Significance
Clock genes have been shown to be important in regulating many key agronomic traits. Therefore, identifying new players in this interconnected clock network will provide novel strategies toward developing new crop varieties. Our study identifies CONSTANS-LIKE 3 (COL3) as a critical protein-binding partner for B-BOX32 (BBX32) action in Arabidopsis. The discovery of the interaction with COL3 provides molecular clues as to how BBX32 exerts its effects on growth and yield. It also implicates COL3 as an integral protein-binding partner that can be used in combination with BBX32 for increased productivity. This regulatory pathway could be applied as an efficient strategy for genetic manipulation in crops for increased agricultural productivity.
Keywords: BBX32, Flowering, Constans-like, Arabidopsis, circadian rhythms
Abstract
Plants have the ability to respond to seasonal environmental variations by monitoring day length to initiate flowering. The transition from vegetative to the reproductive stage is the critical developmental switch in flowering plants to ensure optimal fitness and/or yield. It has been previously reported that B-BOX32 (BBX32) has the potential to increase grain yield when ectopically expressed in soybean. In the present study, we performed a detailed molecular characterization of the Arabidopsis B-box domain gene BBX32. We showed that the circadian clock in Arabidopsis regulates BBX32 and expressed in the early morning. To understand the molecular mechanism of BBX32 regulation, we performed a large-scale yeast two-hybrid screen and identified CONSTANS-LIKE 3 (COL3)/BBX4 as one of its interacting protein partners. Using different genetic and biochemical assays, we have validated this interaction and shown that COL3 targets FT in the presence of BBX32 to regulate the flowering pathway. Based on these findings, we hypothesized that this BBX32-COL3 module could be an additional regulatory mechanism affecting the reproductive development in Arabidopsis that could be translated to crops for increased agricultural productivity.
Many organisms prepare for seasonal changes by integrating day-length and growth and developmental information. Thus, having a robust mechanism for day-length measurement to thrive in the face of seasonal variations and geographic constraints represents an adaptive advantage for plants. For example, in agriculture, premature flowering affects the overall fitness of the plant, which could lead to dramatic downstream events resulting in loss of crop productivity. The mechanism of flowering has been elucidated through studies describing different physiological and genetic phenotypes in the model species Arabidopsis thaliana (1–3). The mechanism for how plants perceive seasonal variations via sensing the light period, and light quality and further coordination of the network of signaling pathways, has been clearly outlined (1, 3–6). Arabidopsis, a facultative long-day plant, flowers earlier in long days than in short days, in coordination with the circadian clock and photoperiod (7–10). The molecular mechanism of day-length measurement is comprised of the circadian regulation of CONSTANS (CO) gene expression and the light regulation of CO protein stability and activity (11). FLOWERING LOCUS T (FT) protein, expressed in long days, acts as a floral integrator, relaying the signal from the light-sensing leaves via the phloem to the shoot apical meristem (SAM), where flowering is initiated (3, 11). This process, however, occurs only under long-day conditions, when the repression of CO by CYCLING DOF FACTORS1 and 2 (CDF1 and CDF2) is relieved. This derepression is mediated primarily by the interaction of the clock-regulated proteins GIGANTEA (GI) and FLAVIN-BINDING, KELCH REPEAT F-BOX1 (FKF1); GI and FKF1 form a light-dependent complex near the end of long days and targets the CDFs to the proteasome via polyubiquitination (10). CO and its interacting partners such as ASYMMETRIC LEAVES 1 (AS1) bind to the FT promoter via the C-terminal CCT [CO, CONSTANS-LIKE (COL), and TIMING OF CAB EXPRESSION1 (TOC1)] domain (12, 13). The TARGET OF EAT1 (TOE1) protein binds to the FT promoter near the CO-binding site, leading to the reduction of CO activity during the morning. TOE1 interacts with FKF1 in the afternoon, competitively interfering with the FKF1–CO interaction and stabilizing CO (11). Taken together, these results suggest that CO may induce FT expression through different regulatory mechanisms at different times of day to fine-tune photoperiodic flowering. The CO-FT module is highly conserved across different crop species such as rice, barley, maize, tomatoes, sunflowers, and sugar beet with a different mode of action (3). However, the mechanism of CO-FT action varies in short days (3). Temperature and geographical locations are also reported to affect the photoperiodic flowering pathway thus affects the seasonal flowering (3). Thus, the integration of geographical and environmental cues makes the whole photoperiodic flowering regulatory framework complex.
In Arabidopsis, zinc finger transcription factors (TFs) account for ∼15% of the total genome and play a critical role in plant growth and development (14, 15). The B-BOX (BBX) family members are zinc finger TFs that contain 32 family members. The TF family is divided into five groups (I–V) based on one or two B-BOX motifs involved in protein–protein interaction and the presence or absence of a CCT domain (15). BBX proteins exist in species from algae, mosses, animals, and plants and are functionally diverse (15, 16). The founding and best-characterized member of the B-box family is CO. Studies have reported that other members of the BBX family such as COL3/BBX4, BBX6, BBX7 (COL9), BBX19, BBX24/SALT TOLERANCE (STO), microProtein1a (MiP1a)/BBX31 and BBX32/EMBRYONIC FLOWER-1 INTERACTING PROTEIN6 (EIP6) also affect flowering with distinct mechanisms (17–22).
It was suggested that BBX32 may play a critical role at the interface of light and the circadian clock in soybean (23). Interestingly, overexpression of the Arabidopsis BBX32 has been shown to significantly increase grain yield in soybean (23), implicating the role of this family of TFs in plant reproductive fitness. In the present study, we dissected the molecular mechanism of BBX32 action in Arabidopsis. We show that BBX32 is regulated by the circadian clock, regulates flowering and hypocotyl growth, and directly interacts with COL3. We provided various supporting genetic and biochemical evidence to establish this interaction and proved that FT is a direct target of COL3. Thus, this study proposes a role for BBX32 in the flowering pathway and a possible mechanism of action.
Results
BBX32 Is Regulated by the Circadian Clock.
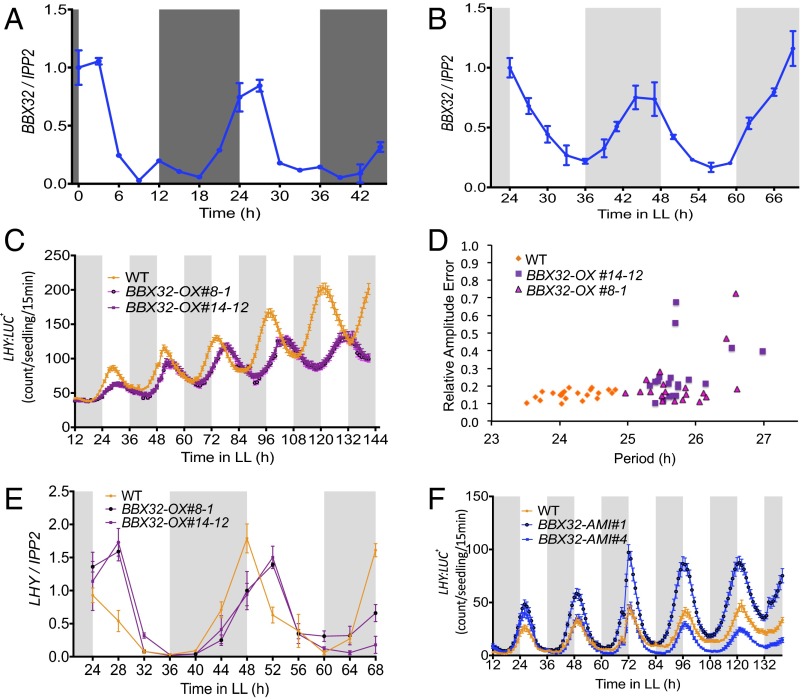
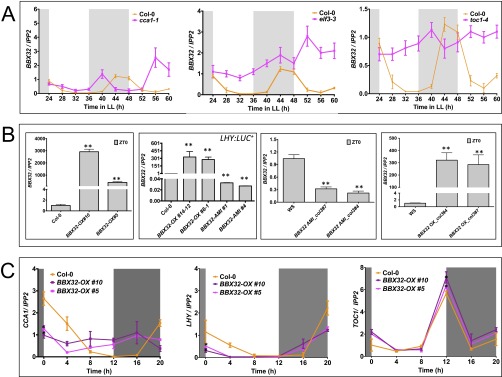
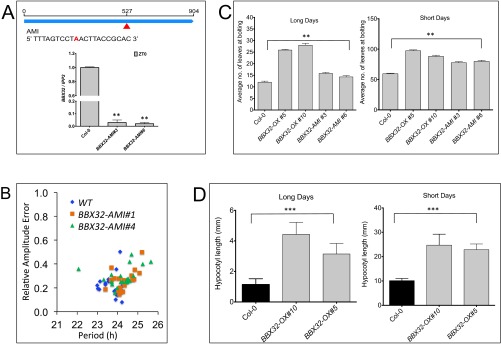
To understand the relationship between the Arabidopsis BBX32 (At3g21150) and the clock, we first evaluated the rhythmic expression pattern of BBX32 in diurnal conditions (12 h light and 12 h dark cycles; 12L:12D) and free-running conditions (continuous light; LL). We performed a time course experiment with samples harvested every 3 h and determined the relative mRNA expression level by using quantitative RT-PCR (qPCR). Our results show that BBX32 expression is rhythmic under both diurnal and free-running conditions with the peak expression in the early morning (Fig. 1 A and B). To confirm BBX32 clock regulation, its expression in mutants of CIRCADIAN CLOCK ASSOCIATED1 (cca1-1), toc1-4, and EARLY FLOWERING3 (elf3-3) was checked (24–26). Relative mRNA expression of samples harvested every 4 h showed expression of BBX32 was altered, indicated by the loss of robust oscillations in elf3-3 and almost arrhythmic with increased expression level in toc1-4 mutants (Fig. S1A). In cca1-1 mutant, expression of BBX32 appears to be altered with a short-period phenotype (Fig. S1A). These observations suggest a quantitative change in the pace of the clock, but to better understand the detail mechanism, further investigation is needed. BBX32 has been shown to alter CCA1, LONG ELONGATED HYPOCOTYL (LHY), and TOC1 gene expression in soybean (23); thus, CCA1, LHY, and TOC1 expression was checked in Arabidopsis plants overexpressing BBX32. Flag-tagged BBX32 overexpression line (BBX32-OX) (Fig. S1B) harvested every 4 h in 12L:12D conditions showed altered and reduced expression of CCA1 and LHY and a modest increase in TOC1 levels in Arabidopsis (Fig. S1C). Bioluminescence assays performed with the BBX32 overexpression line containing a LHY promoter luciferase fusion (LHY::LUC+) under LL showed that when overexpressed, BBX32 lengthens the period by ∼2 h (Fig. 1 C and D). To determine whether this alteration in promoter activity is also evident at the mRNA level, we checked the endogenous expression of LHY in BBX32-OX under LL over 2 d. We observed a shift in the LHY expression compared with wild type, confirming that BBX32 regulates the clock in Arabidopsis (Fig. 1E). We also generated artificial microRNA lines (BBX32-AMI) (Fig. S2A). These BBX32-AMI lines were transformed with the LHY reporter, and bioluminescence assays were performed to assess LHY promoter activity under LL and a modest period lengthening was observed (Fig. 1F and Fig. S2B). Furthermore, clock-regulated phenotypes such as flowering time and hypocotyl growth were examined. BBX32-OX flowered late under both long (16L:8D) and short (8L:16D) day conditions at 22 °C compared with wild type (Fig. S2C). BBX32-AMI plants also have a late flowering phenotype compared with the wild type under both long- and short-day conditions (Fig. S2C). The delayed flowering and circadian phenotypes of BBX32-AMI could be due to the redundancy among the TF family and/or feedback regulation. Also, BBX32-OX showed long hypocotyl growth in long and short days (Fig. S2D). Thus, taken together, these data indicate that BBX32 is a morning gene under clock regulation, and it suggests that BBX32 may regulate the clock and may have a role in flowering and hypocotyl growth.
Fig. 1.
Gene expression and phenotype analysis of BBX32. Relative gene expression analysis of BBX32 under diurnal conditions (12L:12D) (A) and free running conditions (LL) (B) at 22 °C. Wild-type (Col-0) seedlings were grown in 12L:12D for 7 d (A) and 12L:12D for 6 d and transferred to continuous light (LL) and harvested 24–68 h later (B). Error bars represent the SEM of biological triplicates. Experiments were independently repeated three times, each time with two biological replicates per genotype. (C) BBX32-OX:LHY-LUC+ activity under LL. Six-day-old seedlings entrained in 12L:12D were monitored for 5–7 d under LL. Values are shown as mean ± SEM; n = 20. (D) Relative amplitude error of WT and BBX32-OX seedlings imaged for 5 d under LL conditions calculated by using FFT-NLLS; n = 20. (E) Relative mRNA expression of LHY in WT and BBX32-OX. Six-day-old WT and BBX32-OX seedlings were grown in 12L:12D and transferred to continuous light (LL) and harvested 24–68 h later. (F) BBX32-AMI:LHY-LUC+ activity under LL. Six-day-old seedlings entrained in 12L:12D were monitored for 5–7 d under LL. Values are shown as mean ± SEM; n = 20. Experiments were repeated three times independently.
Fig. S1.
Gene expression of BBX32 and phenotypes. (A) Relative gene expression of BBX32 in clock mutants. Wild-type (Col-0) and clock mutant (cca1-1, toc1-4, elf3-3) seedlings were grown for 6 d in 12L:12D and transferred to continuous light (LL) on day 7 and harvested 24–60 h later. Error bars represent the SEM of biological triplicates. Experiments were repeated three times independently. Unshaded and shaded areas represent subjective light and dark periods, respectively. (B) Expression of BBX32 in mutants generated for this study. Wild-type (Col-0 and WS), Flag-tagged BBX32-OX, Flag-tagged BBX32 OX_col3, and BBX32 AMI_col3 seedlings were grown for 6-d in 12L:12D and harvested on day 7 at Zeitgeber Time (ZT) 0. Error bars represent the SEM of biological triplicates. Experiments were repeated three times independently with two biological replicates per genotype. **P ≤ 0.001. (C) Expression analysis of major clock genes in Flag-tagged BBX32-OX. Experiments were performed on 7-d-old seedlings grown in 12L:12D cycles at 22 °C. Error bars represent the SEM of biological triplicates. Experiments were repeated three times independently with two biological replicates per genotype. *P ≤ 0.05. Unshaded and shaded area represents light and dark period, respectively.
Fig. S2.
Gene expression of BBX32-AMI lines and phenotype analysis. (A) Graphical representation of BBX32 amiRNA and its relative gene expression analysis. Upper represents the single point insertion and the amiRNA sequence targeting BBX32. Lower shows the gene expression analysis of the BBX32-AMI. Wild-type (Col-0), BBX32-AMI seedlings were grown for 6 d in 12L:12D at 22 °C and were harvested on day 7 at ZT0. Error bars represent the SEM of biological triplicates. Experiments were repeated three times independently. **P ≤ 0.001. (B) Relative amplitude error of WT and BBX32-OX seedlings imaged for 5 d under LL conditions calculated by using FFT-NLLS; n = 20. (C) Flowering time analysis of Flag-tagged BBX32-OX and BBX32-AMI. Flowering time analysis was performed on plants grown in 16L:8D (Long days) and 8L:16D (Short days) cycles at 22 °C. Data shows average number of leaves at bolting, and error bars represent SE (n = 12). **P ≤ 0.001; unpaired t test. (D) Hypocotyl length of Flag-tagged BBX32-OX. Flag-tagged BBX32-OX show long hypocotyl phenotype in both long (16L:8D) and short days (8L:16D). ***P ≤ 0.0001; unpaired t test.
BBX32 Physically Interacts with COL3.
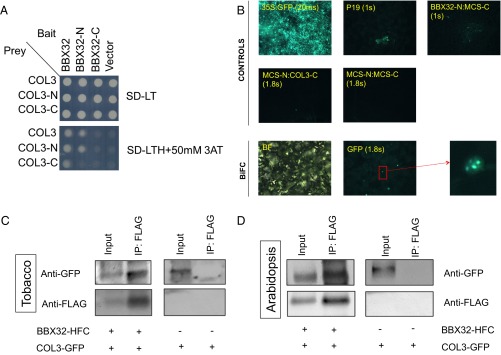
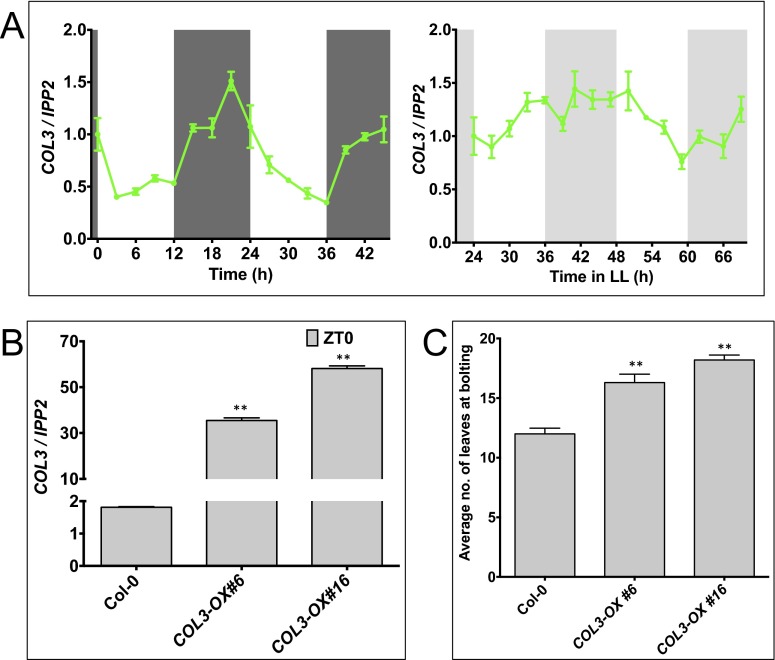
Protein–protein interaction is a key functional aspect of this TF family’s regulatory mechanism. B-box proteins, including BBX32, have been shown to interact with SALT TOLERANCE HOMOLOG2/BBX21 to regulate photomorphogenesis by acting antagonistically on ELONGATED HYPOCOTYL5 (HY5) (27). To gain a deeper insight into the molecular function of BBX32, we performed a high-throughput yeast two-hybrid (Y2H) screen by using BBX32 as bait (Dataset S1). One of the promising interactors in our Y2H screen was COL3 (At2g24790). Further analysis confirms that the N terminus of COL3 interacts with BBX32 (Fig. 2A). COL3 is also a BBX TF family member and is temporally coexpressed with BBX32 (Fig. S3A). GFP-tagged overexpression of COL3 (COL3-OX) (Fig. S3B) also shows a late flowering phenotype in long days compared with the wild type similar to what was observed for BBX32-OX (Fig. S3C).
Fig. 2.
BBX32 physically interacts with COL3. (A) Y2H assay between BBX32 and each of COL3, COL3-N, and COL3-C. These experiments were repeated twice. (B) BiFC in tobacco. COL3-YFPC interacts with BBX32-YFPN in the nucleus and localized to nuclear speckles. 35S:GFP, p19 and empty vector (MCS-N MCS-C) were used as negative controls. B32-N MCS-C represents the fusion of BBX32 on N terminus alone; MCS-N COL3-C represents the fusion of COL3 on C terminus alone (Materials and Methods). Enlarged image of the nuclei shows the size and number of speckles. BF, bright field. (C and D) Co-Immunoprecipitation (Co-IP) in tobacco and Arabidopsis, respectively. Immunoprecipitations (IPs) were performed on tobacco leaves (3 dpi) and 10-d-old Arabidopsis seedlings grown in long days (16L: 8D) at 22 °C. Tissues were harvested 1 h after dawn. IP is performed by using anti-FLAG antibody and COL3 was coimmunoprecipitated with anti-GFP antibody. A 5% input was used. Western blots were performed on 10% (wt/vol) precast gels (Bio-Rad), and experiments were repeated at least five times.
Fig. S3.
Gene expression and flowering time analyses of COL3. (A) Relative gene expression analysis of COL3 under diurnal and free running conditions at 22 °C. Wild-type (Col-0) seedlings were grown in 12L:12D for 7 d for diurnal and 6 d in 12L:12D and transferred to continuous light (LL) and harvested 24–60 h later. Error bars represent the SEM of biological triplicates. Experiments were repeated three times independently with two biological replicates per genotype. Unshaded and shaded areas represent light and dark period (diurnal) and subjective light and dark period (circadian), respectively. (B) Relative expression of COL3 in GFP-tagged COL3-OX lines generated for this study. Wild-type (Col-0), GFP-tagged COL3-OX seedlings were grown for 6 d in 12L:12D at 22 °C and were harvested on day 7 at ZT0. Error bars represent the SEM of biological triplicates. Experiments were repeated three times independently with two biological replicates per genotype. **P ≤ 0.001. (C) GFP-tagged COL3-OX flowers late in long days (16L:8D) at 22 °C. Data show the average number of leaves at bolting. Error bars represent SE (n = 12) **P ≤ 0.001; unpaired t test.
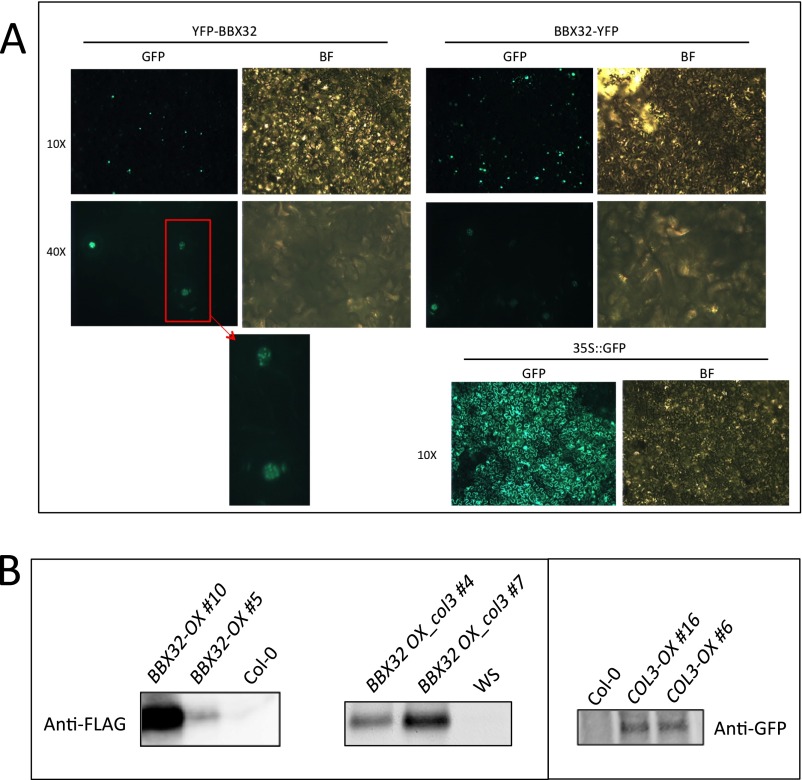
To validate this interaction and evaluate its subcellular localization, we performed a bimolecular fluorescence complementation (BiFC) assay. For this assay, the 35S:BBX32:YFP construct with YFP tag fused either to the N or C terminus were generated. Similar to COL3, BBX32 localized to the nuclear speckles (Fig. S4A) (26). Therefore, to confirm the interaction of BBX32 and COL3 in planta, the N-terminal half of YFP was fused to BBX32, and the C-terminal half of YFP was fused to COL3. Both constructs were then transiently expressed in tobacco (Nicotiana bethamiana). Reconstituted YFP fluorescence was observed in the nucleus, confirming that BBX32 and COL3 are colocalized and interacts in planta (Fig. 2B). Together these results indicate that both proteins possess a similar localization pattern and phenotype, which suggests that these proteins may form a complex to regulate flowering.
Fig. S4.
Subcellular localization of BBX32 and BBX32 and COL3 protein expression. (A) BBX32 is localized to nuclear speckles. Enlarged image of the nuclei shows the size and number of speckles. 35S::GFP was used as a control. BF, bright field. (B) Protein expression analysis of Flag-tagged BBX32-OX; Flag-tagged BBX32 OX_col3, and GFP-tagged COL3-OX lines. Ten-day-old seedlings grown in 12L:12D conditions were harvested at ZT1, and Western blot analysis was performed. Anti-FLAG and anti-GFP antibodies were used for BBX32-OX; BBX32 OX_col3, and COL3-OX, respectively.
To further validate this interaction in vivo, coimmunoprecipitation assays were performed in tobacco and Arabidopsis. For this assay, a BBX32 C-terminal fusion construct (BBX32 OX-HFC) and a COL3-GFP fusion construct (COL3 OX-GFP) were generated (Fig. S4B). COL3-GFP specifically coimmunoprecipitated with BBX32-HFC when both BBX32 OX-HFC and COL3 OX-GFP were expressed in tobacco leaves (Fig. 2C). In Arabidopsis, COL3-GFP also immunoprecipitated with BBX32-HFC, confirming that BBX32 physically interacts with COL3 in vivo (Fig. 2D).
Genetic Interaction and Physiological Characterization of Flowering Time.
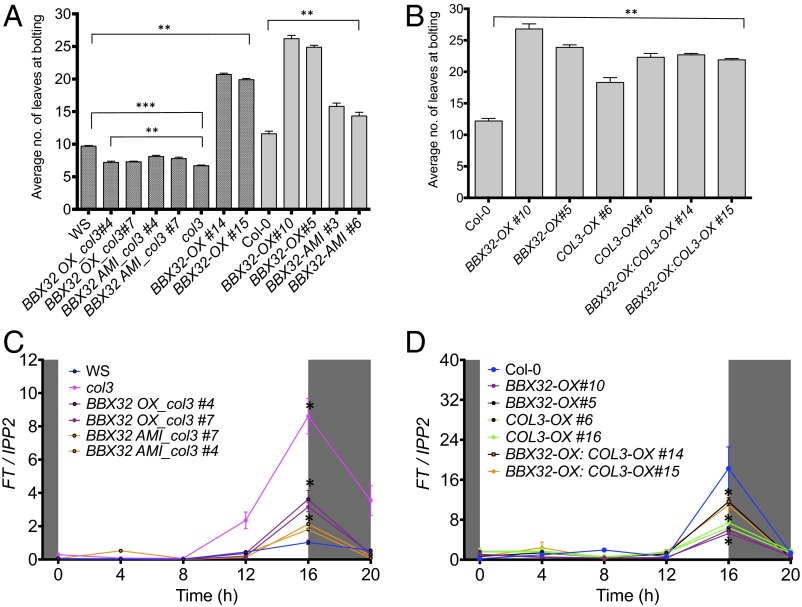
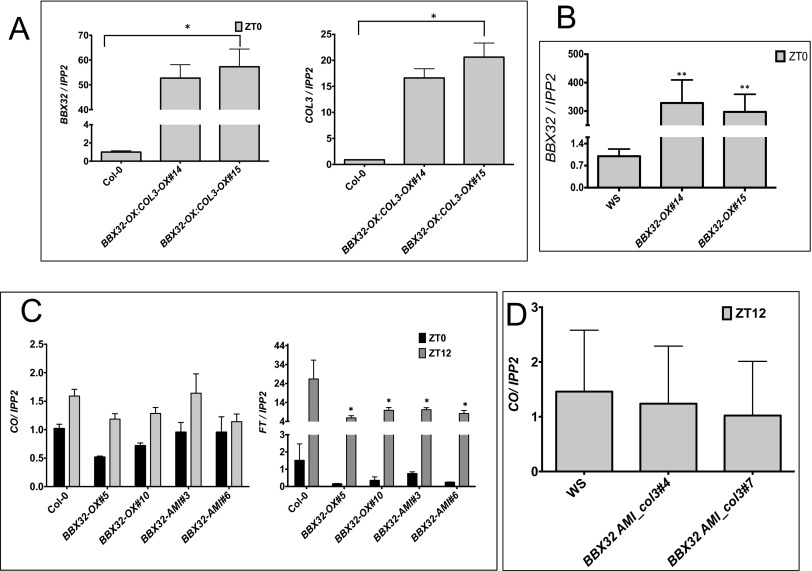
To understand the functional relationship and genetic interaction between BBX32 and COL3 and their role in the regulation of flowering, we investigated whether the late flowering phenotype of BBX32 depends on COL3. We generated BBX32-OX in Wassilewskija (WS) background along with BBX32-OX and BBX32-AMI lines in a col3 mutant background (BBX32-OX_col3 and BBX32-AMI_col3) (Fig. S1B). Homozygous lines were obtained for each construct, and flowering time analysis was performed in long days (16L:8D) at 22 °C (Fig. 3A). Regardless of the genetic background, BBX32 produces a late flowering phenotype when overexpressed (Fig. 3A). Also, COL3 gene is required for alterations in BBX32 mRNA levels to alter flowering time, because both BBX32-OX_col3 and BBX32-AMI_col3 showed an early flowering phenotype similar to col3 (Fig. 3A) compared with wild type (in this case, WS). Moreover, epitope-tagged double mutants overexpressing both BBX32 and COL3 (BBX32-OX: COL3-OX) were generated (Fig. S5A), and flowering time was assessed (Fig. 3B). The BBX32-OX:COL3-OX plants were late flowering compared with wild type (Fig. 3B). Taken together, our data suggest that the late flowering observed in lines with altered levels of BBX32 depends on COL3.
Fig. 3.
Flowering time analysis and expression of FT in overexpression and higher order mutant lines. (A and B) Flowering time analysis was performed on plants grown in 16L:8D cycles at 22 °C for different epitope-tagged single and double transgenic plants. Shaded bars represent mutant lines in WS background. Data shows average number of leaves at bolting, and error bar represents SE (n = 12). ***P ≤ 0.0001; one-way ANOVA (A) and **P ≤ 0.001; unpaired t test (B). (C and D) Relative expression analysis of FT in all epitope-tagged single and double mutants. Experiments were performed on 7-d-old seedlings grown in 16L:8D cycles at 22 °C. Error bars represent the SEM of biological triplicates. Experiments were independently repeated three times, each time with two biological replicates per genotype. Unshaded and shaded area represents light and dark period, respectively.
Fig. S5.
Gene expression analysis of BBX32, CO, and FT. (A) Relative expression of BBX32 and COL3 in BBX32-OX: COL3-OX. Wild-type (Col-0), BBX32-OX:COL3-OX seedlings were grown for 6 d in 12L:12D and harvested on day 7 at ZT0. (B) Relative expression of BBX32 in BBX32-OX in WS background. Wild-type (WS), BBX32-OX seedlings were grown for 6 d in 12L:12D and harvested on day 7 at ZT0. Error bars represent the SEM of biological triplicates. Experiments were repeated three times independently. *P ≤ 0.05. (C) Relative gene expression analysis of CO and FT. Seven-day-old wild-type (Col-0), FLAG-tagged BBX32-OX and BBX32-AMI seedlings grown in long-day conditions (16L:16D) were harvested at ZT0 and ZT12. Error bars represent the SEM of biological triplicates. Experiments were repeated three times independently. (D) Gene expression analysis of CO. Seven-day-old wild-type (WS) and BBX32-AMI_col3 seedlings grown in long-day conditions (16L:16D) were harvested at ZT12. Error bars represent the SEM of biological triplicates. Experiments were repeated three times independently with two biological replicates per genotype.
CO and FT are two major genes regulating the flowering pathway in Arabidopsis under long days. Hence, we checked the expression of these two genes in BBX32-OX and BBX32-AMI lines. No significant change in CO expression was observed in BBX32-OX, BBX32-AMI, and BBX32 AMI_col3 lines (Fig. S5 B and C). However, FT levels were reduced in BBX32-OX and BBX32-AMI lines (Fig. S5B). Thus, we reasoned that the late flowering phenotype could be FT mediated. We next checked the expression of FT in our epitope-tagged single and double mutants of BBX32 and COL3 harvested every 4 h under long-day conditions (16L:8D) at 22 °C. Our data distinctly show that lines that flowered earlier have more FT expression, whereas the late flowering lines have less FT expression (Fig. 3 C and D).
FT Is a Target of COL3.
Because COL3 is a TF and col3 exhibits an early flowering phenotype and increased levels of FT mRNA, we reasoned that the FT promoter could be a target of COL3 regulation. To answer this question, we used a modified yeast-one hybrid (Y1.5H) and chromatin immunoprecipitation (ChIP) assays. In our Y1.5H, we evaluated the binding of COL3 to the FT promoter with or without the presence of BBX32. The concept underlying this assay was to express the protein of interest (BBX32) with an activation domain and evaluate the activation of the candidate DNA (FT) regions fused to a reporter in the presence and/or absence of the interacting partner (COL3) in the yeast system. This assay will allow us to determine whether the interaction between these two proteins is required for the activation of the target.
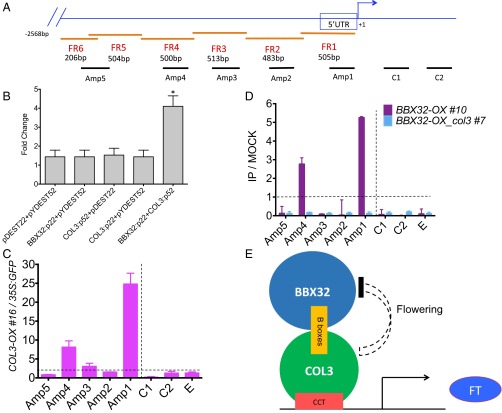
Six fragments (FR1–6) of the FT promoter were integrated into the yeast strain YM4271 (Fig. 4A). Individually, neither BBX32 nor COL3 was sufficient to bind and activate FT. However, coexpressing the BBX32 activation domain fusion with COL3 resulted in successful binding and activation of FT FR1 and FR4. To further quantify this observed activation, we measured the β-galactosidase activity with FR1 and observed a fourfold induction of the reporter gene activity (Fig. 4B). FR4 did not pass our threshold of ≥twofold. These data suggest that COL3 directly binds to FT promoter in the presence of BBX32, at least in a heterologous system.
Fig. 4.
COL3 targets FT to regulate flowering. (A) Schematic representation of the FT gene and amplified promoter regions used for yeast and ChIP assays. FR (1–6) represent fragments cloned in yeast and Amp (1–5) represent amplicons amplified for ChIP assay. C1 and C2 represent regions of locus-control used in ChIP as described (10). (B) β-galactosidase activity in yeast FR1. The fold change of β-galactosidase activity of each construct was calculated by normalizing over control vectors. Bars represent the average of two biological replicates, and error bars represent the SEM. **P < 0.001 (unpaired t test). (C and D) Chromatin immunoprecipitation (ChIP) analysis of GFP-tagged COL3-OX #16 and Flag-tagged BBX32-OX #10 on the FT promoter. The 10-d-old GFP-tagged COL3-OX #16, 35S:GFP, Flag-tagged BBX32-OX #10, and Flag-tagged BBX32 OX_col3 #7 seedlings were grown in 16L:8D cycle at 22 °C and harvested at 2–3 h after dawn. The ratio of specific enrichment in the GFP-tagged COL3-OX #16 samples and that in the 35S:GFP sample is represented (C) whereas FLAG-tagged BBX32-OX #10; FLAG-tagged BBX32 OX_col3 #7 and that in mock (no antibody) (D) in each region were plotted. Experiments were independently repeated three times, each time with two biological replicates. C1 and C2 are the control regions and E (UBQ10) is a nonlocus control as described (10). Dotted line represents no enrichment. (E) Model representing mechanism of action of BBX32 in regulating flowering.
To confirm the binding of COL3 to the FT promoter in vivo, we also performed a ChIP assay by using plants expressing functional GFP-tagged COL3 (COL3-OX) (Fig. S4B) and determined whether COL3-GFP protein was able to immunoprecipitate the FT promoter region of the chromatin (Fig. 4C). The immunoprecipitated DNA was highly enriched in the FT promoter regions denoted as amplicons 1 and 4 in COL3-OX line relative to 35S:GFP (Fig. 4C). These amplicons contain the regions to which CCT domain proteins may associate with the FT promoter (12, 28). Moreover, ChIP experiments were also performed in plants expressing a functional Flag-tagged BBX32 (BBX32-OX) in wild type and (BBX32 OX_col3) in col3 mutant background (Fig. S4B). The immunoprecipitated DNA was enriched in the FT promoter regions (amplicons 1 and 4) in BBX32-OX but not in BBX32-OX_col3 (Fig. 4D). Our ChIP data indicates the presence of COL3 in the region adjacent to the transcriptional start site of FT.
Discussion
We have performed characterization of BBX32, a group V member of BBX TF family, at the molecular level (15). The unique architecture of BBX32 with only one B-box domain known for protein–protein interactions made BBX32 an interesting candidate to study further (21). Also, previous studies in soybean have shown that the Arabidopsis BBX32 has a high potential for increasing crop productivity (23). However, the molecular mechanism of BBX32 regulation in soybean or Arabidopsis was not known. Thus, we performed a detailed molecular study in Arabidopsis and found that BBX32 participates in the regulation of flowering. With enhanced agronomical trait(s) such as flowering, BBX32 can play an important role in plant growth and development, thus contributing as one among other plausible mechanism(s) for the increase in broad acre yield in soybean. We have demonstrated that BBX32 is a clock gene with an early morning expression peak. We have established that COL3 interacts with BBX32, and possess similar expression patterns and phenotypes. Moreover, our data corroborates results from previous studies, which implicated the B-BOX domain of BBX32 in the formation of heterodimers with other members of the family to modulate their activities (27). Hence, this BBX32–COL3 interaction is indispensable in defining its mechanism of action. We have also shown that COL3 targets FT and affects its regulation of flowering time in Arabidopsis. Thus, our proposed model is the interaction of BBX32 with COL3 enables COL3 to bind to the FT promoter and represses its transcription (Fig. 4D), but the detailed mechanism needs further investigation.
The broad acre yield observed in soybean upon ectopic expression of Arabidopsis BBX32 suggests that it might have pleiotropic effects on growth parameters (23). This BBX32-COL3 module suggests they potentially modulate growth parameters via flowering time regulation. Despite the variation of genome complexity and domestication, BBX32-COL3 could hold its merit to explain the enhanced agronomical trait(s) in soybean. It could be an additional regulatory mechanism, which facilitates soybean flowering in a range of photoperiods. Also, BBX32 is in phase with the photoperiod-sensitive gene, COL3. Whether this relation of clock and flowering pathway via BBX32 is direct or indirect, with or without the involvement of CO needs further investigation. The phenotype of the BBX32-OX:COL3-OX suggest that the COL3 could be a part of the bigger complex involving other “modifiers” including COL3 that leads to architectural changes in overall plant growth as observed in soybean. At the same time, the possibility of B-BOX proteins being recruited to their targets via common DNA binding motif facilitating diverse effects in different plants cannot be ruled out either. Also, recent studies with BBX19 and MiP1a (BBX31) showing attenuation of CO activity add a new layer to the flowering regulation by BBX proteins (20, 22).
Thus, the involvement of BBX proteins in flowering suggests their potential use in transgenic crops to obtain desirable agronomic trait. This study describes the molecular function of two B-BOX family members whose physical interaction result in a late flowering phenotype. In fact, this present study provides the direct primary evidence of translational approach on how BBX32 exerts an effect on growth and yield, which could be a profitable strategy to generate early or late flowering plants depending on production requirements or climatic and geographic limitations. Early flowering could be a favorable trait for crops cultivated primarily for seeds whereas late flowering could be an advantage when total biomass is the goal of production (29). This BBX32-COL3 module could apply to other crop species and can potentially result in crop improvement.
Materials and Methods
Plant Materials and Growth Conditions.
Overexpression and artificial microRNA Arabidopsis lines were generated in the Col-0 background, whereas the higher order mutants were generated in col3 background by using A. tumefaciens-mediated transformation (floral dip) (30). BBX32-OX:COL3-OX double transgenic plant was generated by crossing Flag-tagged BBX32-OX to GFP-tagged COL3-OX. The col3 mutant was in WS background (18) hence the WS is used as wild type when single mutant and higher order mutants were used in the experiments. For flowering time analyses, seeds were sown in soil (Sunshine Mix), fertilized once in a month (15–16–17 Peat Lite Special; Everris) and grown in a growth chamber (Conviron) with long-day conditions of 16 h light and 8 h dark (16L:8D) at 22 °C and 50% humidity. The total leaf number was counted at the time of bolting. For hypocotyl measurement, seeds after 3-d stratification were grown on 0.5× MS (no sucrose) vertical plates under short-day conditions (8L:16D) in a growth chamber (Percival) at 80 μE intensity at 22 °C. The hypocotyls were measured by using ImageJ software after scanning the seedlings on day 6. For RNA isolation, 7-d-old seedlings grown in growth chambers (Percival) with lights at 60–80 μE intensity were harvested and immediately frozen in liquid nitrogen and stored in −80 °C. All of the phenotyping experiments were repeated at least three times with similar results, using two independent transgenic lines and data shown for one representative line.
Details.
Details of molecular cloning and constructs, luciferase imaging and data analysis, yeast one-hybrid assay, Y2H, bimolecular fluorescence complementation, coimmunoprecipitation, chromatin immunoprecipitation, and gene expression analyses are provided in SI Materials and Methods and Table S1.
Table S1.
List of primers used in the study
| Gene name | Primer sequence (5′–3′) | Coordinates/positions/size, bp | Notes |
| qPCR | |||
| BBX32 RT-F | TGTGTCTCAAGCTCCGAGCTATCG | — | — |
| BBX32 RT-R | TTTCCCTCCCTCGCGCTCTGTTTA | — | — |
| BBX32-AMI-F | GCGGATGGCATTTTTGTAA | — | — |
| BBX32-AMI-R | CGCCGCTAAGAACACTCTCT | — | — |
| LHY-F | ACCAACGAAACAGGTAAGTGGCG | — | — |
| LHY-R | GTGCACAGTAGTACCATCGTTACC | — | — |
| CCA1 -F | AATGTGACCGGTGGGTTCATGG | — | — |
| CCA1-R | AAGTGCATAGCACGCGCCTTAC | — | — |
| TOC1 -F | CTTCAATCTACTTTTCTTCGGTGCT | — | — |
| TOC1-R | AATAGTAATCCAGCGCAATTTTCTTC | — | — |
| CO-F | AACGACATAGGTAGTGGAGAGAACAAC | — | — |
| CO-R | GCAGAATCTGCATGGCAATACA | — | — |
| FT-F | CAACCCTCACCTCCGAGAATAT | — | — |
| FT-R | TGCCAAAGGTTGTTCCAGTGT | — | — |
| IPP2-F | GTATGAGTTGCTTCTCCAGCAAAG | — | — |
| IPP2-R | GAGGATGGCTGCAACAAGTGT | — | — |
| COL3-F | TACCGTTGGTGCCTGAAAGTGG | — | — |
| COL3-R | ATAACTGCACAGCTGGCGTCTC | — | — |
| Y1.5H | |||
| FT.FR1.1.E-F | CACCCTTATTTTTATTAATAATCTTAC | −508 /−4 | TOPO cloning primer |
| FT.FR1.1-R | CTTTGATCTTGAACAAACAGG | ||
| FT.FR2.1.E-F | CACCGAAGAGAAATAAAACAATTGATTTGG | −975/−493 | TOPO cloning primer |
| FT.FR2.1-R | TATTAATAAAAATAAGAAAAAAGTAGGG | ||
| FT.FR3.1.E-F | CACCCTGAGATGTTACAAA | −1468/−956 | TOPO cloning primer |
| FT.FR3.1-R | CAATTGTTTTATTTCTCTTCTTTTTTTA | ||
| FT.FR4.1.E-F | CACCCAAAAAGATACACTTTT | −1960/−1461 | TOPO cloning primer |
| FT.FR4.1-R | CATCTCAGTAATGTATTCATATTG | ||
| FT.FR5.1.E-F | CACCCGAACATTATGCGA | −2451/−1948 | TOPO cloning primer |
| FT.FR5.1-R | GTGTATCTTTTTGTTTTTTGATTTAAT | ||
| FT.FR6.1.E-F | CACCGTGATGCACTATATG | −2571/−2366 | TOPO cloning primer |
| FT.FR6.1-R | GTTTTGTCTTTATCAACTCCAC | ||
| ChIP | |||
| FT-Amp5-F | CATGCGAAAATCTAGTGGAAGAAA | −2537/−2514 | — |
| FT-Amp5-R | AAGCCGTGATGTTCCTAAACAC | −2335/−2314 | — |
| FT-Amp4-F | GCGACTGCGACCTATTTTTTTC | −1860/−1839 | — |
| FT-Amp4-R | GCTATATGCACTTTTTAACGACTAGC | −1692/−1667 | — |
| FT-Amp3-F | CCTACAGTTGTTAGGCTATGGT | −1354/−1333 | — |
| FT-Amp3-R | CTAACCATCCATTTGCACGAC | −1179/−1159 | — |
| FT-Amp2-F | GACTATTCTCAAATGTCCTGGTTC | −782/−759 | — |
| FT-Amp2-R | GGTACCGCCAAGTTTATAGAGA | −540/−519 | — |
| FT-Amp1-F | GTGTATTAGTGTGGTGGGTT | −297/−278 | — |
| FT-Amp1-R | TGATCTTGAACAAACAGGTG | −26/−7 | — |
| C1-F | GAGACCCTCTTATAGTAAGCAGA | — | Locus related control |
| C1-R | GTATAGAAGTTCCTGAGGTCTTC | — | Locus related control |
| C2-F | TTCTTTTTTCATGAAGATGGACCC | — | Locus related control |
| C2-R | GAACATCAATTTTCTATATCTTTTTGCAG | — | Locus related control |
| UBQ10-F/E-F | TCCAGGACAAGGAGGTATTCCTCCG | — | Locus unrelated control |
| UBQ10-R/E-R | CCACCAAAGTTTTACATGAAACGAA | — | Locus unrelated control |
| Cloning | |||
| BBX32-F | CACCTGTGTCTCAAGCTCCGAGCTATCG | — | TOPO cloning primer |
| BBX32-R | TAAACAGAGCGCGAGGGAGGGAAA | — | Full-length cDNA |
| COL3-F | CACCATGGCGTCGTCGTCAAGACTTTGCG | — | TOPO cloning primer |
| COL3-R | TCAGAAACTCGGAACAACACCG | — | Full-length cDNA |
| Reverse COL3 N-term +Stop | TCAACCGCCATCTTCATCTAC | — | — |
| Reverse COL3 C-term | TCAGAAACTCGGAACAACACCG | — | — |
| BBX32 N term -F | CACCATGGTGAGCTTTTGCGAG | — | TOPO cloning primer |
SI Materials and Methods
Molecular Cloning and Constructs.
For BBX32 OX-HFC and COL3 OX-GFP, the full-length gene was cloned in pENTR/D-TOPO followed by LR Clonase II (Invitrogen) transformation as per the user manual in pB7FWG2 (31) and pMDC83 (32) vectors, respectively. Upon sequencing confirmation, these constructs were transformed in the col3 mutant (BBX32 OX_col3) background by using A. tumefaciens-mediated transformation (floral dip) (29). The artificial microRNA (amiRNA) (TTTAGTCCTAACTTACCGCAC) targeting BBX32 was constructed as described (33). Primers designed using WMD3 Web microRNA Designer (wmd3.weigelworld.org/cgi-bin/webapp.cgi) were used to amplify the amiRNA precursor by overlapping PCR from the pRS300 template. The fragment containing the amiRNA foldback was cloned into pENTR/d-TOPO, sequenced, and subsequently recombined by using Gateway LR Clonase II (Invitrogen) into the pK2GW7 vector (34) for constitutive expression under control of the 35S promoter. This construct was transformed in Col-0 (BBX32 AMI) and col3 mutant (BBXAMI_col3) background. Transformants were selected on appropriate antibiotic and propagated for single-insertion, homozygous plants. At least three independent homozygous lines were generated for each construct, and all experiments were conducted on at least two independent homozygous lines. For yeast assays, pENTR clones of BBX32 and COL3 were recombined by Gateway LR clonase II in pDEST22 (Invitrogen) and pYDEST52 (Invitrogen) vectors. All primers used were listed in Dataset S1.
Luciferase Imaging and Data Analysis.
Six-day-old seedlings were grown on 0.5× MS (no sucrose) in 12L:12D at 22 °C in growth chambers (Percival) transferred to an LL chamber equipped with a custom-made digital CCD system (Princeton Instruments) with a Distagon T 28-mm large-format lens F/2.0 25-mm F/1.4 1 inch (Megapixel) on the seventh day. Bioluminescence was monitored for single seedlings, and 15-min acquisitions were taken every 1 h for 5–7 d. Circadian period estimates were based on the data recorded after the first 24–36 h under constant conditions by FFT-NLLS analysis in the Biological Rhythms Analysis Software System (BRASS 3.0) (2). Imaging experiments were performed more than five times, yielding similar results. Statistical data analysis was performed by using GraphPad Software for one-way ANOVA and unpaired t test calculation.
Yeast One-Hybrid Assay.
FT promoter fragments (FR1–6) from the transcription start site up to 2,568 bp upstream were PCR amplified and cloned into pENTR/d-TOPO. Clones were recombined by using Gateway LR Clonase II (Invitrogen) into the pLacZi vector. Clones were sequenced and integrated into the yeast genome strain YM4271 as described (35). Ortho-nitrophenyl-β-galactoside served as the substrate for measurement of β-galactosidase activity.
Y2H Assay.
The cDNA encoding full-length BBX32 was subcloned into the bait vector pDEST32 by using Gateway LR clonase II (Invitrogen). Y2H screen was performed as described (31). For small-scale Y2H, full-length, N terminus, and C terminus BBX32 and COL3 were cloned into the pACT-GAL4 activation domain (AD) and the pAS-GAL4 DNA-binding domain (DBD) (Clontech) expression vectors by using Gateway LR clonase II (Invitrogen). Y2H assay was performed as described (31).
Bimolecular Fluorescence Complementation.
The full-length coding sequences for the BBX32 and COL3 proteins were cloned into the 35S-SPYNE and 35S-SPYCE Gateway-compatible vectors, respectively. The constructs were transformed in Agrobacterium and infiltrated into Nicotiana benthamiana leaves as described (31). 35S:GFP, p19, and empty vector (MCS-N MCS-C) were used as negative controls. B32-N MCS-C was used as a control to show that BBX32-YFPN coexpressed with YFPC alone does not give a fluorescent signal. Likewise, MCS-N COL3-C control to show that COL3-YFPC with YFPN alone does not give positive interaction. Plants were kept in long-day (16L:8D) conditions at 22 °C for at least 48 h. Leaves were observed under an OLYMPUS IX71 microscope.
Coimmunoprecipitation.
35S:BBX32-HFC and 35:COL3-GFP clones were generated as described above. The gene silencing inhibitor P19, the overexpressors of BBX32 (35S:BBX32-HFC), and COL3 (35S:COL3-GFP) were transformed into GV1301 Agrobacterium cells. Tobacco plants were grown in the long-day (16L:8D) conditions at 22 °C. Infiltration of cells to N. benthamiana leaves and subsequent protein extraction was done as described (36), and leaves are harvested 3 d after infiltration (dpi), 1 h after dawn. For Arabidopsis, 10-d-old homozygous double mutants were grown in a growth chamber (Percival) under long-day (16L:8D) conditions at 22 °C. All samples were collected 1 h after dawn. Anti-FLAG M2 antibody (Sigma) was used for immunoprecipitation as described (30). To examine the interaction between BBX32 and COL3, membranes were cut at ∼50 kDa. The upper portion was used to detect COL3-GFP with anti-GFP (Roche), and the lower portion was used to detect BBX32-HFC with anti-FLAG M2 Peroxidase HRP (Sigma).
Chromatin Immunoprecipitation.
Arabidopsis seedlings for 35S:COL3-GFP, 35S:BBX32-HFC, 35S:BBX32_col3, and 35S:GFP were grown on 0.5× MS plates (no sucrose) in a growth chamber (Percival) for 10 d under long-day conditions at 22 °C. Samples were collected 2–3 h after dawn and processed as described (37). Anti-GFP antibody (Abcam 290) for GFP lines anti-FLAG M2 antibody (Sigma) for Flag-tagged lines were used for immunoprecipitation. Primers are listed in Dataset S1. The qPCR results were normalized to input DNA by using the equation 2^(Ct input − Ct ChIP) × 0.1.
Gene Expression Analyses.
Seven-day-old whole seedlings were harvested after entrainment in a growth chamber (Percival) under a 12L:12D conditions or 6-d entrainment and then transferred to continuous light at 22 °C. Total RNA was extracted by using Gene Jet Thermo RNA kit as per manual instructions (Thermo). cDNA synthesis using iScript (Bio-Rad) was performed followed by qPCR using the Bio-Rad CFX 384 machine. qPCR analysis was performed with CFX Manager 3.1 software. qPCR data represent values from two biological replicates, each with three technical replicates. The graphs illustrate the mean expression of the biological replicates, with error bars denoting the SEM. All of the experiments were repeated at least three times independently with similar results, using two independent transgenic lines. Data for one representative line was shown. All qPCR primers used in the study are listed in Table S1.
Supplementary Material
Acknowledgments
We thank D. H. Nagel, M. A. Nohales, S. E. Sanchez, S. Porco, F. Csukasi, and S. S. Wang for critical reading of the manuscript; J. L. Pruneda-Paz for the help with Y2H screen and analysis; V. Chien for technical assistance; and all laboratory members for helpful discussions. We are also thankful to the late M. Holm for the generous gift of col3 seeds. Research reported in this publication was supported by Monsanto Company (to S.A.K.). Monsanto played a role in design, data analysis, and the decision to publish. Research reported in this publication was also supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Nos. R01GM067837 and R01GM056006 (to S.A.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1616459114/-/DCSupplemental.
References
- 1.Yanovsky MJ, Kay SA. Living by the calendar: How plants know when to flower. Nat Rev Mol Cell Biol. 2003;4(4):265–275. doi: 10.1038/nrm1077. [DOI] [PubMed] [Google Scholar]
- 2.Imaizumi T, Kay SA. Photoperiodic control of flowering: Not only by coincidence. Trends Plant Sci. 2006;11(11):550–558. doi: 10.1016/j.tplants.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Andrés F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 2012;13(9):627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- 4.Song YH, Ito S, Imaizumi T. Similarities in the circadian clock and photoperiodism in plants. Curr Opin Plant Biol. 2010;13(5):594–603. doi: 10.1016/j.pbi.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo H, Yang H, Mockler TC, Lin C. Regulation of flowering time by Arabidopsis photoreceptors. Science. 1998;279(5355):1360–1363. doi: 10.1126/science.279.5355.1360. [DOI] [PubMed] [Google Scholar]
- 6.Mouradov A, Cremer F, Coupland G. Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell. 2002;14(Suppl):S111–S130. doi: 10.1105/tpc.001362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suárez-López P, et al. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410(6832):1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- 8.Yanovsky MJ, Kay SA. Molecular basis of seasonal time measurement in Arabidopsis. Nature. 2002;419(6904):308–312. doi: 10.1038/nature00996. [DOI] [PubMed] [Google Scholar]
- 9.Valverde F, et al. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303(5660):1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- 10.Sawa M, Nusinow DA, Kay SA, Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 2007;318(5848):261–265. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang B, Wang L, Zeng L, Zhang C, Ma H. Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes Dev. 2015;29(9):975–987. doi: 10.1101/gad.251520.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiwari SB, et al. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 2010;187(1):57–66. doi: 10.1111/j.1469-8137.2010.03251.x. [DOI] [PubMed] [Google Scholar]
- 13.Song YH, Lee I, Lee SY, Imaizumi T, Hong JC. CONSTANS and ASYMMETRIC LEAVES 1 complex is involved in the induction of FLOWERING LOCUS T in photoperiodic flowering in Arabidopsis. Plant J. 2012;69(2):332–342. doi: 10.1111/j.1365-313X.2011.04793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riechmann JL, et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science. 2000;290(5499):2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 15.Khanna R, et al. The Arabidopsis B-box zinc finger family. Plant Cell. 2009;21(11):3416–3420. doi: 10.1105/tpc.109.069088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crocco CD, Botto JF. BBX proteins in green plants: Insights into their evolution, structure, feature and functional diversification. Gene. 2013;531(1):44–52. doi: 10.1016/j.gene.2013.08.037. [DOI] [PubMed] [Google Scholar]
- 17.Cheng X-F, Wang Z-Y. Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J. 2005;43(5):758–768. doi: 10.1111/j.1365-313X.2005.02491.x. [DOI] [PubMed] [Google Scholar]
- 18.Datta S, Hettiarachchi GHCM, Deng XW, Holm M. Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell. 2006;18(1):70–84. doi: 10.1105/tpc.105.038182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park H-Y, et al. EMF1 interacts with EIP1, EIP6 or EIP9 involved in the regulation of flowering time in Arabidopsis. Plant Cell Physiol. 2011;52(8):1376–1388. doi: 10.1093/pcp/pcr084. [DOI] [PubMed] [Google Scholar]
- 20.Wang C-Q, Guthrie C, Sarmast MK, Dehesh K. BBX19 interacts with CONSTANS to repress FLOWERING LOCUS T transcription, defining a flowering time checkpoint in Arabidopsis. Plant Cell. 2014;26(9):3589–602. doi: 10.1105/tpc.114.130252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li F, et al. The B-box family gene STO (BBX24) in Arabidopsis thaliana regulates flowering time in different pathways. PLoS One. 2014;9(2):e87544. doi: 10.1371/journal.pone.0087544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graeff M, et al. MicroProtein-mediated recruitment of CONSTANS into a TOPLESS trimeric complex represses flowering in Arabidopsis. PLoS Genet. 2016;12(3):e1005959. doi: 10.1371/journal.pgen.1005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Preuss SB, et al. Expression of the Arabidopsis thaliana BBX32 gene in soybean increases grain yield. PLoS One. 2012;7(2):e30717. doi: 10.1371/journal.pone.0030717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green RM, Tobin EM. Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc Natl Acad Sci USA. 1999;96(7):4176–4179. doi: 10.1073/pnas.96.7.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strayer C, et al. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289(5480):768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- 26.Zagotta MT, et al. The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J. 1996;10(4):691–702. doi: 10.1046/j.1365-313x.1996.10040691.x. [DOI] [PubMed] [Google Scholar]
- 27.Holtan HE, et al. BBX32, an Arabidopsis B-Box protein, functions in light signaling by suppressing HY5-regulated gene expression and interacting with STH2/BBX21. Plant Physiol. 2011;156(4):2109–2123. doi: 10.1104/pp.111.177139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gendron JM, et al. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA. 2012;109(8):3167–3172. doi: 10.1073/pnas.1200355109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gangappa SN, Botto JF. The BBX family of plant transcription factors. Trends Plant Sci. 2014;19(7):460–470. doi: 10.1016/j.tplants.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 30.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 31.Nusinow DA, et al. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475(7356):398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133(2):462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18(5):1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karimi M, Inzé D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7(5):193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 35.Pruneda-Paz JL, et al. A genome-scale resource for the functional characterization of Arabidopsis transcription factors. Cell Reports. 2014;8(2):622–632. doi: 10.1016/j.celrep.2014.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pruneda-Paz JL, Breton G, Para A, Kay SA. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323(5920):1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowler C, et al. Chromatin techniques for plant cells. Plant J. 2004;39(5):776–789. doi: 10.1111/j.1365-313X.2004.02169.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.