As a response to the surrounding environment most organisms regulate their physiology and behavior in rhythmic manner. Such temporal organization is controlled by two global systems, the cell cycle and the circadian clock. The cell cycle controls sequentially recurring phases of cell growth, DNA replication and cell division. The circadian clock, in turn, coordinates the global physiology and behavior with the 24 h periodicity of daily environmental changes associated with the rotation of the earth. Both systems co-exist in many cell types and ought to be coordinated to avoid potentially conflicting harmonics in biological functions. In many differentiated tissues the circadian clock is the dominant pacemaker driving rhythmic metabolism which also has the potential to gate the cell cycle. Many mammalian cells show in vivo and in vitro a close to 24 h period of cell division. In vitro, i.e. in the absence of rhythmic systemic cues, the cell cycle provides sufficiently strong signals to reset the phase of the circadian clock, indicating that both systems are interconnected and mutually affect each other.1
Specific conditions such as embryonic development and tumor growth are often associated with rapid cell proliferation and doubling times that are shorter than 24 h. Since cell growth and metabolism are tightly interrelated, avoiding circadian (down-) regulation of metabolic functions might be advantageous under such circumstances. Indeed, many cancer cell lines and embryonic tissues show weak or no detectable circadian oscillations, implying that the impact of the circadian clock on cellular physiology and metabolism can be blunted or suppressed under certain conditions.2,3 The underling molecular mechanism has been missing. Cancer cells show deregulated expression of oncogenes such as RAS and MYC, which contribute to the malignant development. It has been shown that overexpression of RAS and MYC can impair circadian functions.2,4 In particular MYC is a major regulator of cell growth, which drives metabolic changes supporting proliferation of cancer cells.5
In our recent study we aimed to determine the mechanism by which MYC impacts on the circadian clock. Using inducible overexpression of MYC in rhythmic U2OS cells, we confirmed that high levels of MYC severely attenuate circadian oscillations.6 Circadian oscillations are driven by the bHLH transcription factor BMAL1/CLOCK, which activates via E-boxes expression of circadian repressors (CRYs, PERs and REV-ERBs). CRYs and PERs interact and inhibit the CLOCK/BMAL1 heterodimer, while REV-ERBs repress via RORE elements the transcription of BMAL1 and CLOCK genes. MYC is also a bHLH transcription factor and activates its target genes via E-boxes. ChIP-seq analyses revealed that MYC is recruited to the same binding sites as BMAL1/CLOCK. Hence, MYC could potentially activate transcription of the E-box-controlled negative regulators of the circadian clock. However, we could not detect a significantly increased expression of these genes. Yet, overexpression of MYC resulted in strong reduction of BMAL1 and CLOCK mRNA and protein levels. Since BMAL1 and CLOCK genes are not regulated via E-boxes, we hypothesized that MYC might repress these genes via a different pathway. Indeed, it had been shown that MYC in addition to being an activator also associates with the transcription factor MIZ1 to form a repressive complex. This repressive role of MYC is as crucial as its activating function, since MYC-induced cell proliferation and differentiation are dependent on MYC/MIZ1 complexes that downregulate inhibitors of cyclin-dependent kinases (p15INK4B, p21CIP1, etc.) and transcription factors such as CEBPs.5 In line with this, we found that overexpressed MYC is recruited to MIZ1 binding sites in the promoters of BMAL1 and CLOCK. Mutant variants of MYC (V394D and V393D) that are functional as transcription activators but cannot stably interact with MIZ1 are severely compromised in their capacity to repress the circadian clock. Furthermore, overexpression of the mutant MYC versions did no longer stimulate cell proliferation. On the opposite, RNAi-induced downregulation of MYC reduced the number of dividing U2OS cells and boosted the amplitude of circadian oscillations. Analysis of the transcriptomes of human lymphomas revealed a negative correlation between expression levels of clock genes and MYC.
Thus, our work suggests a role of MYC as a master coordinator of the cell cycle and the circadian clock (Fig. 1).
Figure 1.
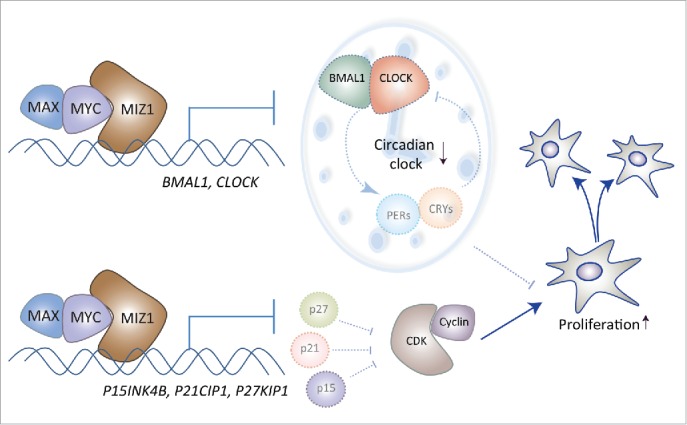
MYC coordinates the cell cycle and the circadian clock. At high levels of MYC, dimers of MYC/MAX form repressive complexes with MIZ1. These complexes inhibit transcription of cyclin-dependent kinase inhibitors (p15, p21, p27), which promotes the cell cycle. These complexes also repress BMAL1 and CLOCK and thereby attenuate clock-driven rhythmic transcriptional programs that might interfere with fast proliferation of cancer cells.
However, our experiments were conducted in vitro using a single rhythmic cancer cell line (U2OS). In these cells the activating potential of overexpressed MYC appeared to be rather low compared to its repressive potential. Particularly, overexpressed MYC did not support sustained elevated expression of the E-box-controlled negative regulators of the clock including REV-ERBα, a potent inhibitor of BMAL1 transcription. Other cell types may differ in the relative contribution of the activating and repressive limbs of MYC. This will determine to which extent the circadian clock is downregulated by MYC/MIZ1-mediated repression of BMAL1 and CLOCK or by MYC-mediated activation of CRYs, PERs and REV-ERBs. In vivo the situation is even more complex, since circadian rhythms are not strictly dependent on a functional cellular oscillator but can be driven by rhythmic systemic cues even when the cell-endogenous clock is blunted (e.g. by downregulation of BMAL1 in liver cells).7 Cancer development often involves a rather complex and heterogeneous environment. Hence, in vivo the constellation of many factors, including rhythmic systemic cues and expression of the corresponding signal transduction components in various cell types, might determine whether and by which mechanism overexpressed MYC will attenuate circadian rhythms in a tumor.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Feillet C, van der Horst GTJ, Levi F, Rand DA, Delaunay F.. Coupling between the circadian clock and cell cycle oscillators: implication for healthy cells and malignant growth. Front Neurol 2015; 6:96; PMID:26029155; http://dx.doi.org/ 10.3389/fneur.2015.00096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Relogio A, Thomas P, Medina-Pérez P, Reischl S, Bervoets S, Gloc E, Riemer P, Mang-Fatehi S, Maier B, Schäfer R, et al.. Ras-Mediated Deregulation of the circadian clock in cancer. PLoS Genet 2014; 10(5):e1004338; PMID:24875049; http://dx.doi.org/ 10.1371/journal.pgen.1004338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Yagita K, Horie K, Koinuma S, Nakamura W, Yamanaka I, Urasaki A, Shigeyoshi Y, Kawakami K, Shimada S, Uchiyama Y.. Development of the circadian oscillator during differentiation of mouse embryonic stem cells in vitro. Proc Natl Acad Sci U S A 2010; 107(8):3846-51; PMID:20133594; http://dx.doi.org/ 10.1073/pnas.0913256107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Altman BJ, Hsieh AL, Sengupta A, Krishnanaiah SY, Stine ZE, Walton ZE, Gouw AM, Venkataraman A, Li B, Goraksha-Hicks P, et al.. MYC disrupts the circadian clock and metabolism in cancer cells. Cell Metab 2015; 22(6):1009-19; PMID:26387865; http://dx.doi.org/ 10.1016/j.cmet.2015.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wiese KE, Walz S, von Eyss B, Wolf E, Athineos D, Sansom O, Eilers M.. The role of MIZ-1 in MYC-dependent tumorigenesis. Cold Spring Harb Perspect Med 2013; 3(12):a014290; PMID:24296348; http://dx.doi.org/ 10.1101/cshperspect.a014290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Shostak A, Ruppert B, Ha N, Bruns P, Toprak UH, ICGC MMML-Seq Project, Eils R, Schlesner M, Diernfellner A, Brunner M.. MYC/MIZ1-dependent gene repression inversely coordinates the circadian clock with cell cycle and proliferation. Nat Commun 2016; 7:11807; PMID:27339797; http://dx.doi.org/ 10.1038/ncomms11807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U.. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLOS Biol 2007; 5(2):e34; PMID:17298173; http://dx.doi.org/ 10.1371/journal.pbio.0050034 [DOI] [PMC free article] [PubMed] [Google Scholar]


