Abstract
Despite the extensive use of Saccharomyces cerevisiae as a platform for synthetic biology, strain engineering remains slow and laborious. Here, we employ CRISPR/Cas9 technology to build a cloning-free toolkit that addresses commonly encountered obstacles in metabolic engineering, including chromosomal integration locus and promoter selection, as well as protein localization and solubility. The toolkit includes 23 Cas9-sgRNA plasmids, 37 promoters of various strengths and temporal expression profiles, and 10 protein-localization, degradation and solubility tags. We facilitated the use of these parts via a web-based tool, that automates the generation of DNA fragments for integration. Our system builds upon existing gene editing methods in the thoroughness with which the parts are standardized and characterized, the types and number of parts available and the ease with which our methodology can be used to perform genetic edits in yeast. We demonstrated the applicability of this toolkit by optimizing the expression of a challenging but industrially important enzyme, taxadiene synthase (TXS). This approach enabled us to diagnose an issue with TXS solubility, the resolution of which yielded a 25-fold improvement in taxadiene production.
INTRODUCTION
The yeast Saccharomyces cerevisiae is an excellent organism for commercial-scale production of biological molecules due to its capacity for high-density fermentation, status as a GRAS (generally regarded as safe) organism, genetic tractability and the availability of tools for genetic engineering (1,2). However, strain development remains slow and laborious because of difficulties anticipating the combined effect of different expression parts and conditions. Although the field of synthetic biology has advanced rapidly in recent years, the assortment of expression contexts currently used for heterologous gene expression is limited and poorly characterized. Native genes, on the other hand, are controlled by a large variety of gene expression parts allowing for dynamic responses to environmental signals. Therefore, the advancement of strain engineering relies on the provision of more diverse and characterized expression parts that can be easily combined so that a large number of expression contexts can be explored.
Plasmid-based systems are routinely used in S. cerevisiae to express synthetic pathways. Specifically, 2-micron plasmids provide a convenient route for overexpression due to their high copy numbers. There are, however, significant problems associated with plasmids such as variability in plasmid copy number between cells and recombinational and segregational instability, which make it difficult to maintain stable unimodal cell populations (1,3). This heterogeneity can result subpopulations of cells that don't express the full pathway at optimal levels. Additionally, plasmid-based systems require selectable markers and media that can be too expensive for industrial applications. Genomic integration is therefore often the best method to ensure pathway stability and homogeneous expression in a population.
The recent refactoring of the type II CRISPR (Clustered Regularly InterSpaced Short Palindromic Repeats) Cas9 (CRISPR-associated protein 9) system for use in genome editing has enabled more efficient and accessible integration into the genome. In yeast, this system works by employing a Streptococcus pyogenes Cas9 endonuclease that is directed to cleave at DNA sequences specified by a non-coding single-guide RNA (sgRNA) that works by base pairing with the DNA target sequence, enabling the endonuclease to introduce a site-specific double-stranded break (DSB) in the DNA (4). The 20-nucleotide guide sequence at the 5′ end of the sgRNA specifies the cut site, and can be designed to target any site in the genome that contains a protospacer adjacent motif NGG sequence. The DSB can be repaired by homologous recombination (HR), allowing for marker-less disruption, deletion or insertion (5).
Several groups have demonstrated high-efficiency chromosomal integration of marker-less donor DNA cassette(s) in S. cerevisiae using Cas9-sgRNA plasmid(s) (1,6–8). A single DSB introduced by a sgRNA-Cas9 complex allows for integration of multiple kilobases of DNA into a locus that can be added as several PCR fragments or oligonucleotides that assemble in vivo by HR (8). Transformation of multiple sgRNAs can be used to integrate into several sites simultaneously, allowing for rapid construction of entire metabolic pathways (7–10). After chromosomal editing, strains can be cured of the Cas9-sgRNA plasmid(s) and used or modified further. Because integrations using Cas9 can be marker-less, there is no need for additional transformations or for marker recycling as is needed in the Cre/LoxP strategy (11), avoiding associated genome instability (12). Additionally, cloning in E. coli can be avoided by using PCR-generated fragments encoding new sgRNA(s) that assemble with a fixed linear backbone by HR (8). Thus, the use of Cas9 greatly reduces the time of strain construction, and vastly increases the capacity for introduction of modifications.
The improvements in strain construction pipelines afforded by CRISPR/Cas9 allow for increased throughput and flexibility of integration sequences. However, high production of target compounds from stable chromosomal integration also requires proper expression contexts to obtain maximal product levels (13–15). Unfortunately, choosing proper expression context in a systematic manner is hindered by scant characterization data, little standardization of parts and measurements (16) and laborious assembly schemes.
In this study, we developed a Cas9-based toolkit to quickly institute genetic changes in S. cerevisiae for optimizing heterologous gene expression. We characterized a range of expression programs, including exponential- or stationary-phase specific expression, subcellular protein localization to specific organelles, and altered protein turnover or solubility afforded by gene expression parts. Our toolkit contains high-efficiency, Cas9-sgRNA plasmids targeting 23 characterized integration loci, 37 promoters, 10 protein tags and a software tool to design integration cassettes. We employed our toolkit to optimize the expression of taxadiene synthase, the first committed enzyme in a pathway for the production of the anti-cancer compound paclitaxel, demonstrating the usefulness of our rapid and expanded workflow for yeast strain engineering.
MATERIALS AND METHODS
Strain and plasmid availability
Escherichia coli and S. cerevisiae strains and DNA sequences reported here (Supplementary Tables S3 and S4), along with their associated information (including annotated sequence files), have been deposited in the public instance of the JBEI Registry (17) (https://public-registry.jbei.org/folders/248) and are physically available from the authors. Because Cas9 has been implemented in many of the S. cerevisiae strains and recent literature cautions the creation of unintended gene drives upon mating of haploid yeast (18), all S. cerevisiae strains will be cured of any Cas9 plasmids prior to distribution. In order to keep the biosafety concern of creating gene drives low in this study, all S. cerevisiae strains were stored and maintained as haploids.
DNA design and construction
DNA cloning was performed using Gibson assembly (19) in E. coli, or homologous recombination in S. cerevisiae. Plasmids were recovered from S. cerevisiae by lysing the cells mechanically using glass beads, followed by plasmid mini-prep (Qiagen) and transformation of the eluted fraction into E. coli.
All pCut plasmids were derived from a pRS426 vector and have a 2 μ origin of replication and a URA3 marker. Cas9 is driven by the ADH1 promoter and CYC1 terminator. The sgRNA is driven by a tyrosine promoter with or without a HDV ribozyme structure, as indicated, and a SNR52 terminator. For the construction of the taxadiene synthase (TXS) library, the URA3 marker of the pCut plasmids was replaced by the LEU2 marker.
For characterizing the effect of promoter and integration locus on the protein expression level of GFP, promoter and terminator sequences were picked from the S. cerevisiae genome, defined as 600 bp upstream of the start codon, or 250 bp downstream of the stop codon, respectively. For the locus studies, a yeast enhanced GFP (yeGFP) cassette, driven by a TEF1 promoter and an ADH1 terminator, was integrated into each of the sites. For the promoter studies, a short-half life UbiM-GFP (20) was integrated into the ARS1021b site, preceded by each of the various promoters and followed by the ADH1 terminator.
Our CASdesigner web tool was used to design primers to create donor DNA fragments with 30–60 bp inter-fragment homology and 1-kbp flanking regions homologous to the respective target site. BY4742 genomic DNA served as a template to generate all flanking regions, promoters and terminator fragments. All other pieces were amplified from plasmids or from genomic DNA of previously integrated and sequence confirmed strains.
Strains, media and cultivation conditions
The parent S. cerevisiae strain used for integrating and characterization of loci, promoters and tags on GFP was BY4742 {MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0}. The parent S. cerevisiae strain used for integrating and characterizing TXS variants was GTy116 {MATa, leu2-3, 112::HIS3MX6-GAL1p-ERG19/GAL10p-ERG8; ura3-52::URA3-GAL1p-MvaSA110G/GAL10p-MvaE (codon optimized); his3Δ1::hphMX4-GAL1p-ERG12/GAL10p-IDI1; trp1-289::TRP1_GAL1p-CrtE(X.den)/GAL10p-ERG20; YPRCdelta15::NatMX-GAL1p-CrtE(opt)/GAL10p-CrtE}.
Promoter and integration locus characterization experiments were conducted in synthetic defined (SD, 0.67% (w/v) yeast nitrogen base without amino acids (VWR International), 0.2% (w/v) complete supplement mixture w/o yeast nitrogen base (Sunrise Science Products)) or standard rich media (YP, 1% (w/v) Bacto yeast extract, 2% (w/v) Bacto peptone) with 2% (w/v) sugar). Pre-cultures for taxadiene production were grown in YP-raffinose (YP supplemented with 2% (w/v) raffinose.). Production of taxadiene was performed in YP-galactose (YP supplemented with 2% (w/v) galactose). E. coli DH10b, used for cloning and plasmid amplification, were grown in LB supplemented with 100 μg/ml carbenicillin.
S. cerevisiae transformation and integration confirmation
S. cerevisiae were transformed by electroporation based on a previously published protocol (21), using ∼2 × 108 yeast cells, 100 ng of plasmid and 1 μg of donor DNA for each transformation. When using multiple pieces of donor DNA, equimolar amounts of the fragments were used to a total of 1 μg. When performing gap-repair plasmid cloning, 100 ng of plasmid backbone and 200 ng of DNA encoding the sgRNA with 30 bp homology to the backbone on either side were used for transformation.
Saturated overnight cultures of S. cerevisiae were diluted to an OD600 of 0.15, and incubated at 30°C, shaking at 250 rpm until cells reached an OD600 of 0.8 to 1.2. The cultures were centrifuged for 3 min at 3000 rpm, and the cells were washed once with 45 ml ice-cold water and once with 45 ml ice-cold yZap buffer (1 M Sorbitol, 10 mM CaCl2), before they were resuspended in 20 ml yCondition buffer (0.1 M LiAc, 1 mM DTT) and incubated for 30 min at 30°C, shaking at 250 rpm. Afterward, the cells were centrifuged again for 3 min at 3000 rpm and washed once more with 45 ml ice-cold yZap buffer. The resulting cell pellets were resupended in n × 200 μl ice-cold yZap buffer (n = number of transformations) and stored on ice until use.
A total of 200 μl of cells were added to microcentrifugation tubes containing the transforming DNA. The DNA – cell suspension was added to pre-chilled 2 mm electroporation cuvettes and electroporated at 2.5 kV, 200 mA and 25 μF. Immediately after electroporation, samples were resuspended in 0.5 ml YPD. After incubation at 30°C at 200 rpm for 30 min, cells were centrifuged at 10 000 rpm for 1 min and the cells were resuspended in 100 μl double distilled H2O and plated on selective media plates. Plates were incubated at 30°C for 2–3 days, until colonies became visible.
Upon appearance of colonies, integration at the intended target site was confirmed via directed colony PCR prior to measurement of GFP or taxadiene levels. Additionally, all taxadiene strains were completely sequenced at the integrated locus to confirm that the integrated cassette(s) contained no mutations.
Flow cytometry
High-throughput flow cytometry experiments were performed using the Accuri C6 flow cytometer equipped with an autosampler (BD). Cells were cultivated in 500 μl YPD, CSM or YP-galactose in 96 deep-well plates at 30°C, 250 rpm for 24 h prior to being diluted to OD600 0.1 in 500 μl of the respective medium and cultivated under the same conditions until fluorescence measurements were performed in biological triplicate. Immediately before starting the fluorescence measurements, 1 drop of SPHEROTM Rainbow Calibration Particles (8 peaks, Spherotech, Catalog No: RCP-30-5A, Lot No: AG01) was added to 100 μl of the respective media in an empty well, dispersed and measured along site the samples to generate a standard curve and allow the calculation of molecules of equivalent Fluorescein (MEFL) values (16). A total of 30 000 events were recorded at a flow rate of 66 μl/min, and a core size of 22 μm. GFP was excited at 478 nm at 70 mW and emission detected at 530 nm. Data acquisition was performed as described in the Accuri C6 Sampler User's Guide. The acquired data were analyzed in the Accuri C6 Sampler software.
Taxadiene production
Strains were grown overnight at 30°C in 200 μl YP-raffinose, and the resulting cultures were used to inoculate (to an OD600nm of 0.05) 5 ml YP-galactose production cultures. The production cultures were overlaid with 10% (v/v) dodecane spiked with an internal standard of 200 mg/l C19 methyl ester (Sigma). A total of 10 μl of the dodecane overlay was mixed with 90 μl of ethyl acetate and analyzed using GC–MS and a previously described method (22) with modifications as follows. An initial temperature of 120°C was maintained for 3 min, followed by ramping to 250°C at a rate of 20°C/min to 250°C, and then held at 250°C for another 3 min. The total flow was set to 8.3 ml/min and helium flow was set to 1 ml/min. Taxadiene production measurements were performed in biological triplicates or quadruplicates. A standard containing known concentrations if the internal standard and taxadiene obtained from Phil Baran was used to determine titer.
Integration efficiency
A total of 100 ng pCut plasmid was co-transformed with 1 μg of the respective donor DNA as a three-piece gap repair (two 300 ng 1 kbp homology regions and 400 ng GFP cassette) in biological triplicate. Unless otherwise indicated, 48 clones from each transformation replicate were analyzed for GFP expression using the Accuri C6 high-throughput flow cytometer as described above, resulting in 144 clones analyzed in total for each integration locus.
Growth rate experiments
Saturated overnight cultures of S. cerevisiae grown in 500 μl YPD (1% (w/v) Bacto Yeast Extract, 2% (w/v) Bacto Peptone, 2% (w/ v) Dextrose) were diluted to an OD600 of 0.1 in 500 μl YPD in a 96 deep-well plate and cultivated at 30°C with shaking at 250 rpm. Samples were taken at 0, 4, 8, 12, 24 and 48 h post dilution. OD600 was monitored using a Synergy 4 (BioTek), preheated to 30°C.
High performance liquid chromatography (HPLC) analysis
Sugar and metabolite concentrations were quantified on a 1200 series HPLC (Agilent Technologies) equipped with an Aminex H column. Samples were filtered through 0.45 μm filters (VWR) to remove cells, and 5 μl of each sample was injected onto the column, preheated to 50°C. The column was eluted with 4 mM H2SO4 at a flow rate of 600 μl/min for 25 min. Sugars and metabolites were monitored by a refractive index detector, and concentrations were calculated by peak area comparison to known standards.
Experiment data availability
All flow cytometry and gas chromatography mass spectrometry data are available from the JBEI Experimental Data Depot (https://public-edd.jbei.org).
CASdesigner implementation, licensing and availability
CASdesigner is implemented in python 3.4 and third party libraries including biopython (http://biopython.org), pandas (http://pandas.org) and Intermine (http://intermine.org). CASdesigner is open-source software under the BSD license (https://opensource.org/licenses/BSD-2-Clause), is freely available from GitHub (https://github.com/JBEI/CASdesigner) and is also available through its web interface on the public CASdesigner webserver (http://casdesigner.jbei.org).
RESULTS
Chromosomal integration using Cas9–sgRNA plasmids and toolkit-generated donor DNA
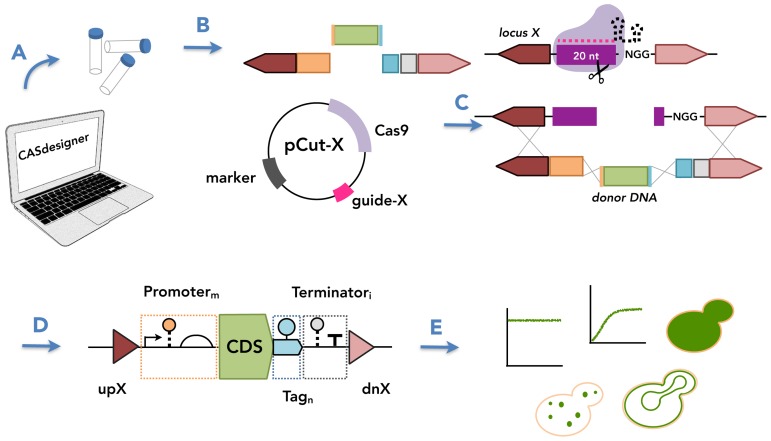
Our approach to chromosomal integration employs marker-less donor DNA and a single high-copy plasmid expressing both Cas9 and a sgRNA, henceforth referred to as pCut-X. In our system, Cas9 is expressed using an ADH1 promoter and a CYC1 terminator; the sgRNA using a tyrosine tRNA promoter (fused to a ribozyme as described in (1)) and an SNR52 terminator. Our CASdesigner web tool (described later) was used to design primers to create donor DNA fragments with 30–60 bp inter-fragment homology and 1-kbp flanking regions homologous to the individual chromosomal target site (Figure 1).
Figure 1.
Cas9-based toolkit for programming gene expression in S. cerevisiae. The toolkit facilitates strain construction by providing characterized genetic parts that can be used to program gene expression. (A) CASdesigner web-based software assists the researcher in selecting genetic parts based on the data reported herein and automatically designs the primers required to (B) generate donor DNA fragments with 30–60 bp inter-fragment and 1 kbp chromosomal homology. The homology regions and a Cas9–sgRNA plasmid (pCut-X) together specify the chromosomal integration site. (C) The sgRNA contains a 20-nt guide sequence also present in the targeted chromosomal locus (cut site). The cut site is cleaved by the sgRNA–Cas9 complex, then repaired by donor DNA via HR. (D) The choice of parts can confer different biological activities, here represented using Synthetic Biology Open Language (SBOL) Visual glyphs (triangles, HR regions; half-circle, ribosome binding site; pentagons pointing to the right, protein-coding sequences; T, terminator; arrow, promoter; circle connected by dashed line, RNA stability element; circle connected by solid line, protein stability element) following established conventions (52). (E) By the choice of genetic parts, a gene of interest can be expressed under various expression programs (level, timing, subcellular localization).
Fluorescence intensity varies ∼2-fold across 23 integration sites
Chromosomal loci are associated with features such as patterns of nucleosome occupancy and histone modification, timing of DNA replication initiation and availability of transcription or other factors with known general effects on gene expression (23,24). Furthermore, the structure of the sgRNA has been shown to influence the success rate of site-specific DSB generated by Cas9 (25). While protein expression from a given site and the Cas9 cleavage efficiency mediated by a specific sgRNA are important aspects in strain engineering, there are few sgRNAs and target chromosomal sites characterized to date, limiting the wider applicability of Cas9 technology.
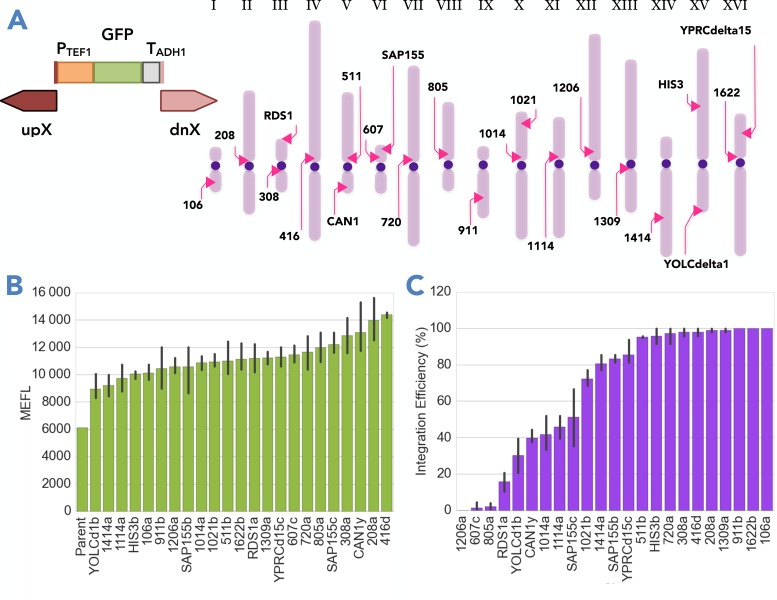
To increase the number of characterized genomic integration sites for stable expression of heterologous genes in S. cerevisiae, we generated a set of pCut plasmids and analyzed integration efficiency and protein expression from a GFP reporter cassette using flow cytometry. We chose 23 sites distributed across the 16 S. cerevisiae chromosomes, allowing flexibility and capacity for large numbers of integrations while minimizing the risk of chromosomal rearrangements when integrating into multiple loci in parallel. Because of a suggested link between high expression and replication initiation, we picked sites near several autonomously replicating sequences (ARSs) with a range of DNA replication initiation times including control sites previously reported to be high-expressing (23,26). The 20-nt guide sequences targeting these sites were chosen from a published list of sequences with minimal off-target effects (6). The sites characterized in this study are named according to the neighboring gene, followed by a letter, with sites near ARSs specified by the ARS number only (e.g. 208a).
Using our pCut plasmids, we integrated a reporter cassette encoding a green fluorescent protein (GFP, expressed using a TEF1 promoter and ADH1 terminator) into each site. The GFP donor DNA was transformed as three pieces (the GFP cassette and two 1-kb flanking homology regions) (Figure 2A), and strains were analyzed for fluorescence using flow cytometry. To extend the applicability of our toolkit for laboratory and industrially relevant conditions, we performed our assays in rich (Yeast extract Peptone Dextrose media, YPD) and minimal (Complete Supplement Media, CSM) media and at two time points representing exponential and (early) stationary phases (8 and 24 h, respectively). Because these growth phases are poorly defined, we assayed optical density at 600 nm (OD600) and extracellular metabolites HPLC for a wild type strain through time in both minimal and rich media to select sampling time points representative of the different cell growth phases (Supplementary Figure S1). In both YPD and CSM, the strains rapidly consumed glucose and were in active growth by 8 h. In YPD, the strains consumed all glucose by 24 h, reaching an OD600 of 8.4. In CSM, the strains grew more slowly and only reached an OD600 of 3.6, but nevertheless consumed the majority of glucose by 24 h. We therefore define the cell growth phase at 8 h of cultivation after back dilution as exponential phase, and 24 h as stationary phase.
Figure 2.
Effect of chromosomal locus upon integration efficiency and reporter protein expression. We integrated a GFP reporter cassette into 23 chromosomal sites of S. cerevisiae to analyze the integration efficiency and reporter protein expression associated with each locus. (A) The GFP reporter cassette (PTEF1-GFP-TADH1) was integrated into each site (indicated by pink arrows) using a Cas9-sgRNA plasmid (pCut).(B) GFP fluorescence levels are reported in NIST-standard molecules of equivalent Fluorescein (MEFL) (16) shown in this plot for strains grown in YPD at 8 h. Data for all other conditions are available in Supplementary Figure S2. Parent is a BY4742 wild-type strain. The bars represent the mean, and the error bars the standard deviation, for three to four biological replicates (independently derived colonies). (C) Integration efficiency for each site was quantified from three separate transformations, using three fragments with 1-kbp homology to the target site and 30–60 bp inter-fragment homology as donor DNA. Bars represent average percent of positive GFP integrants of 48 clones from each transformation replicate; error bars standard deviation, from three separate transformations.
In general, we found minimal variation in fluorescence intensity across all sites within a given condition (Figure 2B and Supplementary Table S1). Overall, the integration of GFP into the various sites led to tight distributions of fluorescence intensities for a given population as well as among different biological replicates (CV < 0.1 among means of different replicates). Within a given medium, we observed a moderate decrease in GFP expression as strains transitioned from exponential into stationary phase (3.7-fold in YPD, 1.9 in CSM). Medium-dependent differences in fluorescence intensity can be observed for the sites in exponential phase: in most cases fluorescence intensity was higher in YPD, however, 607c and 1414a showed the opposite trend, and are the only sites conferring slightly higher fluorescence levels in CSM as compared to YPD. This result could be due to dynamic regulation in response to environmental signals, highlighting the need to characterize integration sites in different conditions. However, several sites stood out as having higher-than-average expression in all or most conditions tested (208a, 1622b, YPRCdelta15c), and include the highest-expression site reported in a previous study, YPRCdelta15 (23).
The slower growth, lower consumption of sugar and overall diminished fluorescence from integrated GFP cassettes in CSM medium supports that CSM may not be a good medium for bioproduction, and reiterates the need for genomic integration of pathway genes so that selective medium is not needed.
Integration efficiency varies by cut site and sgRNA sequence
To explore the impact of the target locus and sgRNA on integration efficiency, we evaluated the rate of positive integrations of a GFP cassette into each site using flow cytometry. For each locus, we performed transformations in triplicate, maintaining the same three-piece design of donor DNA as previously described (Figure 2A), and identified positive integrants through analysis of fluorescence intensity.
We observed high integration efficiencies with the majority of our pCut plasmids (Figure 2C). For 13 sites, integration of the GFP cassette was detected in 80% or more of the colonies tested; for 10 of these, in 95% or more. Several of these sites also conferred higher-than-average GFP expression levels (208a, 308a, 416d, Figure 2 and Supplementary Table S1). However, in five sites GFP expression was detected in fewer than 40% of the colonies tested. Two of these poor-efficiency sites (RDS1a, YOLCdelta1b) are located near telomeres, which are known to be associated with heterochromatin domains that are inaccessible to genomic integration (27). However, the other three low-efficiency sites showed no obvious ominous locus features (607c, 805a, 1206a), leading us to hypothesize that the sgRNAs targeting these sites may be the underlying issue.
The sgRNA expression cassette within pCut contains a tyrosine tRNA promoter fused to a self-cleaving, genomic hepatitis delta virus C ribozyme. The addition of the ribozyme to the sgRNA cassette has been shown to increase sgRNA transcript levels (1). While Ryan et al. achieved high integration efficiencies using a ribozyme fused to the 5′ end of their selected sgRNAs, other groups have shown similar results without a ribozyme (7,8). To test whether the ribozyme fusion negatively impacted the effectiveness of the low-efficiency pCut plasmids in our set, we cloned any plasmid with lower than 40% integration efficiency without the ribozyme (pCut-r, for sites RDS1a, 1206a, 805a, 607c) and performed integration efficiency tests with GFP as the reporter as described earlier. For each of these sites, integration efficiency improved at least 2.5-fold when using pCut-r compared to pCut (Supplementary Figure S3). For site 805a, integration efficiency using pCut-r significantly improved to above 90%. Sites 1206a and 607c, however, remained low-efficiency, suggesting that the guide sequences are ineffective or that the target regions are inaccessible, and therefore should not be used for strain engineering.
While the 23 cut sites were chosen from a BY S. cerevisiae strain background, all 23 sites are also present in both CEN.PK and W303 backgrounds with two exceptions: (i) 208a is mutated at the fourth nucleotide in CEN.PK, and (ii) His3b is missing in W303. Therefore, in addition to the two low efficiency sites, 208a should not be used for strain engineering in a CEN.PK background and His3b should be avoided when working in W303.
Previously, it has been shown that Cas9–sgRNA plasmid(s) can be transformed as a linear backbone and PCR-generated fragment(s) containing the sgRNA(s) (8). To assess the possibility of targeting additional chromosomal sites while maintaining a completely cloning-free toolkit, we compared the integration efficiency of our system using in vivo gap repair of a linear backbone and sgRNA with 30–60 bp homology to that obtained using a pre-cloned pCut plasmid for two of our high-efficiency sites (100% efficiency with pre-cloned pCut, 106a and 911b). While colony numbers decreased by 30% when using in vivo gap repair, integration efficiency only decreased by 15% (Supplementary Figure S4). Because the backbone can be generated ahead of time, and the sgRNA quickly in one round of PCR, this strategy allows for integrating cassettes without cloning any plasmids, and retains our rapid pipeline even for targeting a new chromosomal site.
To assess the impact of integrating a standardized cassette into the different loci characterized in this study on cell physiology, we tested each GFP integrated strain (as previously described above) for growth defects over the course of 48 h in YPD. None of the strains showed any growth deficiencies in YPD media as compared to the parental wild-type strain (data not shown). We also verified that the selected cut sites did not lie in any 5′ or 3′ UTRs of neighboring genes to decrease the risk of unintentionally affecting regulatory processes (28). No cut sites were found in any UTRs, but four sites do cut in genes (208a-ECM15, SAP155b-SAP155, CAN1y-CAN1 and 1114a-YKR05c). The cut sites 208a, and 1114a lie in putative genes or sequences that code for non-essential proteins of unknown function. The SAP155b sgRNA guides Cas9 to cut in a protein that functions with the Sit4 phosphatase. The cut site Can1y is located in an arginine permease gene and was selected as a control because it was studied in a previous investigation (6). To aid users in cut site choice, annotated gene bank files of the 2.5 kb genomic region surrounding each cut site can be found as an attachment to each pCut plasmid file on the JBEI Registry (https://public-registry.jbei.org/folders/248).
Standardized promoters follow a wide range of expression levels at various growth phases and media
In yeast, the term ‘promoter’ is frequently used to refer to sequences upstream of a protein-coding gene including the 5′ untranslated region, by arbitrary demarcation. These regions can affect transcription as well as mRNA subcellular localization (29) and stability (30), and thus are major determinants of gene expression. Native promoters are known to mediate a wide range of expression levels (31,32) and condition-specific activities, and can additionally be easily amplified from genomic DNA. However, there is little agreement on what sequences constitute a promoter and little characterization data in different commonly used media, or in growth phases other than exponential. Because we expected they would allow a wide set of expression programs, we made native yeast promoters the largest set of gene expression parts in the toolkit.
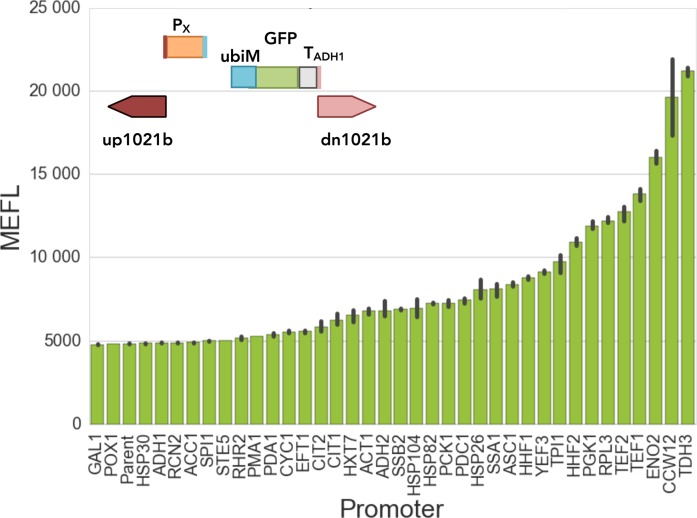
We selected 37 native promoters expected to cover various expression levels and programs (e.g. glycolysis, TCA cycle, stress response) based on the ontological annotations of their genes. We defined each promoter as the 600 bp upstream of the ATG start site of its corresponding coding sequence, observing that these regions generally contain all or most regulatory sequences controlling transcription (33). Following a standardized assembly scheme, we integrated these promoter sequences into a fixed locus (1021b) using a fixed reporter cassette. Because long-lived reporters could mask decreases in transcriptional activity, we used an N-degron GFP (UbiM-GFP) with a protein half-life of ∼1.5 h (versus >36 h for wild type GFP) (20) as our reporter. Expecting possible temporal changes in expression from our promoter strains, we assayed GFP fluorescence via flow cytometry at additional time points (4, 8, 24 and 48 h) compared to the previous experiment (8 and 24 h). We also assayed activity in Yeast extract Peptone Galactose medium (YPG), which is used to induce the widely used PGAL1 promoter, to examine how the latter and other promoters behaved in these conditions.
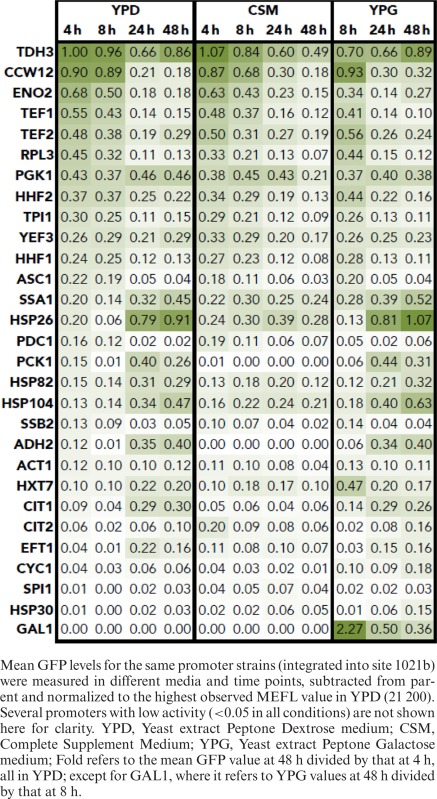
Generally, all promoters led to tight unimodal GFP fluorescence distributions (CV < 0.1 among means of different replicates, <1 among cells within a biological sample) with means spanning a ∼100-fold dynamic range, differing by media and time (Figure 3 and Table 1). In most conditions, PTDH3 (a widely used, high-activity promoter from a glycolytic enzyme) and several other promoters associated with glycolysis and translation (PTEF1, PPGK1, PTEF2, PENO2) produced the highest fluorescence levels, matching previous reports (31). Of these, PPGK1 provided the most constant activity through time and media. The highest fluorescence levels were observed for PGAL1 (natively driving the gene encoding galactokinase), but only in rich galactose medium at 8 h. In these conditions, PGAL1 fluorescence was 2.3-fold higher than that of the second most active promoter (PTDH3). However, fluorescence from PGAL1 rapidly decreased ∼6.3-fold during stationary phase (going from 8- to 48-h). Other promoters with high activity in exponential phase that decreased at later culture times (<0.3-fold from 4- to 48-h) are PCCW12, PTEF1 and PENO2 (from a cell wall mannoprotein, a translation factor and a glycolytic enzyme, respectively). PGAL1 and PTEF1 are often employed in synthetic biology, yet our data show these promoters are minimally active in stationary phase in the media tested. Fortunately, our screen found promoters more appropriate for expression in later growth phases.
Figure 3.
Promoters allow a range of expression levels and profiles across different media and growth phases. We examined 37 promoters driving the expression of a short half-life GFP reporter (20) integrated into a fixed chromosomal site in various conditions. The GFP reporter was fused to an N-degron tag containing ubiquitin (Ubi) followed by a methionine residue. GFP fluorescence levels of promoters in cells growing in YPD medium at exponential phase (4 h after dilution) are reported in NIST-standard MEFL units (16). Data for all other conditions are available in Supplementary Figure S5 and Table 1. The bars represent the mean, and the error bars the standard deviation, for three to four biological replicates.
Table 1. Reporter GFP expression from characterized promoters in different conditions.
|
In stationary phase, we observed several promoters that are induced (>3-fold from 4- to 48-h), including PHSP26, PHSP104 and PSSA1, all annotated to regulate stress-response genes (Table 1). PHSP26, which yielded some of the highest fluorescence we observed in any condition or time (>24 h in rich media), has been observed to mediate cytosolic localization and active translation upon glucose starvation, when most other transcripts are sequestered in non-translating foci (29). To our knowledge, this promoter has never been used in a synthetic biology application. Promoters silenced in stationary phase (<0.3-fold from 4- to 48-h) include PGAL1 (induced only in galactose media), PCCW12, PTEF1 and PENO2 (from a cell wall mannoprotein, a translation factor and a glycolytic enzyme, respectively).
Developing a set of localization and other protein tags using a modular linker
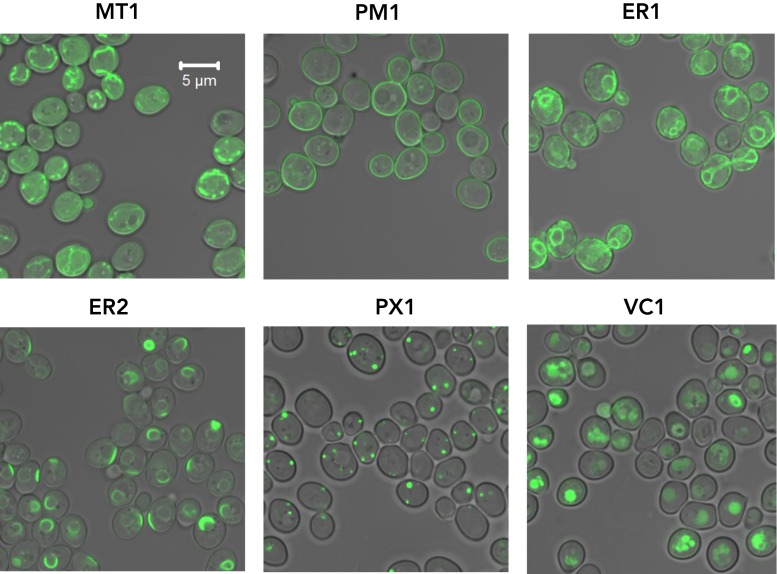
Protein motifs are known to affect important parameters such as stability and subcellular localization. Subcellular targeting, for example, is observed for several native metabolic pathways (e.g. TCA cycle, β-oxidation), exposing an enzyme to different conditions such as concentrations of substrates (or inhibitors), pH or redox cofactors. Localization tags from native genes have been used to target heterologous (or native) enzymes to specific organelles as a way to increase local enzyme concentration and improve titers (34,35). However, fusion tags can be laborious to design and assemble since they require understanding of protein trafficking and are therefore not used to their fullest extent in synthetic biology. To alleviate this obstacle, we developed a modular framework through which a validated tag can be attached, N- or C- terminally, to a target protein using a flexible six-glycine linker (gly6). Based on previous reports, we chose tags targeting various subcellular locations (20,34,36–41) to attach to a reporter GFP to validate their activity (Supplementary Table S2). We constructed these localized-GFP strains using a fixed expression context (site 208a, PTEF1 and TADH1) and visualized localization by confocal microscopy.
Gratifyingly, all but one of our tags led to the expected subcellular localization pattern (Figure 4). The only exception was the vacuolar tag VC1, which appeared to interfere with vesicle trafficking. For tag ER2, we observed GFP localization patterns resembling proliferations of the ER membrane, which have been observed after overexpression of ER proteins such as HMG-CoA reductase (42). Because these tags conferred their native localization on GFPp, the data suggest that they can be used to specify the subcellular localization of heterologous proteins of interest.
Figure 4.
Subcellular localization of GFP can be modified using protein tags. We examined the effect of localization tags fused through a universal six-glycine linker, N- or C- terminally, to a GFP reporter integrated into chromosomal site 208a. Subcellular localization patterns of GFP appended to different localization tags (Supplementary Table S2) as imaged by confocal fluorescence microscopy.
Solubility and N-degron tags change the expression level of GFP
When expressing heterologous proteins, insolubility and accumulation in inclusion bodies resulting in inactivity is a common problem. It has been shown that fusion to a soluble protein can often improve solubility. In E. coli, proteins such as the maltose binding protein (MBP) are routinely fused to heterologous proteins to improve solubility. However, to our knowledge, MBP has not been tried as a solubility tag in yeast. We therefore tested this by attaching a yeast codon-optimized version of the E. coli MBP coding sequence to the C-terminus of GFP. This led to an 18% increase in GFP fluorescence levels as measured by flow cytometry (data not shown).
In addition to solubility, the activity of a heterologous protein is affected by its stability. Earlier we utilized an N-degron tag in our promoter studies that modulates protein degradation rate by the N-end Rule (NER, (43)). Linear fusions of Ubi to target proteins are cotranslationally cleaved by deubiquitylases at the last residue of Ubi, making it possible to dictate the rate of protein degradation by producing, in vivo, different residues at the N-termini of otherwise identical proteins (44). Residues such as glutamate are known to be destabilizing by the NER; ones such as serine stabilizing. These N-degron tags can be used to alter the steady-state expression level of a protein of interest while maintaining the regulatory dynamics afforded by a given promoter or terminator.
CASdesigner web-based software facilitates characterization data visualization and semi-automated donor DNA design and construction
To simplify the workflow of constructing integration cassettes based on our characterization data, we developed a web tool, Computer-Aided Strain designer (CASdesigner, http://casdesigner.jbei.org). Our python-based tool guides users with simple in-line instructions to choose parts and automatically designs the primers required to construct donor DNA. CASdesigner can be used to design cassettes targeting any of our characterized chromosomal sites, using any of our promoters or tags. Terminators or promoters for any native S. cerevisiae gene not included in this study can also be retrieved and incorporated into the design. In all cases, promoters are defined as the 600 bp upstream of the start codon, and terminators as the 250 bp downstream of the stop codon. CASdesigner also allows targeting any native gene by its common name, allowing deletions or replacements. Tools for choosing sgRNA sequences are reported elsewhere (45). See Materials and Methods for CASdesigner implementation details, licensing and availability.
Building an expression context library for a problematic enzyme, taxadiene synthase
Our toolkit can be used to express proteins and pathways with a diverse set of programs, including exponential- or stationary-phase specific expression, subcellular targeting to various organelles and altered protein turnover or solubility. As a demonstration of its utility, we use this context-diversity approach that employs an exploratory set of parts aimed at solving common expression problems to quickly diagnose and address poor activity of an enzyme.
For our test enzyme, we chose TXS that converts the isoprenoid pathway intermediate geranylgeranyl pyrophosphate (GGPP) into taxa-4(5),11(12)-diene, the first committed intermediate in a pathway producing paclitaxel, a powerful and widely used cancer drug (46) (Figure 5A). Both paclitaxel and taxadiene have only been identified in coniferous trees of the genus Taxus. Because tree harvesting is unsustainable, there have been a number of attempts to build paclitaxel pathways in S. cerevisiae over the last decade, although titers remain <10 mg/l (47).
Figure 5.
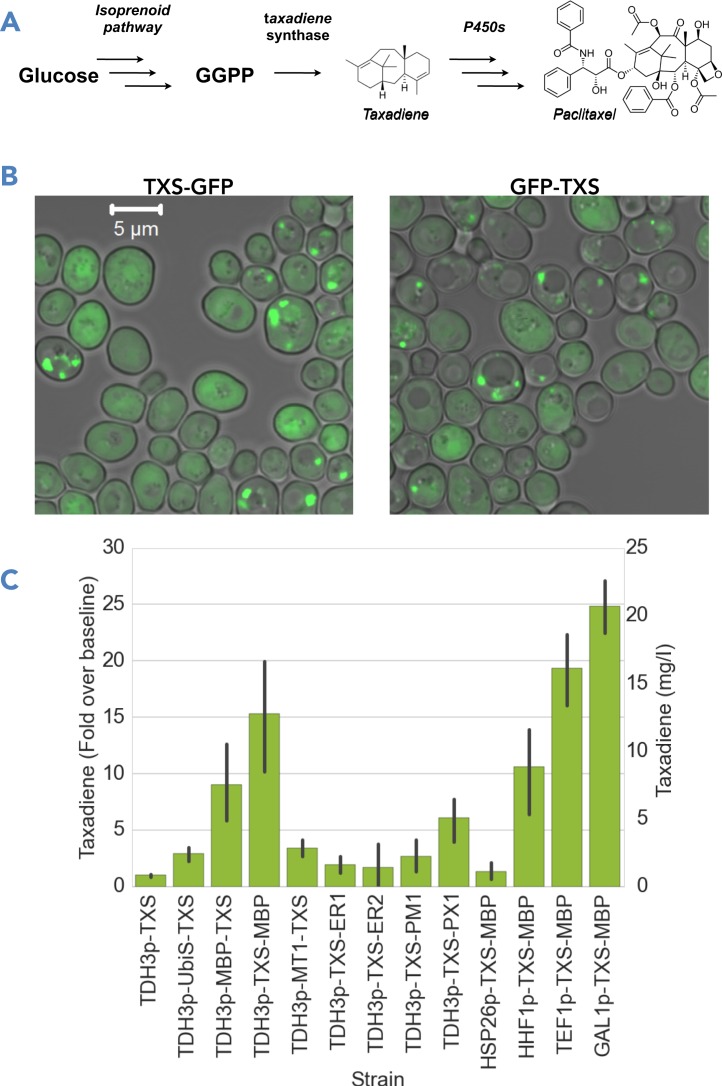
Diversity context library to optimize Taxadiene Synthase activity. (A) Taxadiene synthase (TXS) catalyzes the cyclization of geranylgeranyl-pyrophosphate (GGPP) into taxadiene, an intermediate in a pathway to produce the cancer drug Paclitaxel. (B) C- and N-terminal GFP tags attached to TXS show punctate subcellular localization consistent with poor TXS solubility. (C) Gas chromatography-mass spectroscopy (GC-MS) analysis was used to quantify the production of taxadiene in strains expressing TXS in our diversity-context library. Shown is the fold-change in taxadiene titer over the production of the baseline strain (untagged PTDH3–TXS). The bars represent the mean, and the error bars the standard deviation, for three to four biological replicates.
For our TXS library, we constructed strains using a representative subset of parts that explore a wide set of expression contexts, with 10 protein tags and 5 promoters (Figure 5). These include localization tags (MT1, PM1, ER1, ER2, PM1), a stabilizing UbiS tag, N- and C-terminal MBP tags for solubility and N- and C-terminal GFP tags to visualize subcellular localization. Our tags were fused to TXS using the gly6 linker, expressed from site 1014a, using PTDH3 and TADH1. We also built strains containing untagged TXS expressed from various promoters, including a low-, medium- and high-strength promoter (PHHF1, PTEF1 and PTDH3, respectively), a stress-responsive promoter (PHSP26) and an inducible promoter (PGAL1). The untagged, PTDH3-driven TXS strain served as a baseline level of taxadiene production on which to improve. All TXS cassettes were introduced into GTy116, a strain in which all enzymes in the pathway from acetyl–CoA to GGPP are overexpressed (Supplementary Figure S6).
MBP tags on TXS improve taxadiene production
First, we visualized both C- and N-terminal GFP fusions of TXS to examine protein expression and subcellular localization by confocal microscopy (Figure 5B). Both GFP fusions to TXS formed punctate foci, suggesting poor solubility. To examine if changing expression contexts led to any improvements in taxadiene production, we performed small-scale fermentations in parallel for our TXS strains and analyzed taxadiene levels by gas chromatography-mass spectroscopy (Figure 5C). The parent strain used for the construction of the taxadiene producer strains, GTy116, did not produce any detectable taxadiene (data not shown). In all our TXS strains, we observed a peak, which was not present in our parent strain, that matched the published fragmentation pattern of taxadiene (46) (Supplementary Figure S7). The baseline production level from our untagged TXS was 0.8 mg/l. Among the promoters tested on untagged TXS, none showed significant increases in taxadiene production (data not shown).
Consistent with a hypothesized solubility problem as diagnosed by confocal microscopy, the addition of MBP solubility tags led to significant improvements in titer. The highest taxadiene production was observed for TXS–MBP, at 15-fold higher than the untagged baseline strain. We hypothesized that the promoter library might be effective with a more soluble TXS, so we tested the same promoters again with TXS–MBP. We observed a trend of increasing taxadiene titer with increasing promoter strength (as measured for our GFP promoter strains in YPGal at exponential phase), reaching a 25-fold improvement for the PGAL1-TXS-MBP strain to 20 mg/l taxadiene (Figure 5C and Supplementary Figure S8).
DISCUSSION
In the present study, we describe a methodology and corresponding toolkit that allows for rapid and easy genetic modification for strain engineering in S. cerevisiae. We provide an extensive integration plasmid library allowing for high-efficiency integration into 23 characterized genomic loci, many of which allow for integration efficiencies above 95% (Figure 2C). In vivo assembly of sgRNAs into Cas9 plasmids afforded integration rates just below that of pre-cloned plasmids, allowing users to easily choose their own integration sites without the need for cloning (Supplementary Figure S4).
In our work, most integration sites led to similar GFP expression levels, with only a ∼2-fold range in protein expression (as measured by fluorescence, Figure 2B), in agreement with a recent report examining 11 integration loci (48). A previous study examining reporter protein cassettes integrated at various chromosomal sites report a slightly larger (∼10-fold) range in expression level using an assay based on β-galactosidase activity (23). Other recent reports observe that locus effects carry over to different expression cassettes, with the locus accounting for 35% (as measured by qRT-PCR) or 90% (as measured by fluorescence) of changes in observed expression, making our findings generalizable for other heterologous genes (24,49). The differences in observed expression level of the reporter protein could be accounted for by the different methods of analysis employed in these studies, as well as choice of sites within an individual study (e.g. intergenic versus intragenic), highlighting the need to use standardized measurements units to quantify expression, as we did.
While the majority of the sgRNAs in our pCut library initially resulted in high-efficiency integrations (Figure 2C), removal of the ribozyme structure from the sgRNA resulted in a dramatically increased rate of integration for one of the low-efficiency sites (805a, Supplementary Figure S3). Additionally, two other sgRNAs led to low rates of integration with or without the ribozyme (1206a and 607c). A previous study has examined the chromosomal landscape of nucleosome-dependent gene expression in yeast using a histone depletion assay, and charted expression changes for each gene post depletion (50). We examined the landscape for each of our failed cut sites and found that 1206a is more than 5 kb away from any hypothesized heterochromatic region, while 607c lies in a region of gene upregulation after depletion. These results suggest that the 607c sgRNA may not function because it lacks access to the DNA due to heterochromatic structure, whereas 1206a may have folding or other issues with the sgRNA itself.
Nonetheless, clearly factors beyond off-target effects need to be considered for optimal sgRNA design, such as sgRNA folding, transcription and degradation, which are excellent topics for future investigation. Furthermore, the impact of locus features such as chromatin structure on the efficiency of genomic integration (51) by potentially affecting accessibility to the sgRNA–Cas9p complex and HR repair machinery requires further research and elucidation.
This study is the first to integrate temporal resolution of parts activity across different conditions in a systematic manner. Much of chemical production (especially during batch fermentation) occurs in stationary phase where there is little to no characterization of promoters and other expression parts. Because we tested various growth phases and media, our investigation led to several novel findings. While PGAL1 is considered one of the highest-expressing promoters in S. cerevisiae, its activity drops significantly at later stages of growth (Table 1). Using a GFP reporter cassette, we identified six promoters that resulted in fluorescence levels higher than that of PGAL1 in stationary phase (PHSP26, PSSA1, PPCK1, PHSP82, PHSP104, PADH2). Of particular note is PHSP26, which is barely active in exponential cells yet reaches among the highest expression levels of any condition at stationary phase. Growth phase-specific activity may be desirable in certain synthetic biology designs, including strategies for growth-decoupled stationary phase production, or temporally separating enzyme activities that are incompatible.
Additionally, we identified several other high-strength promoters not examined in other studies, including PENO2, PRPL3, PTPI1 and PYEF3, as well as several stationary phase-induced promoters (PHSP26, PSSA1, PPCK1, PHSP82, PHSP104 and PADH2). A handful of the promoters chosen for this study were also tested in exponential phase in another recent investigation and rank similarly in protein expression strength (PTDH3, PCCW12, PTEF1, PTEF2, PPGK1, PHHF2, PHHF1) as determined by fluorescence (31).
The toolkit described in this work provides a rapid approach to examine multiple gene expression contexts simultaneously, thus increasing the chances of improving activity even in the absence of complete biological understanding. To demonstrate its utility, we applied this context-diversity approach to diagnose and address poor activity for the enzyme TXS (Figure 5A). Yeast TXS activity is especially desirable since many of the paclitaxel-pathway steps downstream of TXS involve cytochrome P450s, notoriously difficult to express in E. coli. Using our toolkit, we were able to show that protein solubility was limiting for TXS activity in yeast (Figure 5). By constructing TXS–MBP fusion proteins, we achieved a 25-fold improvement in taxadiene titer. MBP tags have been previously used in yeast for affinity purification, however, this is the first study to show that the MBP tag can also be used to increase protein solubility in S. cerevisiae. A similar methodology could help to identify and resolve other limiting parameters such as poor intracellular localization or protein degradation for other enzymes, and be a generalizable approach to optimize expression contexts of heterologous enzymes.
Acknowledgments
The authors thank Jake Beal for discussions on flow cytometry and standardization of measurements, as well as for assistance with flow cytometry data processing pipeline access and use. The authors also thank Leonard Katz for suggestions while preparing this manuscript, Hector Plahar, William Morrell and Jacob Coble for technical assistance, and Phil Baran for providing the taxadiene standard. This work was part of the DOE Joint BioEnergy Institute (https://www.jbei.org) and the DOE Joint Genome Institute (https://jgi.doe.gov) supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U. S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, worldwide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
Author contributions: A.R., L.d'E. and M.W. designed and conducted experiments, analyzed data and composed the manuscript. M.W., D.S. and M.G. constructed strains. G.J.T. constructed and characterized GTy116. L.d'E. and R.A.L. carried out taxadiene production runs. L.d'E., O.N, W.Z. and N.H. coded and designed the user interface for the CASdesigner tool. A.M. and J.D.K. guided the scope of the project, provided critical input for the manuscript and provided resources. All authors read and approved the final manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research [contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U. S. Department of Energy]. Funding for open access charge: Lawrence Berkeley National Laboratory [DE-AC02-05CH11231].
Conflict of interest statement. J.D.K. has a financial interest in Amyris, Lygos and LS9.
REFERENCES
- 1.Ryan O.W., Skerker J.M., Maurer M.J., Li X., Tsai J.C., Poddar S., Lee M.E., DeLoache W., Dueber J.E., Arkin A.P., et al. Selection of chromosomal DNA libraries using a multiplex CRISPR system. Elife. 2014 doi: 10.7554/eLife.03703. doi:10.7554/eLife.03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Curran K.A., Karim A.S., Gupta A., Alper H.S. Use of expression-enhancing terminators in Saccharomyces cerevisiae to increase mRNA half-life and improve gene expression control for metabolic engineering applications. Metab. Eng. 2013;19:88–97. doi: 10.1016/j.ymben.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Da Silva N.A., Srikrishnan S. Introduction and expression of genes for metabolic engineering applications in Saccharomyces cerevisiae. FEMS Yeast Res. 2012;12:197–214. doi: 10.1111/j.1567-1364.2011.00769.x. [DOI] [PubMed] [Google Scholar]
- 4.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doudna J.A., Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 6.DiCarlo J.E., Norville J.E., Mali P., Rios X., Aach J., Church G.M. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jakočiūnas T., Rajkumar A.S., Zhang J., Arsovska D., Rodriguez A., Jendresen C.B., Skjødt M.L., Nielsen A.T., Borodina I., Jensen M.K., et al. CasEMBLR: Cas9-facilitated multiloci genomic integration of in vivo assembled DNA parts in Saccharomyces cerevisiae. ACS Synth. Biol. 2015 doi: 10.1021/acssynbio.5b00007. doi:10.1021/acssynbio.5b00007. [DOI] [PubMed] [Google Scholar]
- 8.Horwitz A.A., Walter J.M., Schubert M.G., Kung S.H., Hawkins K., Platt D.M., Hernday A.D., Mahatdejkul-Meadows T., Szeto W., Chandran S.S., et al. Efficient multiplexed integration of synergistic alleles and metabolic pathways in yeasts via CRISPR-Cas. Cell Syst. 2015;1:88–96. doi: 10.1016/j.cels.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Ronda C., Maury J., Jakočiūnas T., Baallal Jacobsen S.A., Germann S.M., Harrison S.J., Borodina I., Keasling J.D., Jensen M.K., Nielsen A.T. CrEdit: CRISPR mediated multi-loci gene integration in Saccharomyces cerevisiae. Microb. Cell Fact. 2015;14:97. doi: 10.1186/s12934-015-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mans R., van Rossum H.M., Wijsman M., Backx A., Kuijpers N.G.A., van den Broek M., Daran-Lapujade P., Pronk J.T., van Maris A.J.A., Daran J.M.G. CRISPR/Cas9: A molecular swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res. 2015;15:1–15. doi: 10.1093/femsyr/fov004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gueldener U., Heinisch J., Koehler G.J., Voss D., Hegemann J.H. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solis-Escalante D., Kuijpers N.G.A., van der Linden F.H., Pronk J.T., Daran J.-M., Daran-Lapujade P., Agmon N., Liefshitz B., Zimmer C., Fabre E., et al. Efficient simultaneous excision of multiple selectable marker cassettes using I-SceI-induced double-strand DNA breaks in Saccharomyces cerevisiae. FEMS Yeast Res. 2014;14:741–754. doi: 10.1111/1567-1364.12162. [DOI] [PubMed] [Google Scholar]
- 13.Kang H.-S., Charlop-Powers Z., Brady S.F. Multiplexed CRISPR/Cas9 and TAR-mediated promoter engineering of natural product biosynthetic gene clusters in yeast. ACS Synth. Biol. 2016 doi: 10.1021/acssynbio.6b00080. doi:10.1021/acssynbio.6b00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Latimer L.N., Lee M.E., Medina-Cleghorn D., Kohnz R.A., Nomura D.K., Dueber J.E. Employing a combinatorial expression approach to characterize xylose utilization in Saccharomyces cerevisiae. Metab. Eng. 2014;25:20–29. doi: 10.1016/j.ymben.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Alonso-Gutierrez J., Kim E.-M., Batth T.S., Cho N., Hu Q., Chan L.J.G., Petzold C.J., Hillson N.J., Adams P.D., Keasling J.D., et al. Principal component analysis of proteomics (PCAP) as a tool to direct metabolic engineering. Metab. Eng. 2015;28:123–133. doi: 10.1016/j.ymben.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 16.Beal J., Haddock-Angelli T., Gershater M., De Mora K., Lizarazo M., Hollenhorst J., Rettberg R., Demling P., Hanke R., Osthege M., et al. Reproducibility of fluorescent expression from engineered biological constructs in E. coli. PLoS One. 2016;11:1–22. doi: 10.1371/journal.pone.0150182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ham T.S., Dmytriv Z., Plahar H., Chen J., Hillson N.J., Keasling J.D. Design, implementation and practice of JBEI-ICE: an open source biological part registry platform and tools. Nucleic Acids Res. 2012;40:e141. doi: 10.1093/nar/gks531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DiCarlo J.E., Chavez A., Dietz S.L., Esvelt K.M., Church G.M. Safeguarding CRISPR-Cas9 gene drives in yeast. Nat. Biotechnol. 2015;33:1250–1255. doi: 10.1038/nbt.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibson D.G., Young L., Chuang R., Venter J.C., Iii C.A.H., Smith H.O., America N. Enzymatic assembly of DNA molecules up to several hundred kilobases. 2009;6:12–16. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 20.Houser J.R., Ford E., Chatterjea S.M., Maleri S., Elston T.C., Errede B. An improved short-lived fluorescent protein transcriptional reporter for Saccharomyces cerevisiae. Yeast. 2012;29:519–530. doi: 10.1002/yea.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benatuil L., Perez J.M., Belk J., Hsieh C.-M. An improved yeast transformation method for the generation of very large human antibody libraries. Protein Eng. Des. Sel. 2010;23:155–159. doi: 10.1093/protein/gzq002. [DOI] [PubMed] [Google Scholar]
- 22.Steen E.J., Kang Y., Bokinsky G., Hu Z., Schirmer A., McClure A., Del Cardayre S.B., Keasling J.D. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature. 2010;463:559–562. doi: 10.1038/nature08721. [DOI] [PubMed] [Google Scholar]
- 23.Flagfeldt D.B., Siewers V., Huang L., Nielsen J. Characterization of chromosomal integration sites for heterologous gene expression in Saccharomyces cerevisiae. Yeast. 2009;26:545–551. doi: 10.1002/yea.1705. [DOI] [PubMed] [Google Scholar]
- 24.Chen X., Zhang J. The genomic landscape of position effects on protein expression level and noise in yeast. Cell Syst. 2016;2:1–8. doi: 10.1016/j.cels.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dang Y., Jia G., Choi J., Ma H., Anaya E., Ye C., Shankar P., Wu H., Mali P., Yang L., et al. Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome Biol. 2015;16:280. doi: 10.1186/s13059-015-0846-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamane S., Yamaoka M., Yamamoto M., Maruki T., Matsuzaki H., Hatano T., Fukui S. Region specificity of chromosome III on gene expression in the yeast Saccharomyces cerevisiae. J. Gen. Appl. Microbiol. 1998;44:275–281. doi: 10.2323/jgam.44.275. [DOI] [PubMed] [Google Scholar]
- 27.Eckert-Boulet N., Lisby M. Regulation of homologous recombination at telomeres in budding yeast. FEBS Lett. 2010;584:3696–3702. doi: 10.1016/j.febslet.2010.05.037. [DOI] [PubMed] [Google Scholar]
- 28.Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., Gerstein M., Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zid B.M., O'Shea E.K. Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature. 2014;514:117–121. doi: 10.1038/nature13578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trcek T., Larson D.R., Moldon A., Query C.C., Singer R.H. Single-molecule mRNA decay measurements reveal promoter regulated mRNA stability in yeast. Cell. 2011;147:1484–1497. doi: 10.1016/j.cell.2011.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee M.E., DeLoache W.C., Cervantes B., Dueber J.E. A highly-characterized yeast toolkit for modular, multi-part assembly. ACS Synth. Biol. 2015;4:975–986. doi: 10.1021/sb500366v. [DOI] [PubMed] [Google Scholar]
- 32.Guo Z., Zhang L., Ding Z., Shi G. Minimization of glycerol synthesis in industrial ethanol yeast without influencing its fermentation performance. Metab. Eng. 2011;13:49–59. doi: 10.1016/j.ymben.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 33.Sharon E., Kalma Y., Sharp A., Raveh-Sadka T., Levo M., Zeevi D., Keren L., Yakhini Z., Weinberger A., Segal E. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat. Biotechnol. 2012;30:521–530. doi: 10.1038/nbt.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Avalos J.L., Fink G.R., Stephanopoulos G. Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols. Nat. Biotechnol. 2013;31:335–341. doi: 10.1038/nbt.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeLoache W.C., Russ Z.N., Dueber J.E. Towards repurposing the yeast peroxisome for compartmentalizing heterologous metabolic pathways. Nat. Commun. 2016;7:11152. doi: 10.1038/ncomms11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun P., Tropea J.E., Waugh D.S. Enhancing the solubility of recombinant proteins in Escherichia coli by using hexahistidine-tagged maltose-binding protein as a fusion partner. Methods Mol. Biol. 2011;705:259–274. doi: 10.1007/978-1-61737-967-3_16. [DOI] [PubMed] [Google Scholar]
- 37.Parlati F., Dominguez M., Bergeron J.J.M., Thomas D.Y. Saccharomyces cerevisiae CNE1 Encodes an Endoplasmic Reticulum (ER) Membrane Protein with Sequence Similarity to Calnexin and Calreticulin and Functions as a Constituent of the ER Quality Control Apparatus. J. Biol. Chem. 1995;270:244–253. doi: 10.1074/jbc.270.1.244. [DOI] [PubMed] [Google Scholar]
- 38.Beilharz T., Egan B., Silver P.A., Hofmann K., Lithgow T. Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:8219–8223. doi: 10.1074/jbc.M212725200. [DOI] [PubMed] [Google Scholar]
- 39.Lewis M.J., Nichols B.J., Prescianotto-Baschong C., Riezman H., Pelham H.R.B. Specific retrieval of the exocytic SNARE snc1p from early yeast endosomes. Mol. Biol. Cell. 2000;11:23–38. doi: 10.1091/mbc.11.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma C., Hagstrom D., Polley S.G., Subramani S. Redox-regulated cargo binding and release by the peroxisomal targeting signal receptor, Pex5. J. Biol. Chem. 2013;288:27220–27231. doi: 10.1074/jbc.M113.492694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reggiori F., Black M.W., Pelham H.R. Polar transmembrane domains target proteins to the interior of the yeast vacuole. Mol. Biol. Cell. 2000;11:3737–3749. doi: 10.1091/mbc.11.11.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wright R., Basson M., D'Ari L., Rine J. Increased amounts of HMG-CoA reductase induce ‘karmellae’: A proliferation of stacked membrane pairs surrounding the yeast nucleus. J. Cell Biol. 1988;107:101–114. doi: 10.1083/jcb.107.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bachmair A., Finley D., Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;523:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 44.Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011;20:1298–1345. doi: 10.1002/pro.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montague T.G., Cruz J.M., Gagnon J.A., Church G.M., Valen E. CHOPCHOP: A CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42:401–407. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wildung M.R., Croteau R. A cDNA clone for taxadiene synthase, the diterpene cyclase that catalyzes the committed step of taxol biosynthesis. J. Biol. Chem. 1996;271:9201–9204. doi: 10.1074/jbc.271.16.9201. [DOI] [PubMed] [Google Scholar]
- 47.Engels B., Dahm P., Jennewein S. Metabolic engineering of taxadiene biosynthesis in yeast as a first step towards Taxol (Paclitaxel) production. Metab. Eng. 2008;10:201–206. doi: 10.1016/j.ymben.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 48.Jessop-Fabre M.M., Jakočiūnas T., Stovicek V., Dai Z., Jensen M.K., Keasling J.D., Borodina I. EasyClone-MarkerFree: A vector toolkit for marker-less integration of genes into Saccharomyces cerevisiae via CRISPR-Cas9. Biotechnol. J. 2016;11:1110–1117. doi: 10.1002/biot.201600147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen M., Licon K., Otsuka R., Pillus L., Ideker T. Decoupling epigenetic and genetic effects through systematic analysis of gene position. Cell Rep. 2013;3:128–137. doi: 10.1016/j.celrep.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wyrick J.J., Holstege F.C., Jennings E.G., Causton H.C., Shore D., Grunstein M., Lander E.S., Young R.A. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature. 1999;402:418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- 51.Smith J.D., Suresh S., Schlecht U., Wu M., Wagih O., Peltz G., Davis R.W., Steinmetz L.M., Parts L., St. Onge R.P. Quantitative CRISPR interference screens in yeast identify chemical-genetic interactions and new rules for guide RNA design. Genome Biol. 2016;17:45. doi: 10.1186/s13059-016-0900-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quinn J.Y., Cox R.S., Adler A., Beal J., Bhatia S., Cai Y., Chen J., Clancy K., Galdzicki M., Hillson N.J., et al. SBOL Visual: a graphical language for genetic designs. PLoS Biol. 2015;13:1–9. doi: 10.1371/journal.pbio.1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]