Abstract
Cells and viruses can utilize internal ribosome entry sites (IRES) to drive translation when cap-dependent translation is inhibited by stress or viral factors. IRES trans-acting factors (ITAFs) are known to participate in such cap-independent translation, but there are gaps in the understanding as to how ITAFs, particularly negative ITAFs, regulate IRES-driven translation. This study found that Lys109, Lys121 and Lys122 represent critical ubiquitination sites for far upstream element-binding protein 2 (KHSRP, also known as KH-type splicing regulatory protein or FBP2), a negative ITAF. Mutations at these sites subsequently reduced KHSRP ubiquitination and abolished its inhibitory effect on IRES-driven translation. We further found that interaction between the Kelch domain of Kelch-like protein 12 (KLHL12) and the C-terminal domain of KHSRP contributed to KHSRP ubiquitination, leading to downregulation of enterovirus IRES-mediated translation in infected cells and increased competition against other positive ITAFs. Together, these results show that ubiquitination can exert control over IRES-driven translation via modification of ITAFs, and to the best of our knowledge, this is the first description of such a regulatory mechanism for IRES-dependent translation.
INTRODUCTION
Most eukaryotic mRNAs undergo cap-dependent translation, but cap-independent translation processes exist as well (1), and may be utilized during viral-host interactions to hijack host translation machinery. Internal ribosome entry sites (IRES) in picornaviruses, including poliovirus (PV) and encephalomycarditis virus (EMCV), were first discovered in 1988 (2,3), and it was shown that the IRES in the 5′ untranslated region (UTR) of the picornavirus RNA genome can direct translation in a cap-independent manner. In cells infected with certain picornaviruses, viral protease 2Apro or Lpro can cleave the translation initiation factor eIF4G, causing rapid termination of most host cap-dependent translation processes (4). However, the cleavage product containing the C-terminal fragment of eIF4G can still bind eIF3 and eIF4A and recruit other eukaryotic translation initiation factors to form the 43S initiation complex, which can then recognize a sequence or RNA structure within the IRES to initiate translation at the authentic initiation codon (5).
Several cellular proteins that can bind and stabilize IRES structures and regulate IRES-driven translation have been reported, and these proteins are known as IRES trans-acting factors (ITAFs) (6). Polypyrimidine tract-binding protein 1 (PTBP1), autoantigen La (LARP), poly(rC) binding protein 2 (PCBP2), upstream of N-ras protein (Unr), serine/arginine-rich splicing factor 3 (SRSF3), nucleolin, heterogeneous nuclear ribonucleoprotein A1 (HNRNPA1), double-stranded RNA binding protein 76 (ILF3), glycyl-tRNA synthetase (GARS), AU-rich element RNA binding factor 1 (HNRNPD), far upstream element-binding protein 1 (FUBP1, also known as FBP1), and far upstream element-binding protein 2 (KHSRP, also known as KH-type splicing regulatory protein, KSRP, FUBP2, FBP2 or P75) are known ITAFs that are functionally important to picornavirus translation (7). Most of these ITAFs are positive regulators of viral translation, but ILF3, HNRNPD, and KHSRP have been found to play negative roles. ILF3 is known to inhibit human rhinovirus type 2 (HRV2) IRES-driven translation (8,9), while HNRNPD can bind to many picornaviruses, such as PV, coxsackievirus B3 (CVB3), HRV2 and enterovirus 71 (EV71) to negatively regulate viral translation (10–12). In a previous study, we also found that KHSRP inhibits EV71 translation (13). However, the exact mechanisms by which ITAFs, particularly negative ITAFs, act to regulate IRES-driven translation is worthy of further investigation, and may yield new insight regarding host-viral interactions and novel anti-viral strategies.
In this study, we used EV71 as a model to explore the negative regulatory mechanism of KHSRP in detail. EV71 is a positive single-strand RNA virus in the Picornaviridae family, and represents a potent emerging threat worldwide (14). EV71 infections normally cause mild diseases, such as hand-foot-and-mouth disease (HFMD) or herpangina. However, children under five years of age are particularly susceptible to the most severe forms of EV71-associated neurological complications, including aseptic meningitis, brainstem and/or cerebellar encephalitis, acute flaccid paralysis (AFP), myocarditis, and rapid fatal pulmonary edema and hemorrhage (15). We previously found that KHSRP is a negative regulator (13), while FUBP1 is a positive regulator of EV71 IRES-dependent translation (16). Moreover, the C-terminal of KHSRP is cleaved upon EV71 infection, and the cleaved form of KHSRP (KHSRP1–503) then becomes a positive regulator of EV71 IRES-driven translation (17). As an important adenosine–uridine element-binding protein (ARE-BP) that can interact with many AREs (18,19), KHSRP contains four K-homologous domains in its central domain, of which KH3 and KH4 are essential to promote mRNA decay (20). In addition, KHSRP is also involved in diverse biological roles, such as regulating the post-transcriptional modification of type I interferon genes to render cells impervious to herpes simplex virus type 1 (HSV1) and vesicular stomatitis virus (VSV) infection (21), or participating in miRNA biogenesis (22). Moreover, KHSRP is a component of both the Drosha and Dicer complexes, and it can bind to the terminal loop sequence of miRNA precursors to promote their maturation (23).
Ubiquitin is a 76-amino acid polypeptide that can be covalently attached to lysine residues in substrate proteins. Ubiquitination involves three enzymes that respectively govern the activation (E1), conjugation (E2) and ligation (E3) of monoubiquitin or polyubiquitin to substrate proteins, thereby altering protein function or activating ubiquitin proteasome pathways (24). An important post-translational process that regulates protein degradation and signaling in gene transcription, nuclear transport, cargo sorting, endocytosis, autophagy, DNA repair and the immune response (25), ubiquitination is known to regulate cap-dependent translation as well (26–28), but the role of ubiquitination in IRES-driven translation has not been studied extensively as yet.
In this study, we sought to understand how KHSRP acts as a negative regulator in EV71 IRES-driven translation, particularly as a similar protein, FUBP1, which differs from KHSRP in its C-terminal domain, is known to act as a positive ITAF. In virus-infected cells, KHSRP is cleaved and the resulting cleavage product (with the C-terminal domain removed) acts as a positive ITAF instead. We utilized iTRAQ-LC-MS/MS analysis to compare the KHSRP-associated proteins that differ from FUBP1 and truncated KHSRP, and thus identified Kelch-like protein 12 (KLHL12), which serves as a substrate adaptor for the cullin 3 (CUL3)-based ubiquitin–protein E3 ligase complex (29–31). Cullin-RING E3 ubiquitin ligases (CRLs) are the largest family of E3 ligases, and seven cullin proteins (CUL1, CUL2, CUL3, CUL4A/4B, CUL5, CUL7 and CUL9) are known to serve as scaffold proteins in CRLs. The C-terminal domain of cullin binds a RING finger protein (RBX1 or RNF7) that can interact with E2 to mediate ubiquitin transfer to the substrate. The N-terminal of cullin binds to different substrate adaptors to recruit different proteins (32).
We found that KLHL12 can differentially interact with KHSRP and promote KHSRP ubiquitination. We then sought to elucidate how KLHL12-mediated KHSRP ubiquitination affects IRES-driven translation. This led us to identify a hitherto undescribed regulatory mechanism of IRES-driven translation involving ubiquitination—namely, the regulation of IRES-dependent translation via ubiquitin modulation of ITAFs. Our findings are expected to have important implications for future research regarding viral-host interactions, and may also help to identify novel drug targets for anti-viral therapies.
MATERIALS AND METHODS
Plasmid construction
The pFLAG-CMV2-FUBP1, KHSRP and KHSRP1–503 plasmids were constructed as follows: cDNA from FUBP1, KHSRP and KHSRP1–503 was amplified by PCR from constructs described previously (16,17). The cDNA of FUBP1 was inserted into the NotI and EcoRV sites of pFLAG-CMV2 vector. The cDNA of KHSRP and KHSRP1–503 were subcloned between the EcoRI and EcoRV sites of pFLAG-CMV2 vector. The KHSRP mutants (residues Lys71, Lys87, Lys109, Lys121, Lys122, Lys251, Lys628, Lys646 and Lys654 were replaced by arginine) were generated by site-directed mutagenesis. KLHL12 was amplified by RT-PCR from the RNA of RD cells. Cloned human KLHL12 cDNA was verified by sequencing and confirmed to be the same as that deposited in GenBank (NM_021633). The cDNA of KLHL12, KLHL12ΔBTB and KLHL12ΔKelch were subcloned to the EcoRI and KpnI sites of the pCMV-HA or pcDNA3.1/myc-His (+) A vector. pcDNA3-HA-Ub (WT) and Ub (KO) were provided by Dr Rei-Lin Kuo and Dr. Chen Zhao. pcDNA3-Myc-CUL3 and pcDNA3-Myc-CUL3ΔRBX1 were gifts from Dr Hsiu-Ming Shih. GST-CUL3 and pcDNA3-3×Myc-RBX1 were gifts from Dr Ruey-Hwa Chen. pCRII-TOPO-EV71 5′ UTR and pGL3-EV71 5′ UTR-FLuc were constructed as previously described (13,16). EV71 replicon 3DD330A was mutated from the EV71 replicon by site-directed mutagenesis as previously described (17).
Antibodies
Anti-FLAG M2 (F3165), anti-Myc (M4439) and anti-HA (H9658) antibodies were purchased from Sigma-Aldrich. Anti-KLHL12 (GTX83228), anti-CUL3 (GTX62065), anti-cyclin B1 (GTX100911), anti-β-catenin (GTX101435) and anti-streptavidin HRP (GTX85912) antibodies were obtained from GeneTex. Anti-actin (MAB1501) antibody was purchased from Merck Millipore. Anti-KHSRP (A302-021A) antibody was purchased from Bethyl Laboratories. Anti-PTBP1 (sc-16547) antibody was obtained from Santa Cruz Biotechnology. Anti-His (OB05) antibody was purchased from CalBioChem. Anti-PCBP2 antibody was a gift from Dr Ian Goodfellow. Anti-EV71 2B antibody was provided by Dr Jim-Tong Horng.
Cell culture and virus infection
Human muscle rhabdomyosarcoma (RD) and 293T cells were cultured at 37°C in Dulbecco's modified Eagle's medium (DMEM; Gibco) containing 10% fetal bovine serum (FBS; Gibco). RD cells with 80–90% confluency were challenged with EV71 (strain Tainan/4643/98) at a multiplicity of infection (MOI) of 10 PFU per cell. After 1 h of adsorption at 37°C in serum-free DMEM, cells were washed with PBS and incubated with DMEM containing 2% FBS. Stable RD cell lines were generated by lentivirus transduction.
Lentiviral vector preparation
The two shRNA constructs used for knockdown of KHSRP are described as follows: the sequence targeting nt 813–837 of human KHSRP mRNA (shKHSRP #1, 5′-CACATTCGTATTCTGAGATCCGTCC-3′) was constructed in the pLKO_TRC005 vector, and shKHSRP #2 (TRCN0000013253) was purchased from the Taiwan National RNAi Core Facility, Academia Sinica. The pLKO.1-shLacZ control plasmid used was TRCN0000072224 (shLacZ). For lentivirus preparation, X-treme GENE transfection reagent (Roche) was used in 293T cells with different shRNA constructs and two helper plasmids, pMD.G and pCMVΔR8.91. The culture supernatants containing the viral particles were harvested, and then the RD cells were transduced with shKHSRP lentivirus for 24 h and selected with puromycin (5 μg/ml). The procedure was conducted in accordance with the recommended protocol on the Taiwan National RNAi Core Facility website (http://rnai.genmed.sinica.edu.tw/webContent/web/protocols/wicket:pageMapName/wicket-0).
Protein identification and iTRAQ LC/LC mass spectrometry
Details are described in the Supplementary Data.
Immunoprecipitation assay
RD or 293T cells were transfected with plasmids from the X-treme GENE transfection reagent (Roche) according to the manufacturer's instructions. After 48 h post-transfection, cells were harvested using lysis buffer (50 mM Tris–HCl, pH 7.4, with 150 mM NaCl, 1 mM EDTA, and 1% Triton-X-100), placed for 30 min on ice, centrifuged at 12 000 × g for 10 min, and subsequently incubated with Anti-FLAG M2 affinity gel (Sigma) for 16 h at 4°C. The Anti-FLAG M2 affinity gel was washed five times with wash buffer (50 mM Tris–HCl, pH 7.4, with 150 mM NaCl). The immunoprecipitation complex was eluted by 2× sample buffer (125 mM Tris–HCl, pH 6.8, with 4% SDS, 20% (v/v) glycerol, and 0.004% bromophenol blue) or in competition with 3× FLAG peptides. Bound proteins were subjected to SDS-PAGE and analyzed by western blotting using specific antibodies. For the in vivo ubiquitination assay, 293T cells were co-transfected with ubiquitin fused with HA tags (HA-Ub), and treated with 20 μM MG132 (Sigma) for 4 h before harvesting. Cells were lysed with lysis buffer containing 5 mM of N-ethylmaleimide (Sigma), and immunoprecipitated with Anti-FLAG M2 affinity gel. The ubiquitinated proteins were detected by western blot using anti-HA antibodies.
In vitro transcription
T7-EV71 5′ UTR was excised from the pCRII-TOPO vector via cleavage with EcoRI. pGL3-EV71 5′ UTR-FLuc was linearized by XbaI as a template for generating EV71 5′ UTR- FLuc RNA. The EV71 replicon, 3DD330A, was linearized by SalI. RNA transcript probes were synthesized using a MEGAscript T7 kit (Thermo Fisher Scientific), according to the protocol recommended by the manufacturer. Biotinylated EV71 5′ UTR RNA probes were synthesized with 1.25 μl 10 mM Biotin-16-UTP (Roche) in a MEGAscript reaction, according to instructions provided by the manufacturer. RNA transcripts were purified using an RNeasy Mini kit (Qiagen).
RNA-protein pull-down assay
RD cell extracts (200 μg), 12.5 pM of a biotinylated EV71 5′ UTR RNA probe, and recombinant PTBP1, FUBP1 and PCBP2 were mixed together, and the mixture (with a final volume of 100 μl), which contained 5 mM HEPES pH 7.1, 40 mM KCl, 0.1 mM EDTA, 2 mM MgCl2, 2 mM DTT and 0.25 mg/ml heparin (RNA mobility shift buffer), was incubated for 15 min at 30°C, and then added to 400 μl of Streptavidin MagneSphere Paramagnetic Particles (Promega) for binding at room temperature for 10 min. The protein-RNA complexes were washed five times with heparin-free RNA mobility shift buffer, after which 30 μl of 2× sample buffer was added to the beads and allowed to incubate for 10 min at room temperature. The sample containing the eluted proteins was incubated at 95°C for 5 min and resolved on a 12% gel by SDS-PAGE. The interactions were detected by western blot using specific antibodies.
Fluorescence microscope analysis
Fluorescence microscope analysis was performed as previously described (16). RD cells were infected with EV71 at 10 MOI. After 6 h post-infection, cells were washed with PBS and fixed with 3.7% formaldehyde for 20 min at room temperature. Cells were permeabilized and immunostained with anti-KHSRP, anti-KLHL12 and anti-EV71 2B antibodies, and then stained with Alexa Fluor 488 goat anti-rabbit (Invitrogen, A11008), Alexa Fluor 647 goat anti-mouse (Invitrogen, A21235) or Alexa Fluor 568 goat anti-rat (Invitrogen, A11077) secondary antibodies. The nucleus was stained with DAPI. The cells were examined under a confocal laser-scanning microscope (Zeiss; LSM 510 NLO).
In vitro ubiquitination assay
In vitro ubiquitination assays were performed using an ubiquitination kit (Enzo Life Science), according to the protocol provided by the manufacturer. FLAG-KHSRP WT and FLAG-KHSRP (K109, 121, 122R) proteins were purified by Anti-FLAG M2 affinity gel and eluted by 3× FLAG peptides. GST-CUL3, 3×Myc-RBX1, and Myc-KLHL12 (or Myc-KLHL12ΔKelch) were co-transfected to 293T cells, and then purified with Glutathione Sepharose 4 Fast Flow beads (GE Healthcare Life Sciences). The KLHL12–CUL3–RBX1 complex (or KLHL12ΔKelch–CUL3–RBX1 complex) bound on the beads was incubated with KHSRP, E1, E2 (UbcH5a), biotin–ubiquitin, Mg-ATP and ubiquitination buffer at 37°C for 1 h. The beads were resuspended in buffer containing 2% SDS, 10 mM Tris–HCl (pH 8.0), 150 mM NaCl and 1 mM DTT at 95°C for 5 min. The supernatant was diluted 5-fold with buffer (10 mM Tris–HCl (pH 8.0), 150 mM NaCl and 1 mM DTT) before immunoprecipitation with anti-FLAG antibodies. The immunoprecipitation samples were eluted by sample buffer and subjected to western blotting, and ubiquitinated proteins were analyzed using anti-streptavidin HRP.
(35S)-methionine labeling assay
shKHSRP knockdown cells were transfected with different FLAG-KHSRP expression vectors. After 24 h, 2.5 × 105 RD cells were seeded into each well of a 12-well plate and incubated at 37°C for an additional 24 h before infection with EV71 at a MOI of 10 PFU per cell. The medium was replaced with methionine-free DMEM and incubation was continued at 37°C for 1 h before labeling with (35S) methionine, conducted by replacing the medium with methionine-free DMEM containing (35S)-Met for labeling (50 μCi/ml). The cells were washed with PBS and lysed with lysis buffer after 1 h of labeling. The cell lysates were centrifuged at 10 000 × g for 10 min at 4°C, and then the supernatants were subjected to SDS-PAGE, transferred to a PVDF membrane, and detected by autoradiography. The same membrane was also used for western blotting.
In vivo and in vitro IRES activity assay
For the in vivo IRES activity assay, pFLAG-CMV2 vector, pFLAG-CMV2-KHSRP or KHSRP (K109, 121, 122R) plasmids were transfected to RD shKHSRP knockdown cells. After 48 h post-transfection, cells were transfected with 0.5 μg EV71 replicon 3DD330A RNA, following which cells were harvested using 5× cell culture lysis buffer (Promega), and then assayed for Firefly luciferase (FLuc) activity. The in vitro IRES activity assay was performed in a final volume of 25 μl containing 0.25 μg RNA, 250 ng KHSRP proteins, 60% volume of RD shKHSRP knockdown cell translation extracts, translation mixture (10 mM creatine phosphate, 50 μg/ml creatine phosphokinase, 79 mM KOAc, 0.5 mM Mg(OAc)2, 2 mM DTT, 0.02 mM hemin, 0.5 mM spermidine), 20 mM HEPES–KOH (pH 7.6), 20 μM amino acid mixture (Promega), 0.4 mM ATP (Promega) and RNase inhibitor. The mixtures were incubated at 30°C for 90 min and measured for FLuc activity by the Luciferase Assay System (Promega).
Preparation of RD cell translation extracts
RD shLacZ and shKHSRP knockdown cells were grown to 90% confluence. The cells were washed and scraped with PBS, and then centrifuged at 300 × g for 10 min at 4°C. The cell pellets were resuspended in 1.5× volume of hypotonic lysis buffer containing 10 mM HEPES–KOH (pH 7.6), 10 mM KOAc, 0.5 mM Mg(OAc)2, 2 mM DTT and 1x protease inhibitor cocktail (Roche), and the mixture was placed on ice for 30 min, then homogenized with a 27-gauge 1/2-in. needle. Cell extracts were centrifuged at 10 000 × g for 20 min at 4°C and stored at −80°C.
Quantitative RT-PCR
Total RNA from the indicated cells was purified using the RNeasy mini Kit (QIAGEN). One μg of the RNA was used as a template to synthesize cDNAs with ReverTra Ace (TOYOBO). The Roche LightCycler® 480 System and KAPA SYBR FAST qPCR Master Mix (Kapa Biosystems) were deployed for quantitative detection of nucleic acids. To detect EV71 5′ UTR by real-time PCR, a set of primers was designed (forward 5′-CCCTGAATGCGGCTAATC-3′; reverse 5′-ATTGTCACCATAAGCAGCCA-3′), and actin was used as an internal control (Primers: forward 5′-GCTCGTCGTCGACAACGGCTC-3′; reverse 5′-CAAACATGATCCTGGGTCATCTTCTC-3′).
Expression and purification of PTBP1, FUBP1 and PCBP2
Statistical analysis
Experimental data were analyzed by Student's two-tailed unpaired t-test using GraphPad Prism 6 software. P-values <0.05 were considered to be statistically significant.
RESULTS
Identification of KHSRP-associated proteins by iTRAQ–LC–MS/MS analysis
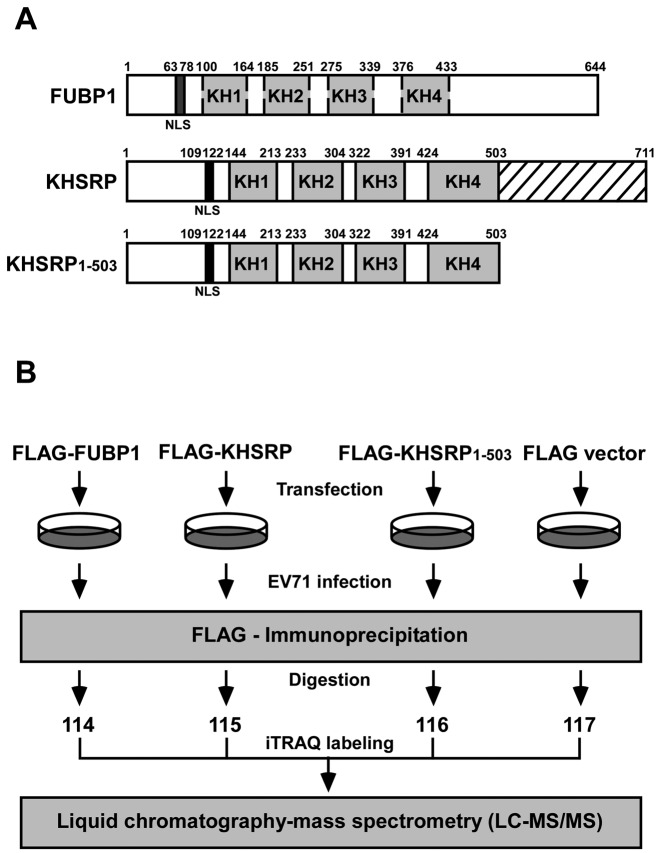
Our previous studies demonstrated that KHSRP is a negative regulator for viral IRES-dependent translation, whereas a similar protein, FUBP1, which differs from KHSRP in its C-terminal domain, is a positive regulator of viral translation (13,16). Moreover, KHSRP is cleaved in EV71-infected cells, and the truncated KHSRP without its C-terminal then acts as a positive regulator (17). Figure 1A illustrates the differences between FUBP1, KHSRP and the truncated form of KHSRP (KHSRP1–503). All of them contain KH1 to KH4 domains in the central region that are responsible for RNA binding. The central region of FUBP1 and KHSRP share approximately 80% sequence homology. The N-terminal domain contains a nuclear localization signal (NLS) that is responsible for protein trafficking. The C-terminal domains of FUBP1 and KHSRP respectively contain three and four repeat tyrosine-rich motifs. Importantly, the C-terminal domains of FUBP1 and KHSRP share only 60% sequence homology (33).
Figure 1.
Identification of KHSRP-associated protein by iTRAQ–LC–MS/MS analysis. (A) Schematic illustration of FUBP1, KHSRP and KHSRP1–503. (B) Flow chart for identification of KHSRP-associated proteins by iTRAQ-based proteomics analysis.
To understand why KHSRP plays a negative role in the regulation of EV71 IRES-dependent translation, we attempted to identify the KHSRP-associated proteins that vary from FUBP1 and KHSRP1–503. FLAG-fused FUBP1, KHSRP or KHSRP1–503 or a vector control, were respectively transfected into human rhabdomyosarcoma (RD cells). The cells were then challenged with EV71 (10 multiplicity of infection, MOI) and cell lysates were collected at 6 hours post-infection. Following FLAG immunoprecipitation, the precipitants underwent digestion with trypsin, and were subsequently labeled with iTRAQ reagent. After liquid chromatography–mass spectrometry (LC–MS/MS) analysis (Figure 1B), we compared the different associated proteins in these four groups. KHSRP/KHSRP1–503 and KHSRP/FUBP1 ratios were normalized to the median and expressed as log2 values. Standard deviations (SD) of log2 ratios were calculated for each data set. Proteins with log2 ratios >2 standard deviations (2SD) from the mean were considered to be upregulated candidates. In the KHSRP/KHSRP1–503 and KHSRP/FUBP1 data sets, 2-fold and 4-fold increases were respectively used as cut-off values for selecting upregulated candidates. Table 1 summarizes the different associated proteins between KHSRP and KHSRP1–503, and Table 2 outlines the differences in associated proteins between KHSRP and FUBP1. Among those proteins, KLHL12 and heterogeneous nuclear ribonucleoprotein C-like 1 protein (HNRNPCL1) appear in both tables. We proceeded to select KLHL12 for further study, as it had a higher ranking in terms of KHSRP/KHSRP1–503 (Table 1) and KHSRP/FUBP1 (Table 2) ratios than the other proteins identified.
Table 1. Comparison of associated proteins between KHSRP and KHSRP1–503.
| Accession no. | Gene symbol | Description | Coverage | Unique peptides | KHSRP/KHSRP1–503 (115/116) |
|---|---|---|---|---|---|
| Q96EP5 | DAZAP1 | DAZ associated protein 1 | 38.82 | 15 | 15.540 |
| Q16637 | SMN1 | Survival of motor neuron 1 | 23.81 | 4 | 4.907 |
| Q53G59 | KLHL12 | Kelch-like protein 12 | 24.65 | 10 | 4.802 |
| Q16630 | CPSF6 | Cleavage and polyadenylation specific factor 6 | 22.87 | 9 | 3.898 |
| O43809 | NUDT21 | Cleavage and polyadenylation specific factor 5 | 50.66 | 11 | 3.549 |
| O60812 | HNRNPCL1 | Heterogeneous nuclear ribonucleoprotein C-like 1 | 37.88 | 2 | 2.919 |
| P09234 | SNRPC | Small nuclear ribonucleoprotein polypeptide C | 25.79 | 5 | 2.389 |
| Q15637 | SF1 | Splicing factor 1 | 12.36 | 7 | 2.342 |
| Q9Y3E5 | PTRH2 | Peptidyl-tRNA hydrolase 2, mitochondrial | 18.44 | 2 | 2.265 |
| P61254 | RPL26 | 60S ribosomal protein L26 | 52.41 | 3 | 2.232 |
| Q8N684 | CPSF7 | Cleavage and polyadenylation specific factor 7 | 41.4 | 17 | 2.161 |
| Q16718 | NDUFA5 | NADH:ubiquinone oxidoreductase subunit A5 | 24.14 | 2 | 2.092 |
| P49756 | RBM25 | RNA binding motif protein 25 | 6.17 | 3 | 2.025 |
| Q13325 | IFIT5 | Interferon-induced protein with tetratricopeptide repeats 5 | 4.98 | 2 | 2.002 |
Table 2. Comparison of associated proteins between KHSRP and FUBP1.
| Accession no. | Gene symbol | Description | Coverage | Unique peptides | KHSRP/FUBP1 (115/114) |
|---|---|---|---|---|---|
| P17096 | HMGA1 | High mobility group AT-hook 1 | 36.45 | 3 | 17.499 |
| Q9BRP8 | PYM1 | Partner of Y14 and Mago | 28.92 | 5 | 11.828 |
| Q9HC36 | MRM3 | Mitochondrial rRNA methyltransferase 3 | 16.67 | 5 | 6.716 |
| Q92945 | KHSRP | Far upstream element-binding protein 2 | 80.73 | 55 | 6.464 |
| Q53G59 | KLHL12 | Kelch-like protein 12 | 24.65 | 10 | 6.406 |
| P98179 | RBM3 | RNA binding motif (RNP1, RRM) protein 3 | 60.51 | 7 | 5.814 |
| O60812 | HNRNPCL1 | Heterogeneous nuclear ribonucleoprotein C-like 1 | 37.88 | 2 | 5.541 |
| O75127 | PTCD1 | Pentatricopeptide repeat domain 1, mitochondrial | 23.29 | 10 | 5.312 |
| Q9ULV0 | MYO5B | Myosin VB | 1.14 | 2 | 4.995 |
| Q9H9L3 | ISG20L2 | Interferon stimulated exonuclease gene 20 like 2 | 12.18 | 3 | 4.986 |
| Q14011 | CIRBP | Cold-inducible RNA-binding protein | 49.42 | 9 | 4.762 |
| Q4U2R6 | MRPL51 | Mitochondrial ribosomal protein L51 | 21.09 | 3 | 4.469 |
| Q8IVS2 | MCAT | Malonyl-CoA-acyl carrier protein transacylase, mitochondrial | 10 | 2 | 4.445 |
| P25398 | RPS12 | 40S ribosomal protein S12 | 73.48 | 10 | 4.304 |
| Q96I51 | RCC1L | Williams-Beuren syndrome chromosome region 16 | 35.99 | 10 | 4.232 |
| Q9BQ75 | CMSS1 | Cms1 ribosomal small subunit | 5.73 | 2 | 4.219 |
| O95900 | TRUB2 | TruB pseudouridine synthase family member 2 | 65.26 | 16 | 4.216 |
| Q7Z7H8 | MRPL10 | Mitochondrial ribosomal protein L10 | 27.2 | 3 | 4.072 |
Confirmation of the interaction between KHSRP and KLHL12
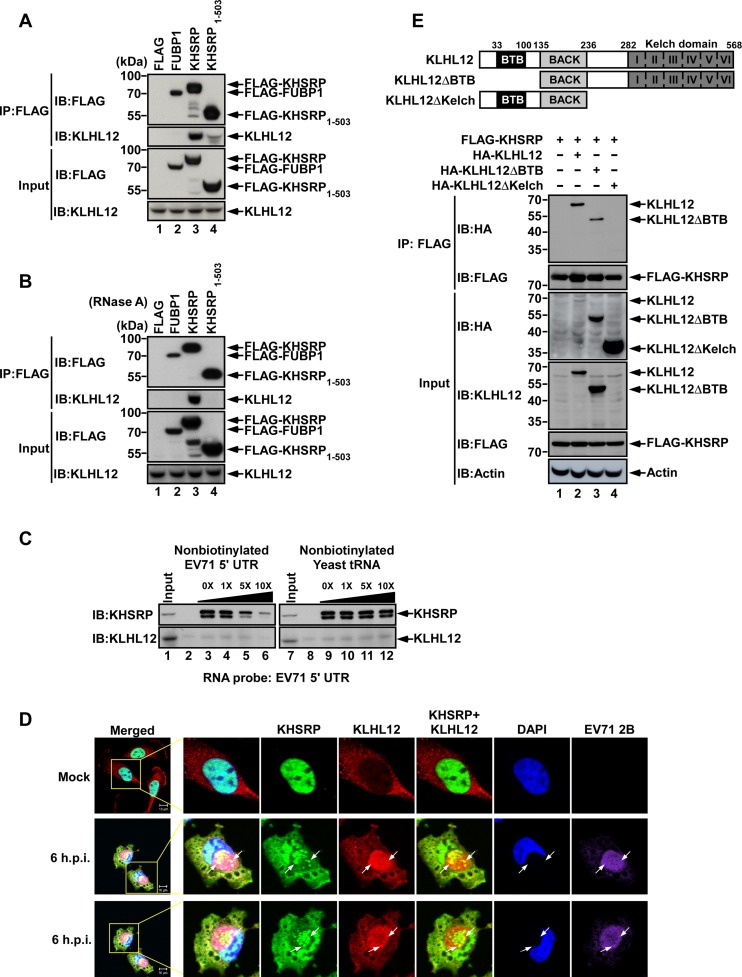
To confirm the interaction between KHSRP and KLHL12, we performed immunoprecipitation (IP) and western blot analysis. FLAG-fused FUBP1, KHSRP, KHSRP1–503, or a vector control, were respectively transfected into RD cells, and then challenged with EV71 (10 MOI). Anti-FLAG M2 affinity gel was used to immunoprecipitate the associated proteins. KLHL12 antibody was then applied in western blot analysis. The results in Figure 2A demonstrate that KLHL12 was present in KHSRP precipitant (lane 3), but not in FUBP1 precipitant (lane 2) or in FLAG precipitant (vector control) (lane 1). There is a weak band in KHSRP1–503 precipitant (lane 4), indicating that truncation of the KHSRP C-terminal domain reduced the interaction with KLHL12. Since KHSRP is an RNA-binding protein, we sought to examine whether the interaction between KHSRP and KLHL12 is dependent on RNA. We conducted a similar experiment as the one for Figure 2A, but further added RNase A to each reaction. The appearance of KLHL12 in KHSRP immunoprecipitant remains quite clear (Figure 2B, lane 3), suggesting that the interaction between KHSRP and KLHL12 can occur without RNA.
Figure 2.
Interaction between KHSRP and KLHL12. (A and B) RD cells were transfected with DNA constructs of FLAG only, FUBP1, KHSRP or KHSRP1–503 fused with FLAG-tags, and cells were infected with 10 MOI EV71 after 48 h; cell extracts were harvested at 6 hours post-infection, and total lysates (A) and RNase A treated lysates (B) were incubated with Anti-FLAG M2 affinity gel. FUBP1, KHSRP and KHSRP1–503 bound proteins were analyzed by SDS-PAGE and western blot. The amount of input was 5% of total IP lysates. (C) EV71 5′ UTR RNA was labeled with biotin and transcribed in vitro. RD cell lysates were incubated with biotinylated RNA probes, increasing amounts of nonbiotinylated EV71 5′ UTR RNA (lanes 2, 4–6) or yeast tRNA (lanes 8, 10–12). The RNA-protein complex was pulled down by streptavidin beads and resolved by SDS-PAGE. Antibodies against KHSRP and KLHL12 were utilized in western blot analysis. (D) KHSRP was observed to co-localize with KLHL12 in EV71-infected cells. RD cells were mock-infected or infected with 10 MOI EV71 for 6 h, fixed and stained with DAPI and antibodies against KHSRP, KLHL12 and EV71 viral protein 2B, and then examined by confocal microscopy. The arrow indicates the colocalization of KHSRP and KLHL12. Scale bar: 10 μm. (E) Schematic diagram of the wild type and truncated forms of KLHL12 protein. RD cells were transfected with FLAG-KHSRP and truncated forms of KLHL12 with HA-tags. Cell lysates were harvested after 48 h post-transfection and were immunoprecipitated with Anti-FLAG M2 affinity gel. The immunoprecipitates were analyzed by western blot using anti-HA and anti-FLAG antibodies. IP, immunoprecipitation. IB, immunoblot.
As KHSRP is an ITAF for the EV71 IRES (13), we wished to ascertain whether KLHL12 associates with the KHSRP-IRES complex. Viral RNA containing the IRES sequence was labeled with biotin, and streptavidin beads were then used to pull down the biotinylated RNA and its associated proteins. Western blot analysis using specific antibodies revealed that KHSRP was present in the complex bound by the EV71 IRES, but KLHL12 may not associate with the EV71 IRES directly (Figure 2C). We further examined the localization of KHSRP and KLHL12 in EV71-infected cells, and found that in mock-infected cells, KHSRP localized in the nucleus; whereas KLHL12 localized in the cytoplasm. However, upon virus infection, KHSRP relocalized from the nucleus to the cytoplasm, where it colocalized with KLHL12 (Figure 2D). KLHL12 is a substrate-specific adapter in the cullin 3 (CUL3)-based ubiquitin–protein E3 ligase complex (29–31). It contains BTB (Broad-Complex, Tramtrack and Bric a brac), BACK (BTB and C-terminal Kelch) and Kelch domains, as illustrated in Figure 2E. It has been known that the BTB domain is required for interacting with CUL3, while the Kelch domain is responsible for substrate binding (34). Therefore, we introduced a deletion mutation into the Kelch domain, and examined the binding ability of the derived mutant. KLHL12 lacking the BTB domain was used as a control. As the results in Figure 2E demonstrate, KLHL12 wild-type and ΔBTB mutant were pulled down by FLAG antibody against FLAG-KHSRP, but not KLHL12ΔKelch, indicating that the Kelch domain is important for the interaction between KHSRP and KLHL12 (Figure 2E).
KLHL12-based CUL3 complex promotes KHSRP ubiquitination
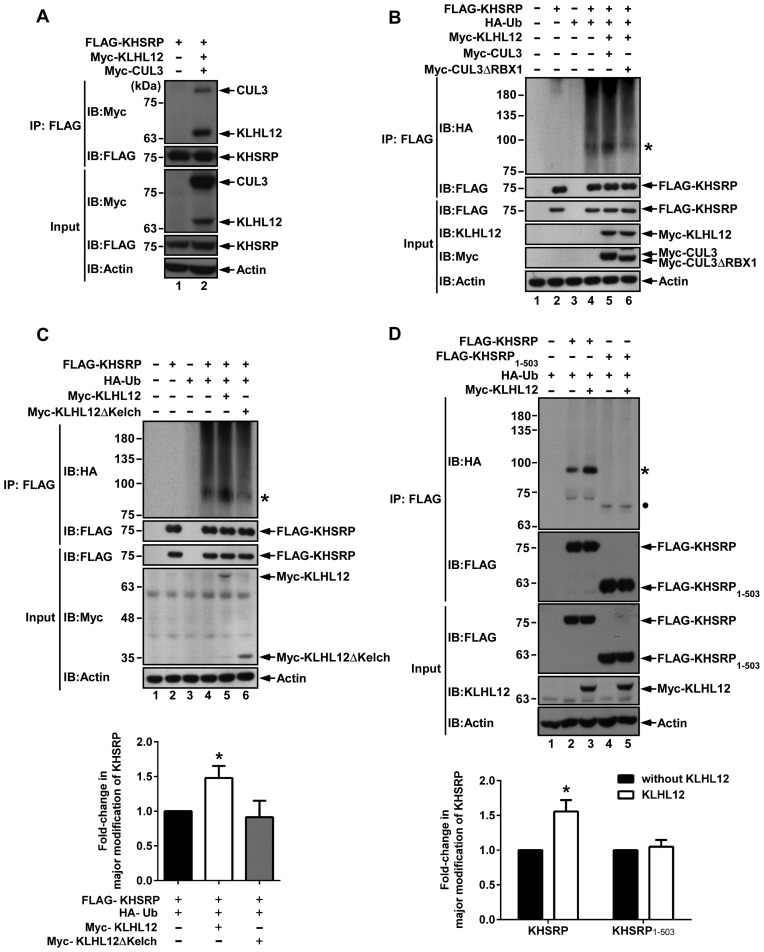
It has been known that KLHL12 serves as a substrate-specific adaptor for the CUL3-based ubiquitin E3 ligase complex. Therefore, we co-transfected FLAG-KHSRP, Myc-KLHL12 and Myc-CUL3 into cells, and then examined the complex formation in the FLAG immunoprecipitant. The results in Figure 3A show that KHSRP associated with KLHL12 and the CUL3 complex. To ascertain whether such interactions affected KHSRP ubiquitination, we co-transfected FLAG-KHSRP and HA-ubiquitin (HA-Ub) into cells, and treated them with MG132 for an additional 4 h before harvesting. Cell lysates were subjected to anti-FLAG IP and western blot, and then HA antibody was used to monitor the ubiquitination of KHSRP (Figure 3B). Ubiquitinated KHSRP was clearly observed when cells were transfected with FLAG-KHSRP, HA-Ub, Myc-KLHL12 and Myc-CUL3 (lane 5). When CUL3 was replaced by CUL3ΔRBX1, a mutant CUL3 without the interacting domain that binds with RBX1 (a component that promotes binding to the E2 conjugating enzyme), ubiquitination was reduced (lane 6). Incidentally, levels of ubiquitinated KHSRP were low in the input, as it was not possible to directly detect the major modification of KHSRP (indicative of ubiquitination) in immunoblots using anti-FLAG antibody, despite increasing input levels to 10% of IP lysates, the maximum loading volume (Supplementary Figure S1).
Figure 3.
KHSRP can be ubiquitinated by the KLHL12-based CUL3 complex. (A) KHSRP was found to form a complex with KLHL12 and CUL3. FLAG-KHSRP, Myc-KLHL12 and Myc-CUL3 expression vectors were co-transfected to 293T cells. The cell lysates were immunoprecipitated with Anti-FLAG M2 affinity gel, and then analyzed by western blot using anti-Myc and anti-FLAG antibodies. (B) 293T cells were co-transfected with FLAG-KHSRP, HA-Ub, Myc-KLHL12, Myc-CUL3 or Myc-CUL3ΔRBX1, and then treated with 20 μM MG132 for 4 h before harvesting. Cell lysates were immunoprecipitated with Anti-FLAG M2 affinity gel and resolved by SDS-PAGE, and KHSRP ubiquitination was detected by anti-HA antibody. The asterisk (*) represents the major modification of KHSRP. KHSRP, KLHL12, CUL3 and CUL3ΔRBX1 were visualized by individual antibodies against FLAG, KLHL12 and Myc. (C) Various constructs expressing KHSRP, Ub, KLHL12 or KLHL12 truncated form (KLHL12ΔKelch) were co-transfected to 293T cells. After 48 h, cells were treated with 20 μM MG132 for 4 h before harvesting, and lysates were then subjected to immunoprecipitation using Anti-FLAG M2 affinity gel. KHSRP ubiqitination was assessed by western blot, and KHSRP, KLHL12 and KLHL12ΔKelch were visualized using anti-FLAG and anti-Myc antibodies. (D) 293T cells were co-transfected with various indicated constructs and subsequently treated with 20 μM MG132 for 4 h before harvesting. Western blots were utilized to analyze ubiquitinated proteins. The asterisk (*) represents the major modification of KHSRP. The circle represents the major modification of KHSRP1-503. For both (C) and (D), levels of KHSRP or KHSRP1–503 with major modification were respectively normalized against levels of FLAG-KHSRP or FLAG-KHSRP1–503, and fold-changes over control reactions without KLHL12 were calculated and plotted.
To further confirm that the interaction between KHSRP and KLHL12 is important for the ubiquitination of KHSRP, we examined cells overexpressing either KLHL12 or KLHL12ΔKelch, a mutant shown in Figure 2E to reduce the interaction between KHSRP and KLHL12. The results in Figure 3C reveal that KLHL12 promotes KHSRP ubiquitination (comparing lanes 4 and 5). Such an effect was not observed in KLHL12ΔKelch overexpressed cells (lane 6). We also overexpressed either KHSRP or KHSRP1–503 with ubiquitin and KLHL12 (Figure 3D). The results show that ubiquitination of KHSRP was reduced in the absence of the C-terminal domain (comparing lanes 2 and 4), and KLHL12 was unable to induce ubiquitination of KHSRP1–503 (comparing lanes 4 and 5). To understand the topology of KHSRP ubiquitination, plasmids encoding FLAG-KHSRP, HA-Ub wild type (WT) or lysine-free (KO) were co-transfected into 293T cells, and cell lysates were immunoprecipitated with anti-FLAG affinity gel. A major modification of KHSRP was observed in cells co-transfected with KHSRP and Ub WT (Supplementary Figure S2, lane 4), but not in cells co-transfected with the Ub KO construct (lane 6). Based on molecular weight, the major modification of KHSRP is speculated to be di-ubiquitination. Taken together, the data demonstrates that the KLHL12-based CUL3 ubiquitin ligase complex promotes KHSRP ubiquitination, in a manner that is dependent on the existence of the Kelch domain of KLHL12 and the C-terminal domain of KHSRP, regions that are also important for KHSRP–KLHL12 interaction.
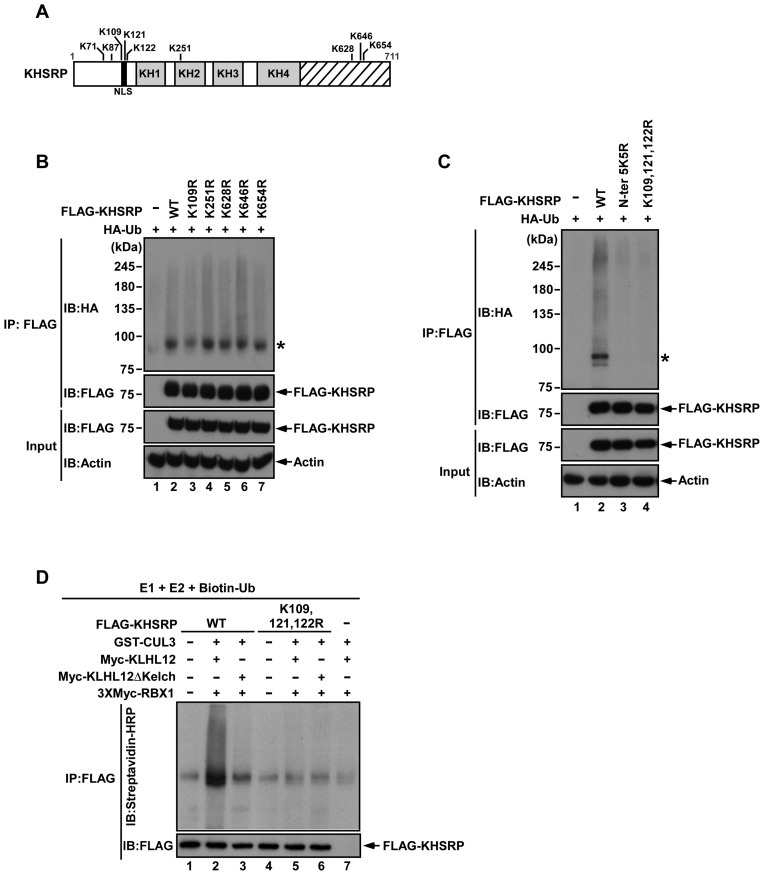
K109, K121, K122 are potential ubiquitination sites in KHSRP
In order to identify the main sites of ubiquitination on KHSRP, we mutated lysine residues to ascertain the variants that could block KHSRP ubiquitination by the KLHL12-based CUL3 ubiquitin ligase complex. Five lysine residues (Lys109, Lys251, Lys628, Lys646 and Lys654) were predicted as ubiquitination sites of KHSRP via proteomics analysis (Figure 4A) (35). These lysine residues were replaced by arginine, and the respective variant constructs were then co-transfected with HA-Ub to 293T cells. The ubiquitination of KHSRP K109R was reduced in comparison to wild type (WT) (Figure 4B, lanes 2 and 3), but this was not seen for the KHSRP K251R, K628R, K646R, or K654R variants (Figure 4B, lanes 4–7). In preparation for the mutation of other lysine residues adjacent to Lys109, five lysine residues were identified in the N-terminal region of KHSRP (Figure 4A). We replaced all five lysine residues with arginine at positions 71, 87, 109, 121, 122 in the N-terminal region of KHSRP (N-ter. 5K5R) and generated a triple mutation variant, KHSRP (K109, 121, 122R). The ubiquitination levels of KHSRP (N-ter. 5K5R) and KHSRP (K109, 121, 122R) were reduced in comparison to KHSRP WT (Figure 4C). In addition, we generated a KHSRP (K121, 122R) variant to assess the contribution of the Lys121 and Lys122 residues to KHSRP ubiquitination (Supplementary Figure S3). The results indicate that KHSRP (K121, 122R) has lower ubiquitination levels as compared to KHSRP WT and KHSRP K109R; however, ubiquitination levels of KHSRP (K109, 121, 122R) remained lower than KHSRP (K121, 122R), as can be compared from immunoblotting results of the major modification of KHSRP (indicated by an asterisk in Supplementary Figure S3).
Figure 4.
Potential ubiquitination sites of KHSRP. (A) A schematic diagram of predicted ubiquitination sites in KHSRP is shown. (B, C) Indicated constructs expressing different KHSRP mutants were co-transfected with HA-Ub to 293T cells, then immunoprecipitated with Anti-FLAG M2 affinity gel and analyzed by western blotting. (D) GST-CUL3, 3×Myc-RBX1 and Myc-KLHL12 (or Myc-KLHL12ΔKelch) were co-transfected to 293T cells, and then pulled down by glutathione Sepharose beads. The E3 ligase complex was incubated with E1, E2, Biotin-Ub, and KHSRP WT or K109, 121, 122R with FLAG tags. The ubiquitination reactions were further subjected to FLAG-immunoprecipitation and were resolved by SDS-PAGE. KHSRP ubiquitination was detected by anti-streptavidin HRP antibody.
An in vitro ubiquitination assay was applied to further confirm the ubiquitination status of KHSRP. FLAG-KHSRP WT or FLAG-KHSRP (K109, 121, 122R) proteins were incubated with KLHL12–CUL3–RBX1 complex (or KLHL12ΔKelch–CUL3–RBX1 complex), E1, E2, and ubiquitin with biotin tags, and were then immunoprecipitated with anti-FLAG antibody. Anti-streptavidin HRP antibody was used to detect ubiquitination of KHSRP. The KLHL12–CUL3–RBX1 complex promoted ubiquitination of KHSRP WT, but not KHSRP (K109, 121, 122R) (Figure 4D, lanes 2 and 5). Ubiquitination of KHSRP was reduced when KLHL12 without the Kelch domain was present in the complex (Figure 4D, lane 3), indicating that KLHL12 is an important mediator between KHSRP and CUL3 for KHSRP ubiquitination. The results in Figure 4 indicate Lys109, Lys121, Lys122 to be likely sites for KLHL12-mediated ubiquitination in KHSRP.
KLHL12 does not affect KHSRP stability
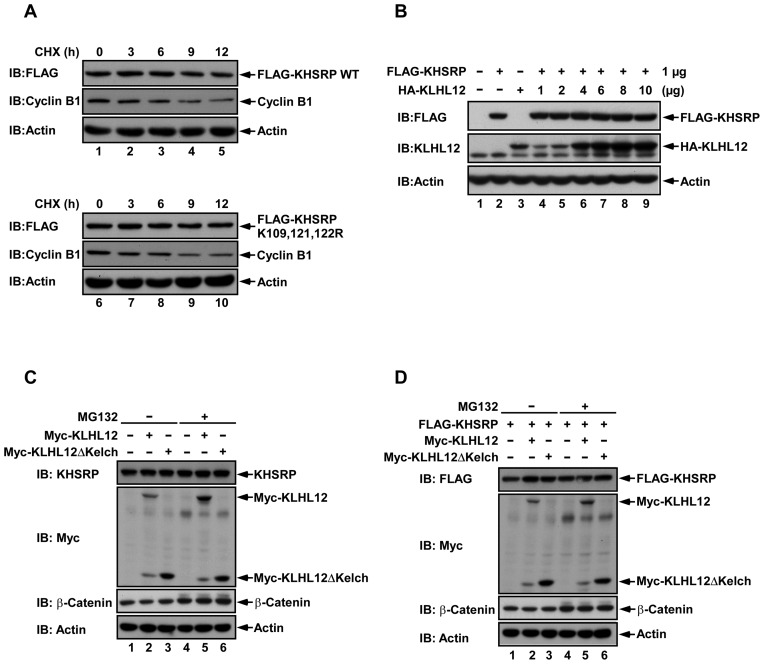
A ubiquitinated protein may undergo proteosome degradation following ubiquitination, and therefore we assessed the half-lives of KHSRP WT and the reduced ubiquitination variant KHSRP (K109, 121, 122R), using the cycloheximide (CHX) chase assay. KHSRP WT or KHSRP (K109, 121, 122R) constructs with FLAG tags were transfected to RD cells, and then treated with CHX for 0, 3, 6, 9 or 12 h before harvesting to derive cell lysates for monitoring of protein levels. Results showed that the half-lives of KHSRP WT and KHSRP (K109, 121, 122R) were not affected (Figure 5A), and we moved on to examine whether KLHL12 affected KHSRP stability. When cells were co-transfected with a fixed amount of KHSRP plasmid (FLAG-KHSRP, 1 μg) and increasing amounts of KLHL12 plasmid (HA-KLHL12, 0–10 μg), expression levels of KHSRP detected by western blot remained the same despite increasing levels of KLHL12 (Figure 5B, lane 2 and lanes 4–9). To further confirm that the interaction between KHSRP and KLHL12 is not related to proteasome degradation, we co-transfected cells with KLHL12 alone, KLHL12ΔKelch alone (Figure 5C), or both of these constructs with KHSRP (Figure 5D), and inhibited proteasome activity with MG132. Endogenous or overexpressed KHSRP protein levels remained the same (Figure 5C and D) regardless of treatment with MG132 or not (comparing lanes 1–3 to 4–6). Taken together, the results in Figure 5 indicate that KLHL12 does not affect KHSRP stability.
Figure 5.
KLHL12 does not affect KHSRP stability. (A) KHSRP WT or KHSRP (K109, 121, 122R) with FLAG tags were transfected to RD cells, and then treated with 100 μg/ml cycloheximide (CHX) for the indicated times. Cyclin B1 was used as a positive control. Cells were lysed and analyzed by western blotting, using antibodies against FLAG, cyclin B1 and actin. (B) RD cells were co-transfected with 1 μg of FLAG-KHSRP and increasing amounts of HA-KLHL12 (0–10 μg). After 48 hours post-transfection, cell lysates were collected, resolved by SDS-PAGE and analyzed by western blot using anti-FLAG and anti-KLHL12 antibodies. (C, D) 293T cells were transfected with the indicated constructs, and then treated with 20 μM MG132 or DMSO for 4 h before cell lysis. β-catenin was used as a positive control. Cell lysates were analyzed by western blotting, using anti-FLAG, anti-Myc and anti-β-catenin antibodies.
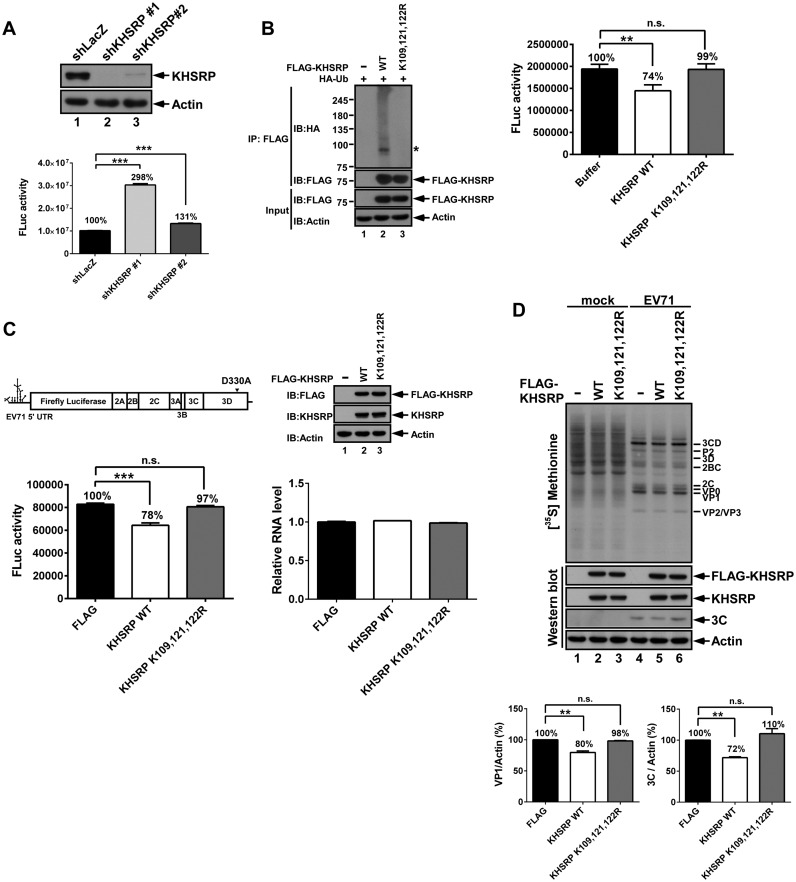
Ubiquitination is crucial for KHSRP downregulation of IRES-driven translation
To investigate the functional impact of KHSRP ubiquitination, we constructed KHSRP knockdown (KD) cells. Both KHSRP shRNA clones 1 and 2 reduced KHSRP protein levels efficiently in comparison to the shLacZ control (Figure 6A). We performed an initial in vitro translation assay to measure EV71 IRES activity in shLacZ cells and KHSRP shRNA clones 1 and 2. The results indicated that EV71 IRES activity increased to 298% in KHSRP shRNA clone 1 cells, as compared to shLacZ cells. However, EV71 IRES activity in KHSRP shRNA clone 2 cells only increased to 131%. As KHSRP shRNA clone 1 demonstrated better knockdown efficiency and induced higher EV71 IRES activity compared to clone 2, we opted to use clone 1 for further studies. To assess the impact of ubiquitinated KHSRP on EV71-driven translation, an in vitro translation assay was applied to measure IRES activity (Figure 6B). KHSRP WT or KHSRP (K109, 121, 122R) were co-transfected with HA-Ub to 293T cells, immunoprecipitated by FLAG antibody, and subsequently eluted using 3× FLAG peptides. Ubiquitination levels of KHSRP WT and KHSRP (K109, 121, 122R) were confirmed by western blot (Figure 6B, left panel). Subsequently, cell translation extracts of shKHSRP clone 1 were used to support the translation of EV71 5′ UTR-FLuc RNA, either in the presence of ubiquitinated KHSRP WT or the reduced ubiquitination variant (KHSRP (K109, 121, 122R)). Ubiquitinated KHSRP WT decreased IRES activity to 74%, whereas the reduced ubiquitination variant (KHSRP (K109, 121, 122R)) demonstrated no significant differences from buffer control (Figure 6B, right panel). We also used shLacZ cells as a control, and EV71 IRES activity showed no significant difference among buffer controls or cells presenting ubiquitinated KHSRP WT or the reduced ubiquitination variant (Supplementary Figure S4). To further confirm the results of this in vitro translation assay, an EV71 replicon bearing a replication-defective 3D polymerase (3DD330A) was used to monitor the impact of ubiquitinated KHSRP on EV71 translation (Figure 6C). We first transfected vector controls or vectors expressing KHSRP WT or KHSRP (K109, 121, 122R) to KHSRP knockdown cells, and then transfected EV71 replicon 3DD330A RNA encoding the firefly luciferase gene. With the equivalent amounts of replicon RNA in all three samples (Figure 6C, right panel), FLuc activity was reduced to 78% in KHSRP WT-expressing cells; however, the reduced ubiquitination variant (KHSRP (K109, 121, 122R)) was not able to diminish IRES activity compared to the FLAG control (Figure 6C, left panel).
Figure 6.
Ubiquitination is crucial for KHSRP downregulation of IRES-driven translation. (A) Cell lysates from KHSRP shRNA knockdown (KD) RD cells were resolved by SDS-PAGE, and then examined using anti-KHSRP and anti-actin antibodies. The cell translation extracts of shLacZ, shKHSRP clone 1, or shKHSRP clone 2 were incubated with EV71 5′ UTR-FLuc RNA, and then the reactions were applied to the Firefly luciferase (FLuc) assay. Experiments were conducted in triplicate, and results were subjected to statistical analysis. Error bars, mean ± SD. ***P < 0.001. (B) KHSRP WT or KHSRP (K109, 121, 122R) vectors were co-transfected with HA-Ub, and then subjected to FLAG immunoprecipitation. The KHSRP WT or KHSRP (K109, 121, 122R) proteins were eluted by 3× FLAG peptides, and then incubated with shKHSRP knockdown RD cell translation extracts and EV71 5′ UTR-FLuc RNA. After 90 minutes, the reactions were assayed with the FLuc assay. Experiments were conducted in triplicate, and results were subjected to statistical analysis. Error bars, mean ± SD. P values were calculated on the basis of luciferase activity. **P < 0.01, Student's two-tailed unpaired t-test. (C) shKHSRP knockdown RD cells expressing FLAG vector, KHSRP WT, or KHSRP (K109, 121, 122R) were grown for 2 days, and then transfected with EV71 replicon 3DD330A RNA. After 6 h post-transfection, cells were lysed or underwent RNA extraction, and lysates were then assayed with FLuc assay, western blotting, and real-time PCR. Experiments were conducted in triplicate, and results were subjected to statistical analysis. Error bars, mean ± SD. ***P < 0.001, Student's two-tailed unpaired t-test. (D) The effect of KHSRP WT and KHSRP (K109, 121, 122R) on viral protein synthesis. shKHSRP knockdown RD cells expressing FLAG vector, KHSRP WT or KHSRP (K109, 121, 122R) were grown for 2 days, and then challenged with EV71 at 10 MOI. Synthesized proteins were labeled with (35S) methionine at 5–6 h post-infection. The lysates from mock-infected and EV71-infected cells were subjected to SDS-PAGE, and then examined by autoradiography (upper panel) and western blot (lower panel). The indicated viral proteins were identified according to protein size. The levels of (35S) methionine-labeled VP1 and 3Cpro were quantified and normalized against actin levels, based on two repeated experiments. Error bars, mean ± SD. **P < 0.01.
In order to investigate how KHSRP ubiquitination affects viral protein synthesis, a (35S)-methionine labeling assay was performed in EV71-infected cells. KHSRP knockdown cells were expressed with FLAG vector, KHSRP WT, or KHSRP (K109, 121, 122R), and then infected with EV71 at a MOI of 10. Newly synthesized viral proteins were diminished in KHSRP WT-expressing cells at 5–6 h post-infection (Figure 6D, lane 5). The reduced ubiquitination variant (KHSRP (K109, 121, 122R)) had comparable viral protein expression to FLAG vector control (Figure 6D, lanes 4 and 6). The quantified results of viral protein VP1 and 3Cpro shown in Figure 6D indicate that VP1 and 3Cpro expression was lower in KHSRP WT-expressing cells as compared to cells expressing FLAG vector and the reduced ubiquitination variant. These results indicate that the ubiquitination of KHSRP is essential for its downregulatory effect on EV71 IRES-driven translation.
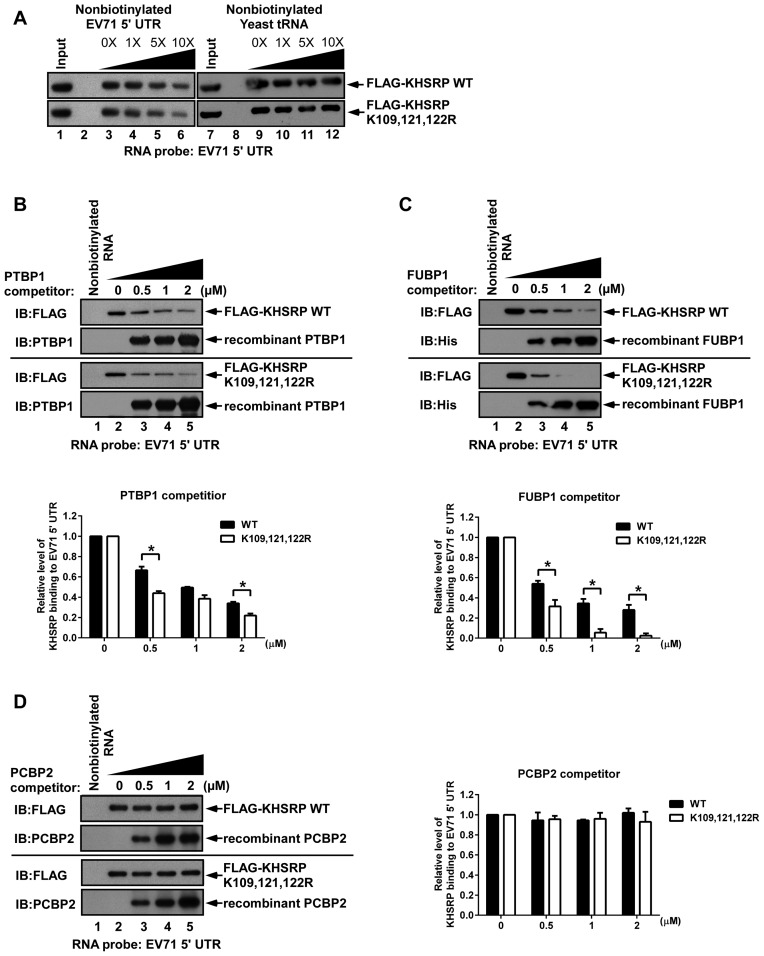
Ubiquitination status affects the capability of KHSRP to compete against FUBP1
To confirm whether ubiquitination affects KHSRP binding to EV71 IRES, lysates from cells overexpressing KHSRP WT or KHSRP (K109, 121, 122R) were incubated with biotinylated EV71 IRES RNA. Increasing amounts of nonbiotinylated EV71 5′ UTR or yeast tRNA probe were added as competitors. Streptavidin beads were used to pull down biotinylated RNA, and it was found that the interaction between KHSRP WT or the KHSRP (K109, 121, 122R) variant with EV71 5′ UTR RNA could be competed by nonbiotinylated EV71 5′ UTR, but not nonbiotinylated yeast tRNA, demonstrating that KHSRP WT and the KHSRP (K109, 121, 122R) variant bind to the EV71 5′ UTR region specifically (Figure 7A). We speculated that although both KHSRP WT and the reduced ubiquitination variant can still associate with the EV71 IRES, the competitive capability of KHSRP against other positive ITAFs may possibly be affected by ubiquitination. To address this question, a competition assay was performed (Figure 7B–D and Supplementary Figure S5). PTBP1, FUBP1 and PCBP2 are known to be positive regulators of picornavirus IRES. Lysates from cells overexpressing KHSRP WT or KHSRP (K109, 121, 122R) were incubated with biotinylated EV71 IRES RNA and increasing amounts of recombinant PTBP1, FUBP1 or PCBP2 proteins, to evaluate the EV71 IRES RNA competition dynamics for KHSRP, reduced ubiquitination variant KHSRP, and these positive ITAFs. The results indicated that increasing amounts of PTBP1 (Figure 7B) and FUBP1 (Figure 7C) weakly compete with both KHSRP WT and the reduced ubiquitination variant for binding to EV71 5′ UTR RNA, while PCBP2 was unable to compete against both forms of KHSRP (Figure 7D). We noted that PTBP1 competed slightly more effectively against reduced ubiquitination variant KHSRP as compared to KHSRP WT, with the differences being statistically significant (Figure 7C, lanes 3–5). In addition, FUBP1 also outcompeted KHSRP (K109, 121, 122R) more readily than KHSRP WT (Figure 7C, lanes 4 and 5). We further used KHSRP WT or KHSRP (K109, 121, 122R) to conduct a reverse competition assay against PTBP1, FUBP1 and PCBP2 (Supplementary Figure S5). Interestingly, increasing levels of KHSRP and KHSRP (K109, 121, 122R) outcompeted FUBP1 for binding to EV71 5′ UTR RNA, but were unable to affect PTBP1 and PCBP2 binding (Supplementary Figure S5B and S5C). However, although 0.5 μM of KHSRP (K109, 121, 122R) led to clearly enhanced competitive capability against FUBP1, competition results remained static despite an increase in KHSRP (K109, 121, 122R) to 2 μM (Supplementary Figure S5C). By contrast, increasing KHSRP WT levels appeared to enhance competitive capability against FUBP1 (Supplementary Figure S5B, lanes 3–5). The ubiquitination status of KHSRP WT and KHSRP (K109, 121, 122R) were assessed by western blot (Supplementary Figure S5A). The results suggest that KHSRP WT can better compete against FUBP1 for binding to EV71 5′ UTR RNA, as compared to KHSRP (K109, 121, 122R), and demonstrate that ubiquitination status may affect the ability of KHSRP to compete against FUBP1 for binding to EV71 IRES RNA.
Figure 7.
Reduction of KHSRP ubiquitination diminishes the competitive capability of KHSRP against FUBP1. (A) RD cells overexpressing KHSRP WT or KHSRP (K109, 121, 122R) were incubated with biotinylated EV71 5′ UTR RNA. Nonbiotinylated EV71 5′ UTR RNA (lanes 2, 4–6) or yeast tRNA (lanes 8, 10–12) were used in the competition assay. Increasing amounts of (B) PTBP1, (C) FUBP1, and (D) PCBP2 recombinant proteins were added to compete with KHSRP WT or KHSRP (K109, 121, 122R) in interacting with biotinylated EV71 5′ UTR. Biotinylated EV71 5′ UTR and associated proteins were pulled down by streptavidin beads, and then analyzed by western blotting, using antibodies against FLAG, PTBP1, His and PCBP2.
DISCUSSION
Translation initiation in eukaryotic mRNA can be triggered by two different mechanisms, cap-dependent translation and IRES-dependent translation. Picornaviruses are known to utilize IRES for translation initiation, and this process is regulated by several cellular proteins, termed ITAFs. Most known ITAFs are positive regulators of picornavirus IRES, but this may be because there is less literature available regarding negative ITAF regulators. Here, we use EV71 as a model to illustrate how KHSRP, a negative ITAF, downregulates EV71 IRES-driven translation. In this study, we utilized iTRAQ-LC-MS/MS analysis to identify KHSRP-associated proteins. We found that KHSRP associates with KLHL12, and stable isotope labeling by amino acids in cell culture (SILAC) confirmed that KLHL12 associates with KHSRP upon EV71 infection (Supplementary Table S1). KLHL12, a BTB-domain containing protein, is a member of the KLHL (Kelch-like) family (34). Several studies have reported that KLHL12 is a substrate adaptor of the CUL3 ubiquitin ligase complex, which can promote Dishevelled (Dsh) ubiquitination and degradation to negatively regulate the Wnt–β-catenin pathway (29). Moreover, the CUL3–KLHL12 ubiquitin ligase complex is known to monoubiquitinate SEC31 to regulate COPII vesicle coat formation (31), and can also promote polyubiquitination of the dopamine D4 receptor, but without causing degradation (30,36). Our results show that KHSRP, KLHL12 and CUL3 can form a complex to promote KHSRP ubiquitination (Figure 3), and this subsequently alters KHSRP regulation of EV71 IRES-driven translation.
Many ITAFs are nuclear-resident proteins, but in the event of picornavirus infection, nuclear pore complexes (NPC) are disrupted to induce redistribution of these factors (7). In previous research, we found that KHSRP, a nuclear protein, redistributed to the cytoplasm upon EV71 infection (13). In this study, we found that KHSRP colocalized in the cytoplasm with KLHL12 in EV71-infected cells (Figure 2D). We also noted that both KLHL12 and viral protein 2B accumulated in specific cellular compartments around the nucleus: KLHL12 is involved in ER-Golgi transport and localizes to the endoplasmic reticulum (ER) (31), while the picornavirus 2B protein also localizes in the ER and Golgi complex (37,38). These findings suggest that KLHL12 and viral protein 2B may colocalize in the ER, and this could be related to formation of the viral replication complex.
Although several ITAFs, such as HNRNPA1 and tumor suppressor programmed cell death 4 (PDCD4), have been reported to act as negative regulators of cellular IRES (39,40), to the best of our knowledge, the question of whether ubiquitination modulates ITAF function with regard to IRES-mediated translation has not been previously addressed. To determine the effects of ubiquitinated KHSRP on EV71 IRES-driven translation, we first sought to identify the ubiquitination sites of KHSRP. Lys109, Lys121 and Lys122 residues, located in the nuclear localization signal (NLS) of KHSRP, represent potential ubiquitination sites. When we replaced all three lysine residues to arginine (K109R, K121R and K122R), we found that the resulting KHSRP variant (K109, 121, 122R) remained in the cytoplasm, but the wild-type form of KHSRP was imported into the nucleus (Supplementary Figure S6). In order to maintain KHSRP WT in the cytoplasm to observe its effect on EV71 IRES-driven translation, we transfected the EV71 replicon into cells, and then forced wild-type KHSRP to relocalize to the cytoplasm (Supplementary Figure S6). The in vitro translation assay and (35S)-methionine labeling assay were also applied to determine the effects of ubiquitinated KHSRP WT and the reduced ubiquitination variant on EV71 IRES-driven translation. The results indicate that ubiquitinated KHSRP serves as a negative regulator in EV71 IRES translation (Figure 6B–D); moreover, the reduced ubiquitination variant could not downregulate EV71 IRES-driven translation despite remaining in the cytoplasm, and this excludes the possibility of cellular localization acting as a differentiating factor. Combined with our results in Figure 2A, we found that the C-terminal domain of KHSRP interacts with KLHL12, leading to ubiquitination of KHSRP at the N-terminal domain. In a previous study, we found that the C-terminal domain is important for KHSRP to play a negative role in EV71 translation (17), and this study further confirms that the C-terminal domain of KHSRP is critical to KHSRP–KLHL12 interaction; without the C-terminal domain (KHSRP1–503), ubiquitination of KHSRP1–503 cannot be enhanced by KLHL12 (Figure 3D). Together, this shows that KHSRP likely interacts with KLHL12 to result in KHSRP ubiquitination and negative regulation of EV71 translation.
Ubiquitination can cause protein degradation in a proteasome-dependent manner, or otherwise alter protein function by changing topology (41). We sought to elucidate whether ubiquitination of KHSRP through the KLHL12-based CUL3 E3 ligase complex leads to degradation or not. Protein expression levels of KHSRP were not significantly affected by increasing levels of KLHL12 or by treatment with the proteasome inhibitor MG132 (Figure 5). However, the observed major modification of KHSRP may alter protein function, and from the results in Supplementary Figure S2, we speculated that this major modification may be di-ubiquitination of KHSRP. We have found that KHSRP is cleaved by caspase activation, proteasome activity, and autophagy at a late point of EV71 infection (10 h.p.i). Moreover, ubiquitination of KHSRP was previously found to be enhanced in RD cells treated with MG132 (17). A different cell line (293T cells) was used to observe ubiquitination of KHSRP in this study, and major modification of KHSRP was seen in 293T cells, but not in RD cells. This suggests that the presence of other ubiquitination sites or different modes of ubiquitination or modification (such as sumoylation) cannot be ruled out for KHSRP. Still, Lys109, Lys121 and Lys122 represent critical sites for KHSRP ubiquitination and regulation of IRES-driven translation. Moreover, we observed KHSRP–KLHL12 interactions at 6 hours post-infection with EV71 (Figure 2), but KHSRP was shown to be cleaved at 10 hours post-infection (17), suggesting that host cells may modulate KHSRP to serve as a negative regulator that suppresses EV71 IRES-driven translation initially. However, EV71 eventually triggers several pathways that result in KHSRP cleavage at a later point after infection.
We hypothesized that ubiquitination may alter the capability of KHSRP to compete against other positive ITAF regulators for binding to the EV71 IRES, and therefore we selected the positive ITAFs, PTBP1, FUBP1 and PCBP2, to conduct a competition assay for binding to EV71 5′ UTR RNA (Figure 7B–D and Supplementary Figure S5). The results showed that PTBP1 and FUBP1 can outcompete KHSRP WT and KHSRP (K109, 121, 122R) (Figure 7B and C), while KHSRP WT and KHSRP (K109, 121, 122R) can outcompete FUBP1 (Supplementary Figure S5B and S5C); however, no competition was observed between PCBP2 and KHSRP WT or KHSRP (K109, 121, 122R) (Figure 7D and Supplementary Figure S5B and S5C), and neither KHSRP WT nor KHSRP (K109, 121, 122R) were able to outcompete PTBP1 (Supplementary Figure S5B and S5C). In a previous study, we found that KHSRP interacts with nt 1–167 (stem loop I-II), 91–228 (stem loop II-III), and 566–745 (stem loop VI to linker region) in the EV71 IRES (13). The binding region of FUBP1 is located at nt 636–745 (linker region) in the EV71 IRES (16), while PTBP1 is thought to interact with stem loop V and its flanking regions in the poliovirus IRES (42), and PCBP2 is known to bind with stem loop IV of the poliovirus and EV71 IRES (5,43,44). It is possible that the KHSRP proteins could not outcompete PTBP1 and PCBP2 because the respective binding regions on the EV71 5′ UTR of these three proteins are all different; however, as both KHSRP and FUBP1 bind to the linker region, competitive activity was observed. FUBP1 weakly competes with KHSRP WT and KHSRP (K109, 121, 122R) (Figure 7C), but KHSRP proteins appear to compete more strongly against FUBP1 (Supplementary Figure S5B and S5C), suggesting that KHSRP may have stronger binding affinity to the EV71 5′ UTR than FUBP1. These results lend support to our hypothesis that ubiquitination primarily serves to improve the competitive binding ability of KHSRP against FUBP1 and possibly other positive ITAFs at the EV71 IRES, rather than altering the binding site of KHSRP on the EV71 IRES.
In summary, we demonstrate that KHSRP acts as a negative regulator in EV71 IRES-mediated translation via ubiquitination by the KLHL12-based CUL3 E3 ubiquitin ligase complex. Although ubiquitination is known to regulate cap-dependent translation, our results elucidate a new regulatory mechanism by which ubiquitination regulates cap-independent IRES-driven translation by modulating ITAFs. Specifically, we found that KHSRP serves as a negative regulator in EV71 translation, and ubiquitination at the Lys109, Lys121 and Lys122 residues enhances the ability of KHSRP to compete against other positive ITAF regulators at the EV71 IRES. This represents a novel mechanism by which viral IRES-driven translation is mediated by ubiquitination of host ITAFs, and may have important implications for the future study of viral–host interactions and the development of anti-viral drug targets.
Acknowledgments
We would like to acknowledge Dr Hsiu-Ming Shih and Dr Ruey-Hwa Chen of the Academic Sinica, Dr Rei-Lin Kuo and Dr Jim-Tong Horng of Chang Gung University, Dr Chen Zhao of the University of Texas at Austin and Dr Ian Goodfellow of the University of Cambridge for generously providing materials, as well as the Proteomics Core Laboratory of Chang Gung University for providing technical support. We also thank Dr Hsiu-Ming Shih, Dr Rei-Lin Kuo, Dr Chen Zhao, Dr Jing-Yi Lin and Dr Chih-Ching Wu for their comments and insight regarding this manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology, Taiwan [104-2632-B-182-002]; Chang Gung Memorial Hospital [CMRPD1D0311-313]; Chang Gung Memorial Hospital [CLRPD190015]. Funding for open access charge: Chang Gung University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Jackson R.J., Hellen C.U., Pestova T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pelletier J., Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 3.Jang S.K., Krausslich H.G., Nicklin M.J., Duke G.M., Palmenberg A.C., Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lloyd R.E. Translational control by viral proteinases. Virus Res. 2006;119:76–88. doi: 10.1016/j.virusres.2005.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sweeney T.R., Abaeva I.S., Pestova T.V., Hellen C.U. The mechanism of translation initiation on Type 1 picornavirus IRESs. EMBO J. 2014;33:76–92. doi: 10.1002/embj.201386124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shih S.R., Stollar V., Li M.L. Host factors in enterovirus 71 replication. J. Virol. 2011;85:9658–9666. doi: 10.1128/JVI.05063-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flather D., Semler B.L. Picornaviruses and nuclear functions: targeting a cellular compartment distinct from the replication site of a positive-strand RNA virus. Front. Microbiol. 2015;6:594. doi: 10.3389/fmicb.2015.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merrill M.K., Gromeier M. The double-stranded RNA binding protein 76:NF45 heterodimer inhibits translation initiation at the rhinovirus type 2 internal ribosome entry site. J. Virol. 2006;80:6936–6942. doi: 10.1128/JVI.00243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merrill M.K., Dobrikova E.Y., Gromeier M. Cell-type-specific repression of internal ribosome entry site activity by double-stranded RNA-binding protein 76. J. Virol. 2006;80:3147–3156. doi: 10.1128/JVI.80.7.3147-3156.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cathcart A.L., Rozovics J.M., Semler B.L. Cellular mRNA decay protein AUF1 negatively regulates enterovirus and human rhinovirus infections. J. Virol. 2013;87:10423–10434. doi: 10.1128/JVI.01049-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong J., Si X., Angeles A., Zhang J., Shi J., Fung G., Jagdeo J., Wang T., Zhong Z., Jan E., et al. Cytoplasmic redistribution and cleavage of AUF1 during coxsackievirus infection enhance the stability of its viral genome. FASEB J. 2013;27:2777–2787. doi: 10.1096/fj.12-226498. [DOI] [PubMed] [Google Scholar]
- 12.Lin J.Y., Li M.L., Brewer G. mRNA decay factor AUF1 binds the internal ribosomal entry site of enterovirus 71 and inhibits virus replication. PLoS One. 2014;9:e103827. doi: 10.1371/journal.pone.0103827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin J.Y., Li M.L., Shih S.R. Far upstream element binding protein 2 interacts with enterovirus 71 internal ribosomal entry site and negatively regulates viral translation. Nucleic Acids Res. 2009;37:47–59. doi: 10.1093/nar/gkn901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown B.A., Pallansch M.A. Complete nucleotide sequence of enterovirus 71 is distinct from poliovirus. Virus Res. 1995;39:195–205. doi: 10.1016/0168-1702(95)00087-9. [DOI] [PubMed] [Google Scholar]
- 15.Ooi M.H., Wong S.C., Lewthwaite P., Cardosa M.J., Solomon T. Clinical features, diagnosis, and management of enterovirus 71. Lancet Neurol. 2010;9:1097–1105. doi: 10.1016/S1474-4422(10)70209-X. [DOI] [PubMed] [Google Scholar]
- 16.Huang P.N., Lin J.Y., Locker N., Kung Y.A., Hung C.T., Lin J.Y., Huang H.I., Li M.L., Shih S.R. Far upstream element binding protein 1 binds the internal ribosomal entry site of enterovirus 71 and enhances viral translation and viral growth. Nucleic Acids Res. 2011;39:9633–9648. doi: 10.1093/nar/gkr682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L.L., Kung Y.A., Weng K.F., Lin J.Y., Horng J.T., Shih S.R. Enterovirus 71 infection cleaves a negative regulator for viral internal ribosomal entry site-driven translation. J. Virol. 2013;87:3828–3838. doi: 10.1128/JVI.02278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Min H., Turck C.W., Nikolic J.M., Black D.L. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- 19.Chen C.Y., Gherzi R., Ong S.E., Chan E.L., Raijmakers R., Pruijn G.J., Stoecklin G., Moroni C., Mann M., Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Mayoral M.F., Hollingworth D., Masino L., Diaz-Moreno I., Kelly G., Gherzi R., Chou C.F., Chen C.Y., Ramos A. The structure of the C-terminal KH domains of KSRP reveals a noncanonical motif important for mRNA degradation. Structure. 2007;15:485–498. doi: 10.1016/j.str.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 21.Lin W.J., Zheng X., Lin C.C., Tsao J., Zhu X., Cody J.J., Coleman J.M., Gherzi R., Luo M., Townes T.M., et al. Posttranscriptional control of type I interferon genes by KSRP in the innate immune response against viral infection. Mol. Cell Biol. 2011;31:3196–3207. doi: 10.1128/MCB.05073-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trabucchi M., Briata P., Garcia-Mayoral M., Haase A.D., Filipowicz W., Ramos A., Gherzi R., Rosenfeld M.G. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trabucchi M., Briata P., Filipowicz W., Ramos A., Gherzi R., Rosenfeld M.G. KSRP promotes the maturation of a group of miRNA precursors. Adv. Exp. Med. Biol. 2011;700:36–42. [PMC free article] [PubMed] [Google Scholar]
- 24.Clague M.J., Heride C., Urbe S. The demographics of the ubiquitin system. Trends Cell Biol. 2015;25:417–426. doi: 10.1016/j.tcb.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Liu F., Walters K.J. Multitasking with ubiquitin through multivalent interactions. Trends Biochem. Sci. 2010;35:352–360. doi: 10.1016/j.tibs.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida M., Yoshida K., Kozlov G., Lim N.S., De Crescenzo G., Pang Z., Berlanga J.J., Kahvejian A., Gehring K., Wing S.S., et al. Poly(A) binding protein (PABP) homeostasis is mediated by the stability of its inhibitor, Paip2. EMBO J. 2006;25:1934–1944. doi: 10.1038/sj.emboj.7601079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murata T., Shimotohno K. Ubiquitination and proteasome-dependent degradation of human eukaryotic translation initiation factor 4E. J. Biol. Chem. 2006;281:20788–20800. doi: 10.1074/jbc.M600563200. [DOI] [PubMed] [Google Scholar]
- 28.Yanagiya A., Suyama E., Adachi H., Svitkin Y.V., Aza-Blanc P., Imataka H., Mikami S., Martineau Y., Ronai Z.A., Sonenberg N. Translational homeostasis via the mRNA cap-binding protein, eIF4E. Mol. Cell. 2012;46:847–858. doi: 10.1016/j.molcel.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Angers S., Thorpe C.J., Biechele T.L., Goldenberg S.J., Zheng N., MacCoss M.J., Moon R.T. The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-beta-catenin pathway by targeting Dishevelled for degradation. Nat. Cell Biol. 2006;8:348–357. doi: 10.1038/ncb1381. [DOI] [PubMed] [Google Scholar]
- 30.Rondou P., Haegeman G., Vanhoenacker P., Van Craenenbroeck K. BTB Protein KLHL12 targets the dopamine D4 receptor for ubiquitination by a Cul3-based E3 ligase. J. Biol. Chem. 2008;283:11083–11096. doi: 10.1074/jbc.M708473200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin L., Pahuja K.B., Wickliffe K.E., Gorur A., Baumgartel C., Schekman R., Rape M. Ubiquitin-dependent regulation of COPII coat size and function. Nature. 2012;482:495–500. doi: 10.1038/nature10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lydeard J.R., Schulman B.A., Harper J.W. Building and remodelling Cullin-RING E3 ubiquitin ligases. EMBO Rep. 2013;14:1050–1061. doi: 10.1038/embor.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis-Smyth T., Duncan R.C., Zheng T., Michelotti G., Levens D. The far upstream element-binding proteins comprise an ancient family of single-strand DNA-binding transactivators. J. Biol. Chem. 1996;271:31679–31687. doi: 10.1074/jbc.271.49.31679. [DOI] [PubMed] [Google Scholar]
- 34.Dhanoa B.S., Cogliati T., Satish A.G., Bruford E.A., Friedman J.S. Update on the Kelch-like (KLHL) gene family. Hum. Genomics. 2013;7:13. doi: 10.1186/1479-7364-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim W., Bennett E.J., Huttlin E.L., Guo A., Li J., Possemato A., Sowa M.E., Rad R., Rush J., Comb M.J., et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rondou P., Skieterska K., Packeu A., Lintermans B., Vanhoenacker P., Vauquelin G., Haegeman G., Van Craenenbroeck K. KLHL12-mediated ubiquitination of the dopamine D4 receptor does not target the receptor for degradation. Cell Signal. 2010;22:900–913. doi: 10.1016/j.cellsig.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 37.de Jong A.S., de Mattia F., Van Dommelen M.M., Lanke K., Melchers W.J., Willems P.H., van Kuppeveld F.J. Functional analysis of picornavirus 2B proteins: effects on calcium homeostasis and intracellular protein trafficking. J. Virol. 2008;82:3782–3790. doi: 10.1128/JVI.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shukla A., Dey D., Banerjee K., Nain A., Banerjee M. The C-terminal region of the non-structural protein 2B from hepatitis A virus demonstrates lipid-specific viroporin-like activity. Sci. Rep. 2015;5:15884. doi: 10.1038/srep15884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liwak U., Thakor N., Jordan L.E., Roy R., Lewis S.M., Pardo O.E., Seckl M., Holcik M. Tumor suppressor PDCD4 represses internal ribosome entry site-mediated translation of antiapoptotic proteins and is regulated by S6 kinase 2. Mol. Cell Biol. 2012;32:1818–1829. doi: 10.1128/MCB.06317-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bevilacqua E., Wang X., Majumder M., Gaccioli F., Yuan C.L., Wang C., Zhu X., Jordan L.E., Scheuner D., Kaufman R.J., et al. eIF2alpha phosphorylation tips the balance to apoptosis during osmotic stress. J. Biol. Chem. 2010;285:17098–17111. doi: 10.1074/jbc.M110.109439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komander D., Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 42.Kafasla P., Morgner N., Robinson C.V., Jackson R.J. Polypyrimidine tract-binding protein stimulates the poliovirus IRES by modulating eIF4G binding. EMBO J. 2010;29:3710–3722. doi: 10.1038/emboj.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blyn L.B., Swiderek K.M., Richards O., Stahl D.C., Semler B.L., Ehrenfeld E. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5′ noncoding region: identification by automated liquid chromatography-tandem mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 1996;93:11115–11120. doi: 10.1073/pnas.93.20.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blyn L.B., Towner J.S., Semler B.L., Ehrenfeld E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 1997;71:6243–6246. doi: 10.1128/jvi.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]