Abstract
RNase H enzymes sense the presence of ribonucleotides in the genome and initiate their removal by incising the ribonucleotide-containing strand of an RNA:DNA hybrid. Mycobacterium smegmatis encodes four RNase H enzymes: RnhA, RnhB, RnhC and RnhD. Here, we interrogate the biochemical activity and nucleic acid substrate specificity of RnhA. We report that RnhA (like RnhC characterized previously) is an RNase H1-type magnesium-dependent endonuclease with stringent specificity for RNA:DNA hybrid duplexes. Whereas RnhA does not incise an embedded mono-ribonucleotide, it can efficiently cleave within tracts of four or more ribonucleotides in duplex DNA. We gained genetic insights to the division of labor among mycobacterial RNases H by deleting the rnhA, rnhB, rnhC and rnhD genes, individually and in various combinations. The salient conclusions are that: (i) RNase H1 activity is essential for mycobacterial growth and can be provided by either RnhC or RnhA; (ii) the RNase H2 enzymes RnhB and RnhD are dispensable for growth and (iii) RnhB and RnhA collaborate to protect M. smegmatis against oxidative damage in stationary phase. Our findings highlight RnhC, the sole RNase H1 in pathogenic mycobacteria, as a candidate drug discovery target for tuberculosis and leprosy.
INTRODUCTION
There is rising interest in the biological impact of ribonucleotides embedded in bacterial chromosomes during DNA replication and repair, and in the pathways of ribonucleotide surveillance that deal with such ‘lesions’ (1). Bacterial polymerases display a range of fidelities with respect to discrimination of dNTP and rNTP substrates. The roster of DNA repair polymerases of the human pathogen Mycobacterium tuberculosis and its avirulent relative M. smegmatis includes four enzymes—LigD-POL, PolD1, PolD2 and DinB2—that have the distinctive properties of low fidelity and of readily incorporating ribonucleotides in lieu of deoxyribonucleotides during primer extension and gap repair in vitro (2–9). We are interested in the connections in mycobacteria between ribonucleotide utilization and replicative quiescence, which is central to the long-term carriage of M. tuberculosis in a clinically dormant state. Cells that are not replicating their DNA may have reduced dNTP pools compared to actively dividing cells. We speculate that DNA repair with a ‘ribo patch’ by polymerase utilization of available rNTPs provides a strategy for quiescent cells to avoid otherwise deadly chromosome damage.
The in vivo impact of ribonucleotide incorporation by mycobacterial polymerases may be obscured by the presence in the M. smegmatis proteome of four ribonuclease H enzymes: MSMEG_5562/RnhA (10); MSMEG_4305/RnhC (11); MSMEG_2442/RnhB (12) and MSMEG_5849 (13). RNase H enzymes incise the RNA strand of RNA:DNA hybrid duplexes; they are classified as type I (H1) or type II (H2 and H3) (14–17). RNase H1 requires an oligoribonucleotide tract and is unable to incise a single ribonucleotide embedded in duplex DNA. RNase H2 is uniquely capable of incising a single embedded rNMP. Of the four M. smegmatis RNase H enzymes, only RnhC has been characterized with respect to its RNA requirements (11), a relevant issue given that the ribo-utilizing mycobacterial polymerases differ with respect to how many sequential ribonucleotides they can embed. The 365-aa RnhC polypeptide consists of two autonomous catalytic domains: an N-terminal 140-aa RNase H1 module and a C-terminal 211-aa acid phosphatase module. [The homologous bifunctional RnhC in M. tuberculosis is Rv2228c (18).] The M. smegmatis RnhC endonuclease is stringently specific for RNA:DNA hybrid duplexes, but RnhC does not selectively recognize and cleave at DNA–RNA or RNA–DNA junctions in duplex nucleic acid. RnhC can incise tracts of four or more ribonucleotides embedded in duplex DNA, leaving two or more residual ribonucleotides at the cleaved 3′-OH end and at least one or two ribonucleotides on the 5′-PO4 end. However, RnhC cannot incise an embedded mono-ribonucleotide or di-ribonucleotide in duplex DNA (11).
These biochemical characteristics of RnhC seem to rule it out as an agent of ribonucleotide excision repair (RER) of single ribonucleotides embedded during DNA replication or repair. This RER function, to the extent that it is operative in mycobacteria, would likely default to one or both of the putative type II RNase H enzymes: RnhB and MSMEG_5849 (referred to henceforth as RnhD). Neither M. smegmatis RnhB (272-aa) nor its M. tuberculosis counterpart Rv2902c (264-aa), both of which are single-domain proteins homologous to Escherichia coli RNase H2, has been characterized biochemically. Deletion of the M. smegmatis rnhB gene has no impact on bacterial growth in liquid culture, sensitivity to hydroxyurea or spontaneous mutation rate (12). Mycobacterium smegmatis RnhD (alias RHII-RSD) is a 567-aa bifunctional enzyme composed of an N-terminal RNase H2-like domain and a C-terminal (p)ppGpp synthetase domain (13). Full-length RnhD, but not an isolated N-terminal domain (aa 1–280), displayed RNase H activity in vitro (13). The counterpart in M. tuberculosis is Rv0776c, a 259-aa protein homologous to the N-terminal segment of M. smegmatis RnhD, but lacking the (p)ppGpp synthetase module.
Mycobacterium smegmatis RnhA (159-aa) was identified in 1995 by Mizrahi et al., who cloned the rnhA gene, produced the protein in E. coli as a maltose-binding protein (MBP)–RnhA fusion, and showed that the affinity-purified MBP–RnhA displayed endoribonuclease activity on an RNA:DNA hybrid substrate (10). Here, we extend their work by purifying and characterizing a His-tagged version of the M. smegmatis RnhA protein. We find that RnhA is a vigorous magnesium-dependent RNase H with strict specificity for cleavage of an RNA:DNA hybrid. It does not cleave an RNA:RNA duplex of otherwise identical primary structure. Like RnhC, RnhA can recognize and efficiently cleave a 4-nucleotide ribo tract embedded in duplex DNA, but not an embedded di-ribonucleotide or mono-ribonucleotide. Thus, RnhA is a type I RNase H. Whereas M. smegmatis has two type I RNase H enzymes in RnhA and RnhC, the M. tuberculosis proteome is devoid of an RnhA homolog.
We extend our biochemical findings by querying the genetics of RNase H in M. smegmatis, via constructing and characterizing deletion mutants of rnhA, rnhB, rnhC and rnhD, singly and in combination. We find that ΔrnhA, ΔrnhB, ΔrnhC and ΔrnhD single mutants are viable, as are ΔrnhA ΔrnhB, ΔrnhB ΔrnhC and ΔrnhB ΔrnhD double mutants, and a ΔrnhA ΔrnhB ΔrnhD triple mutant. We conclude that there is no essential role for M. smegmatis RNase H2-type enzymes under standard laboratory growth conditions. However, we find that ΔrnhA ΔrnhB cells in stationary phase are highly sensitized to killing by hydrogen peroxide and display enhanced sensitivity to UV irradiation.
In agreement with the recent data of Minias et al. (19), we were unable to simultaneously delete the rnhA and rnhC loci, unless a second copy of rnhC or rnhA was introduced elsewhere on the M. smegmatis chromosome. We proceeded to discriminate whether the synthetic lethality reflected the absence of an RNase H1-type protein, an RNase H1 endonuclease activity, or simultaneous absence of an RNase H1 activity and the acid phosphatase activity of RnhC, by complementation with biochemically confirmed ‘catalytic-dead’ mutant alleles of rnhA and rnhC. We show that: (i) RNase H1 activity is essential for viability of M. smegmatis and (ii) the acid phosphatase activity of RnhC is not. We discuss how these findings fortify the case for RNase H1 as a therapeutic target for human mycobacterial diseases.
MATERIALS AND METHODS
Recombinant RnhA proteins
A 480-nt DNA fragment comprising the rnhA open reading frame (ORF) was amplified from M. smegmatis genomic DNA by PCR with primers that introduced a NdeI site at the start codon and a BglII site immediately after the stop codon. The PCR product was digested with NdeI and BglII and inserted between the NdeI and BamHI sites of pET16b-His10 to generate expression plasmid pET-His10•RnhA. Missense mutations E50Q and D72N were introduced into the rnhC ORF by PCR with mutagenic primers. The inserts in each plasmid were sequenced to verify the fusion junctions and to ensure that no unwanted coding changes were introduced during amplification and cloning.
The RnhA expression plasmids were transfected into E. coli BL21 (DE3) cells. Cultures (2-liter) amplified from single transformants were grown at 37°C in Terrific Broth containing 100 μg/ml ampicillin until the A600 reached 0.8. The cultures were chilled on ice for 1 h, adjusted to 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and then incubated for 20 h at 18°C with constant shaking. All subsequent steps of purification were performed at 4°C. Cells were harvested by centrifugation and resuspended in 35 ml of buffer A (50 mM Tris–HCl, pH 7.5, 500 mM NaCl, 20 mM imidazole, 10% glycerol). The cells were lysed by sonication and the insoluble material was removed by centrifugation at 18000 rpm for 45 min. Supernatants were mixed for 1 h with 4 ml of Ni-NTA resin (Qiagen) that had been equilibrated with buffer A. The resins were poured into gravity-flow columns and then washed with 60 ml of buffer A. The adsorbed proteins were step-eluted with 300 mM imidazole in buffer A. The polypeptide compositions of the eluate fractions were monitored by SDS-PAGE and the peak fractions containing each recombinant protein were pooled. RnhA proteins were then subjected to gel filtration through a Superdex-200 column (GE Healthcare) equilibrated in 50 mM Tris–HCl, pH 7.5, 200 mM NaCl, 10% glycerol. Peak monomeric RnhA fractions were pooled, concentrated by centrifugal ultrafiltration to 23 mg/ml [wild-type RnhA], 12 mg/ml [RnhC-E50Q], and 13 mg/ml [RnhC-D72N] and then stored at −80°C. Protein concentrations were determined from the A280 measured with a Nanodrop spectrophotometer (Thermo scientific), applying extinction coefficients calculated with Protparam.
RNase H substrates
RNA and DNA oligonucleotides were purchased from Dharmacon and ThermoFisher Scientific, respectively. Oligonucleotides were 5′ 32P-labeled by reaction with T4 polynucleotide kinase and [γ32P]ATP, then gel-purified. Labeled oligonucleotides were annealed to a 4- to 8-fold molar excess of the unlabeled complementary strands, in buffer containing 0.2 M NaCl, to form the various duplex substrates shown in the figures. Partial alkaline hydrolysates of the 5′ 32P-labeled strands were prepared by incubation in a solution of 40 mM NaHCO3 (pH 8.0), 60 mM Na2CO3 (pH 10.5) for 12 min at 95°C.
M. smegmatis Δrnh mutants
In-frame deletion of rnhB in wild-type M. smegmatis was achieved by a two-step allelic exchange process that uses a suicide vector containing a hygromycin-resistance marker and counterselectable markers sacB and galK as described previously (20). Deletions of rnhA (Mgm4087), rnhC (Mgm4084) and rnhD (Mgm4089), in M. smegmatis were attained by specialized transduction using a temperature-sensitive mycobacteriophage (21). Diagnostic restriction endonuclease digestion and Southern blotting of genomic DNA was performed to confirm each Δrnh knockout using either 5′- or 3′-flanking DNA sequences of the gene as the hybridization probe. The double mutants ΔrnhB ΔrnhA, ΔrnhB ΔrnhC and ΔrnhB ΔrnhD were constructed by transduction of the ΔrnhA, ΔrnhC and ΔrnhD phages in the unmarked ΔrnhB strain. In case of the ΔrnhB ΔrnhA mutant, the hygR marker (flanked by loxP sites) at the disrupted rnhA locus was subsequently excised by expressing Cre recombinase to generate the unmarked ΔrnhB ΔrnhA strain, which was then used as the starting strain to delete rnhD by phage transduction to yield the triple mutant ΔrnhB ΔrnhA ΔrnhD. The ΔrnhC single mutant was unmarked by expressing Cre to provide the starting strain (Mgm4084) for attempts (unsuccessful) to generate a ΔrnhC ΔrnhA double mutant. The double and triple Δrnh mutants were verified by diagnostic restriction endonuclease digestion and Southern blotting of genomic DNA. The M. smegmatis strains used in the study and their genotypes are compiled in Supplementary Table S1.
Growth studies
M. smegmatis strains were revived from frozen glycerol stocks in LB medium (supplemented with 0.5% glycerol, 0.5% dextrose, 0.05% Tween 80) followed by sub-culturing in 7H9 medium (supplemented with 0.5% glycerol, 0.5% dextrose, 0.05% Tween 80). To study growth kinetics, the strains were re-inoculated into fresh 7H9 medium to A600 of 0.1 and incubated at 37°C with constant shaking (150 rpm). Once log phase was achieved, the cultures were repeatedly diluted into fresh medium at regular intervals to maintain growth in log phase. Culture aliquots were removed at these times to measure the A600 and determine viable bacterial counts by serial dilution plating. A change in absorbance or colony counts was plotted against time, and doubling time was calculated using the following formula: doubling time = (tt – t0)/G, where tt is the time at which an aliquot from a growing culture was removed and t0 is the time at which the culture was rediluted into fresh medium to begin the growth curve, and where G (the number of generations) = (log[number of bacteria or A600 at tt]—log[number of bacteria or A600 at t0])/0.301.
UV irradiation sensitivity
M. smegmatis strains to be tested were grown to logarithmic phase (A600 0.3–0.4) or stationary-phase (for 72 h after achieving A600 of 0.5). Serial 10-fold dilutions prepared in phosphate-buffered saline (PBS) with 0.05% Tween 80 were spotted on 7H10 agar plates supplemented with 0.5% glycerol, 0.5% dextrose. UV irradiation at the doses specified in the figures was performed with a Stratalinker (Stratagene) fitted with 254 nm bulbs. Immediately after exposure, the plates were wrapped in foil to prevent repair by photolyase. Surviving colonies were counted after 3 days by using a dissecting microscope, and the percent survival was calculated in comparison to the non-irradiated cells from the same culture. The assays were performed at least twice with biological duplicates for all strains in each experiment. The mean value of percent survival (± standard error of the mean; SEM) for every strain is plotted as a function of UV dose in the figures shown.
Ionizing radiation (IR) sensitivity
M. smegmatis strains grown to logarithmic phase or stationary-phase (as described in the preceding section) were collected by centrifugation and resuspended in PBS with 0.05% Tween 80. Aliquots (200 μl) of the resuspended cells were irradiated with a 137Cs source using a rotating platform to ensure equal exposure to each sample. Following IR exposure, serial 10-fold dilutions were spread on agar media, and surviving colonies were counted after 3 days. Percent survival was calculated after normalization to the colony counts obtained from the unexposed control cells. The assays were performed at least twice with biological duplicates for all strains in each experiment. The mean value of percent survival (±SEM) for every strain is plotted as a function of IR dose in the figures shown.
Hydrogen peroxide sensitivity
Log phase or stationary phase cultures were treated for 2 h with hydrogen peroxide at the concentrations specified in the figures. Serial 10-fold dilutions were plated and percent survival was calculated by determining the colony-forming units (cfu) of the treated versus untreated cultures. The assays were performed at least twice with biological duplicates for all strains in each experiment. The mean value of percent survival (±SEM) for every strain is plotted as a function of peroxide concentration in the figures shown.
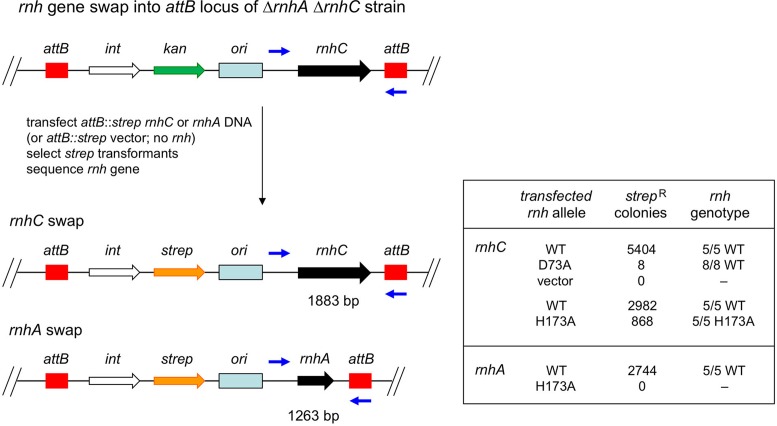
Insertion of rnhC at the M. smegmatis attB locus and tests of RnhC and RnhA function by allelic exchange
To test the essentiality of RNase H1 activity, we constructed a merodiploid strain carrying chromosomal ΔrnhC::loxP ΔrnhA and a complementing copy of rnhC at the attB chromosomal integration site (strain Mgm4083) on a plasmid conferring kanamycin resistance (see schematic in Figure 9). We employed marker swapping with streptomycin resistance-conferring attB integrating plasmids to interrogate the ability of different alleles of rnhC or rnhA to support viability. Streptomycin resistant colonies after transfection with 50 ng of plasmid DNA were enumerated and survivors were genotyped by PCR and sequencing to confirm allelic replacement with the expected rnh allele.
Figure 9.
Genetic testing of the essentiality of RNase H1 endonuclease activity. A schematic representation of the chromosomal attB locus in strain Mgm4083, which has chromosomal deletions of rnhC and rnhA and a copy of rnhC at the attB locus on an integrated plasmid element conferring kanamycin resistance. Because of spontaneous excision of attB integrated plasmids, replacement with a transfected plasmid carrying a streptomycin resistance gene can be efficiently achieved, allowing swapping of the wild type rnhC gene for wild type and mutated versions of rnhC or rnhA. The results of these marker exchange experiments are given in the table as: (i) the number of strepR colonies recovered after transfection with 50 ng of the plasmid carrying the indicated allele and (ii) the results of genotyping of survivors to confirm allelic exchange.
RESULTS
Recombinant RnhA has ribonuclease H activity
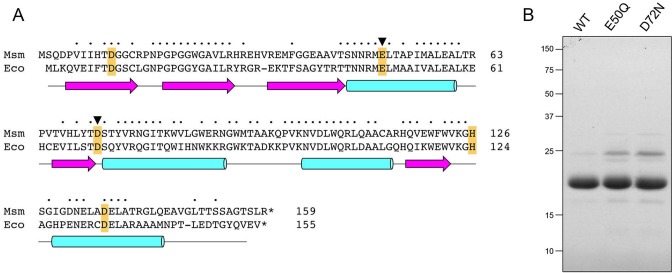
M. smegmatis RnhA (159-aa) is a homolog of E. coli RNase H1 (155-aa), which has been characterized biochemically and structurally (22). Alignment of their amino acid sequences highlights 88 positions of side chain identity/similarity (indicated by • in Figure 1A). Five conserved acidic residues, Asp11, Glu50, Asp72, His126 and Asp136, are predicted to coordinate two catalytic metal ions in the RnhA active site (15). We produced full-length wild-type M. smegmatis RnhA in E. coli as a His10 fusion and isolated the protein from a soluble extract by nickel-agarose chromatography. After gel filtration through Superdex 200, the preparation consisted of a predominant ∼18 kDa RnhA polypeptide, as gauged by SDS-PAGE (Figure 1B). In parallel, we produced and purified two mutants, E50Q and D72N, that have conservative substitutions at the metal binding site (Figure 1B).
Figure 1.
Recombinant RnhA. (A) Primary structure. The amino acid sequence of M. smegmatis (Msm) RnhA is aligned to that of E. coli (Eco) RNase H1. Positions of side chain identity/similarity are denoted by dots above the residues. Gaps in the alignments are denoted by dashes. Five conserved acidic residues—Asp11, Glu50, Asp72, His126 and Asp136 in RnhA—that are predicted to coordinate two catalytic metal ions in the active site are highlighted in gold shading. The Glu49 and Asp72 residues that were mutated are denoted by black arrowheads. The secondary structure elements of the E. coli RNase H1 crystal structure (24; pdb 1RNH) are displayed below the amino acid sequence, with β-strands as magenta arrows and α-helices as cyan cylinders. (B) Purification. Aliquots (5 μg) of recombinant wild-type RnhA (WT) and mutants E50Q and D72N were analyzed by SDS-PAGE. The Coomassie Blue-stained gel is shown. The positions and sizes (kDa) of marker polypeptides are indicated on the left.
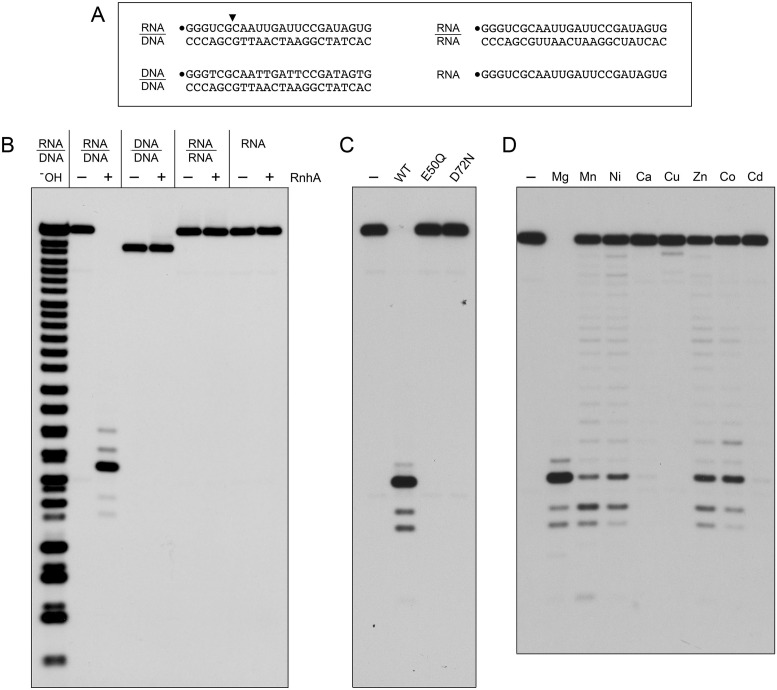
To assay RNase H activity, we annealed a 5′ 32P-labeled 24-mer RNA oligonucleotide to an unlabeled complementary 24-mer DNA strand and reacted the RNA:DNA hybrid (depicted in Figure 2A) with RnhA and magnesium for 20 min at 37°C. The reactions were quenched with EDTA, after which the products were analyzed by urea–PAGE and visualized by autoradiography. RnhA quantitatively incised the RNA:DNA hybrid to yield a shorter end-labeled RNA fragment (Figure 2A). A partial alkaline hydrolysis ladder was analyzed in parallel in Figure 2A, to provide a rough indication of product size. (Note that partial alkaline hydrolysis generates a mixture of 2′-phosphate, 3′-phosphate, and 2′,3′-cyclic phosphate ends; the monophosphate and cyclic phosphate species are resolved as doublets only for the shortest fragments in the ladder. The RNase H cleavage products, which have 3′-OH termini, migrate slower during PAGE than end-labeled oligonucleotides of the same chain length in the alkaline ladder.) As described previously (11), in order to accurately assign the sites of RNA cleavage, we analyzed the reaction products in parallel with a partial digest of the 24-mer 32P-RNA strand by purified M. smegmatis polynucleotide phosphorylase to produce a ladder of 5′ 32P-labeled RNAs with 3′-OH termini. We thereby determined that the predominant 32P-labeled RnhA reaction product derived from incision at the sixth inter-nucleotide phosphodiester (the site indicated by an arrowhead in Figure 2A). No ribonuclease activity was detected when divalent cation was omitted (Figure 2D). Manganese, nickel, zinc and cobalt (at 5 mM concentration) were feeble activators of the RnhA endonuclease; calcium, copper and cadmium (5 mM) were inactive (Figure 2D). Mutations E50Q and D72N abolished the magnesium-dependent nuclease activity (Figure 2C). We conclude that the observed RNase H activity inheres to the recombinant RnhA protein.
Figure 2.
Metal-dependent RNase H activity and nucleic acid substrate specificity of RnhA. (A) Substrates. The 32P-RNA:DNA, 32P-DNA:DNA and 32P-RNA:RNA duplexes and the 32P-RNA single strand substrates are shown, with the 5′ 32P-label denoted by •. The principal site of RnhA incision of the RNA:DNA hybrid is indicated by a black arrowhead above the 32P-labeled RNA strand. (B) Nucleic acid substrate specificity. Reaction mixtures (10 μl) containing 25 mM Tris-HCl (pH 7.5), 50 mM NaCl, 5 mM MgCl2, either 20 nM (200 fmol) 32P-RNA:DNA, 32P-DNA:DNA or 32P-RNA:RNA duplexes or 20 nM (200 fmol) 32P-RNA single strand, and 8 nM (80 fmol) RnhA (where indicated by +) were incubated at 37°C for 20 min. The reactions were quenched with an equal volume of 90% formamide, 50 mM EDTA, 0.3% bromophenol blue. The reaction products were analyzed by electrophoresis through a 40-cm 18% polyacrylamide gel containing 7 M urea in 45 mM Tris-borate, 1 mM EDTA. An alkaline hydrolysis ladder of the 32P-labeled 24-mer RNA strand was analyzed in parallel in lane –OH. The radiolabeled RNAs were visualized by autoradiography. (C) Active site mutations. Reaction mixtures (10 μl) containing 25 mM Tris–HCl (pH 7.5), 50 mM NaCl, 5 mM MgCl2, 20 nM (200 fmol) 32P-RNA:DNA hybrid duplex, and 8 nM (80 fmol) of wild-type RnhA, RnhA-E50Q or RnhA-D72N were incubated at 37°C for 20 min. RnhA was omitted from a control reaction in lane –. (D) Metal cofactor requirement. Reaction mixtures (10 μl) containing 25 mM Tris–HCl, pH 7.5, 50 mM NaCl, 20 nM 32P-RNA:DNA hybrid duplex, 8 nM wild-type RnhA, and 5 mM of the indicated divalent cation (as the chloride salt) were incubated at 37°C for 20 min. Divalent cation was omitted from a control reaction in lane –.
Nucleic acid substrate specificity of the RnhA nuclease
To probe substrate specificity, we prepared a series of 5′ 32P-labeled 24-bp duplex nucleic acids in which the labeled strand was RNA or DNA and the complementary unlabeled strand was either RNA or DNA (Figure 2A). At a level of input RnhA that sufficed for quantitative cleavage of the labeled RNA strand of the RNA:DNA hybrid, we detected no cleavage of a DNA:DNA duplex or an RNA:RNA duplex (Figure 2B). Moreover, RnhA did not incise the 32P-labeled RNA single strand (Figure 2B).
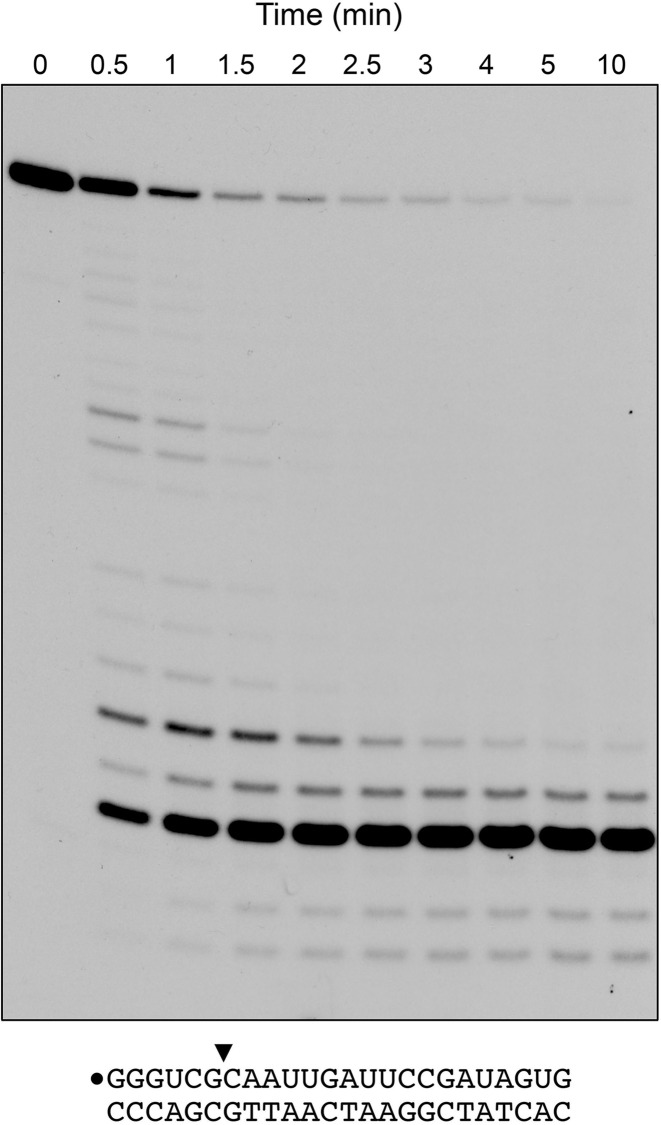
The kinetic profile of a reaction of 8 nM RnhA with 20 nM 24-bp RNA:DNA hybrid showed that, at early times (0.5 min), when not all of the input 24-mer RNA strand had been incised, cleavage generated a mixture of 32P-labeled strands 6- to 21-nucleotides long, reflecting initial incision at multiple different inter-nucleotide phosphodiesters (Figure 3). As the input substrate was consumed, the product distribution shifted over time so that the 6-mer end-labeled cleavage product predominated (Figure 3). By plotting the consumption of the input 24-mer RNA strand with time, we estimated a turnover number of 3.4 min−1. RnhA activity was active over a broad pH range from pH 6.0 to 9.0 (not shown).
Figure 3.
Time course. A reaction mixture containing 25 mM Tris–HCl (pH 7.5), 50 mM NaCl, 5 mM MgCl2, 20 nM 32P-RNA:DNA hybrid duplex (depicted at bottom), and 8 nM RnhA was incubated at 37°C. Aliquots (10 μl) were withdrawn at the times specified and quenched with formamide/EDTA. The reaction products were analyzed by urea–PAGE and visualized by autoradiography.
Cleavage of chimeric RNA–DNA strands by RnhA
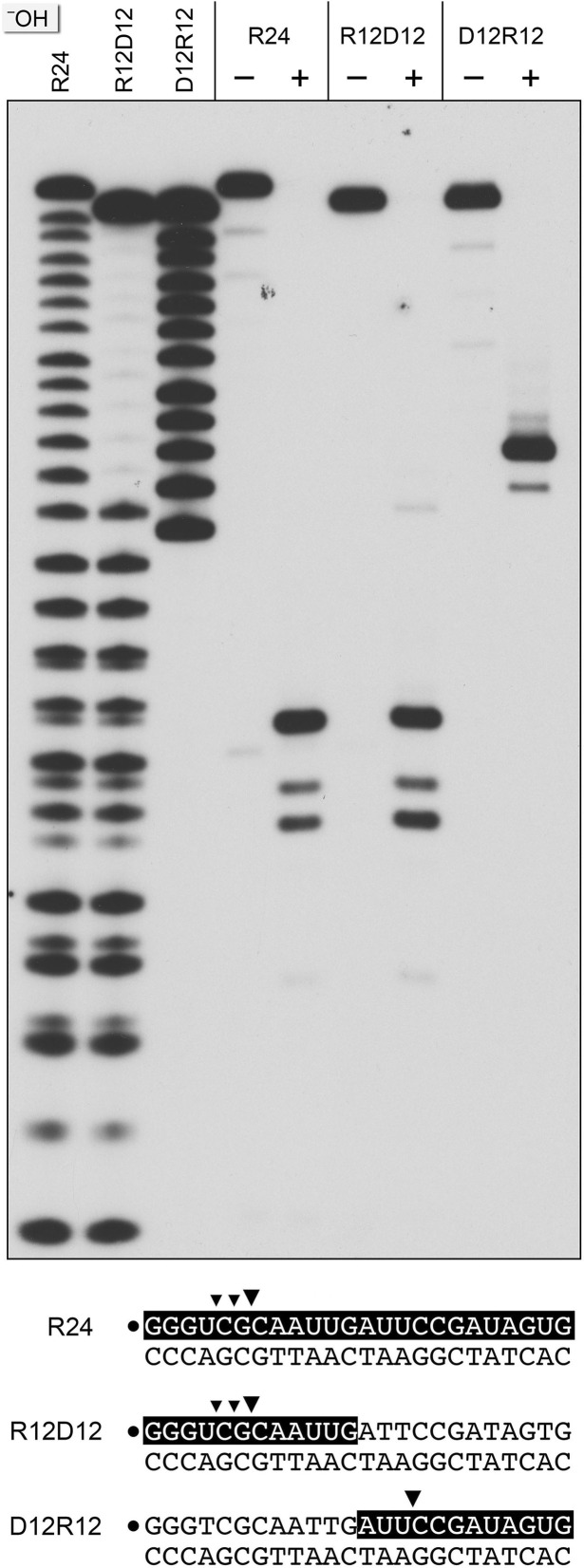
Chimeric duplex substrates were prepared in which the 32P-labeled scissile strand consisted of a 5′ segment of 12 ribonucleotides and a 3′ segment of 12 deoxynucleotides (R12D12) or a 5′ segment of 12 deoxynucleotides and a 3′ segment of 12 ribonucleotides (D12R12) (Figure 4). The R12D12 duplex is analogous to an Okazaki fragment; the D12-R12 duplex is analogous to the fill-in repair reaction product synthesized by M. smegmatis DinB2 in the presence of rNTPs. RnhA was reacted with the chimeric substrates in parallel with an RNA:DNA hybrid with an all-RNA strand (R24) of identical primary structure. Partial alkaline hydrolysis confirmed the chimeric nature of the R12D12 and D12R12 strands (Figure 4).
Figure 4.
Cleavage of chimeric RNA–DNA junction substrates. Reaction mixtures (10 μl) containing 25 mM Tris–HCl (pH 7.5), 50 mM NaCl, 5 mM MgCl2, 20 nM (200 fmol) 32P-labeled 24-mer duplexes R24, R12D12 or D12R12 (shown at the bottom, with the 32P label denoted by • and the ribonucleotides depicted in white on a black background), and 8 nM (80 fmol) RnhA (where indicated by +) were incubated for 20 min at 37°C. The products were resolved by urea-PAGE and visualized by autoradiography. Alkaline hydrolysis ladders of 32P-labeled R24, R12D12 and D12R12 strand were analyzed in parallel in the three lanes on the left (–OH).
Reaction of 8 nM RnhA with 20 nM R12D12 duplex resulted in quantitative incision of the RNA segment of the chimeric strand at the same sites that were cleaved in the R24 control duplex (Figure 4). RnhA cleaved the RNA segment of the D12R12 duplex to yield one major radiolabeled product, which consisted of the proximal DNA segment plus three 3′-terminal ribonucleotides (Figure 4). We surmise that: (i) RnhA does not preferentially cleave the junctions of RNA and DNA segments and (ii) RnhA does not completely remove a ribonucleotide tract installed 3′ of DNA to yield a ‘clean’ DNA3′OH end.
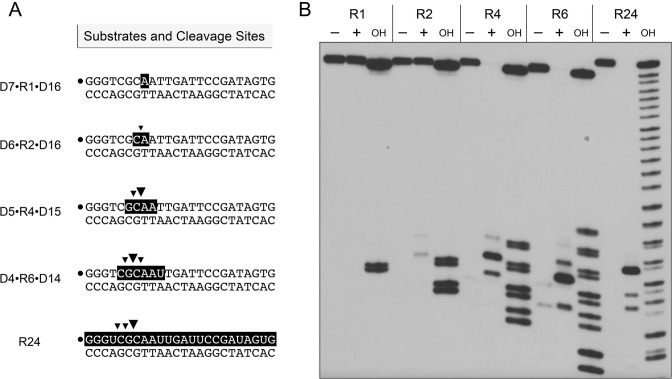
Minimal RNA requirement for RnhA
Further insights to substrate specificity were gleaned by reacting M. smegmatis RnhA with a series of chimeric 24-bp substrates, of otherwise identical primary structure, in which one, two, four, or six ribonucleotides were embedded between 5′ and 3′ flanking DNA segments (Figure 5A). Reactions with the all-RNA:DNA hybrid duplex (R24) were included as positive controls. Partial alkaline hydrolysis of the 32P-labeled strands verified the length and position of the ribonucleotide tracts (Figure 5B). Previous studies of the reaction of these substrates with E. coli RNase H2 revealed that it efficiently and specifically incised the R1, R2, R4 and R6 duplexes at the phosphodiester immediately 5′ of the ribonucleotide at the RNA–DNA junction (11).
Figure 5.
Minimum RNA requirement. (A) Substrates and cleavage sites. The 32P label is denoted by •. Ribonucleotides are depicted in white on a black background. Principal sites of incision by M. smegmatis RnhA are indicated by black arrowheads. (B) Reaction mixtures (10 μl) containing 25 mM Tris–HCl (pH 7.5), 50 mM NaCl, 5 mM MgCl2, 20 nM (200 fmol) duplex substrate as specified, and either no enzyme (lanes –) or 8 nM (80 fmol) RnhA were incubated at 37°C for 20 min. The products were analyzed by urea-PAGE and visualize by autoradiography. Alkaline hydrolysis ladders of the 32P-labeled strands were analyzed in parallel in lanes OH.
Here, we find that M. smegmatis RnhA quantitatively cleaved the R4 and R6 substrates, but failed to incise the R1 duplex and was extremely feeble in cleaving the R2 duplex (5% of the input 24-mer was incised) (Figure 5B). RnhA incised the R4 and R6 duplexes at the positions denoted by black arrowheads in Figure 5A. The RnhA cleavage patterns on the R4 and R6 strands suggest that this enzyme works best when there are at least two ribonucleotides on the ‘upstream’ side of the scissile phosphodiester and at least two ribonucleotides on the ‘downstream’ side: p(rN)p(rN)↓p(rN)p(rN). We conclude from this analysis that RnhA is a canonical type I RNase H enzyme.
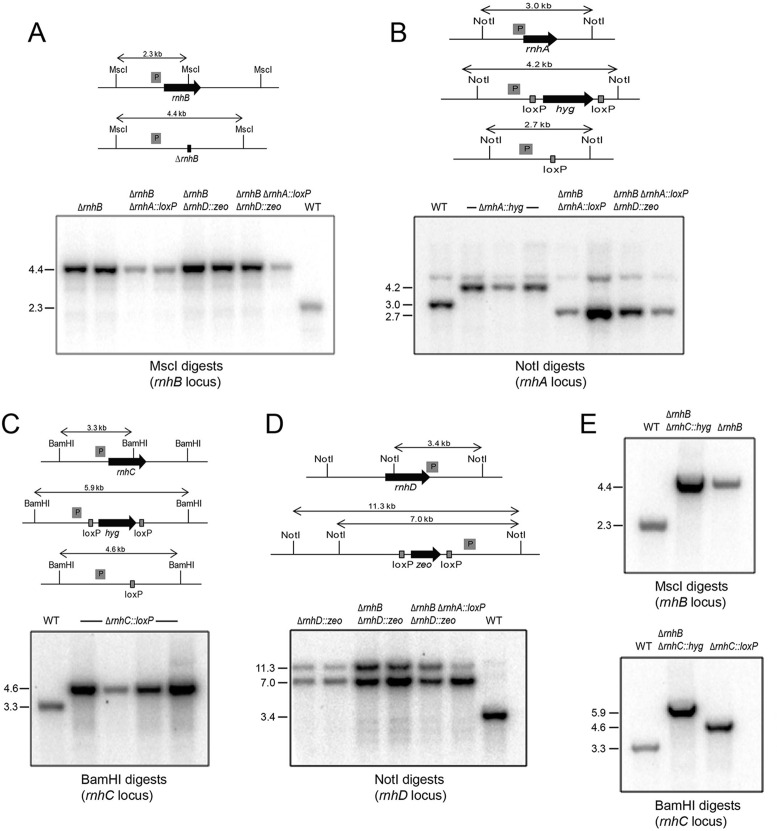
Viable deletion mutants of M. smegmatis rnhA, rnhB, rnhC and rnhD
Single gene deletion strains were constructed as described under Materials and Methods. The genotypes of the Δrnh strains were analyzed by diagnostic restriction endonuclease digestion of genomic DNA and Southern blotting with gene-specific probes, which verified that the fragment containing the native wild-type rnh locus had been replaced with a new restriction fragment corresponding to the intended Δrnh allele (Figure 6A–D). The ΔrnhA, ΔrnhB, ΔrnhC and ΔrnhD single mutants grew as well as wild-type M. smegmatis in liquid culture in 7H9 medium at 37°C, with doubling times calculated from A600 cell density measurements as follows: wild-type (179 min); ΔrnhA (177 min); ΔrnhB (181 min); ΔrnhC (178 min); ΔrnhD (179 min). Doubling times determined by quantitation of viable bacteria were as follows: wild-type (169 min); ΔrnhA (173 min); ΔrnhB (168 min); ΔrnhC (171 min); ΔrnhD (167 min). Double mutants ΔrnhA ΔrnhB, ΔrnhB ΔrnhC, and ΔrnhB ΔrnhD and a triple mutant ΔrnhA ΔrnhB ΔrnhD were generated by sequential knockout maneuvers and the genotypes of these strains were verified by restriction and Southern blotting of genomic DNA (Figure 6A–E). Their doubling times by A600 were: ΔrnhA ΔrnhB (186 min); ΔrnhB ΔrnhC (182 min); ΔrnhB ΔrnhD (183 min); ΔrnhA ΔrnhB ΔrnhD (185 min). Their doubling times by viable bacterial counts were: ΔrnhA ΔrnhB (177 min); ΔrnhB ΔrnhC (170 min); ΔrnhB ΔrnhD (171 min); ΔrnhA ΔrnhB ΔrnhD (180 min). Two salient points emerge from these gene deletions: (i) M. smegmatis does not require an RNase H2-type enzyme, viz., the ΔrnhB ΔrnhD strain is viable and has no growth defect and (ii) M. smegmatis is viable when RnhC is the only RNase H enzyme available.
Figure 6.
Genotyping of Δrnh mutants. For each panel, schematic representations of the wild type and Δrnh genetic loci are shown with probe location (marked by a grey box ‘P’) and restriction endonuclease sites. Predicted hybridization products are indicated for wild type and deletion alleles. Below these schematics are autoradiograms of Southern blots of restriction endonuclease-digested chromosomal DNA hybridized with the indicated radiolabelled DNA probe from candidate recombinant strains in either wild type or Δrnh backgrounds, as indicated above each lane, along with chromosomal DNA derived from wild type M. smegmatis (WT). (A) Deletion of rnhB in WT (strain Mgm4085), ΔrnhA (strain Mgm4088), ΔrnhD (strain Mgm4090) and ΔrnhA ΔrnhD (strain Mgm4091). (B) Deletion of rnhA in WT (strain Mgm4087), ΔrnhB (strain Mgm4088), and ΔrnhB ΔrnhD (strain Mgm4091). (C) Deletion of rnhC in WT background after removal of the hygR marker via Cre recombinase (strain Mgm4084). (D) Deletion of rnhD in WT (strain Mgm4089), ΔrnhB (strain Mgm4090), ΔrnhA ΔrnhB (strain Mgm4091). (E) Confirmation of the ΔrnhB ΔrnhC double mutant (Mgm4086). Deletion of rnhB in the ΔrnhC background (top panel, see A for schematic) and rnhC in the ΔrnhB background (see C for schematic). In both top and bottom panels, the single mutants ΔrnhB (strain Mgm4085) and ΔrnhC::loxP (strain Mgm4084) were used as the controls in the last lanes, respectively.
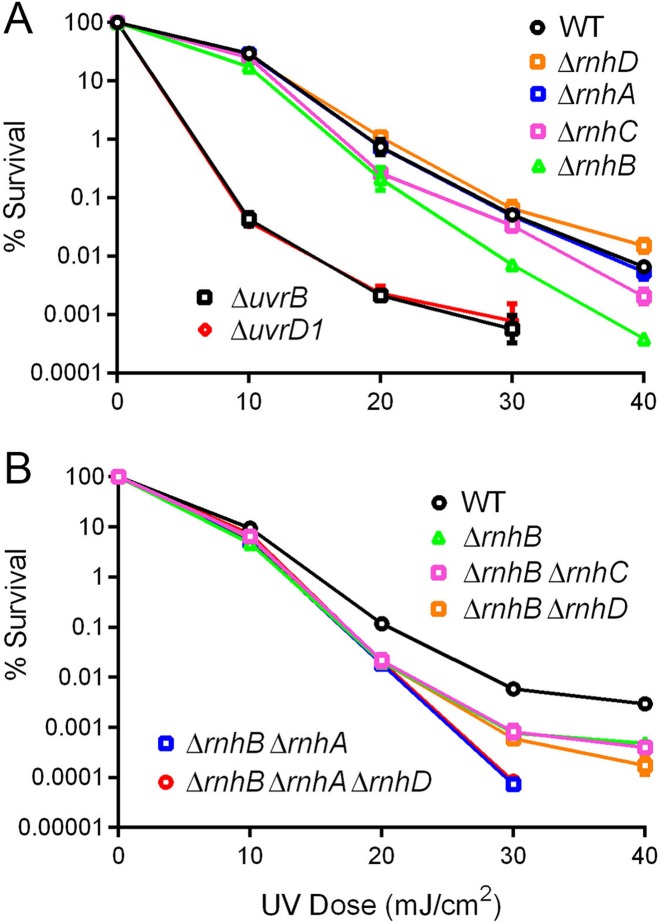
Are Δrnh mutants sensitized to DNA radiation damage?
We tested the clastogen sensitivity of Δrnh strains by exposing them to ultraviolet radiation (UV) or ionizing radiation (IR). Logarithmically growing ΔrnhA, ΔrnhB, ΔrnhC, and ΔrnhD cells were no more sensitive than wild-type M. smegmatis to killing by escalating doses of UV or IR (Supplementary Figure S1A and B). We reasoned that non-replicating cells might experience a greater load of embedded ribonucleotides during DNA repair by ribonucleotide-utilizing polymerases. Therefore, we proceeded to test the radiation sensitivity of cells in stationary phase. Whereas stationary wild-type, ΔrnhA, ΔrnhB, ΔrnhC and ΔrnhD cells were equally sensitive to IR (Supplementary Figure S2), we observed that the ΔrnhB strain, but not the other Δrnh single-mutants, displayed enhanced killing by UV irradiation, i.e. 10-fold lower survival than wild-type after a 40 mJ/cm2 dose (Figure 7A). The ΔrnhB stationary phase UV phenotype was much milder than that of a stationary phase ΔuvrB strain that lacks the UV damage recognition protein UvrB (23) or a stationary phase ΔuvrD1 strain that lacks the UvrD1 helicase component of the mycobacterial nucleotide excision repair pathway (Figure 7A) (24). The ΔrnhA ΔrnhB double mutant was more sensitive than ΔrnhB to killing by UV in stationary phase (Figure 7B). By contrast, the ΔrnhB ΔrnhC and ΔrnhB ΔrnhD double mutants behaved like ΔrnhB with respect to UV sensitivity in stationary phase (Figure 7B). The triple mutant ΔrnhA ΔrnhB ΔrnhD was no more sensitive than the ΔrnhA ΔrnhB strain (Figure 7B). These results implicate RnhB in the repair of UV damage in non-replicating M. smegmatis and suggest a backup role for RnhA when RnhB is missing.
Figure 7.
Deletion of rnhB sensitizes M. smegmatis to UV irradiation in stationary phase. (A and B) Strains of the indicated genotypes were grown to stationary phase and exposed to escalating doses of UV irradiation as described in Materials and Methods. Strain survival was quantified by culturing serial 10-fold dilutions of exposed bacteria and normalizing to an unexposed sample from the same culture. When error bars are not visible they are within the plotted symbol.
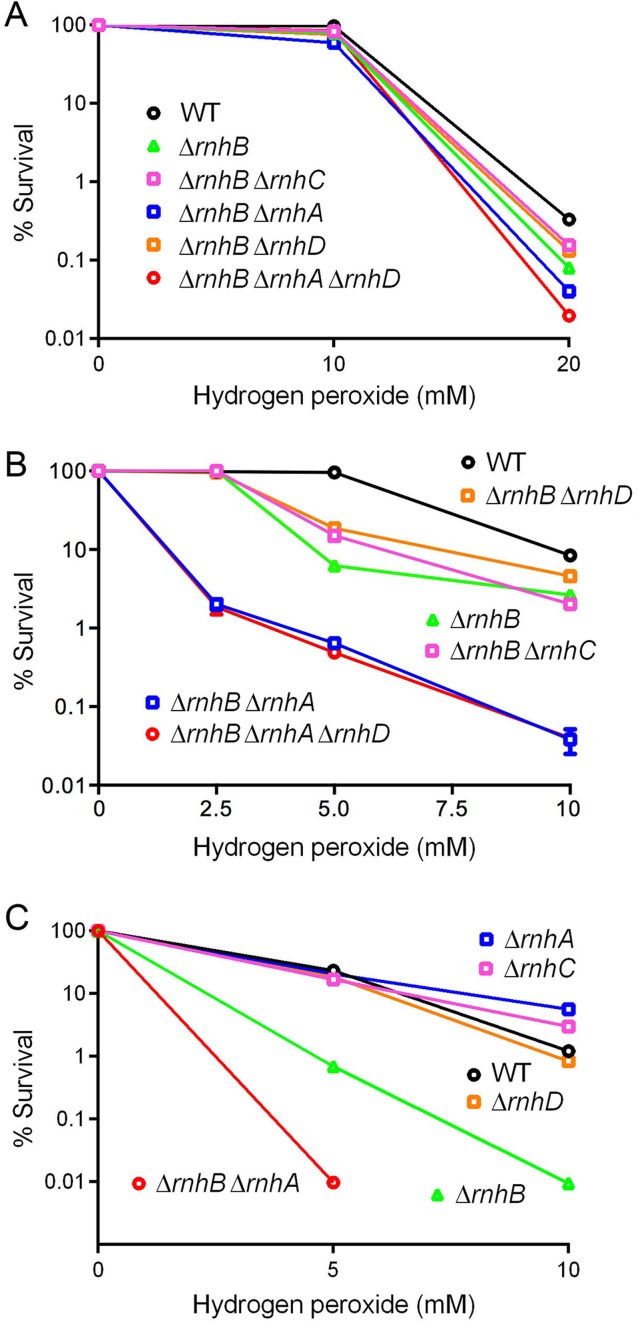
ΔrnhA ΔrnhB cells in stationary phase are sensitized to killing by hydrogen peroxide
We next surveyed the collection of ΔrnhB strains for their tolerance to exposure to increasing concentrations of hydrogen peroxide. Logarithmically growing wild-type M. smegmatis was unaffected by 10 mM H2O2 but was highly susceptible 20 mM H2O2, which elicited a ∼300-fold decrement in survival (Figure 8A). The ΔrnhA ΔrnhB ΔrnhD mutant was ∼17-fold more sensitive than wild-type to killing in log phase by 20 mM H2O2. The other ΔrnhB strains in log phase displayed intermediate sensitivities to 20 mM H2O2 (Figure 8A).
Figure 8.
RnhB, aided by RnhA, protects against killing by hydrogen peroxide in stationary phase. Strains of the indicated genotypes either in logarithmic phase (A) or stationary phase (B and C) were exposed to escalating doses of hydrogen peroxide (indicated on the x-axis) for 2 h as described in Materials and Methods. Survival was quantified by culturing serial 10-fold dilutions of exposed bacteria and normalizing to an unexposed sample from the same culture. When error bars are not visible they are within the plotted symbol.
Different peroxide effects were seen in stationary phase, whereby survival of the ΔrnhA ΔrnhB ΔrnhD and ΔrnhA ΔrnhB strains was reduced by factors of 50- and 200-fold by exposure to 2.5 and 5 mM H2O2, concentrations that had no effect on wild-type stationary phase cells (Figure 8B). The stationary phase ΔrnhA ΔrnhB ΔrnhD and ΔrnhA ΔrnhB strains were 200-fold more sensitive than wild-type to killing by 10 mM H2O2 (Figure 8B). By contrast, the ΔrnhB single mutant and the ΔrnhB ΔrnhC and ΔrnhB ΔrnhD double mutants were unaffected by 2.5 mM H2O2 and displayed more modest sensitization to 5 and 10 mM H2O2 (Figure 8B). Stationary phase ΔrnhA, ΔrnhC, and ΔrnhD single mutants were no more sensitive than wild-type M. smegmatis to killing by 5 to 10 mM H2O2 (Figure 8C), whereas the ΔrnhB single mutant was acutely sensitive to killing by 5 to 10 mM H2O2 and the ΔrnhA ΔrnhB double mutant was even more sensitive than the ΔrnhB strain. These results highlight a key function for RnhB in guarding against oxidative damage in non-replicating M. smegmatis and a crucial ancillary role for RnhA in this process when RnhB is missing.
ΔrnhA and ΔrnhC are synthetically lethal
Attempts to sequentially delete the chromosomal rnhC and rnhA loci in M. smegmatis were unsuccessful, suggesting that these two type I RNase H enzymes play an essential but functionally redundant role in mycobacterial survival. To fortify this conclusion genetically, we integrated a copy of the rnhC gene (under the control of the putative ‘native’ rnhC promoter, i.e. the promoter that drives expression of genes MSMEG_4307, MSMEG_4306 and MSMEG_4305 (rnhC) that are arranged as an operon at the endogenous chromosomal locus) at the attB site of the M. smegmatis chromosome in the ΔrnhC strain. This maneuver enabled the deletion of the chromosomal rnhA gene, as verified by Southern blotting of NotI-digested genomic DNA from independent isolates of the ΔrnhA::hygR ΔrnhC::loxP attB::rnhC strain (data not shown). Thus, either RnhC or RnhA is necessary for the viability of M. smegmatis.
RNase H1 catalytic activity is essential for growth of M. smegmatis
Synthetic lethality of rnhC and rnhA gene deletions does not constitute proof that their respective RNase H1 enzymatic activities are essential, i.e. it is conceivable that: (i) the absence of the RnhC and RnhA proteins is lethal for reasons unrelated to RNase H catalysis (for example if RnhC and RnhA are essential but functionally redundant structural components of the mycobacterial replication/repair machinery) or (ii) lethality arises because of the combined loss of RNase H1 and the acid phosphatase activity inherent in the C-terminal domain of RnhC.
To address these issues, we established a genetic complementation test in the ΔrnhA ΔrnhC attB::rnhC strain whereby we attempted to replace the attB::kan rnhC locus marked with a gene conferring kanamycin resistance with a homology-directed attB::strep rnhC locus with a marker gene conferring resistance to streptomycin (see Figure 9). We tested in parallel the efficacy of allelic replacement (gauged by recovery of strepR integrants) by transfected attB::rnhC constructs in which rnhC was wild-type (positive control), the RNase H catalytic-dead mutant rnhC-D73A, or the acid phosphatase catalytic-dead mutant rnhC-H173A (11). An attB strepR vector lacking the rnhC gene was used as a negative control. We recovered 5404 strepR integrants from the positive control transformation, 0 strepR colonies from the vector control (affirming that rnhC cannot be deleted in a ΔrnhA background), and only 8 strepR integrants from the transfection with rnhC-D73A. Whereas all eight of the strepR integrants from the transformation with rnhC-D73A were sensitive to kanamycin (affirming the marker swap at the attB site), sequencing of the rnhC locus after PCR amplification with flanking primers (denoted by blue arrows in Figure 9, yielding a 1883 bp PCR product) showed that the wild-type rnhC allele was present in every case, signifying that the recombination event at attB entailed genetic exchange distal to the mutated D73A codon in the transfected rnhC gene. Put simply, the rnhC-D73A mutation was lethal in a ΔrnhA background, indicating that the RNase H activity of RnhC is essential when RnhC is the only type I RNase H available.
In parallel experiments, we recovered 2982 strepR integrants from the positive control transfection with wild-type attB::rnhC and 868 strepR integrants from the transfection with attB::rnhC-H173A. Analysis of five independent strepR attB::rnhC-H173A isolates showed that they were sensitive to kanamycin and that the H173A mutation was present in the rnhC gene in all five isolates. Thus, the acid phosphatase activity of RnhC is dispensable for its essential function in the ΔrnhA background.
We extended the complementation analysis in the ΔrnhA ΔrnhC attB::rnhC strain by testing for replacement of the kanR-marked attB::rnhC locus with a strepR-marked attB integrating plasmid carrying the rnhA gene (with rnhA expression driven by the native rnhA promoter) in which rnhA was wild-type (positive control) or the RNase H catalytic-dead mutant rnhA-E50Q. We recovered 2744 strepR integrants from the wild-type rnhA transformation and 0 strepR isolates from the transformation with rnhA-E50Q. PCR amplification with flanking primers confirmed the exchange of the original rnhC ORF with transfected rnhA, i.e. PCR yielded a 1263-bp product containing the wild-type rnhA gene sequence. We conclude that the RNase H activity of RnhA is essential when RnhA is the only type I RNase H present in vivo.
DISCUSSION
M. smegmatis has a rich roster of four different RNase H enzymes. The present biochemical characterization of recombinant RnhA, and previous studies of recombinant RnhC (11), establish that both are magnesium-dependent RNase H1 enzymes, i.e. they incise RNA:DNA hybrids containing a run of four or more consecutive ribonucleotides. Whereas the substrate specificities of RnhB and RnhD have not been reported, it is presumed that they are members of the RNase H2 clade, and therefore likely to be capable of incising DNA with a single embedded ribonucleotide. Here we gained genetic insights to the division of labor among mycobacterial RNase H1 and H2 enzymes, by deleting the rnhA, rnhB, rnhC and rnhD genes, individually and in various combinations. The salient conclusions are that: (i) RNase H1 activity is essential for mycobacterial growth and can be provided by either RnhC or RnhA; (ii) the RNase H2 proteins RnhB and RnhD are dispensable for mycobacterial growth under laboratory conditions and (iii) RnhB and RnhA collaborate to protect M. smegmatis against oxidative damage in stationary phase. The implications of our findings for mycobacterial physiology and anti-bacterial drug discovery are discussed below.
RNase H1 activity is essential for mycobacterial growth
The properties of bacterial RNase H enzymes implicate them in the metabolism of the RNA primers of Okazaki fragments formed during lagging strand DNA replication and/or the RNA strands of R-loops formed during transcription. In the model bacterium E. coli, which has a single RNase H1 (RnhA) and a single RNase H2 (RnhB), a ΔrnhA ΔrnhB double mutant is viable (25), signifying that E. coli can survive without an RNase H enzyme in an otherwise wild-type background. Bacillus subtilis has four RNase H enzymes: YpdQ, YpeP, RnhB and RnhC (25–27). Initial inferences that B. subtilis ΔrnhB and ΔrnhC mutations were synthetically lethal (25) have been superceded by the isolation of a viable ΔrnhB ΔrnhC double mutant as well as a viable quadruple mutant in which all four B. subtilis RNase H genes were disrupted (27). Thus, B. subtilis can survive without an RNase H enzyme in an otherwise wild-type background. As we show here, the situation is different in M. smegmatis, where the simultaneous loss of the two RNase H1 enzymes RnhC and RnhA is lethal. Moreover, by testing complementation with catalytically defective mutants of RnhC and RnhA, we affirm that RNase H1 endonuclease activity is essential in M. smegmatis. (This contrasts with the acid phosphatase activity of RnhC, which is inessential.) To our knowledge, this is the first instance in which a bacterium requires RNase H1 activity for growth. It is worth pointing out that M. smegmatis requires RNase H1 notwithstanding that it has a potential alternative biochemical route to process Okazaki fragment RNA primers via the 5′ exonuclease activity of mycobacterial DNA polymerase I (28,29).
RnhC as a therapeutic target for tuberculosis and leprosy
The distinctive essentiality of RNase H1 activity in M. smegmatis highlights RNase H1 inhibition as strategy to interdict pathogenic mycobacteria (potentially without affecting other microbial flora). However, the RNase H1 landscape in M. smegmatis (an avirulent model mycobacterium) is pharmacologically daunting, insofar as one would have to simultaneously inhibit both RnhA and RnhC to achieve growth arrest or bacterial killing. The case for targeting RNase H1 would be simplified if the mycobacterial pathogens of interest had only one RNase H1 enzyme and it was the same RNase H1 enzyme in every case. PSI-BLAST searches of the NCBI database (taxid mycobacteria) with M. smegmatis RnhA, RnhC, and RnhB (as control) revealed that: (i) all mycobacterial species encode an RnhB enzyme (size range 223 to 279 aa); (ii) all mycobacterial species encode a bifunctional RNase H1/acid phosphatase RnhC enzyme (size range 352 to 383 aa); and (iii) only a subset of mycobacterial species encode an RnhA enzyme (size range 148–186 aa). The members of the RnhA-plus group of mycobacteria (comprising M. smegmatis and at least 33 other taxa) are listed in Supplementary Table S2. The RnhA-minus group of mycobacteria (at least 34 species) is compiled in Supplementary Table S3. The RnhA-minus clade includes the major human pathogens M. tuberculosis and M. leprae as well as several animal pathogens (M. bovis, M. avium) and ‘minor’ human pathogens (M. ulcerans, M. lepromatosis, M. canettii, M. kansasii, M. marinum, et al.). The lack of RnhA, and the presence of RnhC as the sole RNase H1 in clinically relevant mycobacterial pathogens, fortifies the case for inhibitors of the RnhC endonuclease as potential anti-tuberculosis and anti-leprosy agents. [Prior attention to RNase H as a drug target has focused primarily on inhibitors of the RNase H component of HIV reverse transcriptase as potential antivirals for treatment of AIDS (30–32).] The M. smegmatis Δrnh strains we have constructed, which rely on only RnhC or RnhA for growth, are suitable for cell-based screening for specific inhibitors of RnhC, i.e., by selecting small molecules that arrest growth of a rnhC+ ΔrnhA strain but do not affect an rnhA+ ΔrnhC strain.
A role for RnhB in protection against UV and oxidative damage in stationary phase
Surveying the single Δrnh mutants for clastogen sensitivity phenotypes revealed modest sensitization of stationary phase ΔrnhB cells to UV irradiation and a more profound sensitization to killing by transient exposure to hydrogen peroxide (a trigger of oxidative damage). It is notable that the ΔrnhD strain was not UV sensitive in stationary phase and that the ΔrnhB ΔrnhD double mutant lacking both RNase H2 enzymes phenocopied the ΔrnhB strain with respect to UV, in light of the recent report of Krishnan et al. (33) that expression of the M. smegmatis rnhD gene (as gauged by qRT-PCR) increases in response to UV stress, whereas expression of rnhB and rnhA is unchanged after UV exposure. Our data indicate that the absence of RnhD has no effect on UV damage survival (in logarithmic or stationary phase cells) and they raise the prospect that the UV stress induction of RnhD expression (33) may have more to do with up-regulating its (p)ppGpp synthetase activity than exerting an effect on ribonucleotide surveillance. Similarly, our results that ΔrnhD does not affect peroxide killing, by itself or when combined with ΔrnhB, establishes that RnhB and RnhD are functionally distinct.
We envision that the enhanced killing by peroxide of ΔrnhB cells in stationary phase reflects both oxidative damage to the DNA genome (especially the generation of 8-oxoguanine) and oxidation of the cellular NTP pool to generate oxo-rGTP and oxo-dGTP as substrates for repair polymerases, among which M. smegmatis DinB2 stands out as highly adept at using ribonucleotides, including oxo-rGTP, for templated synthesis and lesion bypass in vitro (3,4). This scenario predicts the accumulation of sporadically embedded ribonucleotides (and oxo-rGMP, a potent mutagen) in the genome of peroxide-treated stationary phase cells, and thus the reliance on an RNase H2 activity (specifically RnhB) to initiate ribonucleotide excision repair.
Why is RnhB particularly relevant to peroxide sensitivity in stationary phase? Are there not other means of preventing or dealing with oxidized ribo lesions? Mycobacteria have MutT-type enzyme systems that can detoxify oxo-dGTP and oxo-rGTP in the NTP pool, by converting them to oxo-dGMP and oxo-rNMP (34,35). Whether the M. smegmatis MutT systems are operative in stationary phase at a level sufficient to cope with peroxide stress on the guanine nucleotide pool is not clear. Mycobacteria have a MutM glycosylase that that excises the oxoG nucleobase at an oxo-dG:dC pair and a MutY glycosylase that excises the dA nucleobase at an oxo-dG:dA pair (36,37), but it is not reported whether the mycobacterial MutM and MutY enzymes can act at the corresponding ribo base pair lesions oxo-rG:dC and oxo-rG-dA. It is noteworthy that a M. smegmatis ΔmutM strain is sensitized to inhibition of logarithmic growth by peroxide (36), whereas a ΔmutY strain is not peroxide sensitive (37). How well MutM and MutY operate in stationary phase is untested, to our knowledge.
The intriguing observation here is that the ΔrnhA mutation, which has no effect per se on clastogen sensitivity, synergizes with ΔrnhB with respect to peroxide sensitivity in stationary phase. Yet, the loss of RnhC, which is functionally redundant to RnhA with respect to mycobacterial survival, does not synergize with ΔrnhB anent peroxide killing, signifying that RnhC and RnhA perform differently in stationary phase cells suffering oxidative DNA damage. This could reflect an inherent biochemical distinction between RnhA and RnhC (e.g., unique protein-protein interactions) or differences in the expression of RnhA and RnhC in stationary phase mycobacteria. [In that vein, it is notable that rnhC gene expression is repressed ∼5-fold in response to UV irradiation, whereas rnhA expression is unaffected (33).] The open question is how an RNase H1 enzyme (RnhA) cooperates with an RNase H2 enzyme (RnhB) to protect M. smegmatis against peroxide damage. The biochemical characterization of M. smegmatis RnhA presented here does not point to a direct role as an initiator of excision repair at a single embedded ribo. However, recent studies of E. coli RnhA unveiled a manganese-dependent RNA:DNA junction-cleaving capacity in vitro that, acting in tandem with E. coli RnhB, can lead to clean excision of a single ribonucleotide embedded in duplex DNA (38). It is not clear whether (or in what DNA damage situation) this dual-RNase H excision pathway might operate in E. coli in vivo. With respect to mycobacterial RNases H, the present study provides direction for future analyses that should address: (i) the biochemical specificities of the RNase H2 proteins RnhB and RnhD and (ii) the enzymes and repair pathway responsible for ribonucleotide stress on the stationary phase mycobacterial genome in response to peroxide.
Footnotes
Present address: Richa Gupta, Department of Natural Sciences, LaGuardia Community College, Long Island City, NY 11101, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
U.S. National Institutes of Health Grants [AI64693, P30CA008748]. Funding for open access charge: U.S. National Institutes of Health Grant [AI64693].
Conflict of interest statement. None declared.
REFERENCES
- 1.Schroeder J.W., Randall J.R., Mattews L.A., Simmons L.A. Ribonucleotides in bacterial DNA. Crit. Rev. Biochem. Mol. Biol. 2014;50:181–193. doi: 10.3109/10409238.2014.981647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu H., Bhattarai H., Yan H., Shuman S., Glickman M. Characterization of Mycobacterium smegmatis PolD2 and PolD1 as RNA/DNA polymerases homologous to the POL domain of bacterial DNA ligase D. Biochemistry. 2012;51:10147–10158. doi: 10.1021/bi301202e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ordonez H., Uson M.L., Shuman S. Characterization of three Mycobacterial DinB (DNA polymerase IV) paralogs highlights DinB2 as naturally adept at ribonucleotide incorporation. Nucleic Acids Res. 2014;42:11056–11070. doi: 10.1093/nar/gku752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ordonez H., Shuman S. Mycobacterium smegmatis DinB2 misincorporates deoxyribonucleotides and ribonucleotides during templated synthesis and lesion bypass. Nucleic Acids Res. 2014;42:12722–12734. doi: 10.1093/nar/gku1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma A., Nair D.T. MsDpo4—a DinB homolog from Mycobacterium smegmatis—is an error-prone DNA polymerase than can promote G;T and T:G mismatches. J. Nucleic Acids. 2012;2012:285481. doi: 10.1155/2012/285481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu H., Shuman S. Novel 3′-ribonuclease and 3′-phosphatase activities of the bacterial non-homologous end-joining protein, DNA ligase D. J. Biol. Chem. 2005;280:25973–25981. doi: 10.1074/jbc.M504002200. [DOI] [PubMed] [Google Scholar]
- 7.Yakovleva L., Shuman S. Nucleotide misincorporation, 3′-mismatch extension, and responses to abasic sites and DNA adducts by the polymerase component of bacterial DNA ligase D. J. Biol. Chem. 2006;281:25026–25040. doi: 10.1074/jbc.M603302200. [DOI] [PubMed] [Google Scholar]
- 8.Pitcher R.S., Brissett N.C., Picher A.J., Andrade P., Juarez R., Thompson D., Fox G.C., Blanco L., Doherty A.J. Structure and function of a mycobacterial NHEJ DNA repair polymerase. J. Mol. Biol. 2007;366:391–405. doi: 10.1016/j.jmb.2006.10.046. [DOI] [PubMed] [Google Scholar]
- 9.Zhu H., Nandakumar J., Aniukwu J., Wang L.K., Glickman M.S., Lima C.D., Shuman S. Atomic structure and NHEJ function of the polymerase component of bacterial DNA ligase D. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1711–1716. doi: 10.1073/pnas.0509083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawes S.S., Crouch R.J., Morris S.L., Mizrahi V. Cloning, sequence analysis, overproduction in Escherichia coli and enzymatic characterization of the RNase HI from Mycobacterium smegmatis. Gene. 1995;165:71–75. doi: 10.1016/0378-1119(95)00523-9. [DOI] [PubMed] [Google Scholar]
- 11.Jacewicz A., Shuman S. Biochemical characterization of Mycobacterium smegmatis RnhC (MSMEG_4305), a bifunctional enzyme composed of autonomous N-terminal type I ribonuclease H and C-terminal acid phosphatase domains. J. Bacteriol. 2015;197:2489–2498. doi: 10.1128/JB.00268-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minias A.E., Brzostek A.M., Minias P., Dziadek J. The deletion of rnhB in Mycobacterium smegmatis does not affect the level of RNase HII substrates or influence genome stability. PLoS One. 2015;10:e0115521. doi: 10.1371/journal.pone.0115521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murdeshwar M.S., Chatterji D. MS_RHII-RSD, a dual-function RNase HII-(p)ppGpp synthetase from Mycobacterium smegmatis. J. Bacteriol. 2012;194:4003–4014. doi: 10.1128/JB.00258-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tadokoro T., Kanaya S. Ribonuclease H: molecular diversities, substrate binding domains, and catalytic mechanism of the prokaryotic enzymes. FEBS J. 2009;276:1482–1493. doi: 10.1111/j.1742-4658.2009.06907.x. [DOI] [PubMed] [Google Scholar]
- 15.Nowotny M., Gaidamakov S.A., Crouch R.J., Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 16.Rychlik M.P., Chon H., Cerritelli S.M., Klimek P., Crouch R.J., Nowotny M. Crystal structures of RNase H2 in complex with nucleic acid reveal the mechanism of RNA–DNA junction recognition and cleavage. Mol. Cell. 2010;40:658–670. doi: 10.1016/j.molcel.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Figiel M., Nowotny M. Crystal structure of RNase H3-substrate complex reveals parallel evolution of RNA/DNA hybrid recognition. Nucleic Acids Res. 2014;42:9285–9294. doi: 10.1093/nar/gku615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watkins H.A., Baker E.N. Structural and functional characterization of an RNase HI domain from the bifunctional protein Rv2228c from Mycobacterium tuberculosis. J. Bacteriol. 2010;192:2878–2886. doi: 10.1128/JB.01615-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minias A.E., Brzostek A.M., Korycka-Machala M., Dziadek B., Minias P., Rajagopalan M., Madiraju M., Dziadek J. RNase HI is essential for survival of Mycobacterium smegmatis. PLoS One. 2015;10:e0126260. doi: 10.1371/journal.pone.0126260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barkan D., Stallings C.L., Glickman M.S. An improved counterselectable marker system for mycobacterial recombination using galK and 2-deoxy-galactose. Gene. 2011;470:31–36. doi: 10.1016/j.gene.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glickman M.S., Cox J.S., Jacobs W.R. A novel mycolic acid cyclopropane synthetase is required for cording, persistence, and virulence of Mycobacterium tuberculosis. Mol. Cell. 2000;5:717–727. doi: 10.1016/s1097-2765(00)80250-6. [DOI] [PubMed] [Google Scholar]
- 22.Yang W., Hendrickson W.A., Crouch R.J., Satow Y. Structure of ribonuclease H phased at 2 Å resolution by MAD analysis of the selenomethionyl protein. Science. 1990;249:1398–1405. doi: 10.1126/science.2169648. [DOI] [PubMed] [Google Scholar]
- 23.Mazloum N., Stegman M.A., Croteau D.L., Van Houten B., Kwon N.S., Ling Y., Dickinson C., Venugopal A., Towheed M.A., Nathan C. Identification of a chemical that inhibits the mycobacterial UvrABC complex in nucleotide excision repair. Biochemistry. 2011;50:1329–1335. doi: 10.1021/bi101674c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinha K.M., Stephanou N.C., Gao F., Glickman M.S., Shuman S. Mycobacterial UvrD1 is a Ku-dependent DNA helicase that plays a role in multiple DNA repair events, including double-strand break repair. J. Biol. Chem. 2007;282:15114–15125. doi: 10.1074/jbc.M701167200. [DOI] [PubMed] [Google Scholar]
- 25.Itaya M., Omori A., Kanaya S., Crouch R.J., Tanaka T., Kondo K. Isolation of RNase H genes that are essential for growth of Bacillus subtilis 168. J. Bacteriol. 1999;181:2118–2123. doi: 10.1128/jb.181.7.2118-2123.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohtani N., Haruki M., Morikawam M., Crouch R.J., Itaya M., Kanaya S. Identification of the genes encoding Mn2+-depednent RNAase HII and Mg2+-dependent RNAse HIII from Bacillus subtilis: classification of RNases H into three families. Biochemistry. 1999;38:605–618. doi: 10.1021/bi982207z. [DOI] [PubMed] [Google Scholar]
- 27.Fukushima S., Itaya M., Kato H., Ogasawara N., Yoshikawa H. Reassessment of the in vivo functions of DNA polymerase I and RNase H in bacterial cell growth. J. Bacteriol. 2007;189:8575–8583. doi: 10.1128/JB.00653-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huberts P., Mizrahi V. Cloning and sequence analysis of the gene encoding the DNA polymerase I from Mycobacterium tuberculosis. Gene. 1995;164:133–136. doi: 10.1016/0378-1119(95)00453-d. [DOI] [PubMed] [Google Scholar]
- 29.Gordhan B.G., Andersen S.J., De Meyer A.R., Mizrahi V. Construction by homologous recombination and phenotypic characterization of a DNA polymerase domain polA mutant of Mycobacterium smegmatis. Gene. 1996;178:125–130. doi: 10.1016/0378-1119(96)00350-2. [DOI] [PubMed] [Google Scholar]
- 30.Ilina T., Labarge K., Sarafianos S.G., Ishima R., Parniak M.A. Inhibitors of HIV-1 reverse transcriptase-associated ribonuclease H activity. Biology. 2012;1:521–541. doi: 10.3390/biology1030521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Himmel D.M., Myshakina N.S., Ilina T., Van Ry A., Ho W.C., Parniak M.A., Arnold E. Structure of a dihydroxycoumarin actiove-site inhibitor in complex with the RNase H domainsof HIV-1 reverse transcripase and structure-activity analysis of inhibitor analogs. J. Mol. Biol. 2014;426:2617–2631. doi: 10.1016/j.jmb.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corona A., Di Leva F.S., Thierry S., Pescatori L., Cuzzucoli Crucitti G., Subra F., Delelis O., Esposito F., Rigogliuso G., Costi R., et al. Dentification of highly conserved residues involved in inhibition of HIV-1 RNase H function by diketo acid derivatives. Antimicrob. Agents Chemother. 2014;58:6101–6110. doi: 10.1128/AAC.03605-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krishnan S., Petchiappan A., Singh A., Bhatt A., Chatterji D. R-loop induced stress response by second (p)ppGpp synthetase in Mycobacterium smegmatis: functional and domain interdependence. Mol. Microbiol. 2016;102:168–182. doi: 10.1111/mmi.13453. [DOI] [PubMed] [Google Scholar]
- 34.Patil A.G., Sang P.B., Govindan A., Varshney U. Mycobacterium tuberculosis MutT1 (Rv2985) and ADPRase (Rv1700) proteins constitute a two-stage mechanism of 8-oxo-dGTP and 8-oxoGTP detoxification and adenosine to cytidine mutation avoidance. J. Biol. Chem. 2013;288:11252–11262. doi: 10.1074/jbc.M112.442566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sang P.B., Varshney U. Biochemical properties of MutT2 proteins from Mycobacterium tuberculosis and M. smegmatis and their contrasting antimutator roles in Escherichia coli. J. Bacteriol. 2013;195:1552–1560. doi: 10.1128/JB.02102-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jain R., Kumar P., Varshney U. A distinct role of formamidopyrimidine DNA glycosylase (MutM) in down-regulation of accumulation of G, C mutations and protections against oxidiative stress in mycobacteria. DNA Repair. 2007;6:1774–1785. doi: 10.1016/j.dnarep.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 37.Kurthkoti K., Srinath T., Kumar P., Malshetty V., Sang P.B., Jain R., Manjunath R., Varshney U. A distinct physiological role of MutY in mutation prevention in mycobacteria. Microbiology. 2010;156:88–98. doi: 10.1099/mic.0.033621-0. [DOI] [PubMed] [Google Scholar]
- 38.Tannous E., Kanaya E., Kanaya S. Role of RNase H1 in DNA repair: removal of single ribonucleotide misincorporated into DNA in collaboration with RNase H2. Sci. Rep. 2015;5:9969. doi: 10.1038/srep09969. [DOI] [PMC free article] [PubMed] [Google Scholar]