Abstract
The potential use of G-quadruplex (GQ) stabilizing small molecules as anti-cancer drugs has created a flurry of activity on various aspects of these molecules. Telomestatin and oxazole telomestatin derivatives (OTD) are some of the most prominent of such molecules, yet the underlying dynamics of their interactions with GQ and the extent of heterogeneities in these interactions are not known. We performed single molecule measurements to study binding kinetics, rotational freedom, and dwell time distributions of a Cy5-labeled OTD (L1Cy5–7OTD) as it interacted with several different GQ structures. Our measurements show that L1Cy5–7OTD dwells on more stable GQ for longer times and binds to such GQ with higher frequency. The dwell times showed a broad distribution, but were longer than a minute for a significant fraction of molecules (characteristic dwell time τ = 192 ± 15 s and τ = 98 ± 15 s for the more and less stable GQ, respectively). In addition, L1Cy5–7OTD might be able to bind to GQ in at least two different primary orientations and occasionally transition between these orientations. The dwell time in one of these orientations was significantly longer than that in the other one, suggesting different stabilities for different binding orientations.
INTRODUCTION
G-quadruplex (GQ) structures are non-canonical nucleic acid secondary structures that form in guanine-rich regions of the genome (1,2). Inability to unfold these structures is known to retard the replication machinery and give rise to elevated levels of DNA breaks and genomic instability (3,4). At the telomeric context, the GQs formed at the 3′ overhang can interfere with telomerase activity and prevent telomere elongation (1,2). Genome-wide computational studies identified several hundred thousand potentially GQ forming sequences (PQS) in the human genome, with promoters being particularly enriched in these sequences (5,6). GQs were used as specific drug targets to modulate gene expression (7,8) or to induce synthetic lethality or radiosensitivity in cancer cells (9,10). As this general picture illustrates, being able to modulate GQ stability with small molecules is a potentially effective mechanism for slowing down tumor proliferation by inhibiting telomerase or the replication machinery or regulating gene expression by modulating the stability of promoter site GQs.
Because of their medical and technological potential, identifying, synthesizing, and improving the capabilities of such molecules have been active research fields with a number of exciting discoveries in the recent years. In addition to a large body of work that is focused on identifying and synthesizing GQ stabilizing small molecules (11–15), a flurry of activity has been observed in terms of improving the affinity, specificity and multi-functionality of these molecules (16–19). Despite these exciting developments, the single molecule work on GQ stabilizing small molecules has been limited (20–22). In this study, we employed single molecule Förster Resonance Energy Transfer (smFRET) to study binding kinetics of a single L1Cy5–7OTD (16) to several GQ structures.
MATERIALS AND METHODS
DNA constructs and oxazole telomestatin derivative
All the unlabeled and site-specific labeled DNA oligonucleotides were purchased as PAGE or HPLC purified from Integrated DNA Technologies (Coralville, IA, USA). The DNA sequences are given in Table 1. The partial duplex DNA (pdDNA) constructs were formed by heating the corresponding ssDNA strands at 90°C for 3 min, followed by a slow cool down to room temperature over a couple hours. A particular convention was followed for naming the DNA sequences used in the manuscript. To illustrate, one of the names used is pd-hGQ12T, which essentially has three parts: pd, hGQ, and 12T. pd designates a partial duplex construct which has a duplex stem, hGQ designates a GQ with human telomeric sequence, and 12T designates the ssDNA overhang which has 12 thymines (T). Similarly, pd-12ThGQ has the same ingredients but the 12T overhang is placed between the duplex stem and hGQ. In the constructs pd-3Ly1Lp12T an d pd-12T3Ly1Lp similar convention is followed with the phrase ‘3Ly1Lp’ replacing ‘hGQ’. The GQ forming part 3Ly1Lp has the sequence GGGTGGGTGGGTGGG which forms a GQ with 3-layers (3Ly) and has 1T loops (1Lp). L1Cy5–7OTD was dissolved and diluted to 10 mM concentration in 100% dimethyl sulfoxide (DMSO) solution before use. The relevant data on purity and characterization of this OTD is given in reference (16).
Table 1. Sequences of the DNA constructs used in this study.
| Construct | Sequence (5′ to 3′) | pd-Comp |
|---|---|---|
| pd-3Ly1Lp12T | TGGCGACGGCAGCGAGGC TT GGGTTAGGGTTAGGGTTAGGG T12-Cy3 | Strand 1 |
| pd-hGQ12T | Cy3-T12GGGTTAGGGTTAGGGTTAGGG TT TGGCGACGGCAGCGAGGC | Strand 2 |
| pd-12T3Ly1Lp | Cy3-TT GGGTGGGTGGGTGGG T12TGGCGACGGCAGCGAGGC | Strand 2 |
| pd-12ThGQ | Cy3-TT GGGTTAGGGTTAGGGTTAGGG T12TGGCGACGGCAGCGAGGC | Strand 2 |
| pd-Cy312T3Ly1Lp | TT GGGTGGGTGGGTGGG T12TGGCGACGGCAGCGAGGC | Strand 3 |
| Strand 1 | GCC TCG CTG CCG TCG CCA-Biotin | |
| 12ThGQ | T12GGGTTAGGGTTAGGGTTAGGG TT | |
| 12T3Ly1Lp | T12GGGTGGGTGGGTGGG TT | |
| Strand 2 | Biotin- GCC TCG CTG CCG TCG CCA | |
| Strand 3 | Biotin- GCC TCG CTG CCG TCG CCA-Cy3 |
The GQ forming sections are underlined. The parts forming the duplex stem are in bold. Strand 1 and Strand 2 are complementary to the bold sections of other strands. The column name ‘pd-Comp’ refers to partial duplex complement.
Circular dichroism (CD) assay
CD measurements were performed using a Jasco J-810 spectrophotometer at room temperature in a 1 mm wide cuvette. The DNA concentration was 4 μM for all samples. The measurements were performed in 10 mM Tris (pH 7.5), 150 mM KCl and 2 mM MgCl2 and the samples were prepared (annealing and storage) using an identical protocol to that used for the smFRET assay. The measurements were performed in triplicates, and the average is reported.
Single molecule FRET setup and assay
Total internal reflection fluorescence (TIRF) microscopy was used for smFRET imaging. An Olympus IX71 microscope equipped with an Andor IXON EMCCD camera (IXON DV-887 EMCCD, Andor Technology, CT, USA) was used in our prism-based TIRF setup. A green laser (SpectraPhysics Excelsior) with λ = 532 nm wavelength was used as the excitation source. The sample chamber was formed by sandwiching a double sided tape between a quartz slide and a glass coverslip. The inner surfaces of the sample chamber were passivated by polyethylene glycol (PEG) coating (Laysan Bio). One percent of PEG molecules were tagged with biotin to provide attachment points for biotinylated DNA molecules, which bind to biotin-PEG via neutravidin. Detailed protocols of chamber preparation and surface passivation are described in reference (23).
The smFRET imaging buffer contained Tris base (50mM, pH 7.5), 2 mM Trolox, 0.8 mg/ml glucose, 0.1 mg/ml bovine serum albumin (BSA), 0.1 mg/ml glucose oxidase, 0.02 mg/ml catalase, 2 mM MgCl2, and indicated concentrations of KCl or NaCl. Glucose, glucose oxidase, and catalase form an oxygen scavenging system that delays photobleaching of fluorophores. Trolox increases brightness of the fluorophores by quenching their dark triplet state. MgCl2 is added to better mimic the physiological ionic conditions. In the rest of the manuscript, only the KCl (or NaCl) concentration will be explicitly stated however, it should be assumed that 2 mM Mg2+ is always present along with the monovalent cations. BSA is added in order to patch any regions of the surface that might have imperfections in the PEG coating.
The FRET efficiency is calculated using the expression: EFRET = IA/(IA + ID), where IA is the acceptor intensity and ID is the donor intensity. Unless otherwise specified, all quoted errors are standard error of the mean. Except for dwell time measurements, 100 ms/frame integration time was used in all measurements. Dwell time measurements reported in the paper were performed at 2.5 s integration time/frame to compensate for the low laser excitation power. 1000–2000 frame movies were recorded and smFRET traces were generated for each molecule. All smFRET data presented in this manuscript were selected from traces that showed at least one L1Cy5–7OTD binding event. Those that did not show any binding event contributed to a donor-only (DO) peak, as the DNA molecules only have the donor fluorophore (the acceptor fluorophore is on L1Cy5–7OTD). The DO peak was subtracted from the histograms and the FRET scale was corrected using a standard protocol (as we described in supplementary materials of reference (24)). The smFRET traces (Figures 2C and 3A, and Supplementary Figure S5) are shown without the DO correction as this correction is made after individual traces are collected within a histogram. Because of this, EFRET ≈ 0.12 after acceptor dissociates from GQ in these traces, which is the DO FRET level ( ). After DO correction this FRET level will be shifted to EDO ≈ 0.0 and the scale will normalized accordingly.
). After DO correction this FRET level will be shifted to EDO ≈ 0.0 and the scale will normalized accordingly.
Figure 2.
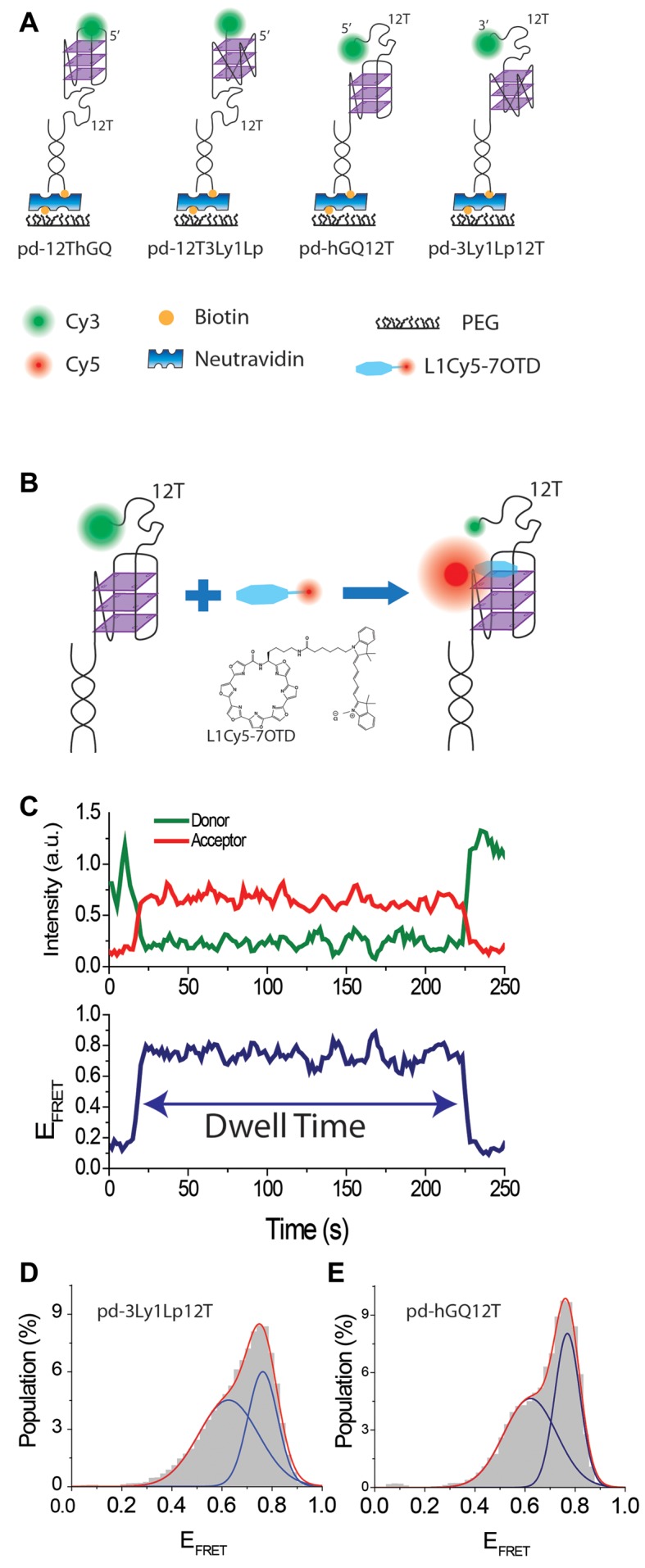
(A) Schematics of DNA constructs. (B) Schematic of L1Cy5–7OTD binding to GQ and structure of L1Cy5–7OTD. As the DNA construct does not have an acceptor, the FRET is at donor only level before binding of L1Cy5–7OTD. (C) A sample smFRET trace showing L1Cy5–7OTD binding and how dwell time is measured. (D) smFRET histogram for L1Cy5–7OTD binding to pd-3Ly1Lp12T. The blue lines are Gaussian fit peaks and the red line is the cumulative of two peaks. (E) Similar histogram and fits to pd-hGQ12T construct.
Figure 3.
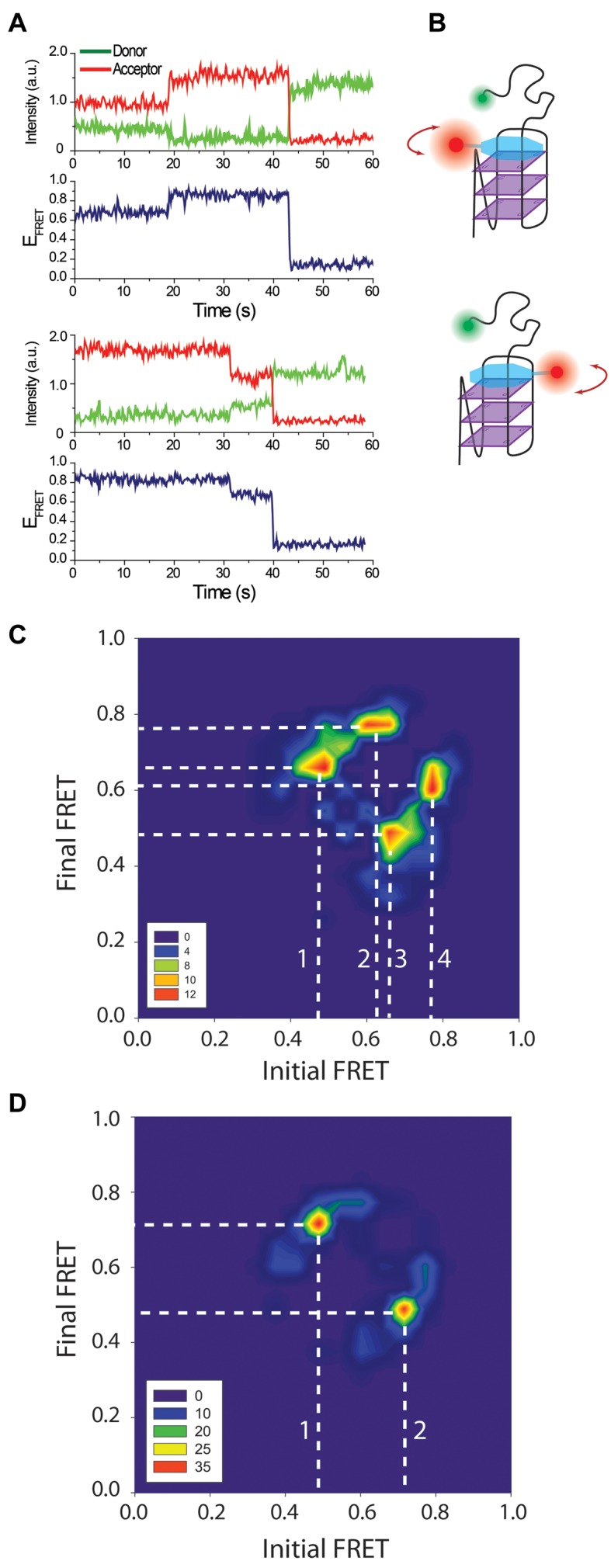
(A) smFRET traces showing a transition from EF-Low to EF-High (top), and from EF-High to EF-Low (bottom). (B) A schematic of the model proposing different binding orientations for L1Cy5–7OTD, which we propose is the source of the observed FRET levels. (C) TDP constructed from pd-3Ly1Lp12T and pd-12T3Ly1Lp traces. Data were combined to accumulate enough statistics as the transitions are rare. (D) TDP constructed from pd-hGQ12T and pd-12ThGQ traces (data combined). The white dashed lines indicate the transitions between prominent FRET states. These transitions are marked with numbers to facilitate their identification. The TDP in both (C) and (D) are symmetric indicating that transitions from EF-High to EF-Low and vice versa are possible.
The L1Cy5–7OTD concentration was kept at 50 nM in all assays except the dwell time measurements, where 100 nM L1Cy5–7OTD was used in some measurements. This concentration was high enough to collect large enough data sets to perform statistical analysis but low enough to maintain a low fluorescent background and insignificant non-specific binding of L1Cy5–7OTD to the surface.
RESULTS AND DISCUSSION
Circular dichroism measurements
GQ formation and conformation could not be directly probed using smFRET assay since the DNA molecules were labeled only with a donor fluorophore. Therefore, we performed CD measurements, under ionic conditions identical to those used in smFRET measurements, to investigate GQ folding and conformation. DNA constructs that lack the donor fluorophore but are otherwise identical to smFRET constructs (pd-3Ly1Lp12T, pd-hGQ12T, pd-12T3Ly1Lp and pd-12ThGQ) were used for the CD measurements. As the constructs are in partial duplex form, the contribution of duplex stem needs to be subtracted from CD spectra. Therefore, we also measured the CD spectrum of just the duplex stem formed by Strand 1 and its complementary (Table 1). Figure 1 shows these data after subtraction of duplex stem spectrum. The pd-3Ly1Lp12T and pd-12T3Ly1Lp constructs show a peak at 265 and 266 nm, respectively (see Supplementary Figure S1 for the peak fitting analysis). In both spectra, a trough at ≈240 nm accompanies the peaks, which are characteristics of parallel conformation. On the other hand, hGQ constructs demonstrate a broad distribution which can be decomposed into two peaks at 269 and 288 nm for pd-12ThGQ and 272 nm and 291 nm for pd-hGQ12T, which are characteristics of a mix of parallel and anti-parallel, or hybrid conformation. These spectra is similar to that reported on a telomeric sequence with a 11T overhang (25). In order to probe the possible influence of the duplex stem on GQ conformation, we also measured the CD spectra of ssDNA (without duplex stem) with identical sequence to overhang of partial duplex constructs. These data, overlaid with spectra from the respective partial duplex counterparts, are shown in Supplementary Figure S1. The peak positions for the ssDNA construct 12T3Ly1Lp is 266 nm, which is identical to partial duplex constructs. Similarly, the peak positions for 12ThGQ construct are 267 nm and 286 nm, which are in agreement for partial duplex versions of hGQ with only minor variations.
Figure 1.
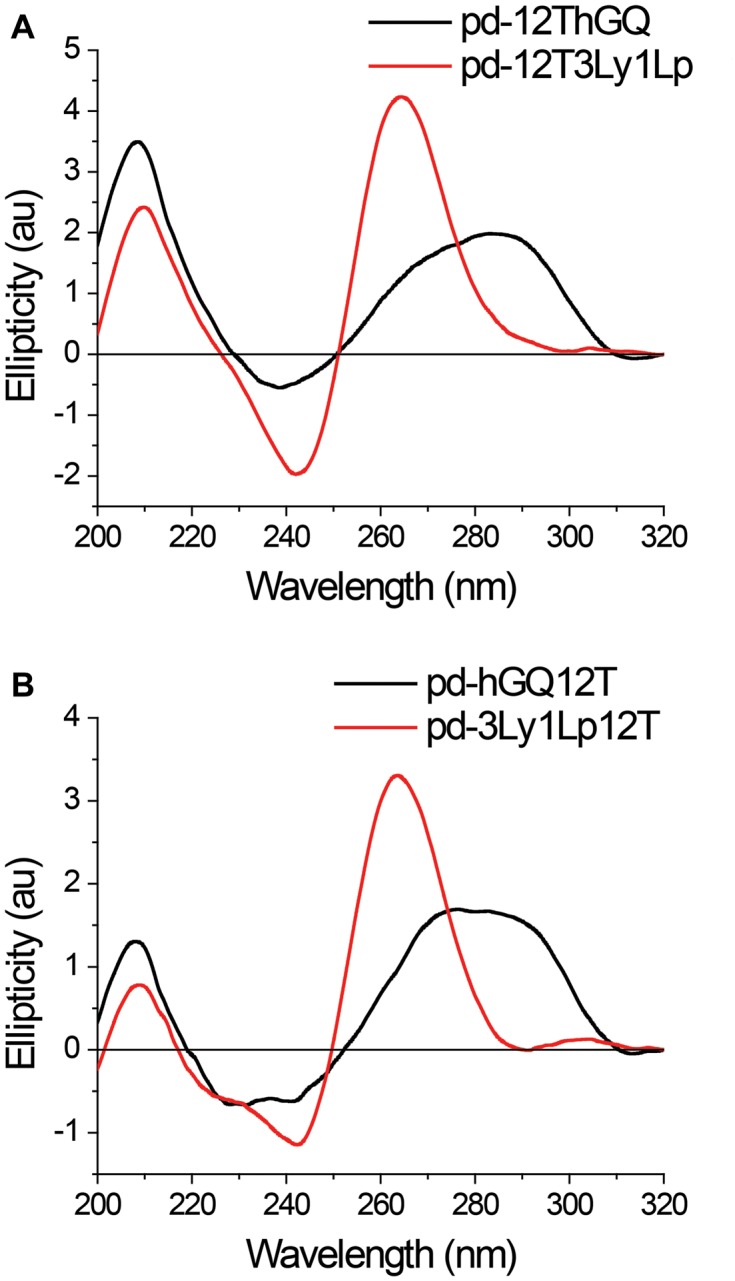
Circular dichroism measurements were performed at room temperature in 150 mM KCl on (A) pd-12ThGQ and pd-12T3Ly1Lp; (B) pd-3Ly1Lp12T and pd-hGQ12T constructs. The signal from the duplex stem was measured separately and subtracted from the spectra of partial duplex constructs to isolate the signal from the GQ. The spectra of pd-3Ly1Lp12T and pd-12T3Ly1Lp are consistent with parallel GQ conformation while the telomeric constructs (pd-hGQ12T and pd-12ThGQ) demonstrate a mix of conformations.
In addition to differences in their folding conformations, the hGQ constructs (pd-hGQ12T and pd-12ThGQ) and 3Ly1Lp constructs (pd-3Ly1Lp12T and pd-12T3Ly1Lp) also demonstrate significant differences in terms of their stability. In particular, the (GGGT)3GGG sequence, which is the GQ forming section of 3Ly1Lp constructs, has been utilized in different applications due to its extreme stability (26,27). The thermal melting temperature of this sequence is Tm = 84°C at 5 mM KCl, and Tm > 94°C at 15 mM KCl (26). In comparison, the thermal melting temperature of hGQ with 11T overhang has been measured to be Tm = 57.9°C at 50 mM KCl (25). Therefore, these two sets of constructs enable probing how variations in stability and conformation influence binding of L1Cy5–7OTD to GQs.
Binding of L1Cy5–7OTD to GQ
A schematic of the DNA constructs and smFRET assay are shown in Figure 2A–B, respectively. An acceptor (Cy5) labeled OTD, L1Cy5–7OTD of reference (16), and donor (Cy3) labeled DNA constructs were used for smFRET measurements. We ensured GQ formation by incubating the DNA samples in 150 mM K+ for 30 min, twice as long as our earlier work and CD measurements (Figure 1) have demonstrated to be adequate (28,29). An imaging buffer that contained 50 nM L1Cy5–7OTD and 150 mM K+ was added to chamber before data acquisition started. Figure 2C demonstrates a sample smFRET trace showing the FRET state that results upon binding of L1Cy5–7OTD to the GQ. Histograms of such smFRET traces are shown in Figure 2D for pd-3Ly1Lp12T and in Figure 2E for pd-hGQ12T. Gaussian fits to smFRET distributions result in two FRET peaks for each construct, which we will call EF-High and EF-Low: EF-Low = 0.63 ± 0.12 and EF-High = 0.76 ± 0.06 for pd-3Ly1Lp12T construct and EF-Low = 0.62 ± 0.10 and EF-High = 0.77 ± 0.05 for pd-hGQ12T construct.
We did not observe L1Cy5–7OTD binding for DNA constructs that do not form GQ, such as those with just poly-thymine overhang. Electrophoretic mobility shift assay (EMSA) measurements also show L1Cy5–7OTD selectively binds to GQ but not double stranded DNA (dsDNA) or single stranded DNA (ssDNA) (16). NMR data (30) showed that an OTD binds to a particular G-tetrad, ‘top G-tetrad’ rather than both ‘top’ and ‘bottom G-tetrad’, at low OTD concentrations, such as those used in this manuscript. Given this, why two peaks are observed in our data needs clarification. Binding of L1Cy5–7OTD to different GQ conformations (parallel, anti-parallel, or hybrid) or its binding to the top or bottom G-tetrad are potential explanations we will consider first. Since pd-3Ly1Lp12T folds only into the parallel conformation (Figure 1) yet it shows two peaks in smFRET histograms, we rule out the conformations as the source of these different peaks. The similarity of the smFRET histograms for pd-3Ly1Lp12T and pd-hGQ12T constructs, which are very different in terms of their conformations also argues against this possibility. On the other hand, binding of L1Cy5–7OTD to the top (or bottom) G-tetrad would result in very similar donor-acceptor separations for pd-3Ly1Lp12T and pd-hGQ12T since the donor fluorophore in both constructs is separated from the G-tetrad layers by a 12T overhang. However, elucidating whether the two peaks are due to binding to top and bottom G-tetrads required a deeper inspection of smFRET traces which provided insights that argue against this explanation. In ∼5% of all smFRET traces that showed L1Cy5–7OTD binding, we observed binding at one level followed by a transition to another level, without dissociation of L1Cy5–7OTD from GQ (Figure 3A). In all such traces, dissociation (or photobleaching) of L1Cy5–7OTD takes place in a single step, suggesting a single L1Cy5–7OTD molecule is responsible for both FRET states. As the transition takes place without loss of L1Cy5–7OTD signal, it is unlikely that L1Cy5–7OTD can switch from one tetrad to another in this manner. We also tested this idea by switching the Cy3 position on the DNA construct, and observed further evidence against the plausibility of this model (see Supplementary Figure S2).
Two possible explanations that would be consistent with our measurements could be conceived. The less interesting possibility is the two peaks being due to the Cy5 molecule in L1Cy5–7OTD stacking to different parts of the GQ by using the long and flexible linker connecting Cy5 to OTD (see Supplementary Figure S3 for a schematic of this model). Even though we cannot rule it out, we do not think this non-specific stacking scenario is likely, primarily due to the consistency and reproducibility of the peak positions for different GQ constructs. Instead, we propose the two FRET peaks to be due to stacking of L1Cy5–7OTD to a G-tetrad in at least two different orientations (see Figure 3B for a schematic of this model). In this model, the transitions between the FRET states are due to L1Cy5–7OTD rotating from one orientation to the other while it remains stacked on the G-tetrad. The significant width of the histograms, especially the EF-Low peaks which are twice as broad as the EF-High peaks, suggests that the L1Cy5–7OTD molecules may bind in ‘bands’ of possible orientations. However, the clearly distinguishable peaks suggest these bands are distinct from each other. Even though larger width of the EF-Low peaks is partially due to this peak being in the more sensitive part of FRET (∼18% more sensitive based on a comparison of the derivative of EFRET = 1/[1 + (R/RFörster)6] at EFRET = 0.63 compared to EFRET = 0.76), the 2-fold broader peak suggests there may be a wider band of angles that the molecule binds around the EF-Low peak. It is also possible that this lower FRET peak is composed of two peaks with significant overlap. It should be mentioned that two FRET peaks do not necessarily represent just two primary orientations. For fluorescent molecules attached via flexible linkers, as in our constructs, the FRET efficiency depends on the separation between the donor-acceptor fluorophores, which could be similar for different binding orientations of L1Cy5–7OTD. Therefore, we propose L1Cy5–7OTD to have ‘at least’ two different primary orientations.
In order to gain further insight about these transitions, we performed a Hidden–Markov modeling based analysis (vbFRET) on traces that showed such transitions (31). We constructed a transition density plot (TDP) for data combined from pd-3Ly1Lp12T and pd-12T3Ly1Lp traces (Figure 3C, N = 77 molecules) and a TDP for data combined from pd-hGQ12T and pd-12ThGQ traces (Figure 3D, N = 112 molecules). Only smFRET traces that demonstrate a transition between different FRET states are included in the TDP. The x-axis of this histogram represents the initial FRET state before the transition and the y-axis represents the final FRET state after the transition. To illustrate, if a molecule transitions from EFRET ≈ 0.60 to EFRET ≈ 0.83, it will contribute to the contour plot at (x, y) = (0.60, 0.83), x being the horizontal axis and y the vertical axis. On the other hand, a transition from EFRET = 0.83 to EFRET = 0.60 will contribute to the contour plot at (x, y) = (0.83, 0.60). The color coding on the plot represents how often such transitions are observed. The symmetry of the contour plot in TDP suggests that L1Cy5–7OTD can bind to either EF-High or EF-Low and then transition to the other state. Figure 3C shows transitions between three primary states (EFRET≈0.47, EFRET≈0.63-0.66, and EFRET≈0.77), while Figure 3D shows transitions between two primary states (EFRET ≈ 0.47 and EFRET ≈ 0.72). These transitions are shown with white dashed lines which are marked with numbers. These numbers represent transitions between the following FRET states: 1: 0.47 to 0.66; 2: 0.63 to 0.77; 3: 0.66 to 0.47; and 4: 0.77 to 0.62 in Figure 3C. Transition 1 and 3 are back and forth transitions between EFRET ≈ 0.54 and EFRET ≈ 0.71, while 2 and 4 are back and forth transitions between EFRET ≈ 0.68 and EFRET ≈ 0.80. The EFRET = 0.62, 0.63, and 0.66 are close enough that they are considered to represent the same state. Whether the three prominent states in Figure 3C represent three different orientations is not clear at this point. The relatively small dataset that resulted from rarity of these transitions, which also required combining of data on two different constructs, could also be a reason for the fragmented distribution. However, there is in principle no reason for why L1Cy5–7OTD could not bind in more than two primary orientations. The prominent transitions in Figure 3D are 1: 0.47 to 0.72 and 2: 0.72 to 0.47, which are essentially back and forth transitions between the two prominent states.
Dwell time analysis
We also measured the dwell time of L1Cy5–7OTD, which is a measure of how long L1Cy5–7OTD remains bound on the G-tetrad before it dissociates (Figure 2C). The DNA constructs pd-hGQ12T and pd-3Ly1Lp12T were used. Binding of 50–100 nM L1Cy5–7OTD was studied in 150 mM K+. As photobleaching of Cy5 and dissociation of L1Cy5–7OTD are indistinguishable in this assay, it is important to minimize photobleaching. Therefore, the laser power should be low enough to ensure the characteristic time for photobleaching to be significantly longer than the typical dwell time. We performed a systematic measure of the dwell time as a function of laser power (see Supplementary Figure S4) and identified 2.5 mW laser excitation power, which is ∼16x lower than typical power used in these assays, to satisfy this condition. Therefore, the dwell time histograms were constructed from data obtained with 2.5 mW or lower laser excitation power. The image integration time was increased from 0.1 to 2.5 s to compensate for the reduced laser power. Figure 2C was acquired under these conditions.
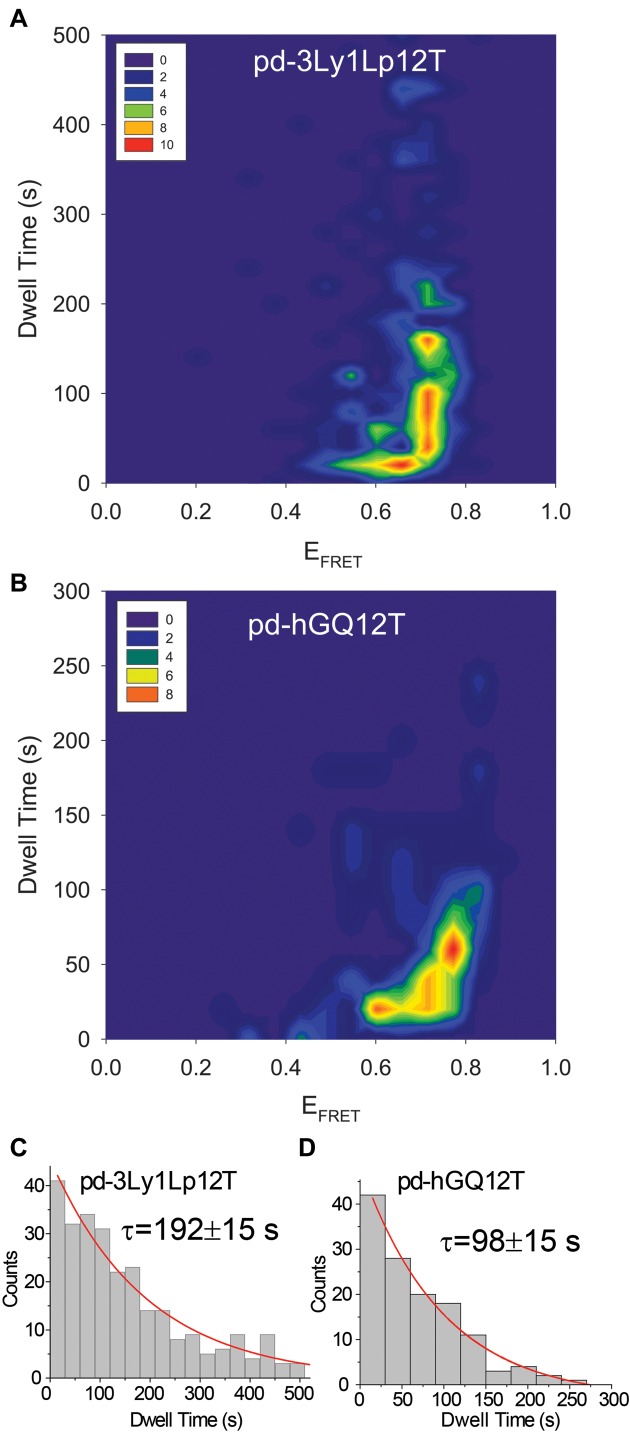
In general, we observe four types of traces upon stacking of L1Cy5–7OTD to GQ: (i) L1Cy5–7OTD was already bound to the GQ when imaging started; (ii) L1Cy5–7OTD did not dissociate from GQ when imaging ended; (iii) multiple non-overlapping binding events were observed for the same DNA molecule during the imaging time; (iv) a single binding event that started and ended during the imaging period was observed. Examples of these are shown in Supplementary Figure S5. In categories (i) and (ii) the dwell time would be underestimated as we miss some part of the binding event either at the beginning in (i) or at the end in (ii). Category (iii) should in principle be able to demonstrate an accurate dwell time however, it is challenging in such traces to distinguish between different binding events and a single binding event that undergoes an extended transition to a dark state (‘blinking of acceptor or donor’). Category (iv) traces, on the other hand, should provide the most accurate measurement of the dwell time with the disadvantage that a significant number of molecules had to be eliminated from the analysis. Figure 4 shows a contour plot of dwell times as a function of EFRET for traces in category (iv) with the low laser power. In order to determine a characteristic dwell time for each DNA construct, we build dwell time histograms as shown in Figure 4C and D (number of molecules N = 282 for pd-3Ly1Lp12T and N = 129 for pd-hGQ12T). Exponential fits to these histograms result in a characteristic (decay) time of τ = 192 ± 15 s for pd-3Ly1Lp12T and τ = 98 ± 15 s for pd-hGQ12T constructs. The majority of L1Cy5–7OTD molecules remain stacked on GQ for tens of seconds, with significantly longer dwells observed for the more stable pd-3Ly1Lp12T compared to pd-hGQ12T. In addition, the state with higher EFRET demonstrates a much broader distribution with a significantly higher characteristic dwell time. The lack of a clear boundary in EFRET between the two states prevents a quantitative comparison of the dwell times but is approximately several fold based the distributions in Figure 4A and B. At the typical laser powers used for other assays, 40 mW, we did not observe any difference between the two GQ constructs since the analysis was dominated by photobleaching of Cy5, which is similar for all conditions (see Supplementary Figure S6 for these data). Therefore, we believe the observed differences in dwell times reflect genuine variations in L1Cy5–7OTD dynamics as it interacts with different GQs or binds in different orientations. Since photobleaching cannot be completely eliminated even at such low laser powers, the quoted dwell times maybe somewhat smaller than the real dwell times, and should be considered as a lower limit compared to the case when photobleaching is completely eliminated.
Figure 4.
Contour plot of dwell time versus EFRET in 150 mM K+ for (A) pd-3Ly1Lp12T; and (B) pd-hGQ12T. Low laser power and 2.5 s integration time were used to minimize the influence of photobleaching. Significantly longer dwell times are observed for the more stable pd-3Ly1Lp12 compared to pd-hGQ12T. Also, the dwell times are significantly longer for EF-High levels compared to EF-Low levels. Dwell time histogram of (C) pd-3Ly1Lp12T and (D) pd-hGQ12T constructs. An exponential decay fit (red curves) to these histograms results in a decay parameter (characteristic dwell time) of τ = 192 ± 15 s for pd-3Ly1Lp12T and τ = 98 ± 15 s for pd-3Ly1Lp12T.
Binding frequency
How frequency of L1Cy5–7OTD binding depends on the stability of GQ is another aspect we investigated. To quantify the frequency of binding, we determined the percentile of DNA molecules with respect to the total number of DNA molecules, in which we observed at least one L1Cy5–7OTD binding during our imaging time. The imaging time was kept constant for all DNA constructs for this analysis. The molecules that did not show any L1Cy5–7OTD binding essentially only showed donor-only FRET level. The results of this analysis are shown in Figure 5.
Figure 5.
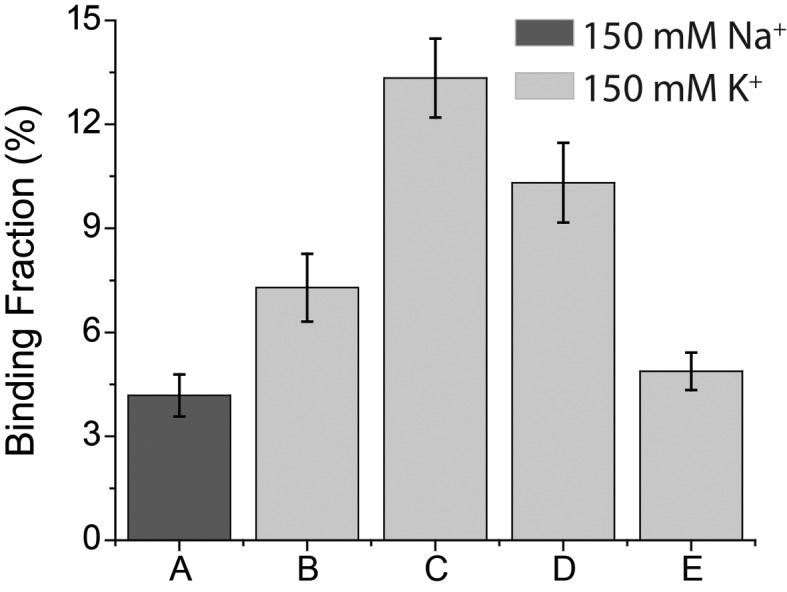
The fraction of GQ molecules showing L1Cy5–7OTD binding for different DNA constructs. (A) pd-hGQ12T in 150 mM Na+; (B–E) were in 150mM K+. (B) pd-hGQ12T; (C) pd-3Ly1Lp12T; (D) pd-12T3Ly1Lp; (E) pd-12ThGQ.
In one case we compared the binding frequency for pd-hGQ12T in 150 mM Na+ vs. 150 mM K+ based on the large body of work that has demonstrated human telomeric GQ to be more stable in K+ compared to Na+ (32). As shown in Figure 5, L1Cy5–7OTD binding frequency is 1.7 times higher in K+ compared to Na+. In the second case, we compared the binding frequency of L1Cy5–7OTD, at 150 mM K+, to four different GQ constructs we introduced earlier (see Figure 2A for schematics of these constructs). Of particular interest here is the stability of these constructs, and GQs with short loops, as pd-12T3Ly1Lp and pd-3Ly1Lp12T, are known to be more stable (33). Figure 5 shows that L1Cy5–7OTD binds 1.8±0.3 times more frequently to pd-3Ly1Lp12T construct than to pd-hGQ12T construct. A similar ratio, 2.1 ± 0.3, is observed between the binding frequency to pd-12T3Ly1Lp and to pd-12ThGQ. These results suggest that L1Cy5–7OTD binds more frequently to more stable GQ. Another comparison that we performed is whether having a 12T (pd-3Ly1Lp12T and pd-hGQ12T) or 2T overhang (pd-12T3Ly1Lp and pd-12ThGQ) downstream of GQ has any influence on binding frequency. Although less prominent, the binding frequency is higher for 12T overhang constructs compared to 2T overhang constructs. The binding frequency is 1.3 ± 0.2 fold higher for pd-3Ly1Lp12T compared to pd-12T3Ly1Lp, and 1.5 ± 0.3 fold higher for pd-hGQ12T compared to pd-12ThGQ. This increase could be due to the longer overhang being more permissive to L1Cy5–7OTD binding, possibly because the Cy3 fluorophore is further displaced from the binding site of L1Cy5–7OTD.
Our studies with L1Cy5–7OTD demonstrate that interactions of this small molecule with GQ are dynamic in terms of binding kinetics and possibly in terms of rotational freedom. The Cy5 fluorophore has enabled monitoring and quantifying binding and dissociation dynamics of a single L1Cy5–7OTD molecule to GQ, which to our knowledge has not been demonstrated for any GQ stabilizing small molecule before. We show that most L1Cy5–7OTD molecules remain bound to GQ for tens of seconds, with significantly higher dwell times for more stable GQ. Furthermore, frequency of L1Cy5–7OTD binding also shows significant correlations with GQ stability, with more frequent events taking place for more stable GQ. In addition, L1Cy5–7OTD possibly has at least two preferred primary binding orientations and is able to occasionally transition between these orientations while it remains stacked on a G-tetrad. The binding is significantly more stable in one of these orientations compared to the other for both GQ constructs studied.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [1R15GM109386 to H.B.]; Grant-in-Aid for Scientific Research (B) from JSPS [26282214 to K.N.]; Grant-in-Aid for Challenging Exploratory Research from JSPS [16K13094 to K.N.]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Blackburn E.H., Greider C.W., Szostak J.W. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat. Med. 2006;12:1133–1138. doi: 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- 2.Fletcher T.M., Sun D., Salazar M., Hurley L.H. Effect of DNA secondary structure on human telomerase activity. Biochemistry. 1998;37:5536–5541. doi: 10.1021/bi972681p. [DOI] [PubMed] [Google Scholar]
- 3.Johnson J.E., Cao K., Ryvkin P., Wang L.S., Johnson F.B. Altered gene expression in the Werner and Bloom syndromes is associated with sequences having G-quadruplex forming potential. Nucleic Acids Res. 2010;38:1114–1122. doi: 10.1093/nar/gkp1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paeschke K., Capra J.A., Zakian V.A. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huppert J.L., Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Todd A.K., Johnston M., Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balasubramanian S., Hurley L.H., Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat. Rev. Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morris M.J., Negishi Y., Pazsint C., Schonhoft J.D., Basu S. An RNA G-Quadruplex Is Essential for Cap-Independent Translation Initiation in Human VEGF IRES. J. Am. Chem. Soc. 2010;132:17831–17839. doi: 10.1021/ja106287x. [DOI] [PubMed] [Google Scholar]
- 9.McLuckie K.I.E., Antonio M.D., Zecchini H., Xian J., Caldas C., Krippendorff B.-F., Tannahill D., Lowe C., Balasubramanian S. G-Quadruplex DNA as a Molecular Target for Induced Synthetic Lethality in Cancer Cells. J. Am. Chem. Soc. 2013;135:9640–9643. doi: 10.1021/ja404868t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merle P., Gueugneau M., Teulade-Fichou M.P., Muller-Barthelemy M., Amiard S., Chautard E., Guetta C., Dedieu V., Communal Y., Mergny J.L., et al. Highly efficient radiosensitization of human glioblastoma and lung cancer cells by a G-quadruplex DNA binding compound. Sci. Rep. 2015;5:16255. doi: 10.1038/srep16255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diveshkumar K.V., Sakrikar S., Harikrishna S., Dhamodharan V., Pradeepkumar P.I. Targeting promoter G-quadruplex DNAs by indenopyrimidine-based ligands. ChemMedChem. 2014;9:2754–2765. doi: 10.1002/cmdc.201402394. [DOI] [PubMed] [Google Scholar]
- 12.Iida K., Nagasawa K. Macrocyclic polyoxazoles as G-quadruplex ligands. Chem. Rec. 2013;13:539–548. doi: 10.1002/tcr.201300015. [DOI] [PubMed] [Google Scholar]
- 13.Muller S., Laxmi-Reddy K., Jena P.V., Baptiste B., Dong Z., Godde F., Ha T., Rodriguez R., Ba.lasubramanian S., Huc I. Targeting DNA G-quadruplexes with helical small molecules. Chembiochem. 2014;15:2563–2570. doi: 10.1002/cbic.201402439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rahman K.M., Tizkova K., Reszka A.P., Neidle S., Thurston D.E. Identification of novel telomeric G-quadruplex-targeting chemical scaffolds through screening of three NCI libraries. Bioor. Med. Chem. Lett. 2012;22:3006–3010. doi: 10.1016/j.bmcl.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 15.Ohnmacht S.A., Varavipour E., Nanjunda R., Pazitna I., Di Vita G., Gunaratnam M., Kumar A., Ismail M.A., Boykin D.W., Wilson W.D., et al. Discovery of new G-quadruplex binding chemotypes. Chem. Commun. (Camb.) 2014;50:960–963. doi: 10.1039/c3cc48616h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iida K., Nakamura T., Yoshida W., Tera M., Nakabayashi K., Hata K., Ikebukuro K., Nagasawa K. Fluorescent-ligand-mediated screening of G-quadruplex structures using a DNA microarray. Angew. Chem. Int. Ed. Engl. 2013;52:12052–12055. doi: 10.1002/anie.201305366. [DOI] [PubMed] [Google Scholar]
- 17.Largy E., Granzhan A., Hamon F., Verga D., Teulade-Fichou M.P. Visualizing the quadruplex: from fluorescent ligands to light-up probes. Top. Curr.Chem. 2013;330:111–177. doi: 10.1007/128_2012_346. [DOI] [PubMed] [Google Scholar]
- 18.Verga D., Hamon F., Poyer F., Bombard S., Teulade-Fichou M.P. Photo-cross-linking probes for trapping G-quadruplex DNA. Angew. Chem. Int. Ed. Engl. 2014;53:994–998. doi: 10.1002/anie.201307413. [DOI] [PubMed] [Google Scholar]
- 19.Iida K., Tera M., Hirokawa T., Shin-ya K., Nagasawa K. G-quadruplex recognition by macrocyclic hexaoxazole (6OTD) dimer: greater selectivity than monomer. Chem. Commun. (Camb). 2009:6481–6483. doi: 10.1039/b910242f. [DOI] [PubMed] [Google Scholar]
- 20.Jena P.V., Shirude P.S., Okumus B., Laxmi-Reddy K., Godde F., Huc I., Balasubramanian S., Ha T. G-quadruplex DNA bound by a synthetic ligand is highly dynamic. J. Am. Chem. Soc. 2009;131:12522–12523. doi: 10.1021/ja903408r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kreig A., Calvert J., Sanoica J., Cullum E., Tipanna R., Myong S. G-quadruplex formation in double strand DNA probed by NMM and CV fluorescence. Nucleic Acids Res. 2015;43:7961–7970. doi: 10.1093/nar/gkv749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yangyuoru P.M., Di Antonio M., Ghimire C., Biffi G., Balasubramanian S., Mao H. Dual binding of an antibody and a small molecule increases the stability of TERRA G-Quadruplex. Angew. Chem. Weinheim. Bergstr. Ger. 2015;127:924–927. doi: 10.1002/ange.201408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joo C., Ha T. Single-molecule FRET with total internal reflection microscopy. Cold Spring Harbor Protoc. 2012;2012 doi: 10.1101/pdb.top072058. doi:10.1101/pdb.top072058. [DOI] [PubMed] [Google Scholar]
- 24.Ray S., Bandaria J.N., Qureshi M.H., Yildiz A., Balci H. G-quadruplex formation in telomeres enhances POT1/TPP1 protection against RPA binding. Proc. Natl. Acad. Sci. U.S.A. 2014;111:2990–2995. doi: 10.1073/pnas.1321436111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Viglasky V., Bauer L., Tluckova K., Javorsky P. Evaluation of human telomeric g-quadruplexes: the influence of overhanging sequences on quadruplex stability and folding. J. Nucleic Acids. 2010 doi: 10.4061/2010/820356. doi:10.4061/2010/820356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathias J., Okyere R., Lomidze L., Gvarjaladze D., Musier-Forsyth K., Kankia B. Thermal stability of quadruplex primers for highly versatile isothermal DNA amplification. Biophys. Chem. 2014;185:14–18. doi: 10.1016/j.bpc.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Jing N., Marchand C., Liu J., Mitra R., Hogan M.E., Pommier Y. Mechanism of inhibition of HIV-1 integrase by G-tetrad-forming oligonucleotides in Vitro. J. Biol. Chem. 2000;275:21460–21467. doi: 10.1074/jbc.M001436200. [DOI] [PubMed] [Google Scholar]
- 28.Budhathoki J.B., Stafford E.J., Yodh J.G., Balci H. ATP-dependent G-quadruplex unfolding by Bloom helicase exhibits low processivity. Nucleic Acids Res. 2015;43:5961–5970. doi: 10.1093/nar/gkv531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Budhathoki J.B., Ray S., Urban V., Janscak P., Yodh J.G., Balci H. RecQ-core of BLM unfolds telomeric G-quadruplex in the absence of ATP. Nucleic Acids Res. 2014;42:11528–11545. doi: 10.1093/nar/gku856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung W.J., Heddi B., Tera M., Iida K., Nagasawa K., Phan A.T. Solution structure of an intramolecular (3 + 1) human telomeric G-quadruplex bound to a telomestatin derivative. J. American Chem. Soc. 2013;135:13495–13501. doi: 10.1021/ja405843r. [DOI] [PubMed] [Google Scholar]
- 31.Bronson J.E., Fei J., Hofman J.M., Gonzalez R.L., Jr, Wiggins C.H. Learning rates and states from biophysical time series: a Bayesian approach to model selection and single-molecule FRET data. Biophys. J. 2009;97:3196–3205. doi: 10.1016/j.bpj.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tran P.L., Mergny J.L., Alberti P. Stability of telomeric G-quadruplexes. Nucleic Acids Res. 2011;39:3282–3294. doi: 10.1093/nar/gkq1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guedin A., Gros J., Alberti P., Mergny J.L. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010;38:7858–7868. doi: 10.1093/nar/gkq639. [DOI] [PMC free article] [PubMed] [Google Scholar]