Abstract
NusG, the only universally conserved transcription factor, comprises an N- and a C-terminal domain (NTD, CTD) that are flexibly connected and move independently in Escherichia coli and other organisms. In NusG from the hyperthermophilic bacterium Thermotoga maritima (tmNusG), however, NTD and CTD interact tightly. This closed state stabilizes the CTD, but masks the binding sites for the interaction partners Rho, NusE and RNA polymerase (RNAP), suggesting that tmNusG is autoinhibited. Furthermore, tmNusG and some other bacterial NusGs have an additional domain, DII, of unknown function. Here we demonstrate that tmNusG is indeed autoinhibited and that binding to RNAP may stabilize the open conformation. We identified two interdomain salt bridges as well as Phe336 as major determinants of the domain interaction. By successive weakening of this interaction we show that after domain dissociation tmNusG-CTD can bind to Rho and NusE, similar to the Escherichia coli NusG-CTD, indicating that these interactions are conserved in bacteria. Furthermore, we show that tmNusG-DII interacts with RNAP as well as nucleic acids with a clear preference for double stranded DNA. We suggest that tmNusG-DII supports tmNusG recruitment to the transcription elongation complex and stabilizes the tmNusG:RNAP complex, a necessary adaptation to high temperatures.
INTRODUCTION
Transcription is catalyzed by the enzyme RNA polymerase (RNAP) in all domains of life. RNAP is highly regulated by numerous transcription factors, amongst them the N-utilization substance (Nus) factor NusG, the only universally conserved transcription factor (called Spt5 in archaea and eukarya) (1,2). Escherichia coli (E. coli) NusG (ecNusG) comprises an N-terminal and a C-terminal domain (NTD, CTD) that are connected by a flexible 15 amino acid linker (3). ecNusG-NTD binds to the clamp of RNAP by making bridging contacts between the β’ clamp helices (β’CH) and the β gate loop (βGL) (4,5). This interaction is proposed to lock the clamp in a closed state to keep RNAP processive and to suppress pauses (5). ecNusG-CTD is the target of several interaction partners. It either binds to termination factor Rho, supporting Rho-dependent termination or it interacts with protein S10 of the 30S subunit of the ribosome, thus coupling transcription and translation (6,7). S10 is identical to antitermination factor NusE, and as such it can form a complex with NusB. This heterodimer is part of the multiprotein antitermination complex in which RNAP is modified to read through termination signals, a process necessary for the transcription of ribosomal RNA operons as well as DNA of lambdoid phages (8–10).
The NTD and CTD of NusG from the hyperthermophilic bacterium Thermotoga maritima (T. maritima, tmNusG) tightly interact with each other, in striking contrast to the corresponding domains of ecNusG (11,12). As the binding sites for RNAP, NusE and Rho are masked in this closed state of tmNusG, the respective interactions are supposed to be repressed (11). Compared with ecNusG, tmNusG comprises an additional domain, DII, that is inserted into tmNusG-NTD (11). tmNusG-DII is composed of two subdomains and interacts neither with tmNusG-NTD nor with tmNusG-CTD. An insertion is also present in NusG from Aquifex aeolicus (aaNusG) which, however, is significantly smaller than tmNusG-DII and shows no sequential or structural homology to tmNusG-DII (11,13–14). Although non-sequence specific binding to nucleic acids has been demonstrated for the additional domain in both tmNusG and aaNusG, the functions of these domains remain unclear (13,15).
NusE/S10 is an important interaction partner of NusG. In E. coli, NusE (ecNusE) is a monomeric, globular protein with a ribosome binding loop. This loop has no function in antitermination but is important for its insertion into the 30S subunit, where it folds into a two-stranded β-sheet (16). In solution, this loop is unstructured and causes severe protein instability. Even a deletion variant, in which this loop (aa 46–67) is replaced by a single Ser residue (ecNusEΔ), is only stable in complex with ecNusB (16). During antitermination the ecNusE:ecNusB complex binds a highly conserved RNA sequence (boxA) (8,17). While NusE appears to be the active transcription factor, the function of NusB is to recruit NusE to RNA, as NusB's affinity to boxA-RNA is significantly stronger than that of NusE (16,18).
So far, the intramolecular domain interaction in tmNusG is unique among NusG proteins. Here we used nuclear magnetic resonance (NMR) spectroscopy and targeted amino acid substitutions to demonstrate that tmNusG is indeed autoinhibited and to identify the major determinants of the tight domain interaction. Furthermore, NMR and fluorescence spectroscopic studies were carried out to elucidate possible roles for tmNusG-DII.
MATERIALS AND METHODS
Cloning
The codon optimized gene encoding tmNusEΔ (residues 1, Gly, 3–45, Ser, 68–102) fused to the codons corresponding to an N-terminal His6 tag and a Tobacco Etch Virus (TEV) protease cleavage site was obtained from GenScript (Piscataway, NJ, USA) and cloned into pET29b (Novagen, Madison, WI, USA) via NdeI and BamHI restriction sites.
The plasmid pETM11-tmNusGΔR275A,R279A coding for tmNusGΔR275A,R279A (11) was used as template for site-directed mutagenesis to generate tmNusGΔR275A,R279A,F336A with the following primers: 5′-gta aac gta act ata gcc gga cgt gaa act cc-3′, 5′- gg agt ttc acg tcc ggc tat agt tac gtt tac -3′. Point mutations were introduced according to the QuikChange kit (Agilent Technologies, Santa Clara, CA, USA) and confirmed by sequencing (GATC Biotech, Köln, Germany). tmNusG-DII (encoding residues 42–233 of tmNusG) was cloned into pETGB1a (provided by Gunter Stier, EMBL Heidelberg, Germany) via NcoI and BamHI restriction sites using pET22b-tmNusG (provided by Markus Wahl, FU Berlin, Germany) as template and the primers 5′-cat gcc atg gct gaa gag gta gtt ttg gac-3′ and 5′-cgg gat ccc tac ggg aag agt ttt ctt ctt g-3′. The resulting protein carried a His6 tag followed by the 56 amino acid immunoglobulin binding domain B1 of streptococcal protein G (GB1) and a TEV protease cleavage site at its N-terminus.
The codon optimized tmNusB gene was obtained from GenScript (Piscataway, NJ, USA) and cloned into pET16b (Novagen, Madison, WI, USA) using NcoI and BamHI restriction sites.
The codon optimized gene encoding tmRho was obtained from GenScript (Piscataway, NJ, USA) and cloned into the E. coli expression vector pET101/D-TOPO with the Champion™ pET101 Directional TOPO© Expression Kit (Invitrogen, Carlsbad, CA, USA).
Genes encoding the β, β’, and ω subunits of T. maritima RNAP (tmRNAP) were amplified by polymerase chain reaction (PCR) from genomic T. maritima DNA (DSMZ, Braunschweig, Germany) with the following primers:
tmrpoB: 5′-gga att cca tat gaa aga gat ctc ttg cgg tag g-3′, 5′-cgg gat cct cag tac ttg tcg ata tct atc tcg-3′
tmrpoC: 5′-gga att cca tat gcc aat gtc ctc ttt caa gag g-3′, 5′-acg cgt cga ccg cga gtt ctt ctt cca ctg c-3′
tmrpoZ: 5′-gga att cca tat gga aaa aat tgt gaa gtt cg-3′, 5′-ggg gta cct cac ttc acc ttc gga atg-3′
tmrpoB and tmrpoC were both cloned into pET29b (Novagen, Madison, WI, USA) using NdeI and BamHI or NdeI and SalI restriction sites, respectively. This allowed the production of the β’ subunit with a C-terminal nine amino acid linker followed by a His6 tag. The gene encoding the ω subunit was cloned into the multiple cloning site 2 of pACYCDuet-1 (Novagen, Madison, WI, USA) via NdeI and KpnI restriction sites.
Gene expression and protein purification
tmNusEΔ
His6-tmNusEΔ was expressed in E. coli Rosetta (DE3) pLysS (Novagen, Madison, WI, USA). Cells were grown in lysogeny broth (LB) medium supplemented with 30 μg/ml kanamycin and 34 μg/ml chloramphenicol at 37°C until an optical density at 600 nm (OD600) of 0.6–0.8 was reached. Gene expression was induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and cells were harvested 4 h after induction by centrifugation (9 000 × g, 15 min, 4°C). Cells were resuspended in buffer A (50 mM Tris/HCl, pH 7.5, 300 mM NaCl, 10 mM imidazole) and lysed using a microfluidizer (Microfluidics, Newton, MA, USA). After centrifugation (12 000 × g, 30 min, 4°C) the crude extract was applied to a HisTrap FF column (GE Healthcare, Munich, Germany). Elution was performed with a step gradient with increasing imidazole concentrations (10–500 mM in buffer A). His6-tmNusEΔ containing fractions were combined and cleaved during overnight dialysis at 4°C (50 mM Tris/HCl, pH 7.5, 300 mM NaCl; molecular weight cut-off (MWCO): 1 000 Da) by TEV protease. TEV protease and the cleaved off GB1 tag were removed by a second nickel affinity chromatography using the same conditions as above. Pure tmNusEΔ was in the wash fraction as it exhibits weak and non-specific binding to the HisTrap FF column. It was dialyzed against the required buffer, concentrated by ultrafiltration (Vivascience, Hannover, Germany; MWCO: 1000 Da), frozen in liquid nitrogen, and stored at −80°C.
tmNusB
E. coli BL21(DE3) cells (Novagen, Madison, WI, USA) were transformed with pET16b_tmNusB and grown in LB medium containing 100 μg/ml ampicillin. Overexpression was induced by 1 mM IPTG at an OD600 of 0.6–0.8. 4 h after induction cells were harvested by centrifugation (9 000 × g, 15 min, 4°C) and resuspended in buffer B (75 mM potassium phosphate, pH 7.0). After addition of half a protease inhibitor cocktail tablet (cOmplete, EDTA-free, Roche Diagnostics GmbH, Mannheim, Germany), 0.05 g lysozyme (Roche Diagnostics GmbH, Mannheim, Germany) and DNase I (Roche Diagnostics GmbH, Mannheim, Germany), the cell suspension was stirred on ice for 30 min. Cells were lysed with a microfluidizer (Microfluidics, Newton, MA, USA) and the lysate centrifuged (12 000 × g, 30 min, 4°C). The supernatant was incubated at 80°C for 30 min and centrifuged (12 000 × g, 30 min, 4°C). Subsequently, nucleic acids were precipitated by addition of 0.6 % (v/v) polyethylenimine and 15 min stirring on ice. After centrifugation (12 000 × g, 30 min, 4°C) (NH4)2SO4 was added to the supernatant to a final concentration of 1.1 M, and the protein solution was applied to a HiTrap HP Butyl column (GE Healthcare, Munich, Germany). After washing with buffer B supplemented with 1 M KCl and 1 M (NH4)2SO4, elution was performed using a step gradient with buffer B. tmNusB containing fractions were combined and dialyzed over night against buffer C (50 mM Tris/HCl, pH 7.5, 50 mM NaCl). The protein solution was applied to a HiTrap Heparin HP column (GE Healthcare, Munich, Germany) and elution was performed by a step gradient with increasing NaCl concentrations (50 mM-1 M in buffer C). Fractions containing pure tmNusB were combined, dialyzed against the desired buffer, concentrated by ultrafiltration (Vivascience, Hannover, Germany; MWCO: 5000 Da), frozen in liquid nitrogen and stored at −80°C.
tmNusG
E. coli BL21 (DE3) cells (Novagen, Madison, WI, USA) containing pET22b-tmNusG were grown in LB medium in the presence of 100 μg/ml ampicillin at 37°C. At an OD600 of 0.6–0.8 expression was induced with 1 mM IPTG and cells were harvested 3 h after induction by centrifugation (9 000 × g, 15 min, 4°C). Cells were resuspended in buffer D (50 mM Tris/HCl, pH 7.5, 100 mM NaCl), lysed with a microfluidizer (Microfluidics, Newton, MA, USA) and the lysate was heated to 85°C for 20 min. After centrifugation (12 000 × g, 30 min, 4°C), the supernatant was applied to a HiTrap Heparin HP column (GE Healthcare, Munich, Germany). Elution was performed with a step gradient with increasing NaCl concentrations (100 mM–1 M in buffer D). Fractions containing pure tmNusG were combined, dialyzed against the required buffer, concentrated by ultrafiltration (MWCO = 10 000 Da), frozen in liquid nitrogen and stored at −80°C.
tmNusG-NTD, tmNusG-CTD, tmNusGΔ
The proteins were produced and purified as described (11). The protocol for tmNusGΔ was also used for all its variants.
tmNusG-DII
The gene encoding tmNusG-DII fused to a His6-GB1 tag was expressed in E. coli Rosetta (DE3) (Novagen, Madison, WI, USA) harboring the plasmid pETGB1a-tmNusG-DII. Gene expression, cell lysis and purification were carried out as described for tmNusEΔ, with an MWCO of 3.5 or 5 kDa for dialysis or ultrafiltration, respectively.
tmRho
E. coli BL21 (DE3) cells (Novagen, Madison, WI, USA) were transformed with pET101/D-TOPO_tmRho and grown at 37°C in the presence of 100 μg/ml ampicillin. After induction with 0.4 mM IPTG at an OD600 of 0.6–0.8 cells were grown for 3 h, harvested by centrifugation (9 000 × g, 15 min, 4°C) and resuspended in buffer E (100 mM potassium phosphate, pH 7.6, 100 mM KCl). The crude extract was prepared as described for tmNusEΔ and then heated to 90°C for 15 min in a water bath. Subsequently, nucleic acids were precipitated by addition of 0.2 % (v/v) polyethylenimine and 15 min stirring on ice. After centrifugation (12 000 × g, 30 min, 4°C) (NH4)2SO4 was added to the supernatant to a final concentration of 1.1 M, and the protein solution was applied to a HiTrap HP Butyl column (GE Healthcare, Munich, Germany). After washing with buffer E supplemented with 1 M (NH4)2SO4, elution was performed using a step gradient with buffer E containing 10 % (v/v) 2-propanole. tmRho containing fractions were combined, dialyzed over night against buffer C, and concentrated by ultrafiltration (MWCO = 10 000 Da). The protein was applied to a Superdex 200 column (Tricorn 10/300; GE Healthcare, Munich, Germany), and the fractions containing pure tmRho were combined. tmRho dialysis and storage were performed as above (MWCO = 10 000 Da). Concentrations of tmRho always refer to the hexamer.
β subunit of tmRNAP
The gene coding for the β subunit of tmRNAP was expressed in E. coli Rosetta (DE3) pLysS (Novagen, Madison, WI, USA). Cells were grown at 37°C supplemented with 30 μg/ml kanamycin and 34 μg/ml chloramphenicol. At an OD600 of ∼0.8 expression was induced by addition of 0.1 mM IPTG. Cells were harvested 3 h after induction by centrifugation (9 000 × g, 15 min), resuspended in 50 mM Tris/HCl (pH 7.3), 50 mM NaCl and lysed as described for tmNusEΔ. After centrifugation (30 min, 4°C, 12 000 × g) the pellet was dissolved in 1 mM ethylenediaminetetraacetic acid (EDTA), pH 8.0, supplemented with 1 mg/ml deoxycholic acid sodium salt, 20 mM dithiotreitol (DTT) and 0.2 mg/ml lysozyme, and again centrifuged (30 min, 4°C, 12 000 × g). The pellet was washed three times with the same solution, three times with 50 mM Tris/HCl, pH 8.0, 50 mM NaCl, 10 mM EDTA, 5 mM DTT and once with H2O. Finally, the pellet was resuspended in 50 mM Tris/HCl, pH 7.3, 8 M urea, 500 mM NaCl and stirred for 1 h at room temperature. Urea was removed by dialysis against 50 mM Tris/HCl, pH 7.3, 5 % (v/v) glycerol, 500 mM NaCl, 0.5 mM EDTA, 1 mM DTT at 4°C for 3 h and the same buffer without NaCl overnight. The dialysate was centrifuged (30 min, 4°C, 12 000 × g) and the supernatant was applied to a Q Sepharose FF column (GE Healthcare, Munich, Germany). After washing with buffer F (50 mM Tris/HCl, pH 7.5, 5 % (v/v) glycerol, 0.5 mM EDTA, 1 mM DTT) elution was performed using a constant NaCl gradient (0-1 M NaCl) in buffer F. Fractions containing pure tmβ were combined, dialyzed against the required buffer, concentrated by ultrafiltration (MWCO = 10 000 Da) and stored at −80°C after freezing in liquid nitrogen.
The complex of β’ and ω subunits of tmRNAP
The genes of β’ and ω subunits of tmRNAP (tmβ’, tmω) were coexpressed from individual plasmids in E. coli BL21 (DE3) cells (Novagen, Madison, WI, USA) to prevent tmβ’ from degradation in LB medium in the presence of 30 μg/ml kanamycin and 34 μg/ml chloramphenicol. At an OD600 of 0.6-0.8 expression was induced with 0.1 mM IPTG and cells were harvested (9 000 × g, 15 min, 4°C) after 4 h. Cells were resuspended in buffer G (50 mM Tris/HCl, pH 7.5, 500 mM NaCl, 10 mM imidazole) and the crude extract was prepared as described for tmNusEΔ. The supernatant was applied to a HisTrap HP column (GE Healthcare, Munich, Germany). The column was washed with buffer G, and the tmβ’:tmω complex was eluted using a step gradient with increasing imidazole concentrations (10–500 mM imidazole in 50 mM Tris/HCl, pH 7,5, 500 mM NaCl). Fractions containing tmβ’:tmω were combined and treated as tmβ above.
E. coli RNAP (ecRNAP)
The protein was produced and purified as described (19).
Isotopic labeling
15N- and 15N-, 13C-labeled proteins were obtained from E. coli cells grown in M9 minimal medium (20,21) with (15NH4)2SO4 (Campro Scientific, Berlin, Germany) and 13C D-glucose (Spectra Stable Isotopes, Columbia, MD, USA), respectively, as only nitrogen and carbon source. Expression and purification were as described for proteins produced in LB medium.
NMR spectroscopy
NMR measurements were performed on Bruker Avance 600 MHz, 700 MHz and 800 MHz spectrometers, the latter two equipped with cryogenically cooled probes. All spectra were recorded at 323 K and the initial sample volume was always 550 μl, if not stated otherwise. Standard heteronuclear double and triple resonance experiments were conducted for resonance assignment at 323 K (tmNusEΔ: backbone and side chain; tmNusG-DII: backbone) (22,23). 15N- and 13C-edited 3D nuclear Overhauser-effect spectroscopy (NOESY) experiments were recorded with mixing times of 120 ms for obtaining distance restraints.
For hydrogen/deuterium (H/D) exchange measurements the 15N-labeled proteins were in 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.5, 50 mM NaCl. After lyophilization proteins were dissolved in D2O (99.98 %) and the decay of signal intensities was observed in a series of [1H,15N]-heteronuclear single quantum coherence (HSQC) spectra. Exchange rates were determined by fitting the signal decay to a monoexponential curve, and the protection factors (PFs) were calculated by dividing the experimental exchange rates by the intrinsic exchange rates calculated from the amino acid sequence and experimental conditions with tabulated parameters (24,25).
15N longitudinal and transverse relaxation rates of tmNusGΔR275A,R279A,F336A were determined using standard methods at 600.2 MHz 1H frequency and a calibrated temperature of 323 K (buffer: 25 mM HEPES, pH 7.5, 50 mM NaCl) (26). The relaxation rates (R1, R2) were fitted to monoexponential decays using the program CURVE FIT (A. G. Palmer, Department of Biochemistry and Molecular Biophysics, Columbia University, USA). The rotation correlation times were calculated with TENSOR2 (27).
For the interaction studies of tmNusG variants and tmNusEΔ proteins were in 25 mM HEPES, pH 7.5, 50 mM NaCl. To evaluate titration experiments we calculated the normalized chemical shift changes Δδnorm according to Equation (1).
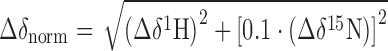 |
(1) |
where Δδ is the resonance frequency difference in ppm.
The dissociation constant (KD) was calculated from [1H,15N]-HSQC titrations by analyzing the chemical shift changes and fitting a two-state model with Equations (1) and (2) to the chemical shift change of residues showing fast exchange in the chemical shift timescale.
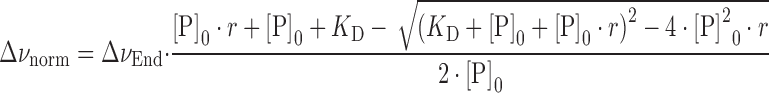 |
(2) |
where Δνnorm is the normalized resonance frequency difference in Hz, ΔνEnd the normalized resonance frequency difference between free and fully bound labeled protein in Hz, KD the dissociation constant, r the ligand:labeled protein ratio and [P]0 the total concentration of labeled protein. KD and ΔνEnd were used as fitting parameters. The reduction of [P]0 due to dilution was accounted for during fitting.
Interaction studies between 15N-tmNusG variants and tmRho, tmRNAP subunits or E. coli RNAP (ecRNAP) were carried out with 20–30 μM labeled protein and the signal intensity in one-dimensional (1D) [1H,15N] HSQC spectra after addition of the binding partner was analyzed. Intensities were normalized by concentration and number of scans. As pulse lengths changed less than 1 % upon addition of the potential binding partner, the influence of these changes on signal intensity was neglected. Binding to tmRho was measured in 100 mM potassium phosphate (pH 7.6), 100 mM KCl at 298 K, binding to the tmRNAP subunits in 10 mM potassium phosphate, pH 7.5, 50 mM NaCl, 5 % (v/v) glycerol, 0.5 mM EDTA at 323 K, and binding to ecRNAP in 10 mM potassium phosphate, pH 7.5, 50 mM NaCl, 5 % (v/v) glycerol, 0.5 mM EDTA, 2 mM DTT at 298 K.
Backbone assignments of tmNusG-CTD, tmNusGΔ, tmNusGΔR279A tmNusGΔR275A,R279A were taken from a previous study (11) and used to assign tmNusGΔR275A,R279A,F336A. Spectra of tmNusG variants were recorded with proteins in 25 mM HEPES, pH 7.5, 50 mM NaCl.
Structure calculation of tmNusEΔ
tmNusEΔ was in 25 mM HEPES, pH 7.5, 50 mM NaCl and all spectra were recorded at 323 K. Distance restraints for structure calculation were derived from 15N-edited NOESY and 13C-edited NOESY spectra. NOESY cross peaks were classified according to their relative intensities and converted to distance restraints with upper limits of 3.0 Å, strong, 4.0 Å, medium, 5.0 Å, weak and 6.0 Å, very weak. For ambiguous distance restraints the r−6 summation over all assigned possibilities defined the upper limit. Experimental NOESY spectra were validated semi-quantitatively against back-calculated spectra to confirm the assignment and to avoid bias of upper distance restraints by spin diffusion. Dihedral restraints were taken from analysis of chemical shifts by the TALOS software package (28). Hydrogen bonds were included for backbone amide protons in regular secondary structure, when the amide proton does not show a water exchange cross peak in the 15N-edited NOESY spectrum.
Structure calculations were performed with the program XPLOR-NIH 1.2.1 with a three-step simulated annealing protocol with floating assignment of prochiral groups including a conformational database potential (29). The 20 structures (out of a total of 120 structures) showing the lowest values of the target function excluding the database potential were further analyzed with X-PLOR, PROCHECK and PyMOL (29–31).
Molecular modeling
In order to test the possibility of an intramolecular interdomain interaction of ecNusG and aaNusG analogous to the closed conformation in tmNusG, we explored closed states of ecNusG and aaNusG by molecular dynamics simulations. Structural models of ecNusG and aaNusG with domain interaction were constructed by fitting the structures of the individual ecNusG or aaNusG domains to the tmNusG structure. During the simulation using a standard XPLOR protocol (XPLOR-NIH 2.1.2) for solution structure calculation, backbone coordinates were held fixed for all residues except the linker region between NTD and CTD (ecNusG: residues 116–127, aaNusG: residues 183–192) and the flexible region (residues 45–64) to reduce steric clashes between NTD and CTD in the initial structure. No further restraints were applied. This approach does not include attractive forces, but is used only for testing possible conformations that do not sacrifice typical bond geometries.
Programs
Graphical representations of protein structures as well as superimpositions were created with PyMOL (31). Sequence alignments were calculated with ClustalW (32) and edited with GeneDoc (http://www.psc.edu/biomed/genedoc). Electrostatic surface potentials were calculated with APBS (33).
Fluorescence anisotropy
Fluorescence anisotropy titrations were performed on a Fluorolog-Tau-3 spectrofluorometer (HORIBA Jobin Yvon GmbH, Unterhaching, Germany). 6-carboxyfluorescein (6-FAM) labeled nucleic acids (Supplementary Table S1) were titrated with tmNusG-NTD, tmNusG-DII or tmNusG-CTD at 25°C. Double stranded (ds) DNA was generated by mixing the single stranded (ss) oligonucleotides in equimolar concentrations, heating them to 95°C for 1 min, followed by incubation for 10 min at 60°C and cooling them to room temperature. Titrations were carried out in 1.3 ml 25 mM HEPES, pH 7.5, 50 mM NaCl for DNA or 25 mM Tris/HCl, pH 7.7, 50 mM NaCl for RNA in a 10 × 4 mm quartz cuvette (Hellma, Müllheim, Germany) using 5 or 50 nM of nucleic acids. The sample was excited at 492 nm, and emission was recorded at 515–517 nm. The slit widths were set to 3 and 4 nm for excitation and emission, respectively, when 50 nM nucleic acid were used and to 7 and 8 nm for 5 nM nucleic acid. After sample equilibration, each data point was collected six times with an integration time of 0.8 s. The anisotropic data was fitted to a two-state binding model (Equations 3 and 4) for determination of the KD values using GraFit 5.0 (Erithacus Software).
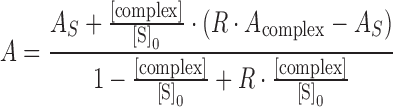 |
(3) |
with
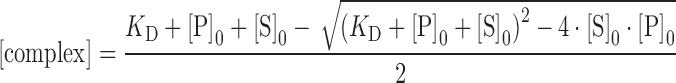 |
(4) |
where A is the measured anisotropy, AS the anisotropy of free nucleic acid, Acomplex the anisotropy of the complex, [complex] the concentration of the complex, [S]0 and [P]0 the total nucleic acid and protein concentrations, respectively, KD the dissociation constant, and R the ratio of the fluorescence intensities of fully bound and free substrate. Each titration was measured three times.
RESULTS AND DISCUSSION
Solution structure of tmNusEΔ
ecNusE interacts with ecNusG-CTD (6). To test whether a similar interaction also occurs in T. maritima, we first solved the solution structure of tmNusE. Initial attempts to study the full length protein, however, failed due to its structural instability after purification, a situation already known from ecNusE (16,34). Substitution of the ribosomal binding loop (residues 46–67) by a single Ser residue, tmNusEΔ, resulted in samples suitable for NMR spectroscopic studies, even in the absence of tmNusB. The [1H,15N]-HSQC spectra of tmNusEΔ showed the large chemical shift dispersion typical for globular, folded proteins, even up to 80°C (Supplementary Figure S1A–C). This is in stark contrast to ecNusEΔ, which exists in a structured state only in the NusEΔ:NusB complex or when it is part of the ribosome (16,35).
With multidimensional heteronuclear NMR spectroscopy, resonances were assigned and the solution structure of tmNusEΔ was determined based on 1341 distance and 110 dihedral restraints derived from multiple NMR experiments (Table 1 and Figure 1A). Free tmNusEΔ is composed of a four-stranded, antiparallel β-sheet, flanked by two roughly antiparallel α-helices on one side. The structure superimposes well with that of ecNusEΔ in the ecNusEΔ:ecNusB complex (Figure 1B, root mean square deviation for backbone atoms: 1.0 Å). Characteristic structural features of ecNusEΔ, e.g. the cis-conformation of Pro39 in the ecNusEΔ:ecNusB interface, are also present in tmNusEΔ, suggesting that these characteristics do not result from ecNusEΔ:ecNusB complex formation. Titration of 15N-tmNusEΔ with tmNusB indicated protein interaction (Supplementary Figure S1D). However, due to exchange processes close to or in the intermediate range of the NMR timescale, 15N-tmNusEΔ signals disappear upon tmNusB addition. Although finally the signals of the complex reappear, tracking of signal shifts during the titration and resonance assignment of the complex was not possible. 15N-tmNusEΔ signals with similar chemical shifts in the free and complex form are most likely from residues that are not directly involved in tmNusB binding. Comparison of tmNusEΔ with ecNusEΔ in the ecNusEΔ:ecNusB complex shows that these residues are located opposite of the ecNusB binding surface in ecNusEΔ, suggesting a similar type of interaction of the E. coli and the T. maritima proteins. These data and the high structural similarity to ecNusEΔ may indicate that tmNusEΔ fulfills the same functions as its E. coli homolog.
Table 1. Experimental constraints for structure calculation and statistics of tmNusEΔ.
| Experimental derived restraints | ||
|---|---|---|
| distance restraints | ||
| NOE | 1265 | |
| intraresidual | 467 | |
| sequential | 321 | |
| medium range | 182 | |
| long range | 295 | |
| hydrogen bonds | 37 | |
| dihedral restraints | 110 | |
| restraint violation | ||
| average distance restraint violation (Å) | 0.0029 +/− 0.0002 | |
| distance restraint violation > 0.1 Å | 0 | |
| average dihedral restraint violation (°) | 0.05 +/− 0.02 | |
| dihedral restraint violation > 1° | 0 | |
| deviation from ideal geometry | ||
| bond length (Å) | 0.00043 +/− 0.00001 | |
| bond angle (°) | 0.08 +/− 0.002 | |
| coordinate precisiona,b | ||
| backbone heavy atoms (Å) | 0.44 | |
| all heavy atoms (Å) | 0.94 | |
| Ramachandran plot statisticsc (%) | 93.8/ 5.1/ 0.1/ 1.0 | |
aThe precision of the coordinates is defined as the average atomic root mean square difference between the accepted simulated annealing structures and the corresponding mean structure calculated for the given sequence regions.
bCalculated for residues Gly3-Val78.
cRamachandran plot statistics are determined by PROCHECK (30) and noted by most favored/additionally allowed/generously allowed/disallowed.
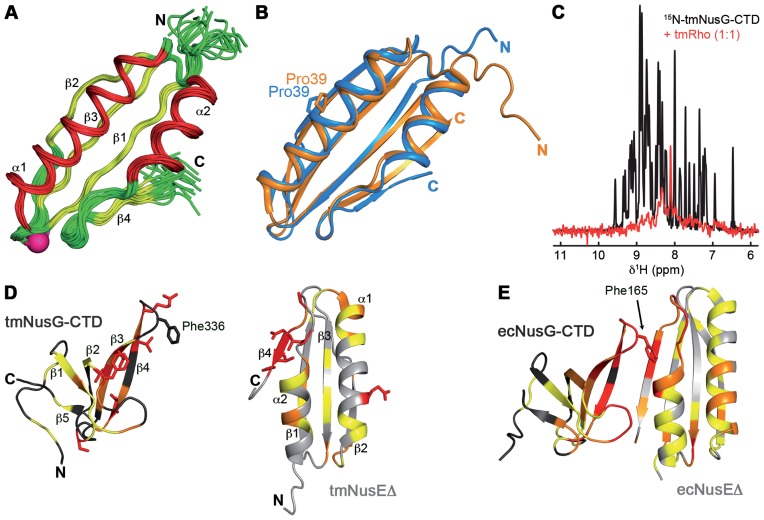
Figure 1.
tmNusEΔ and tmRho are interaction partners of tmNusG-CTD. (A) Solution structure of tmNusEΔ. Superposition of 20 accepted structures of tmNusEΔ in ribbon representation. α-helices, red; β-strands, yellow; loops, green. Secondary structure elements and termini are labeled. The Ser replacing the ribosome binding loop is shown as pink sphere. (B) Superposition of tmNusEΔ, blue and ecNusEΔ in the ecNusEΔ:ecNusB complex, orange (Protein Data Bank (PDB) ID: 3D3B). Selected amino acids are shown as sticks and labeled. (C) 1D [1H,15N]-HSQC spectra of 338 μM 15N-tmNusG-CTD, black, and of 27 μM 15N-tmNusG-CTD in the presence of tmRho in equimolar concentrations, red. (D) Interaction of tmNusEΔ with tmNusG-CTD. tmNusG-CTD (dark gray; PDB ID: 2LQ8) and tmNusEΔ (light gray) are in ribbon representation. Normalized chemical shift changes of the [1H,15N]-HSQC titrations of 15N-tmNusEΔ with tmNusG-CTD and 15N- tmNusG-CTD with tmNusEΔ are mapped on the structures. Δδnorm > 0.2 ppm, red; 0.2 ppm > Δδnorm > 0.1 ppm, orange; 0.1 ppm > Δδnorm > 0.04 ppm, yellow. Highly affected amino acids are in sticks representation. (E) Structure of the ecNusEΔ:ecNusG-CTD complex (PDB ID: 2KVQ). Representation as in (D). ecNusG-CTD, dark gray; ecNusEΔ, light gray. Normalized chemical shift changes of [1H,15N]-HSQC titrations are taken from Ref. (6).
tmNusEΔ and tmRho are targets of tmNusG-CTD
As NusE and Rho are both interaction partners of NusG-CTD in E. coli, we asked whether tmNusG-CTD had the analogous targets in T. maritima (6). First, 1D [1H,15N]-HSQC spectra of 15N-tmNusG-CTD in the absence and in the presence of tmRho were recorded (Figure 1C). 15N-tmNusG-CTD signals decreased significantly upon addition of tmRho, indicating interaction of these proteins as complex formation dramatically increases the molecular mass (MM) of tmNusG-CTD (MMtmNusG-CTD: 7 kDa, MMtmRho: 290 kDa), which, in turn, leads to faster magnetization relaxation and finally results in significant line broadening. Only some signals corresponding to amino acids in random coil areas remain visible. Weak or no binding to tmRho was detected for tmNusG-NTD and tmNusG-DII, respectively (Supplementary Figure S2). The weak tmNusG-NTD:tmRho interaction is probably unspecific and might be attributed generally to the hydrophobic area of tmNusG-NTD responsible for RNAP binding. This confirms that tmNusG interacts specifically with tmRho via its CTD, just like ecNusG, suggesting that NusG:Rho binding is conserved in bacteria.
To test if tmNusG-CTD binds to tmNusEΔ we performed [1H,15N]-HSQC NMR titrations in which either 15N-labeled tmNusEΔ was titrated with unlabeled tmNusG-CTD or vice versa (Supplementary Figure S3A and B). In both cases signals of the 15N-labeled protein shifted significantly or disappeared upon stepwise addition of the unlabeled partner due to tmNusG-CTD:tmNusEΔ complex formation. The normalized changes of the chemical shifts (Δδnorm) were plotted against the amino acid sequences and mapped on the structures to identify the binding surfaces (Supplementary Figure S3C and D; Figure 1D). In tmNusEΔ strongly affected residues are predominantly found at the C-terminus of β-strand β4 as well as in the preceding loop. Binding of tmNusEΔ had an effect on two regions of tmNusG-CTD in particular that involve β-strands β3 and β4 as well as the loop between β1 and β2. The binding interface between tmNusEΔ and tmNusG-CTD is highly similar to that of the ecNusEΔ:ecNusG-CTD complex (Figure 1E) (6). In ecNusG-CTD, Phe165 is a key residue for NusE binding (6) and the corresponding residue in tmNusG-CTD, Phe336, is also located in the loop between β3 and β4 (Figure 1D and E). This suggests that Phe336 is also involved in tmNusEΔ binding, although direct evidence could not be found as we could not assign this residue in the [1H,15N]-HSQC spectra. Since overlapping ecRho and ecNusEΔ binding sites have been proposed for ecNusG-CTD, this might also be true for tmNusG-CTD as suggested by the structural similarity of ecNusG-CTD and tmNusG-CTD (6,11).
From the chemical shift perturbations a KD-value of around 13 μM was estimated for the tmNusEΔ:tmNusG-CTD interaction (Supplementary Figure S3E and F), indicating tighter binding than in the E. coli system (50 μM) (6). Moreover, owing to the high protein concentrations required for NMR experiments, the KD-values in the low micromolar range rather represent an upper limit. Although the titrations were carried out at 50°C, a temperature close to the growth conditions of T. maritima (55–90°C), the KD might still be different at the optimal growth temperature of 80°C (36). Neither for tmNusG-NTD nor for tmNusG-DII an interaction with tmNusEΔ was detected (Supplementary Figure S4). The high similarity of the binding interfaces of NusG-CTD and NusE in T. maritima and E. coli in combination with the fact that T. maritima and E. coli are evolutionary distant (37) suggests a conserved binding mode for NusG-CTD and NusE in bacteria, an interaction important for both transcription:translation coupling and antitermination.
Domain interaction renders tmNusG autoinhibited
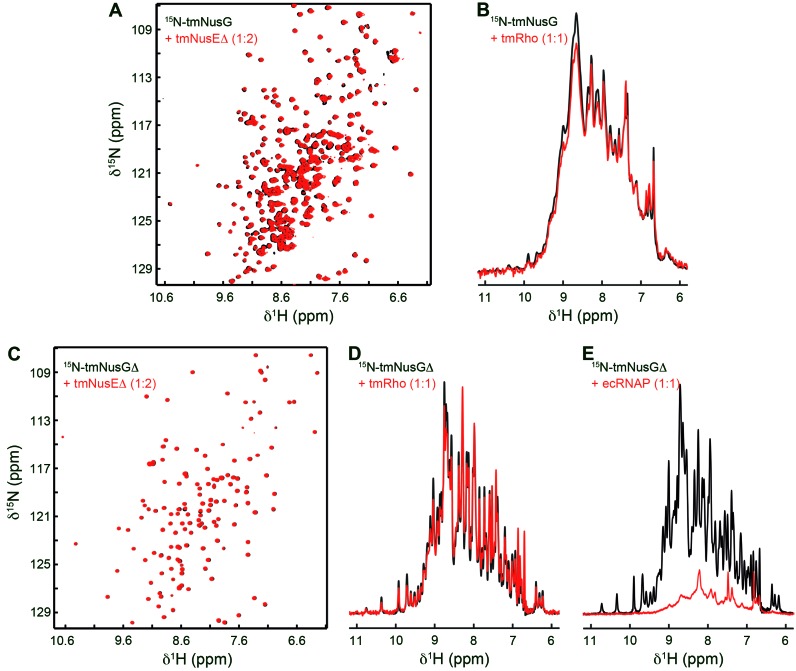
Since tmNusG-CTD has interaction partners analogous to those of ecNusG-CTD, we suggest that it also carries out analogous functions. In the closed state, however, the binding sites for tmRho and tmNusE on tmNusG-CTD and that for RNAP on tmNusG-NTD are masked, implying an autoinhibitory regulation for tmNusG (11). To test whether full length tmNusG was able to bind tmNusEΔ or tmRho, we recorded 1D or two-dimensional (2D) [1H,15N]-HSQC spectra of 15N-tmNusG in the absence and presence of these factors (Figure 2A and B). However, neither addition of tmNusEΔ nor addition of tmRho changed the corresponding spectrum, indicating that no complex formation occurs. Repeating the experiments with 15N-tmNusGΔ, a variant in which tmNusG-DII is replaced by the linker of ecNusG (11), led to identical results (Figure 2C and D). These data indicate that the closed conformation of tmNusG is indeed a silent state. The open and closed conformation are in dynamic equilibrium with the majority of tmNusG being in the closed form (11). Neither tmNusEΔ nor tmRho can shift this equilbrium toward the open state.
Figure 2.
tmNusG and tmNusGΔ are autoinhibited. (A) 2D [1H,15N]-HSQC spectra of 150 μM 15N-tmNusG, black, and of 89 μM 15N-tmNusG in the presence of tmNusEΔ in a twofold molar excess, red. (B) 1D [1H,15N]-HSQC spectra of 278 μM 15N-tmNusG, black, and of 30 μM 15N-tmNusG in the presence of tmRho in equimolar concentrations, red. (C) 2D [1H,15N]-HSQC spectra of 100 μM 15N-tmNusGΔ, black, and of 74 μM 15N-tmNusGΔ in the presence of tmNusEΔ in a twofold molar excess, red. (D) 1D [1H,15N]-HSQC spectra of 348 μM 15N-tmNusGΔ, black, and of 27 μM 15N-tmNusGΔ in the presence of tmRho in equimolar concentrations, red. (E) 1D [1H,15N]-HSQC spectra of 348 μM 15N-tmNusGΔ, black, and of 25 μM 15N-tmNusGΔ in the presence of ecRNAP in equimolar concentrations, red.
Next we tested whether 15N-tmNusGΔ interacts with RNAP. Since RNAP from T. maritima (tmRNAP) was not available, we used RNAP from E. coli (ecRNAP; Figure 2E). In the 1D [1H,15N]-HSQC spectrum 15N-tmNusGΔ signals decreased significantly upon ecRNAP addition, suggesting complex formation. This implies that ecRNAP stabilizes the open conformation by interacting with tmNusG-NTD. Although we cannot exclude that the autoinhibited form interacts non-specifically with ecRNAP, RNAP may be the signal to promote NusG domain dissociation.
Determinants of the domain interaction in tmNusGΔ
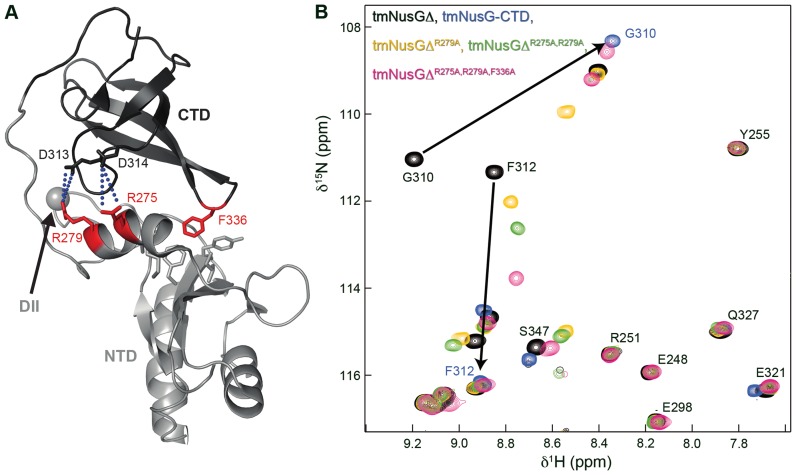
The intramolecular interaction in tmNusG strongly increases the stability of tmNusG-CTD (ΔΔG = 10.3 kJ/mol at 50°C) and is so far unique for NusG proteins (11). Two interdomain salt bridges, Arg275:Asp314 and Arg279:Glu313, were suggested to contribute to the tmNusG-NTD:tmNusG-CTD interaction. However, their elimination results only in partial domain opening, indicating that additional interactions are responsible for the stabilization of the autoinhibited state (11). Another important determinant of the domain interaction may be Phe336 in tmNusG-CTD as it points into a hydrophobic pocket of the RNAP binding site on tmNusG-NTD (Figure 3A) (11). Due to the similarity between the NusG-CTD:NusEΔ binding sites in E. coli and T. maritima (Figure 1D and E) and the fact that EcNusEΔ and EcRho interaction surfaces on EcNusG-CTD overlap (6), Phe336 in tmNusG-CTD is probably involved in the interaction of tmNusG-CTD with tmRho and tmNusE.
Figure 3.
Determinants of the domain interactions in tmNusGΔ. (A) Cartoon representation of tmNusGΔ (PDB ID: 2LQ8). tmNusG-NTD, light gray; tmNusG-CTD, dark gray. Amino acids involved in domain interaction are shown as sticks with residues which were exchanged by alanines colored in red. The gray sphere marks the position where tmNusG-DII is integrated into tmNusG-NTD. Salt bridges are indicated by blue dots. (B) Section of the superposition of 2D [1H,15N]-HSQC spectra of 15N-tmNusGΔ (100 μM), black, 15N-tmNusG-CTD (150 μM), cyan, 15N-tmNusGΔR279A (100 μM), yellow, 15N-tmNusGΔR275A,R279A (100 μM), green and 15N-tmNusGΔR275A,R279A,F336A (420 μM), pink. Arrows indicate how tmNusG-CTD signals of tmNusGΔ shift toward the signals of isolated tmNusG-CTD upon successive amino acid exchanges.
To quantify the contribution of the salt bridges and Phe336 to the domain interaction, we stepwise exchanged Arg275, Arg279 and Phe336 by Ala in tmNusGΔ and measured H/D exchange. The lyophilized, 1H,15N-labeled proteins were dissolved in D2O and H/D exchange was monitored at 323 K via the decay of signal intensities in a series of 2D [1H,15N]-HSQC spectra. Based on the amide proton exchange rates the PF was calculated, which is a measure for the stabilization of a conformation compared to the unfolded state (Supplementary Figure S5A). In tmNusGΔ the PFs are significantly higher than those of the isolated tmNusG-CTD, corresponding to the increased stability of the CTD in tmNusGΔ (11). Successive disruption of both salt bridges and the Phe336Ala exchange gradually lowered the PFs, finally resulting in PF values similar to those of the isolated tmNusG-CTD (Supplementary Figure S5A). This is exemplified by the PF of Gly317: 2.4 × 108 in tmNusGΔ, 2.8 × 107 in tmNusGΔR279A, 7.4 × 106 in tmNusGΔR275A,R279A, 3.6 × 106 in tmNusGΔR275A,R279A,F336A, 5.2 × 106 in tmNusG-CTD (11). The decrease in PFs correlates with a weakening of the domain interaction. Thus, both tmNusGΔR275A,R279A and tmNusGΔR275A,R279A,F336A were not stabilized as compared to isolated tmNusG-CTD. Weakening of the domain interaction was confirmed by 2D [1H,15N]-HSQC spectra of the 15N-labeled tmNusGΔ variants (Figure 3B and Supplementary Figure S5B). With every amino acid exchange the CTD signals gradually shifted from the position typical for tmNusGΔ toward the resonances of isolated tmNusG-CTD. Hence, the tmNusGΔ variants appear to reflect intermediate stages in domain opening of tmNusG, and confirming Phe336 as important factor for domain interaction (see signal shift of Phe312 or Glu339). In [1H,15N]-HSQC spectra of 15N-labeled wild type NusG proteins from other bacteria signals of the isolated CTD and of the CTD in full length protein superimpose perfectly, demonstrating the absence of a closed conformation (ecNusG (3), Thermus thermophilus NusG (ttNusG) (38), Mycobacterium tuberculosis NusG (mtNusG) (39)). The small chemical shift differences between the spectra of tmNusGΔR275A,R279A,F336A and isolated tmNusG-CTD thus suggest still an observable contribution of the closed conformation to the NMR parameters.
To confirm these results we conducted spin relaxation experiments to analyze the relative movements of the NTD and the CTD in tmNusGΔR275A,R279A,F336A, which, in turn, characterizes the overall tumbling of the protein. Uniform rotational tumbling of a multidomain protein corresponds to tight domain interaction, whereas a multidomain protein with non-interacting domains requires an individual description of the rotational tumbling for each domain. Analysis of the ratio of the transverse relaxation rate R2 and the longitudinal relaxation rate R1 offers an elegant method to detect relative domain movements on a timescale faster than the overall molecular tumbling (40). This ratio shows a uniform distribution for tmNusGΔ, confirming the association of tmNusG-NTD and tmNusG-CTD (11). The R2/R1 distribution in tmNusGΔR275A,R279A,F336A (Supplementary Figure S5C) exhibits a slightly bimodal characteristic, reflecting a contribution of individual rotational behavior of the two domains to the overall tumbling. The effective rotation correlation times are 15.5 and 13.4 ns for tmNusG-NTD and tmNusG-CTD, respectively, suggesting that tmNusGΔR275A,R279A,F336A is not in a fully closed conformation. As the differences in the apparent rotational correlation times of the domains are not as big as in ecNusG (12), NTD and CTD in tmNusGΔR275A,R279A,F336A still affect each other during rotation, and the open and the closed conformation are in an equilibrium on the time scale of molecular rotation. This is consistent with the small chemical shift differences of isolated tmNusG-CTD as compared to tmNusGΔR275A,R279A,F336A. Thus, Arg275, Arg279 and Phe336 are major determinants of the autoinhibited state in tmNusG, although further interactions are involved.
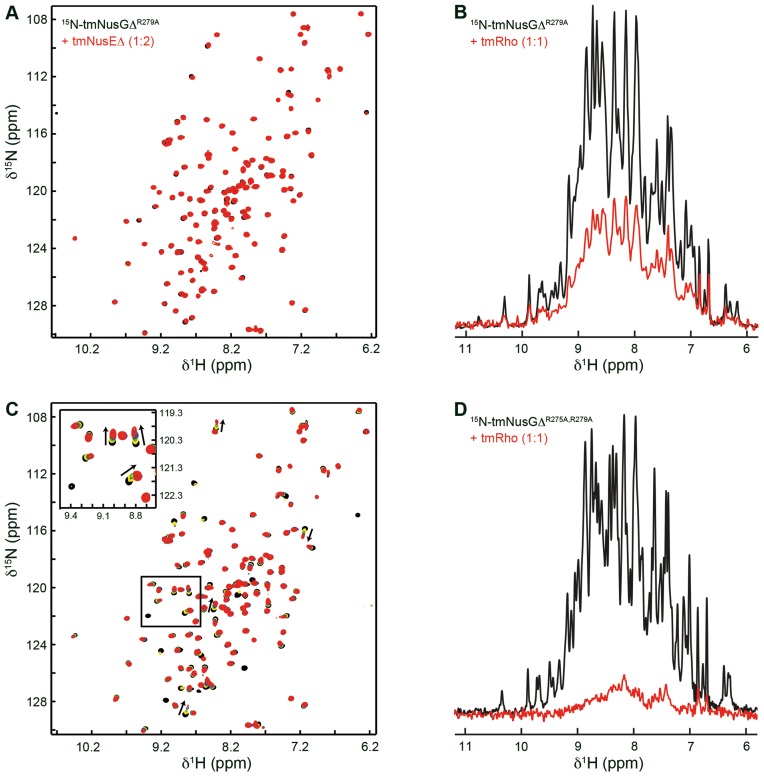
Domain opening of tmNusG allows binding to tmNusEΔ and tmRho
In contrast to tmNusGΔ isolated tmNusG-CTD is able to bind tmRho and tmNusEΔ. As RNAP is the potential trigger to promote domain separation, we used the tmNusGΔ variants with weakened domain interaction to demonstrate that tmNusG can execute the same functions as ecNusG as soon as the autoinhibited state has been released. 1D or 2D [1H,15N]-HSQC spectra of 15N-tmNusGΔR279A recorded in the absence or presence of tmNusEΔ or tmRho clearly show no or only weak binding, respectively. This is in accordance with the H/D exchange experiments since domain association is only slightly reduced (Figure 4A and B). In contrast, significant chemical shift changes occurred when tmNusEΔ was titrated to 15N-tmNusGΔR275A,R279A, and signal intensity of 15N-tmNusGΔR275A,R279A decreased considerably upon addition of tmRho (Figure 4C and D), indicating domain opening. Although the additional elimination of Phe336 further decreased domain interaction (Figure 3B), tmNusGΔR275A,R279A,F336A binds neither to tmNusEΔ nor to tmRho (Supplementary Figure S6), confirming that Phe336 is essential for these interactions, just like the corresponding Phe165 in ecNusG (6).
Figure 4.
Interaction of tmNusGΔ variants with tmNusEΔ and tmRho. (A) 2D [1H,15N]-HSQC spectra of 150 μM 15N-tmNusGΔR279A, black, and of 89 μM 15N-tmNusGΔR279A in the presence of tmNusEΔ in a 2-fold molar excess, red. (B) 1D [1H,15N]-HSQC spectra of 172 μM 15N-tmNusGΔR279A, black, and of 30 μM 15N-tmNusGΔR279A in the presence of tmRho in equimolar concentrations, red. (C) 2D [1H,15N]-HSQC spectra of the titration of 15N-tmNusGΔR275A,R279A with tmNusEΔ. tmNusEΔ (stock concentration: 438 μM) was added to 150 μM 15N-tmNusGΔR275A,R279A (molar ratios 1:0, black; 1:0.5, yellow; 1:1, green; 1:2, blue; 1:5, red). The insert shows a blow-up of the boxed region. Arrows indicate changes of the chemical shifts during the titration. (D) 1D [1H,15N]-HSQC spectra of 71 μM 15N-tmNusGΔR275A,R279A, black, and of 26 μM 15N-tmNusGΔR275A,R279A in the presence of tmRho in equimolar concentrations, red.
Autoinhibition of tmNusG probably parallels its thermostability. While intermolecular interactions between NTD and CTD of bacterial NusG proteins have been reported, the tight intramolecular domain interaction in tmNusG is unique (11–12,14). As demonstrated, two salt bridges and Phe336 are important contributors to the domain interaction. Phe336 is highly conserved in bacterial NusG proteins (Supplementary Figure S7), probably since it is essential for NusE and Rho binding. However, the presence of Phe336 is sufficient to cause partial autoinhibition only in tmNusG. Therefore, Phe336 and its binding pocket in tmNusG-NTD are optimized with respect to tighter binding compared to other NusGs. The amino acid combinations allowing the formation of the two salt bridges are not conserved. Only in ttNusG an Arg and an Asp residue are present at the positions corresponding to Arg275 and Asp314 in tmNusG, but no NTD:CTD interaction was observed (38). tmNusG has a longer linker than several other NusGs. The linker of ttNusG is three amino acids shorter, that of ecNusG five and that of aaNusG even seven amino acids. Molecular modeling showed that the linker of ecNusG and even aaNusG are sufficient to allow a closed state similar to tmNusG. However, in either case the linker must adopt an unlikely, nearly extended conformation. Compared to tmNusG, mtNusG contains a significantly longer linker, but did not show any domain interaction (39) suggesting that linker length is not the pivotal factor for intramolecular domain interaction. Although transient intra- or intermolecular NTD:CTD interactions might occur in all NusG proteins, only tmNusG has developed additional features to stabilize the autoinhibited state. However, in contrast to the ecNusG paralog RfaH, autoinhibition in tmNusG appears to merely support thermostability and has no regulatory role (11,41–42).
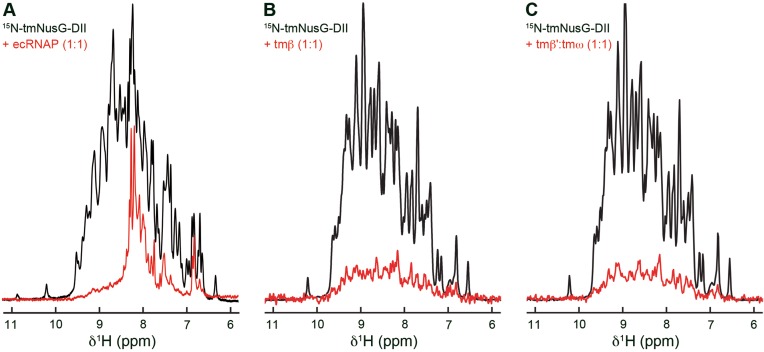
tmNusG-DII binds to RNAP
Aside from non-specific binding to nucleic acids (15), functions of tmNusG-DII are unknown. Since tmNusGΔ interacts with RNAP (Figure 2E) we analyzed if also tmNusG-DII binds to RNAP to uncover possible roles of this domain. As for tmNusGΔ we recorded 1D [1H,15N]-HSQC spectra of 15N-tmNusG-DII in the absence and presence of ecRNAP (Figure 5A). 15N-tmNusG-DII signals decreased significantly upon addition of ecRNAP and residual signal intensity could only be found in regions typical for unstructured parts, indicating complex formation. As NusG-NTD binds to the β and β’ subunits (4,5), tmNusG-DII will probably also interact with one of these.
Figure 5.
Binding of tmNusG-DII to ecRNAP and tmRNAP subunits. 1D [1H,15N]-HSQC spectra of 375 μM 15N-tmNusG-DII, black, and of 15N-tmNusG-DII in the presence, red, of (A) ecRNAP (25 μM each), (B) tmβ (30 μM each) and (C) tmβ’:tmω (30 μM each). Differences between the spectra of free 15N-tmNusG-DII in (A) and (B and C) are due to different temperatures in the experiments.
We used a method previously established for the E. coli system to identify the interacting RNAP subunit (19). tmRNAP subunit genes coding for the β and β’ subunit (tmβ, tmβ’) were expressed separately. While tmβ could be purified individually, tmβ’ was only stable in complex with the ω subunit (tmω). The integrity of tmβ and tmβ’:tmω was checked by testing their ability to bind to known interaction partners with ecRNAP as reference (Supplementary Figure S8). The 1D [1H,15N]-HSQC spectra revealed an interaction of 15N-tmNusG-NTD with both tmβ and tmβ’:tmω whereas 15N-tmNusG-CTD did not bind either subunit.
The interaction of tmNusG-DII with tmRNAP subunits was tested using the same approach. Addition of tmβ and tmβ’:tmω to 15N-tmNusG-DII led to a drastic signal decrease (Figure 5B and C), suggesting that tmNusG-DII binds to both the tmβ subunit and the tmβ’:tmω complex.
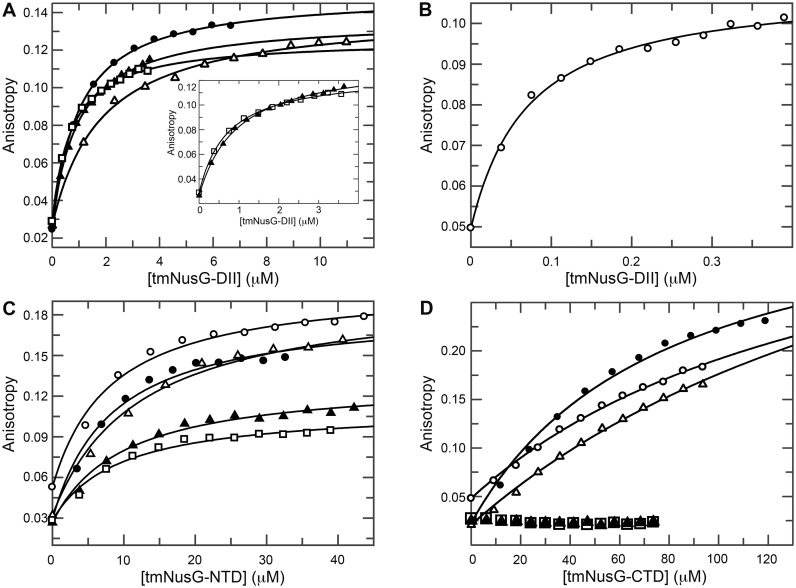
tmNusG-DII binds preferably dsDNA
The ability of tmNusG-DII to bind non-specifically to nucleic acids has been demonstrated qualitatively by gel shift assays and electron microscopy using long oligonucleotides (0.7-5.2 kb) (15). These data suggested a preference for dsDNA and RNA over ssDNA. We used fluorescence anisotropy measurements to further elucidate these interactions by titrating 6-FAM labeled ssDNA, dsDNA and ssRNA of about 20 nucleotides (nt) in length with tmNusG-DII (Figure 6A and B; Table 2). The domain had a similar low micromolar affinity for ssDNA as for ssRNA, but exhibited a clear preference for dsDNA with a KD value of 40 nM. Different sequences for each type of nucleic acid resulted in similar KD values, suggesting that the interaction is sequence-independent.
Figure 6.
Fluorescence anisotropy titrations of nucleic acids with tmNusG domains. Titrations of (A) ssDNA 1 (filled circles), ssDNA 2 (open triangles), ssRNA 1 (filled triangles) and ssRNA 2 (open squares) and (B) dsDNA (open circles) with tmNusG-DII. The insert in (A) shows a blow-up of the titration with ssRNAs 1 and 2. (C) Titration of ssDNA 1, ssDNA 2, ssRNA 1, ssRNA 2 and dsDNA with tmNusG-NTD (symbols as in (A and B)). (D) Titration of ssDNA 1, ssDNA 2, ssRNA 1, ssRNA 2 and dsDNA with tmNusG-CTD (symbols as in (A and B)). Solid lines show the best fit to Equation (3). Nucleic acids were labeled with 6-FAM.
Table 2. Affinities of tmNusG domains for nucleic acids. n.d.: not detectable.
| tmNusG domain | dsDNA | ssDNA 1 | ssDNA 2 | ssRNA 1 | ssRNA 2 |
|---|---|---|---|---|---|
| DII | 0.04 μM | 1 μM | 1.5 μM | 0.8 μM | 0.5 μM |
| NTD | 4 μM | 4 μM | 3 μM | 3.5 μM | 4 μM |
| CTD | >100 μM | >50 μM | >150 μM | n.d. | n.d. |
As non-specific nucleic acid binding has also been reported for a tmNusG construct lacking the DII domain, similar to the tmNusGΔ variant, we repeated the titrations with isolated tmNusG-NTD and tmNusG-CTD (Figure 6C and D) (15). While tmNusG-CTD showed only very weak binding to ssDNA and dsDNA and no binding to ssRNA, tmNusG-NTD interacted with all nucleic acids with a similar affinity of 3–4 μM. These data are consistent with the finding that Bacillus subtilis NusG, ttNusG and E. coli RfaH contact the non-template DNA strand when bound to the RNAP in the TEC (43–45). This seems to be a feature conserved in all NusG proteins. The main nucleic acid binding ability of tmNusG, however, can be attributed to tmNusG-DII, which preferentially binds dsDNA.
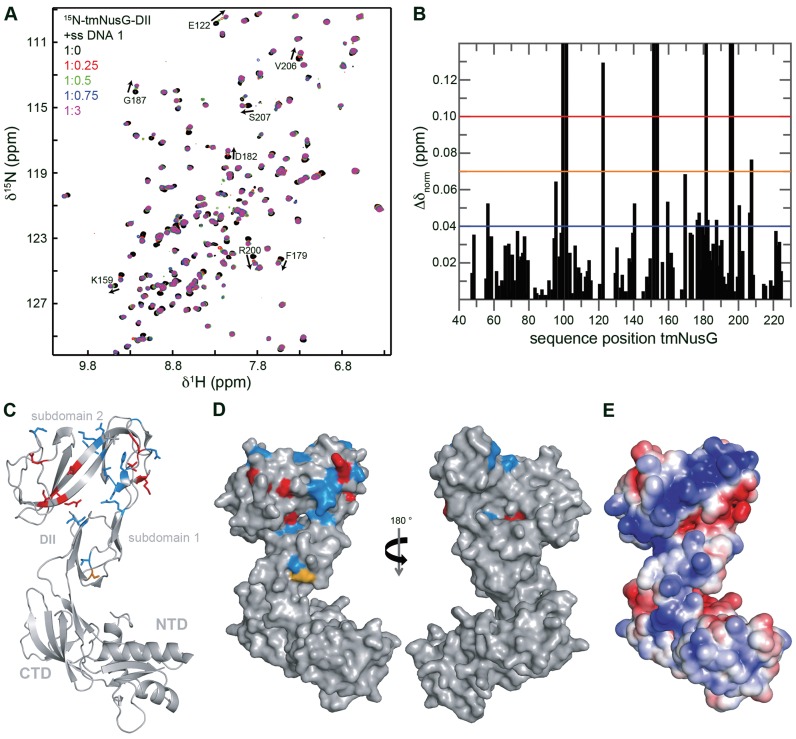
To determine the nucleic acid binding site of tmNusG-DII, we conducted a [1H,15N]-HSQC titration with 15N-tmNusG-DII and ssDNA. Chemical shift changes as well as disappearing signals could be observed, confirming complex formation (Figure 7A). The normalized chemical shift changes of affected residues were mapped on the structure of tmNusG-DII, revealing that especially the part of tmNusG-DII that comprises subdomain 2 is involved in ssDNA binding. Although the affected residues do not form a completely continuous patch, the electrostatic surface potential reveals that the determined binding site superimposes with a positively charged area, suggesting that we indeed identified the nucleic acid binding site.
Figure 7.
The DNA binding site of tmNusG-DII. (A) [1H,15N]-HSQC spectra of the titration of 15N-tmNusG-DII with ssDNA 1. ssDNA 1 (stock concentration: 1 mM) was added to 150 μM 15N-tmNusG-DII (molar ratios 1:0, black; 1:0.25, red; 1:0.5, green; 1:0.75, blue: 1:3, magenta). Selected signals are assigned, arrows indicate changes of the chemical shifts. (B) Normalized chemical shift changes derived from the HSQC titration in (A) versus tmNusG-DII sequence position. The significance levels are indicated by horizontal lines. Δδnorm = 0.04 ppm, blue; Δδnorm = 0.07 ppm, orange; Δδnorm = 0.1 ppm, red. Gaps represent prolines and not assigned amino acids. (C and D) Mapping of the normalized chemical shift changes on the structure of tmNusG (gray, PDB ID: 2XHC) in ribbon (C) and surface (D) representation. Δδnorm > 0.1 ppm, red; 0.1 ppm > Δδnorm > 0.07 ppm, orange; 0.07 ppm > Δδnorm > 0.04 ppm, blue. (E) Electrostatic surface potential of tmNusG calculated with the program APBS (33), colored from -3 kT/e− (red) to +3 kT/e− (blue).
Conclusions: tmNusG-DII recruits tmNusG to the TEC and stabilizes the tmNusG:TEC complex
The interaction of NTD and CTD in tmNusG masks the binding sites for tmNusE, tmRho and tmRNAP, preventing these interactions and rendering tmNusG silent. We showed that domain opening is essential for tmNusG to accomplish the functions known from ecNusG. The closed and the open state are in dynamic equilibrium with 98 % being in the closed conformation, even at temperatures close to the optimal growth conditions of T. maritima (11). Neither tmNusEΔ nor tmRho are able to shift the equilibrium toward the open state.
We suggest that several factors might contribute to release the autoinhibition. As shown by NMR spectroscopy interaction of tmNusG with RNAP might be the driving force to promote domain opening. Under physiological conditions, however, tmNusG binds to the TEC and not to RNAP alone. We demonstrated that tmNusG-NTD interacts non-specifically with nucleic acids, suggesting that tmNusG-NTD, once bound to the TEC, might additionally interact with the non-template strand in the transcription bubble to increase the overall affinity of tmNusG. Similar situations are known from other NusG proteins. B. subtilis NusG, for example, recognizes a specific sequence in the non-template strand within the paused transcription bubble in certain operons (43) and also the NTD of RfaH, a paralog of NusG, interacts with a specific DNA element in the non-template strand during its recruitment to the TEC (46). Moreover, tmNusG-DII interacts with the tmβ subunit and the tmβ’:tmω complex. This interaction might again increase the overall affinity of tmNusG for the TEC, facilitating its recruitment.
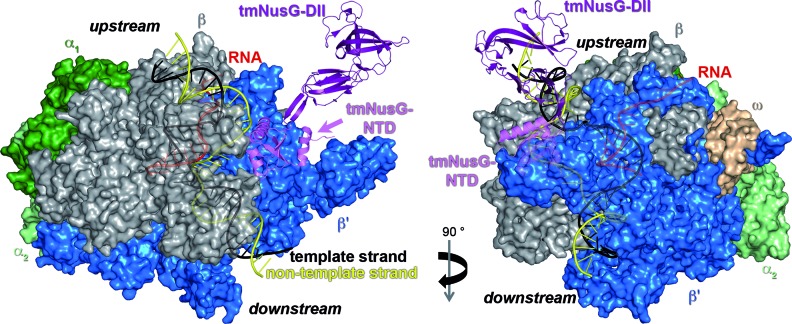
Based on the complex structure of Pyrococcal Spt4/5 and the clamp domain (4) we generated a model of tmNusG bound to elongating RNAP from T. thermophilus (ttRNAP) (Figure 8). Fluorescence spectroscopic titrations of nucleic acids with tmNusG-DII showed a clear preference of this domain for dsDNA. In the model tmNusG-DII is in close proximity to upstream dsDNA and separated from downstream dsDNA by parts of the β’ subunit, suggesting that tmNusG-DII interacts with upstream dsDNA. The binding of tmNusG-DII to dsDNA might further stabilize the tmNusG:TEC complex at the high temperatures at which T. maritima lives. A. aeolicus is a hyperthermophilic organism that grows at temperatures between 67 and 95°C with an optimum at 85°C (47). Like tmNusG, aaNusG has an additional domain that binds non-specifically ssDNA, dsDNA and RNA, but that is much smaller than tmNusG-DII and exhibits no sequential similarity to tmNusG-DII (11,13). aaNusG also differs from tmNusG as aaNusG-NTD and aaNusG-CTD do not interact and aaNusG is not autoinhibited (13,14). Thus, the additional domain found in NusG proteins in some (hyper)thermophilic organisms might be an adaption to the high temperatures of the natural habitats as it allows the stabilization of the TEC by either interaction with nucleic acids or with RNAP or both.
Figure 8.
Model of tmNusG bound to elongating ttRNAP. Surface representation of ttRNAP with nucleic acids shown as ribbons (PDB ID: 2O5I; α1, dark green; α2, light green; β, gray; β’, blue; ω, light orange; DNA template strand, black; DNA non-template strand, yellow; RNA, red). tmNusG (PDB ID: 2HXC; ribbon representation; tmNusG-NTD, violet; tmNusG-DII, purple; tmNusG-CTD, not shown; termini are labeled) was modeled to ttRNAP according to the Pyrococcus furiosus Spt4/5 complex bound to the RNAP clamp domain (PDB ID: 3QQC) (4) by superimposition of tmNusG-NTD on Spt5-NTD.
ACCESSION NUMBERS
The structure coordinates and chemical shift assignments of tmNusEΔ have been deposited in the PDB and the Biological Magnetic Resonance Data Bank with accession numbers 2MEW and 19533, respectively.
Acknowledgments
We thank Ramona Heißmann and Ulrike Persau for excellent technical support and Dr Max Gottesman (Columbia University, USA) for helpful discussions. We also thank Dr. Gunter Stier (EMBL Heidelberg, Germany) and Dr. Markus Wahl (FU Berlin, Germany) for kindly providing pETGB1a and pET22b-tmNusG, respectively.
Footnotes
Present address: Christin Schneider, Lehrstuhl für Biochemie und Molekulare Medizin, Friedrich-Alexander-Universität Erlangen-Nürnberg, Fahrstraße 17, 91054 Erlangen, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [Ro 617/17-1, Ro 617/21-1 to P.R.]; Columbia University Medical Center Ludwig-Schaefer-Scholarship 2015 (to P.R.). Funding for open access charge: German Research Foundation (DFG) and the University of Bayreuth in the funding programme Open Access Publishing.
Conflict of interest statement. None declared.
REFERENCES
- 1.Werner F. A nexus for gene expression-molecular mechanisms of Spt5 and NusG in the three domains of life. J. Mol. Biol. 2012;417:13–27. doi: 10.1016/j.jmb.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.NandyMazumdar M., Artsimovitch I. Ubiquitous transcription factors display structural plasticity and diverse functions: NusG proteins—shifting shapes and paradigms. Bioessays. 2015;37:324–334. doi: 10.1002/bies.201400177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mooney R.A., Schweimer K., Rösch P., Gottesman M.E., Landick R. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J. Mol. Biol. 2009;391:341–358. doi: 10.1016/j.jmb.2009.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martinez-Rucobo F.W., Sainsbury S., Cheung A.C., Cramer P. Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J. 2011;30:1302–1310. doi: 10.1038/emboj.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sevostyanova A., Belogurov G.A., Mooney R.A., Landick R., Artsimovitch I. The β subunit gate loop is required for RNA polymerase modification by RfaH and NusG. Mol. Cell. 2011;43:253–262. doi: 10.1016/j.molcel.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burmann B.M., Schweimer K., Luo X., Wahl M.C., Stitt B.L., Gottesman M.E., Rösch P. A NusE:NusG complex links transcription and translation. Science. 2010;328:501–504. doi: 10.1126/science.1184953. [DOI] [PubMed] [Google Scholar]
- 7.Cardinale C.J., Washburn R.S., Tadigotla V.R., Brown L.M., Gottesman M.E., Nudler E. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science. 2008;320:935–938. doi: 10.1126/science.1152763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nodwell J.R., Greenblatt J. Recognition of boxA antiterminator RNA by the E. coli antitermination factors NusB and ribosomal protein S10. Cell. 1993;72:261–268. doi: 10.1016/0092-8674(93)90665-d. [DOI] [PubMed] [Google Scholar]
- 9.Friedman D.I., Olson E.R., Georgopoulos C., Tilly K., Herskowitz I., Banuett F. Interactions of bacteriophage and host macromolecules in the growth of bacteriophage lambda. Microbiol. Rev. 1984;48:299–325. doi: 10.1128/mr.48.4.299-325.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torres M., Balada J.M., Zellars M., Squires C., Squires C.L. In vivo effect of NusB and NusG on rRNA transcription antitermination. J. Bacteriol. 2004;186:1304–1310. doi: 10.1128/JB.186.5.1304-1310.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drögemüller J., Stegmann C.M., Mandal A., Steiner T., Burmann B.M., Gottesman M.E., Wöhrl B.M., Rösch P., Wahl M.C., Schweimer K. An auto-inhibited state in the crystal structure of Thermotoga maritima NusG. Structure. 2013;21:365–375. doi: 10.1016/j.str.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burmann B.M., Scheckenhofer U., Schweimer K., Rösch P. Domain interactions of the transcription-translation coupling factor Escherichia coli NusG are intermolecular and transient. Biochem. J. 2011;435:783–789. doi: 10.1042/BJ20101679. [DOI] [PubMed] [Google Scholar]
- 13.Steiner T., Kaiser J.T., Marinkovic S., Huber R., Wahl M.C. Crystal structures of transcription factor NusG in light of its nucleic acid- and protein-binding activities. EMBO J. 2002;21:4641–4653. doi: 10.1093/emboj/cdf455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knowlton J.R., Bubunenko M., Andrykovitch M., Guo W., Routzahn K.M., Waugh D.S., Court D.L., Ji X. A spring-loaded state of NusG in its functional cycle is suggested by X-ray crystallography and supported by site-directed mutants. Biochemistry. 2003;42:2275–2281. doi: 10.1021/bi0272508. [DOI] [PubMed] [Google Scholar]
- 15.Liao D., Lurz R., Dobrinski B., Dennis P.P. A NusG-like protein from Thermotoga maritima binds to DNA and RNA. J. Bacteriol. 1996;178:4089–4098. doi: 10.1128/jb.178.14.4089-4098.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo X., Hsiao H.H., Bubunenko M., Weber G., Court D.L., Gottesman M.E., Urlaub H., Wahl M.C. Structural and functional analysis of the E. coli NusB-S10 transcription antitermination complex. Mol. Cell. 2008;32:791–802. doi: 10.1016/j.molcel.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burmann B.M., Luo X., Wahl M.C., Rösch P., Gottesman M.E. Fine tuning of the E. coli NusB:NusE complex affinity to BoxA RNA is required for processive antitermination. Nucleic Acids Res. 2010;38:314–326. doi: 10.1093/nar/gkp736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greive S.J., Lins A.F., von Hippel P.H. Assembly of an RNA-protein complex. Binding of NusB and NusE (S10) proteins to boxA RNA nucleates the formation of the antitermination complex involved in controlling rRNA transcription in Escherichia coli. J. Biol. Chem. 2005;280:36397–36408. doi: 10.1074/jbc.M507146200. [DOI] [PubMed] [Google Scholar]
- 19.Drögemüller J., Strauss M., Schweimer K., Wohrl B.M., Knauer S.H., Rösch P. Exploring RNA polymerase regulation by NMR spectroscopy. Sci. Rep. 2015;5:10825–10835. doi: 10.1038/srep10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J., Russel D.W. Molecular CLONING—a Laboratory Manual. NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 21.Meyer O., Schlegel H.G. Biology of aerobic carbon monoxide-oxidizing bacteria. Annu. Rev. Microbiol. 1983;37:277–310. doi: 10.1146/annurev.mi.37.100183.001425. [DOI] [PubMed] [Google Scholar]
- 22.Bax A., Grzesiek A. Methodological advances in protein NMR. Acc. Chem. Res. 1993;26:131–138. [Google Scholar]
- 23.Sattler M., Schleucher J., Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution empolying pulsed field gradients. Prog. NMR Spectrosc. 1999;34:93–158. [Google Scholar]
- 24.Bai Y., Milne J.S., Mayne L., Englander S.W. Protein stability parameters measured by hydrogen exchange. Proteins. 1994;20:4–14. doi: 10.1002/prot.340200103. [DOI] [PubMed] [Google Scholar]
- 25.Bai Y., Milne J.S., Mayne L., Englander S.W. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kay L.E., Torchia D.A., Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: Application to staphylococcal nuclease. Biochemistry (N. Y. ) 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 27.Dosset P., Hus J.C., Blackledge M., Marion D. Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J. Biomol. NMR. 2000;16:23–28. doi: 10.1023/a:1008305808620. [DOI] [PubMed] [Google Scholar]
- 28.Cornilescu G., Delaglio F., Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 29.Schwieters C.D., Kuszewski J.J., Tjandra N., Clore G.M. The xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:66–74. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 30.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 31.Schrödinger L. The PyMOL Molecular Graphics System, Version 1.3. Mannheim: Schrödinger, LLC; 2010. [Google Scholar]
- 32.Higgins D.G., Thompson J.D., Gibson T.J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das R., Loss S., Li J., Waugh D.S., Tarasov S., Wingfield P.T., Byrd R.A., Altieri A.S. Structural biophysics of the NusB:NusE antitermination complex. J. Mol. Biol. 2008;376:705–720. doi: 10.1016/j.jmb.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wimberly B.T., Brodersen D.E., Clemons W.M.J., Morgan-Warren R.J., Carter A.P., Vonrhein C., Hartsch T., Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 36.Huber R., Langworthy T.A., König H., Thomm M., Woese C.R., Sleytr U.B., Stetter K.O. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90°C. Arch. Microbiol. 1986;144:324–333. [Google Scholar]
- 37.Woese C.R. Bacterial evolution. Microbiol. Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reay P., Yamasaki K., Terada T., Kuramitsu S., Shirouzu M., Yokoyama S. Structural and sequence comparisons arising from the solution structure of the transcription elongation factor NusG from Thermus thermophilus. Proteins. 2004;56:40–51. doi: 10.1002/prot.20054. [DOI] [PubMed] [Google Scholar]
- 39.Strauss M., Schweimer K., Burmann B.M., Richter A., Guttler S., Wöhrl B.M., Rösch P. The two domains of Mycobacterium tuberculosis NusG protein are dynamically independent. J. Biomol. Struct. Dyn. 2016;34:352–361. doi: 10.1080/07391102.2015.1031700. [DOI] [PubMed] [Google Scholar]
- 40.Horstmann M., Ehses P., Schweimer K., Steinert M., Kamphausen T., Fischer G., Hacker J., Rösch P., Faber C. Domain motions of the Mip protein from Legionella pneumophila. Biochemistry. 2006;45:12303–12311. doi: 10.1021/bi060818i. [DOI] [PubMed] [Google Scholar]
- 41.Burmann B.M., Knauer S.H., Sevostyanova A., Schweimer K., Mooney R.A., Landick R., Artsimovitch I., Rösch P. An α-helix to β-barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell. 2012;150:291–303. doi: 10.1016/j.cell.2012.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knauer S.H., Rösch P., Artsimovitch I. Transformation: The next level of regulation. RNA Biol. 2012;9:1418–1423. doi: 10.4161/rna.22724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yakhnin A.V., Murakami K.S., Babitzke P. NusG is a sequence-specific RNA polymerase pause factor that binds to the non-template DNA within the paused transcription bubble. J. Biol. Chem. 2016;291:5299–5308. doi: 10.1074/jbc.M115.704189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sevostyanova A., Artsimovitch I. Functional analysis of Thermus thermophilus transcription factor NusG. Nucleic Acids Res. 2010;38:7432–7445. doi: 10.1093/nar/gkq623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Artsimovitch I., Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- 46.Belogurov G.A., Vassylyeva M.N., Svetlov V., Klyuyev S., Grishin N.V., Vassylyev D.G., Artsimovitch I. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol. Cell. 2007;26:117–129. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huber R., Wilharm T., Huber D., Trincone A., Burggraf S., König H., Reinhard R., Rockinger I., Fricke H., Stetter K.O. Aquifex pyrophilus gen. nov. sp. nov., represents a novel group of marine hyperthermophilic hydrogen-oxidizing bacteria. Syst. Appl. Microbiol. 1992;15:340–351. [Google Scholar]