Abstract
Transposons are highly abundant in eukaryotic genomes, but their mobilization must be finely tuned to maintain host organism fitness and allow for transposon propagation. Forty percent of the human genome is comprised of transposable element sequences, and the most abundant cut-and-paste transposons are from the hAT superfamily. We found that the hAT transposase TcBuster from Tribolium castaneum formed filamentous structures, or rodlets, in human tissue culture cells, after gene transfer to adult mice, and ex vivo in cell-free conditions, indicating that host co-factors or cellular structures were not required for rodlet formation. Time-lapsed imaging of GFP-laced rodlets in human cells revealed that they formed quickly in a dynamic process involving fusion and fission. We delayed the availability of the transposon DNA and found that transposition declined after transposase concentrations became high enough for visible transposase rodlets to appear. In combination with earlier findings for maize Ac elements, these results give insight into transposase overproduction inhibition by demonstrating that the appearance of transposase protein structures and the end of active transposition are simultaneous, an effect with implications for genetic engineering and horizontal gene transfer.
INTRODUCTION
Transposition is an ancient and influential force on the genome of nearly every organism, reshaping chromosomes by an inherently mutagenic process (1). Transposons commonly make up the majority of eukaryotic genomes, with maize having the most transposon-rich genome (>85%) (2). The transposase protein coding genes are the most abundant and ubiquitous genes in nature (3). Using these diverse transposases as genetic starting material, hosts have tamed them into “domesticated transposases” with diverse and crucial functions (4).
Class II DNA transposons cut-and-paste DNA sequences in the genome and are useful for gene transfer applications. Transposons with a linear dsDNA intermediate (5) propagate mainly by replication of the host genome, in the process also replicating the transposon. In this study, we report data on two DD(E/D)-transposons of insect origin: Tribolium castaneum Buster (TcBuster; (6) and piggyBac (7). TcBuster is a newly discovered member of the hAT (hobo, Ac, Tam3) transposon superfamily that includes the transposon Hermes which has a known crystal structure (8,9). PiggyBac is the namesake for its own family (10). We found that TcBuster has a high level of activity in human tissue culture cells but displayed an overproduction inhibition/negative dosage effect—meaning that high doses of transposase resulted in suboptimal transposition (11). In fact, in a careful review of the literature the majority of DD(E/D) transposons tested displayed overproduction inhibition (12), indicating that the cause of overproduction inhibition must be: (i) independent of the details of transposase structure or exact mechanism of DNA cleavage; (ii) present in all transposons and (iii) independent of the host.
The overproduction inhibition/negative dosage effect was first described by McClintock (13). This effect has presented the need for optimization for the successful creation of transposon vectors for genome modification, most notably for the Sleeping Beauty transposon (12,14,15). Despite the difficulty this presents in genome engineering (16), in nature transposon self-regulation is expected so that suicidal autointegration events are limited (17). Additionally, since the health of the host is crucial for transposon replication, transposons cannot act as purely ‘selfish’ genes: this adjective implies unmitigated harm to the host on which the transposon relies (18). DNA transposons keep their activity in check through an assortment of mechanisms, including complementary regulatory RNA (19), repressors (20,21), and concentration-dependent negative feedback mechanisms (22). The relationship between transposase concentration and the rate of transposition could be thought of as taking three possible courses: a linear or exponential increase in activity, overproduction inhibition in which activity decreases above a certain transposase concentration, and saturated transposition activity by transposase overconcentration where the increasing amount of transposase beyond a certain point does not affect the rate of transposition. In 33 out of 50 manuscripts reviewed by Bire et al., classic overproduction inhibition was reported; therefore, the most common outcome of a surplus of transposase protein across a wide variety of DNA cut-and-paste transposons from different families is a decrease in transposition rate (12). Our data expand upon previous studies that linked overproduction inhibition or negative dosage effect to high transposase protein concentration by linking this inhibition to the creation of host-independent structures. To further investigate these rodlet structures, we made several tagged versions of TcBuster transposase and discovered they were created in a dynamic process in human tissue culture cells, in cell-free solutions ex vivo, and in mice. We also demonstrated that transposition occurred prior to the detection of transposase rodlets and that the presence of aggregates for both TcBuster and piggyBac signaled the end of a window of transposition.
MATERIALS AND METHODS
Plasmid constructs
Plasmids were prepared by endotoxin-free maxiprep (Qiagen, Valencia, CA, USA) or midiprep (Zymo Research Corp., Irvine, CA, USA). The filler DNA plasmid pUC19 (Invitrogen, Carlsbad, CA, USA) is commercially available. The construction of the piggyBac plasmids pCMV-piggyBac (23), pCMV-HA-piggyBac (24), pTpB (23), and pTpBCAGLuc (referred to as pT-CAGLuc in (25) and the TcBuster plasmids pCMV-TcBuster and pTcBNeo (11) have been described elsewhere.
pCMV-HA-TcBuster was created concurrently with pCMV-TcBuster (11), the only difference being the forward primer SpeITCB2 (GCTGACTAGTATGATGCTGAATTGGCTGAAAAGC) used to clone the insert into the SpeI site of pCMV-HA-piggyBac to allow retention of the HA tag at the N-terminus to create pCMV-HA-TcBuster.
pCMV-Flag-TcBuster was created by PCR of pCMV-TcBuster with the primers TcB-Flag-F (CCCCGCGGATGGACTACAAAGACCACGACGGCGATTATAAAGATCACGATATCGATTACAAGGATGACGATGACAAGATGATGCTGAATTGGCTGAAAAG) and TcB-Flag-R (CTATGCGTCGGCTGATAGTG) to create a Flag-tagged insert fragment that was digested and cloned into the SacII and BstBI sites of pCMV-TcBuster.
To create pCMV-TcBuster-GFP, a similar PCR of pCMV-TcBuster with the primers TcB-c3GFP-for (GCCGCCGCGATGATGCTGAATTGGCTGAAAAG) and TCB-c3GFP-rev (GGTGAGATTTCTGGGCCTGCTTG) synthesized an insert containing the TcBuster reading frame that was TOPO cloned via the pCDNA3.1/CT-GFP-TOPO kit (Invitrogen), which was also transformed alone to create the pCMV-GFP empty vector control. To create pCMV-GFP-TcBuster, a PCR of pCMV-TcBuster was performed with the primers TcB-c3GFP-for and RevKpnITcB2 (GCCGGGTACCTCAGTGAGATTTCTGGGCCTGC) and TOPO cloned into pCDNA3.1/NT-GFP-TOPO (Invitrogen). The same linker (GGSGGSGGSGGSGTS) we have previously employed (15,26) was introduced via annealed oligos into the AgeI site in pCMV-GFP-TcBuster and the XbaI site in pCMV-TcBuster-GFP.
The luciferase-expressing TcBuster transposon pTcBCAGLuc was created by digest of the pTpBCAGLuc plasmid with BsaI, SpeI, and BamHI to liberate the CAGLuc cassette. This insert was then cloned into the TcBuster transposon pXL-TcB-D-GFP/Bsd (27) digested with BglII and SpeI.
Tissue culture
HEK-293 and HeLa cells (ATCC, Manassas, VA) were cultured in cellgro Minimum Essential Medium, Alpha 1× with Earle's Salts, ribonucleosides, deoxyribonucleosides and l-glutamine (Mediatech, Inc., Manassas, VA) to which 10% fetal bovine serum (Thermo Scientific, Logan, UT) and 1% penicillin/streptomycin (Invitrogen) were added.
Immunofluorescent staining and live cell imaging of tissue culture cells
For staining of the HA tag, HEK-293 cells were plated onto poly-d-lysine-coated 12 mm glass coverslips (BD Biosciences, San Jose, CA) or HeLa cells were plated onto uncoated 18 mm glass coverslips. Cells were transfected via FuGene 6 (Roche, Indianapolis, IN) and fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS; USB Corporation, Cleveland, OH) for 20 min. Fixation was followed by five washes of PBS, permeabilization in PBST (PBS with 0.1% Triton-X 100) for 10 min, blocking in PBSBT (PBS with 3% bovine serum albumin, 0.1% Triton-X 100 and 0.02% sodium azide) for 30 min, incubation in primary rat α-HA antibody (3F10, Roche) or rat IgG (Santa Cruz Biotechnology, Dallas, TX) diluted 1:100 in PBSBT for 45 min, three washes of PBSBT, and incubation in secondary goat α-rat Alexa Fluor 594 antibody (Invitrogen) diluted 1:1000 in PBSBT for 30 min. After three washes of PBSBT, the coverslips were mounted in ProLong Gold Antifade Reagent plus DAPI (Invitrogen). The following day, the coverslips were sealed with nail polish.
For co-staining of the Flag and HA tags, the above procedure was carried out with the addition of a Flag staining step after blocking and before incubation with α-HA. Either mouse IgG (Santa Cruz Biotechnologies) or mouse M2 monoclonal α-Flag (Sigma, St. Louis, MO) were diluted 1:500 in PBSBT, clarified, and incubated on the cells for 45 min followed by three washes in PBSBT. The secondary antibody was goat α-mouse Alexa Fluor 488 (Invitrogen) for 30 minutes followed by 3 washes in PBSBT.
For imaging of GFP and DAPI together, cells grown on coverslips were mounted directly in ProLong Gold Antifade Reagent with DAPI. For live cell imaging of GFP-tagged rodlets, HEK-293 cells were grown and transfected in 35 mm dishes with glass windows (MatTek Corporation, Ashland, MA) in media without phenol red. They were maintained in a heated incubator affixed to an inverted confocal microscope and imaged every 10 min overnight.
Images were acquired on an Olympus BX51, Nikon Eclipse 80i, LSM510 Meta Inverted, or a DeltaVision Image Restoration Microscope. Images were merged in Photoshop (Adobe, San Jose, CA). For quantification of rodlet number, five pictures per coverslip were obtained and picture names were coded. We then counted number of DAPI+ nuclei, the number of nuclei that have rodlets, and the total number of rodlets per picture in a blinded fashion. Data were analyzed and lines of best fit determined in Excel (Microsoft, Seattle, WA).
Colony counts
HEK-293 cells seeded onto six-well plates were transfected at 50–80% confluence in triplicate with FuGene6 (Roche) at a 3:1 ratio of FuGene6 to DNA according to the manufacturer's instructions. Two days later, the cells were trypsinized, reconstituted in 1 ml of media, and 13.33 μl was transferred onto 10 cm plates containing 10 ml of media containing 1 mg/ml geneticin (Invitrogen) for a 1:750 dilution (unless noted differently in the figure legend). Two weeks later, the cells were fixed and stained with 50% methanol (VWR, Radnor, PA) and 1% methylene blue (Invitrogen). After 1 h, the plates were washed twice with PBS and colonies were counted by hand following drying. Error bars represent the standard error of the mean.
Protein purification and cell-free chamber system
We transfected eight 10 cm plates of HEK-293 cells with 8 μg of pCMV-Flag-TcBuster plasmid each 48 h prior to protein lysis in 125 μl of CellLytic M buffer (Sigma) with mini cOmplete ULTRA protease inhibitor cocktail tablet (Roche) per plate. The Flag M Purification Kit was used to absorb the Flag-TcBuster protein onto ANTI-FLAG M2 affinity gel followed by elution by competition with the supplied 3xFlag peptide (Sigma). A clean flow chamber was created as previously described (28). A solution containing 32% purified protein in modified PEM buffer (90 mM PIPES, 1.1 mM ethylene glycol tetraacetic acid (EGTA) and 0.1 mM MgCl2, pH 6.9) with or without 0.4 ng/μl pTcBNeo was flowed into the chamber. After evaporation overnight at 37°C, 10 mM PIPES buffer was introduced into the chamber which was sealed with wax and the rodlets were imaged by confocal differential interference contrast (DIC) microscopy on a LSM 510 Meta Inverted microscope.
Western blots
For Figure 1, our procedure for obtaining cytoplasmic and nuclear protein fractions has been described previously (29). To obtain the insoluble protein fraction, the pellet that was not soluble in either the hypertonic or nuclear lysis buffers was resuspended in the same volume of cell lysis buffer as used to obtain soluble protein fractions (550 μl). The 10% Bis–Tris NuPage Gel (Invitrogen) was loaded with 20 μg of soluble protein and 40 μl of insoluble suspension, transferred via iBLOT (Invitrogen) to a nitrocellulose membrane, immunoblotted for HA and β-actin, and imaged as previously described (29) except that rat α-HA (3F10, Roche) diluted 1:1000 and goat anti-rat 800 diluted 1:15 000 were used to detect HA. For Figure 7, cells were harvested by scraping in PBS, washed once in PBS, and frozen. RIPA (Sigma) supplemented with PhosSTOP (Roche) and cOmplete ULTRA Tablets (Roche) inhibitors was added to each thawed cell pellet and cells were further lysed by freeze-thaw. Protein lysates were analyzed by Pierce BCA Protein Assay Kit (Thermo Scientific) and 25 μg protein was combined with NuPAGE sample buffer and reducing agent (Life Technologies), loaded onto 4–12% Bis–Tris gels (Life Technologies), and processed further as described above. Image Studio software (LiCOR) was used for quantification and further analysis was performed in Excel and GraphPad Prism.
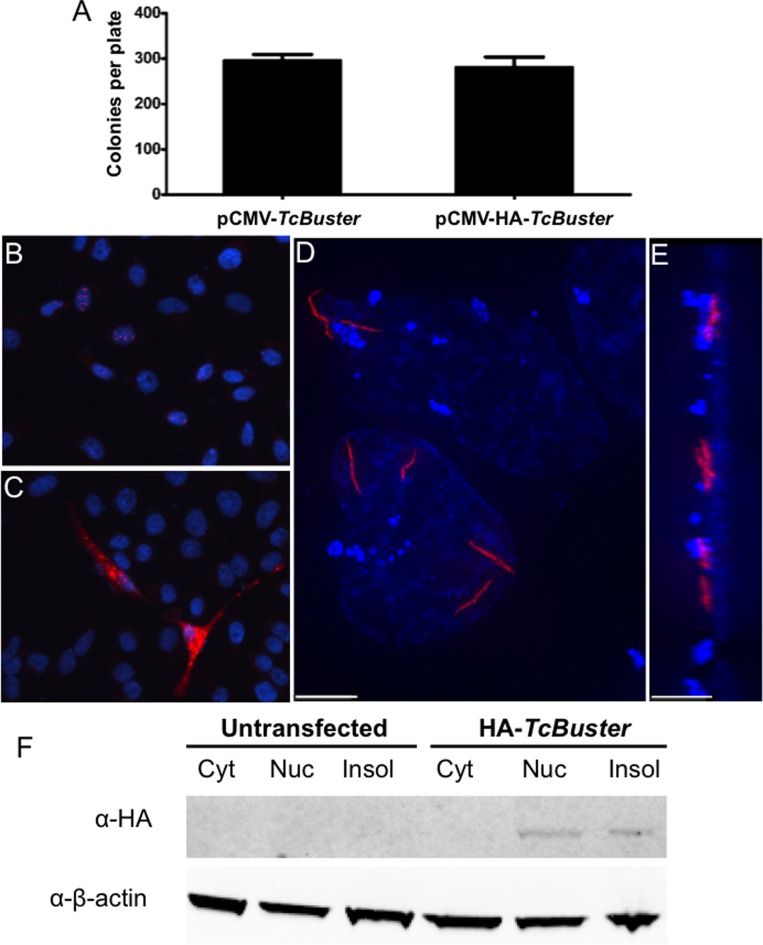
Figure 1.
TcBuster transposase is localized to rodlet structures in human tissue culture cells. (A) To test the activity of the HA-tagged TcBuster transposase, HEK-293 cells were transfected with the transposase plasmid indicated and the pTcBNeo neomycin-resistance transposon plasmid at a 1:5 ratio, n = 3 per group. After 48 h, the cells were split into selection media and colonies were counted after two weeks of selection. To localize the transposase protein, HeLa cells were transfected with 1 μg of pCMV-HA-TcBuster plasmid. The following day, the cells were fixed with formaldehyde and stained to detect the HA tag (red) and DNA with DAPI (blue). Representative image shows nuclear rodlet inclusions (B), non-representative image showing extreme overexpression phenotype with cytoplasmic inclusions that were observed rarely (C). No red staining could be found at this exposure on IgG only or untagged controls (not shown). Imaging of HA-TcBuster was performed on a deconvolution microscope (D). In (E), the image shown in (D) has been rotated 90° such that the left is the top of the cells and the right is the bottom of the cells. The scale bar represents 5 μm in (D) and (E). (F) Cytoplasmic (Cyt), nuclear (Nuc), and insoluble (Insol) protein lysates prepared from transfected HEK-293 cells were subjected to immunoblotting for the HA-tagged transposase (top row) and the β-actin loading control (bottom row). The transposase was detected at the expected size in the nuclear and insoluble fractions.
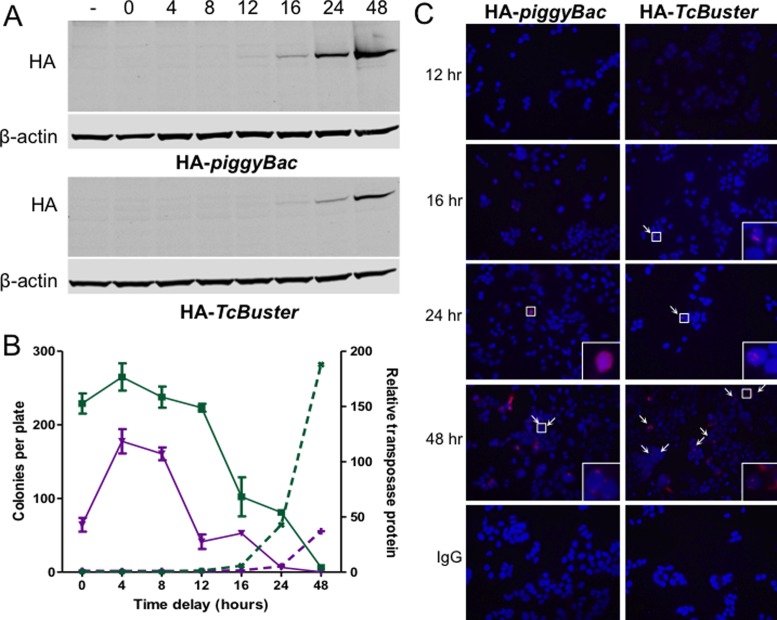
Figure 7.
The presence of transposase structures signals the end of active transposition. (A) HEK-293 cells in 10 cm plates were transfected with 800 ng of pCMV-HA-piggyBac or pCMV-HA-TcBuster and 6.7 μg of pUC19 by FuGene6. Equal amounts of the lysates obtained at various timepoints after transfection as indicated above the immunoblots (in hours). The two NuPAGE gels were run, immunoblotted, and imaged simultaneously for rat α-HA and mouse α-β-actin. (−), untransfected HEK-293 cells were the negative control. (B) To test the ability of piggyBac and TcBuster transposases to integrate transposons at various timepoints after expression, HEK-293 cells were transfected by FuGene 6 with 100 ng of pCMV-piggyBac (green squares, solid lines) or pCMV-TcBuster (purple inverted triangles, solid lines) transposase plasmid and 900 ng of pUC19 filler DNA. Cells were transfected with 1 μg of the corresponding neomycin-resistance transposon plasmid, either pTpB or pTcBNeo, at the indicated time delay after the first transfection. After 56 h the cells were split into selection media (1:2000) and colonies were counted after two weeks of selection (plotted on the left y-axis). The bands from the immunoblots shown in (A) were quantified in LiCOR Image Studio. The HA signal was divided by the β-actin signal to normalize, then each fraction was divided by the signal in the untransfected lane (−) to give the relative protein abundance over time for each transposase (piggyBac, green x's, dashed lines; TcBuster, purple +'s, dashed lines; plotted on the right y-axis). (C) Timecourse of transposase expression. HEK-293 cells were seeded onto poly-L-lysine coated coverslips and transfected via FuGene6 with 100 ng of the indicated transposase plasmid and 900 ng of pUC19 filler DNA. The coverslips were harvested by formaldehyde fixation at the indicated timepoint after transfection (12, 16, 24 or 48 h). Cells were stained for HA or with rat IgG for the negative control (red) and DNA by DAPI (blue). White arrows point to rodlets or inclusions; areas indicated by white boxes are magnified 16x in the lower right corners. Representative images are shown.
For detection of Flag, the buffers and washes were Tris and milk-based as described elsewhere (30). The amount of unpurified protein sample loaded was 30 μl while the amount of purified protein sample loaded was 20 μl. The primary rabbit α-Flag antibody was diluted 1:1000 (Cell Signalling, Danvers, MA) and the secondary antibody was goat anti-rabbit HRP diluted 1:10 000 (Bio-Rad, Hercules, CA). The membrane was developed with SuperSignal West Pico Chemiluminescent Substrate (Thermo Pierce) and imaged on a ChemiDoc XRS+ (Bio-Rad). The membrane was stained with Ponceau S Staining Solution (Tocris Biosciences, Bristol, UK).
Gene delivery in vivo, live animal imaging, and liver immunofluorescence
Plasmid DNA was prepared by endo-free maxiprep (Qiagen). For long-term luciferase imaging, female FVB 8-week-old mice obtained from Baylor College of Medicine (Houston, TX) received hydrodynamic tail vein injections of TransIT-QR hydrodynamic delivery solution (Mirus Bio, Madison, WI) containing 25 μg of transposon DNA with or without transposase DNA (2.5 μg for pCMV-TcBuster or 5 μg for pCMV-piggyBac). Mice were imaged periodically under isoflurane on an IVIS machine (Perkin Elmer, Waltham, MA) as previously described (25). Mice that were sacrificed at 24 h were 4-month-old male FVB mice obtained from Charles River. Hydrodynamic tail vein injections contained 10 μg of pTcBCAGLuc and either 2.5 μg or 25 μg of pCMV-HA-TcBuster (n = 3 per group) in 2 ml of TransIT-QR solution. Luciferase positive mice (n = 2 per group) were sacrificed 24 h post-injection and their livers fixed in 10% formalin at 4°C overnight. Samples were stained for the HA antigen as previously described (29). The Institutional Care and Use Committees of Baylor College of Medicine and Vanderbilt University approved all animal experiments. Error bars represent the standard error of the mean.
RESULTS
TcBuster transposase localization in human cells
We initially sought to evaluate the subcellular localization of TcBuster transposase in human cells to determine if the transposase was able to localize to the nucleus. We constructed a plasmid to add the hemagglutinin (HA) tag to the N-terminus of the transposase, pCMV-HA-TcBuster, to permit detection of the protein with antibodies to the tag. We tested the relative activity of HA-TcBuster transposase by drug-resistant colony assay to measure the integration efficiency of a transposon carrying the neomycin-resistance gene (pTcBNeo) and found it to be indistinguishable from untagged TcBuster transposase (Figure 1A). By immunofluorescent staining of transfected HEK-293 cells, we observed rod-shaped structures (rodlets) that colocalized with the 4′,6-diamidino-2-phenylindole (DAPI)-stained genomic DNA (Figure 1B). The vast majority of expressing cells have several rodlets between 1–5 μm in length with their localization limited to the nuclei. Rarely, cells could be found that were overexpressing the protein with bright inclusions throughout the cytoplasm (Figure 1C). To better visualize the nuclear rodlet structures, we used deconvolution microscopy to remove the light scatter emanating from the fluorescent particles. The rodlets were shown by this method to be thin and filamentous (Figure 1D and E). To confirm the localization observed by immunofluorescent imaging, we also obtained cytoplasmic and nuclear protein fractions along with the insoluble pellet from cells transfected with pCMV-HA-TcBuster and subjected these fractions to immunoblotting. We found that the TcBuster transposase was not detectable in the cytoplasm but was present in the nuclear fraction with a substantial portion remaining in the insoluble pellet (Figure 1F). We concluded that TcBuster transposase formed nuclear rodlet protein structures in the nucleus.
The number of rodlets increases with transposase plasmid dose
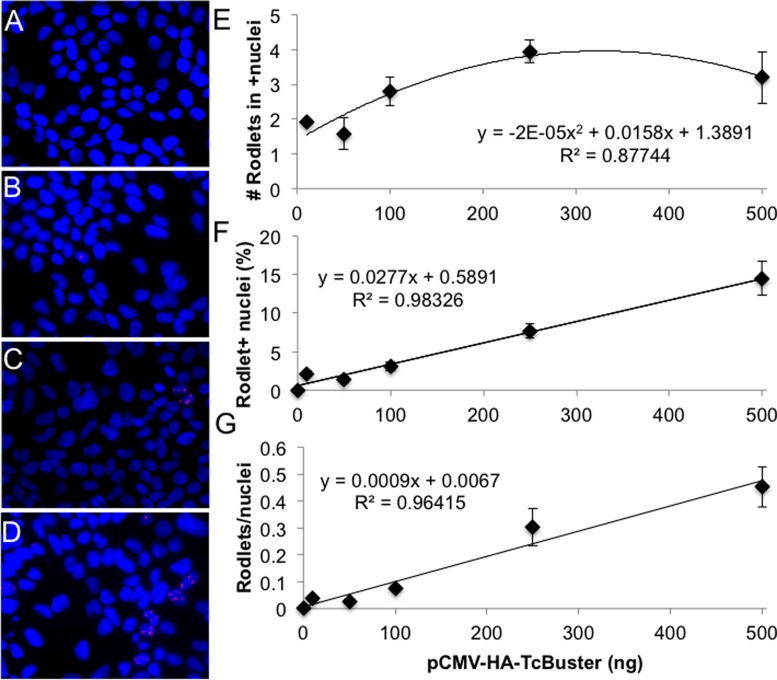
We previously found a non-linear relationship between transposase dose and colony count when the transposon amount was held constant (11). With increasing transposase dose at low concentrations, the number of colonies increased steadily. When 100 ng of transposase was transfected, the maximal number of colonies was reached and thereafter the number of colonies decreased with a 500 ng dose of transposase producing approximately half the number of colonies as the 100 ng dose (11). To determine the number of rodlets present at various doses, we repeated the transfections done for the colony assay (11) with pCMV-HA-TcBuster on HEK-293 cells seeded onto coverslips. The total number of nuclei, total number of rodlets and the number of nuclei with rodlets were counted blindly (Figure 2). Representative images of cells given 0 ng (Figure 2A), 50 ng (Figure 2B), 100 ng (Figure 2C) and 500 ng (Figure 2D) of transposase plasmid along with 500 ng of transposon plasmid are shown. The number of rodlets inside of positive nuclei increased modestly from 1–2 to 3–4 with increasing dosage (Figure 2E). The percentage of nuclei that had rodlets (Figure 2F) and the total number of rodlets per nuclei (Figure 2G) increased linearly. For instance, at the 100 ng transposase dose, peak transpositional activity declined after transposase concentrations became high enough for visible transposase rodlets to appear in >5% of cells (Figure 2F) (11). Therefore, we found that the presence of rodlets coincided with the inhibition of activity at high transposase doses. These data suggest that the negative dosage effect was related to the presence of the rodlets.
Figure 2.
TcBuster transposase rodlet number increases with increasing plasmid DNA doses. HEK-293 cells on coverslips in 6-well plates were transfected by FuGene 6 with 500 ng of transposon (pTcBNeo) and varying doses of pCMV-HA-TcBuster (0 ng, 10 ng, 50 ng, 100 ng, 250 ng or 500 ng), and pUC19 as needed to increase the DNA amount to 1000 ng into HEK-293 cells in six-well dishes. Representative images of cells given 0 ng (A), 50 ng (B), 100 ng (C) or 500 ng (D) of pCMV-HA-TcBuster are shown (DAPI-stained DNA is in blue; HA-TcBuster transposase is stained in red). At each dose, two coverslips were stained and three images per coverslip were taken of random of fields containing at least one rodlet. The average and standard error of the six images taken based on data from blinded counts are plotted (E–G). (E) The number of rodlets per HA-TcBuster positive nuclei is shown for various transposase plasmid doses. (F) The percentage of nuclei with rodlets was calculated at each DNA dose. (G) The number of rodlets as a fraction of all nuclei. Lines of best fit are displayed with the corresponding equation and R2 value for each (E–G).
The dynamics of rodlet formation
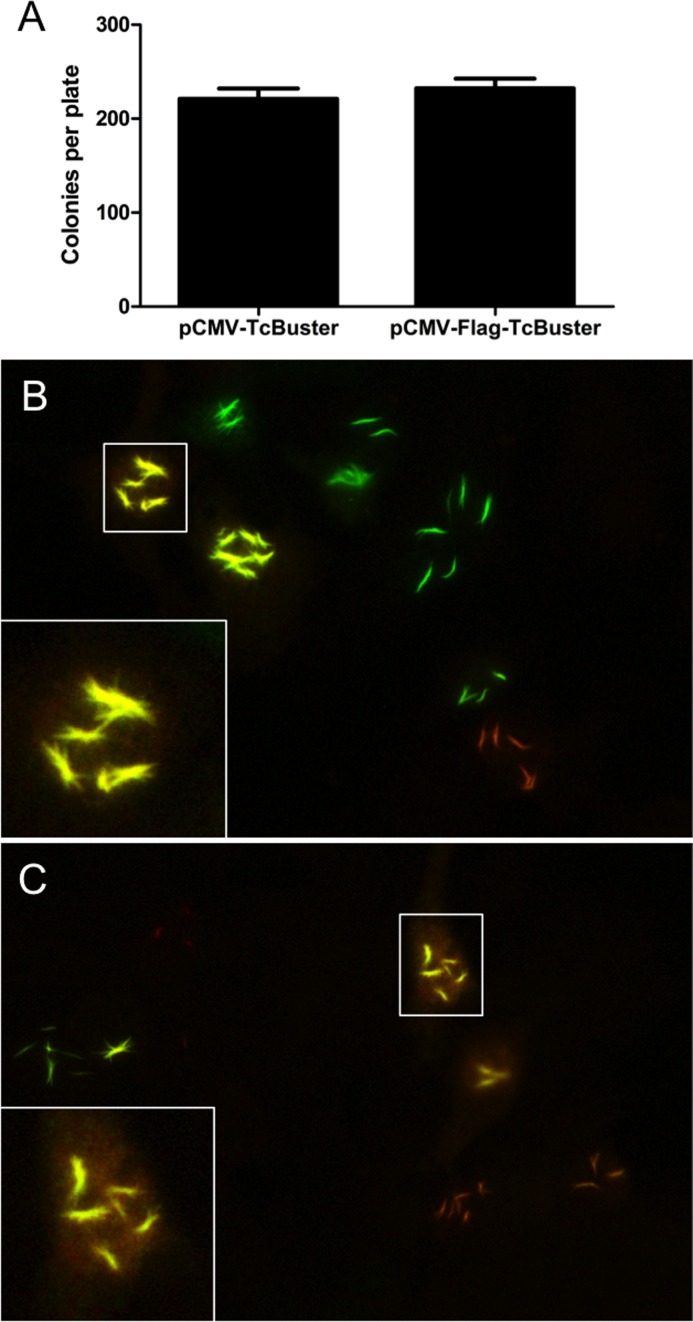
We created a N-terminally Flag-tagged version of TcBuster transposase (pCMV-Flag-TcBuster) and found that it retained activity when compared to untagged TcBuster in a colony assay (Figure 3A). If the rodlets were formed slowly over time, we would expect a ‘seeding’ to take place during which the rodlet would form from a static starting location, with polymerization continuing out from this initial seed either from one or both ends. To test this hypothesis, we transfected HEK-293 cells with either first the HA, then the Flag-tagged transposase, or vice versa, 6 h apart and stained the following day (Figure 3B and C). We found cells with rodlets that stained for only HA (red rodlets, Figure 3B and C) or Flag (green rodlets, Figure 3B and C), presumably because they were transfected in only one of the two transfections. Many cells expressed both HA and Flag at varying ratios (yellow rodlets, Figure 3B and C), but all of the yellow rodlets examined appeared at this resolution to co-stain throughout the rodlet for both HA and Flag (inset, Figure 3B and C), suggesting that the rodlets may not be slowly formed from a starting seed but rather are a dynamic structure that is flexible in composition.
Figure 3.
Colocalization within rodlets following delayed transfection of differently tagged transposase constructs. (A) To test the activity of the Flag-tagged TcBuster transposase, HEK-293 cells were transfected with the transposase plasmid indicated and the pTcBNeo neomycin-resistance transposon plasmid at a 1:9 ratio, n = 3 per group. After 48 h, the cells were split into selection media and colonies were counted after two weeks of selection. To examine the dynamics of rodlet formation, HEK-293 cells grown on coverslips in six-well plates were transfected by FuGene 6 with pCMV-TcBuster (500 ng) and pCMV-HA-TcBuster (500 ng) then 6 h later with pCMV-TcBuster (500 ng) and pCMV-Flag-TcBuster (500 ng) (B) or vice versa (C). The following day, the cells were double-stained for Flag-tagged TcBuster transposase (green) and HA-tagged TcBuster transposase (red). Cells transfected with one of the transfections produced red or green rodlets only, whereas cells transfected with both produced only uniformly yellow rodlets as shown in the representative images including a 4× magnification of a nuclei with rodlets costaining for Flag-TcBuster and HA-TcBuster, inset.
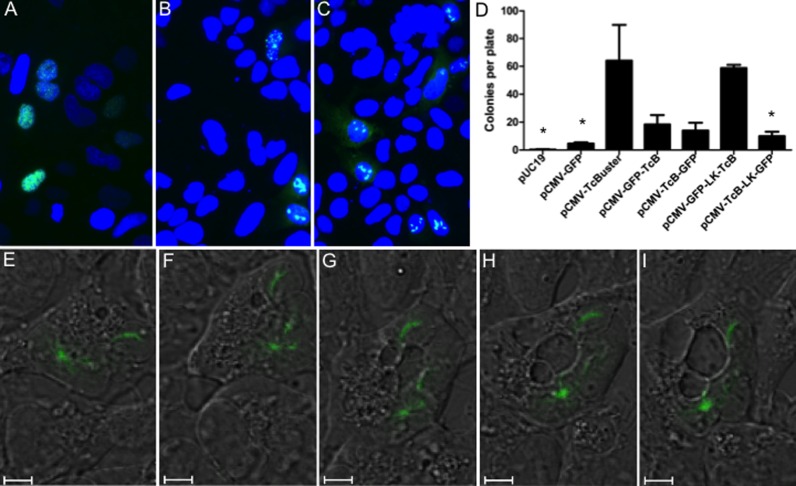
In order to examine how rodlets form in real time, we created a series of GFP fusions to TcBuster. All of the GFP fusions changed the rodlet phenotype from a rodlet to diffuse speckled inclusions, as shown for pCMV-TcBuster-GFP in Figure 4A (GFP images for each fusion are shown in the supplementary data, Figure S1A). By introducing untagged or HA-tagged TcBuster transposase along with the GFP-tagged transposase, we were able to ‘lace’ the rodlets with GFP to permit live cell imaging. GFP-positive cells exhibited a spectrum of phenotypes ranging from cells having speckles without rodlets to cells with rodlets indistinguishable from those seen when HA or Flag tags were used to detect them. This spectrum likely represents the many random ratios of plasmids that could occur during a lipid transfection of both the GFP-tagged and untagged transposase. The average cell exhibited an intermediate phenotype where the rodlets were shortened and increased in number (Figure 4B and C).
Figure 4.
Live cell imaging of rodlet formation. HEK-293 cells plated onto coverslips in 6-well dishes were transfected with either 1 μg of pCMV-TcBuster-GFP (A), 500 ng of pCMV-TcBuster-GFP + 500 ng of pCMV-TcBuster (B) or 500 ng of pCMV-TcBuster-GFP + 500 ng of pCMV-HA-TcBuster (C). Cells were mounted directly with media containing DAPI to stain the nuclei (blue) to observe GFP expression (green). (D) To test the activity of the GFP-tagged TcBuster transposase (TcB) constructs, with and without a linker (LK), HEK-293 cells were transfected with the transposase plasmid indicated and the pTcBNeo neomycin-resistance transposon plasmid at a 1:9 ratio, n = 3 per group. After 48 h, the cells were split into selection media and colonies were counted after two weeks. Asterisk (*) indicates this group was significantly different than pCMV-TcBuster by Tukey post-test analysis. (E–I) Rodlets were imaged in HEK-293 cells by confocal live cell imaging of DIC (black and white) and GFP (green) 32 h after transfection with 500 ng each of pCMV-TcBuster and pCMV-GFP-LK-TcBuster. A cell is shown at 32:00 (E), 32:30 (F), 33:00 (G), 33:30 (H) and 34:00 (I).
Although still slightly above the background pCMV-GFP, GFP fusion to either the N- or C-terminus severely blunted the activity of TcBuster transposase as measured by colony assay (Figure 4D). It should be noted that since the pCMV-GFP helper plasmid vector contains a neomycin resistance cassette and some will randomly integrate, more colonies were found when using pCMV-GFP alone even without transposase present. To increase the activity of the protein, we introduced a linker region that we have previously used to retain transposase activity when fused to DNA-binding domains (15,26). We found that the N-terminal pCMV-GFP-LK-TcBuster was similarly active to untagged TcBuster (Figure 4D), so we used this construct to study GFP-labeled rodlets by live imaging (Figure S1B). The GFP-laced rodlets were generally confined to the nucleus, colocalizing with H2B-mCherry fusion protein (31) to define the nucleus (Figure S1C).
We proceeded to carry out time-lapse live cell DIC and fluorescent confocal imaging at a 1:1 ratio of pCMV-TcBuster to pCMV-GFP-LK-TcBuster in HEK-293 cells. Snapshots beginning at 32 h post-transfection are shown in Figure 4E–I in 30-min increments. In Figure 4E, a HEK-293 cell is shown containing one rodlet in the upper right portion of the cell and one round inclusion at the bottom left. In Figure 4F, the upper rodlet changed position by ∼90° and the lower round inclusion split off into three short rodlets. In Figure 4G, the middle rodlet elongated whilst the lower two remained in close proximity. In Figure 4H, the two lowest inclusions reformed a larger, brighter round inclusion and the middle rodlet faded, and was hardly visible in Figure 4I. Therefore, the transposase rodlets were created in a fast, dynamic process involving fission and fusion.
Ex vivo formation of TcBuster rodlets
When we first found them, we wondered if the rodlets were labeling a pre-existing nuclear structure. Slowly we began to acquire mounting evidence that rodlets were generated by the transposase alone. In support of this hypothesis are the following points: (i) the TcBuster rodlets are unique structures that only look similar to those filamentous structures others have observed for Ac transposase (32); (ii) we have found the TcBuster rodlets in animal cells while Ac rodlets were observed in plant cells (32), which are different kingdoms of life; (iii) the rodlet length was decreased dramatically through fusion to GFP (Figure 4A); (iv) the rodlet length could be manipulated in a linear fashion depending on the ratio of GFP fusion protein to untagged protein provided to the cells (Figure 4B) and (v) transposon DNA was not required for rodlets to form.
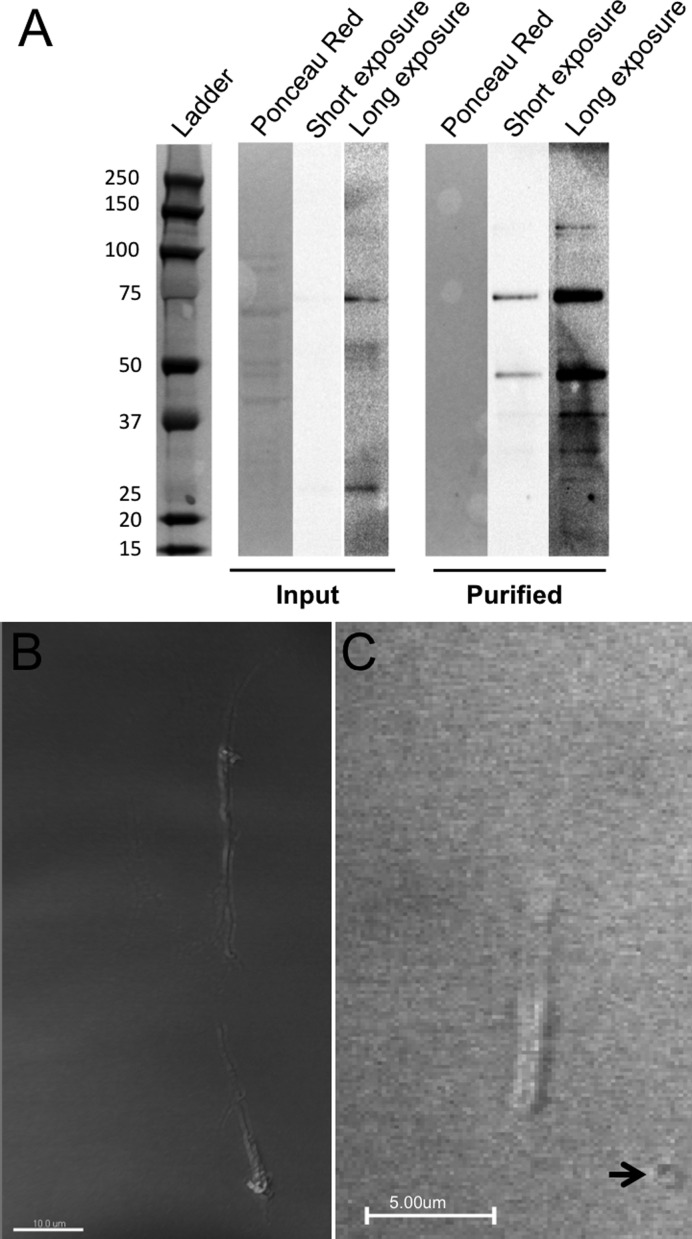
To further test the hypothesis that the rodlet structures are formed from TcBuster protein rather than the transposase protein being drawn to a pre-existing cellular structure, we purified Flag-TcBuster from HEK-293 cells that were overexpressing the protein at 48 h post-transfection. The Ponceau Red staining for nonspecific protein revealed many more bands in the Input lane and none in the Purified protein lane as expected (Figure 5A). A band at the expected size (75.8 kDa) was weakly present in the Input lane after a long exposure and gamma manipulation of the image; this band was enriched in the purified protein lane (Figure 5A). A smaller 50 kDa band present in the purified protein lane was not present in the input or in other samples analyzed (Figures 1 and 7), therefore, we conclude the band is either an endogenous protein that cross-reacted with the Flag epitope or a cleavage product generated during protein purification.
Figure 5.
Formation of rodlets from purified transposase ex vivo. Protein was purified with a Flag affinity column from HEK-293 cells transfected via FuGene 6 with pCMV-Flag-TcBuster 48 h prior. (A) Input (unpurified protein lysate) and purified protein were run together, transferred and Flag was detected by chemiluminescent western blot followed by staining with Ponceau Red to determine overall protein amount in each lane. Both a short, unmodified exposure and long exposure optimized for sensitivity are shown. Expected running size for Flag-TcBuster is 75.8 kDa. The size of each band in the ladder is given in kDa. (B and C) Purified Flag-TcBuster formed rodlets in cell-free conditions both in the absence (B) and presence (C) of transposon DNA in a cell-free buffer system. Slides were DIC imaged on a confocal microscope. Larger circular structures (arrow) were present only when DNA was added.
To observe rodlet formation ex vivo we followed microtubule polymerization protocols (28). Chamber slides were prepared from cleaned coverslips and a solution of ∼1/3 protein in a PEM-like buffer was flowed into the chamber. After incubation at 37°C, rodlets formed both in the absence (Figure 5B) and presence (Figure 5C) of transposon DNA. The rodlets were thin (average diameter 0.86 μm; 95% confidence interval of 0.81–0.99 μm) and measured several microns in length, indistinguishable from those that appear in cells. The rodlets in the presence of DNA (Figure 5C) were somewhat thicker than those without (Figure 5B). Additionally there were small ring-like structures visible only in preparations that included DNA (Figure 5C). In the absence of DNA, the ends of the rodlets appear somewhat frayed (Figure 5B), an effect that we also saw in cells more frequently for Flag-TcBuster rodlets than the HA-TcBuster rodlets (Figure 3).
TcBuster activity and localization in mouse liver in vivo
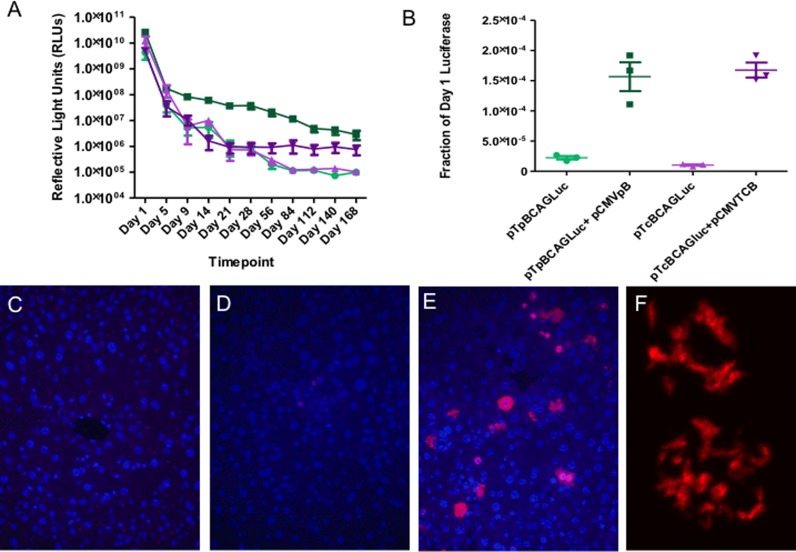
To test the utility of TcBuster for liver-directed gene transfer, we compared TcBuster to piggyBac for long-term gene expression after hydrodynamic injection of mice. First, we created pTcBCAGLuc, a plasmid carrying TcBuster IRs and the same luciferase expression cassette as pTpBCAGLuc (25). The luciferase expression cassette consists of the CAGGS chicken β-actin enhancer with a minimal CMV promoter driving expression of the firefly luciferase protein. The transposon alone or transposon plus transposase for piggyBac or TcBuster were introduced into the mouse liver by hydrodynamic tail vein injection, a commonly used method to transfect mouse hepatocytes in vivo (33,34). Luciferase expression was measured in reflective light units by live animal imaging over a 168-day time-course (Figure 6A). The difference in the groups was statistically significant by one-way analysis of variance. To better compare the two systems, the last three time-points (Day 112, Day 140, and Day 168) representing stable long-term gene expression were normalized to Day 1 to account for differences in transfection efficiency between groups and the promoter activity within the 5′ inverted repeat of piggyBac (35)(Figure 6B). For both systems, the mice given transposase had higher levels of gene expression at the later timepoints than mice receiving transposon alone (piggyBac, P < 0.01; TcBuster, P < 0.0005). However, no significant difference was found between the TcBuster and piggyBac groups. The mice were monitored for gross evidence of cancer or other illness and remained healthy until sacrifice. We used PCR to confirm that TcBuster excision occurred in the mouse liver in vivo. Three mice were given hydrodynamic injection and plasmid DNA was purified from their livers the following day and used as a template for excision PCR. Excision PCR products of the correct size were sequenced and confirmed TcBuster activity in vivo (Figure S2). TcBuster transposition in mouse liver successfully produced long-term gene expression in vivo.
Figure 6.
The TcBuster transposon system is active in mouse liver at a low dose and forms rodlets at a high dose. (A) Mice were given hydrodynamic tail vein injections of either piggyBac transposon alone (25 μg pTpBCAGLuc; light green circles, n = 5), piggyBac transposon plus piggyBac transposase (25 μg pTpBCAGLuc + 5 μg pCMV-piggyBac; dark green squares, n = 5), TcBuster transposon alone (25 μg pTcBCAGLuc; light purple triangles, n = 4), or TcBuster transposon plus TcBuster transposase (25 μg pTcBCAGluc + 2.5 μg pCMV-TcBuster; dark purple inverted triangles, n = 5). Mice were imaged at the indicated timepoints to quantify the amount of liver-specific luciferase expression in Reflective Light Units (RLU). One-way analysis of variance indicates that the groups were significantly different (Kruskal-Wallis test, P = 0.01). (B) To compare the abilities of piggyBac with that of TcBuster to mediate long-term gene expression, the Day 112, Day 140 and Day 168 RLU values were normalized to Day 1 RLU values for each group and then graphed on a dot-plot. Unpaired two-tailed t-tests comparing the late-timepoint normalized RLU values for each system were significantly higher with transposase than without (piggyBac, P < 0.01; TcBuster, P < 0.0005), while the two transposon systems were not significantly different from each other. (C–F) Mice were hydrodynamically injected and livers harvested after 24 h were stained for the HA-tagged TcBuster transposase (red) and DNA by DAPI (blue). (C) IgG control. (D) Non-representative image of mice injected with 10 μg pTcBCAGluc and 2.5 μg pCMV-TcBuster showing the only cells with detectable HA staining in the section. (E) Representative image of mice injected with 10 μg pTcBCAGluc and 25 μg pCMV-HA-TcBuster. (F) Confocal image of representative nuclei from mice shown in (E).
To assess if rodlet formation is also found upon transposase introduction to a living organism, we performed hydrodynamic tail vein injection of the pCMV-HA-TcBuster plasmid in low (2.5 μg) and high (25 μg) doses along with the 10 μg of the pTcBCAGLuc plasmid to monitor injection efficacy by live animal luciferase imaging. Immunofluorescent staining for the HA-TcBuster protein was performed on 24-h samples of liver obtained from mice that received a successful injection. No positive cells were present on the IgG control (Figure 6C). Only a few positive cells could be found in sections from the mice receiving 2.5 μg of pCMV-HA-TcBuster (Figure 6D). Other mice received 10 times this dose, 25 μg, which is a standard hydrodynamic dose for the transgene to be integrated but high for the transposase DNA. Sections from these mice were abundant in positive cells, many of which contained rodlets. However, some cells had cytoplasmic inclusions only, nuclear and cytoplasmic inclusions, or more diffuse staining patterns without obvious inclusions (Figure 6E). Confocal microscopy of hepatocyte nuclei (Figure 6F) revealed a similar rodlet pattern to that observed in transfected tissue culture cells (Figure 1D). These results demonstrate that the formation of TcBuster transposase into rodlets occurred in somatic tissues and that overall the properties of TcBuster transposase rodlets were consistent regardless of the setting (cell-free, tissue culture cells or live mice).
Transposition is complete when transposase rodlets appear
To examine protein expression over time, HEK-293 cells were transfected with a low amount of either transposase and cells were harvested at 0, 4, 8, 12, 16, 24 or 48 h post-transfection. Immunoblotting to detect the HA-tagged transposases revealed a greater overall RIPA-soluble protein amount for HA-piggyBac than for HA-TcBuster (Figure 7A). Protein began to be detectable by western blot at 12 h for HA-piggyBac and 16 h for HA-TcBuster, with the transposase protein abundance increasing greatly between 24 and 48 h post-transfection for both (Figure 7B, right y-axis).
We have previously compared the activity of the TcBuster and piggyBac systems and found that at low doses of transposase plasmid the systems appear comparable in activity whereas at high transposase doses piggyBac is able to yield a higher number of drug-resistant colonies (11). We transfected HEK-293 cells with 100 ng of the pCMV-piggyBac or pCMV-TcBuster transposase plasmids. After 0, 4, 8, 12, 16, 24 or 48 h, cells were transfected a second time with 1 μg of the matching transposon carrying a neomycin-resistance cassette to produce drug-resistant colonies. We found that the cells are transposition-competent for a much shorter time for TcBuster than for piggyBac (Figure 7B, left y-axis). For TcBuster, transposition was greatly attenuated between 8 and 12 h following transfection of the pCMV-TcBuster plasmid, with transposition completely ceased at 24 h (Figure 7B). For piggyBac, transposition dropped sharply between 12 and 16 h following transfection with pCMV-piggyBac plasmid, with transposition over by 48 h after transfection (Figure 7B). Therefore, one reason that piggyBac is more active than TcBuster simply because it retains the ability to transpose for a longer time.
To examine the localization of the transposases at the same timepoints and doses as the colony assay in Figure 7A, HEK-293 cells were transfected with 100 ng of either pCMV-HA-piggyBac or pCMV-HA-TcBuster. Interestingly, the number of positive cells at each timepoint was approximately equal for either transposase. No positive cells were found at 0, 4, 8 or 12 h (data not shown; Figure 7C), and very few positive cells were seen at 16 or 24 h (Figure 7C). At 48 h, the number of positive cells was higher with many cells having bright positive staining, consistent with the immunoblots (Figure 7A and B). While the number of positive cells was approximately the same for the different transposases, localization differed considerably. At 24 h, the HA-piggyBac samples had diffuse nuclear staining while at 48 h there were a variety of localization phenotypes present including cytoplasmic, cytoplasmic and nuclear, or inclusions (white arrows) in the nucleus. Additionally, at 48 h most cells had the characteristic HA-TcBuster rodlets (white arrows), but some had cytoplasmic non-nuclear staining. In both cases there was a diversity of localization phenotypes present at 48 h that we did not observe at 24 h, when all HA-piggyBac-expressing cells had diffuse nuclear staining and all HA-TcBuster cells had nuclear rodlets. Therefore, we conclude that the end of active transposition is signaled by the presence of structures formed by the transposase.
DISCUSSION
We found that TcBuster localized to long nuclear rodlet structures. Rodlet formation was not affected by staining parameters such as the fixation method (formaldehyde versus methanol, data not shown), cell type (HEK-293 (Figure 1B) versus HeLa (Figure 1D), or the presence of transposon plasmid DNA (absent, Figure 1B versus present, Figure 2D). Similarities between Hermes and TcBuster transposase domains and amino acid sequences (9) might suggest that TcBuster forms similar octamers to Hermes (8), but at this point we have no information as to whether the active octameric structures are present within the rodlet structures as subunits or if the protein forms another conformation in the rodlets. If anything, the inhibitory aspect of the rodlets suggests the later. The published octameric ring structure of Hermes transposase is only 20 nm in diameter (8), while the TcBuster rodlets are much larger structures (∼860 nm diameter by ∼5000 nm in length) that, for reference, are similar in size to the primary cilia. Herein, we could not detect signs of a seed and growth mechanism (Figures 3 and 4). Rodlet formation was disrupted by addition of the GFP sequence, and the rodlet phenotype was partially rescued when untagged TcBuster transposase was introduced to create GFP-laced rodlets of approximately half the length. This suggests that rodlets are formed by a polymeric mechanism of some sort that depends on the strength of protein-protein interactions between transposase molecules: in the case of fusion proteins, such as to GFP, these interactions are weakened, resulting in shorter rodlets. Rodlets branched off from a large inclusion (Figure 4E and F) and also combined to become one large inclusion (Figure 4G and H). Therefore, the TcBuster transposase rodlets exhibit a propensity for fission and fusion. Rodlets form without host cofactors, a conclusion supported by the cross-species nature of rodlet formation (Figures 6 and 7). By the time transposase rodlets were detectable by immunofluorescence in some of the cells, transposition was essentially terminated (Figure 7).
We first observed similarities between TcBuster and the classic hAT element Ac in the negative dose response that they exhibit (11,36). Now, further parallels between TcBuster and Ac transposase can be drawn with regard to the unique filamentous structures to which the transposase protein is localized. Similar rodlets were found in Ac localization experiments from synthetic constructs that were carried out in a number of diverse species: insect (37), petunia (32), maize (32,38), tobacco (22) and zebrafish (39). The recombinant transposases can be difficult to purify due to protein insolubility (40–42). It is possible that smaller, precursor aggregated forms are inhibitory rather than the larger aggregates we visualized; however, such hypothetical structures are beyond detection with the techniques we utilized. A detailed review of the association between transposase concentration and activity for different transposons has been published recently in which there are several transposases (Mos1, POKEY, mPB, SB10 and HSB16) that when overexpressed as GFP fusion proteins (12) exhibited punctate staining similar to our TcBuster GFP fusions (Figure 4, Figure S1A). It is possible that these punctate structures may, like TcBuster, be modulated in shape when untagged transposases are introduced to ‘lace’ the rodlets with GFP rather than expressing only the GFP fusions.
Yeast two-hybrid studies on truncated and mutated forms of Ac transposase identified the hAT ‘C-terminal dimerization domain (38)’. This highly conserved region of the hAT transposases is now known from the Hermes crystal structure to be critical for maintaining the overall structure of the transposase but, surprisingly, was not directly involved in the dimerization interfaces of the transpososome in octameric form (8,9,38). One dominant negative mutant in a highly conserved residue within this region of Ac transposase, R733A, formed aggregates that resembled hexagonal crystals rather than filamentous rodlets (38). Therefore, it is possible that the ‘dimerization domain’ could play a role in the oligomerization that may occur in the context of hAT transposase rodlet formation, rather than in the direct transposase–transposase interfaces of the active complex. For Ac transposase, filamentous aggregation is thought to limit transposition and has been proposed as a protective mechanism (32). Scofield et al. put forward the hypothesis that Ac transposase accumulates during a phase in which transposition is encouraged by monomers that bind to the transposon to form the DNA-protein transpososome, but eventually protein levels exceed some threshold, at which time the transposition reaction becomes inhibited by protein-protein interactions (22). However, this hypothesis does not address the complete cessation of transposition that we observed (Figure 7).
The termination of transposition after rodlets appear suggests a window or timer effect. This timer begins when transposase is made in undetectable amounts in the cells and ends when an “off switch” is flipped by transposase aggregation. It is striking how complete the mechanism is (Figure 7) and that the length of the timer correlated perfectly with well-established overall levels of activity since piggyBac is more active than TcBuster (11,27) and on a longer timer (Figure 7). We found an inverse relationship between colonies formed and the transposase concentration in the cell at any given timepoint for both transposons (Figure 7). For TcBuster, transposition was essentially over by the time the protein was detectable by western blot while for piggyBac, detectable protein expression overlapped with colony formation at the 12, 16 and 24 h timepoints. We previously analyzed piggyBac protein expression over the week following transfection and found protein expression slowly dropped after the 48-h peak (43). Different transposases are expected to reach this ‘point of no return’ earlier or later due to their solubility properties, providing an important factor by which we may be able to increase transposase activity in our favor for gene transfer or mutagenesis. Higher activity transposase mutants or fusions could be identified with an extended active window that is achieved through greater solubility rather than changing the transposase-DNA interaction.
As with other filamentous proteins, purification of TcBuster monomers together with advanced imaging techniques may allow us to further elucidate the mechanism of rodlet formation. TcBuster transposase expression in the red flour beetle should be studied to understand if rodlets form and attenuate transposition in the native setting. Fusion of other domains to the N-terminus of TcBuster transposase with a linker may allow the enzyme to maintain activity while directing it with a DNA-binding domain to direct transposition to particular sites in the genome. Finally, we hope to find TcBuster mutants in which rodlet formation is disrupted such that the negative dosage effect can be mitigated to increase transposition for gene transfer applications.
Acknowledgments
We thank Nancy Craig, Peter Atkinson, Reinhard Kunze, Joseph Gall, Allison Hickman and Fred Dyda for helpful discussions regarding the TcBuster rodlets. We thank the Baylor College of Medicine Integrated Microscopy Core and the Vanderbilt Cell Imaging Shared Resource for use of their microscopes. H2B-mCherry was a gift from Robert Benezra (Addgene plasmid #20972). We thank Ruth Ann Veach and Felisha M. Williams for their technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [5T32DK062706 and 2T32DK060445-11]; a fellowship from Dr and Mrs Harold Seltzman; Career Development Award from the Department of Veterans Affairs [BX002797 to L.E.W.]; Vanderbilt Student Research Training Program in Diabetes and Obesity, Kidney Disease and Digestive Disease funded by the Vanderbilt Short Term Research Training Program for Medical Students from the National Institutes of Health [DK007383]; Vanderbilt Diabetes Research and Training Center [DK20593 to L.M.D.]; Baylor College of Medicine Integrative Molecular and Biomedical Sciences (to Y.C.L.); Baylor College of Medicine SMART PREP program funded by the National Institutes of Health [GM069234-2003-2014 to E.S.T.]; National Institutes of Health [DK093660]; Department of Veterans Affairs [BX002190]; Vanderbilt Center for Kidney Disease (to M.H.W.). Funding for open access charge: VA and NIH funding.
Conflict of interest statement. None declared.
REFERENCES
- 1.Biemont C. A brief history of the status of transposable elements: from junk DNA to major players in evolution. Genetics. 2010;186:1085–1093. doi: 10.1534/genetics.110.124180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schnable P.S., Ware D., Fulton R.S., Stein J.C., Wei F., Pasternak S., Liang C., Zhang J., Fulton L., Graves T.A., et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 3.Aziz R.K., Breitbart M., Edwards R.A. Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res. 2010;38:4207–4217. doi: 10.1093/nar/gkq140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Volff J.N. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays. 2006;28:913–922. doi: 10.1002/bies.20452. [DOI] [PubMed] [Google Scholar]
- 5.Piegu B., Bire S., Arensburger P., Bigot Y. A survey of transposable element classification systems–a call for a fundamental update to meet the challenge of their diversity and complexity. Mol. Phylogenet. Evol. 2015;86:90–109. doi: 10.1016/j.ympev.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 6.Arensburger P., Hice R.H., Zhou L., Smith R.C., Tom A.C., Wright J.A., Knapp J., O'Brochta D.A., Craig N.L., Atkinson P.W. Phylogenetic and functional characterization of the hAT transposon superfamily. Genetics. 2011;188:45–57. doi: 10.1534/genetics.111.126813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cary L.C., Goebel M., Corsaro B.G., Wang H.G., Rosen E., Fraser M.J. Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology. 1989;172:156–169. doi: 10.1016/0042-6822(89)90117-7. [DOI] [PubMed] [Google Scholar]
- 8.Hickman A.B., Ewis H.E., Li X., Knapp J.A., Laver T., Doss A.L., Tolun G., Steven A.C., Grishaev A., Bax A., et al. Structural basis of hAT transposon end recognition by Hermes, an octameric DNA transposase from Musca domestica. Cell. 2014;158:353–367. doi: 10.1016/j.cell.2014.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atkinson P.W. hAT transposable elements. Microbiol. Spectr. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0054-2014. doi:10.1128/microbiolspec.MDNA3-0054-2014. [DOI] [PubMed] [Google Scholar]
- 10.Sarkar A., Sim C., Hong Y.S., Hogan J.R., Fraser M.J., Robertson H.M., Collins F.H. Molecular evolutionary analysis of the widespread piggyBac transposon family and related "domesticated" sequences. Mol. Genet. Genomics. 2003;270:173–180. doi: 10.1007/s00438-003-0909-0. [DOI] [PubMed] [Google Scholar]
- 11.Woodard L.E., Li X., Malani N., Kaja A., Hice R.H., Atkinson P.W., Bushman F.D., Craig N.L., Wilson M.H. Comparative analysis of the recently discovered hAT transposon TcBuster in human cells. PLoS One. 2012;7:e42666. doi: 10.1371/journal.pone.0042666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bire S., Casteret S., Arnaoty A., Piegu B., Lecomte T., Bigot Y. Transposase concentration controls transposition activity: myth or reality. Gene. 2013;530:165–171. doi: 10.1016/j.gene.2013.08.039. [DOI] [PubMed] [Google Scholar]
- 13.McClintock B. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. U.S.A. 1950;36:344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yant S.R., Meuse L., Chiu W., Ivics Z., Izsvak Z., Kay M.A. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat. Genet. 2000;25:35–41. doi: 10.1038/75568. [DOI] [PubMed] [Google Scholar]
- 15.Wilson M.H., Kaminski J.M., George A.L., Jr Functional zinc finger/sleeping beauty transposase chimeras exhibit attenuated overproduction inhibition. FEBS Lett. 2005;579:6205–6209. doi: 10.1016/j.febslet.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 16.Ivics Z., Li M.A., Mates L., Boeke J.D., Nagy A., Bradley A., Izsvak Z. Transposon-mediated genome manipulation in vertebrates. Nat. Methods. 2009;6:415–422. doi: 10.1038/nmeth.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y., Wang J., Devaraj A., Singh M., Jimenez Orgaz A., Chen J.X., Selbach M., Ivics Z., Izsvak Z. Suicidal autointegration of sleeping beauty and piggyBac transposons in eukaryotic cells. PLoS Genet. 2014;10:e1004103. doi: 10.1371/journal.pgen.1004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doolittle W.F., Kirkwood T.B., Dempster M.A. Selfish DNAs with self-restraint. Nature. 1984;307:501–502. doi: 10.1038/307501b0. [DOI] [PubMed] [Google Scholar]
- 19.Simons R.W., Kleckner N. Translational control of IS10 transposition. Cell. 1983;34:683–691. doi: 10.1016/0092-8674(83)90401-4. [DOI] [PubMed] [Google Scholar]
- 20.Misra S., Rio D.C. Cytotype control of Drosophila P element transposition: the 66 kd protein is a repressor of transposase activity. Cell. 1990;62:269–284. doi: 10.1016/0092-8674(90)90365-l. [DOI] [PubMed] [Google Scholar]
- 21.Chou J., Casadaban M.J., Lemaux P.G., Cohen S.N. Identification and characterization of a self-regulated repressor of translocation of the Tn3 element. Proc. Natl. Acad. Sci. U.S.A. 1979;76:4020–4024. doi: 10.1073/pnas.76.8.4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scofield S.R., English J.J., Jones J.D. High level expression of the Activator transposase gene inhibits the excision of Dissociation in tobacco cotyledons. Cell. 1993;75:507–517. doi: 10.1016/0092-8674(93)90385-4. [DOI] [PubMed] [Google Scholar]
- 23.Wilson M.H., Coates C.J., George A.L., Jr PiggyBac transposon-mediated gene transfer in human cells. Mol. Ther. 2007;15:139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- 24.Doherty J.E., Huye L.E., Yusa K., Zhou L., Craig N.L., Wilson M.H. Hyperactive piggyBac gene transfer in human cells and in vivo. Hum. Gene Ther. 2012;23:311–320. doi: 10.1089/hum.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saridey S.K., Liu L., Doherty J.E., Kaja A., Galvan D.L., Fletcher B.S., Wilson M.H. PiggyBac Transposon-based Inducible Gene Expression In Vivo After Somatic Cell Gene Transfer. Mol. Ther. 2009;17:2115–2120. doi: 10.1038/mt.2009.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kettlun C., Galvan D.L., George A.L., Jr, Kaja A., Wilson M.H. Manipulating piggyBac transposon chromosomal integration site selection in human cells. Mol. Ther. 2011;19:1636–1644. doi: 10.1038/mt.2011.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X., Ewis H., Hice R.H., Malani N., Parker N., Zhou L., Feschotte C., Bushman F.D., Atkinson P.W., Craig N.L. A resurrected mammalian hAT transposable element and a closely related insect element are highly active in human cell culture. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E478–E487. doi: 10.1073/pnas.1121543109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gell C., Bormuth V., Brouhard G.J., Cohen D.N., Diez S., Friel C.T., Helenius J., Nitzsche B., Petzold H., Ribbe J., et al. Microtubule dynamics reconstituted in vitro and imaged by single-molecule fluorescence microscopy. Methods Cell Biol. 2010;95:221–245. doi: 10.1016/S0091-679X(10)95013-9. [DOI] [PubMed] [Google Scholar]
- 29.Doherty J.E., Woodard L.E., Bear A.S., Foster A.E., Wilson M.H. An adaptable system for improving transposon-based gene expression in vivo via transient transgene repression. FASEB J. 2013;27:3753–3762. doi: 10.1096/fj.13-232090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keravala A., Lee S., Thyagarajan B., Olivares E.C., Gabrovsky V., Woodard L.E., Calos M.P. Mutational derivatives of phiC31 integrase with enhanced efficiency and specificity. Mol. Ther. 2009;17:112–120. doi: 10.1038/mt.2008.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nam H.S., Benezra R. High levels of Id1 expression define B1 type adult neural stem cells. Cell Stem Cell. 2009;5:515–526. doi: 10.1016/j.stem.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinlein M., Brattig T., Kunze R. In vivo aggregation of maize Activator (Ac) transposase in nuclei of maize endosperm and Petunia protoplasts. Plant J. 1994;5:705–714. doi: 10.1111/j.1365-313x.1994.00705.x. [DOI] [PubMed] [Google Scholar]
- 33.Liu F., Song Y., Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 34.Zhang G., Budker V., Wolff J.A. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum. Gene Ther. 1999;10:1735–1737. doi: 10.1089/10430349950017734. [DOI] [PubMed] [Google Scholar]
- 35.Cadinanos J., Bradley A. Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 2007;35:e87. doi: 10.1093/nar/gkm446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scofield S.R., Harrison K., Nurrish S.J., Jones J.D. Promoter fusions to the activator transposase gene cause distinct patterns of Dissociation excision in tobacco cotyledons. Plant Cell. 1992;4:573–582. doi: 10.1105/tpc.4.5.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hauser C., Fusswinkel H., Li J., Oellig C., Kunze R., Muller-Neumann M., Heinlein M., Starlinger P., Doerfler W. Overproduction of the protein encoded by the maize transposable element Ac in insect cells by a baculovirus vector. Mol. Gen. Genet. 1988;214:373–378. doi: 10.1007/BF00330469. [DOI] [PubMed] [Google Scholar]
- 38.Essers L., Adolphs R.H., Kunze R. A highly conserved domain of the maize activator transposase is involved in dimerization. Plant Cell. 2000;12:211–224. doi: 10.1105/tpc.12.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emelyanov A., Gao Y., Naqvi N.I., Parinov S. Trans-kingdom transposition of the maize dissociation element. Genetics. 2006;174:1095–1104. doi: 10.1534/genetics.106.061184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hickman A.B., Perez Z.N., Zhou L., Musingarimi P., Ghirlando R., Hinshaw J.E., Craig N.L., Dyda F. Molecular architecture of a eukaryotic DNA transposase. Nat. Struct. Mol. Biol. 2005;12:715–721. doi: 10.1038/nsmb970. [DOI] [PubMed] [Google Scholar]
- 41.Iida A., Tachibana A., Hamada S., Hori H., Koga A. Low transposition frequency of the medaka fish Tol2 element may be due to extranuclear localization of its transposase. Genes Genet. Syst. 2004;79:119–124. doi: 10.1266/ggs.79.119. [DOI] [PubMed] [Google Scholar]
- 42.Michel K., Atkinson P.W. Nuclear localization of the Hermes transposase depends on basic amino acid residues at the N-terminus of the protein. J. Cell Biochem. 2003;89:778–790. doi: 10.1002/jcb.10554. [DOI] [PubMed] [Google Scholar]
- 43.Saha S., Woodard L.E., Charron E.M., Welch R.C., Rooney C.M., Wilson M.H. Evaluating the potential for undesired genomic effects of the piggyBac transposon system in human cells. Nucleic Acids Res. 2015;43:1770–1782. doi: 10.1093/nar/gkv017. [DOI] [PMC free article] [PubMed] [Google Scholar]