Abstract
Co-translational protein targeting and membrane protein insertion is a fundamental process and depends on the signal recognition particle (SRP). In mammals, SRP is composed of the SRP RNA crucial for SRP assembly and function and six proteins. The two largest proteins SRP68 and SRP72 form a heterodimer and bind to a regulatory site of the SRP RNA. Despite their essential roles in the SRP pathway, structural information has been available only for the SRP68 RNA-binding domain (RBD). Here we present the crystal structures of the SRP68 protein-binding domain (PBD) in complex with SRP72-PBD and of the SRP72-RBD bound to the SRP S domain (SRP RNA, SRP19 and SRP68) detailing all interactions of SRP72 within SRP. The SRP72-PBD is a tetratricopeptide repeat, which binds an extended linear motif of SRP68 with high affinity. The SRP72-RBD is a flexible peptide crawling along the 5e- and 5f-loops of SRP RNA. A conserved tryptophan inserts into the 5e-loop forming a novel type of RNA kink-turn stabilized by a potassium ion, which we define as K+-turn. In addition, SRP72-RBD remodels the 5f-loop involved in ribosome binding and visualizes SRP RNA plasticity. Docking of the S domain structure into cryo-electron microscopy density maps reveals multiple contact sites between SRP68/72 and the ribosome, and explains the role of SRP72 in the SRP pathway.
INTRODUCTION
The universally conserved signal recognition particle (SRP) mediates co-translational targeting of proteins to membranes by recognizing hydrophobic N-terminal signal sequences of its client proteins emerging from ribosomes (1). In eukaryotes, SRP—ribosome nascent chain complexes (RNCs) are targeted to the endoplasmic reticulum by guanosine-5′-triphosphate- (GTP-) dependent interaction with the membrane-bound SRP receptor (SRαβ) and the RNC is transferred to a vacant translocation channel (Sec 61 complex) (2,3). The heterodimeric targeting complex formed by the SRP GTPases SRP54 and SRα constitutes the core of the SRP system and regulates the entire process (4–6). After the RNC is handed over to the translocation channel and translation resumes, the targeting complex relocates on SRP RNA and the SRP GTPases are stimulated by a regulatory ‘distal site’ of the RNA. This is so far only shown for Escherichia coli (7,8) despite the conservation of the targeting complex and its binding sites on the SRP RNA. Upon GTP-hydrolysis, SRP dissociates from the SR and the SRP targeting cycle is closed.
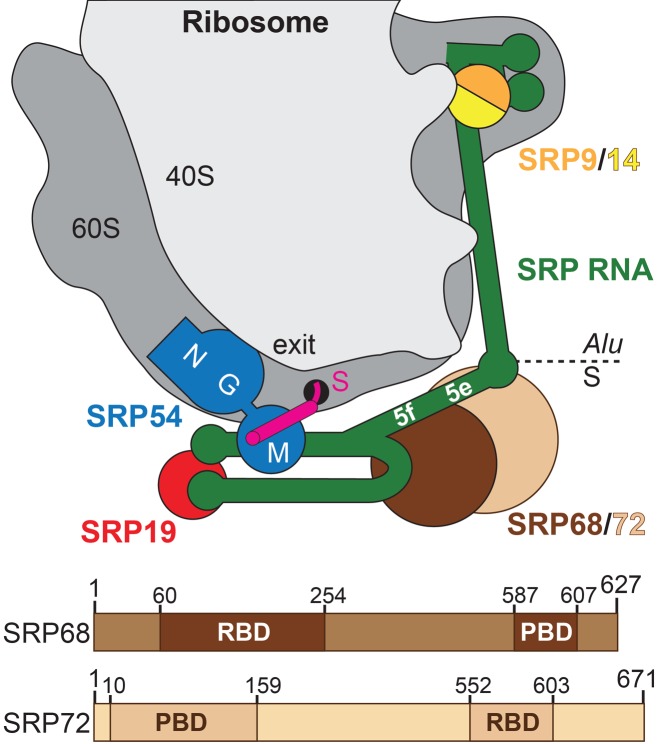
Human SRP is a ribonucleoprotein complex comprising the highly base-paired SRP RNA (also referred to as 7SL RNA) of 300 nucleotides and six proteins (SRP9, SRP14, SRP19, SRP54, SRP68 and SRP72) (Figure 1) (9). It is divided into the Alu domain (SRP9/14 and 5′/3′ ends of SRP RNA) responsible for elongation retardation (10) and the S domain, which recognizes the signal sequence of the ribosome nascent chain and binds to the SR. Both S domain functions are performed by the highly conserved SRP54 protein while SRP19 is a scaffolding protein clamping helices 6 and 8 of SRP RNA (11,12). SRP19 binding to the apical tetra-nucleotide loops (tetraloops) of helices 6 and 8 prepares the flexible internal ‘asymmetric loop’ within helix 8 for SRP54 binding. The two largest α-solenoidal proteins SRP68 and SRP72 (60% of SRP protein mass) belong to the S domain and form a stable heterodimer essential for SRP function (13). Reconstituted SRP with alkylated SRP68/72 or without the heterodimer is not functional in translocation assays (14). The SRP68/72 heterodimer is assembled into the pre-SRP particle in the nucleus and its presence is necessary for export to the cytoplasm. Knockout of the heterodimer in yeast leads to an accumulation of pre-SRP in the nucleus (15) and gene silencing of SRP68/72 with RNAi in Trypanosoma brucei causes cell death due to the toxicity of nuclear SRP-accumulation (16). SRP72 is cleaved during apoptosis by caspases and a 6 kDa C-terminal fragment that is phosphorylated on serine residues is released (17). Proteolytic cleavage of this peptide is connected with the development of autoantibodies against SRP72 triggering the formation of autoimmune diseases, like systemic lupus erythematosus (17,18). Mutations in the SRP72 gene are causative for familial aplastic anemia and an increased risk of acute myeloid leukemia (19).
Figure 1.
SRP68/72 in context of the SRP-RNC. Upper panel: Scheme of human SRP bound to a ribosome—nascent chain complex (gray). Exit: polypeptide tunnel exit; S: signal sequence (magenta); NGM: domains of SRP54 (blue); 5e/5f: loops of SRP RNA (green); SRP9/14 (orange/yellow); SRP19 (red); SRP68 (brown); and SRP72 (sand). Lower panel: Domain architecture of human SRP68/72 (RBD: RNA-binding domain; PBD: protein-binding domain).
The subunits of the SRP68/72 heterodimer (Figure 1) bind individually to SRP RNA (20–22). The N-terminal SRP68 RNA-binding domain (RBD) locates to a central RNA three-way junction between RNA helices 5, 6 and 8 (22). SRP68 binding to the RNA kinks the S domain RNA and remodels the nearby 5f-loop (23). Kinking is important for ribosome interaction at the previously described ‘C4-contact’ (24), while 5f-loop remodeling might activate the targeting complex as described in E. coli (7,8). Here, a conserved guanine base complements the interface of the targeting complex and activates GTP hydrolysis. SRP72 binding to SRP RNA was pinpointed to the adjacent 5e-loop (21). The C-terminal SRP72-RBD, preceding the caspase cleavage and phosphorylation sites of SRP72 (17), comprises a lysine-rich cluster and a linear sequence motif (SRP72-Pfam) harboring a conserved tryptophan residue (21) (Supplementary Figure S1). Heterodimer formation with SRP68 occurs via the N-terminal protein-binding domain (SRP72-PBD), which is predicted to consist of four tetratricopeptide repeats (TPRs) (25). TPRs consist of 34 amino acids, which form two antiparallel helices with a conserved residue pattern. TPR domains contain 3 to 16 repeats and form right handed α-solenoid structures (26). Usually, small peptides bind to the concave side of the TPR domain in a linear mode ranging from an extended coil to α-helical conformation (27). The TPRs of SRP72-PBD bind to a highly conserved C-terminal peptide of SRP68 (SRP68-PBD). However, despite the importance of SRP72 for SRP biogenesis and function and despite the progress with recent high-resolution cryo-EM structures of SRP-RNC complexes (28,29), it remained the last mammalian SRP protein without any structural information available. To close this gap, we determined the structures of the human SRP68/72-PBD complex and SRP72-RBD bound to SRP RNA in context of a quaternary S domain complex, providing important insights into SRP RNA plasticity and revealing all relevant interactions of SRP72 within SRP.
MATERIALS AND METHODS
DNA manipulations and plasmids construction
All constructs used in this study are listed in Supplementary Table S1. Total human mRNA was extracted from HEK cells (SV Total RNA Isolation System, Promega), reversed-transcribed into cDNA (Transcription First Strand cDNA Synthesis Kit with dT primers, Roche), and used for cloning in this study. The DNA sequence encoding human SRP72-RBD (512–662) fused to a C-terminal His6-tag was cloned into the pET24d vector (Novagen) digested NcoI/BamHI. DNA encoding for human SRP72-PBD (8–166) was cloned into the N-terminal His6-Sumo fusion vector pCA528 (30) using the BsaI/BamHI restriction sites. Untagged human SRP68-PBD (546–614) was cloned into the NcoI/BamHI restriction sites of pET16b (Novagen) and into pETtrx1a (31) containing an N-terminal thioredoxin. Short human SRP72-RBD peptides P1 (560–605), P2 (557–605) and P3 (549–605) were cloned into the NcoI/BamHI restriction sites of the pETtrx1a vector.
For yeast two-hybrid analyses, human SRP72-PBD (1–159) was fused to an N-terminal GAL4 binding (BD) and activation (AD) domain using the plasmids pG4ADN111/pG4BDN22 (32), respectively. Human SRP68-PBD (585–607) was fused to a C-terminal GAL4-BD and GAL4-AD using the plasmids pG4BDC22/pG4ADC111 (32). Point mutations were introduced according to the QuikChange Lightning kit (Agilent) site-directed mutagenesis protocol.
The 145 nucleotides long (105–249) human SRP RNA gene was amplified from human genomic DNA, fused to a 3′-hammerhead ribozyme and cloned under the control of the T7 RNA polymerase promotor into pSP64 vector (Promega) digested EcoRI/HindIII. Numbering and sequences correspond to UniProtKB o76094 (human SRP72), UniProtKB q9uhb9 (human SRP68) and GenBank accession no. X04248.1 (human SRP RNA).
Protein/RNA production and purification
The human SRP72-RBD constructs were transformed into BL21(DE3) E. coli cells (Novagen) and proteins were expressed by auto-induction for 16 h at 25°C (33). Sumo-fused human SRP72-PBD and non-tagged SRP68-PBD were co-expressed by auto-induction for 16 h at 25°C. Seleno-methionine labeled SRP72-RBD as well as SRP72-PBD (L31M/L65M) were expressed in BL21 (DE3) E. coli cells grown in M9 medium supplemented with seleno-methionine as described in (34). Cells were grown at 37°C to an OD600 of 0.6 and induced with of 0.5 mM IPTG for 16 h at 22°C.
Purification of SRP72-RBD, SRP19 and SRP68-RBD were performed as follows: the cell pellet was resuspended in lysis buffer containing 40 mM Tris/HCl pH 8.0, 1 M NaCl, 5 mM MgCl2, 5 mM KCl, 20 mM imidazole, 10% (v/v) glycerol, and 0.02% (v/v) 1-thioglycerol. Cells were lysed using an M-110L Microfluidizer (Microfluidics), the cleared lysate was loaded onto a Ni-NTA column (GE Healthcare), the column was washed with the lysis buffer and the protein was eluted by an imidazole gradient (20–400 mM). The eluate was further purified on a size exclusion chromatography (SEC) column Superdex 75 26/60 (GE Healthcare) equilibrated with a buffer containing 20 mM Hepes pH 7.5, 500 mM NaCl, 5 mM MgCl2, 5 mM KCl, 5% (v/v) glycerol and 0.02% (v/v) 1-thioglycerol.
Purification of SRP72-PBD, SRP68-PBD (both proteins individually or in complex) and SRP72-RBD P1, P2 and P3 were performed as described above using buffers containing 200 mM NaCl. In order to remove the fusion constructs, SRP72-PBD and SRP72-RBD P1, P2 and P3 were treated with Ulp1 Sumo- and Tev-protease, respectively, after the first Ni-NTA elution. The proteins were dialyzed against a buffer containing 20 mM Tris/HCl pH 8.0, 200 mM NaCl, 5 mM MgCl2, 5 mM KCl, 10% (v/v) glycerol and 0.02% (v/v) 1-thioglycerol for 16 h at 4°C. The proteases and cleaved fusion tags were bound to a reverse Ni-NTA column and the flow through with the containing proteins was purified via SEC. All proteins were concentrated using Amicon Centrifugal filters (Millipore), frozen in liquid nitrogen and stored at -80°C.
In vitro run-off transcription of the 145 nucleotides long SRP RNA construct was performed according to a modified protocol described previously (35). In vitro transcription was performed at 37°C for 3 h. The RNA was purified by urea-polyacrylamide gel electrophoresis and extracted by crush-and-soak using 0.3 M sodium acetate followed by isopropanol precipitation and desalting (PD10, GE Healthcare).
Reconstitution of the human quaternary S domain
SRP RNA was refolded using a snap cool protocol. The RNA was heated to 90°C for 1 min, cooled immediately on ice for 1 min, diluted with 6× folding buffer containing 120 mM Tris/HCl pH 8.0, 1.2 M NaCl, 60 mM KCl, 60 mM MgCl2, 52% (v/v) glycerol and incubated for 10 min at 37°C. SRP19, SRP68-RBD and SRP72-RBD were added to the RNA, the salt concentration was adjusted to 200 mM and the complex was incubated at 25°C for 10 min. The reconstituted S domain was purified by SEC using a Superdex S200 16/60 column (GE Healthcare) equilibrated with 20 mM HEPES/NaOH pH 7.5, 150 mM NaCl, 5 mM MgCl2, and 5 mM KCl. SRP19, SRP68-RBD and SRP72-RBD were dialyzed two times against a buffer containing 20 mM HEPES/NaOH pH 7.5, 500 mM NaCl, 5 mM MgCl2, 5 mM CsCl, 5% (v/v) glycerol and 0.02% (v/v) 1-thioglycerol for 2 h at 4°C to replace potassium for cesium ions. Folding buffer and SEC buffer for S domain reconstitution were prepared with CsCl using the respective KCl concentrations. S domain reconstitution was performed as described above.
Crystallization and data collection
Crystals of the SRP68/72-PBD complex (native and seleno-methionine labeled SRP68/72-PBD L31M/L65M) were grown by automated crytallization in sitting drops containing 0.2 M K2SO4 and 20% (w/v) PEG3350 at 18°C. The concentration of the complex was adjusted to 12 mg/ml and drops were set in a 1:1 ratio of protein solution to crystallization buffer. Crystals of the quaternary S domain (native, seleno-methionine labeled SRP72-RBD either L562M, I598M, or cesium containing S domain) grew at 4°C in hanging drops manually set containing 17–20% (w/v) PEG3350 and 0.2 M KF. The complex had a concentration of 10 mg/ml and drops were set in 1:1 ratio. Crystals were cryoprotected in 20% (v/v) glycerol added to the mother liquor and were frozen in liquid nitrogen. Data were collected at the European Synchrotron Radiation Facility (ESRF, Grenoble). Single-wavelength anomalous dispersion (SAD) data were collected for the seleno-methionine labeled samples or cesium containing crystals at anomalous peak wavelengths.
Structure determination and refinement
Data processing was performed with XDS (36) and the CCP4i-implemented program AIMLESS (37) for all collected datasets. The structure was solved using SAD data at 3.0 Å resolution using the Phenix suite (38,39). The structure was refined against the native dataset using Phenix.refine (40) and model building was done in COOT (41). The structure was solved by molecular replacement with the program Phenix.phaser (42) using the human ternary S domain complex (4p3e (23)) as a search model. The structure was refined with Phenix and built in iterative cycles in COOT with the help of the anomalous signal for methionine of SRP72-RBD L562M or I598M. Surface potentials were calculated by APBS (43) in PyMol (44). Superposition of structures was performed in COOT. Figures of the structures and electron density maps were created using PyMol and Chimera (45), respectively.
Isothermal titration calorimetry (ITC)
ITC experiments were performed using a MicroCal PEAQ-ITC (Malvern) in a buffer containing 20 mM HEPES/NaOH pH 7.5, 200 mM NaCl, 10 mM MgCl2, 10 mM KCl and 10% (v/v) glycerol (for RNA binding) or 5% (v/v) glycerol for the protein interaction. The titrations were conducted in 19 injection of 2 μl aliquots at 25°C, a reference power of 10 μcal/s and a stirring speed of 750 rpm. Typical concentrations of the titrants were between 100–200 μM (RNA-binding domains) and 150–200 μM (SRP68-PBD), the concentrations in the cell were between 7–17 μM (S domain) and 20–30 μM (SRP72-PBD). The measurements were analyzed using the MicroCal PEAQ-ITC Analysis Software. ITC experiments were performed at least in triplicates (N).
Multi-angle light scattering (MALS)
A total of 150 μl of the SRP68/72-PBD complex (2.5 mg/ml) was injected onto a Superdex 75 10/300 column (GE Healthcare) coupled to a MALS system (Dawn Heleos II 8+ and Optilab T-rEX, Wyatt Technology). Data was analyzed using Astra 6 software (Wyatt Technology).
Pull-down assay
A total of 500 μl of 40 μM His6-SRP68-PBD were loaded on spin-columns with 100 μl Ni-NTA beads equilibrated with the SRP68/72-PBD SEC buffer (buffer A) and incubated for 5 min at 4°C on a slow stirring wheel. The beads were washed once with 500 μl buffer A and once with 100 μl buffer A containing 50 mM imidazole (buffer B). A total of 500 μl of 40 μM SRP72-PBD were loaded on the Ni-NTA beads and incubated for 5 min at 4°C. The beads were washed twice with 100 μl buffer B before elution of the complex with 100 μl buffer A containing 500 mM imidazole (buffer C). As control, SRP72-PBD was loaded without His6-SRP68-PBD to 100 μl Ni-NTA beads and incubated for 5 min at 4°C. The column was washed with 500 μl buffer A, twice with 100 μl buffer B and once with 100 μl buffer C. All flow through samples were analyzed on a 15% sodium dodecyl sulphate-polyacrylamide gel electrophoresis.
Yeast two-hybrid assays
The Saccharomyces cerevisiae strain PJ69–4A was co-transformed with a bait and prey plasmid. Double transformants were selected on SDC medium lacking leucine and tryptophan. For each transformation, four colonies were arrayed in 96-well microtiter plates. The colonies were diluted to an OD600 of 3.0 in water followed by 10-fold serial dilutions and spotted onto SDC-Leu-Trp, SDC-Leu-Trp-His and SDC-Leu-Trp-Ade plates. Growth was determined after 3 days incubation at 30°C. Transactivation controls were systematically performed for each construct with the opposite vector without insert.
RESULTS
Structure of the SRP68/72-PBD complex
In order to characterize the SRP68/72 interaction, the SRP72-PBD was co-expressed and co-purified with SRP68-PBD. Stoichiometric complex formation was confirmed by pull-down assays and analytical SEC coupled with multi-angle light scattering (Supplementary Figure S2). The complex was crystallized in space group C2 with one molecule per asymmetric unit. A native dataset was collected to 1.6 Å resolution. However, the structure could not be solved by molecular replacement. Therefore, the double mutant SRP72-PBD (L31M/L65M) was prepared for seleno-methionine based de novo single-wavelength anomalous dispersion (SAD) phasing. The final model was refined to an Rwork (Rfree) of 14.4% (18.6%) and Ramachandran statistics show 100% of the residues in allowed regions (Table 1 and Supplementary Figure S3).
Table 1. Data collection and refinement statistics for the SRP68/72-PBD complex and the quaternary S domain complex.
| SRP68/72-PBD | quaternary S domain | |||
|---|---|---|---|---|
| native | L31M/L65M | native | cesium | |
| Data collection | ||||
| Space group | C 1 2 1 | C 1 2 1 | P 21 21 21 | P 21 21 21 |
| Cell dimensions | ||||
| a, b, c (Å) | 66.6 52.3 62.9 | 67.0 52.2 63.0 | 134.6 139.3 152.2 | 130.0 141.4 170.6 |
| α, β, γ (°) | 90 118.2 90 | 90 90 90 | 90 90 90 | 90 90 90 |
| Resolution (Å) | 38.0-1.6 (1.66-1.6) | 39.0-3.0 (3.2-3.0) | 48.4-3.4 (3.5-3.4) | 48.9-6.0 (6.7-6.0) |
| Rpim (%) | 2.8 (27.9) | 2.1 (4.0) | 7.2 (68.5) | 7.4 (83.9) |
| Wilson B-factor (Å2) | 21.0 | 95.8 | ||
| I/σ(I) | 14.2 (3.0) | 29.8 (15.5) | 8.7 (1.3) | 7.5 (1.2) |
| CC1/2 | 99.8 (93.3) | 100 (91.2) | ||
| Completeness (%) | 99.4 (98.9) | 93.3 (68.1) | 99.7 (99.4) | 99.8 (100) |
| Redundancy | 5.0 (4.6) | 12.3 (9.4) | 13.4 (13.8) | 25.3 (25.8) |
| Heavy Atom | Se | Cs | ||
| Wavelength (Å) | 0.97264 | 2.06640 | ||
| Anom. Completeness (%) | 92.7 (66.7) | 99.9 (100) | ||
| Anom. Redundancy | 6.3 (4.8) | 13.7 (13.6) | ||
| Refinement | ||||
| No. reflections | 25061 (2507) | 39632 (3863) | ||
| Rwork (%) | 14.2 (19.1) | 23.7 (36.7) | ||
| Rfree (%)* | 18.4 (25.4) | 28.0 (42.0) | ||
| No. atoms | 1582 | 12128 | ||
| Protein | 1409 | 11987 | ||
| Ligand/ion | 23 | 141 | ||
| Protein residues | 171 | 984 | ||
| B-factors (Å2) | 36.1 | 137.5 | ||
| Protein | 34.6 | 137.5 | ||
| Ligands | 65.8 | 137.6 | ||
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.005 | 0.011 | ||
| Bond angles (°) | 0.73 | 1.57 | ||
| Ramachandran allowed (%) | 100 | 97.8 | ||
*for 5% of all data.
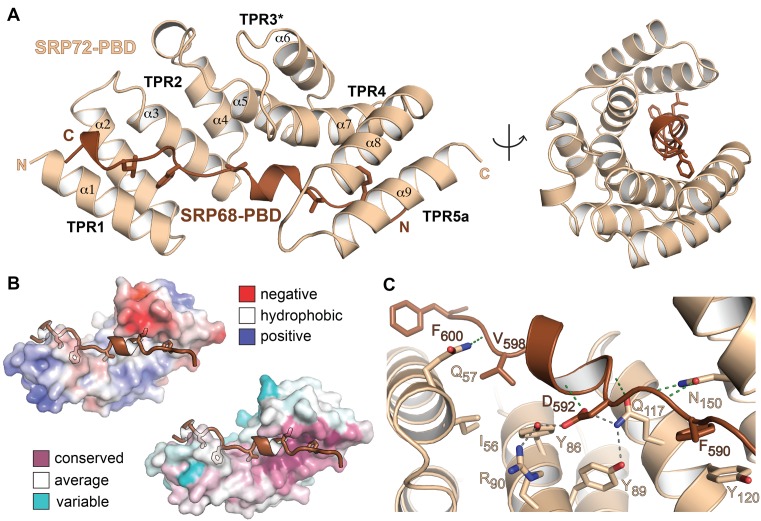
SRP72-PBD (residues S10 to S159) is composed of nine anti-parallel α-helices arranged in a right-handed α-solenoid (Figure 2A). SRP72-PBD comprises three classical TPRs (TPR1: 11–44, TPR2: 45–78, TPR4: 109–142). Residues 79–108 form two anti-parallel α-helices, which also possess the characteristics of a TPR but are shorter in length (Supplementary Figure S4). We define these helices as TPR3*. The C-terminal helix 9 forms a cap and probably corresponds to the first helix of a following TPR5. SRP72-PBD accommodates the TPR-ligand SRP68-PBD in a conserved groove on the concave side of the α-solenoid, as typical for TPR domains (Figure 2B). The tight interaction was confirmed by isothermal titration calorimetry (ITC; KD of 33 nM; Table 2 and Supplementary Figure S5). SRP68-PBD (residues K587 to L607) adopts an extended structure with a central short α-helix (D592 to H597) and a C-terminal helical turn (E604 to L607) predicted to continue until K612. Superposition of SRP72-PBD with its nearest structural homolog (PDB 3vty (46); r.m.s.d. of 2.8 Å for 148 Cα-atoms) indicates a closed α-solenoid (Supplementary Figure S4). Closure is induced by a tilt of TPR4 and TPR5a toward the groove, which is enabled by the shortening of TPR3*. The contact surface between SRP68 and SRP72 is 1200 Å2, which is 43% of the whole surface of SRP68-PBD (47). The interface is larger than for common TPR–ligand complexes with extended peptide conformation like the Hop/Hsp70-Hsp90 multichaperone machine (48) (peptide interfaces less than 10 residues and below 700 Å2, KD in the μM range). The difference in affinity reflects the stability of the SRP68/72 complex in contrast to the diverse and reversible Hop interactions.
Figure 2.
Structure of the human SRP68/72-PBD complex. (A) Structure of the SRP68/72-PBD complex in side and front views of the solenoid with labeled TPRs and α-helices. Hydrophobic sidechains of SRP68-PBD are shown. (B) Electrostatic surface potential map (top, ±5 kBT/e) and sequence conservation (bottom) of SRP72-PBD. The SRP68-binding groove is mainly hydrophobic and the interface is highly conserved. (C) Details of the interface with the hydrogen-bonding network around D592 including the ‘amide ladder’.
Table 2. Average KD values and thermodynamic parameters of ITC measurements.
| syringe | cell | KD in nM | N | n | ΔH kcal mol−1 | TΔS kcal mol−1 | ΔG kcal mol−1 |
|---|---|---|---|---|---|---|---|
| SRP68-PBD | SRP72-PBD | 33 (±11) | 6 | 0.67 | −15.7 (±0.5) | 5.3 (±0.7) | −10.2 (±0.2) |
| SRP68-RBD | SRP RNA + SRP19 | 2.3 (±1.1) | 3 | 1.07 | −9.3 (±1.5) | −5.8 (±1.7) | −15.0 (±2.4) |
| SRP68-RBD | SRP RNA + SRP19 + SRP72-RBD | 2.1 (±0.3) | 3 | 1.07 | −11.0 (±1.0) | −1.4 (±1.4) | −12.5 (±0.7) |
| SRP72-RBD | SRP RNA + SRP19 | 427 (±105) | 7 | 0.99 | −29.9 (±3.1) | 21.1 (±3.4) | −8.7 (±0.1) |
| SRP72-RBD | SRP RNA + SRP19 + SRP68-RBD | 181 (±51) | 10 | 0.89 | −30.6 (±3.5) | 21.2 (±3.4) | −9.2 (±0.2) |
| SRP72-RBD | SRP RNA(U110C) + SRP19 + SRP68-RBD | 427 (±96) | 5 | 1.11 | −26.4 (±3.1) | 17.6 (±2.9) | −8.7 (±0.2) |
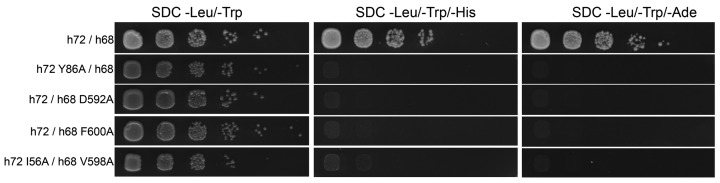
The SRP68/72 interaction is rather hydrophobic and the complex is stable in high salt conditions (1 M). Most prominent, the aromatic rings of the phenylalanines F590 and F600 within SRP68-PBD are anchored in hydrophobic pockets formed in between TPR4 and 5a, and TPR1 and 2, respectively (Figure 2C). The SRP68-PBD helix forms the center of the interface and is clamped in between TPR2, TPR3* and TPR4. Hydrogen bonding to the SRP68-PBD backbone are restricted to amide sidechains as typically found for ‘amide ladders’ in other TPR or Armadillo-repeat proteins (49). To test for the importance of individual residues on the interaction we performed yeast-two-hybrid assays (Figure 3). We show that the region P585-L607 of SRP68 is sufficient for SRP72 binding. Single alanine mutations of the strictly conserved residues Y86 and D592 abolished the binding completely, consistent with previous interaction studies (25) and underlining the central role of the intertwined hydrogen-bonding network between residues Y86, R90, Q117 and D592 (Figure 2C and Supplementary Figure S6). Similarly, the modification of the hydrophobic patch by the single mutant F600A and the double alanine mutant I56A/V598A resulted in a complete loss of binding.
Figure 3.
Yeast two-hybrid (Y2H) analysis of the SRP68/72-PBD complex. The Y2H is tested in both combinations of activating domain (AD) and binding domain (BD), but only BD-SRP72 and AD-SRP68 are shown. In the first panel, the interaction of the wild-type SRP72-PBD with SRP68-PBD shows the binding of the two domains. Single mutations Y86A of SRP72 (second panel), D592A or F600A of SRP68 (third and fourth panel), as well as the double mutation I56A(SRP72)—V598A(SRP68) (fifth panel) lead to a complete loss of binding. The Y2H of the inverted AD/BD combination is shown in Supplementary Figure S6B.
Structure of the human S domain with SRP72-RBD
While interaction between the PBDs of SRP68 and SRP72 results in the formation of a stable heterodimer, both proteins can bind to SRP RNA individually via their RBDs. Previously, we showed that the SRP68-RBD remodels the SRP RNA using a S domain RNA construct of 125 nucleotides (23). In order to characterize now the RNA-binding of SRP72, we reconstituted a quaternary human SRP sub-complex including the S domain RNA (using a longer RNA variant of 145 nucleotides from 105 to 249), SRP19, SRP68-RBD and SRP72-RBD. ITC measurements were performed to determine binding affinities of SRP68-RBD and SRP72-RBD to SRP RNA (Table 2 and Supplementary Figure S5). SRP19 was prebound in all experiments to SRP RNA in order to clamp helices 6 and 8 together and thereby to provide the binding platform for SRP68 and SRP72. SRP68-RBD binds strongly to SRP RNA (KD of 2 nM) independent of the presence of SRP72-RBD. In contrast, the binding affinity for SRP72-RBD is lower by two orders of magnitude with a twofold increase in presence of SRP68 (KD values of 181 and 427 nM, respectively). This indicates that RNA binding of SRP72 is enhanced by interaction with SRP68 or that a SRP68-induced conformational change in the SRP RNA facilitates recruitment of SRP72. Interestingly, while SRP68-RBD binding to SRP RNA is enthalpy-driven, SRP72-RBD binding is entropy-driven suggesting folding upon binding.
To answer this question, we set out to determine the structure of the human quaternary S domain including SRP72-RBD. Crystals could be obtained belonging to space group P212121 with two molecules in the asymmetric unit. The structure was solved by molecular replacement at 3.4 Å resolution (Figure 4A). The final model was refined to an Rwork (Rfree) of 23.7% (28.0%) and Ramachandran statistics show 97.8% of the residues in allowed regions (Table 1 and Supplementary Figure S3). SRP72-RBD and metal ion assignments were validated by mutational analyses and anomalous difference Fourier techniques (Table 1, Supplementary Figure S7 and Supplementary Table S2). The structure reveals the fold of the entire S domain RNA including the 5e-loop, that so far was only modeled (21,50). SRP19 and SRP68-RBD structures as well as the way they interact with SRP RNA are identical as in a ternary S domain complex solved previously (23) (Supplementary Figure S8). Especially the 20° kink at the RNA three-way junction as induced by SRP68-RBD and the insertion of its arginine-rich motif (ARM) into the major groove at the 5f-loop are maintained. Comparison of the RNA-binding site for SRP54 (asymmetric loop of helix 8) shows that it adopts a different structure than previously observed (23,51,52). The difference is due to a concerted helix-bending induced by crystal packing (Supplementary Figure S8) and reflects the plasticity of this RNA site. However, asymmetric loop flexibility has no influence on SRP72-RBD binding.
Figure 4.
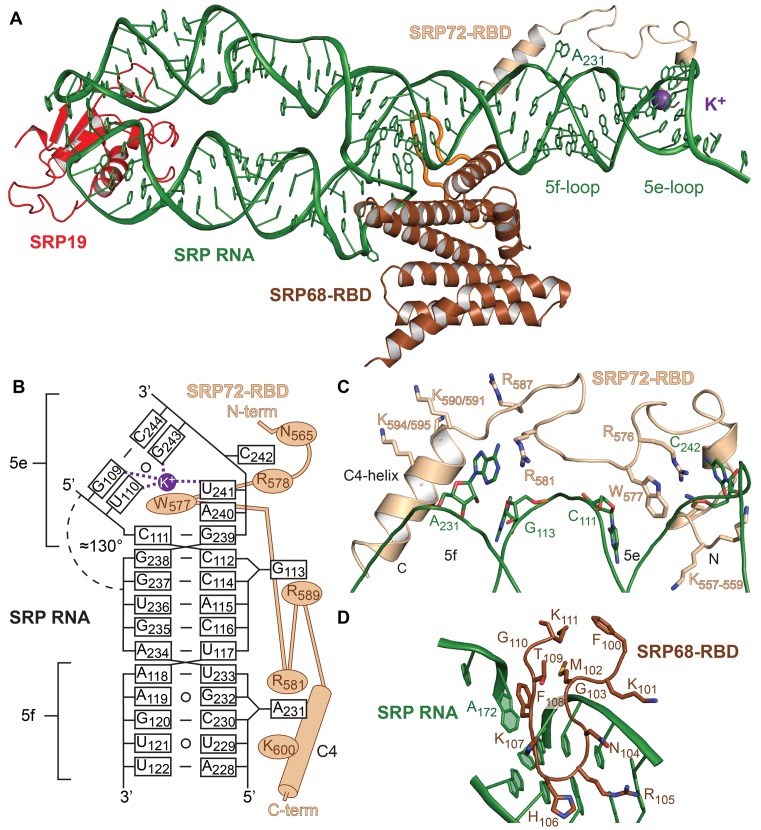
Structure of the human SRP S domain. (A) The quaternary SRP S domain complex. SRP72-RBD (sand) crawls along the 5e- and 5f-loops of SRP RNA (green). A potassium ion (purple sphere) is coordinated in the 5e-loop. The 5f-loop is indicated by the bulged-out adenine A231. The extended loop of SRP68-RBD (brown) in the RNA three-way junction is indicated in orange. (B) Scheme of SRP72-RBD binding to SRP RNA. Non-Watson Crick base pairs are indicated by open circles. (C) Detailed view of SRP72-RBD and its binding to SRP RNA. Residues R576 and W577 are clamped between nucleotides C111 and C242. Adenine A231 stacks onto R581. The lysine-rich region (K557-K559) binds to the 5e-loop and the C4-helix aligns with the 5f-loop. (D) The extended loop of SRP68-RBD (100-111) forms a β-hairpin motif and binds mainly to the backside of the 5f-loop and the SRP RNA three-way junction. The interaction includes a π-stacking between phenylalanine F108 and adenosine A172.
The SRP72-RBD (K557-Q603) appears as a linear peptide that crawls along the ‘distal site’ of SRP RNA at the 5e- and 5f-loop (Figure 4B and C). At the N-terminus, the lysine-rich motif involved in RNA-binding (21) is only partially ordered (K557-559) probably due to the limited length of our RNA construct (Supplementary Figure S9). Binding studies showed that the lysine-rich motif is needed for SRP72 binding to the SRP RNA (Supplementary Figure S9). The SRP72-Pfam motif (572PDPERWLPMRER583) is central to the interaction. The strictly conserved 572PDP574 motif is highly flexible and electron density is weak. Most prominent, the strictly conserved tryptophan (W577) inserts into the 5e-loop, which introduces a kink into SRP RNA (Figure 4B and C). Together with the neighboring strictly conserved arginine R576, the two residues form a continuous stack with the nucleobases of C111 and C242 thus stabilizing the 5e-loop geometry. Arginine R581 stabilizes nucleotide A231 of the 5f-loop in a bulged-out conformation. The 5f-loop is opened by the ARM of SRP68-RBD (23), suggesting an allosteric stimulation of SRP72-RBD binding (2-fold) as shown by our ITC data. Interestingly, while in the S domain structure without SRP72-RBD two nucleotides (A231 and G232) were found to be bulged-out, in the structure with SRP72-RBD guanine G232 is base-pairing with A119 underlining the previously observed plasticity of the 5f-loop (23). At the C-terminus following the Pfam motif, a positively charged helix (C4-helix) aligns with the 5f-loop region (Figure 4C) and together with nucleotide G113 mediates a crystal contact with SRP RNA (Supplementary Figure S8). A strictly conserved 603QG604 motif locates to the C-terminal end of the helix. The very C-terminus beyond the C4-helix and the caspase cleavage site (D614) is intrinsically disordered and not visible in the quaternary complex. The C-terminus of SRP72-RBD is located close to an extended SRP68-RBD loop (F100-K111) that forms a β-hairpin inserting in the three-way junction of SRP RNA connecting helices 5, 6 and 8 (Figure 4A and D). An exposed phenylalanine (F108) within this loop stacks onto an RNA U-turn as predicted (23).
The 5e-loop constitutes a potassium-binding RNA kink-turn
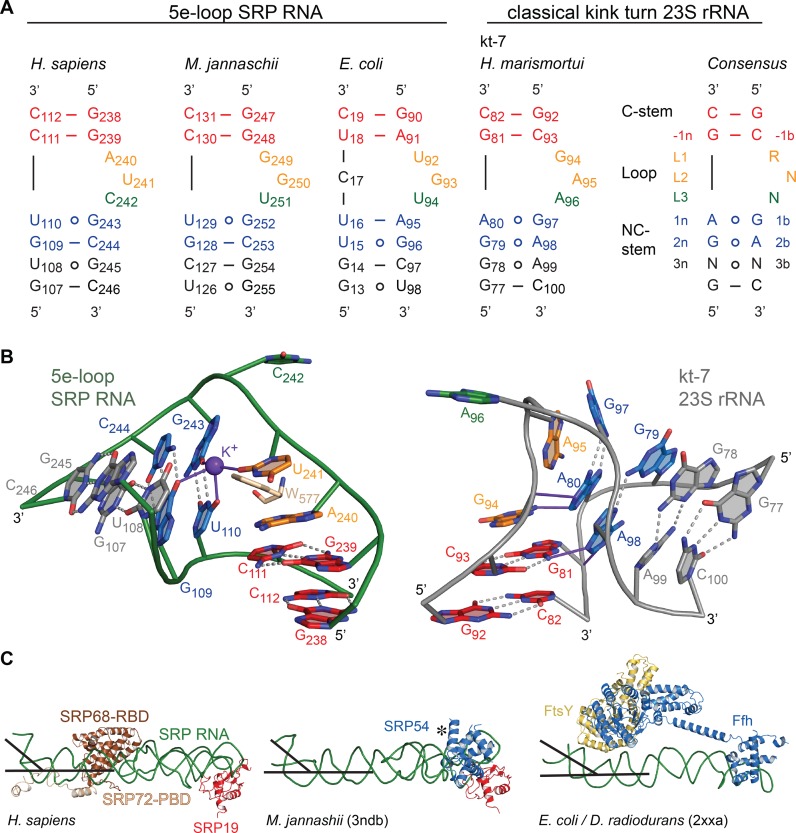
The 5e-loop has been described as a putative ‘kink-turn’ (K-turn) motif (21) that represents an extremely widespread RNA-folding unit present in ribosomal RNAs and riboswitches (53). Classical K-turns comprise a three-nucleotide bulge flanked by two highly conserved sheared A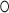 G pairs on the 3′ side (non-canonical NC-stem) and a Watson-Crick helix (canonical C-stem) at the 5′ side of the loop (Figure 5A) (54). The 5e-loop does not contain the A
G pairs on the 3′ side (non-canonical NC-stem) and a Watson-Crick helix (canonical C-stem) at the 5′ side of the loop (Figure 5A) (54). The 5e-loop does not contain the A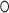 G pairs, and although resembling the canonical motif with three unpaired nucleotides (A240, U241 and C242), it significantly differs. Most strikingly, the 5e-loop is stabilized by the presence of a central potassium ion (Figure 5B), validated by cesium replacement (Supplementary Figure S7), which is not present in classical K-turns. We therefore define the 5e-loop geometry as ‘K+-turn’. The potassium locates to a central G
G pairs, and although resembling the canonical motif with three unpaired nucleotides (A240, U241 and C242), it significantly differs. Most strikingly, the 5e-loop is stabilized by the presence of a central potassium ion (Figure 5B), validated by cesium replacement (Supplementary Figure S7), which is not present in classical K-turns. We therefore define the 5e-loop geometry as ‘K+-turn’. The potassium locates to a central G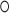 U wobble base pair starting the C-stem (U110
U wobble base pair starting the C-stem (U110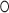 G243; -1b and -1n position in K-turn nomenclature (54)) and is coordinated by four oxygens of G109, U110, U241 and G243. The potassium bridges the NC- to the C-stem as in classical K-turns performed by the central adenines of the A
G243; -1b and -1n position in K-turn nomenclature (54)) and is coordinated by four oxygens of G109, U110, U241 and G243. The potassium bridges the NC- to the C-stem as in classical K-turns performed by the central adenines of the A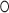 G pairs that are involved in A-minor motifs. C-stem superposition of the K+-turn with a classical K-turn indicates a diametric opposed kinking of the RNA (Figure 5B). Although different in shape, most K-turns serve as protein binding sites with varying structures (53). In order to test the influence of the potassium ion for SRP72 binding, we exchanged the central G
G pairs that are involved in A-minor motifs. C-stem superposition of the K+-turn with a classical K-turn indicates a diametric opposed kinking of the RNA (Figure 5B). Although different in shape, most K-turns serve as protein binding sites with varying structures (53). In order to test the influence of the potassium ion for SRP72 binding, we exchanged the central G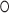 U wobble for a standard G-C base pair (U110C mutant) to impair the potassium coordination sphere. ITC experiments revealed a twofold reduced binding affinity compared to the wild-type RNA (KD of 427 nM, Table 2 and Supplementary Figure S5).
U wobble for a standard G-C base pair (U110C mutant) to impair the potassium coordination sphere. ITC experiments revealed a twofold reduced binding affinity compared to the wild-type RNA (KD of 427 nM, Table 2 and Supplementary Figure S5).
Figure 5.
Comparison of K-turns with SRP RNA 5e-loops. (A) The consensus sequence for classical K-turns, the K-turn kt-7 of Haloarcula marismortui and SRP RNA 5e-loops of representative members of all three kingdoms of life are shown. In the classical K-turn, the C-stem is formed by two G-C basepairs (red) and lies on the 5′ end of the loop composed of three bulged-out nucleotides (orange, green). Following on the 3′ site a conserved GA/AG (blue) pair forms the NC-stem, which is the most structurally conserved motif in K-turns and which is absent in the 5e-loop of the SRP RNAs. (B) Zoom into the human 5e-loop (green) and and the classical K-turn kt-7 of H. marismortui (54) (PDB 1jj2, gray). The conserved tryptophan W577 (sand) inserts into the 5e-loop and hydrogen bonds are formed in presence of a potassium ion between L2, 1b, 1n and 2n (purple). In the classical K-turn, the two adenines of the NC-stem form hydrogen bonds (purple) with the C-stem (-1n and L1) leading to a kink of approximately 120°. In contrast the SRP RNA 5e-loop kinks to the diametral opposite side of the RNA with approximately 50°. In both cases L3 (green) is bulged-out completely. (C) SRP RNA kinking at the 5e-loop region is present in all kingdoms of life (PDB 2xxa and 3ndb) (55,56). The similar kinking is highlighted by black lines. The kink provides an essential docking platform for the activated targeting complex consisting of the SRP GTPases Ffh (SRP54) and FtsY (SRα). The flexible NG domain (indicated by *) in 3ndb was removed for clarity.
Strikingly, a respective SRP RNA kink of similar extent (∼50°) is also present in bacteria and archaea (55,56) despite the absence of SRP68/72 (Figure 5C). In an archaeal structure, the three unpaired nucleotides are stacked into the loop (56) as typical for other SRP RNA bulge-loops in the absence of protein (23,51). The ligand sphere of potassium including the G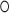 U wobble is present in archaea and according to sequence comparisons, the K+-turn might be a general feature of archaeal, eukaryotic, as well as Gram-positive bacterial SRP RNA (57). In Gram-negative bacteria the 5e-loop corresponds to loop E, which has been determined as ‘primary docking site’ for the targeting complex formed by Ffh (SRP54 in archaea and eukaryotes) and its receptor FtsY (SRα in eukaryotes) (7) (Figure 5C). The integrity of this ‘distal site’ has been found to be crucial for the GTPase-activating function of SRP RNA on the SRP GTPase heterodimer in E. coli (7,8). Our data showing a conserved SRP RNA kink indicate the general conservation of targeting complex activation by RNA. Such mechanism is supported by the conservation of the respective RNA interaction sites in the targeting complexes from all kingdoms of life (6).
U wobble is present in archaea and according to sequence comparisons, the K+-turn might be a general feature of archaeal, eukaryotic, as well as Gram-positive bacterial SRP RNA (57). In Gram-negative bacteria the 5e-loop corresponds to loop E, which has been determined as ‘primary docking site’ for the targeting complex formed by Ffh (SRP54 in archaea and eukaryotes) and its receptor FtsY (SRα in eukaryotes) (7) (Figure 5C). The integrity of this ‘distal site’ has been found to be crucial for the GTPase-activating function of SRP RNA on the SRP GTPase heterodimer in E. coli (7,8). Our data showing a conserved SRP RNA kink indicate the general conservation of targeting complex activation by RNA. Such mechanism is supported by the conservation of the respective RNA interaction sites in the targeting complexes from all kingdoms of life (6).
The SRP72-RBD in the interface of SRP/RNC complexes
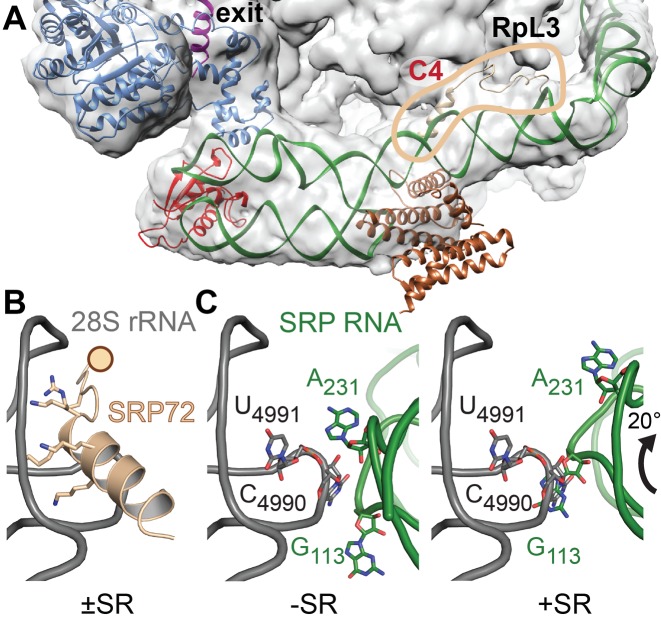
In order to analyze the function of SRP72-RBD for RNC interactions, the quaternary S domain structure was fitted into electron density maps of eukaryotic SRP-RNC and SRP/SR-RNC complexes previously obtained by cryo-electron microscopy (24,58) (Figure 6A). In these structures, SRP binds across the ribosomal surface, spanning from the polypeptide exit tunnel on the 60S subunit to the elongation factor binding site in the ribosomal subunit interface. The placement of our S domain structure suggests an intriguing function for SRP68-RBD and SRP72-RBD in guiding and modulating the SRP-RNC interaction at the C4-contact. The β-hairpin of SRP68-RBD in the SRP RNA three-way junction is in close contact with the 28S rRNA, an interaction that due to limited local resolution of the cryo-EM structure escaped previous analyses. SRP72-RBD is not resolved in the cryo-EM structure, but threads between SRP and the ribosome and contributes to the interaction with its PDP motif and the C-terminal C4-helix. The PDP motif is in proximity to an extended β-hairpin of the ribosomal protein RpL3 and apparently confers rigidity to the interaction. The C4-helix contains multiple positive charges (K590/591, K594/595) and is involved in a protein–RNA contact in our S domain crystal (Figure 4C and Supplementary Figure S8). On the ribosome, it reads out the 3′-terminal three-way junction of 28S rRNA (Figure 6B). However, rigid-body docking of the SRP onto the ribosome would result in a clash of the C4-helix with 28S rRNA (Supplementary Figure S10) suggesting a flexible tethering of the C4-helix within the S domain. Upon SR binding, eukaryotic SRP rotates at the C4-contact in respect to the ribosome (58). The rotation places the C4-helix within the ribosomal three-way junction in a very similar way as in our protein–RNA crystal contact. Therefore, we assume this contact to mimick the physiological C4-contact. The bulged-out nucleotide A231, which is stabilized by R581, contributes to the C4-contact possibly forming a base pair with the 28S rRNA (Figure 6C). SR dependent rotation of SRP induces a switch of this RNA–RNA contact and results in replacement of nucleotide A231 by G113. Thus, the C4-contact includes protein–protein, protein–RNA and RNA–RNA interactions (Supplementary Figure S10). It is maintained upon SR interaction, but adjusts during the SRP cycle. Of note, all ribosomal contacts locate to eukaryotic extensions not present in bacterial ribosomes, which do not establish a stable C4-contact with SRP (59).
Figure 6.
SRP–RNC interactions at the C4-contact. (A) Fit of the quaternary S domain complex into the cryo-EM density of a mammalian SRP–RNC complex (EMD-3037 (29)). SRP72-RBD, highlighted by the circle in sand, tethers the S domain on the ribosome (28S rRNA and protein RpL3 at the C4-contact. Large parts of SRP68/72 including the PBDs are flexible and therefore, without density. Color coding as in Figure 1. (B) The interaction of the SRP72-RBD C4-helix with the 3′-terminal three-way junction of 28S rRNA (gray) based on cryo-EM data for SRP/SR-RNC (58) and the human ribosome (PDB 4ugo). The rotation of SRP on the ribosome upon SR interaction requires flexible tethering of the ribosome-bound C4-helix (indicated by a sphere). (C) Rotation of SRP RNA induced by SR binding coincides with a switch of ribosomal contacts from the 5f-loop A231 (left panel) to G113 (right panel). Putative ribosomal base-pairing partners are indicated (C4990 and U4991). In the SR bound state the 5f-loop is exposed, ready to activate the targeting complex. SRP72 is omitted for clarity.
DISCUSSION
SRP72, the largest SRP protein, forms a stable heterodimer with SRP68 and is essential for SRP function. It is the only SRP protein, which is post-translationally modified by MAPK phosphorylation and contains a C-terminal caspase cleavage site. Caspase cleavage during apoptosis is discussed as a regulatory step for SRP-dependent protein secretion. Even though SRP72 is biochemically and pathologically well described, its structure and its functional implications have stayed enigmatic.
The presented crystal structures close this gap and show in detail how the N-terminal TPR domain of SRP72 enfolds a conserved C-terminal peptide of SRP68 and how SRP72 interacts with the potassium-bound 5e-loop of SRP RNA. Strikingly, SRP72 binding to the 5e-loop influences the adjacent 5f-loop, with a bulged-out nucleotide providing a binding platform for the ribosomal C4-contact. Docking of the S domain structure into cryo-EM densities of entire SRP/SR–RNC complexes reveals the C4-contact to be maintained during the SRP cycle and that SRP68, SRP72 and SRP RNA contribute to the ribosome contact. In addition the kink-turn of the 5e-loop seems to be present in all domains of life providing a binding platform for the targeting complex. On the basis of this work we propose the following scenario for the human SRP targeting cycle (Figure 7): Without SR, SRP72-RBD presents the 5f-loop (A231) to ribosomal RNA and the flexibly linked C4-helix tethers SRP onto the ribosome. Upon SR binding and docking of the targeting complex to the 5e-loop, SRP rotates and the 5f-loop becomes available for SRP GTPase activation as shown for the bacterial system (7,8) potentially involving two-bulged out nucleotides (A231 and G232) (23). The bulged-out nucleotide (G113) of SRP RNA takes over the RNA–RNA tertiary interaction and the SRP-ribosome contact is maintained in contrast to Gram-negative bacteria (59). This might explain why ribosomes are essential for the GTPase activation in eukaryotes in contrast to prokaryotes (60). Although the C4-helix of SRP72 stays in place, its conserved C-terminus might aid in GTP-hydrolysis. This idea is supported by the activation of an SRP GTPase dimer by a glutamine residue located in an α-helix of a protein partner as observed for the third SRP GTPase FlhF (involved in flagella biosynthesis) (61,62).
Figure 7.
Model for SRP72 function in the SRP/SR-RNC complex interface. Scheme for the interaction and repositioning of SRP in context of the RNC±SR (in orthogonal views). The 5e-loop is kinked (potassium ion shown as purple sphere) and forms the docking platform for the targeting complex. The targeting complex is embedded by the SRP68/72 solenoids. The rotation of SRP places the 5f-loop in the targeting complex interface. The C-terminal tail of SRP72 (dashed lines indicate intrinsic disorder, red line indicates caspase cleavage site) locates to the same region and may interact with the targeting complex.
The caspase cleavage and phosphorylation sites within SRP72 immediately follow the C4-helix and would come in close proximity to the re-localized targeting complex. Positively-charged surface patches on the targeting complex (6) are readily available to bind to phosphorylated serine residues within the SRP72 tail. As caspase cleavage removes the C-terminal tail, SRP targeting would be clearly impaired. This model matches with data on SRP72 cleavage in apoptosis, where protein secretion is indeed decreased (17,63) and in disease by generating an SRP72 epitope for autoimmune response finally resulting in myopathies or lupus (17,18).
In summary, although SRP is a conserved and ancient machinery in all kingdoms of life, its complexity in structure and regulation increased significantly during evolution. Our data underline that all players need to be carefully characterized on the atomic level to finally understand the mechanism of the whole machinery.
ACCESSION NUMBERS
Coordinates and structure factors are deposited in the RCSB protein data bank (PDB) with the accession numbers 5M72 and 5M73.
Acknowledgments
We acknowledge Friederike Schreiter and Jonathan Paulitz for technical assistance. We thank Yvonne Hackmann for human cDNA preparation; Gunter Stier for providing the pETTrx1a vector and Bernd Bukau for providing the pCA528 plasmid. We are grateful to Ed Hurt for sharing plasmids pG4ADN111, pG4ADC111, pG4BDN22 and pG4BDC22. We thank Jürgen Kopp and Claudia Siegmann from the BZH/Cluster of Excellence:CellNetworks crystallization platform. We acknowledge access to the beamlines at the European Synchrotron Radiation Facility (ESRF) in Grenoble and the support of the beamline scientists. I.S. is an investigator of the Cluster of Excellence:CellNetworks.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (DFG) [SFB638 to I.S., K.W., GRK1188 and the Leibniz programme to I.S.]. Funding for open access charge: DFG (Leibniz Programme).
Conflict of interest statement. None declared.
REFERENCES
- 1.Elvekrog M.M., Walter P. Dynamics of co-translational protein targeting. Curr. Opin. Chem. Biol. 2015;29:79–86. doi: 10.1016/j.cbpa.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nyathi Y., Wilkinson B.M., Pool M.R. Co-translational targeting and translocation of proteins to the endoplasmic reticulum. Biochim. Biophys. Acta Mol. Cell Res. 2013;1833:2392–2402. doi: 10.1016/j.bbamcr.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 3.Voorhees R.M., Hegde R.S. Toward a structural understanding of co-translational protein translocation. Curr. Opin. Cell Biol. 2016;41:91–99. doi: 10.1016/j.ceb.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Egea P.F., Shan S.-O., Napetschnig J., Savage D.F., Walter P., Stroud R.M. Substrate twinning activates the signal recognition particle and its receptor. Nature. 2004;427:215–221. doi: 10.1038/nature02250. [DOI] [PubMed] [Google Scholar]
- 5.Focia P.J., Shepotinovskaya I., Seidler J., Freymann D.M. Heterodimeric GTPase core of the SRP targeting complex. Science. 2004;303:373–377. doi: 10.1126/science.1090827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wild K., Bange G., Motiejunas D., Kribelbauer J., Hendricks A., Segnitz B., Wade R.C., Sinning I. Structural basis for conserved regulation and adaptation of the signal recognition particle targeting complex. J. Mol. Biol. 2016;428:2880–2897. doi: 10.1016/j.jmb.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 7.Shen K., Wang Y., Hwang Fu Y.H., Zhang Q., Feigon J., Shan S.O. Molecular mechanism of GTPase activation at the signal recognition particle (SRP) RNA distal end. J. Biol. Chem. 2013;288:36385–36397. doi: 10.1074/jbc.M113.513614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voigts-Hoffmann F., Schmitz N., Shen K., Shan S. ou, Ataide S.F., Ban N. The structural basis of FtsY recruitment and GTPase activation by SRP RNA. Mol. Cell. 2013;52:643–654. doi: 10.1016/j.molcel.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walter P., Gilmore R., Blobel G. Protein translocation across the endoplasmic reticulum. Cell. 1984;38:5–8. doi: 10.1016/0092-8674(84)90520-8. [DOI] [PubMed] [Google Scholar]
- 10.Wolin S.L., Walter P. Signal recognition particle mediates a transient elongation arrest of preprolactin in reticulocyte lysate. J. Cell Biol. 1989;109:2617–2622. doi: 10.1083/jcb.109.6.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wild K., Sinning I., Cusack S. Crystal structure of an early protein-RNA assembly complex of the signal recognition particle. Science. 2001;294:598–601. doi: 10.1126/science.1063839. [DOI] [PubMed] [Google Scholar]
- 12.Oubridge C., Kuglstatter A., Jovine L., Nagai K. Crystal structure of SRP19 in complex with the S domain of SRP RNA and its implication for the assembly of the signal recognition particle. Mol. Cell. 2002;9:1251–1261. doi: 10.1016/s1097-2765(02)00530-0. [DOI] [PubMed] [Google Scholar]
- 13.Lütcke H., Prehn S., Ashford A.J., Remus M., Frank R., Dobberstein B. Assembly of the 68- and 72-kD proteins of signal recognition particle with 7S RNA. J. Cell Biol. 1993;121:977–985. doi: 10.1083/jcb.121.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siegel V., Walter P. Each of the activities of signal recognition particle (SRP) is contained within a distinct domain: analysis of biochemical mutants of SRP. Cell. 1988;52:39–49. doi: 10.1016/0092-8674(88)90529-6. [DOI] [PubMed] [Google Scholar]
- 15.Grosshans H., Deinert K., Hurt E., Simos G. Biogenesis of the signal recognition particle (SRP) involves import of SRP proteins into the nucleolus, assembly with the SRP-RNA, and Xpo1p-mediated export. J. Cell Biol. 2001;153:745–761. doi: 10.1083/jcb.153.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lustig Y., Goldshmidt H., Uliel S., Michaeli S. The trypanosoma brucei signal recognition particle lacks the Alu-domain-binding proteins: purification and functional analysis of its binding proteins by RNAi. J. Cell Sci. 2005;118:4551–4562. doi: 10.1242/jcs.02578. [DOI] [PubMed] [Google Scholar]
- 17.Utz P.J., Hottelet M., Le T.M., Kim S.J., Geiger M.E., Van Venrooij W.J., Anderson P. The 72-kDa component of signal recognition particle is cleaved during apoptosis. J. Biol. Chem. 1998;273:35362–35370. doi: 10.1074/jbc.273.52.35362. [DOI] [PubMed] [Google Scholar]
- 18.Hengstman G.J.D., ter Laak H.J., Vree Egberts W.T.M., Lundberg I.E., Moutsopoulos H.M., Vencovsky J., Doria A., Mosca M., van Venrooij W.J., van Engelen B.G.M. Anti-signal recognition particle autoantibodies: marker of a necrotising myopathy. Ann. Rheum. Dis. 2006;65:1635–1638. doi: 10.1136/ard.2006.052191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirwan M., Walne A.J., Plagnol V., Velangi M., Ho A., Hossain U., Vulliamy T., Dokal I. Exome sequencing identifies autosomal-dominant SRP72 mutations associated with familial aplasia and myelodysplasia. Am. J. Hum. Genet. 2012;90:888–892. doi: 10.1016/j.ajhg.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iakhiaeva E., Bhuiyan S.H., Yin J., Zwieb C. Protein SRP68 of human signal recognition particle: identification of the RNA and SRP72 binding domains. Protein Sci. 2006;15:1290–1302. doi: 10.1110/ps.051861406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iakhiaeva E., Iakhiaev A., Zwieb C. Identification of amino acid residues in protein SRP72 required for binding to a kinked 5e motif of the human signal recognition particle RNA. BMC Mol. Biol. 2010;11:83. doi: 10.1186/1471-2199-11-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menichelli E., Isel C., Oubridge C., Nagai K. Protein-induced conformational changes of RNA during the assembly of human signal recognition particle. J. Mol. Biol. 2007;367:187–203. doi: 10.1016/j.jmb.2006.12.056. [DOI] [PubMed] [Google Scholar]
- 23.Grotwinkel J.T., Wild K., Segnitz B., Sinning I. SRP RNA remodeling by SRP68 explains its role in protein translocation. Science. 2014;344:101–104. doi: 10.1126/science.1249094. [DOI] [PubMed] [Google Scholar]
- 24.Halic M., Becker T., Pool M.R., Spahn C.M.T., Grassucci R.A., Frank J., Beckmann R. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427:808–814. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- 25.Iakhiaeva E., Hinck C.S., Hinck A.P., Zwieb C. Characterization of the SRP68/72 interface of human signal recognition particle by systematic site-directed mutagenesiss. Protein Sci. 2009;18:2183–2195. doi: 10.1002/pro.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D'Andrea L.D., Regan L. TPR proteins: The versatile helix. Trends Biochem. Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 27.Zeytuni N., Zarivach R. Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure. 2012;20:397–405. doi: 10.1016/j.str.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 28.Beckert B., Kedrov A., Sohmen D., Kempf G., Wild K., Sinning I., Stahlberg H., Wilson D.N., Beckmann R. Translational arrest by a prokaryotic signal recognition particle is mediated by RNA interactions. Nat. Struct. Mol. Biol. 2015;22:767–773. doi: 10.1038/nsmb.3086. [DOI] [PubMed] [Google Scholar]
- 29.Voorhees R.M., Hegde R.S. Structures of the scanning and engaged states of the mammalian SRP-ribosome complex. Elife. 2015;4:1–21. doi: 10.7554/eLife.07975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andréasson C., Fiaux J., Rampelt H., Mayer M.P., Bukau B. Hsp110 is a nucleotide-activated exchange factor for Hsp70. J. Biol. Chem. 2008;283:8877–8884. doi: 10.1074/jbc.M710063200. [DOI] [PubMed] [Google Scholar]
- 31.Bogomolovas J., Simon B., Sattler M., Stier G. Screening of fusion partners for high yield expression and purification of bioactive viscotoxins. Protein Expr. Purif. 2009;64:16–23. doi: 10.1016/j.pep.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 32.Thoms M., Thomson E., Baßler J., Gnädig M., Griesel S., Hurt E. The Exosome Is Recruited to RNA Substrates through Specific Adaptor Proteins. Cell. 2015;162:1029–1038. doi: 10.1016/j.cell.2015.07.060. [DOI] [PubMed] [Google Scholar]
- 33.Studier F.W. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 34.Hendrickson W. A., Horton J.R., LeMaster D.M. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price S.R., Ito N., Oubridge C., Avis J.M., Nagai K. Crystallization of RNA-protein complexes. I. Methods for the large-scale preparation of RNA suitable for crystallographic studies. J. Mol. Biol. 1995;249:398–408. doi: 10.1006/jmbi.1995.0305. [DOI] [PubMed] [Google Scholar]
- 36.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans P.R., Murshudov G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams P.D., Afonine P. V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terwilliger T.C., Adams P.D., Read R.J., McCoy A.J., Moriarty N.W., Grosse-Kunstleve R.W., Afonine P. V., Zwart P.H., Hung L.W. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Afonine P. V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schrödinger LLC. PyMOL Molecular Graphics System. 2010.
- 45.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 46.Zeytuni N., Baran D., Davidov G., Zarivach R. Inter-phylum structural conservation of the magnetosome-associated TPR-containing protein, MamA. J. Struct. Biol. 2012;180:479–487. doi: 10.1016/j.jsb.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 47.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 48.Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F.U., Moarefi I. Structure of TPR domain-peptide complexes: Critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 49.Jinek M., Rehwinkel J., Lazarus B.D., Izaurralde E., Hanover J.A., Conti E. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat Struct Mol Biol. 2004;11:1001–1007. doi: 10.1038/nsmb833. [DOI] [PubMed] [Google Scholar]
- 50.Iakhiaeva E., Wower J., Wower I.K., Zwieb C. The 5e motif of eukaryotic signal recognition particle RNA contains a conserved adenosine for the binding of SRP72. RNA. 2008;14:1143–1153. doi: 10.1261/rna.979508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wild K., Bange G., Bozkurt G., Segnitz B., Hendricks A., Sinning I. Structural insights into the assembly of the human and archaeal signal recognition particles. Acta Crystallogr. D Biol. Crystallogr. 2010;66:295–303. doi: 10.1107/S0907444910000879. [DOI] [PubMed] [Google Scholar]
- 52.Kuglstatter A., Oubridge C., Nagai K. Induced structural changes of 7SL RNA during the assembly of human signal recognition particle. Nat. Struct. Biol. 2002;9:740–744. doi: 10.1038/nsb843. [DOI] [PubMed] [Google Scholar]
- 53.Huang L., Lilley D.M.J. The kink turn, a key architectural element in RNA structure. J. Mol. Biol. 2016;428:790–801. doi: 10.1016/j.jmb.2015.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klein D.J., Schmeing T.M., Moore P.B., Steitz T.A. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ataide S.F., Schmitz N., Shen K., Ke A., Shan S., Doudna J. A., Ban N. The crystal structure of the signal recognition particle in complex with its receptor. Science. 2011;331:881–886. doi: 10.1126/science.1196473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hainzl T., Huang S., Merilainen G., Brannstrom K., Sauer-Eriksson A.E. Structural basis of signal-sequence recognition by the signal recognition particle. Nat. Struct. Mol. Biol. 2011;18:389–391. doi: 10.1038/nsmb.1994. [DOI] [PubMed] [Google Scholar]
- 57.Andersen E.S., Rosenblad M.A., Larsen N., Westergaard J.C., Burks J., Wower I.K., Wower J., Gorodkin J., Samuelsson T., Zwieb C. The tmRDB and SRPDB resources. Nucleic Acids Res. 2006;34:D163–D168. doi: 10.1093/nar/gkj142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Halic M., Gartmann M., Schlenker O., Mielke T., Pool M.R., Sinning I., Beckmann R. Signal recognition particle receptor exposes the ribosomal translocon binding site. Science. 2006;312:745–747. doi: 10.1126/science.1124864. [DOI] [PubMed] [Google Scholar]
- 59.Jomaa A., Boehringer D., Leibundgut M., Ban N. Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon. Nat. Commun. 2016;7:1–9. doi: 10.1038/ncomms10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bacher G., Lütcke H., Jungnickel B., Rapoport T. A., Dobberstein B. Regulation by the ribosome of the GTPase of the signal-recognition particle during protein targeting. Nature. 1996;381:248–251. doi: 10.1038/381248a0. [DOI] [PubMed] [Google Scholar]
- 61.Bange G., Kummerer N., Grudnik P., Lindner R., Petzold G., Kressler D., Hurt E., Wild K., Sinning I. Structural basis for the molecular evolution of SRP-GTPase activation by protein. Nat. Struct. Mol. Biol. 2011;18:1376–1380. doi: 10.1038/nsmb.2141. [DOI] [PubMed] [Google Scholar]
- 62.Bange G., Sinning I. SIMIBI twins in protein targeting and localization. Nat. Struct. Mol. Biol. 2013;20:776–780. doi: 10.1038/nsmb.2605. [DOI] [PubMed] [Google Scholar]
- 63.Breckenridge D.G., Germain M., Mathai J.P., Nguyen M., Shore G.C. Regulation of apoptosis by endoplasmic reticulum pathways. Oncogene. 2003;22:8608–8618. doi: 10.1038/sj.onc.1207108. [DOI] [PubMed] [Google Scholar]