Abstract
The functional relevance of the inverted repeat structure (IR/DR) in a subgroup of the Tc1/mariner superfamily of transposons has been enigmatic. In contrast to mariner transposition, where a topological filter suppresses single-ended reactions, the IR/DR orchestrates a regulatory mechanism to enforce synapsis of the transposon ends before cleavage by the transposase occurs. This ordered assembly process shepherds primary transposase binding to the inner 12DRs (where cleavage does not occur), followed by capture of the 12DR of the other transposon end. This extra layer of regulation suppresses aberrant, potentially genotoxic recombination activities, and the mobilization of internally deleted copies in the IR/DR subgroup, including Sleeping Beauty (SB). In contrast, internally deleted sequences (MITEs) are preferred substrates of mariner transposition, and this process is associated with the emergence of Hsmar1-derived miRNA genes in the human genome. Translating IR/DR regulation to in vitro evolution yielded an SB transposon version with optimized substrate recognition (pT4). The ends of SB transposons excised by a K248A excision+/integration- transposase variant are processed by hairpin resolution, representing a link between phylogenetically, and mechanistically different recombination reactions, such as V(D)J recombination and transposition. Such variants generated by random mutation might stabilize transposon-host interactions or prepare the transposon for a horizontal transfer.
INTRODUCTION
DNA recombination inherently involves breakage and joining of distant DNA sites. The recombination reactions require two major functional components: a recombinase protein and specific DNA sites at which the recombinase binds and executes recombination. A highly conserved catalytic domain, containing a DDE signature, commonly characterizes many recombinases (1). The DDE superfamily is widespread from prokaryotes to humans, including transposases encoded by the bacterial IS elements and the Tc1/mariner family of eukaryotic DNA-transposons, retroviral integrases and the RAG1 recombinase of V(D)J recombination, a transposition-derived process (2) that generates the immunglobulin repertoire of the adaptive immune system in vertebrates. The growing numbers of solved crystal structures of various recombinases (3–7) reveal that although these enzymes catalyse similar chemical reactions, there are also important differences in how the different elements process the reaction.
Namely, although all DDE transposases initiate the DNA cleavage reaction with a single-stranded nick at the end of the transposon, second-strand-processing can proceed in three different ways (reviewed in (8)). Cleavage of the second strand can be achieved via a hairpin intermediate that forms either on the transposon end (e.g. Tn5) or at the ends of the cleaved genomic DNA (hAT transposons, V(D)J recombination (reviewed in (8)). In contrast, the mariner elements and Sleeping Beauty (SB), members of the Tc1/mariner family, do not transpose via a hairpin intermediate (9,10), indicating that double-strand cleavage is the result of two sequential hydrolysis reactions by the transposase (11).
In transpositional DNA recombination reactions, the DNA sites, between which the recombination reaction occurs, are strictly defined in order to limit risks on genome stability posed by inter-chromosomal recombination between unlinked transposons scattered around the genome. This strict definition is provided by a requirement for a synapsis of the two ends of the same transposon before any catalytic step can commence. Yet, occasional cleavage events initiated at seemingly unsynapsed sites were observed in RAG1-mediated recombination, as well as in mariner, piggyBac or SB transposition at different frequencies (3,9,12–16). Such events likely result from bimolecular pairing of transposon ends, when synapsis occurs between two separate transposon molecules (16).
Enforcement of synapsis of the transposon ends varies among recombinases. In the transposition of the bacterial elements Mu, Tn5 and Tn10, the synaptic complex has a trans architecture, that is, the monomeric transposase must bind to the other transposon end before dimerization and catalysis to occur. Thus, the trans architecture of the synaptic complex couples the initiation of catalysis and synapsis, thereby suppressing non-canonical reactions. In contrast, RAG1 recombinase and the mariner transposase bind the recognition site as dimers, capable of performing catalysis without synapsis (reviewed in (8)), suggesting that non-canonical recombination events need to be suppressed by other regulatory mechanisms. In V(D)J recombination, unpaired reaction products are filtered out by a highly controlled, ordered assembly process, assisted by a cellular factor, HMGB1 (17–19). In mariner transposition, a topological filter suppresses promiscuous synapses of unlinked transposon ends (14). In these reactions a conformational change of the transposase couples synapsis and cleavage that helps to filter out aberrant recombination products (3,9,11,13,14).
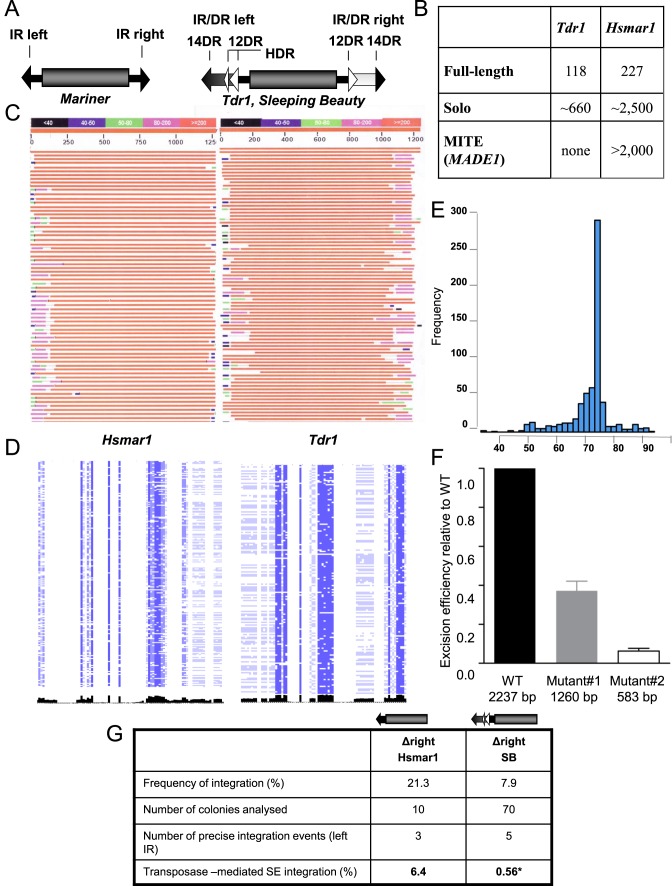
In addition to the transposase, the transposon terminal inverted repeats (IRs) can also potentially contribute a regulatory role to synaptic complex assembly. While mariners have short IRs with one transposon binding site at each transposon end, SB belongs to the inverted repeat/direct repeat (IR/DR) subfamily of transposons, possessing two transposase binding sites (represented by direct repeats, DRs) at each transposon end (reviewed in (20) (Figure 1A). The left IR contains an additional motif (HDR) that acts as an enhancer in SB transposition (21). While the IR/DR is a strict requirement of SB transposition (22), our understanding of its role in the transposition process is limited.
Figure 1.
Genome wide structure of Tc1/mariner elements. (A) Structural comparison of the representatives of two subfamilies of the Tc1/mariner transposons. The transposase coding sequence (gray cylinder) is flanked by either simple- or IR/DR-type inverted repeats (IRs). In mariners, the simple terminal IRs 30 bp) contain a single recognition motif per IRs. The IR/DR elements possessing longer (230 bp) terminal IRs (arrows), with two recognition sequences (30 bp) per IRs, repeated twice in a directly repeated form (DRs). The left IR carries a transpositional enhancer motif (HDR) (13 bp) (21). (B) Genomic structure of the zebrafish Tdr1 (IR/DR) and the human Hsmar1 (IR) elements. For the definition of the categories see Materials and Methods. (C) The structure of genomic copies of the human Hsmar1 (left) and the zebrafish Tdr1 (right) elements. A graphical overview of a representative collection of a BLAST search is shown. Note the polymorphic regions at the ends of the IRs (‘self inflicted wounds’). (D) Genomic alignments of the ends of Hsmar1 and Tdr1 sequences. Pairwise alignment of left and right terminal sequences, concatenated from full-length Hsmar1 and Tdr1 (30 and 60 bp), respectively. Similarity scores are visualized in black. Note that compared to Tdr1, left and right terminal sequences are less similar to each other in Hsmar1. (E) Amplification of a specific structure of Hsmar1-derived MITEs. The genomic frequencies of various Hsmar1-derived sequences are shown. Distances between the left IR start site and right IR end site are plotted. A 78 bp long MITE variant, containing a 6 bp long intervening sequence between the 36 bp long terminal IRs (annotated as MADE1) has been amplified. Note that the number of MADE1-like sequences, including solo configuration (5 kb window) is much higher (∼8000, not shown). Here, only copies having a 100% identity to the Repbase sequence are accounted. (F) Effect of internal deletions on the frequency of Sleeping Beauty (SB) transposon excision. Wild type and two internally deleted transposon versions were subjected to an excision assay. The assay measures the restoration of the open reading frame of GFP upon transposon excision by FACS. (G) Estimating transposon-mediated single ended transposition events. Transposition frequency of Hsmar1 or SB (25,26) transposons, lacking the right IRs is shown relative to the respective wild type transposons. Wild type and mutated transposons lacking the right IRs were subjected to transposition assay in HeLa cells with active (Hsmar1-Ra; SB) or inactive (Hsmar1-cons; SB-D3) transposase versions. Integration frequencies were calculated as a ratio of colony numbers obtained with active versus inactive transposases. To identify transposon-mediated single ended (SE) integration events, the surrounding of transposon integration sites were analysed by linker-mediated PCR in the indicated number of colonies. Only precise left IR integrations (flanked by TA) to the canonical transposon target site were interpreted as single ended integration. *Data of single ended SB integration is based on (16). Δ signs refer to deletion.
No active members of the Tc1/mariner family have been isolated from vertebrate genomes. In an attempt to decipher how simple IR- and IR/DR-type elements are regulated, we first analysed genomic copies generated by past activities of Tdr1 (IR/DR type) and Hsmar1 (simple IR) transposons in the zebrafish and human genome, respectively. In addition, we compared the transposition reactions of resurrected, active versions of Hsmar1 (simple IR) and SB (IR/DR) elements. In a systematic approach, we dissected both the transposon and the transposase of SB to small, functional domains, and addressed their contribution to the transposition process. Our data obtained by using a combination of in vivo, in vitro and in silico approaches suggest that the IR/DR structure might have evolved to promote an ‘ordered assembly’ process of transposon-transposase complexes. This tight regulation suppresses abnormal recombination activities, and more efficiently inhibits the mobilization of short, internally deleted copies of the IR/DR subfamily of transposons, including SB.
Our mechanistic studies combined with molecular evolution resulted in a sequence variant of the transposon IR with improved substrate recognition by the SB transposase. Furthermore, we identified a genetic variant of the SB transposase that can trigger a transition between mechanistically different recombination reactions, such as V(D)J recombination or transposition. Unusually, this transposase variant liberates the transposon by hairpin resolution, and generates an extrachromosomal product. Such variants generated by random mutation might stabilize transposon-host interactions or prepare the transposon for horizontal transfer.
The inactivation process included the generation of mutagenized IRs, known as ‘self-inflicted wounds’ (23), disabling both types of transposons from remobilization. Furthermore, mariners, but not the IR/DR elements seem to give rise to internally deleted, shorter copies that are mobilized more frequently than full-length elements (26). These shorter variants eventually outcompete the transposition of autonomous, transposase encoding elements, and contribute to the inactivation process. In the human genome, this process is associated with the emergence of Hsmar1-derived MITEs that gave rise to a large number of miRNA genes.
MATERIALS AND METHODS
Plasmid constructs
Prokaryotic vectors pET-21a/N57, pET-21a/58-123 and pET-21a/N123 expressing hexahistidine-tagged (HIS) subdomains of the SB DNA-binding domain, PAI, RED and N123 respectively, has been described previously (21); pET28/HMGB1 expressing HIS-tagged version of HMGB1 was kindly provided by M. Bianchi, Milan, Italy (described in (24)). For expression of the SB transposase in HeLa cells a pCMV-SB10 (25) and pCMV-SBD3 (D3), a catalytic mutant (E278D) of SB, has been used; pCMV-Hsmar1 and pCMV-Hsmar1-Ra express the inactive and active versions of Hsmar1 transposase, respectively (26). As donor plasmids in in vivo assays the following constructs have been used: pT/neo described previously (25); pHsmar1-neo and pHsmar1-neo-left lacking the right IR (26). The pre-cleaved transposon substrate, pTBsaXI contains identical left IRs generated by amplifying the transposon using the primer of T-prclvd: ACGTCAGCTCCTACCCTACAGTTGAAGTCGGAAGTTTACATACAC.
Cloning
The mutated SB transposon ends were created by PCR-mediated mutagenesis (for details see Table 1).
Table 1. List of primer sequences and cloning strategies.
| Constructs | Primer sequences | Template of the PCR | Cloning strategy |
|---|---|---|---|
| Construct 2 | 5′-tacagtgacgaccccaagtgtacatacacgcgccccaaatacat-3′ | pT/neo | Ligate to SmaI site of pUC19 |
| 5′-tacagtgacgaccccaagtgtacatacacgcgccttggagtcatta-3′ | |||
| Construct 3 | 5′-gtacatacacgcgcttagtatttggtagcattgccttta-3′ | pT/neo | Ligate the PCR products |
| 5′-gtacatacacgcgcttgactgtgcctttaaacagcttgg-3′ | |||
| 5′-acttggggtcgtcaccaattgtgatacagtgaattataagtg-3′ | pT/neo | ||
| 5′-acttggggtcgtcaccgaatgtgatgaaagaaataaaagc-3′ | |||
| Construct 4 | 5′-gtacatacacgcgcttagtatttggtagcattgccttta-3′ | pT/neo | Ligate the PCR products |
| 5′-gtacatacacgcgcttgactgtgcctttaaacagcttgg-3′ | |||
| 5′-acttggggtcgtcaccaattgtgatacagtgaattataagtg-3′ | Construct2 | ||
| 5′-acttggggtcgtcaccgaatgtgatgaaagaaataaaagc-3′ | |||
| Construct 5 | 5′-acttccgacttcaactgtaggggatcctctagagtcgacctg-3′ | pT/neo | Ligate the PCR products |
| 5′-acttccgacttcaactgtagggtaccgagctcgaattcactg-3′ | |||
| 5′-gtacatacacgcgccccaaatacatttaaactcactttttc-3′ | pT/neo | ||
| 5′-gtacatacacgcgccttggagtcattaaaactcgtttttc-3′ | |||
| Construct 6 (pT4) | 5′-acttctgacccactgggaatgtgatgaaagaaataaaagc-3′ | pT/neo | Ligate the PCR products |
| 5′-acttctgacccactggaattgtgatacagtgaattataagtg-3′ | |||
| 5′-gtacatacacgcgcttagtatttggtagcattgccttta-3′ | pT/neo | ||
| 5′-gtacatacacgcgcttgactgtgcctttaaacagcttgg-3′ | |||
| Construct 7 | 5′-gtacatacacgcgcttagtatttggtagcattgccttta-3′ | pT/neo | Ligate the PCR products |
| 5′-gtacatacacgcgcttgactgtgcctttaaacagcttgg-3′ | |||
| 5′-acttctgacccactgggaatgtgatgaaagaaataaaagc-3′ | Construct 5 | ||
| 5′-acttctgacccactggaattgtgatacagtgaattataagtg-3′ |
Protein expression and purification
Expression and purification of His-tagged PAI and RED subdomains were conducted as described in (21). The His-tagged HMGB1 was expressed in Escherichia coli BL21(DE3) upon addition of 0.8 mM IPTG at OD (A600) ∼0.6 and growth at 30°C for 4 h. The bacterial pellet was resuspended in lysis buffer (50 mM NaH2PO4 pH 7.8, 300 mM NaCl and 10 mM imidazole) containing 1 COMPLETE Mini Tablet (Roche) and sonicated. Purification was done on Ni-NTA Spin Columns (QIAGEN) according to the manufacturer's protocol.
Electromobility shift assay (EMSA)
Double-stranded oligonucleotides corresponding to either 12 or 14DRs were end-labeled using [α-32P]dCTP and Klenow fragment. The DNA probe containing the left IR was a EcoRI fragment of the pT/neo, end-labeled with [α-32P]dATP. Following the Klenow reaction, the labeled DNA was purified on MicroSpin G-25 Columns as described by the manufacturer. Binding reactions were performed in 20 mM HEPES (pH 7.5), 0.1 mM EDTA, 1 mM DTT, 20 000–50 000 cpm labeled DNA probe and various concentrations of the proteins (as noted in the figs) were added in a total volume of 10 μl, and incubated 10 min on ice. After addition of 3 μl of loading dye (containing 50% glycerol and bromophenol blue) the samples were loaded onto a 4% or 6% polyacrylamide gel. The electrophoresis was carried out in Tris-glycine buffer pH 8.3 at 25 mA for 2–3 h. The gels were dried for 45 minutes using the gel dryer from BIORAD. After overnight exposure the gels were scanned with Fujifilm FLA-3000 and analysed with AIDA program.
Sequence of probes used in the experiments:
14DR; 12DR;
CAST-2-S 5′-acatacaccctggtgtatgtaaagatcggacggccggttgg-3′
CAST2-AS 5′-gactccaaccggccgtccgatctttacatacaccagggtgtatgt-3′;
CAST-5-S 5′-acatacaggcgcgtgtatgtacacttggggtcgtcacttgg-3′
CAST-5-AS 5′-gactccaagtgacgaccccaagtgtacatacacgcgcctgtatgt-3′
CAST-9-S 5′-acatacagcaccatgtacttaaatctctgacctgggcttgg-3′
CAST-9-AS 5′-gactccaagcccaggtcagagatttaagtacatggtgctgtatgt-3′
CAST-20-S 5′-acatacacgtaagtgtacatactgtgtacacaaagacttgg-3′
CAST-20-AS 5′-gactccaagtctttgtgtacacagtatgtacacttacgtgtatgt-3′
Chemical crosslinking
Reactions were performed using the bis(sulfosuccinimidyl) substrate (BS3, Pierce Biotechnology, USA) according to manufacturer's recommendations, and as in (21). Briefly, SB derivative RED (N58-123, 3μM) was incubated on ice in 20 mM HEPES (pH 7.5), 5 mM MgCl2, 100 mM NaCl and 2.5 mM BS3 (bis(sulfosuccinimidyl), Pierce), in a final volume of 15 μl for 2 h. The reactions were stopped by adding Tris–HCl pH 7.5 to a final concentration of 50 mM and incubating 10 min at room temperature. Then the Laemmli buffer (125 mM Tris–HCl pH 6.8, 5% SDS, 10% ß-mercaptoethanol, 25% glycerol and bromophenol blue) was added and samples were loaded on 15% SDS-PAGE and analysed by Western blotting using polyclonal anti-SB antibody (R&D Systems, USA) and anti-goat IgG (Pierce Biotechnology, USA).
CASTing experiment
The CASTing was performed based on the method described in (Wright, Binder et al. 1991). Oligonucleotides with random 35 bp long core SB-DOL: 5′-GCG GGA TCC ACT CCA GGC CGG ATG CT (N)35 CAC CAG GGT GTA AGG CGG ATC CCG C-3′ were synthesized and made double-stranded in a PCR reaction with primers complementary to the sequences flanking the core. The nucleoprotein complexes formed during 1 h incubation of 2 μg of the oligonucleotides with 0.15 μg of the purified His-tagged SB transposase (SBFl-6H) (21) were recovered using the Ni-NTA resin (QIAGEN). The bound oligonucleotides were enriched by extensive washing steps. The selected oligonucleotides were extracted and amplified by primers A, 5′-GCG GGA TCC GCC TTA CAC CCT GGT G-3′ and B, 5′-GCG GGA TCC ACT CCA GGC CGG ATG CT-3′ and subjected to additional rounds of the CASTing cycle to increase the specificity of the method. The oligonucleotides obtained from sixth round were sequenced and tested in binding and transposition assays.
Cell culture
HeLa cells were grown in DMEM (GIBCO BRL, Germany) supplemented with 10% Fetal Calf Serum Gold (FCS Gold) (PAA, Germany) and 1% antimycotic antibiotic (Invitrogen, Germany). One day prior transfection cells were seeded onto six-well plates. Cells were transfected with Qiagen purified DNA (Qiaprep spin miniprep kit, Qiagen) using jetPEI RGD transfection reagent (Polyplus Transfection, France). Two days posttransfection cells were harvested for excision assay and/or were plated out on 10 cm plates for selection using 1 mg/ml G418 (Biochrom, Germany). After 3 weeks of selection, colonies were stained and counted as described in (25).
PCR-based excision assay
In order to analyze excision sites of the PCR-based excision assay, plasmid DNA was isolated from harvested cells using the QIAprep Spin Miniprep protocol with small modifications. Instead of the P2 buffer, 1.2% SDS and 0.1 μg/μl Proteinase K was added. DNA was eluted in 50 μl Elution Buffer and 2 μl was used as template for PCR. The PCR was performed with Taq polymerase and 10 pmol of primers aligning to the pUC19 backbone sequence in order to obtain the excision site (primer sequences 5′-cagtaagagaattatgcagtgctgcc-3′ and 5′-cctctgacacatgcagctcccgg-3′). The PCR product was diluted 1:100 and 1 μl was subjected to another round of PCR with nested primers (primer sequences 5′-gcgaaagggggatgtgctgcaagg-3′ and 5′-cagctggcacgacaggtttcccg-3′). The PCR program used in the assay is: 94°C 5 min; 30× (94°C 30 s, 65°C 30 s, 72°C 30 s); 72°C 5 min, 4°C ∞. To normalize the PCR conditions for excision assay, a PCR for ampicillin gene (5′-tgcacgagtgggttacatcgaact-3′ and 5′-ttgttgccattgctacaggcatcg-3′) was performed using PCR program: 94°C, 5 min 15× (94°C 30 s, 68°C 30 s 72°C 20 s); 72°C 5 min 4°C ∞. The products were visualized on 1.2% agarose gel.
Genomic DNA isolation
Cells collected from 12-well or 6-well plates were washed with PBS, then lysed in lysis buffer (10 mM Tris pH 8.5, 5 mM EDTA, 0.5% SDS, 200 mM NaCl and 100 μg/ml Proteinase K) by incubating them at 37°C overnight. Next the DNA was ethanol precipitated and the pellets were dissolved in TE buffer (10 mM Tris pH 8.0, 1 mM EDTA) containing 0.1 mg/ml RNase.
Genomic analysis of transposon integration sites
For SB transposon. 1 μg of genomic DNA was digested with BglII and BclI restriction enzymes, then after precipitation the DNA fragments were circularized by ligation overnight at 16°C in a large volumes to facilitate self-ligation. After precipitation the DNA samples were dissolved in 30 μl TE buffer and 2 μl was used as template for PCR reactions. Nested PCRs were conducted with primers aligning to the neomycin resistance gene (neo) and SB inverted repeat sequences (IRs). Sequence of the PCR primers are the following; neo 5′- ccttgcgcagctgtgctcgacg-3′, cgtcgagcacagctgcgcaagg in the 1. PCR: SB primer 5′-ctcatcaatgtatcttatcatgtctgg-3′, in the 2. PCR: Nested SB primer 5′-cttgtgtcatgcacaaagtagatgtcc-3′. For the Hsmar1 transposon. 1 μg of genomic DNA was digested with FspBI enzyme and ligated to annealed double stranded FspBI linkers (FspBI linker (+) 5′-gtaatacgactcactatagggctccgcttaagggac-3′; FspBI linker (–) 5′ P-tagtcccttaagcggag-amino 3′). One-fifth part of the ligation reaction was used as template for nested PCR reactions carried out with Q5 DNA polymerase. The primers are hybridizing to the linker and to the transposon cargo sequence downstream to the left Hsmar1 transposon end. Sequence of the PCR primers are the following; in the 1. PCR: Linker primer 5′-gtaatacgactcactatagggc-3′, SVpA Rev1 5′-gtggtttgtccaaactcatcaatgt-3′; in the 2. PCR: Nested primer 5′-agggctccgcttaagggac-3′, SVpA Rev2 5′-tcttatcatgtctggatcgggt-3′. The PCR products were extracted from the gel and sequenced.
Sleeping Beauty transposon excision assay using GFP reporter
To evaluate the effects of the length of the internal sequence of the SB transposon on excision efficiency, 977- and 1654-bp sequences (containing partial SV40-neo) were removed from the pCMV(CAT)-GFP/pT2Neo reporter, to derive alternative excision reporters with shorter internal sequences (1260 and 583 bp, respectively). In detail, the reporter construct pCMV(CAT)-GFP/pT2Neo (7) was digested with RsrII and NsiI to remove a 977 bp internal sequence between the inverted repeats, and religated (Mutant#1,1200 bp). Mutant#2 was generated by partially digesting the pCMV(CAT)-GFP/pT2Neo with HindIII to remove 1654 bp internal sequence, and religated. The three transposon constructs were purified using the Qiagen plasmid midi kit. The plasmid DNA was transfected into HeLa cells with the transposase-expressing plasmid pCMV(CAT)SB100X (27) using jetPEI (Polyplus transfection, for mammalian cells) according to instructions of manufacturer. Three days later we estimated the number of GFP-positive cells by FACS.
Genome-wide analyses of the Tdr1 (zebrafish) and Hsmar1 (human)
Genome wide alignments
Hsmar1 and Tdr1 coordinates were filtered from their respective repeat masked genomes, downloaded from UCSC table browsers (https://genome.ucsc.edu/cgi-bin/hgTables). We employed bedtools https://bedtools.readthedocs.io/en/latest/ to fetch their sequences from Human Reference Sequence (hg19/ GRCh37) and Genome Reference Consortium Zebrafish Build 10 (danRer10/GRCz10), respectively. We aligned sequences with their reference sequence from Repbase (http://www.girinst.org/repbase). The sequences were aligned using MUSCLE (28) sequence length and visualized by the jalview tool (http://www.jalview.org/).
Genome-wide analysis of transposon ends
We used full-length Hsmar1 and Tdr1 sequences from Repbase. We concatenated sequences from left and right terminal inverted repeats (30 bp from each end) to build in silico MITE sequence. Alignments were by visualized by the jalview tool.
Identifying solo and MITE structures
We used BLAT (https://users.soe.ucsc.edu/∼kent) to identify genomic sequences on both strands that match a full-length element, solo terminal IRs and MITEs of Hsmar1 and Tdr1 from human and zebrafish genomes, respectively. We analysed genomic coordinates with 90% or higher identity scores further. Genomic coordinates were post-processed to filter in solo terminal Inverted Repeats (solo IRs) and MITEs. For accounting solo terminal IRs, we used a 5 kb window (an IR sequence is not in the proximity of 5 kb with a previous one.) Post-processing included a cross-check with RepeatMasker (http://www.repeatmasker.org/) sequences of Hsmar1 and Tdr1. Upon identifying Hsmar1-derived sequences in the human genome, we noticed that most of solo IRs and MITEs are annotated as MADE1 repeat family as Hsmar1 dependent Tc1/mariner (http://www.repeatmasker.org/cgi-bin/ViewRepeat?id=MADE1). MADE1 is represented by ∼8000 copies in human genome, from which 660 are 100% identical to the Repbase sequence (http://www.girinst.org/repbase).
RESULTS
Genome-wide comparative analysis of simple-IR- and IR/DR-type elements
We analyzed genomic copies of two transposon families, possessing either simple or IR/DR-type inverted repeats with respect to copy number, primary DNA sequence and subfamily structure. Since the transpositionally active, synthetic Sleeping Beauty is resurrected from various fish genomes (25), we have chosen a related IR/DR-type transposon, Tdr1 (29) from the zebrafish genome as a model, and compared it to genomic copies of the simple-IR-type human mariner element Hsmar1 (26,30) (Figure 1A). Genome-wide analysis revealed that both transposons accumulated large copy numbers in their respective hosts (Figure 1B), however neither family is mobile at present, due to inactivating mutations (26,29). The most obvious feature of both Tdr1 and Hsmar1 transposon copies is a high degree of mutational damage of their IRs, especially at the very ends of the transposons (Figure 1C). These ‘self-inflicted wounds’, originally described for the mariner elements (23), are generated by transposase nicking at the transposon IRs. ‘Self-inflicted wounds’ are clearly detectable among the Tdr1 copies but, in contrast to Hsmar1, seem to occur more symmetrically at both ends of the transposons (Figure 1C). Our genome-wide analysis supports this observation, suggesting that the process generating mutations at the ends of the Tdr1 transposon occurs in a more concerted manner when compared to Hsmar1 s (Figure 1D).
The efficiency of both mariner and SB transposition correlates negatively with increasing transposon size (22,31), suggesting that internally deleted transposon copies may have an advantage in transposition, and therefore would be predicted to accumulate higher copy numbers in the genome over time. Alignment of genomic transposon copies suggests that the structure of the internally deleted copies of Tdr1 and Hsmar1 are different. While the truncated Tdr1 copies always carried sequences of various lengths between the IR/DRs (over 400 bps, Supplementary Figure S1), variants lacking the entire internal part, and basically consisting only of the IRs are detectable only in the Hsmar1 population (Supplementary Figure S2). These structures are known as miniature inverted repeat transposable elements or MITEs, and seem to be generally associated with mariners (30,32,33). Curiously, while various Hsmar1-derived MITE-like structures are detectable in the human genome, one particular variant, consisting from two, 37-bp IRs linked by a 6-bp intervening sequence appears to have been preferentially amplified (Figure 1E). Some of these MITEs are annotated as MADE1 (30), and are processed by the RNA interference enzymatic machinery to form 22-nt mature miRNA sequences (hsa-mir-548) (34). Six of these a ‘domesticated’ Hsmar1-derived miRNA genes have been implicated to have a primate specific, cancer-related regulatory role (34). Our analysis identified around 300 MADE1 MITEs with a 100% sequence identity to the consensus sequence in Repbase (35) (Figure 1E), predicting that the number (and the impact) of Hsmar1-derived miRNA genes is much higher than previously estimated. Intriguingly, despite the fact that MITEs (of different origin) are present in the zebrafish genome (36), no Tdr1-derived MITE sequences could be identified, suggesting that the transposition of IR/DR-type elements might not support the amplification of MITE-like structures. Finally, Hsmar1-derived solo IRs greatly outnumber the full-length copies (∼2500 copies versus ∼200 copies, respectively, a 12.5× ratio) (37) in the human genome, while we estimate this ratio significantly lower for Tdr1 in the zebrafish genome (660 versus 118 copies, 5.6×) (Figure 1B).
In sum, both Hsmar1 and Tdr1 accumulated high copy numbers in their respective host genomes during their active evolutionary life cycle. The process of generating of ‘self-inflicted wounds’, thought to be associated with transposase-dependent nicking (23), likely contributed to the inactivation of both transposon families. In contrast, internally deleted versions, MITEs and solo elements accumulated to different extents, suggesting transposon-specific processes.
Sleeping Beauty does not mobilize truncated substrates efficiently
We next probed experimentally the mechanistic differences between simple and IR/DR-type transposons and their contribution to their different genome-wide landscapes. The Hsmar1 transposase was previously reported to prefer the Hsmar1-derived MADE1 MITEs (also called MiHsmar1, (30)) over full-length elements as substrates (26), thereby providing a possible explanation for how Hsmar1-derived MITEs accumulated in the human genome.
To see how an active IR/DR transposase handles internally truncated transposons, we generated two versions of the SB transposon, by deleting either 977 or 1654 bp internally. The Mutant#2 construct consists almost entirely of the IRs (2 × 230 bp), thereby mimicking a MITE. The two internally deleted mutants were subjected to a transposon excision assay that measures fluorescence produced by a restored open reading frame of GFP upon transposon excision. Interestingly, both internally deleted transposon substrates were mobilized less frequently in comparison to the full-length element (Figure 1F). Curiously, the MITE-like substrate was mobilized only at a ∼10% efficiency of the full-length element. Thus, in contrast to Hsmar1, SB does not prefer internally deleted or MITE-like structures for mobilization. This observation might also explain why MITEs are not associated with the related Tdr1 (IR/DR) transposon in the zebrafish genome.
In addition to recombination between genomic copies, solo-IR transposon copies could be resulting from single-ended transposition events, and single-ended transposons of mariner (38), SB and piggyBac (16) were reported to occasionally serve as substrates for transposition. We compared the frequency of single-ended substrate utilization of Hsmar1 (simple IR) versus SB (IR/DR) transposition, by measuring mobilization of truncated substrates that lack one of their IRs in a cell culture-based transposition assay (25) (Figure 1G). After discarding non-transposase-mediated genomic integration events, we estimate that single-ended substrates were integrated more frequently in Hsmar1 (6.4%) versus SB (0.54%) (16) transposition (Figure 1G), suggesting that the more complex inverted repeat structure (IR/DR) could be a contributing factor to the fidelity of SB transposition.
The PAI subdomain of the Sleeping Beauty transposase mediates primary substrate contact
The DRs of the IR/DR have a composite structure, recognized by a composite DNA-binding domain of the transposase (21). The DNA-binding domain of the SB transposase consists of two helix-turn-helix (HTH) motifs, referred to as PAI and RED, based on their resemblance to the PAIRED domain present in the PAX family of transcription factors (21,39). Both subdomains are involved in sequence-specific DNA-binding: PAI binds the 3′- and RED interacts with the 5′-part of the bipartite transposase binding sites represented by the DRs (21,40). In addition to DNA binding, PAI was previously shown to encode a protein-protein interaction function (21). Notably, the four DRs of SB are not identical, as the DRs at the transposon ends are longer by 2 bp (14DRs versus 12DRs in Figure 1A).
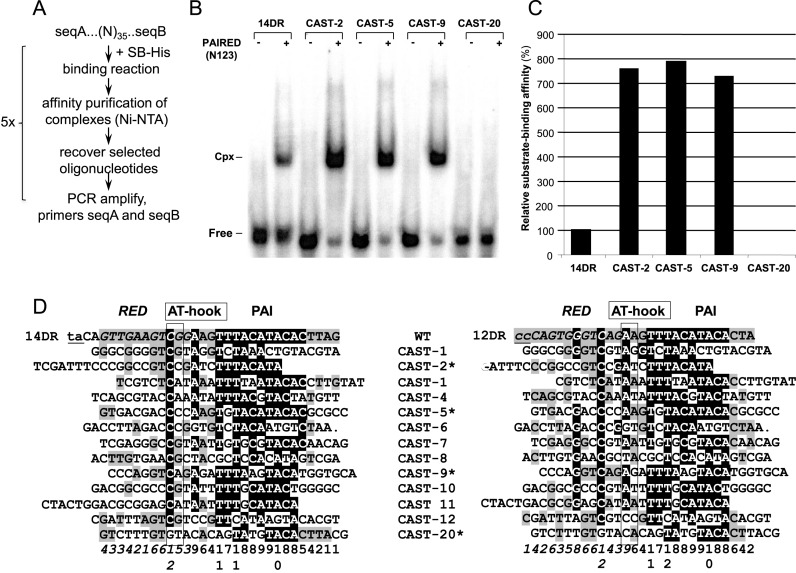
Although the binding site occupied by the PAIRED domain of SB has been determined by footprinting (21,25), this approach is not informative regarding the relative contributions and specificities of DNA-binding by the PAI and RED subdomains. To answer this question, we have used a CASTing approach that was originally developed to identify optimal binding sites for DNA-binding proteins (41) (Figure 2A). CASTing selects preferentially bound sequences out of complex libraries based on sequential enrichment of DNA sequences by affinity purification and PCR amplification. Thus, a CASTing approach should (i) identify high affinity binding sites, and (ii) map sequence motifs that are preferentially involved in primary substrate recognition by the composite DNA-binding domain. Based on footprinting data of SB transposase binding (25), a 35-bp random oligonucleotide library was exposed to binding by recombinant SB transposase in vitro. Oligonucleotides selected after six CASTing cycles were sequenced and tested in electromobility shift assay (EMSA) using the full (PAIRED) DNA-binding domain of the transposase. Some of the CASTing-selected sequences were bound up to eight-fold stronger than the wild-type 14DR sequence (Figure 2B and C). Curiously, the CASTing-selected, high-affinity binding sites had only limited similarity to the wild-type DRs, and sequence similarity concentrated mainly to the PAI recognition motif (Figure 2D). Thus, while the PAI subdomain seems to specify primary substrate recognition (21,40), RED is marginally involved in this process. The sequences captured by the CASTing strategy suggest that DNA-interactions mediated by PAI and RED have distinct functions, and protein-DNA interaction by RED might take place at a later step. Furthermore, CASTing did not appear to be selective for either 12DR or 14DR, suggesting that there is no significant distinction between 12DR (inner) versus 14DR (outer) (Figure 2D) binding sites during the ‘first contact’ between the transposon and transposase.
Figure 2.
Selection of optimal binding sites for the SB transposase by a molecular evolutionary strategy. (A) Flow chart of the optimization strategy (CASTing). (B) Oligonucleotides selected in six CASTing cycles were sequenced and tested in electromobility shift assay (EMSA), using the PAIRED-like DNA-binding domain of the SB transposase, N123 (21). Binding affinities were compared to the wild type 14DR motif of the SB left IR. The position of complexed (Cpx) and uncomplexed (Free) DNA probes. (C) Quantification of nucleo-protein complexes shown on Figure 2B by calculating the relative substrate-binding affinity values. (D) Alignment of CASTing selected, optimized binding sites to wild-type DR motifs. Various CAST sequences are aligned to either 14DR (left panel) or 12DR (right panel) of the left IR of the SB transposon. Binding regions aligning to RED and PAI are shown in italic and in plain, respectively. The AT-hook motifs are boxed. TA target site flanking the transposon and the corresponding nucleotides in the 12DR are in lowercase and underlined. The identity scores are shown below. Identical nucleotides are in shaded background (black – above 50%; gray – below 50%). Note that binding sites selected by the CASTing strategy are more similar to PAI versus RED (70% versus 20%). Selected CAST sequences, tested in EMSA (Figure 2B) are labeled with a star.
The RED subdomain of the Sleeping Beauty transposase mediates the distinction between 12DR versus 14DR
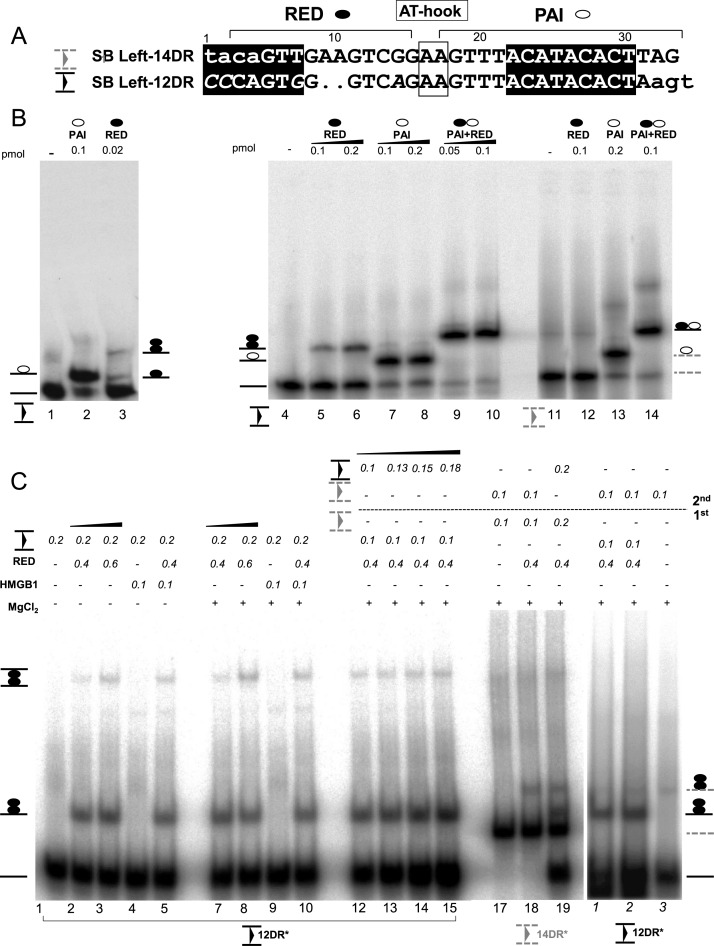
The sequence recognized by either RED or PAI differs between 12DRs and 14DRs (Figure 3A). Notably, the RED binding site overlaps with a sequence that is 2-bp shorter in 12DR (21) (Figure 3A), suggesting that RED might be involved in distinguishing between the inner (12DR) and outer (14DR) binding sites of the transposase. To test this assumption, double-stranded oligonucleotides representing the 12- and 14DRs were subjected to EMSA, using either the PAI (1-57 aa) or the RED (58-123 aa) subdomains of the SB transposase. As shown in Figure 3B, PAI equally bound to both DRs (lanes 2, 7, 8 and 13). In contrast, RED had a clear preference for 12DR, and no significant binding was detected using the 14DR substrate (Figure 3B, lanes 3, 5, 6 and 12). Thus, RED can distinguish between 12- versus 14DRs that might occur by recognizing sequence variation or difference in length. In order to distinguish between these possibilities, the EMSA was repeated with a 12DR-like oligonucleotide filled with 2 nucleotides having the same length as 14DR. Incorporation of two nucleotides into the 12DR abolished specific DNA binding (Supplementary Figure S3A, lanes 6 and 7) by RED, but left binding by PAI unaffected (Supplementary Figure S3A, lane 8). These results indicated that RED distinguishes between inner and outer DRs by length and not sequence. The above data support the hypothesis that selective recognition of the inner (12DRs) versus outer (14DRs) transposase binding sites is guided by length difference between the 12- and 14DRs, recognized by the RED subdomain of the SB transposase. Curiously, RED does not recognize 14DR, located at the end of the inverted repeat in this experimental setup.
Figure 3.
Modeling second-end-capture (SEC) of Sleeping Beauty in gel mobility shift assay (EMSA). (A) Alignment of the 14DR (outer, filled triangle between two lines) and 12DR (inner) of the left inverted repeat (IR). The nucleotides involved in DNA-protein interaction (21,25), are shown in uppercase, while the nonidentical nucleotides are in italics. The nucleotides recognized by PAI (empty circle) or RED (black circle) subdomains, and the AT-hook (framed) are indicated (21). The nucleotides resembling the ‘heptamer’ and ‘nonamer’ motifs of the RAG1 (50) are highlighted in black boxes. The length of the spacer between motifs is 12 bp, or 14 bp in the inner and outer DR, respectively. The 2 bp differences between inner and outer DRs are dotted. (B) DNA binding properties of RED (N58-123, black circle), PAI (N157, empty circle) or the full N-terminal DNA binding domain (PAI+RED) were characterized by EMSA. Upper panels: labeled oligonucleotides corresponding to the 12DR (black triangle, plain lines), the 14DR (gray triangle, dashed lines) or the 12+AA DR (dotted, black) were used as DNA substrates. The schematic of the predicted nucleoprotein complexes are depicted. Uncomplexed DNA—plain line. Complexes formed with the full N-terminal DNA binding domain (N123, PAIRED) were used as size markers (∼2×RED). The complexes were separated on 4% native gels. Note that monomeric RED–12DR complexes can be detected only at low protein concentration (0.02 versus 0.1 pmol, left panel, lane 3). (C) The 12DR/12DR rule (a selective capture of 12DR by the 12DR–RED complex) of Sleeping Beauty second-end-capture (SEC). HMGB1 promotes RED–12DR formation in a Mg2+ independent manner (lanes 1–10). Nucleoprotein complexes of 12DR–RED formed on 12DR (0.2 pmol) at various concentrations of RED indicated in the absence or presence of HMGB1 (μM). See also Supplementary Figure S3C. Asterisks indicate radioactively labeled substrate. Note that the formation of the low mobility band does not require the presence of MgCl2 (5 mM). Investigating SEC by staged EMSA (first–second). (First) 12DR substrate was pre-incubated with RED, (second) followed by the addition of labeled 12DR (lanes 12–15), or 14DR partner (right panel, 1–3), respectively (15 min, followed by 10 min on ice). Binding reactions were performed at constant RED concentration (0.4 pmol). Note that detectable SEC formation was neither detected when 14DR was offered as a partner to 12DR (right panel, 1–3), or in a reciprocal experiment, when complex formation was initiated on 14DR (lanes 17–19). The complexes were separated on 4% native gels.
In addition to 12/12DR distinction, RED is involved in protein-protein interactions
Although the PAI and RED subdomains are of similar size (57 and 66 amino acids, respectively), their nucleoprotein complexes migrate differently in EMSA (Figure 3B). Based on mobility, PAI seems to bind both the 12- and 14DRs as a monomer. In contrast, using similar concentrations, the dominant nucleoprotein complex formed between RED and 12DR migrates slower, consistent with the complex containing two molecules of RED (Figure 3B, lanes 3, 5 and 6). Notably, the complex formed by a RED monomer could be detected at a reduced protein concentration (20-fold less) in the binding reaction (Figure 3B, lane 3). This observation suggests that RED readily forms dimers upon binding to the 12DR, suggesting that similarly to PAI (21) the RED subdomain might be involved in both protein-DNA and protein-protein interactions. To test this, the RED peptide was subjected to chemical crosslinking followed by western blotting. Bands corresponding to dimeric, tetrameric and even higher order multimeric structures of RED were identified, both in the presence (Supplementary Figure S3B) and absence of DNA substrate (not shown). These results indicate that similarly to PAI (21), the RED subdomain is able to homodimerize. In sum, although both the PAI (21) and RED subdomains are proficient in protein-protein interaction, only RED, but not PAI forms dimers upon DNA binding.
The 12/12DR rule of synaptic complex formation during Sleeping Beauty transposition
We have shown previously that HMGB1 is also required for SB transposition, and enhances preferential binding of the SB transposase to the 12DR (42). To test whether HMGB1 is affecting substrate binding of RED, purified human HMGB1 protein was included in the binding reactions (Figure 3C). Increasing the concentration of RED resulted in an additional band of lower mobility, in a process that did not require Mg2+ (Figure 4C, lanes 1–10). The presence of HMGB1 enhanced the intensity of this low mobility band at various RED concentrations (0.2–0.6 pmol) (Supplementary Figure S3C). Notably, HMGB1 was not incorporated stably in the RED–12DR complex, as no supershift was detectable in the EMSA (Supplementary Figure S3C).
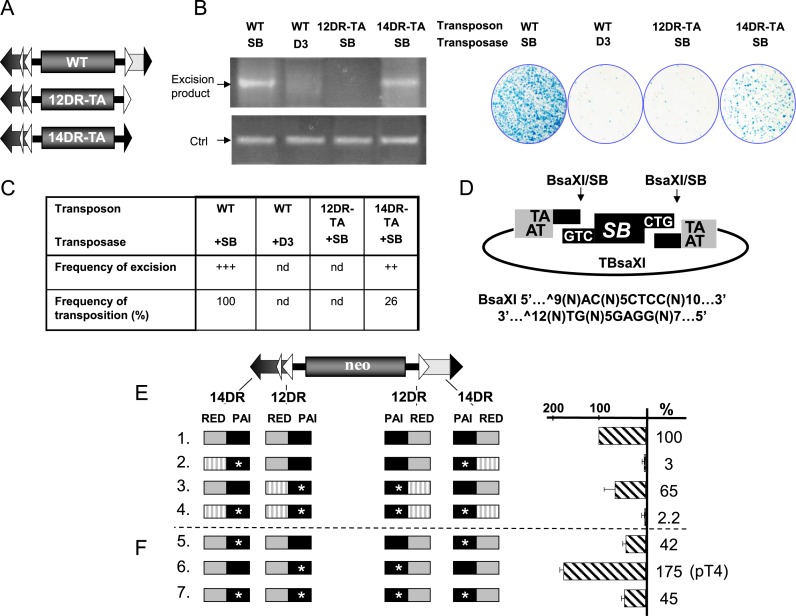
Figure 4.
Enhanced binding affinity at the inner 12DR, resistant for cleavage, improves Sleeping Beauty transposition. (A) Schematic representation of wild type and two single-DR transposon mutants, possessing either the 12DR (white triangle) or the 14DR (black triangle), flanked by the canonical TA dinucleotide (named as 12DR-TA and 14DR-TA, respectively). (B) Excision of Sleeping Beauty is inhibited at the 12DR. Excision (left panel) and integration properties (right panel) of the transposon variants. Semi-quantitative PCR-based excision assay (left panel) was performed on low molecular weight DNA isolated from HeLa cells transfected with the wt or mutant transposon constructs and active (SB) or inactive (D3) transposase versions, as indicated. The excision primers anneal to the plasmid backbone surrounding the transposon and amplify a 322 bp product only if the transposon excised. As an internal control (Ctrl), the ampicillin gene present on the backbone of the transposon constructs is amplified. Note that although the PCR-based excision assay is not quantitative, it is suitable for roughly estimating the excision rate of the transposon from a donor regardless of the reintegration frequencies. (Right panel) Result of a representative transposition assay in HeLa cells (25) transfected with the same transposon and transposase constructs as in the excision assay. (C) Diagram shows frequency of excision and transposition estimated from three independent experiments. (D) Linear DNA, mimicking the excised transposon is not recognized as transposon substrate in SB transposition. BsaXI cleavage produces identical ends as SB excision from the construct called TBsaXI. No detectable transposition events were identified in a colony based transposition assay performed with BsaXI cleaved, linearized transposon fragment in HeLa cells (not shown). (E) Enhanced binding affinity at the inner 12DR improves Sleeping Beauty transposition. On the left, schematics of various neo-marked, mutated transposon constructs are shown. DRs are represented by triangles (DR, large triangles; HDR, narrow triangle). On the right, the respective transpositional activities determined by a colony-based transposition assay (25) are shown in comparison to wild type transposon (construct #1), set as 100% (n = 4). Transposition activity of transposon variants, where the DRs of the transposons were changed to enhanced binding affinity (CAST-5) sites. PAI (black box) or the RED (gray box) recognition motifs into a high-affinity binding site (CAST-5) selected by CASTing (marked by stars at the PAI and stripped at the RED-like recognition motifs). (F) As in Figure 4E, but the CAST-5 sequence was used to replace only the PAI recognition motif, while the rest of the DR was wild type (n = 4). The optimized construct #6 is named as pT4.
Initial formation of a 12DR–RED complex is likely followed by incorporation of additional DNA sites during synaptic complex assembly. To address this, we used staged EMSA. In the first step, a fixed concentration of RED (0.4 pmol) was allowed to bind labeled 12DR. In the second step, the RED–12DR complex was exposed to various amounts of labeled 12DR or 14DR (Figure 3C, lanes 12–15 and 1–3). When labeled 12DR was added to the pre-complex, a lower mobility complex has appeared in the EMSA (Figure 3C, lanes 12–15), while no lower mobility complex was detectable when 14DR was offered as a second DNA substrate (Figure 3C, lanes 1–3). In the reciprocal experiment, no significant low mobility complex formation was detectable, when 14DR was used in the first step of the staged EMSA (Figure 3C, lanes 17–19). Thus, the 12DR–RED pre-complex was able to capture an additional 12DR substrate molecule. In contrast, 14DR was rejected as a partner, suggesting that the low mobility complex consists of 12DR–RED–12DR.
In sum, these data support a model where a RED–12DR complex selectively captures a second 12DR, and not a 14DR, thereby establishing a 12/12DR rule of paired–end complex (PEC) formation in SB transposition. The assembly process is facilitated by HMGB1. Thus, the SB transposase preferentially binds to the inner 12DR located distantly from the end of the transposon, dimerizes via the RED subdomain, and bridges to the second 12DR in the other IR of the transposon.
Cleavage is inhibited at the 12DR
In mariner transposition the recombinase dimer bound to its binding site is catalytically active, and cleaves the transposon end (9). Similarly, a 12DR-transposase-12DR complex holding both arms of the transposon is assumed to be catalytically active. Still, cleavage is not expected to occur, since the TA dinucleotide required for the cleavage (43) is not present next to the 12DR motif (Figure 3A). To see if the transposase could cleave at the internal 12DRs, two versions of mutant transposons were created. In both constructs, the left IR was wild-type, while the right IR was truncated at the internal DR, and was either modified to end with a TA (12DR-TA) or changed to 14DR that has the canonical TA dinucleotide (14DR-TA) (Figure 4A). The two mutant transposons were subjected to both excision and transposition assays (Supplementary Figure S4 and Figure 4B). Despite being truncated, detectable cleavage occurs using 14DR-TA (Figure 4B, left panel). In contrast, almost no cleavage products could be identified by 12DR-TA (Figure 4B, left panel), indicating that regardless of the presence of a TA, 12DR is not compatible with cleavage. In contrast to cleavage, neither of the truncated transposon versions transposed efficiently. Compared to the wild type, the transposition assay estimated ∼26% transposition frequencies using 14DR-TA (Figure 4B, right panel) (22), suggesting that the 12/12DR rule is not absolute. No detectable transposition occurred using 12DR-TA (Figure 4B, right panel), indicating that the inner position is resistant to cleavage even if it is flanked by a TA dinucleotide.
To challenge the ordered assembly process, we asked if a pre-cleaved transposon substrate could be incorporated into the transposition process. We generated a wild type transposon cleaved by BsaXI, a restriction enzyme that similarly to the SB transposase removes three nucleotides from the end of the transposon (Figure 4D). The pre-processed substrate was subjected to a transposition assay. Surprisingly, no significant transposition could be detected using the pre-processed transposon (not shown), suggesting that a pre-cleaved transposon substrate is not accepted by the SB transposase.
The above experiments suggest that SB transposition is a delicately controlled process, where the reaction proceeds in a defined order of distinct steps coupled to quality control. For example, an aberrant transposon can be excised, but still filtered out before the integration step occurs. Pre-cleaved substrates, not validated by the quality control cannot be incorporated in the reaction. The ordered assembly process of SB promotes paired-end complex formation at the inner DRs that are located distantly from the ends of the transposon. The paired complex would then be guided to cleave the transposon ends next to the outer DRs.
IR/DR governs an ‘ordered assembly’ process
Altering the affinity of the binding sites might also challenge the ‘ordered assembly’ process. Thus, a series of transposon versions were constructed where 12DR and/or 14DR motifs were replaced by CASTing-selected, high affinity binding sites shown in Figure 2, and the various constructs were subjected to transposition assays (Figure 4E). Surprisingly, replacing wild type motifs with the high-affinity CAST-5 sequence did not improve transposition frequencies. On the contrary, replacing either 12DRs or 14DRs with the CAST-5 motif resulted in 65% and 3% of wild type activities, respectively (Figure 4E). Similarly, changing all four DRs to CAST-5 had a severe negative effect on transposition (2.2%), suggesting that an enhanced DNA-binding affinity at either DR position might compromise SB transposition. Alternatively, the negative effect of CAST-5 on transposition could, at least partially, be explained by preferential selection for PAI binding, while compromising binding by RED. Indeed, the CASTing sequences are predicted to be sub-optimal for RED interaction, thereby compromising the ability of the SB transposase to distinguish between inner versus outer positions (Figure 2). To distinguish between the two scenarios, we generated CAST-5/wt hybrids, in which CAST-5-derived sequences replaced only the PAI interaction motif and keeping the rest of the transposase binding site wild type. Again, we tested the impact of the hybrid motifs on transposition in various combinations. The high-affinity, CAST-5/wt hybrid motifs were still affecting transposition negatively at the outer and the combined inner/outer positions (Figure 4F). However, the CAST-5/wt motif improved transposition (175%), when replacing 12DRs at the inner positions (pT4) (Figure 4F). In conclusion, the DNA-binding affinity of the DRs at the inner versus outer positions cannot be freely changed, indicating that substrate recognition occurs in well-defined steps at different phases of the transposition reaction, directed by the IR/DR structure.
The K248A exc+/int− mutant liberates transposon via a hairpin intermediate
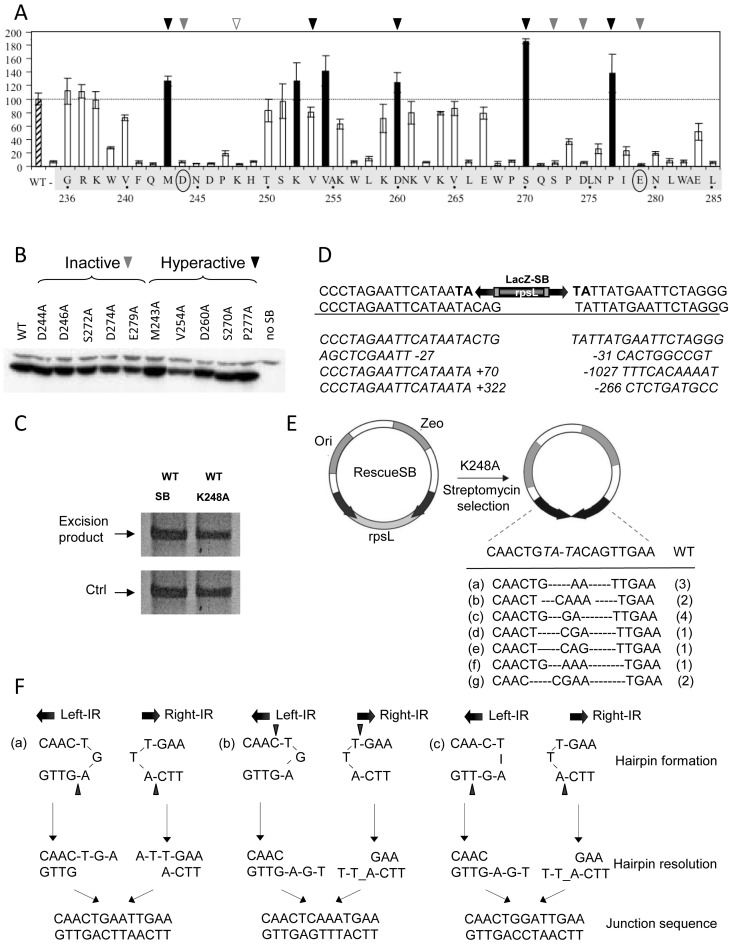
The process generating ‘self-inflicted wounds’, observed for both Hsmar1 and Tdr1 is assumed to be ‘open-and-shut’ cleavage that does not liberate the transposon. Nevertheless, the process seems to require pairing of the IRs of IR/DR elements before cleavage (Figure 1D). ‘Self-inflicted wounds’ are assumed to be generated by transposase variants that cannot process the transposon ends appropriately (44), for example because the first nick at the non-transferred strand is not followed by double-strand cleavage. To identify such SB transposase mutants, we performed systematic alanine-scanning mutagenesis in the catalytic domain. The transposase mutants were tested for both excision (Supplementary Figure S4) and integration activities (Figure 5A). The majority of the single amino acid mutations resulted in decreased activity or were even inactivating, while some of the mutations enhanced activities. The hyperactive mutations (up to 180%) included M243A, K252A, V254A, D260A, S270A and P277A (Figure 5A and B). In general, the mutations affected both excision and transposition in a comparable manner with the exception of K248A. While K248A was able to excise the transposon (Figure 5C), the reintegration of the excised molecule was severely impaired (8% versus wt) (Figure 5A), suggesting that in K248A transposition the excision and reintegration steps are not coupled. The excision footprints generated by K248A were not typical products of nonhomologous end joining (NHEJ) (Figure 5D), but more compatible with being generated by homology-dependent repair (HDR) (10). Alternatively, the footprint structure could be explained by aberrant cleavage that did not occur at the ends of the IRs.
Figure 5.
Liberation by the K248A variant of Sleeping Beauty is resolved by hairpin resolution. (A) Transposition frequencies of transposase variants mutagenized in the catalytic domain of the Sleeping Beauty transposase. Amino acids in the catalytic domain, between positions 236–285 of the catalytic domain were mutagenized by alanine scan mutagenesis. HeLa cells were cotransfected with a plasmid encoding a neomycin-marked transposon (pT/nori) (51) together with a plasmid encoding no transposase (–), the SB transposase (SB10) (25), or a transposase missense mutants. Relative transposition activity of SB and mutated SB transposases are shown (wild type activity is set to 100%; n = 3). The hyperactive mutants are shown as black columns. Certain hyperactive (black triangle) and inactive (gray triangle) mutants (five of each) were subjected to comparative expression analysis shown in B. Numbers indicate residue positions. (B) Immunoblot analysis of SB10 and representative mutant transposase proteins expressed by plasmid transfection of HeLa cells. Protein extracts were prepared from cells 40 h post-transfection and subjected to electrophoresis and electroblotting, and the 39-kDa transposase was detected with a polyclonal rabbit antibody to the SB protein. (C) Semi-quantitative PCR-based excision assay as in Figure 4B. (D) Excision footprints generated by K248A using the substrate of LacZ-SB (16). The canonical footprints generated by SB are ‘cag’ or ‘ctg’. TA target duplication in the donor construct are shown in bold. Representative excision sites generated by K248A show either deletions at both sites flanking the transposon or insertion at the left side and deletion at the right side. The length of the insertion and deletion sequences are shown by numbers. (E) Analysis of the extrachromosomal molecules, generated by K248A by using the reporter, RescueSB (16). RescueSB contains an SB transposon carrying an origin of replication (ori) and a zeocin gene (Zeo). The backbone DNA encodes a streptomycin gene rpsL gene, rendering bacteria sensitive to streptomycin. Black arrows: inverted repeat (IRs). Following transposon excision and circularization of the excised transposon, the rpsL is lost, thereby rendering bacteria StrepR. Extrachromosomal, non-integrated transposon molecules could be recovered from low molecular weight DNA preparation. The sequence of the junction region has been determined. Certain variants were recovered repeatedly (numbers of identical events recovered from total number of 14). (F) Three examples (a–c, from E) of junction sequences generated by hairpin resolution. The position of nicking is indicated by black triangle. Find analysis of further cases on Supplementary Figure S5.
To distinguish between these two possibilities, we followed the fate of the excised transposon by using a reporter construct, RescueSB (16). RescueSB carries a replication origin inside and a gene encoding rpsL outside of the transposon (Figure 5E). Using negative selection provided by rpsL, excised but not integrated circular transposon molecules can be recovered and sequenced. Analysis of the recovered transposons detected no large deletions; instead, they were lacking a few nucleotides (mostly) from the right IR (Figure 5E). These changes would render these circularized transposon molecules unable to reintegrate, in agreement with the exc+int−-like phenotype of K248A. Curiously, the sequences at the junction sites were reminiscent of DNA repair following hairpin resolution (Figure 5F and Supplementary Figure S5). The hairpin intermediate is indicative of single-strand cleavage. Our data are consistent with K248A not being able to perform second strand cleavage effectively. Nevertheless, single-strand cleavage at both IRs of the transposon by K248A could liberate the transposon via a hairpin intermediate. The hairpin intermediate is generated when the 3′-OH of nicked DNA attacks the opposing second strand in a direct trans-esterification reaction resulting in a double-strand break. The reaction is completed by a simple resolution of the hairpin structure on the transposon end. Hairpin resolution by nicking at the tip of the hairpin yields a blunt transposon end, but would diversify the DNA sequence when occurs at other positions (45), disabling the reintegration step.
DISCUSSION
In order to avoid the generation of aberrant side products and potentially genotoxic transposition events, the synapsis of the two transposon ends should precede the catalytic step. Because in the Tc1/mariner family a catalytically active transposase dimer can be formed on a single IR, a mechanism that controls synapsis of the ends is critical. In mariners, a conformational change couples synapsis and cleavage, and suppresses premature cleavage at unsynapsed transposon ends (3,9,11,13,14,44). This constraint provides effective regulation of mariner transposition at low transposase concentrations, but can be challenged by changing the transposase/transposon ratio, or by enhanced activity of the transposase (44).
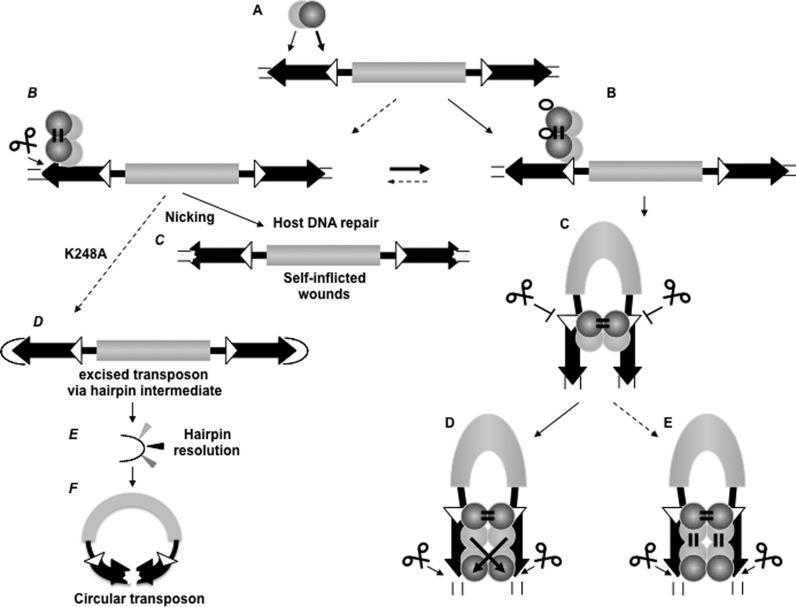
Here we provide evidence that the IR/DR structure, characteristic to a subgroup of the family of Tc1/mariner transposons, provides an extra layer of regulation to enforce paired-end formation before cleavage to occur. Our data are compatible with a model of SB transposition, in which the distinct steps of synaptic complex assembly are orchestrated by the interplay between the IR/DR structure and the composite PAIRED-like domain of the transposase. Both PAI and RED subdomains possess both DNA binding and protein-interaction functions that play important and differential roles at different phases of the transposition reaction. The DRs located at either ends of the IRs are distinguished by their size (14DR versus 12DR). We propose a model of IR/DR-governed complex assembly (Figure 6), in which DNA binding and protein-protein interaction are used alternatively in a defined order. The specific primary DNA recognition is conducted by PAI, and the contribution of RED to ‘first contact’ is limited. RED distinguishes between 12 versus 14DRs, and is involved in specifying complex assembly at the inner 12DR. Upon binding to the 12DR, the transposase readily forms dimers (Figure 3B) (alternatively, the transposase binds DNA as a dimer) through protein–protein interaction via the RED-RED interface. The complex captures the second IR as naked DNA, following the rule of 12/12DR, while the combination of 12/14DR is not accepted (Figure 3C). Synapsis is not accompanied by cleavage at the inner 12DRs. In the following steps further (possibly two) transposase molecules are recruited via the PAI–PAI interface, and finally the 14DRs are incorporated in the complex. While RED does not recognize 14DR in the early phase of the reaction (Figure 2), it is required to complete the assembly process, and prepare the complex for cleavage/reintegration performed by the catalytic domain. This process is assisted by the host-encoded factor HMGB1 that is recruited by the transposase (42). The role of HMGB1 is potentiate DNA binding at the inner 12DR (42). The DNA-bending activity of the HMGB1 might facilitate second-end-capture, and perhaps contributes at later steps of the complex assembly process as well. Nevertheless, the requirement for HMGB1 might not be absolute as the transposase is capable of capturing the second IR even in its absence (Figure 3C). Thus, HMGB1 is likely to improve the fidelity of the reaction.
Figure 6.
The ordered assembly of Sleeping Beauty transposition enforces paired-end complex formation (model). (A) Primary contact, substrate recognition by the transposase. Left IR is preferred over the right one. The transposase is able to recognize both the inner and outer DRs primarily by PAI (light gray ball), while RED (dark gray ball) is involved in the primary substrate identification. (B) RED is able to differentiate between 12DR (white triangle) versus 14DR (black triangle), and transposase binding is guided to 12DR. In the presence of HMGB1 (empty ellipse), the preference for 12DR-binding is further emphasized. The transposase dimerizes rapidly upon binding to 12DR (or binds as a dimer) via RED homo-dimerization. (C) The dimerized transposase captures the right IR in a naked form, governed by the 12/12DR rule. The second end capture complex (SEC) involves the 12DRs, and cleavage is strictly inhibited at this stage of the reaction. SEC formation is promoted by the presence of HMGB1, but Mg2+ is not required (not shown). (D and E) 14DRs are positioned in the complex for cleavage, presumably involving PAI-homo-dimerization and 14DR–RED interaction. The configuration of the complex can be trans-crossed (D) or trans-parallel (E) (43). PAI-enhancer interaction (21) is predicted to stabilize the pre-integration complex (not shown). Note: Since both PAI and RED are involved in dual function, conformational changes might coordinate the DNA-binding and protein dimerization activities (not shown). Left panel: (B) Dotted arrow indicates that the under suboptimal condition and/or by transposase variants (hexagons) generated by genetic drift, nicking occurs at 14DRs (instead of double strand cleavage). Nicking is also preceded by paired end formation, and might liberate the transposon (as in F). (C) The DNA repair machinery of the host diversifies the DNA sequence around the nick (‘self-inflicted wounds’). (D) K248A liberates the transposons via hairpin resolution. (E) Hairpin resolution diversifies the IRs. (F) Hairpin resolution seals the transposon to form an extrachromosomal circle. Circular transposon molecules have markedly reduced integration ability, but might be ideal for horizontal transfer. Note: For simplicity, the catalytic domain of the transposase and the enhancer motif are not shown, and HMGB1 is shown only in B. Possible conformational changes between steps are not shown.
The ordered assembly process provides quality control, and filters out aberrant intermediates by aborting the reaction, and inhibits the formation of MITEs. How IR/DR elements inhibit the accumulation of MITEs is not clear. It is unlikely to assume a requirement of an internal motif for transposition, since SB-based gene vectors only contain the IR/DRs and lack internal transposon sequences. Instead, the nucleoprotein complex formation might require an optimal distance between the IRs, and the connecting DNA cannot be too short. Notably, transposition is not efficient either if the ‘tips’ of the IRs are too close to each other (e.g. on a circular molecule (22)). Perhaps, a certain length of the DNA around the IR/DRs might be necessary to properly accommodate the multimeric transposase and its host factors (e.g. HMGB1) during complex formation.
The highly regulated nature of IR/DR transposition makes it challenging to convert improved binding affinity to enhance the transposition reaction as a whole. Increasing binding affinities at the IRs uniformly or at the outer 14DRs disturbs the delicately regulated ordered assembly process, and results in reduced transposition frequencies (Figure 4E). Nevertheless, enhancing binding affinity at the inner 12DRs could affect the entire transposition process positively (Figure 4F). Notably, the enhancement is not directly proportional with the optimized binding affinity, indicating that the IR/DR structure governs a delicately regulated process that does not tolerate drastic changes. Nevertheless, the attempt to decipher the role of the IR/DR structure in combination of molecular evolutionary approaches could be translated to significantly improve the efficiency of SB transposition for genetic applications (pT4).
Despite regulatory constraints, under suboptimal conditions transposases of the Tc1/mariner family or variants generated by mutations seem to catalyze infrequent, unconstrained transposition reactions (38,44). Besides providing insight into the mechanism of transposition, these events have phylogenetically significant aspects. For example, generating ‘self-inflicted wounds’ represent a suicidal form of autoregulation, assumed to support a stable transposon-host coexistence (44). This reaction initiates by nicking, but the reaction aborts, and the gap is resealed by the host DNA repair machinery (Figure 6C). Similarly, certain RAG1 mutants are able to ‘nick only’ without proceeding with the cleavage reaction further (46). Notably, ‘self-inflicted wounds’ of the IR/DR elements are generated in a fairly concerted manner (Figure 1D), suggesting that the process involves paired-end-formation.
Alternatively, the mutated transposon copies might be integration events of excised transposons. Indeed, the nicks generated by K248A are not resealed, and the transposon gets liberated. The majority of K248 excised transposon molecules do not reintegrate efficiently, but accumulate as circular extrachromosomal DNA, thereby exhibiting an exc+int- phenotype. Thus, in the K248A-mediated reaction the excision and integration functions of the transposase are clearly disconnected. Intriguingly, besides switching between integrating and extrachromosomal mode, K248A demonstrates a yet unprecedented scenario when the reaction converts from hydrolysis to a transesterification mode. The free DNA strand, generated by nicking, attacks the opposite DNA strand. The resulting double stranded cut liberates the transposon via a hairpin intermediate. Resolution via hairpin intermediate is considered as a mechanistically different mode of transposition, characterizing the bacterial Tn5 Tn10 elements or V(D)J recombination. The hairpin resolution diversifies the transposon IRs, explaining the exc+int- phenotype. Excised but not reintegrated molecules were observed during transposition of Tc1, Tc3 or Minos elements (47,48); these extrachromosomal transposon-derived molecules have long been debated to be either transposition intermediates, or side products. We do not consider K248A-generated extrachomosomal circles as natural transposition products, however they might have a phylogenetic relevance. It is noteworthy that a switch between the two modes (integration versus extrachromosomal) is relatively simple. It can be generated by a mutation or perhaps by suboptimal conditions. A single amino acid change in K248 can apparently convert the tranposase to generate extrachromosomal copies, where the second strand is processed, and diversified by hairpin resolution. Although the extrachomosomal circles integrate poorly, they might be optimal vehicles for horizontal transfer, and establish transposition in a new naïve genome.
The analogy between SB transposition and V(D)J recombination is multiple, underpinning the mechanistic relatedness. The sequences recognised by the recombinases are clearly related (Figure 3A), and both processes are assisted by HMGB1 (17,18,49). The IR/DR regulation resembles the ‘ordered assembly’ process of V(D)J recombination (19). An important difference is that during paired end formation the naked DNA captures a heterologous (12/23) partner in V(D)J recombination, while in SB transposition the captured partner is homologous (12/12). Strikingly, while V(D)J recombination was evolved from transposition (2), our current work demonstrates that transposition could be converted by a single amino acid change to a V(D)J-like process.
Acknowledgments
Z. Iz. and Z. Iv. conceived and designed the study, collected and analyzed the data, and drafted the manuscript. D.P.D. C.D.K. Y.W. E.E.N. J.W. has designed and performed the EMSA, hairpin, transposon optimization, transposition, deletion mutagenesis experiments, respectively. M.S. performed bioinformatics analysis. M.A.K. and S.Y. designed and performed the alanine screen. The authors thank A.D. for technical assistance.
Footnotes
Present address:
Yongming Wang, School of Life Sciences, Fudan University, Shanghai 200438, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Z. Iz. is funded by European Research Council, ERC Advanced [ERC-2011-AdG 294742]. Funding for open access charge: European Research Council, ERC Advanced [ERC-2011-AdG 294742].
Conflict of interest statement. None declared.
REFERENCES
- 1.Craig N.L. Unity in transposition reactions. Science. 1995;270:253–254. doi: 10.1126/science.270.5234.253. [DOI] [PubMed] [Google Scholar]
- 2.Huang S., Tao X., Yuan S., Zhang Y., Li P., Beilinson H.A., Zhang Y., Yu W., Pontarotti P., Escriva H., et al. Discovery of an active RAG transposon illuminates the origins of V(D)J recombination. Cell. 2016;166:102–114. doi: 10.1016/j.cell.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richardson J.M., Colloms S.D., Finnegan D.J., Walkinshaw M.D. Molecular architecture of the Mos1 paired-end complex: the structural basis of DNA transposition in a eukaryote. Cell. 2009;138:1096–1108. doi: 10.1016/j.cell.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hare S., Gupta S.S., Valkov E., Engelman A., Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010;464:232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuypers M.G., Trubitsyna M., Callow P., Forsyth V.T., Richardson J.M. Solution conformations of early intermediates in Mos1 transposition. Nucleic Acids Res. 2013;41:2020–2033. doi: 10.1093/nar/gks1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hickman A.B., Ewis H.E., Li X., Knapp J.A., Laver T., Doss A.L., Tolun G., Steven A.C., Grishaev A., Bax A., et al. Structural basis of hAT transposon end recognition by Hermes, an octameric DNA transposase from Musca domestica. Cell. 2014;158:353–367. doi: 10.1016/j.cell.2014.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voigt F., Wiedemann L., Zuliani C., Querques I., Sebe A., Mates L., Izsvak Z., Ivics Z., Barabas O. Sleeping Beauty transposase structure allows rational design of hyperactive variants for genetic engineering. Nat. Commun. 2016;7:11126. doi: 10.1038/ncomms11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hickman A.B., Chandler M., Dyda F. Integrating prokaryotes and eukaryotes: DNA transposases in light of structure. Crit. Rev. Biochem. Mol. Biol. 2010;45:50–69. doi: 10.3109/10409230903505596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dawson A., Finnegan D.J. Excision of the Drosophila mariner transposon Mos1. Comparison with bacterial transposition and V(D)J recombination. Mol. Cell. 2003;11:225–235. doi: 10.1016/s1097-2765(02)00798-0. [DOI] [PubMed] [Google Scholar]
- 10.Izsvak Z., Stuwe E.E., Fiedler D., Katzer A., Jeggo P.A., Ivics Z. Healing the wounds inflicted by sleeping beauty transposition by double-strand break repair in mammalian somatic cells. Mol. Cell. 2004;13:279–290. doi: 10.1016/s1097-2765(03)00524-0. [DOI] [PubMed] [Google Scholar]
- 11.Richardson J.M., Dawson A., O'Hagan N., Taylor P., Finnegan D.J., Walkinshaw M.D. Mechanism of Mos1 transposition: insights from structural analysis. EMBO J. 2006;25:1324–1334. doi: 10.1038/sj.emboj.7601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu K., Lieber M.R. The nicking step in V(D)J recombination is independent of synapsis: implications for the immune repertoire. Mol. Cell. Biol. 2000;20:7914–7921. doi: 10.1128/mcb.20.21.7914-7921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipkow K., Buisine N., Lampe D.J., Chalmers R. Early intermediates of mariner transposition: catalysis without synapsis of the transposon ends suggests a novel architecture of the synaptic complex. Mol. Cell. Biol. 2004;24:8301–8311. doi: 10.1128/MCB.24.18.8301-8311.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Claeys Bouuaert C., Liu D., Chalmers R. A simple topological filter in a eukaryotic transposon as a mechanism to suppress genome instability. Mol. Cell. Biol. 2011;31:317–327. doi: 10.1128/MCB.01066-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Claeys Bouuaert C., Walker N., Liu D., Chalmers R. Crosstalk between transposase subunits during cleavage of the mariner transposon. Nucleic Acids Res. 2014;42:5799–5808. doi: 10.1093/nar/gku172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y., Wang J., Devaraj A., Singh M., Jimenez Orgaz A., Chen J.X., Selbach M., Ivics Z., Izsvak Z. Suicidal autointegration of sleeping beauty and piggyBac transposons in eukaryotic cells. PLoS Genet. 2014;10:e1004103. doi: 10.1371/journal.pgen.1004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Gent D.C., Hiom K., Paull T.T., Gellert M. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agrawal A., Eastman Q.M., Schatz D.G. Transposition mediated by RAG1 and RAG2 and its implicacltions for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 19.Jones J.M., Gellert M. Ordered assembly of the V(D)J synaptic complex ensures accurate recombination. EMBO J. 2002;21:4162–4171. doi: 10.1093/emboj/cdf394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plasterk R.H., Izsvak Z., Ivics Z. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 1999;15:326–332. doi: 10.1016/s0168-9525(99)01777-1. [DOI] [PubMed] [Google Scholar]
- 21.Izsvak Z., Khare D., Behlke J., Heinemann U., Plasterk R.H., Ivics Z. Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in Sleeping Beauty transposition. J. Biol. Chem. 2002;277:34581–34588. doi: 10.1074/jbc.M204001200. [DOI] [PubMed] [Google Scholar]
- 22.Izsvak Z., Ivics Z., Plasterk R.H. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J. Mol. Biol. 2000;302:93–102. doi: 10.1006/jmbi.2000.4047. [DOI] [PubMed] [Google Scholar]
- 23.Lohe A.R., Timmons C., Beerman I., Lozovskaya E.R., Hartl D.L. Self-inflicted wounds, template-directed gap repair and a recombination hotspot. Effects of the mariner transposase. Genetics. 2000;154:647–656. doi: 10.1093/genetics/154.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aidinis V., Bonaldi T., Beltrame M., Santagata S., Bianchi M.E., Spanopoulou E. The RAG1 homeodomain recruits HMG1 and HMG2 to facilitate recombination signal sequence binding and to enhance the intrinsic DNA-bending activity of RAG1-RAG2. Mol. Cell. Biol. 1999;19:6532–6542. doi: 10.1128/mcb.19.10.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivics Z., Hackett P.B., Plasterk R.H., Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 26.Miskey C., Papp B., Mates L., Sinzelle L., Keller H., Izsvak Z., Ivics Z. The ancient mariner sails again: transposition of the human Hsmar1 element by a reconstructed transposase and activities of the SETMAR protein on transposon ends. Mol. Cell. Biol. 2007;27:4589–4600. doi: 10.1128/MCB.02027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mates L., Chuah M.K., Belay E., Jerchow B., Manoj N., Acosta-Sanchez A., Grzela D.P., Schmitt A., Becker K., Matrai J., et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 28.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Izsvak Z., Ivics Z., Hackett P.B. Characterization of a Tc1-like transposable element in zebrafish (Danio rerio) Mol. Gen. Genet. 1995;247:312–322. doi: 10.1007/BF00293199. [DOI] [PubMed] [Google Scholar]
- 30.Robertson H.M., Zumpano K.L. Molecular evolution of an ancient mariner transposon, Hsmar1, in the human genome. Gene. 1997;205:203–217. doi: 10.1016/s0378-1119(97)00472-1. [DOI] [PubMed] [Google Scholar]
- 31.Lampe D.J., Grant T.E., Robertson H.M. Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics. 1998;149:179–187. doi: 10.1093/genetics/149.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wallau G.L., Capy P., Loreto E., Hua-Van A. Genomic landscape and evolutionary dynamics of mariner transposable elements within the Drosophila genus. BMC Genomics. 2014;15:727. doi: 10.1186/1471-2164-15-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang G., Fattash I., Lee C.N., Liu K., Cavinder B. Birth of three stowaway-like MITE families via microhomology-mediated miniaturization of a Tc1/Mariner element in the yellow fever mosquito. Genome Biol. Evol. 2013;5:1937–1948. doi: 10.1093/gbe/evt146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piriyapongsa J., Jordan I.K. A family of human microRNA genes from miniature inverted-repeat transposable elements. PLoS One. 2007;2:e203. doi: 10.1371/journal.pone.0000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bao W., Kojima K.K., Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Izsvak Z., Ivics Z., Shimoda N., Mohn D., Okamoto H., Hackett P.B. Short inverted-repeat transposable elements in teleost fish and implications for a mechanism of their amplification. J. Mol. Evol. 1999;48:13–21. doi: 10.1007/pl00006440. [DOI] [PubMed] [Google Scholar]
- 37.Liu D., Bischerour J., Siddique A., Buisine N., Bigot Y., Chalmers R. The human SETMAR protein preserves most of the activities of the ancestral Hsmar1 transposase. Mol. Cell. Biol. 2007;27:1125–1132. doi: 10.1128/MCB.01899-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sinzelle L., Jegot G., Brillet B., Rouleux-Bonnin F., Bigot Y., Auge-Gouillou C. Factors acting on Mos1 transposition efficiency. BMC Mol. Biol. 2008;9:106. doi: 10.1186/1471-2199-9-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Czerny T., Schaffner G., Busslinger M. DNA sequence recognition by Pax proteins: bipartite structure of the paired domain and its binding site. Genes Dev. 1993;7:2048–2061. doi: 10.1101/gad.7.10.2048. [DOI] [PubMed] [Google Scholar]
- 40.Carpentier C.E., Schreifels J.M., Aronovich E.L., Carlson D.F., Hackett P.B., Nesmelova I.V. NMR structural analysis of Sleeping Beauty transposase binding to DNA. Protein Sci. 2014;23:23–33. doi: 10.1002/pro.2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wright W.E., Binder M., Funk W. Cyclic amplification and selection of targets (CASTing) for the myogenin consensus binding site. Mol. Cell. Biol. 1991;11:4104–4110. doi: 10.1128/mcb.11.8.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zayed H., Izsvak Z., Khare D., Heinemann U., Ivics Z. The DNA-bending protein HMGB1 is a cellular cofactor of Sleeping Beauty transposition. Nucleic Acids Res. 2003;31:2313–2322. doi: 10.1093/nar/gkg341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cui Z., Geurts A.M., Liu G., Kaufman C.D., Hackett P.B. Structure-function analysis of the inverted terminal repeats of the sleeping beauty transposon. J. Mol. Biol. 2002;318:1221–1235. doi: 10.1016/s0022-2836(02)00237-1. [DOI] [PubMed] [Google Scholar]
- 44.Liu D., Chalmers R. Hyperactive mariner transposons are created by mutations that disrupt allosterism and increase the rate of transposon end synapsis. Nucleic Acids Res. 2014;42:2637–2645. doi: 10.1093/nar/gkt1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhasin A., Goryshin I.Y., Reznikoff W.S. Hairpin formation in Tn5 transposition. J. Biol. Chem. 1999;274:37021–37029. doi: 10.1074/jbc.274.52.37021. [DOI] [PubMed] [Google Scholar]
- 46.Lee G.S., Neiditch M.B., Salus S.S., Roth D.B. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- 47.Radice A.D., Emmons S.W. Extrachromosomal circular copies of the transposon Tc1. Nucleic Acids Res. 1993;21:2663–2667. doi: 10.1093/nar/21.11.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Luenen H.G., Colloms S.D., Plasterk R.H. The mechanism of transposition of Tc3 in C. elegans. Cell. 1994;79:293–301. doi: 10.1016/0092-8674(94)90198-8. [DOI] [PubMed] [Google Scholar]
- 49.West R.B., Lieber M.R. The RAG-HMG1 complex enforces the 12/23 rule of V(D)J recombination specifically at the double-hairpin formation step. Mol. Cell. Biol. 1998;18:6408–6415. doi: 10.1128/mcb.18.11.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hesse J.E., Lieber M.R., Mizuuchi K., Gellert M. V(D)J recombination: a functional definition of the joining signals. Genes Dev. 1989;3:1053–1061. doi: 10.1101/gad.3.7.1053. [DOI] [PubMed] [Google Scholar]
- 51.Yant S.R., Meuse L., Chiu W., Ivics Z., Izsvak Z., Kay M.A. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat. Genet. 2000;25:35–41. doi: 10.1038/75568. [DOI] [PubMed] [Google Scholar]