Abstract
Spinal Muscular Atrophy (SMA) is a neuromuscular disorder caused by insufficient levels of the Survival of Motor Neuron (SMN) protein. SMN is expressed ubiquitously and functions in RNA processing pathways that include trafficking of mRNA and assembly of snRNP complexes. Importantly, SMA severity is correlated with decreased snRNP assembly activity. In particular, the minor spliceosomal snRNPs are affected, and some U12-dependent introns have been reported to be aberrantly spliced in patient cells and animal models. SMA is characterized by loss of motor neurons, but the underlying mechanism is largely unknown. It is likely that aberrant splicing of genes expressed in motor neurons is involved in SMA pathogenesis, but increasing evidence indicates that pathologies also exist in other tissues. We present here a comprehensive RNA-seq study that covers multiple tissues in an SMA mouse model. We show elevated U12-intron retention in all examined tissues from SMA mice, and that U12-dependent intron retention is induced upon siRNA knock-down of SMN in HeLa cells. Furthermore, we show that retention of U12-dependent introns is mitigated by ASO treatment of SMA mice and that many transcriptional changes are reversed. Finally, we report on missplicing of several Ca2+ channel genes that may explain disrupted Ca2+ homeostasis in SMA and activation of Cdk5.
INTRODUCTION
Spinal Muscular Atrophy (SMA) is a devastating neuromuscular disorder that often causes death during infancy and invariably leads to progressive loss of muscle strength due to loss of motor neurons (1,2). The disorder is caused by deletions or mutations in the SMN1 gene (3,4), while SMN2, a secondary gene capable of expressing an identical Survival of Motor Neuron (SMN) protein, expresses only a low amount of SMN due to predominant skipping of exon 7. A point mutation at position 6 in exon 7 of SMN2 eliminates an exonic splicing enhancer (ESE) recognized by serine/arginine-rich splicing factor 1 (SRSF1) (5,6) and creates an exonic splicing silencer (ESS) recognized by heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) (7). This ESS acts synergistically with other ESS motifs that bind to hnRNP A1 and Sam68, and repress exon 7 inclusion (8–11). This leads to the production of a truncated and unstable protein (12).
SMN itself is a ubiquitously expressed protein best characterized as a protein involved in the assembly of small nuclear ribonucleic protein (snRNP) complexes, both the catalytically active components of the spliceosome (13–15), and the U7 snRNP (16), which is involved in the processing of histone pre-mRNA (17).
SMN has also been reported to be directly involved in intra-cellular transport of mRNA (reviewed in (18)), to regulate actin (19) and to function as a general regulator of translation (20). Thus, SMN functions in many different contexts, and the SMA phenotype may be the outcome of the disruption of SMN function in one or several of these.
The question of whether or not SMA is entirely or partially caused by a perturbation in the generation of the snRNP repertoire, or by the loss of axonal-specific activity remains unresolved. Several studies support the involvement of specific axonal activities of SMN in SMA pathology and motor neuron function, in particular mRNA packaging and trafficking (19,21–24), but the best characterized and most well-established function of SMN is in the biogenesis and maintenance of snRNPs (13,25). Additionally, an SMN missense mutant that retains snRNP-assembly activity is able to rescue the SMA phenotype in a mouse model expressing the human SMN2 transgene (26), while co-injection of purified snRNP complexes from HeLa cells into fish embryos rescues motor axon development defects in a zebrafish SMA model (27).
Widespread defects in splicing have been reported from studies employing exon arrays, where it was observed that the level of aberrant splicing increases with disease progression (28,29). In particular, the minor spliceosomal snRNPs are affected by low levels of SMN (27,30,31), and SMN-dependent alternative splicing of a few U12-dependent introns has been reported previously (31). Moreover, alternative splicing of a U12-dependent intron in the Stasimon gene was recently reported to result in defects in the neuronal circuitry of Drosophila, and expression of the human orthologue in a zebrafish SMA model rescued motor axon defects (32).
While motor neurons are the cells most severely affected by low levels of SMN, there is also evidence indicating that SMA pathology may affect other cell types in other tissues as well. In both human patients with severe SMA (33) and severe SMA mouse models (34–36), non-motor neuron central nervous system (CNS) pathologies and disrupted enteric nervous system (ENS) signaling that may be related to gastrointestinal dysfunction (37) have been reported. Additionally, liver defects have been reported in several mouse models (38,39), while cardiac defects have been reported in both human patients (40,41) and mouse models (38,42,43).
Furthermore, systemic antisense oligonucleotide (ASO) therapy with subcutaneous injection is necessary for long-term rescue of type I SMA mice (44), while restricting ASO therapy to peripheral tissue reduces tissue necrosis and improves motor function and survival (45). Treatment with a 2′-O-(2-methoxyethyl)-modified ASO targeting ISS-N1 (9) designated ASO10-27/SMNRx/Nusinersen represents a promising treatment for SMA and has recently completed its clinical trials and is now in the process of FDA and EMA approval. Recently, a microarray study on an induced adult SMA mouse model demonstrated that this ASO rescues the gene expression changes observed in adult SMA mice (46), but little is known about the global effects of SMN restoration with an ASO on the splicing dysregulation, in particular intron retention, in severe SMA mice, although pharmacological restoration of SMN levels with a small-molecule drug has been shown to restore snRNA levels and correct a few known aberrant splicing events (47). Importantly, ASO therapy also offers the possibility to study the effect of a postnatal increase in SMN protein levels to identify changes specifically related to the postnatal state versus those that may arise due to delayed maturation during embryogenesis.
In the present study, we have used RNA-seq to investigate global changes in gene expression and alternative splicing in multiple tissues at an early pre-symptomatic stage and at a later symptomatic stage in an SMA mouse model. Furthermore, for the first time we use RNA-seq to examine the transcriptomic changes following ASO treatment targeting ISS-N1 and in particular how U12-dependent intron splicing is corrected in treated severe SMA mice.
MATERIALS AND METHODS
Antisense oligonucleotide treatment
The synthesis and purification of ASO10–29 (5′-ATTCACTTTCATAATGCTGG-3′) MOE-modified oligonucleotide with phosphorothioate backbone and all 5-methylcytosines were performed as described (48). The oligonucleotides were dissolved in 0.9% saline. Mice treated with either saline or 100 μg/g body-weight ASO10-29 were injected twice, once on postnatal day 0 (PND0) and once on postnatal day 1 (PND1). The oligonucleotide or empty saline solutions were injected subcutaneously into the upper back with a 5 μl syringe and 33-gauge custom removable needle (Hamilton) as previously described (48).
Mouse tissue preparation
Mice used in the study were ‘Taiwanese’ SMA model mice generated from an SMA type III model (44,49,50) with a mean lifespan of ∼10–11 days. Mouse tissues were collected on postnatal day 1 (PND1) and postnatal day 5 (PND5). Mice were sacrificed with CO2 asphyxiation and organs/tissues were rinsed in saline, snap-frozen in liquid N2 and stored at −80°C until processing. For extraction of RNA samples, tissues were pulverized in liquid N2 with a mortar and pestle, and total RNA was purified with an RNeasy Mini Kit and treated with RNase-free DNase I. When possible, samples were gender-matched to include two males and two females in each group.
SMN1 knockdown in HeLa cells
HeLa cells were transfected three separate times with SMN1 siRNA (L-011108-00-0005, Dharmacon) using RNAiMAX (Thermo Fisher Scientific) according to manufacturer's instructions. Forty-eight hours post transfection RNA was extracted using Isol-RNA lysis reagent (5 PRIME). Two replicate knockdowns were used in RNA-sequencing. Knockdown was confirmed by Western blotting using anti-SMN (SC-32313, Santa Cruz Biotechnology) and anti-SRSF3 (SC-13510, Santa Cruz Biotechnology) as a loading control.
RNA-seq library preparation and sequencing
RNA purity, integrity and concentration were determined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., USA). Only RNA with a RIN value of 8.0 or higher and a 28s/18s ratio ∼1.8 was taken forward for sample preparation. Mouse spinal cord RNA samples were processed for library construction following the manufactory instructions (Illumina TruSeq RNA sample preparation v2 Guide, Part #15026495 Rev.B February 2012 – ‘Low sample protocol’). In brief, 1 μg of total RNA from each mouse was used for library preparation. mRNA was purified, chemically fragmented for 8 min at 94°C, and processed for first strand cDNA synthesis and then second strand synthesis. Following several purification steps, the cDNA was then end-repaired, purified, adenylated at the 3′-ends, and purified before adding the indexed adaptor sequences. TruSeq RNA sample prep kit v2, set A, cat-RS-122-2001 was used for indexing. Each library preparation was then enriched by 10 cycles of PCR, purified and finally validated in regard to size and concentration. For sizing, libraries were analyzed on the Agilent 2100 Bioanalyzer using a DNA 1000 kit from Agilent Technologies. The Libraries were quantified by qPCR using the KaPa Library quantification Kits (KaPa Biosystems, Cat KK4824) and a final concentration of 15 pM denatured libraries were used for paired-end 75 bp sequencing on an Illumina HiSeq1500.
Mouse brain, liver, muscle, ASO treated tissue samples, and HeLa RNA samples were prepped for library sequencing following Illumina TruSeq Stranded Total RNA Samples Preparation Guide, Part#15031048 Rev.E October 2013—‘Low sample protocol’. 0.5 μg of total RNA from each sample was depleted for cytoplasmic rRNA using the Ribo-Zero ribosomal reduction chemistry and subsequently purified before fragmentation and processing similar to the protocol above. However, during second strand synthesis, the incorporation of dUTP instead of dTTP quenches the second strand during amplification, because the polymerase does not amplify past this nucleotide, and thereby the library becomes strand specific. For indexing, the TruSeq Stranded LT Kit Index set A was used. Samples were pooled in sets of 12 libraries, and a final concentration of 16 pM denatured libraries were used for paired-end 100 bp sequencing using an Illumina HiSeq1500.
Raw sequencing data have been deposited in the ArrayExpress database under accession E-MTAB-3664.
cDNA synthesis
0.5 or 1.0 μg of total RNA was treated with DNase I (Invitrogen, cat. 18068-015), and then reverse transcribed into cDNA using the Superscript VILO cDNA kit (Invitrogen, cat. 11754-050). A reaction volume of 10 μl was used. cDNA synthesis was performed according to manufactory instruction, but with an extended synthesis at 42°C for 120 min. Subsequently, the cDNA was added 50 μl DEPC-water and cDNA concentration was measured by absorbance readings at 260, 280 and 230 nm (NanoDrop™1000 Spectrophotometer; Thermo Scientific, CA, USA).
qPCR
Each cDNA (50–100 ng) was used in triplicates as template for in a reaction volume of 8 μl containing 3.33 μl Fast Start Essential DNA Green Master (2×) (Roche Diagnostics, Hvidovre, Denmark), 0.33 μl primer premix (containing 10 pmol of each primer), and PCR grade water to a total volume of 8 μl. The qPCR was performed in a Light Cycler LC480 (Roche Diagnostics, Hvidovre, Denmark): 1 cycle at 95°C/5 min followed by 45 cycles at 95°C/10 s, 59–64°C (primer dependent)/10 s, 72°C/10 s. Primers used for qPCR are listed in Supplementary Table S9. Threshold values were determined by the Light Cycler software (LCS480 1.5.1.62 SP1) using Absolute Quantification Analysis/2nd derivative maximum. Each qPCR assay included; a standard curve of nine serial dilution (2-fold) points of a cDNA mix of all the samples (250 to 0.97 ng), and a no-template control. PCR efficiency ( = 10(-1/slope) − 1) were ≥70% and r2 = 0.96 or higher. The specificity of each amplification was analyzed by melting curve analysis. Quantification cycle (Cq) was determined for each sample and the comparative method was used to detect relative gene expression ratio (2−Cq) normalized to the reference gene Vps29 in spinal cord, brain, and liver samples, and E430025E21Rik in the muscle samples. In HeLA samples, TBP was used as reference. Reference genes were chosen based on their observed stability across conditions. Significance was ascertained by the two-tailed Student's t-test.
Bioinformatics analysis
Each sample was aligned using STAR (51) with the following additional parameters: ‘–outSAMstrandField intronMotif –outFilterType BySJout’. The gender of each sample was confirmed through Y chromosome coverage and RT-PCR of Y-chromosome-specific genes (data not shown).
Gene-expression analysis
HTSeq (52) was used to obtain gene-counts using the Ensembl v.67 (53) annotation as reference. The Ensembl annotation had prior to this been restricted to genes annotated as protein-coding. Gene counts were subsequently used as input for analysis with DESeq2 (54,55) using R (56). Prior to analysis, genes with fewer than four samples containing at least one read were discarded. Samples were additionally normalized in a gene-wise manner using conditional quantile normalization (57) prior to analysis with DESeq2. Gene expression was modeled with a generalized linear model (GLM) (58) of the form: expression ∼ gender + condition. Genes with adjusted P-values <0.1 were considered significant, equivalent to a false discovery rate (FDR) of 10%.
Differential splicing analysis
Exon-centric differential splicing analysis was performed using DEXSeq (59) with RefSeq (60) annotations downloaded from UCSC, Ensembl v.67 (53) annotations downloaded from Ensembl, and de novo transcript models produced by Cufflinks (61) using the RABT approach (62) and the Ensembl v.67 annotation. We excluded the results of the analysis of endogenous Smn, as the SMA mice only express the human SMN2 transgene correctly, but not the murine Smn gene, which has been disrupted. Ensembl annotations were restricted to genes determined to be protein-coding. To focus the analysis on changes in splicing, we removed significant exonic regions that represented start or end regions within the transcript models, and were therefore not likely to be caused by changes in splicing, but rather by alternative transcriptional initiation or 3′-end formation. We did this by removing the regions that shared either their start position with a start position of a reference transcript, or their end position with an end position of one of the reference transcripts. We divided exonic regions into classes by the following criteria, in descending order: regions that fully matched known exons were classified as cassette exons; regions that were spanned by an upstream exonic region and a downstream intron were classified as alternative 5′ss; regions that were spanned by a downstream exonic region and an upstream intron were classified as alternative 3′ss; regions that had a start position matching a known 5′ss and an end position matching a known 3′ss were classified as retained introns; and regions that did not match any of these criteria were classified as uncategorized. Exon expression was modeled with a generalized linear model of the form: expression ∼ gender + condition. DEXSeq was run with independent filtering turned off in the DESeq2 package, as we found that this filter resulted in loss of detection of SMN2 skipping in some analyses, despite clear skipping. Cufflinks v. 2.1.1 was run using the following parameters: ‘–max-multiread-fraction 0.5 -j 0.1′. Cuffcompare v. 2.1.1 was used to assemble transcripts from all samples into a non-redundant set. A custom Perl script was used to sanitize the resulting GTF file before use with DEXSeq by removing duplicate gene annotations on different chromosomes or strands. U12-intron retention analysis was performed using DEXSeq and RefSeq annotations supplemented with a non-redundant list of 835 U12-dependent introns generated by combining U12DBv2 (63) annotations with introns in the Ensembl v.67 annotation that matched the 5′ss-3′ss consensus AT-AC or GTATCCT-AG. The set of genes analyzed was restricted to only those genes with at least one U12-intron. Similarly, for hg38 we generated a list of 1057 introns in 764 genes using U12DBv2 and Ensembl hg38 annotation version 79.
Pathway analysis
GO pathway analysis was performed using GOseq (64) while significantly expressed KEGG pathways were identified using Gage (65) and visualized using Pathview (66). Pathways with adjusted P-values below 0.1 were considered significant.
Coverage profiles
To compare coverage profiles between groups, we used Gviz (67) to plot the average of the four samples in each group. Prior to this, coverage of each sample had been normalized using size factors calculated by cqn.
RESULTS
We first collected tissue samples from severe SMA mice (68) sacrificed on postnatal day 1 (PND1) and postnatal day 5 (PND5). These time points were selected to allow us to compare an early pre-symptomatic stage with a later symptomatic stage. Additionally, at PND5, treatment with ASO therapy can only weakly delay disease progression (44,48), indicating that by this time point the disease has reached a critical stage. We collected tissue samples from spinal cord, brain, liver, and skeletal muscle, extracted total RNA and performed RNA-seq on 4 SMA mice and 4 litter-matched heterozygous control mice from each time point.
Few changes in gene expression at postnatal day 1
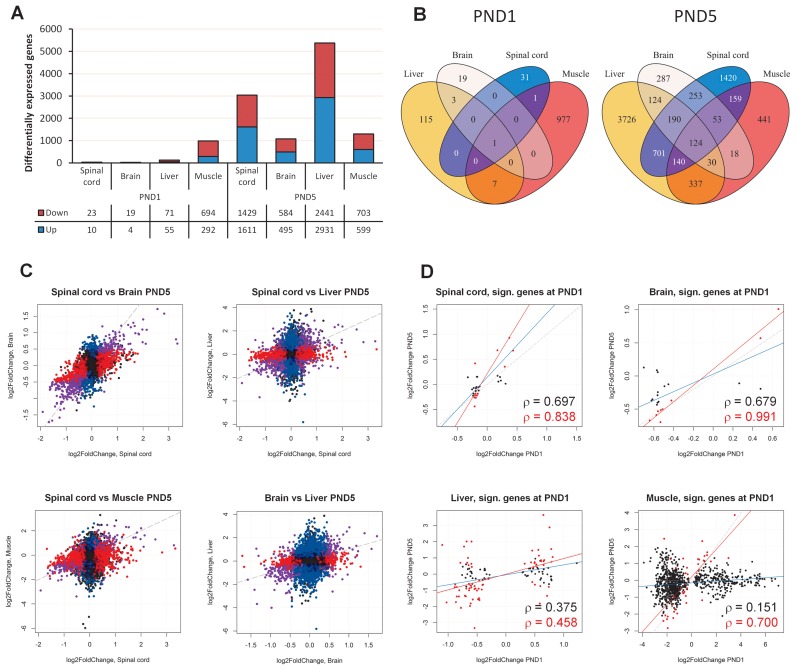
We first examined changes in gene expression between SMA mice and heterozygous mice at PND1 and PND5. At PND5, principal component analysis (PCA) indicated large differences between the two groups, and also that male and female SMA mice respond differently, as indicated by gender grouping in the spinal cord and liver samples (Supplementary Figures S1 and S2). We therefore accounted for the effect of gender in subsequent analyses (see Materials and Methods). At PND1, we identified only few differentially expressed genes at false-discovery rate (FDR) less than 10% or 0.1 (Figure 1A and Supplementary Figures S4 and S5, Table S7 and S8). By contrast, we identified several thousand genes up- or down-regulated at FDR < 0.1 in the SMA mice at PND5 (Figure 1A, Supporting data S1). These numbers indicate that among the tissues examined, the spinal cord and liver are affected more severely or that they are responding more strongly to the decrease in levels of SMN.
Figure 1.
Gene expression analysis. (A) Barplot showing the number of differentially expressed genes in SMA-like mice at PND1 and PND5 in spinal cord, brain, liver and muscle. The number of down- and up-regulated genes is indicated below the barplot. (B) Venn diagrams of the overlap of significant genes in different tissues at PND1 and PND5. (C) Scatterplots of log2 fold-change estimates in spinal cord, brain, liver and muscle. Genes that were significant in both conditions are indicated in purple, genes that were significant only in the condition on the x axis are indicated in red, genes significant only in the condition on the y axis are indicated in blue. (D) Scatterplots of log2 fold-changes of genes in the indicated tissues that were statistically significantly different at PND1 versus the log2 fold-changes at PND5. Genes that were also statistically significantly different at PND5 are indicated in red. The dashed grey line indicates a completely linear relationship, the blue line indicates the linear regression model based on the genes significant at PND1, and the red line indicates the linear regression model based on genes that were significant at both PND1 and PND5. Pearsons rho is indicated in black for all genes significant at PND1, and in red for genes significant at both time points.
We identified only few differences at PND1 in the spinal cord, brain and liver samples (Figure 1A) and PCA also indicated little overall difference between SMA mice and their heterozygous littermates (Supplementary Figure S1A and C, S2A and C).
Comparison of differentially expressed genes across tissues showed considerable overlap at PND5, however, only one gene at PND1 was differentially expressed in all tissues. This could be in part because there were generally few differentially expressed genes at PND1 (Figure 1B). The gene, Snrpa1, was up-regulated in all tissues both at PND1 and at PND5, and was also observed to be up-regulated at P7 in the Δ7 mouse model in an earlier microarray study (29). It is especially interesting since it encodes the A’ polypeptide that is part of the mature U2 snRNP (69), indicating a possible effect on splicing.
When we compared the log2 fold-changes at PND5 in spinal cord versus the log2 fold-changes in other tissues, it was clear that the direction of change was generally maintained, pointing to activation and deactivation of similar pathways in the different tissues (Figure 1C). There was a common overlap of 124 genes between all tissues, and 110 of these were either up-regulated in all tissues, or down-regulated in all tissues. Gene ontology (GO) analysis identified several down-regulated GO terms, in particular angiogenesis (Supporting data S3), indicating that limited supply of growth factors and nutrients may be a contributing factor to SMA pathogenesis in several tissues.
We further examined the degree of agreement between PND1 and PND5 by comparing the log2 fold-change estimates of the differentially expressed genes at PND1 with their log2 fold-changes at PND5 (Figure 1D, Supporting data S2). In the spinal cord, there were 33 significantly altered genes at PND1, and linear regression analysis on these genes showed overall agreement between the two time points. Similarly, in the brain there was a high correlation between the two time points particularly for those genes that were significantly changed at both time-points. Correlation was also observed between the overlap of genes in muscle and liver. Taken together, these data indicate that the changes of gene expression levels represent a consistent response to decreasing levels of SMN protein.
Cell division is down-regulated in SMA mice at PND5 while stress-genes are up-regulated
To identify the overall cellular pathways that are significantly altered in the tissues of SMA mice, we performed GO enrichment analysis on the significant genes (Supporting data S4–7). This analysis indicated that pathways and processes associated with cell-division were significantly down-regulated in the spinal cord at PND5, in particular mitotic-phase genes (Supporting data S4). In a recent study using an inducible adult SMA mouse model, reduced cell division was reported as one of the primary affected pathways that could be reversed with ASO treatment (46). In particular, up-regulation of Cdkn1a and Hist1H1C were reported as the most significant genotype-driven changes and similarly we observe the same up-regulation in spinal cord at PND5. There were no significantly enriched GO terms when we analyzed the up-regulated genes, but we did observe an up-regulation of Mt1 and Mt2 (Figure 2B), which are metal-binding proteins up-regulated in cells under stress (70,71). These two genes are also among the genes that were up-regulated in all tissues at PND5 and, notably, they were also up-regulated at PND1 in several tissues (Figure 2C). This indicates that while there were few overall differences at PND1 between SMA and heterozygous mice, increased cellular stress was apparent at the pre-symptomatic stage. Furthermore, GO terms associated with angiogenesis were down-regulated, and we observed the same at PND5 in the brain, where these were among the most significantly down-regulated GO terms (Supporting data S5). Likewise, angiogenesis seemed to be affected in the liver and the muscles (Supporting data S6 and S7), and in the muscles the cell cycle seemed to be down-regulated. The PI3K–Akt signaling pathway was down-regulated in the liver at PND5, also indicating impaired cellular growth (Supporting data S6) and in line with the adult induced SMA mouse model (46).
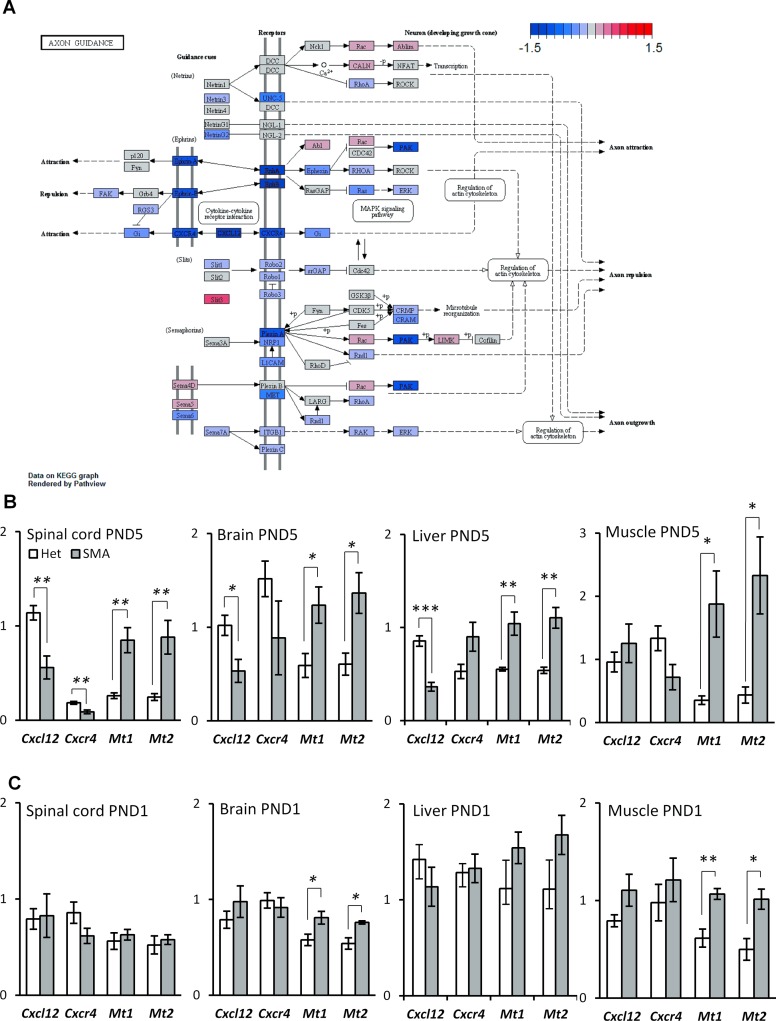
Figure 2.
Expression of axon guidance genes is down-regulated in SMA-like mice at PND5 while stress genes are up-regulated. (A) Schematic depiction of the axon guidance pathway in mice from the KEGG database. Gene regulation is indicated by a color gradient going from down-regulated (blue) to up-regulated (red) with the extremity thresholds of log2 fold-changes set to −1.5 and 1.5, respectively. (B) qPCR validation of differentially expressed genes in SMA-like mice at PND5. (C) qPCR validation of differentially expressed genes in SMA-like mice at PND1. Error bars indicate SEM, n ≥ 3, **P-value < 0.01, *P-value < 0.05. White bars indicate heterozygous control mice, grey bars indicate SMA-like mice.
Axon guidance genes Cxcl12 and Cxcr4 are down-regulated at PND5
We observed significantly altered KEGG pathways (Kyoto Encyclopedia of Genes and Genomes; Supporting data S4–7) and significant down-regulation of the axon-guidance pathway in spinal cord at PND5 (Tables 1 and 2). Some of the most strongly down-regulated genes were Cxcl12 and its receptor, Cxcr4 (Figure 2A, Table 2). When we examined expression of these genes across all samples by qPCR, we found Cxcl12 to be down-regulated in brain and liver as well, but not at PND1 (Figure 2B), while Cxcr4 also appeared to be down-regulated in several tissues.
Table 1. The significantly down-regulated KEGG pathways in spinal cord at PND5.
| Pathway | P-value | q-value | No. of genes in pathway |
|---|---|---|---|
| mmu04110 Cell cycle | 1.31E−07 | 3.60E−05 | 122 |
| mmu04360 Axon guidance | 6.90E−06 | 0.000953 | 128 |
| mmu04114 Oocyte meiosis | 0.000354 | 0.032538 | 106 |
| mmu04914 Progesterone-mediated oocyte maturation | 0.001139 | 0.078583 | 86 |
| mmu05206 MicroRNAs in cancer | 0.001614 | 0.089089 | 140 |
Table 2. Significantly differentially expressed genes in the axon guidance pathway at PND5 in the spinal cord.
| Ensembl ID | Symbol | Description | log2FoldChange | P-value | padj |
|---|---|---|---|---|---|
| ENSMUSG00000029710 | Ephb4 | Eph receptor B4 [Source:MGI Symbol;Acc:MGI:104757] | −0.61541736 | 3.59E−10 | 2.33E−08 |
| ENSMUSG00000045382 | Cxcr4 | chemokine (C-X-C motif) receptor 4 [Source:MGI Symbol;Acc:MGI:109563] | −0.982131677 | 1.16E−09 | 6.75E−08 |
| ENSMUSG00000020099 | Unc5b | unc-5 homolog B (C. elegans) [Source:MGI Symbol;Acc:MGI:894703] | −0.498514056 | 6.92E−08 | 2.86E−06 |
| ENSMUSG00000061353 | Cxcl12 | chemokine (C-X-C motif) ligand 12 [Source:MGI Symbol;Acc:MGI:103556] | −1.129675774 | 9.65E−08 | 3.84E−06 |
| ENSMUSG00000026259 | Ngef | neuronal guanine nucleotide exchange factor [Source:MGI Symbol;Acc:MGI:1858414] | −0.370710424 | 4.43E−07 | 1.57E−05 |
| ENSMUSG00000027852 | Nras | neuroblastoma ras oncogene [Source:MGI Symbol;Acc:MGI:97376] | −0.277599616 | 2.52E−06 | 7.19E−05 |
| ENSMUSG00000032562 | Gnai2 | guanine nucleotide binding protein (G protein), alpha inhibiting 2 [Source:MGI Symbol;Acc:MGI:95772] | −0.325593742 | 7.26E−06 | 0.000180106 |
| ENSMUSG00000030539 | Sema4b | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B [Source:MGI Symbol;Acc:MGI:107559] | 0.375467998 | 3.07E−05 | 0.000614462 |
| ENSMUSG00000027954 | Efna1 | ephrin A1 [Source:MGI Symbol;Acc:MGI:103236] | −0.624353799 | 3.38E−05 | 0.000662811 |
| ENSMUSG00000029095 | Ablim2 | actin-binding LIM protein 2 [Source:MGI Symbol;Acc:MGI:2385758] | 0.237941574 | 0.000103113 | 0.001693553 |
| ENSMUSG00000056427 | Slit3 | slit homolog 3 (Drosophila) [Source:MGI Symbol;Acc:MGI:1315202] | 0.507131782 | 0.000143304 | 0.002233933 |
| ENSMUSG00000031217 | Efnb1 | ephrin B1 [Source:MGI Symbol;Acc:MGI:102708] | −0.503547146 | 0.000180732 | 0.002694972 |
| ENSMUSG00000031391 | L1cam | L1 cell adhesion molecule [Source:MGI Symbol;Acc:MGI:96721] | −0.300231258 | 0.000318591 | 0.004288775 |
| ENSMUSG00000029168 | Dpysl5 | dihydropyrimidinase-like 5 [Source:MGI Symbol;Acc:MGI:1929772] | −0.399882167 | 0.00033427 | 0.004474746 |
| ENSMUSG00000028664 | Ephb2 | Eph receptor B2 [Source:MGI Symbol;Acc:MGI:99611] | −0.320932658 | 0.000447098 | 0.005638681 |
| ENSMUSG00000026640 | Plxna2 | plexin A2 [Source:MGI Symbol;Acc:MGI:107684] | −0.292021681 | 0.000631786 | 0.007465337 |
| ENSMUSG00000022048 | Dpysl2 | dihydropyrimidinase-like 2 [Source:MGI Symbol;Acc:MGI:1349763] | −0.196633206 | 0.000719297 | 0.008222821 |
| ENSMUSG00000025810 | Nrp1 | neuropilin 1 [Source:MGI Symbol;Acc:MGI:106206] | −0.304492143 | 0.001188028 | 0.012042472 |
| ENSMUSG00000059921 | Unc5c | unc-5 homolog C (C. elegans) [Source:MGI Symbol;Acc:MGI:1095412] | −0.305730412 | 0.002565788 | 0.022064245 |
| ENSMUSG00000031398 | Plxna3 | plexin A3 [Source:MGI Symbol;Acc:MGI:107683] | −0.265106665 | 0.002827641 | 0.023693623 |
| ENSMUSG00000003070 | Efna2 | ephrin A2 [Source:MGI Symbol;Acc:MGI:102707] | −0.222009429 | 0.003008549 | 0.024887771 |
| ENSMUSG00000021904 | Sema3g | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G [Source:MGI Symbol;Acc:MGI:3041242] | −0.396876592 | 0.004839997 | 0.035346239 |
| ENSMUSG00000005958 | Ephb3 | Eph receptor B3 [Source:MGI Symbol;Acc:MGI:104770] | −0.239324667 | 0.005089011 | 0.036761342 |
| ENSMUSG00000033220 | Rac2 | RAS-related C3 botulinum substrate 2 [Source:MGI Symbol;Acc:MGI:97846] | 0.422992771 | 0.005092864 | 0.036770732 |
| ENSMUSG00000052133 | Sema5b | sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5B [Source:MGI Symbol;Acc:MGI:107555] | 0.194938034 | 0.005121675 | 0.036904754 |
| ENSMUSG00000059810 | Rgs3 | regulator of G-protein signaling 3 [Source:MGI Symbol;Acc:MGI:1354734] | −0.276075209 | 0.006178853 | 0.042220205 |
| ENSMUSG00000030084 | Plxna1 | plexin A1 [Source:MGI Symbol;Acc:MGI:107685] | −0.187031083 | 0.006338726 | 0.042964265 |
| ENSMUSG00000019647 | Sema6a | sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A [Source:MGI Symbol;Acc:MGI:1203727] | −0.179636461 | 0.010602229 | 0.062109349 |
| ENSMUSG00000052504 | Epha3 | Eph receptor A3 [Source:MGI Symbol;Acc:MGI:99612] | −0.418474638 | 0.012227601 | 0.068266031 |
| ENSMUSG00000074785 | Plxnc1 | plexin C1 [Source:MGI Symbol;Acc:MGI:1890127] | −0.29161526 | 0.012574303 | 0.069634854 |
| ENSMUSG00000028289 | Epha7 | Eph receptor A7 [Source:MGI Symbol;Acc:MGI:95276] | −0.25772596 | 0.012614666 | 0.069702846 |
| ENSMUSG00000020121 | Srgap1 | SLIT-ROBO Rho GTPase activating protein 1 [Source:MGI Symbol;Acc:MGI:2152936] | −0.146573343 | 0.013646582 | 0.073758114 |
| ENSMUSG00000029674 | Limk1 | LIM-domain containing, protein kinase [Source:MGI Symbol;Acc:MGI:104572] | 0.149975362 | 0.015313998 | 0.079827523 |
| ENSMUSG00000029765 | Plxna4 | plexin A4 [Source:MGI Symbol;Acc:MGI:2179061] | −0.206671388 | 0.017069775 | 0.086229278 |
| ENSMUSG00000053646 | Plxnb1 | plexin B1 [Source:MGI Symbol;Acc:MGI:2154238] | 0.180639586 | 0.019246017 | 0.094148523 |
| ENSMUSG00000022781 | Pak2 | p21 protein (Cdc42/Rac)-activated kinase 2 [Source:MGI Symbol;Acc:MGI:1339984] | −0.192645617 | 0.020665462 | 0.098401758 |
The genes are sorted by their P-value.
Expression of spurious transcripts via novel splice sites in SMA mice
The hypothesis that SMA pathogenesis is linked to aberrant splicing has been proposed in the past (15,27) and by this reasoning, it is likely that SMA cells produce aberrant transcripts that are not observable in normal cells. To investigate this scenario, we examined alternative splicing using increasingly comprehensive and diverse transcript annotations to distinguish aberrant splicing from regulated alternative splicing. RNA-seq is particularly well suited for the analysis of aberrant splicing, such as intron retention, relative to exon-arrays, which generally fail to detect any signal from expressed intronic sequences.
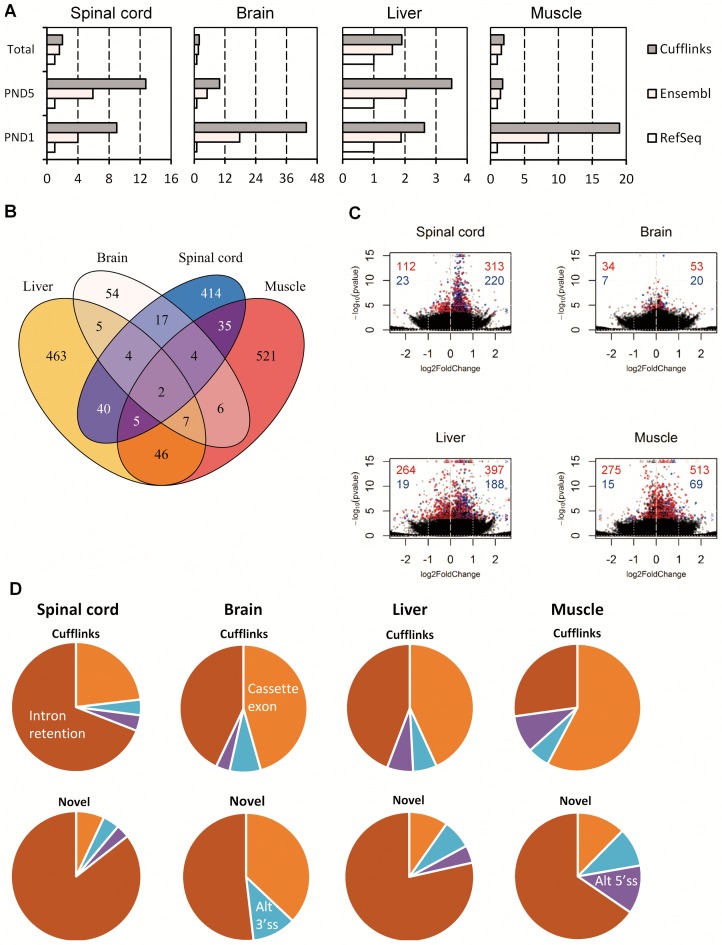
When we analyzed alternative splicing in the spinal cord with the RefSeq annotation (60) and compared it with the Ensembl annotation (53), we found an increase in the number of detected alternative splicing events that was approximately 4–6 times higher than expected from the increase in annotation size alone (Figure 3A, Supporting data S8 and S9). The increase in the number of differentially expressed regions was higher at PND5, indicating that the increase was not only due to the increased number of regions. Similarly, in brain, liver, and muscle, there was an increased number of differentially regulated exonic regions when using the Ensembl reference (Table 3). In muscle, the increase was not as pronounced as in the other tissues and was in fact in line with the increase in annotation complexity (Figure 3A, Table 3), but we identified fewer down-regulated exonic regions with the Ensembl annotation. We also observed the largest number of alternatively regulated exonic regions with the RefSeq annotation in the muscle, which explains why the relative increase is lower.
Figure 3.
Aberrant splicing with increased U2-intron retention in SMA-like mice. (A) Barplots showing the number of regions relative to the RefSeq annotation, either the total set of annotated regions, or those significantly alternatively spliced at PND1 or PND5 in the indicated tissues. (B) Venn diagram showing the overlap of genes with alternative splicing between tissues at PND5. (C) Volcano plots of exonic regions in spinal cord, brain, liver and muscle at PND5. Significantly differentially expressed regions overlapping known transcripts are indicated in red, novel significant regions are indicated in blue. Values exceeding chart limits are plotted at the corresponding edge and indicated by either up or downward facing triangle, or left/right facing arrow heads. Number of known and novel regions that are up or down-regulated are indicated in red (known) and blue (novel) in either the upper-left (down) or upper-right (up). (D) Piecharts of observed alternative splicing patterns in spinal cord, brain, liver and muscle at PND5 in either the full Cufflinks annotation or only the novel regions not overlapping previously annotated transcripts.
Table 3. Number of differentially expressed exonic regions at PND1 and PND5.
| Spinal cord | Brain | Liver | Muscle | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Reference | Regulation | PND1 | PND5 | PND1 | PND5 | PND1 | PND5 | PND1 | PND5 |
| Refseq | Up | 0 | 30 | 1 | 17 | 4 | 124 | 13 | 387 |
| Down | 3 | 50 | 1 | 14 | 4 | 124 | 1 | 285 | |
| Ensembl | Up | 1 | 296 | 2 | 43 | 8 | 331 | 19 | 477 |
| Down | 6 | 92 | 3 | 31 | 7 | 175 | 4 | 279 | |
| Cufflinks | Up | 1 | 533 | 8 | 73 | 11 | 585 | 30 | 582 |
| Down | 8 | 135 | 2 | 41 | 10 | 283 | 10 | 290 | |
| Novel (Cufflinks) | Up | 1 | 220 | 4 | 20 | 2 | 188 | 8 | 69 |
| Down | 2 | 23 | 0 | 7 | 5 | 19 | 0 | 15 | |
While the Ensembl annotation likely contains many transcripts that are non-functional and therefore can be described as aberrant, it does not contain all possible transcripts that can be expressed from all genes, and in particular it lacks transcripts that are expressed in cells with dysfunctional splicing due to a pathological condition. In order to detect these spurious transcripts, we used Cufflinks (61,62), to assemble the RNA-seq reads into novel transcripts using the reference-guided algorithm with the Ensembl annotation as reference. By this approach, we identified 9 differentially expressed exonic regions in seven genes in the spinal cord at PND1, and 668 exonic regions in 517 genes at PND5, at 10% FDR (Table 3, Supporting data S10). Again, the relative increase in the number of differentially expressed regions at PND5 between the Ensembl and Cufflinks derived annotations (72.6%) surpasses the relative increase in total exonic regions (21.1%) and because most of the differentially expressed regions were up-regulated (533), we conclude that spurious and likely aberrant transcripts are preferentially up-regulated in the spinal cord of SMA mice at PND5. Further corroborating this hypothesis, we observed that of the 668 differentially expressed regions, 243 (36.5%) did not overlap any of the known exonic regions in the RefSeq and Ensembl annotations, and of these, 220 (90.2%) were up-regulated in SMA mice, whereas only 23 (9.8%) were down-regulated. In the brain, liver and muscle, the results were similar with large increases at both PND1 and PND5 in the number of detected alternative splicing, and most of the de-novo exonic regions were up-regulated in the SMA mice (Figure 3A, Table 3, Supporting data S10).
A comparison of the genes with alternative splicing using the Cufflinks annotation between tissues (Figure 3B, Supplementary Figure S8) shows that most of the aberrant splicing was tissue-specific, as there was a relatively small overlap in the number of genes alternatively spliced in multiple tissues. Only two genes were alternatively spliced in all tissues at significant levels, albeit in different ways: Srsf5 and Srsf10, both of which are SR proteins that regulate splicing. We speculate that expression of SR proteins may be particularly sensitive to altered splicing fidelity, as they are known to regulate themselves and other SR proteins through alternative splicing (72–75). Changes in the splicing of splicing regulatory genes was also observed in another recent microarray study of SMA mice (76).
U2-dependent introns are retained in SMA mice
We then examined the patterns of alternative splicing by categorizing the exonic regions based on their overlap with the overall transcript structure (see Materials and Methods). The most prevalent type of missplicing detected with the RefSeq annotation was alternative cassette exon splicing (Table 4, Supplementary Figure S9), whereas analysis with the Ensembl annotation indicated that not only alternative 3′ and 5′ splice sites were being activated, but also that increased intron retention was prevalent in SMA mice. When we analyzed alternative splicing using the Cufflinks-derived annotation, pronounced intron retention became more evident, particularly in the spinal cord tissue, where the number of alternative splicing events increased considerably. More specifically, the aberrantly spliced transcripts were generally up-regulated in SMA mice (Figure 3C) and the majority was of the intron-retention type (Figure 3D). This pattern was even more pronounced when we examined only the novel regions, where more than three out of four novel splicing events were intron-retention. The same was true for the other tissues, indicating that increased intron retention is a tissue-wide phenomenon in SMA mice.
Table 4. Categorization of differentially expressed exonic regions using different annotations.
| Spinal cord | Brain | Liver | Muscle | |||||
| RefSeq | RefSeq | RefSeq | RefSeq | |||||
| Region | PND1 | PND5 | PND1 | PND5 | PND1 | PND5 | PND1 | PND5 |
| Cassette exon | 2 | 69 | 2 | 28 | 8 | 225 | 12 | 642 |
| Alt 3′ss | 0 | 2 | 0 | 0 | 0 | 7 | 1 | 7 |
| Alt 5′ss | 1 | 3 | 0 | 3 | 0 | 5 | 1 | 10 |
| Intron retention | 0 | 6 | 0 | 0 | 0 | 11 | 0 | 13 |
| Uncategorized | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ensembl | Ensembl | Ensembl | Ensembl | |||||
| PND1 | PND5 | PND1 | PND5 | PND1 | PND5 | PND1 | PND5 | |
| Cassette exon | 3 | 134 | 2 | 41 | 9 | 255 | 15 | 507 |
| Alt 3′ss | 0 | 11 | 1 | 5 | 1 | 34 | 0 | 36 |
| Alt 5′ss | 1 | 12 | 1 | 3 | 1 | 37 | 1 | 62 |
| Intron retention | 3 | 230 | 1 | 25 | 4 | 180 | 7 | 147 |
| Uncategorized | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| Cufflinks | Cufflinks | Cufflinks | Cufflinks | |||||
| PND1 | PND5 | PND1 | PND5 | PND1 | PND5 | PND1 | PND5 | |
| Cassette exon | 4 | 154 | 3 | 52 | 10 | 372 | 23 | 499 |
| Alt 3′ss | 0 | 26 | 1 | 9 | 1 | 52 | 1 | 48 |
| Alt 5′ss | 1 | 26 | 1 | 4 | 1 | 57 | 3 | 82 |
| Intron retention | 4 | 460 | 5 | 49 | 9 | 381 | 13 | 235 |
| Uncategorized | 0 | 2 | 0 | 0 | 0 | 6 | 0 | 8 |
| Novel (Cufflinks) | Novel (Cufflinks) | Novel (Cufflinks) | Novel (Cufflinks) | |||||
| PND1 | PND5 | PND1 | PND5 | PND1 | PND5 | PND1 | PND5 | |
| Cassette exon | 1 | 17 | 0 | 10 | 2 | 20 | 3 | 10 |
| Alt 3′ss | 0 | 10 | 0 | 3 | 0 | 15 | 0 | 8 |
| Alt 5′ss | 0 | 8 | 1 | 0 | 0 | 9 | 0 | 10 |
| Intron retention | 1 | 208 | 3 | 14 | 4 | 161 | 5 | 53 |
| Uncategorized | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 3 |
U12-dependent intron retention occurs in all examined tissues at PND5
Defective splicing of U12-dependent introns has previously been hypothesized to be a disease mechanism in SMA and has also been reported several times in SMA cells (31,32), and we therefore focused our splicing analysis on this class of introns. We noticed that some of the intron-retention events identified with the Cufflinks annotation were in fact U12-dependent introns (Supplementary Figure S9). This analysis only included U12-dependent introns that were identified by Cufflinks. Therefore, to analyze U12-dependent introns more generally we generated a set of U12-dependent introns by extracting 546 U12-dependent introns contained in the U12DBv2 database (63) and by extracting all introns in the Ensembl annotation that contained either an AT at the donor and an AC at the acceptor, or a GTATCCT sequence at the donor and an AG at the acceptor, both of which are splice-site combinations specific to U12-dependent introns (63). We then combined these annotations into a non-redundant set of 835 U12-dependent introns in the mm9 genome.
We realize that this approach most likely does not identify all U12-dependent introns in the mouse genome. Notably those that are not in the U12DBv2 database or have a different donor site than the AT or GTATCCT consensus that we selected, in addition to those that are not annotated as introns in the Ensembl annotation will be missed. However, we chose to be restrictive in order to avoid including by mistake any non-U12 introns in the analysis.
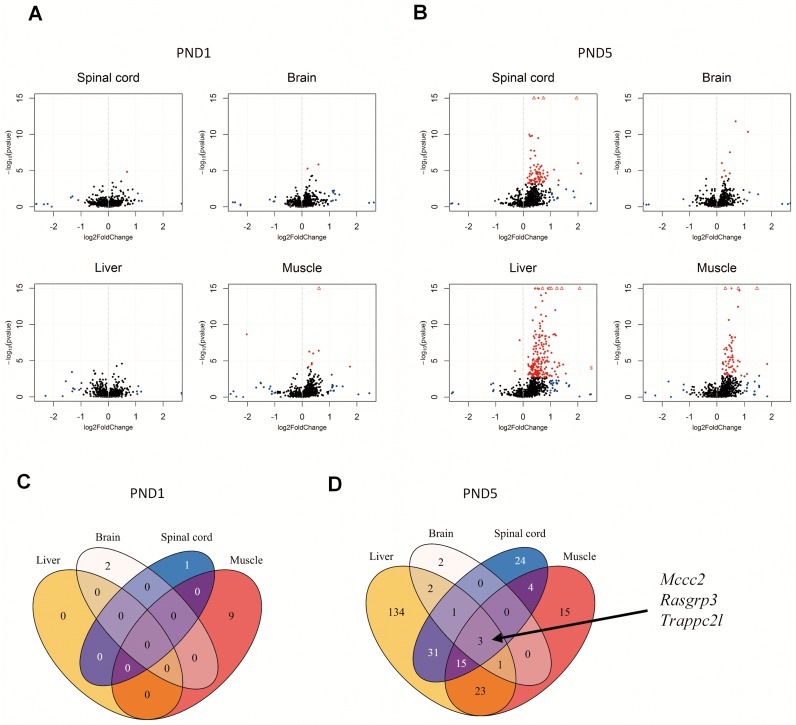
Next, we constructed reference gene annotations with transcripts containing these introns, as well as their normal transcripts from the RefSeq annotation. We further restricted our analysis to genes with U12-dependent introns, leaving us with 701 genes with one or more U12-dependent introns. We then analyzed both PND1 and PND5 samples, and using an FDR cut-off value of 0.1, we identified very few significant changes at PND1, but observed marked increases in the degree of retention of U12-dependent introns at PND5 in the SMA samples (Figure 4A and B, Supporting data S11). Notably, these events were almost exclusively up-regulated in the SMA samples, indicating increased retention of these U12-dependent introns.
Figure 4.
U12-intron retention increases with disease progression. (A) Volcano plots of U12-intron retention SMA-like mice at PND1 in spinal cord, brain, liver and muscle. Significantly differentially expressed introns are indicated in red. Non-significant introns with foldchanges > 2 are indicated in blue. Values exceeding chart limits are plotted at the corresponding edge and indicated by either up or downward facing triangle, or left/right facing arrow heads. (B) Volcano plots of U12-intron retention in SMA-like mice at PND5 in spinal cord, brain, liver and muscle. Significantly differentially expressed introns are indicated in red. Non-significant introns with fold-changes >2 are indicated in blue. Values exceeding chart limits are plotted at the corresponding edge and indicated by either up or downward facing triangle, or left/right facing arrow heads. (C) Venn diagram of the overlap of common significant alternative U12-intron retention across tissue at PND1. (D) Venn diagram of the overlap of common significant alternative U12-intron retention across tissue at PND1.
The cell cycle regulator Rasgrp3 is down-regulated in all tissues and its U12-dependent intron 4 is retained in SMA mice
The overlap of genes with significant U12-dependent intron retention at PND5 included a set of three genes, and of these, the U12-dependent intron in Rasgrp3 was among the most highly up-regulated in all tissues (Figure 4D, Supporting data S11). Rasgrp3 is a regulator of the cell cycle through activation of Ras proteins (77), and since down-regulation of the cell cycle was a general feature of the SMA tissues, aberrant intron 4 retention in Rasgrp3 may be one of the events underlying this change. We validated the increase in retention of the U12-dependent intron and the decrease of the levels of spliced Rasgrp3 by specific qPCR assays in all tissues, at both PND1 and PND5 (Figure 5 and 6).Since retention of the intron could lead to degradation of the transcript via the NMD pathway due to a premature termination codon (PTC) in the U12-dependent intron (Supplementary Figure S10), our observations point out that aberrant retention of the U12-dependent intron in the Rasgrp3 gene might be an underlying mechanism contributing to deregulation of the cell cycle in SMA mice.
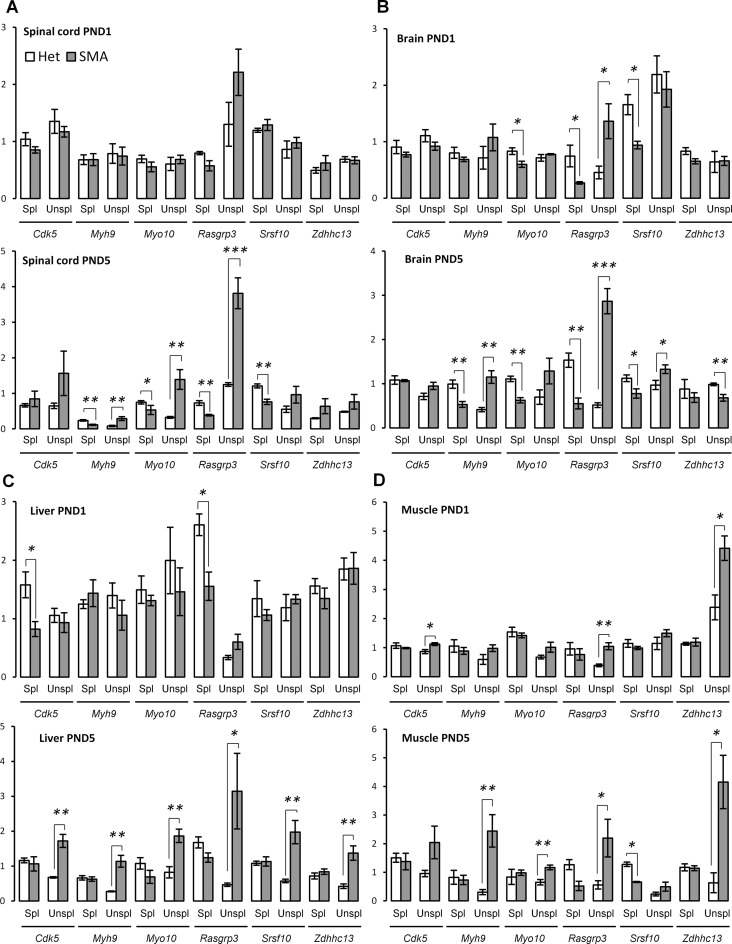
Figure 5.
Increased U12-dependent intron retention in SMA mice. (A) qPCR validation of U12-dependent intron retention at PND1 and PND5 in spinal cord. (B) qPCR validation of U12-dependent intron retention at PND1 and PND5 in brain. (C) qPCR validation of U12-dependent intron retention at PND1 and PND5 in liver. (D) qPCR validation of U12-dependent intron retention at PND1 and PND5 in muscle. Error bars indicate SEM, n ≥ 3, ***P-value < 0.001, **P-value < 0.01, *P-value < 0.05. White bars indicate heterozygous control mice, gray bars indicate SMA-like mice. Spl = spliced, Unspl = unspliced/retained intron.
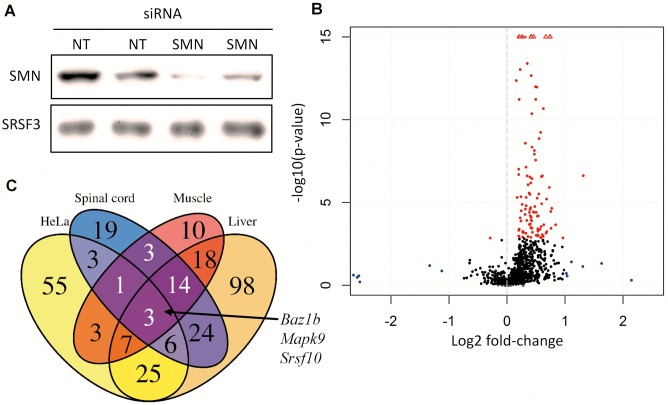
Figure 6.
Knockdown of SMN in HeLa cells recapitulate the findings in SMA mice. (A) Western blot of SMN-targeting siRNA (SMN) versus non-targeting siRNA (NT) samples showing reduced expression of SMN protein. (B) Volcano-plot of U12-dependent intron retention in SMN-deficient HeLa cells. Significantly differentially retained introns are indicated in red. Non-significant introns with fold-changes >2 are indicated in blue. Values exceeding chart limits are plotted at the corresponding edge and indicated by upward facing triangle. (C) Venn diagram showing overlap of genes with U12-dependent intron retention in SMA mice at PND5 and SMN depleted HeLa cells.
U12-dependent intron retention in genes important for neuronal function
Loss of Myo10 has recently been shown to inhibit axon outgrowth (78,79), and our RNA-seq data indicated that the U12-dependent intron 6 in Myo10 is retained, although not to a statistically significant degree. However, qPCR analysis showed that the U12-dependent intron 6 in Myo10 was in fact retained more in SMA mice than in their control littermates, and we observed significant intron retention at PND5 in spinal cord, liver, and muscle (Figure 6) and a significant decrease of spliced Myo10 in spinal cord at PND5 and in brain at both PND1 and PND5. These data suggest that Myo10 missplicing could play a role in SMA pathology.
Similarly, with qPCR we validated the up-regulation of U12-dependent intron retention in the Cdk5, Srsf10, and Zdhhc13 genes, which have all been linked to neuronal development and function (80–83). Curiously, hyperactivity of Cdk5 was recently reported to increase phosphorylation of tau in SMA neurons (84). We observed increased retention of a U12-dependent intron in Cdk5 in both muscle and liver at PND5, while it was slightly more retained in the spinal cord, but at a very low level (Supporting data S11, Supplementary Figure S11). Analysis using specific qPCR assays confirmed up-regulation of the intron in liver and muscle (Figure 6A and B) and also indicated down-regulation of the spliced transcript in liver at PND1 (Figure 6A), while there was only minor intron retention in muscle at PND1 (Figure 6B).
We also observed retention of the U12-dependent intron in the Stasimon/Tmem41b gene that was previously reported as misspliced (32), although at very low expression levels. Since these earlier results indicated that Tmem41b is spliced more aberrantly in some neurons than others, this may result in overall small changes when examining total spinal cord and also indicates that small changes may reflect highly biologically relevant changes in some cell populations.
In the spinal cord, a U12-dependent intron in Myh9 was retained, together with down-regulation of Myh9 transcripts in general. As in the case of Rasgrp3, this may be due to a PTC being present within the U12-dependent intron (Supplementary Figure S13). Validation with qPCR confirmed the down-regulation of spliced Myh9 and up-regulation of unspliced Myh9 in both spinal cord and brain (Figure 5A and B), whereas there was increased retention of the U12-dependent intron in liver and muscle (Figure 5C and D). This could indicate that the NMD pathway is more effective in the CNS, and perhaps explain why we detected more U12-dependent intron retention events in peripheral tissue than in the CNS.
We therefore examined the relationship between U12-dependent intron retention and gene expression (Supplementary Figure S14). There was very little correlation in the liver, followed by moderate correlation in spinal cord and muscle. However, in the brain there was a clear inverse linear relationship, indicating that NMD is an important pathway in the brain. This may explain why we observed fewer aberrant splicing events in this tissue compared the other tissues. These results also corroborate the hypothesis that increased retention of U12-dependent introns leads to degradation of the transcripts.
Knock-down of SMN in HeLa cells recapitulates the U12-dependent intron retention of the SMA mouse model
If SMA pathology is indeed related to a generalized defective splicing of U12-dependent introns, this defect should be present in human cells as well. To examine this, we performed siRNA mediated knock-down of SMN protein in HeLa cells. After 48 hours we harvested the cells and conducted RNA-seq on knock-down samples and controls prepared with a non-targeting siRNA (Figure 6A).
As in the mice, there was a similar striking increase in U12-dependent intron retention in the knock-down samples (Figure 6B), and many of the genes with affected U12-dependent introns in the mice were also affected in HeLa cells (Figure 6C, Supplementary Figure S15B). In total, 103 genes had U12-dependent intron retention in one or more introns. There are likely several more introns in other genes affected by the decrease in SMN levels. These early changes in U12-dependent intron retention following reduced SMN levels indicate that the effect on splicing is an early effect of SMN decrease.
Overall, we observed relatively few significant changes in gene expression compared to what we observed in mice, only 110 genes had significantly altered expression (Supplementary Figure S15A, Supporting data S1). Of note, however, there was an increase in the expression of several mitochondrial genes, indicating an increased number of mitochondria in the cells. We did not detect changes in the Mt1 and Mt2 homologues nor in CXCL12 or CXCR4, indicating that the same stress mechanisms observed in the mouse tissues are not activated in the HeLa cell culture. Therefore, the U12-dependent intron retention is independent of the stress response otherwise activated in SMA mice.
Supporting that splicing differences are more pronounced than gene expression changes, we found splicing changes in 778 genes when we used the same methodology as previously described to examine splicing of known and novel transcripts by Cufflinks and DEXSeq (Supporting data S10). Of these, 152 were intron retention events even though this method does not specifically test for intron retention, either U2 or U12-dependent, but relies on Cufflinks including them in the set of transcripts. The overlap of genes alternatively spliced in the SMA mice and HeLa SMN knockdown was generally small, only two genes were consistently alternatively spliced in all tissues and in HeLa, Srsf5 and Srsf10 (Supplementary Figure S15D). In the case of Srsf5, the splicing events were different from what we observed in the SMA mice; where we observed up-regulation of a distal 5′ss and alternative splicing of an intron and a cassette exon, we observed down-regulation of a distal 3′ss in the HeLa SMN knockdown. However, in the case of Srsf10 the region alternatively spliced in the HeLa SMN knockdown overlaps the dual U12-dependent introns that were detected as retained by the Cufflinks/DEXSeq approach in the SMA mice.
Another gene with an intron consistently affected by decreased SMN levels both in SMA mice and the HeLa knock-down experiment was Baz1b, a transcription factor and regulator of chromatin structure recently shown to regulate the transcription of neurodevelopmental genes (85). Furthermore, Baz1b is involved in DNA damage response (86), and DNA damage is increased in SMA mouse models, including the ‘Taiwanese’ model used in this study (87). This suggests that Baz1b missplicing could lead to a reduction in DNA damage response. Furthermore, Baz1b also regulates the maintenance of correct chromatin structure during DNA replication (88), suggesting that Baz1b missplicing might also contribute to impaired cell division.
Similarly, we detected increased U12-dependent intron retention in MYO10 and SRSF10, and also in TMEM41B, but not in RASGRP3, MYH9, ZDHHC13 or CDK5. With the exception of MYH9, this is likely due to low levels of expression that preclude a reliable detection of intron retention. That we could not detect missplicing of MYH9 could be due to NMD degradation of the aberrant transcripts, but it could also indicate that MYH9 missplicing might not be as pronounced in HeLa cells or even in humans. Further validation is required to establish whether this is true.
Restoration of SMN by ASO therapy reverses the expression profile of SMA mice and mitigates intron retention
One important question is whether the restoration of SMN levels by correcting the splicing of SMN2 exon 7 with an antisense oligonucleotide (ASO) can reverse the splicing deficiency in the SMA mice along with the other transcriptional characteristics. The effects of small-molecule restoration of SMN were recently examined in a few specific cases of aberrant splicing (47), but effects on the global scale and with ASO technology have not yet been characterized. To address this we treated SMA mice with either saline or 100 μg ASO10–29 per gram of body weight using subcutaneous injections in two separate doses on PND0 and PND1, and took out tissue samples for RNA-seq from spinal cord, brain and liver. This ASO has previously been shown to rescue splicing of SMN2 exon 7 (89).
First, we confirmed that ASO treatment restored levels of SMN2 exon 7 inclusion in all tissues (Figure 7A, Supplementary Figure S16A) to similar levels as previous studies using SC injections of the ISS-N1 blocking ASO10-27 (44). Then, we analyzed the gene expression and compared ASO treated SMA mice with saline treated SMA mice to identify genes that responded to the treatment. In spinal cord there was a high degree of overlap between genes that respond to ASO treatment and genes, which are deregulated in SMA mice. Between those two groups, there was an inverse correlation indicating reversal of the gene expression profile consistent with a reversal of the phenotype (Figure 7B) and with earlier microarray findings (46). The same was true in brain and liver, although the effects were less pronounced (Supplementary Figure S16B and C). Similarly, when we analyzed splicing changes with the Cufflinks annotation, we found that there was an inverse correlation between splicing changes in SMA mice and those in ASO treated mice, indicating a reversal of the alternative splicing (Supplementary Figure S17).
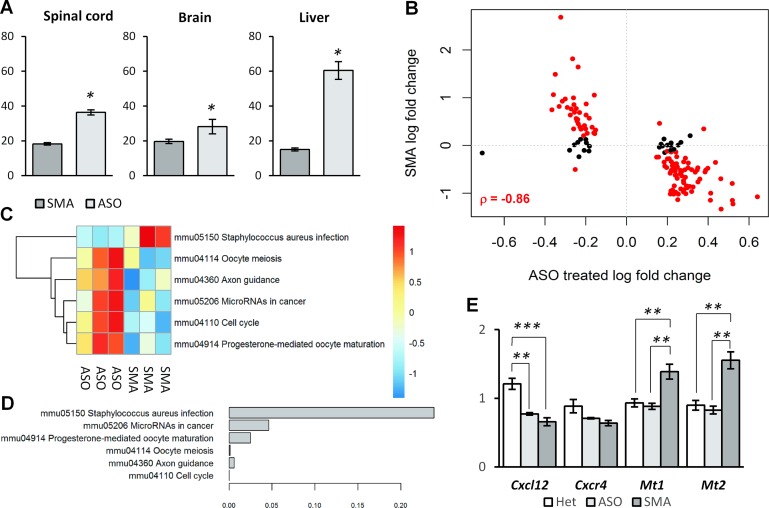
Figure 7.
ASO treatment reverses the expression and splicing profiles of SMA. (A) ASO treatment increases inclusion of SMN2 exon 7. Inclusion percentage were estimated from RT-PCR product sizes quantitated on a fragment analyzer. (B) Scatter plot of log2 fold-changes of significant genes in spinal cord of ASO treated mice relative to saline treated SMA mice and log2 fold-changes of the same genes in SMA mice relative to heterozygous controls. Genes that are significant in both comparisons are indicated in red and Pearson's correlation is indicated for these genes. (C) Heatmap of the average gene expression of genes in pathways significantly altered in the spinal cord of SMA mice relative to healthy controls. Gene expression values are derived from regularized log values and the normalized across samples setting the average to zero. (D) Barplot showing unadjusted P-values for the testing of reversal of the pathway in ASO treated mice relative to untreated SMA mice. (E) qPCR measurements of genes previously found to be deregulated in the spinal cord of SMA mice.
We then compared the pathways previously observed to be significantly altered in SMA mice, and found that the majority were reversed in the ASO treated SMA mice (Figure 7C and D). Importantly, cellular stress seemed to be alleviated as the expression levels of the stress markers Mt1 and Mt2 were down-regulated to heterozygous levels in the ASO treated SMA mice (Figure 7E). Furthermore, the axon guidance pathway activity was increased in the ASO treated SMA mice relative to saline treated SMA mice. Although we observed a trend indicating up-regulation, the increase in Cxcl12 and Cxcr4 levels did not reach significance in the qPCR validation (Figure 7E, Supplementary data S1). This could indicate that the phenotype is not completely reversed, or that the axon guidance pathway may also be down-regulated through factors other than Cxcl12 and Cxcr4.
Next, we analyzed the effect of ASO treatment on U12-dependent intron retention and found that increasing SMN levels through ASO treatment reverses the aberrant intron retention in SMA mice and promotes correct splicing of U12-dependent introns (Figure 8A). While we did not observe correction of splicing in the brain, this may be due to lower ASO efficacy in the brain as indicated by the observed levels of SMN2 exon 7 splicing correction (Figure 7A).
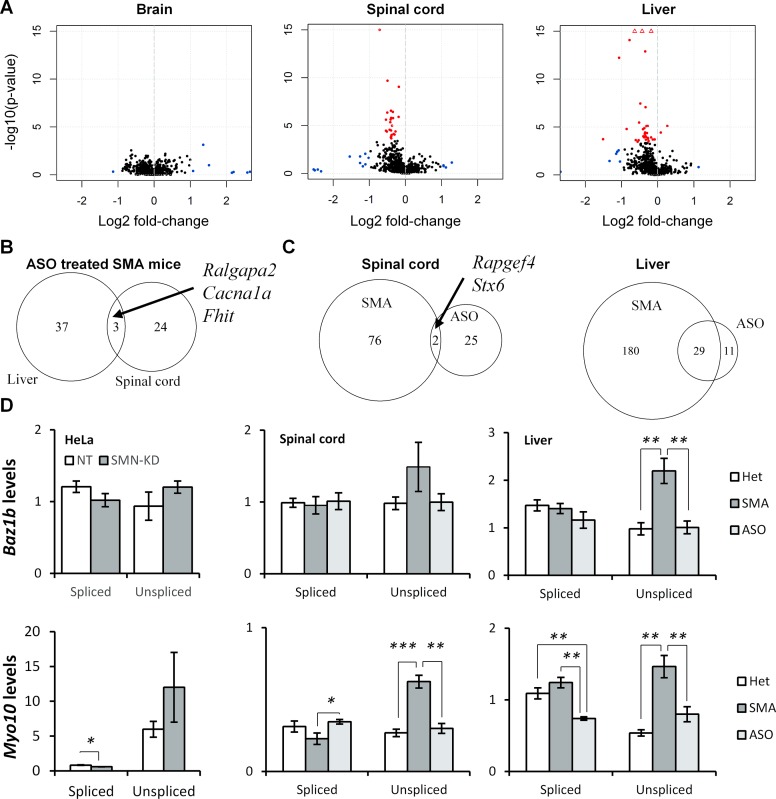
Figure 8.
ASO treatment restores correct splicing of U12-dependent introns in SMA mice. (A) Volcano plots of U12-dependent introns in brain, spinal cord and liver of ASO treated SMA mice. Significantly differentially retained introns are indicated in red. Non-significant introns with fold-changes >2 are indicated in blue. Values exceeding chart limits are plotted at the corresponding edge and indicated by upward facing triangle. (B) Overlap of introns with significantly improved splicing in liver and spinal cord of ASO treated SMA mice. SMA = saline treated SMA mice, ASO = ASO treated SMA mice. (C) Overlap of introns significantly retained in SMA mice and introns significantly improved in ASO treated SMA mice in spinal cord and liver. (D) qPCR analysis of BAZ1B/Baz1b and MYO10/Myo10 U12-dependent intron splicing in HeLa SMN-KD samples, and spinal cord and liver in ASO treated SMA mice.
The degree of overlap between introns that were corrected in spinal cord versus liver was relatively small (Figure 8B), especially considering that more than half of the aberrantly retained U12-dependent introns in spinal cord were also aberrantly retained in the liver (Figure 4C). Similarly, there was little overlap of introns corrected in spinal cord of ASO treated SMA mice and those aberrantly spliced relative to heterozygous controls (Figure 8C). In the liver however, almost three out of four corrected introns had previously been detected as aberrantly retained, including the U12-dependent intron in Rasgrp3. This indicates an overall improvement in the splicing of U12-dependent introns in the liver, which is consistent with the observed higher correction of SMN2 exon 7 inclusion.
Using qPCR we specifically confirmed that ASO treatment restores splicing of the U12-dependent intron in Baz1b to levels equivalent to those in heterozygous control mice (Figure 8D). Likewise, Myo10 splicing is restored in the spinal cord of ASO treated SMA mice and the retention is ameliorated in liver as well (Figure 8D).
Interestingly, among the genes with rescued U12-dependent intron retention in spinal cord there were several encoding the pore-forming subunit in voltage-gated Ca2+ channels: Cacna1a, Cacna1b, Cacna1c, Cacna1e and Cacna1h. Disruption of Ca2+ channel clustering and cellular Ca2+ homeostasis is one of the hallmarks of SMA (90,91), and elevated Ca2+ level leads to activation of Cdk5 through cleavage of p35 (92). We therefore suggest that aberrant splicing of U12-dependent introns in genes encoding Ca2+ channels contribute to disrupted Ca2+ homeostasis leading to axon terminal dysfunction and Cdk5 hyperactivity.
DISCUSSION
SMA has been known for long to be caused by decreased levels of SMN protein, but the molecular pathways involved in the manifestation of the clinical symptoms are not clearly understood. Among important questions raised, is the extent of the involvement of diverse functions of the SMN protein in the disease pathology, in particular the question of the role of aberrant splicing has been raised, but without a clear answer emerging.
Here, we used RNA-seq to investigate the consequences of low levels of SMN on the transcriptome in multiple tissues in SMA mice, and found evidence to support aberrant splicing as one of the contributing disease mechanisms in SMA.
In particular, we found that stress was elevated in several tissues and that in the spinal cord the axon guidance pathway was down-regulated. Cell division pathways were also significantly down-regulated across all tissues. Treatment of SMA mice with an ASO that restores splicing of SMN2 exon 7 resulted in decreased stress and reactivation of the axon guidance pathway as well as cell division pathways, consistent with the remarkable reversal of the phenotype previously observed (44). Interestingly, up-regulation of the stress-genes Mt1 and Mt2 was also reported in an earlier microarray study of gene expression in the spinal cord in the Δ7 SMA model at PND13 (29), indicating that the stress is characteristic of SMA. Similarly, angiogenesis seemed to be inhibited across all tissues examined in SMA mice, possibly limiting the supply of nutrients, growth factors and oxygen to developing cells. Consistent with a hypoxic stress response, hypoxia-inducible factor Hif-3α was up-regulated app. 2.5–4-fold in all tissues at PND5 in SMA mice, which was also observed to be up-regulated in a previous microarray study of spinal cord (29). However, unlike Zhang et al. (28) we did not observe alternative splicing of Hif-3α. Elevated ER stress specific to SMA motor neurons has been reported (93), but in our data from total spinal cord we did not find evidence for up-regulation of this pathway, likely because this stress may be highly specific to the motor neurons.
It could be speculated that many of the transcriptomic changes identified in this study are activated by cellular stress, especially by the hypoxic stress response activated in SMA mice. However, upon examining RNA-seq data from a study on hypoxic stress (94), we found no indication of increased U12-dependent intron retention as a response to hypoxia or siRNA targeting HIF-1α or HIF-2α (Supplementary Figure S18). Additionally, up-regulation of p38MAPK levels following stress has been reported to modulate U12-dependent introns and increase their splicing (95), but we only detected changes in the expression of p38MAPK kinases in the liver and muscle at PND5. In the liver Mapk11 and Mapk12 were down-regulated, whereas Mapk13 was up-regulated, and in the muscle Mapk11 was also down-regulated. Similarly, an earlier study reported that the phosphorylation state of p38MAPK isoforms is not altered in SMA mice (96). Thus, while we cannot rule out that stress is involved in the observed increase in U12-dependent intron retention, we cannot find evidence to suggest that it is the primary cause of aberrant U12-dependent intron processing. Considering the established role of SMN in snRNP assembly, it seems more likely that decreased SMN levels directly influence the aberrant processing of U12-dependent introns. The stress-related changes may be a secondary effect of the SMA pathology, but as SMN deficiency has been shown to sensitize cells to stress (97), stress may further exacerbate the symptoms. Treatments that minimize cellular stress may thus be beneficial to SMA patients.
In addition to elevated cellular stress, cell proliferation pathways were also down-regulated across the tissues examined in this study. Specifically, we noted missplicing of the U12-dependent introns in Baz1b in most tissues, and in Rasgrp3 in all tissues, and down-regulation of correctly spliced Rasgrp3 in tissues at both PND1 and PND5. Rasgrp3 is an activator of Ras, and suppression of Rasgrp3 inhibits cell proliferation, whereas over-expression enhances cell proliferation (77). Similar to others (29,46,98), we also observed up-regulation of the Cdk1 inhibitor Cdk1na involved in p53-mediated cell-cycle arrest and of several histone cluster 1 genes (Supplementary data S1). Additionally, U7 snRNP levels are decreased when SMN levels are low (16), and this may also contribute directly to a decrease in cell proliferation. SMN may thus directly and through several distinct pathways influence cell proliferation in addition to any stress related inhibition of cell growth.
Comparative analysis of our results from the analysis of spinal cord at PND5 with two previous microarray studies that analyzed spinal cord expression in SMA mice vs. normal and heterozygous controls (28,29) shows similar expression profiles, but at different time-points (Supplementary Figure S19). Apart from differences in sensitivities between RNA-seq and microarrays, one explanation for the temporal difference could be that the former studies examined gene expression in the Δ7 mouse model (99), while we used the ‘Taiwanese’ model (68). This model has a shorter life span and appears to have an expression profile at PND5 that is more similar to the expression profile of the Δ7 model at PND11–13, than at PND7. One possible explanation is that the general intracellular stress and therefore expression profile of motor neuron dysfunction persists throughout a longer period, and consistent with a shorter lifespan, the cellular stress is more severe at an earlier time-point in the ‘Taiwanese’ model, than in the Δ7 model.
Similar to previous studies (28,29,76) we also observed alternative splicing of Uspl1 across all tissues and we also noticed alternative splicing of genes associated with U snRNPs, such as Snrpa1 and Rnpc3, that were likewise observed in previous exon-array studies (28,29). Although the change in splicing of Uspl1 in the spinal cord was apparent from the RNA-seq data it was not statistically significant, but we were able to confirm it in a PCR reaction (data not shown). Uspl1 was recently reported to interact with snRNA loci, and knock-down of Uspl1 resulted in decreased snRNP levels (100). The exon which is up-regulated in Uspl1 would seemingly lead to more production of the Uspl1 protein, and in liver and muscle, we observed up-regulation of Uspl1 transcripts. Similarly, it is noteworthy that we observed significant up-regulation of Snrpa1 in all tissues at both PND1 and PND5, while we observed decreased intron retention in Snrpa1, which could lead to the increased expression, as fewer transcripts would be NMD-sensitive. These changes do not appear to be alternative splicing caused by defective splicing, but instead may be regulated splicing of genes as a response to the low SMN levels. These alternative splicing events may also be a specific response to altered snRNP levels and using primers from (28), we examined snRNA levels in treated and untreated SMA mice at PND5, as well as HeLa cells with SMN-KD. In the liver, we observed down-regulation of several snRNAs in SMA mice, which was corrected by ASO treatment, and we also observed trends towards decreased snRNA levels in the HeLa cells (Supplementary Figure S20). Surprisingly, we did not observe the same in spinal cord and brain, in contrast with earlier studies (26,28,30). These conflicting results in relation to earlier studies may be due to differences in the analysis methods employed, such as specific immuno-precipitation of mature snRNP complexes vs total RNA, and cDNA synthesis using specific snRNA primers versus random primers. Tissue differences between liver and CNS may be due to a number of reasons such as differences in snRNA turnover, differences in RNA sample stability, and the proportion of cells affected within the sample, which may be higher in the liver. Therefore, although we did not detect differences in snRNA levels in all tissues, there may still be a significant perturbation in the levels of active snRNP complexes in the affected tissues, as indicated by the altered expression and splicing of several snRNP associated proteins.
It is possible that the alternative splicing of splicing-regulatory proteins, such as SR proteins, is also a case of regulated alternative splicing in response to the disruption of the splicing machinery, rather than being directly caused by decreased splicing fidelity. However, if SR proteins are more sensitive to decreased splicing fidelity, increasing missplicing of SR proteins may explain in part the increasing levels of aberrant splicing with the SMA stage in animal models, as suggested previously (76). In general, there is little overlap between the detected splice changes in this study and a recent exon array study (Supplementary Figure S21), which may be explained by the differences in assay technology and animal model. Thus, we did not detect alternative splicing of Chodl or Uba1, which have been investigated previously (29,101,102), while we detected minor skipping in the Nrxn2 gene of an exon different from the previously reported (103). We did however detect down-regulation of the Uba1 gene in spinal cord, and also aberrant retention of U12-dependent introns in the Uba3, Ubeh2, and Ubl5 genes that support disruption of the ubiquitin homeostasis as previously suggested (101). We also detected missplicing of U12-dependent introns in UBA3 and UBEH2 in HeLa SMN-KD experiments, indicating that disruption of this pathway through U12-dependent intron retention may be present in human patients as well.
In a recent RNA-seq study of micro-dissected motor neurons, Zhang et al. (104) reported up-regulation of the C1q genes in SMA motor neurons, and consistent with this, we observed up-regulation of the C1qa, C1qb and C1qc genes in the spinal cord at PND5 (Supporting data S1). This indicates that it is possible to detect changes at PND5 that may be causative at PND1, but that changes are more pronounced at PND5 and therefore more easily detected. This underscores the relevance of studying mice at later stages in the disease progression. Zhang and coworkers also reported prominent skipping of the Z exons in agrin, but we did not find any evidence of missplicing in the Agrn gene in the spinal cord or brain samples taken at PND1 or PND5. It is possible that this skipping is activated in a limited cell population and therefore not apparent in our total spinal cord samples.
One major issue raised by this and other studies, is the lack of transcriptomic changes at PND1 prior to symptoms, which makes it difficult to differentiate between primary changes directly caused by SMN loss and secondary changes caused by a general cell dysfunction, when examining the transcriptome at a later symptomatic point, such as PND5. One explanation could be that a reduced SMN level is not critical during embryonic development and that it does not manifest significantly until a time-point later than PND1. Decreased SMN protein levels are caused by skipping of exon 7 and it is possible that during the embryonic stage SMN2 exon 7 is included at sufficient levels due to altered expression of splicing factors. During the postnatal period the SMN protein levels are then slowly decreased after which cellular dysfunction becomes apparent. Importantly, both we and others detect missplicing of splicing factors in SMA mice (76), including Tial1, which activates SMN2 exon 7 (105). Furthermore, it has been demonstrated that there is a feedback loop where exon 7 skipping leads to progressively increased skipping of SMN2 exon 7 (106,107). Even a modestly higher SMN2 exon 7 inclusion level during embryonic development may thus result in a substantially improved SMN protein amount, which may be sufficient to ensure normal embryonic development, but postnatally SMN levels drop significantly due to the negative feedback loop of increased exon 7 skipping.
Despite this, we do observe changes in the expression of some genes at PND1 that are correlated with changes observed at PND5, and also missplicing of some U12-dependent introns. It is likely that the SMN decrease is more severe in some cells than in others, and this can explain the lower levels of missplicing when examining the total tissue. It also indicates that U12-dependent intron retention is part of the early response to decreased SMN levels, which is confirmed by the analysis on SMN knockdown in HeLa cells. However, because of NMD clearing of aberrantly spliced transcripts this may be very difficult to detect, and therefore underreported.
In this study, we identified U12-dependent intron retention in all examined tissues (Table 5), indicating that all cells experience some level of dysfunction during SMN deficiency, and this was confirmed in the non-neuronal HeLa cell line. Many of the genes were affected in multiple tissues while others were completely unaffected, indicating that some genes have an increased sensitivity to aberrant U12-dependent intron splicing. In order to characterize what might sensitize some U12-dependent introns to decreased SMN levels, we examined their properties. We found that low GC-content of the introns was associated with intron retention (Supplementary Figure S22). In most tissues, the introns had a statistically significant lower GC-content than introns that were not affected. Interestingly, in the case of the U12-dependent introns responding to ASO treatment in the spinal cord, there was an inverse relationship between the amount of correction and the GC-content (Supplementary Figure S22). While we cannot rule out that sequencing itself biases this analysis, as sequences with lower GC-content are more likely to be amplified during library construction, we did not observe the same pattern for the U2-dependent introns where the significantly retained U2-introns were more likely to have higher GC-content (data not shown). GC-content may thus be one of the factors that differentiate sensitive U12-dependent introns from robustly spliced U12-dependent introns.
Table 5. Genes with U12-dependent intron retention validated with qPCR at PND5.
| Gene | Brain | Spinal cord | Liver | Muscle | Gene description | Related biological processes and functions |
|---|---|---|---|---|---|---|
| Baz1b | R,Q | R,Q | R,Q | R* | Bromodomain Adjacent To Zinc Finger Domain, 1B | Chromatin remodelling, DNA damage response |
| Cdk5 | R | R | R,Q | R | Cyclin-dependent kinase 5 | Synaptic plasticity, neuronal migration |
| Myh9 | R,Q | R,Q | R,Q | R,Q | Myosin, heavy chain 9 | Cytokinesis, cell shape, cytoskeleton reorganization, beta-actin associated |
| Myo10 | Q | Q | R,Q | R,Q | Myosin X | Axon guidance and growth, neuronal migration, actin binding, vesicle transport |
| Rasgrp3 | R,Q | R,Q | R,Q | R,Q | RAS Guanyl Releasing Protein 3 | Cell growth, cell migration |
| Srsf10 | R,Q | R,Q | R,Q | R,Q | Serine/Arginine-Rich Splicing Factor 10 | Regulation of mRNA splicing, neuronal development |
| Zdhhc13 | R,Q | R | R,Q | R,Q | Huntingtin-Interacting Protein 14-Related Protein | Neuronal, bone development |
R = significant in RNA-seq, Q = significant in qPCR. * = not tested with qPCR in the tissue.
Several lines of evidence support the involvement of aberrant U12-dependent intron splicing in SMA pathology rather than missplicing by the major spliceosome. Firstly, several studies have reported on specific decreases in the minor snRNPs rather than in the major snRNPs (27,30,31). Secondly, SMN depletion results in motor neuron defects in a number of species that are evolutionary distinct and vary greatly in size, from fruit flies to humans. The defect therefore seems to be highly conserved and U12-dependent introns do indeed tend to be evolutionary conserved (108). Missplicing of a few key exons or retention of U2-dependent introns spliced by the major spliceosome would be unlikely to be conserved across species so evolutionary distinct as fruit flies and humans. This indicates that while missplicing by the major spliceosome might also contribute to specific pathologies in various animal models and in humans, the common pathology of SMA is more likely to be affected by impaired minor spliceosome activity.
The question therefore remains how increased U12-dependent intron retention is particularly detrimental to the function of motor neurons. Earlier studies have indicated that retention of the U12-dependent intron in Tmem41b in proprioceptive neurons is higher than in motor neurons and that it is the disruption of the interplay between neuronal populations that motor neurons are particular sensitive to (32). Furthermore, motor neuron cell-nonautonomous rescue of SMA mice (45) strongly indicates that motor neuron function is disrupted by peripheral dysfunction as well as motor neuron specific dysfunction. Even so, our data here points to several U12-dependent introns being misspliced in spinal cord tissue that may directly influence neuronal function. Myo10 showed increased U12-dependent intron retention and decreases of correctly spliced Myo10 in brain and spinal cord, while there was less aberrant splicing in the liver and muscle. Myo10 regulates axonal growth and neuronal migration (78,79,109) and has additionally been reported to regulate vesicle transport along tunneling nanotubes (TNT) in neuronal cells (110). As the changes in splicing of Myo10 seem to be more pronounced in the CNS than in peripheral tissue, missplicing of Myo10 may represent a crucial neuron-specific event that can explain some of the neuron-specific characteristics of SMA. Additionally, missplicing of U12-dependent introns in the voltage gated Ca2+ channel subunit genes could lead to the disruption of Ca2+ channel clusters at axon terminals and cellular Ca2+ homeostasis previously reported as hallmarks of SMA (90,91). These U12-dependent introns are additionally conserved and are be found in Ca2+ channel genes in SMA animal models. Since changes in Ca2+ levels can lead to hyperactivation of Cdk5 (92), we propose that aberrant splicing of U12-dependent introns in genes encoding subunits in the Ca2+ channels contribute to disrupted Ca2+ homeostasis leading to axon terminal dysfunction and Cdk5 hyperactivity, which in turn may lead to motor neuron death. Interestingly, the increased oxidative stress as indicated by elevated Hif-3α, Mt1, and Mt2 expression may also influence Ca2+ levels, which might contribute further to the downstream effects of disrupted Ca2+ homeostasis. These genes may be particularly sensitive to aberrant U12-dependent intron splicing as they contain two distinct U12-dependent introns that each may be affected. Crucially, very recently aberrant processing of U12-dependent introns was reported in Amyotrophic Lateral Sclerosis (ALS) associated FUS mutants (111), and among the affected genes were CACNA1B, CACNA1C, CACNA1E and MYO10. Both SMA and ALS are characterized by loss of motor neuron function and disrupted Ca2+ homeostasis and these genes may therefore represent interesting candidates as therapeutic targets in both SMA and ALS.
Acknowledgments
We thank Lone Sundahl, Mette Bebe Hansen, and Simon Toftholm Jakobsen for assistance with experimental assays. We are grateful to Frank Rigo and C. Frank Bennett (Ionis Pharmaceuticals, Carlsbad, CA) for the generous gift of ASO10-29. We also thank all the members of Dr Andresen's lab for critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Danish Medical Research Council [08-045449 to B.S.A.]; Lundbeck Foundation [R54-A5706 to B.S.A., R118-A11714 to B.S.A.]; National Institutes of Health [R37-GM42699 to A.R.K. and Y.H]. Funding for open access charge: University of Southern Denmark.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pearn J. Incidence, prevalence, and gene frequency studies of chronic childhood spinal muscular atrophy. J. Med. Genet. 1978;15:409–413. doi: 10.1136/jmg.15.6.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crawford T.O., Pardo C.A. The neurobiology of childhood spinal muscular atrophy. Neurobiol. Dis. 1996;3:97–110. doi: 10.1006/nbdi.1996.0010. [DOI] [PubMed] [Google Scholar]
- 3.Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 4.Lefebvre S., Burlet P., Liu Q., Bertrandy S., Clermont O., Munnich A., Dreyfuss G., Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 5.Lorson C.L., Hahnen E., Androphy E.J., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. U.S.A. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cartegni L., Krainer A.R. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 7.Kashima T., Manley J.L. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet. 2003;34:460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 8.Bruun G.H., Doktor T.K., Borch-Jensen J., Masuda A., Krainer A.R., Ohno K., Andresen B.S. Global identification of hnRNP A1 binding sites for SSO-based splicing modulation. BMC biology. 2016;14:54. doi: 10.1186/s12915-016-0279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh N.K., Singh N.N., Androphy E.J., Singh R.N. Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron. Mol. Cell. Biol. 2006;26:1333–1346. doi: 10.1128/MCB.26.4.1333-1346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doktor T.K., Schroeder L.D., Vested A., Palmfeldt J., Andersen H.S., Gregersen N., Andresen B.S. SMN2 exon 7 splicing is inhibited by binding of hnRNP A1 to a common ESS motif that spans the 3′ splice site. Hum. Mutat. 2011;32:220–230. doi: 10.1002/humu.21419. [DOI] [PubMed] [Google Scholar]
- 11.Pedrotti S., Bielli P., Paronetto M.P., Ciccosanti F., Fimia G.M., Stamm S., Manley J.L., Sette C. The splicing regulator Sam68 binds to a novel exonic splicing silencer and functions in SMN2 alternative splicing in spinal muscular atrophy. EMBO J. 2010;29:1235–1247. doi: 10.1038/emboj.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorson C.L., Strasswimmer J., Yao J.M., Baleja J.D., Hahnen E., Wirth B., Le T., Burghes A.H., Androphy E.J. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat. Genet. 1998;19:63–66. doi: 10.1038/ng0598-63. [DOI] [PubMed] [Google Scholar]
- 13.Liu Q., Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer U., Liu Q., Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 15.Pellizzoni L., Yong J., Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298:1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- 16.Tisdale S., Lotti F., Saieva L., Van Meerbeke J.P., Crawford T.O., Sumner C.J., Mentis G.Z., Pellizzoni L. SMN is essential for the biogenesis of U7 small nuclear ribonucleoprotein and 3′-end formation of histone mRNAs. Cell Rep. 2013;5:1187–1195. doi: 10.1016/j.celrep.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mowry K.L., Steitz J.A. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA's. Science. 1987;238:1682–1687. doi: 10.1126/science.2825355. [DOI] [PubMed] [Google Scholar]
- 18.Sleeman J. Small nuclear RNAs and mRNAs: linking RNA processing and transport to spinal muscular atrophy. Biochem. Soc. Trans. 2013;41:871–875. doi: 10.1042/BST20120016. [DOI] [PubMed] [Google Scholar]
- 19.Rossoll W., Jablonka S., Andreassi C., Kroning A.K., Karle K., Monani U.R., Sendtner M. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J. Cell Biol. 2003;163:801–812. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez G., Dury A.Y., Murray L.M., Biondi O., Tadesse H., El Fatimy R., Kothary R., Charbonnier F., Khandjian E.W., Cote J. A novel function for the survival motoneuron protein as a translational regulator. Hum. Mol. Genet. 2013;22:668–684. doi: 10.1093/hmg/dds474. [DOI] [PubMed] [Google Scholar]
- 21.Fallini C., Zhang H., Su Y., Silani V., Singer R.H., Rossoll W., Bassell G.J. The survival of motor neuron (SMN) protein interacts with the mRNA-binding protein HuD and regulates localization of poly(A) mRNA in primary motor neuron axons. J. Neurosci. 2011;31:3914–3925. doi: 10.1523/JNEUROSCI.3631-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossoll W., Kroning A.K., Ohndorf U.M., Steegborn C., Jablonka S., Sendtner M. Specific interaction of Smn, the spinal muscular atrophy determining gene product, with hnRNP-R and gry-rbp/hnRNP-Q: a role for Smn in RNA processing in motor axons. Hum. Mol. Genet. 2002;11:93–105. doi: 10.1093/hmg/11.1.93. [DOI] [PubMed] [Google Scholar]
- 23.Akten B., Kye M.J., Hao le T., Wertz M.H., Singh S., Nie D., Huang J., Merianda T.T., Twiss J.L., Beattie C.E., et al. Interaction of survival of motor neuron (SMN) and HuD proteins with mRNA cpg15 rescues motor neuron axonal deficits. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10337–10342. doi: 10.1073/pnas.1104928108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hubers L., Valderrama-Carvajal H., Laframboise J., Timbers J., Sanchez G., Cote J. HuD interacts with survival motor neuron protein and can rescue spinal muscular atrophy-like neuronal defects. Hum. Mol. Genet. 2011;20:553–579. doi: 10.1093/hmg/ddq500. [DOI] [PubMed] [Google Scholar]
- 25.Liu Q., Fischer U., Wang F., Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- 26.Workman E., Saieva L., Carrel T.L., Crawford T.O., Liu D., Lutz C., Beattie C.E., Pellizzoni L., Burghes A.H. A SMN missense mutation complements SMN2 restoring snRNPs and rescuing SMA mice. Hum. Mol. Genet. 2009;18:2215–2229. doi: 10.1093/hmg/ddp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winkler C., Eggert C., Gradl D., Meister G., Giegerich M., Wedlich D., Laggerbauer B., Fischer U. Reduced U snRNP assembly causes motor axon degeneration in an animal model for spinal muscular atrophy. Genes Dev. 2005;19:2320–2330. doi: 10.1101/gad.342005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z., Lotti F., Dittmar K., Younis I., Wan L., Kasim M., Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baumer D., Lee S., Nicholson G., Davies J.L., Parkinson N.J., Murray L.M., Gillingwater T.H., Ansorge O., Davies K.E., Talbot K. Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet. 2009;5:e1000773. doi: 10.1371/journal.pgen.1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gabanella F., Butchbach M.E., Saieva L., Carissimi C., Burghes A.H., Pellizzoni L. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS One. 2007;2:e921. doi: 10.1371/journal.pone.0000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boulisfane N., Choleza M., Rage F., Neel H., Soret J., Bordonne R. Impaired minor tri-snRNP assembly generates differential splicing defects of U12-type introns in lymphoblasts derived from a type I SMA patient. Hum. Mol. Genet. 2011;20:641–648. doi: 10.1093/hmg/ddq508. [DOI] [PubMed] [Google Scholar]
- 32.Lotti F., Imlach W.L., Saieva L., Beck E.S., Hao le T., Li D.K., Jiao W., Mentis G.Z., Beattie C.E., McCabe B.D., et al. An SMN-dependent U12 splicing event essential for motor circuit function. Cell. 2012;151:440–454. doi: 10.1016/j.cell.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu C.F., Chen C.Y., Yuh Y.S., Chen Y.H., Hsu Y.T., Zimmerman R.A. MR findings of Werdnig-Hoffmann disease in two infants. AJNR Am. J. Neuroradiol. 1998;19:550–552. [PMC free article] [PubMed] [Google Scholar]
- 34.Harding B.N., Kariya S., Monani U.R., Chung W.K., Benton M., Yum S.W., Tennekoon G., Finkel R.S. Spectrum of neuropathophysiology in spinal muscular atrophy type I. J. Neuropathol. Exp. Neurol. 2015;74:15–24. doi: 10.1097/NEN.0000000000000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wishart T.M., Huang J.P., Murray L.M., Lamont D.J., Mutsaers C.A., Ross J., Geldsetzer P., Ansorge O., Talbot K., Parson S.H., et al. SMN deficiency disrupts brain development in a mouse model of severe spinal muscular atrophy. Hum. Mol. Genet. 2010;19:4216–4228. doi: 10.1093/hmg/ddq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu H., Beauvais A., Baker A.N., Tsilfidis C., Kothary R. Smn deficiency causes neuritogenesis and neurogenesis defects in the retinal neurons of a mouse model of spinal muscular atrophy. Dev Neurobiol. 2011;71:153–169. doi: 10.1002/dneu.20840. [DOI] [PubMed] [Google Scholar]
- 37.Gombash S.E., Cowley C.J., Fitzgerald J.A., Iyer C.C., Fried D., McGovern V.L., Williams K.C., Burghes A.H., Christofi F.L., Gulbransen B.D., et al. SMN deficiency disrupts gastrointestinal and enteric nervous system function in mice. Hum. Mol. Genet. 2015;24:3847–3860. doi: 10.1093/hmg/ddv127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sahashi K., Ling K.K., Hua Y., Wilkinson J.E., Nomakuchi T., Rigo F., Hung G., Xu D., Jiang Y.P., Lin R.Z., et al. Pathological impact of SMN2 mis-splicing in adult SMA mice. EMBO Mol. Med. 2013;5:1586–1601. doi: 10.1002/emmm.201302567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vitte J.M., Davoult B., Roblot N., Mayer M., Joshi V., Courageot S., Tronche F., Vadrot J., Moreau M.H., Kemeny F., et al. Deletion of murine Smn exon 7 directed to liver leads to severe defect of liver development associated with iron overload. Am. J. Pathol. 2004;165:1731–1741. doi: 10.1016/S0002-9440(10)63428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rudnik-Schoneborn S., Heller R., Berg C., Betzler C., Grimm T., Eggermann T., Eggermann K., Wirth R., Wirth B., Zerres K. Congenital heart disease is a feature of severe infantile spinal muscular atrophy. J. Med. Genet. 2008;45:635–638. doi: 10.1136/jmg.2008.057950. [DOI] [PubMed] [Google Scholar]
- 41.Menke L.A., Poll-The B.T., Clur S.A., Bilardo C.M., van der Wal A.C., Lemmink H.H., Cobben J.M. Congenital heart defects in spinal muscular atrophy type I: a clinical report of two siblings and a review of the literature. Am. J. Med. Genet. A. 2008;146:740–744. doi: 10.1002/ajmg.a.32233. [DOI] [PubMed] [Google Scholar]
- 42.Heier C.R., Satta R., Lutz C., DiDonato C.J. Arrhythmia and cardiac defects are a feature of spinal muscular atrophy model mice. Hum. Mol. Genet. 2010;19:3906–3918. doi: 10.1093/hmg/ddq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bevan A.K., Hutchinson K.R., Foust K.D., Braun L., McGovern V.L., Schmelzer L., Ward J.G., Petruska J.C., Lucchesi P.A., Burghes A.H., et al. Early heart failure in the SMNDelta7 model of spinal muscular atrophy and correction by postnatal scAAV9-SMN delivery. Hum. Mol. Genet. 2010;19:3895–3905. doi: 10.1093/hmg/ddq300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hua Y., Sahashi K., Rigo F., Hung G., Horev G., Bennett C.F., Krainer A.R. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478:123–126. doi: 10.1038/nature10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hua Y., Liu Y.H., Sahashi K., Rigo F., Bennett C.F., Krainer A.R. Motor neuron cell-nonautonomous rescue of spinal muscular atrophy phenotypes in mild and severe transgenic mouse models. Genes Dev. 2015;29:288–297. doi: 10.1101/gad.256644.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staropoli J.F., Li H., Chun S.J., Allaire N., Cullen P., Thai A., Fleet C.M., Hua Y., Bennett C.F., Krainer A.R., et al. Rescue of gene-expression changes in an induced mouse model of spinal muscular atrophy by an antisense oligonucleotide that promotes inclusion of SMN2 exon 7. Genomics. 2015;105:220–228. doi: 10.1016/j.ygeno.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 47.Zhao X., Feng Z., Ling K.K., Mollin A., Sheedy J., Yeh S., Petruska J., Narasimhan J., Dakka A., Welch E.M., et al. Pharmacokinetics, pharmacodynamics, and efficacy of a small-molecule SMN2 splicing modifier in mouse models of spinal muscular atrophy. Hum. Mol. Genet. 2016 doi: 10.1093/hmg/ddw062. doi:10.1093/hmg/ddw062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hua Y., Sahashi K., Hung G., Rigo F., Passini M.A., Bennett C.F., Krainer A.R. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010;24:1634–1644. doi: 10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gogliotti R.G., Hammond S.M., Lutz C., Didonato C.J. Molecular and phenotypic reassessment of an infrequently used mouse model for spinal muscular atrophy. Biochem. Biophys. Res. Commun. 2010;391:517–522. doi: 10.1016/j.bbrc.2009.11.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riessland M., Ackermann B., Forster A., Jakubik M., Hauke J., Garbes L., Fritzsche I., Mende Y., Blumcke I., Hahnen E., et al. SAHA ameliorates the SMA phenotype in two mouse models for spinal muscular atrophy. Hum. Mol. Genet. 2010;19:1492–1506. doi: 10.1093/hmg/ddq023. [DOI] [PubMed] [Google Scholar]
- 51.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anders S., Pyl P.T., Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Flicek P., Ahmed I., Amode M.R., Barrell D., Beal K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fairley S., et al. Ensembl 2013. Nucleic Acids Res. 2013;41:D48–55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.R Core Team. R Foundation for Statistical Computing. 2013. [Google Scholar]
- 57.Hansen K.D., Irizarry R.A., Wu Z. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics. 2012;13:204–216. doi: 10.1093/biostatistics/kxr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCullagh P., Nelder J.A. Generalized Linear Models. 2nd edn. London; NY: Chapman and Hall; 1989. [Google Scholar]
- 59.Anders S., Reyes A., Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22:2008–2017. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pruitt K.D., Tatusova T., Brown G.R., Maglott D.R. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roberts A., Pimentel H., Trapnell C., Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27:2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- 63.Alioto T.S. U12DB: a database of orthologous U12-type spliceosomal introns. Nucleic Acids Res. 2007;35:D110–D115. doi: 10.1093/nar/gkl796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luo W., Friedman M.S., Shedden K., Hankenson K.D., Woolf P.J. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luo W., Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29:1830–1831. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hahne F., D S., Ivanek R., Mueller A., Lianoglou S., Tan G., Parsons L. Gviz: Plotting Data and Annotation Information along Genomic Coordinates. R package version 1.12.0 ed. [Google Scholar]
- 68.Hsieh-Li H.M., Chang J.G., Jong Y.J., Wu M.H., Wang N.M., Tsai C.H., Li H. A mouse model for spinal muscular atrophy. Nat. Genet. 2000;24:66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- 69.Sillekens P.T., Beijer R.P., Habets W.J., van Verooij W.J. Molecular cloning of the cDNA for the human U2 snRNA-specific A' protein. Nucleic Acids Res. 1989;17:1893–1906. doi: 10.1093/nar/17.5.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karin M., Andersen R.D., Herschman H.R. Induction of metallothionein mRNA in HeLa cells by dexamethasone and by heavy metals. Eur. J. Biochem. 1981;118:527–531. doi: 10.1111/j.1432-1033.1981.tb05551.x. [DOI] [PubMed] [Google Scholar]
- 71.Karin M., Slater E.P., Herschman H.R. Regulation of metallothionein synthesis in HeLa cells by heavy metals and glucocorticoids. J. Cell Physiol. 1981;106:63–74. doi: 10.1002/jcp.1041060108. [DOI] [PubMed] [Google Scholar]
- 72.Sureau A., Gattoni R., Dooghe Y., Stevenin J., Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001;20:1785–1796. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lareau L.F., Inada M., Green R.E., Wengrod J.C., Brenner S.E. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 74.Ni J.Z., Grate L., Donohue J.P., Preston C., Nobida N., O'Brien G., Shiue L., Clark T.A., Blume J.E., Ares M., Jr Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun S., Zhang Z., Sinha R., Karni R., Krainer A.R. SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nat. Struct. Mol. Biol. 2010;17:306–312. doi: 10.1038/nsmb.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huo Q., Kayikci M., Odermatt P., Meyer K., Michels O., Saxena S., Ule J., Schumperli D. Splicing changes in SMA mouse motoneurons and SMN-depleted neuroblastoma cells: evidence for involvement of splicing regulatory proteins. RNA Biol. 2014;11:1430–1446. doi: 10.1080/15476286.2014.996494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang D., Tao J., Li L., Kedei N., Toth Z.E., Czap A., Velasquez J.F., Mihova D., Michalowski A.M., Yuspa S.H., et al. RasGRP3, a Ras activator, contributes to signaling and the tumorigenic phenotype in human melanoma. Oncogene. 2011;30:4590–4600. doi: 10.1038/onc.2011.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Raines A.N., Nagdas S., Kerber M.L., Cheney R.E. Headless Myo10 is a negative regulator of full-length Myo10 and inhibits axon outgrowth in cortical neurons. J. Biol. Chem. 2012;287:24873–24883. doi: 10.1074/jbc.M112.369173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu H., Wang N., Ju X., Yang Y., Sun D., Lai M., Cui L., Sheikh M.A., Zhang J., Wang X., et al. PtdIns (3,4,5) P3 recruitment of Myo10 is essential for axon development. PLoS One. 2012;7:e36988. doi: 10.1371/journal.pone.0036988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fu A.K., Ip F.C., Fu W.Y., Cheung J., Wang J.H., Yung W.H., Ip N.Y. Aberrant motor axon projection, acetylcholine receptor clustering, and neurotransmission in cyclin-dependent kinase 5 null mice. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15224–15229. doi: 10.1073/pnas.0507678102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sutton L.M., Sanders S.S., Butland S.L., Singaraja R.R., Franciosi S., Southwell A.L., Doty C.N., Schmidt M.E., Mui K.K., Kovalik V., et al. Hip14l-deficient mice develop neuropathological and behavioural features of Huntington disease. Hum. Mol. Genet. 2013;22:452–465. doi: 10.1093/hmg/dds441. [DOI] [PubMed] [Google Scholar]
- 82.Parker J.A., Metzler M., Georgiou J., Mage M., Roder J.C., Rose A.M., Hayden M.R., Neri C. Huntingtin-interacting protein 1 influences worm and mouse presynaptic function and protects Caenorhabditis elegans neurons against mutant polyglutamine toxicity. J. Neurosci. 2007;27:11056–11064. doi: 10.1523/JNEUROSCI.1941-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu L., Lin J.J., Chen X., Liu X., Xu P. Neural expression and regulation of NSSR1 proteins. Neuroreport. 2003;14:1847–1850. doi: 10.1097/00001756-200310060-00018. [DOI] [PubMed] [Google Scholar]
- 84.Miller N., Feng Z., Edens B.M., Yang B., Shi H., Sze C.C., Hong B.T., Su S.C., Cantu J.A., Topczewski J., et al. Non-aggregating tau phosphorylation by cyclin-dependent kinase 5 contributes to motor neuron degeneration in spinal muscular atrophy. J. Neurosci. 2015;35:6038–6050. doi: 10.1523/JNEUROSCI.3716-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lalli M.A., Jang J., Park J.H., Wang Y., Guzman E., Zhou H., Audouard M., Bridges D., Tovar K.R., Papuc S.M., et al. Haploinsufficiency of BAZ1B contributes to Williams syndrome through transcriptional dysregulation of neurodevelopmental pathways. Hum. Mol. Genet. 2016;25:1294–1306. doi: 10.1093/hmg/ddw010. [DOI] [PubMed] [Google Scholar]
- 86.Xiao A., Li H., Shechter D., Ahn S.H., Fabrizio L.A., Erdjument-Bromage H., Ishibe-Murakami S., Wang B., Tempst P., Hofmann K., et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 2009;457:57–62. doi: 10.1038/nature07668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fayzullina S., Martin L.J. Skeletal muscle DNA damage precedes spinal motor neuron DNA damage in a mouse model of Spinal Muscular Atrophy (SMA) PLoS One. 2014;9:e93329. doi: 10.1371/journal.pone.0093329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bozhenok L., Wade P.A., Varga-Weisz P. WSTF-ISWI chromatin remodeling complex targets heterochromatic replication foci. EMBO J. 2002;21:2231–2241. doi: 10.1093/emboj/21.9.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Porensky P.N., Mitrpant C., McGovern V.L., Bevan A.K., Foust K.D., Kaspar B.K., Wilton S.D., Burghes A.H. A single administration of morpholino antisense oligomer rescues spinal muscular atrophy in mouse. Hum. Mol. Genet. 2012;21:1625–1638. doi: 10.1093/hmg/ddr600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jablonka S., Beck M., Lechner B.D., Mayer C., Sendtner M. Defective Ca2+ channel clustering in axon terminals disturbs excitability in motoneurons in spinal muscular atrophy. J. Cell Biol. 2007;179:139–149. doi: 10.1083/jcb.200703187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ruiz R., Casanas J.J., Torres-Benito L., Cano R., Tabares L. Altered intracellular Ca2+ homeostasis in nerve terminals of severe spinal muscular atrophy mice. J. Neurosci. 2010;30:849–857. doi: 10.1523/JNEUROSCI.4496-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee M.S., Kwon Y.T., Li M., Peng J., Friedlander R.M., Tsai L.H. Neurotoxicity induces cleavage of p35 to p25 by calpain. Nature. 2000;405:360–364. doi: 10.1038/35012636. [DOI] [PubMed] [Google Scholar]
- 93.Ng S.Y., Soh B.S., Rodriguez-Muela N., Hendrickson D.G., Price F., Rinn J.L., Rubin L.L. Genome-wide RNA-Seq of Human Motor Neurons Implicates Selective ER Stress Activation in Spinal Muscular Atrophy. Cell Stem Cell. 2015;17:569–584. doi: 10.1016/j.stem.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Choudhry H., Schodel J., Oikonomopoulos S., Camps C., Grampp S., Harris A.L., Ratcliffe P.J., Ragoussis J., Mole D.R. Extensive regulation of the non-coding transcriptome by hypoxia: role of HIF in releasing paused RNApol2. EMBO Rep. 2014;15:70–76. doi: 10.1002/embr.201337642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Younis I., Dittmar K., Wang W., Foley S.W., Berg M.G., Hu K.Y., Wei Z., Wan L., Dreyfuss G. Minor introns are embedded molecular switches regulated by highly unstable U6atac snRNA. Elife. 2013;2:e00780. doi: 10.7554/eLife.00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Genabai N.K., Ahmad S., Zhang Z., Jiang X., Gabaldon C.A., Gangwani L. Genetic inhibition of JNK3 ameliorates spinal muscular atrophy. Hum. Mol. Genet. 2015;24:6986–7004. doi: 10.1093/hmg/ddv401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zou T., Yang X., Pan D., Huang J., Sahin M., Zhou J. SMN deficiency reduces cellular ability to form stress granules, sensitizing cells to stress. Cell. Mol. Neurobiol. 2011;31:541–550. doi: 10.1007/s10571-011-9647-8. [DOI] [PubMed] [Google Scholar]
- 98.Maeda M., Harris A.W., Kingham B.F., Lumpkin C.J., Opdenaker L.M., McCahan S.M., Wang W., Butchbach M.E. Transcriptome profiling of spinal muscular atrophy motor neurons derived from mouse embryonic stem cells. PLoS One. 2014;9:e106818. doi: 10.1371/journal.pone.0106818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Le T.T., Pham L.T., Butchbach M.E., Zhang H.L., Monani U.R., Coovert D.D., Gavrilina T.O., Xing L., Bassell G.J., Burghes A.H. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum. Mol. Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 100.Hutten S., Chachami G., Winter U., Melchior F., Lamond A.I. A role for the Cajal-body-associated SUMO isopeptidase USPL1 in snRNA transcription mediated by RNA polymerase II. J. Cell Sci. 2014;127:1065–1078. doi: 10.1242/jcs.141788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wishart T.M., Mutsaers C.A., Riessland M., Reimer M.M., Hunter G., Hannam M.L., Eaton S.L., Fuller H.R., Roche S.L., Somers E., et al. Dysregulation of ubiquitin homeostasis and beta-catenin signaling promote spinal muscular atrophy. J. Clin. Invest. 2014;124:1821–1834. doi: 10.1172/JCI71318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sleigh J.N., Barreiro-Iglesias A., Oliver P.L., Biba A., Becker T., Davies K.E., Becker C.G., Talbot K. Chondrolectin affects cell survival and neuronal outgrowth in in vitro and in vivo models of spinal muscular atrophy. Hum. Mol. Genet. 2014;23:855–869. doi: 10.1093/hmg/ddt477. [DOI] [PubMed] [Google Scholar]
- 103.See K., Yadav P., Giegerich M., Cheong P.S., Graf M., Vyas H., Lee S.G., Mathavan S., Fischer U., Sendtner M., et al. SMN deficiency alters Nrxn2 expression and splicing in zebrafish and mouse models of spinal muscular atrophy. Hum. Mol. Genet. 2014;23:1754–1770. doi: 10.1093/hmg/ddt567. [DOI] [PubMed] [Google Scholar]
- 104.Zhang Z., Pinto A.M., Wan L., Wang W., Berg M.G., Oliva I., Singh L.N., Dengler C., Wei Z., Dreyfuss G. Dysregulation of synaptogenesis genes antecedes motor neuron pathology in spinal muscular atrophy. Proc. Natl. Acad. Sci. U.S.A. 2013;110:19348–19353. doi: 10.1073/pnas.1319280110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Singh N.N., Seo J., Ottesen E.W., Shishimorova M., Bhattacharya D., Singh R.N. TIA1 prevents skipping of a critical exon associated with spinal muscular atrophy. Mol. Cell. Biol. 2011;31:935–954. doi: 10.1128/MCB.00945-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jodelka F.M., Ebert A.D., Duelli D.M., Hastings M.L. A feedback loop regulates splicing of the spinal muscular atrophy-modifying gene, SMN2. Hum. Mol. Genet. 2010;19:4906–4917. doi: 10.1093/hmg/ddq425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ruggiu M., McGovern V.L., Lotti F., Saieva L., Li D.K., Kariya S., Monani U.R., Burghes A.H., Pellizzoni L. A role for SMN exon 7 splicing in the selective vulnerability of motor neurons in spinal muscular atrophy. Mol. Cell. Biol. 2012;32:126–138. doi: 10.1128/MCB.06077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lin C.F., Mount S.M., Jarmolowski A., Makalowski W. Evolutionary dynamics of U12-type spliceosomal introns. BMC Evol. Biol. 2010;10:47. doi: 10.1186/1471-2148-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sousa A.D., Berg J.S., Robertson B.W., Meeker R.B., Cheney R.E. Myo10 in brain: developmental regulation, identification of a headless isoform and dynamics in neurons. J. Cell Sci. 2006;119:184–194. doi: 10.1242/jcs.02726. [DOI] [PubMed] [Google Scholar]
- 110.Gousset K., Marzo L., Commere P.H., Zurzolo C. Myo10 is a key regulator of TNT formation in neuronal cells. J. Cell Sci. 2013;126:4424–4435. doi: 10.1242/jcs.129239. [DOI] [PubMed] [Google Scholar]
- 111.Reber S., Stettler J., Filosa G., Colombo M., Jutzi D., Lenzken S.C., Schweingruber C., Bruggmann R., Bachi A., Barabino S.M., et al. Minor intron splicing is regulated by FUS and affected by ALS-associated FUS mutants. EMBO J. 2016 doi: 10.15252/embj.201593791. doi:10.15252/embj.201694763. [DOI] [PMC free article] [PubMed] [Google Scholar]