Abstract
The 2-methylthio (ms2) modification at A37 of tRNAs is critical for accurate decoding, and contributes to metabolic homeostasis in mammals. However, the regulatory mechanism of ms2 modification remains largely unknown. Here, we report that cysteine hydropersulfide (CysSSH), a newly identified reactive sulfur species, is involved in ms2 modification in cells. The suppression of intracellular CysSSH production rapidly reduced ms2 modification, which was rescued by the application of an exogenous CysSSH donor. Using a unique and stable isotope-labeled CysSSH donor, we show that CysSSH was capable of specifically transferring its reactive sulfur atom to the cysteine residues of ms2-modifying enzymes as well as ms2 modification. Furthermore, the suppression of CysSSH production impaired insulin secretion and caused glucose intolerance in both a pancreatic β-cell line and mouse model. These results demonstrate that intracellular CysSSH is a novel sulfur source for ms2 modification, and that it contributes to insulin secretion.
INTRODUCTION
tRNAs undergo a wide variety of post-transcriptional modifications that are essential for accurate and efficient decoding in all living organisms (1,2). The 2-methylthio (ms2) modification is an evolutionarily conserved modification found at position 37 of tRNAs (2). In mammalian cells, two forms of ms2-modified nucleotide have been identified: 2-methylthio-N6-threonylcarbamoyadenosine (ms2t6A) and 2-methylthio-N6-isopentenyladenosine (ms2i6A) (3,4). Cdk5 regulatory subunit associated protein 1-like-1 (CDKAL1) catalyzes the methylthiolation of t6A to form ms2t6A at A37 of cytosolic tRNALys(UUU) (5). On the other hand, Cdk5 regulatory subunit-associated protein 1 (CDK5RAP1) catalyzes the methythiolation of i6A to form ms2i6A at A37 of four mitochondrial tRNAs: mt-tRNASer(UCN), mt-tRNATrp, mt-tRNAPhe and mt-tRNATyr (4). The ms2 modification stabilizes codon–anticodon binding through a cross-strand interaction that contributes to accurate decoding (6). Indeed, ms2 deficiency in cytosolic tRNALys(UUU) impairs proinsulin biosynthesis and causes the development of type 2 diabetes (5). Similarly, ms2 deficiency in ms2i6A impairs mitochondrial protein synthesis and leads to the development of myopathy in mice and mitochondrial diseases in humans (4).
Given the essential role of ms2 modification in regulating protein synthesis, the regulatory mechanism of ms2 modification has attracted wide attention. All ms2-modifying enzymes share similar functional domains: UPF0004, radical S-adenosylmethionine (SAM) and tRNA-binding domains (3). The UPF0004 and radical SAM domains form [4Fe-4S] clusters through conserved Cys residues in each domain (7). The methyl group in ms2 is apparently derived from SAM (7), whereas the origin of the sulfur atom in ms2 has remained unknown for decades. Recently, Forouhar et al. showed that the UPF0004 domain contained exogenous sulfide species that might provide the sulfur atom for ms2 modification using a defined reconstitution system (8). These findings have shed light on the molecular origin of sulfur in ms2. However, whether the exogenous sulfide species also exist in living cells and how these sulfides are involved in ms2 modification remain largely unexplored.
The ms2 modification requires the conversion of a C–H bond to C–S bond, which is a challenging reaction (3). It is thus predicted that reactive sulfur species might be required to initiate the conversion. Cells contain various sulfur species including cysteine hydropersulfide (CysSSH) and hydrogen sulfide (H2S)(9–12). These sulfur species are mainly produced by cystathionine beta-synthase (CBS) and cystathionine gamma-lyase (CTH) (12). CysSSH is highly reactive due to its marked nucleophilicity (12,13). More importantly, it can readily initiate thiol redox exchange, leading to protein Cys-polythiolation (12). Given its ability to transfer sulfur, CysSSH could potentially participate in ms2 modification. In the present study, we investigated the potential role of the CysSSH in the regulation of ms2 modification in tRNAs and its physiological relevance in vivo.
MATERIALS AND METHODS
Reagents
The oxidized CysSSH donor was chemically synthesized as described previously (12). Briefly, 10 mM cysteine hydrochloride was reacted with 10 mM Na2S or Na34SH in 50 ml of 10 mM NaOH solution in the presence of 5% I2 at room temperature. CysS-S2-SCys and CysS-34S2-SCys were purified by high pressure (or high performance) liquid chromatography. Monobromobimane, β-Cyano-L-alanine and Deferoxamine were purchased from Sigma. Predesigned siRNAs against CBS and CTH (siRNA for CBS: s63475, siRNA for CTH: CTHHSS102445) were purchased from Invitrogen and Ambion, respectively. Control siRNA was purchased from Invitrogen. Methylsulfonyl benzothiazole (MSBT) and biotinylated α-cyano ester (CN-biotin) were synthesized as described previously (12). DAPI solution and 3′,6′-dil(O-thiosalicyl) fluorescein (SSP4) were purchased from Dojindo, Japan. Hexadecyltrimethylammonium Bromide (CTAB) was purchased from Tokyo Chemical Industry, Japan.
Mass spectrometric analysis of ms2 modification
For mass spectrometric analysis of ms2 in HeLa cells, total RNA was purified by TRIzol reagent. For bacterial RNA, TOP10 competent cells were transformed with pET21a-CDKAL1 plasmids. A single colony was cultured in LB medium with constant shaking until the OD600 reached 0.7. Next, 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG, WAKO) was added to the culture, which was then incubated overnight. Total RNA from the bacteria was purified using a TRIzol Max Bacterial RNA Isolation Kit (Invitrogen) following the manufacturer's instructions. Twenty micrograms of RNA were digested with 2.5 U of Nuclease P1 (WAKO) and 0.2 U of bacterial alkaline phosphatase (Takara) in 5 mM ammonium acetate and 20 mM HEPES-KOH, pH 7.0, for 3 h at 37°C. The samples were subjected to mass spectrometry (Agilent 6460). The ms2 modification was detected by a multiple reaction monitoring (MRM) method using the positive mode. The MRM parameters for ms2t6A were as follows: precursor ion: m/z 459.4, product ion: m/z 326.8, collision energy: 14, Fragmentor 125 V.
Mass spectrometry for examination of polysulfide in cells
Cysteine polysulfide (CysSSH) was detected using monobromobimane (Invitrogen), as described previously (12). Monobromobimane efficiently and specifically reacts with CysSSH to form a CysS-S-bimane adduct. HeLa cells were grown on a 10-cm dish, washed with phosphate-buffered saline and directly treated with 200 μl of 5 mM monobromobimane dissolved in methanol. Cells were collected, sonicated and incubated at 37°C for 15 min. Insoluble materials were removed by brief centrifugation at 10 000 × g for 10 min. Supernatants were diluted with distilled water and subjected to mass spectrometry using the MRM method in the positive ion mode. The MRM parameters for bimane adducts were as described previously (12).
Imaging of intracellular polysulfides by SSP4
Endogenous reactive sulfur species including CysSSH were observed using the sulfane sulfur-specific fluorescent probe SSP4, as described previously (12) SSP4 is a modified version of SSP2 that was developed for bioimaging sulfane sulfurs in living cells (14). HeLa cells were cultured on 35-mm glass-bottomed dishes (IWAKI) for polysulfide imaging. The cells were washed with serum-free Dulbecco's modified Eagle's medium (DMEM), followed by the addition of 20 μM SSP4 in serum-free DMEM containing 500 μM CTAB at 37°C for 15 min. The cells were then washed twice with phosphate buffered saline (PBS) and counterstained with DAPI in PBS at room temperature for 10 min. Fluorescence was observed using an FV1000 confocal microscope (OLYMPUS). The average fluorescence was quantified using the software FLUOVIEW Ver. 4.2 (OLYMPUS).
Polysulfide-specific biotin-labeling assay
HeLa cells transfected with the pCMV-Myc-CDKAL1 plasmid vector were homogenized in lysis buffer (10 mM Tris-HCl, 1% NP-40 and 150 mM NaCl, pH 7.4) containing a protease inhibitor cocktail (Roche) and immediately incubated with 2 mM MSBT at 37°C for 30 min. Lysates were subsequently reacted with 4 mM CN-biotin at 37°C for 30 min. After the insoluble materials were removed by a brief centrifugation at 10 000 × g for 10 min, the biotin-labeled proteins were purified using streptavidin Sepharose beads (GE Healthcare) at 4°C for 3 h. After an extensive wash with lysis buffer, the biotin-labeled proteins were eluted by the addition of SDS sample buffer (50 mM Tris-HCl, 2% SDS, 6% 2-mercaptoethanol, 10% glycerol and 0.005% BPB). Cypolythiolation of CDKAL1 was detected by Western blotting using anti-Myc antibody (Wako).
Mass spectrometric analysis of cysteine polysulfides in the CDKAL1 protein
HeLa cells were transfected with the pCMV-Myc-CDKAL1 vector for 48 h, followed by treatment with 100 μM CysS-S2-SCys or CysS-34S2-SCys for 6 h. The cells were then homogenized in lysis buffer (10 mM Tris-HCl, 1% NP-40 and 150 mM NaCl, pH 6.8) and immediately reacted with 1 mM monobromobimane at 37°C for 15 min. The lysates were incubated with anti-Myc antibody at 4°C for 1 h. Myc-CDKAL1 protein was then precipitated by Dynabeads Protein G (Life Technologies). After an extensive wash with lysis buffer, the Myc-CDKAL1 proteins captured on the Dynabeads were digested with 0.5 mg/ml pronase (Calbiochem) in sodium phosphate buffer (pH 6.0) at 37°C for 2 h. The supernatants were directly subjected to mass spectrometric analysis for the detection of CysS-S-bimane adducts, as described above.
Measurement of insulin secretion
The mouse pancreatic beta cell line was seeded in 24-well plates and treated with 500 μM BCA for 24 h. After incubation, the cells were washed with Krebs–Ringer bicarbonate buffer (KRB buffer: 115 mM NaCl, 5 mM KCl, 10 mM NaHCO3, 2.5 mM MgCl2, 2.5 mM CaCl2 and 20 mM HEPES, pH 7.4, 0.1% bovine serum albumin) containing 2.8 mM glucose, followed by incubation in the same buffer for 1 h at 37°C. The cells were stimulated with KRB buffer containing 2.8 mM glucose (low glucose) for 30 min, followed by stimulation with KRB buffer containing 16.7 mM glucose (high glucose). The insulin levels in the incubation buffers were measured using an ELISA kit (Shibayagi) following the manufacturer's instructions.
Animals
Cth-deficient mice on C57BL6/6J background were established by Dr. Ishii (15). Animals were housed at 25°C with 12-h light/12-h dark cycles. High-fat chow (D12451, 45% kcal% fat) was purchased from Research Diets. All animal procedures were approved by the Animal Ethics Committee of Kumamoto University (approval ID: A27-037R1).
Cell culture
HeLa cells established from a human cervical cancer cell line were cultured in DMEM high-glucose medium (GIBCO) supplemented with 10% fetal bovine serum (FBS, HyClone) at 37°C and 5% CO2. Two mouse pancreatic β-cell lines was established from insulinoma that developed in transgenic mice expressing the SV40 antigen under the control of the insulin promoter (16). Male mice at the age of 8 weeks were sacrificed and insulinoma tissues were hand-picked under a microscope. The insulinoma was minced and cultured in DMEM high-glucose medium (GIBCO) supplemented with 10% FBS (HyClone) at 37°C and 5% CO2.
Quantitative polymerase chain reaction (PCR) analysis of tRNA modification
Total RNA was extracted from the lysates of cultured cells using TRIzol reagent (Invitrogen) following the manufacturer's instructions. The ms2 modification levels were analyzed using a quantitative PCR-based method as described previously (17). The levels of ms2-modified tRNALys(UUU) were normalized to the total tRNALys(UUU) (17). In all experiments, the ms2 modification levels in control cells are expressed as 100%. The modification levels in compound-treated cells are expressed as levels relative to those in control cells. The sequences of primers were as follows:
Primers for detecting ms2t6A in human and mouse cells
tRNALys_forward primer: GTCGGTAGAGCATCAGACTT
tRNALys_reverse primer r1: CCTGGACCCTCAGATTAAAA
tRNALys_reverse primer r1: GAACAGGGACTTGAACCCTG
Gene expression analysis
RNA was extracted from the cells using TRIzol reagent (Invitrogen) following the manufacturer's instructions. A PrimerScript RT Reagent Kit (TAKARA) was used to generate cDNA. Quantitative real-time PCR was performed using SYBR Premix Ex Taq (TAKARA). The results were normalized to beta-2 microglobulin.
Plasmids
For overexpression of the CDKAL1 protein in HeLa cells, cDNA encoding CDKAL1 was subcloned into the pCMV-Myc vector (Clontech). For the expression of CDKAL1 in E. coli, CDKAL1 was subcloned into the pET21a vector (Novagen). For the construction of CDKAL1 carrying Cys-to-Ala mutations, Cys 83, Cys 109 and Cys 138 in the UPF0004 domain and Cys 214, Cys 218 and Cys 221 in the radical SAM domain were mutated to Ala using QuikChange II Site-Directed Mutagenesis Kits (Agilent Technologies). Plasmids were used to transfect HeLa cells with Lipofectamine 2000 (Invitrogen).
Measurement of blood glucose and plasma insulin levels
The glucose tolerance test was performed in either male mice fed normal chow at the age of 6 weeks old or in male mice fed a high-fat diet for 20 weeks. Briefly, mice were fasted for 20 h (6:00 pm to 14:00 am), followed by the intraperitoneal injection of glucose (1 g/kg). The plasma glucose level was determined by a glucometer (ACCU-CHEK, Aviva; Roche). Plasma insulin levels were determined using an ELISA kit (Shibayagi, Tokyo, Japan) following the manufacturer's instructions.
Statistical analysis
All data are presented as the mean ± SEM unless otherwise indicated. Statistical analyses were performed using Prism 6 Software (GraphPad Software). An unpaired Student's t-test was used to test the differences between two groups. Analysis of variance (one-way ANOVA) was used to test the differences among multiple groups, followed by the Bonferroni procedure to calculate the P-value between two groups. A two-tailed P-value of 0.05 was considered significant.
RESULTS
Regulation of endogenous CysSSH by CBS and CTH
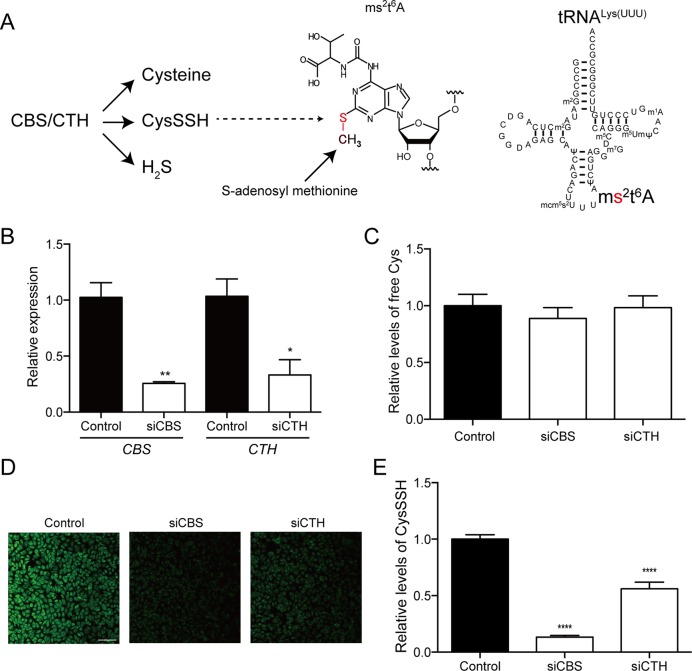
CBS and CTH are key enzymes in sulfur biology (Figure 1A). In addition to the classic cysteine biogenesis pathway, CBS/CTH have recently been implicated in the production of various sulfur species including CysSSH and H2S (12, Figure 1A). To investigate the biological source of the sulfur atom in ms2 modification, we silenced CBS or CTH in Hela cells using specific siRNAs and examined the levels of free cysteine and CysSSH (Figure 1B). The siRNAs successfully downregulated CBS or CTH, but the free cysteine level was unchanged in the siRNA-treated cells, when compared with control siRNA-treated cells (Figure 1C). To examine the endogenous CysSSH level, we applied a sulfane sulfur-specific fluorescent probe, SSP4, in HeLa cells (12). The reactive sulfur atom in CysSSH, but not cysteine or glutathione, is capable of reacting with the SSP4 probe to release its fluorophore (14). There was a significant decrease of SSP4 fluorescence in CBS- and CTH-silenced cells, when compared with control cells (Figure 1D and E). These results demonstrate that the downregulation of CBS or CTH selectively reduced the CysSSH level in HeLa cells.
Figure 1.
Regulation of endogenous cysteine hydropersulfide (CysSSH) by cystathionine beta synthase (CBS) and cystathionine gamma lyase (CTH). (A) Potential molecular origins of ms2 modification of tRNALys(UUU). (B) The efficiency of siRNAs against CBS (siCBS) and CTH (siCTH) in HeLa cells. n = 4, *P < 0.05, **P < 0.01. (C) The endogenous free cysteine levels in cells treated with control siRNA and siRNAs against CBS (siCBS) or CTH (siCTH) were determined by mass spectrometry and normalized to endogenous phenylalanine levels. n = 3 each. (D) The endogenous CysSSH levels in cells treated with control siRNA and siRNAs against CBS (siCBS) or CTH (siCTH) were visualized by the SSP4 probe. Bar = 0.1 mm. (E) Relative levels of CysSSH were estimated by quantification of the intensity of SSP4 in each sample. Results are expressed as the intensity relative to that in control cells. n = 13–20, ****P < 0.0001 versus control sample.
Regulation of ms2 modification by CysSSH in living cells
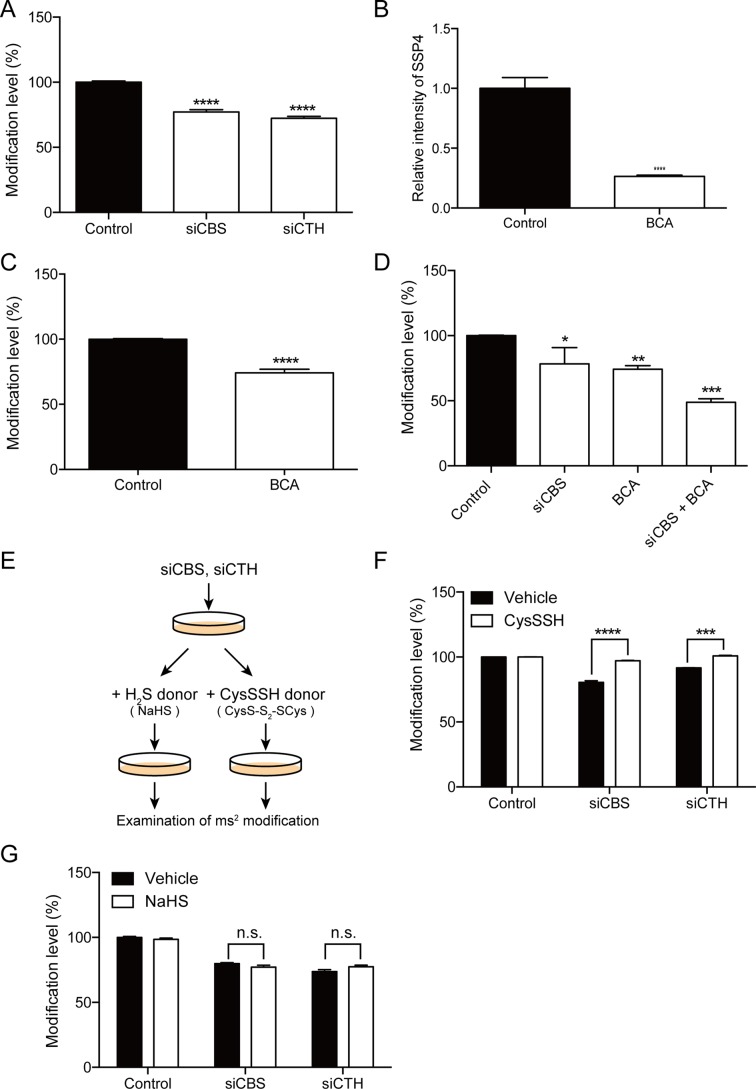
Next, we investigated whether CysSSH is involved in the regulation of ms2 modification of tRNAs. CBS and CTH were silenced by siRNAs, and the levels of ms2 modification in cytosolic tRNALys(UUU) were examined using quantitative PCR (17). There was a significant decrease in the ms2 levels in CBS- and CTH-silenced cells, when compared with control cells (Figure 2A). In addition to the treatment with the siRNAs, we also chemically downregulated CysSSH using β-cyano-L-alanine (BCA), a classic inhibitor of CTH (18). BCA markedly reduced the intracellular CysSSH level, as indicated by the reduction of SSP4 fluorescence (Figure 2B). Accordingly, the ms2 modification was significantly decreased in BCA-treated cells (Figure 2C). Furthermore, co-application of BCA and CBS-targeting siRNA synergistically reduced the ms2 modification level (Figure 2D).
Figure 2.
(A) Regulation of ms2 modification by CysSSH in intact cells. Silencing of CBS and CTH significantly reduced the ms2 level of tRNALys(UUU). n = 4, ****P < 0.0001. (B) Cells treated with 500 μM BCA exhibited a significant decrease in endogenous CySSSH levels. n = 8–9, ****P < 0.0001. (C) Treatment with 500 μM BCA significantly decreased the ms2 modification levels of tRNALys(UUU). n = 4, ****P < 0.0001. (D) Inhibition of CBS and CTH with siCBS and BCA synergistically suppressed endogenous CysSSH levels. n = 4, *P < 0.05, **P < 0.01, ***P < 0.001. (E) Experimental design for rescuing ms2 modification by the exogenous CysSSH donor or H2S donor. (F) The CysSSH donor, but not (G) H2S donor, effectively reversed the suppression of ms2 modification levels of tRNALys(UUU) by silencing CBS or CTH. n = 4, ****P < 0.0001. n.s.: not significant.
To further demonstrate that CysSSH is involved in the regulation of ms2 modification, we aimed to modulate ms2 modification by supplementing exogenous CysSSH. We chemically synthesized a CysSSH donor, CysS-S2-SCys. Upon uptake by cells, CysS-S2-SCys is rapidly broken down to CysSSH due to the reductive cellular environment. Indeed, the application of 100 μM of the CysSSH donor to HeLa cells for 1 h resulted in a marked increase of the intracellular CysSSH level (Supplementary Figure S1, Vehicle: 3.8 nM, CysSSH: 71 nM). HeLa cells were treated with specific siRNAs against CBS and CTH, followed by application of the CysSSH donor (Figure 2E). As expected, CysSSH donor supplementation successfully reversed the ms2 modification, which was downregulated by silencing CBS or CTH (Figure 2F).
Both CBS and CTH are capable of producing H2S, which is also actively involved in diverse biological activities (19). Therefore, to investigate the potential contribution of H2S to ms2 modification, we applied NaHS, a donor of H2S, to HeLa cells with silencing of CBS or CTH (Figure 2E). However, NaHS was not able to reverse the ms2 modification level of tRNALys(UUU) in CBS-silenced or CTH-silenced cells (Figure 2G). Taken together, these results suggest that CBS/CTH-mediated CysSSH production is selectively involved in the regulation of ms2 modification.
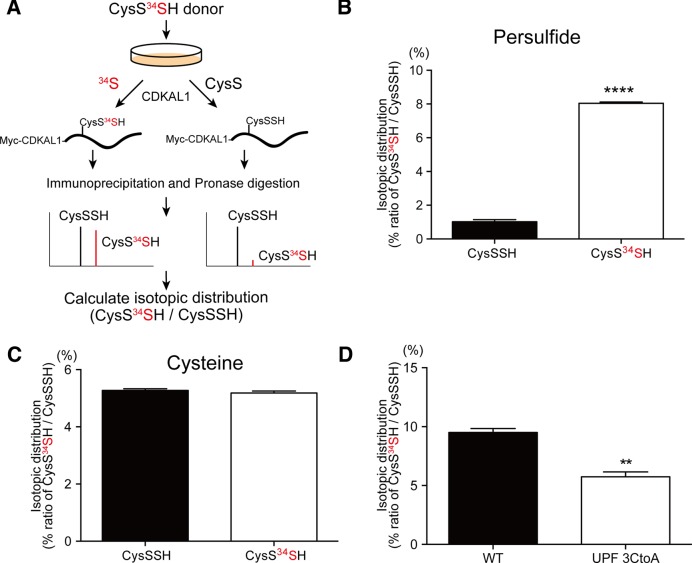
Selective chasing of reactive sulfur atom by stable isotope labeling
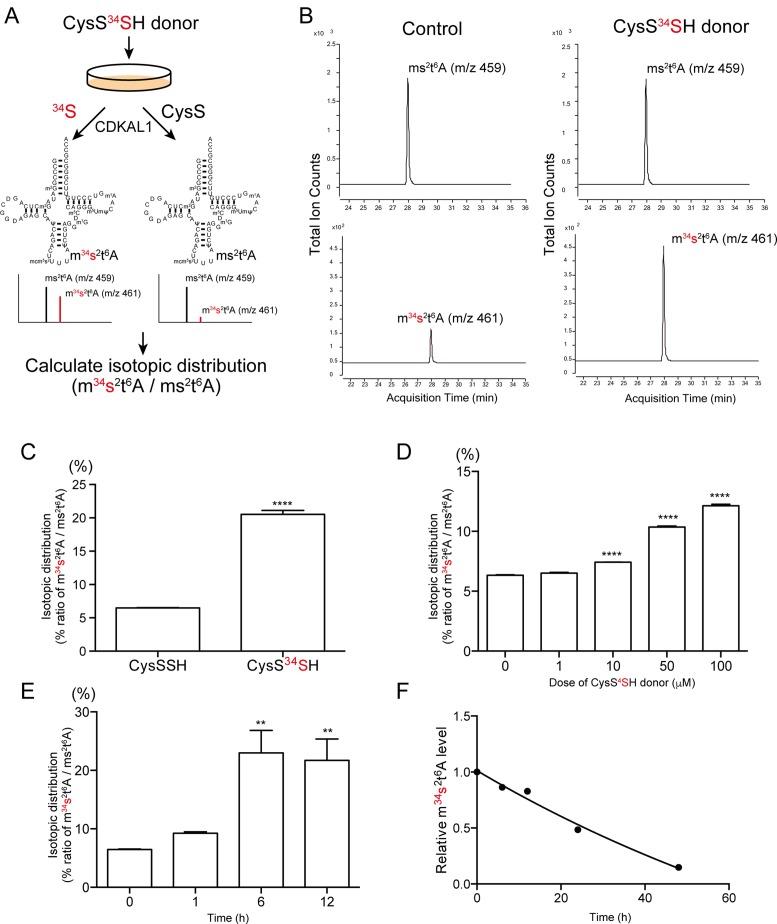
To clarify which sulfur atom in CysSSH is utilized for ms2 modification, we synthesized a ‘heavy’ CysSSH donor (CysS-34S2-SCys), in which reactive sulfur atoms were selectively and stably labeled with heavy isotope 34S instead of natural 32S (Figure 3A). The heavy CysSSH donor markedly and selectively induced the intracellular CysS34SH level as quickly as 1 h after application (Supplementary Figure S2A and B). Notably, application of the CysS34SH donor did not give rise of S34-labeled free cysteine (Supplementary Figure S2C and D). These results clearly demonstrate that the CysS34SH donor enables us to selectively track the mobilization of reactive sulfur atoms by the differential molecular mass.
Figure 3.
Incorporation of reactive sulfur atom in ms2 modification. (A) Experimental design to identify the sulfur source for ms2 in intact cells. The red letters represent the stable isotope-labeled reactive sulfur atoms of CysSSH. (B) HeLa cells were treated with 100 μM of CysSSH or the CysS34SH donor. Subsequently, total RNA was purified and subjected to mass spectrometry. Representative mass chromatograms of ms2t6A and m34s2t6A are shown. The CysS34SH donor markedly increased the m34s2t6A level. (C) The isotopic distribution (relative abundance of m34s2t6A to ms2t6A) of ms2t6A in cells treated with vehicle (CysSSH donor) or the CysS34SH donor. n = 4 each, ****P < 0.0001. The isotopic distribution of ms2t6A in response to different (D) doses or (E) periods of treatment with 100 μM of the CysS34SH donor. **P < 0.01, ****P < 0.0001, n = 4 each. (F) Pulse-chasing of m34s2t6A showing the relatively slow rate of decrease in the amount of m34s2t6A in cells. Data points represent the relative change in the isotopic distribution of ms2t6A from four replicates.
We applied the CysS34SH donor to HeLa cells and examined whether CysS34SH is capable of providing a ‘heavy’ sulfur atom for ms2 modification (Figure 3A). All chemical species contain a trace of the isotope, and exhibit a fixed isotopic distribution pattern. Theoretically, the abundance of naturally occurring m34s2t6A (m/z 461) only accounts for 8.19% of the abundance of ms2t6A (m/z 459). If the heavy 34S of CysS34SH is actively incorporated in the sulfur atom of ms2t6A, there will be an unnatural increase of the abundance of m34s2t6A (Figure 3A). However, if ms2 modification utilizes unlabeled sulfur atoms of cysteine, the abundance of m34s2t6A would remain unchanged (Figure 3A). This isotopic distribution can be monitored by the ratio of the abundance of m34s2t6A to the abundance of ms2t6A (m34s2t6A/ ms2t6A), which thus reflects the incorporation of the reactive 34S atom. When Hela cells were treated with 100 μM of the CysSSH donor, the abundance of m34s2t6A accounted for 6.49% of the abundance of ms2t6A, which is close to the theoretical ratio (Figure 3B and C). Upon treatment with 100 μM of the CysS34SH donor, there was a marked increase of m34s2t6A; the abundance of m34s2t6A in CysS34SH donor-treated cells accounted for 20.5% of the abundance of ms2t6A, which is far beyond the natural distribution (Figure 3B and C). This evidence strongly suggests that a substantial portion of ms2 modification is likely to be derived from the reactive sulfur atom of CysSSH.
We examined the efficacy of the CysS34SH donor by subjecting cells to various conditions. The incorporation of 34S was dose-dependent, with an optimal concentration of 100 μM (Figure 3D). The incorporation of 34S peaked 6 h after treatment (Figure 3E). The utilization of CysS34SH donor allowed us to examine the turnover rate of ms2 modification in vivo. HeLa cells were labeled with the CysS34SH donor for 6 h, followed by various chasing periods. The 34S in m34s2t6A decreased slowly and had mostly disappeared at 48 h after treatment (Figure 3F). The slow turnover rate of ms2-containing tRNA is in agreement with the half-life of cytosolic tRNAs in vivo (20), and reflects the partial labeling efficiency by the CysS34SH donor. Taken together, these results clearly demonstrate that the reactive sulfur atom of CysSSH is transferred for the ms2 modification of tRNA under physiological conditions.
Polysulfidation of CDKAL1 in intact cells
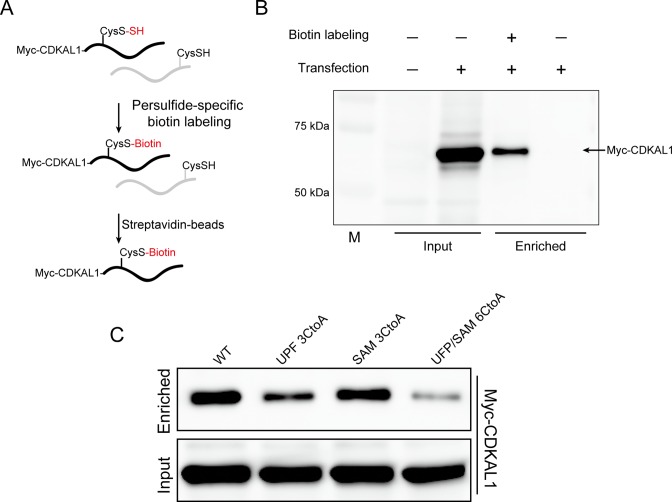
A previous study of an in vitro reconstituted bacterial ms2-modifying enzyme, MiaB, revealed the existence of exogenous sulfur atoms in the enzyme (8). It has been proposed that the enzyme-conjugating polysulfide might be utilized for ms2 modification. We speculated that CDKAL1 may undergo CysSSH-mediated protein polysulfidation, and this is subsequently utilized for ms2 modification. To investigate the potential polysulfidation of CDKAL1, we performed a polysulfide-specific biotin-labeling assay in HeLa cells (12). This unique method enables the selective conjugation of biotin to the reactive sulfur residue present in polysulfide-containing proteins (Figure 4A). The biotin-labeled protein can be specifically enriched by streptavidin-beads and subjected to the downstream applications (Figure 4A). HeLa cells expressing Myc-CDKAL1 were subjected to the biotin-labeling assay. Subsequently, the enriched polysulfide-containing proteins were examined by Western blotting using anti-Myc antibody. As expected, Myc-CDKAL1 was clearly detected in the polysulfide-containing protein fraction (Figure 4B).
Figure 4.
Detection of polysulfidation in CDKAL1. (A) Illustration of the polysulfide-specific biotin labeling assay used to identify the polysulfidation in proteins. (B) HeLa cells overexpressing Myc-CDKAL1 were subjected to a polysulfide-specific biotin labeling assay. Myc-CDKAL1 was detected in enriched proteins using streptavidin beads. (C) The polysulfidation levels in wild-type CDKAL1 (WT) or mutant CDKAL1 carrying Cys-to-Ala mutations in the UPF0004 domain (UPF 3CtoA), radical SAM domain (SAM 3CtoA) or UPF0004/radical SAM domains (UPF/SAM 6CtoA) were evaluated using a polysulfide-specific biotin labeling assay.
The UPF0004 domain of bacterial MiaB protein forms a cluster [4Fe–4S] and has also been proposed to form a polysulfide moiety (3,8). Consistent with the bacterial model, mutation of the Cys residues in the UPF0004 domain of CDKAL1 completely eliminated its activity (Supplementary Figure S3). To examine whether the polysulfidation of CDKAL1 occurred in the UPF0004 domain of intact cells, CDKAL1 in which the Cys residues were mutated to Ala was subjected to the polysulfide-specific biotin labeling assay (Figure 4C). Compared with wild-type CDKAL1, CDKAL1 carrying Cys-to-Ala mutations in the radical SAM domain exhibited a slight decrease in polysulfidation. Intriguingly, the polysulfidation level was markedly decreased in CDKAL1 carrying Cys-to-Ala mutations in the UPF0004 domain. Mutations in both the UPF0004 and radical SAM domains almost eliminated the polysulfidation from the CDKAL1 protein. These results suggest that the UPF0004 domain is the major polysulfidation site in CDKAL1.
CysSSH-mediated polysulfidation of CDKAL1
To investigate whether the polysulfidation of CDKAL1 is also mediated by the reactive sulfur atom of CysSSH, we treated HeLa cells with the CysS34SH donor, and examined 34S-containing polysulfidation in CDKAL1 (Figure 5A). In analogy with the experiment that aimed to detect 34S incorporation in ms2 modification shown in Figure 3A, the formation of 34S-containing polysulfide in Cys residues of CDKAL1 would result in an increased abundance of CysS34SH, which would lead to a shift in the isotopic distribution of CysSSH (Figure 5A). HeLa cells expressing Myc-CDKAL1 were treated with the CysS34SH donor for 1 h. Subsequently, Myc-CDKAL1 was immunoprecipitated with anti-Myc antibody. The immunoprecipitaed proteins were digested by pronase and subjected to mass spectrometry. In control cells treated with the CysSSH donor, abundance of natural occurring CysS34SH accounted for 1% of the abundance of CysSSH in Myc-CDKAL1 (Figure 5B). After application of the CysS34SH donor, the abundance of CysS34SH accounted for as high as 8% of the abundance of CysSSH (Figure 5B). In contrast, the CysS34SH donor did not change the isotopic distribution of cysteine residue (Cys34SH/CysSH) in CDKAL1 protein (Figure 5C). Furthermore, we applied the CysS34SH donor to HeLa cells expressing Myc-CDKAL1 with or without mutations in the UPF0004 domain, and examined polysulfidation by mass spectrometry. There was a significant reduction of 34S-containing polysulfidation in mutant CDKAL1, when compared with the wild-type (Figure 5D). These results strongly suggest that CysSSH selectively transferred the reactive sulfur atom to Cys residues of CDKAL1, and induced polysulfidation in the UPF0004 domain of CDKAL1.
Figure 5.
CysSSH-mediated polysulfidation of CDKAL1. (A) Experimental design to detect 34S-labeled polysulfide in the CDKAL1 protein. (B and C) HeLa cells overexpressing Myc-CDKAL1 were treated with vehicle (CysSSH donor) or the CysS34SH donor for 1 h. Myc-CDKAL1 was immunoprecipitated and subjected to mass spectrometry to determine the isotopic distribution of polysulfidation in Cys residues. The isotopic distribution of CysSSH (abundance of CysS34SH relative to that of CysSSH) is shown in (B). The isotopic distribution of the Cys residue (abundance of CysS34H relative to that of CysSH) is shown in (C). ****P < 0.0001, n = 4. (D) HeLa cells expressing wild-type Myc-CDKAL1 or mutant CDKAL1 carrying Cys-to Ala mutations in the UPF0004 domain were treated with the CysS34SH donor, followed by mass spectrometric analysis. The mutant CDKAL1 had less 34S incorporation in Cys residues in CDKAL1, when compared with the wild-type CDKAL1. n = 3 each, **P < 0.01.
In addition, we aimed to exclude the possibility that the [4Fe-4S] clusters might non-specifically transfer sulfur atoms to cysteine residues of CDKAL1 protein during sample preparation under oxidized conditions. HeLa cells were treated with an iron-chelating reagent, deferoxamine, to degenerate [4Fe-4S] clusters and then subjected to mass spectrometry. The polysulfidation in the CDKAL1 protein was not affected by deferoxamine (DFOM) (Supplementary Figure S4A–D), whereas the reagent significantly impaired CDKAL1 activity (Supplementary Figure S4A–E). Taken together, these results suggest that cysteine residues in UPF0004 of CDKAL1 undergo CysSSH-mediated polysulfidation and the sulfur of the polysulfidated Cys residues is ultimately transferred for ms2 modification.
Regulation of ms2 modification and insulin secretion by CysSSH in pancreatic β-cells
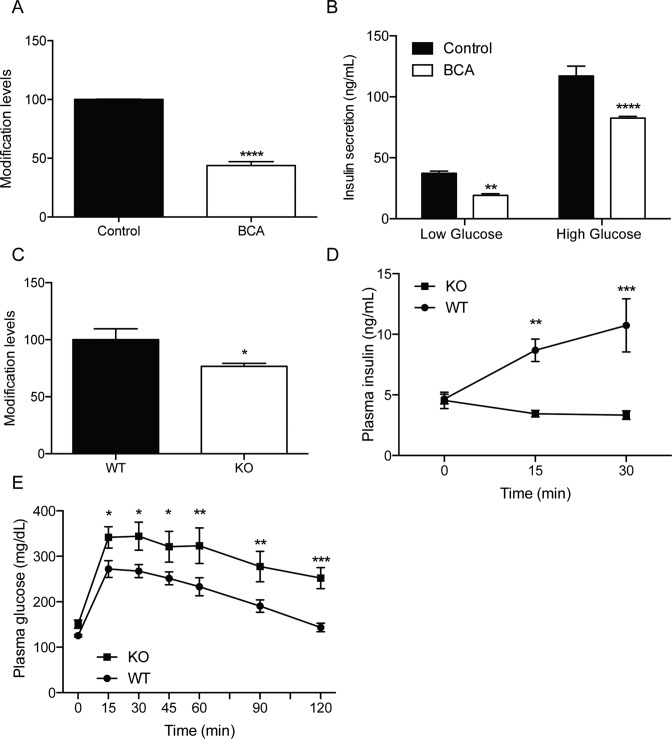
CDKAL1-medated ms2 modification regulates insulin biosynthesis in pancreatic β-cells, and has been implicated in the development of type 2 diabetes (5). Given the important role of CysSSH in ms2 modification, we aimed to investigate whether CysSSH contributes to insulin secretion through the regulation of ms2 modification in pancreatic β-cells. We established two pancreatic β-cell-derived cell lines and applied BCA to reduce the intracellular CysSSH levels. The suppression of CysSSH production by BCA significantly decreased the ms2 modification level in both β-cell lines (Figure 6A and Supplementary Figure S5A). We treated β-cell lines with BCA and then stimulated the cells with 2.8 mM (low) glucose and 16.7 mM (high) glucose. BCA significantly decreased glucose-stimulated insulin secretion in these cells (Figure 6B and Supplementary Figure S5B).
Figure 6.
Regulation of insulin secretion by CySSH. (A) Suppression of CysSSH by 500 μM BCA significantly decreased ms2 levels in the pancreatic β-cell line #1. n = 4, ****P < 0.0001. (B) The inhibition of Cth by BCA significantly impaired glucose-stimulated insulin secretion in the pancreatic β-cell line. n = 8, **P < 0.01, ****P < 0.0001. (C) Wild-type (WT) mice and Cth-deficient mice (KO) were fed a high-fat diet for 20 weeks. The ms2 levels of tRNALys(UUU) in pancreatic islets were examined. n = 4–5, *P < 0.05. (D) WT and KO mice were fed a high-fat diet for 20 weeks and challenged with glucose (1 g/Kg). (D) Plasma insulin levels and (E) blood glucose levels at indicated time-points are shown. n = 4–5, *P < 0.05, **P < 0.01, ***P < 0.001.
Next, we investigated whether or not modulation of the CysSSH level affects ms2 modification and glucose metabolism in vivo. Cth-deficient mice developed normally but exhibited high-fat diet-induced metabolic defects (15,21). Indeed, when the mice were fed normal chow, the hepatic CysSSH level in Cth-deficient mice was comparable with that of wild-type mice (Supplementary Figure S6A). To accelerate the diabetic phenotype, we fed the mice a high-fat diet for 20 weeks. The hepatic CysSSH level of Cth-deficient mice was then significantly lower than that of wild-type mice (Supplementary Figure S6B). Accordingly, there was a significant reduction of ms2 modification in pancreatic islets of Cth-deficient mice, when compared with wild-type mice (Figure 6C). These results suggest that CysSSH is associated with ms2 modification in vivo.
To examine whether a decrease of ms2 modification affects insulin secretion, mice were injected with glucose and the blood insulin level was examined. The Cth-deficient mice fed a high-fat diet exhibited impaired insulin secretion, when compared with wild-type mice (Figure 6D). To examine whether the impairment of insulin secretion was associated with a decrease of glucose metabolism, the mice were injected with glucose and the blood glucose levels were measured. The blood glucose level in Cth-deficient mice was significantly higher than that in control mice (Figure 6E). Taken together, these results suggest that CysSSH contributes to ms2 modification and glucose metabolism in vivo.
DISCUSSION
The present study provides direct evidence that the intracellular CysSSH is closely involved in the regulation of ms2 modifications in mammalian tRNAs. Using the unique CysSSH donor in combination with precision mass spectrometry-based analytic methods, our results clearly demonstrate that the reactive sulfur of CysSSH rapidly initiates protein polysulfidation in ms2-modifying enzymes and mediates the sulfur insertion of ms2 modification. Furthermore, the suppression of CysSSH production resulted in a decreased ms2 level, which ultimately led to the impairment of insulin secretion in vivo.
The regulatory mechanism of ms2 modification in mammalian cells has remained largely unknown. In general, all ms2-modifying enzymes require [4Fe-4S] clusters for their catalytic activities. Based on studies of biotin synthase, a [4Fe-4S] cluster-containing thiotransferase, it was previously assumed that the sulfur atom in ms2 might be derived from the sacrifice of the sulfur atom from its own [4Fe–4S] cluster (22). Recently, a structural study of bacterial MiaB protein questioned this self-sacrifice model, and proposed that the sulfur atom of ms2 might be derived from an extra sulfur group conjugated to the enzyme (8). However, the extra sulfur-regulated ms2 modification could be formed as a byproduct during the in vitro reconstitution of [4Fe–4S] clusters. Using chemically defined CysS34SH donors, we were able to provide direct evidence that the extra sulfur group of CDKAL1 was indeed formed in intact mammalian cells, and that the extra sulfur was derived from the reactive sulfur of CysSSH. In addition, the majority of the polysulfidation was found in the UPF0004 domain of CDKAL1 that was also in agreement with a previous prediction (8). Taken together, these results suggest that CysSSH-mediated protein polysulfidation is a physiological event that contributes to ms2 modification.
Sulfur atoms are widely incorporated into tRNAs during a number of essential modifications, including 2-thiouridine, 4-thiouridine and 2-thiocythidine (2). To date, cysteine has been considered as the only sulfur source for these modifications (23). In the general model, cysteine desulfurase captures a sulfur atom from cysteine, and forms enzyme-bound polysulfide as the first step of the reactions (24). The activated sulfur atom is ultimately relayed to tRNAs by various enzymes. In contrast to the general model, our study showed that ms2 modification and CDKAL1 contained a substantial portion of sulfur atoms, which were derived from the reactive sulfur atoms of CysSSH, but not cysteine. These results thus challenge the classical model, and suggest that there are multiple sulfur sources for tRNA thiolation, including the reactive CysSSH. The CysSSH probes and analytic methodologies utilized in this study would provide unique biochemical tools for studying the molecular mechanisms of these sulfur transfers in the future.
Oxidative stress impairs insulin secretion and subsequently induces glucose intolerance. Because CysSSH is highly susceptible to intracellular reactive oxygen species (12), excess oxidative stress might downregulate the CysSSH level, which in turn impairs ms2 modification as well as insulin secretion. Consistent with this view, the CysSSH level was markedly impaired in Cth-deficient mice in a stress-dependent manner. Furthermore, the decrease of the CysSSH level was associated with a decrease of ms2 modification as well as impaired insulin secretion in Cth-deficient mice. Nevertheless, it is conceivable that the high-fat-induced oxidative stress might also affect the redox state of [4Fe–4S] clusters of CDKAL1.
In summary, our results show that the reactive CysSSH is a novel sulfur source for ms2 modification of tRNA and CDKAL1 in intact cells and in vivo. Suppression of the CysSSH level leads to the reduction of ms2 modification and the impairment of insulin signaling.
Acknowledgments
The authors thank Nobuko Maeda for technical support during the animal experiments.
Author contributions: N.T. performed all experiments and wrote the manuscript; Y.O. and T.S. synthesized the persulfide donors; S.W. performed the animal experiments and analyzed the data; T.A. provided reagents for polysulfide-specific biotin-labeling assay; I.I. established and provided Cth-deficient mice; A.F., T.K. and H.N. contributed to the discussion; and F.Y.W. and K.T. designed the experiments and wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan; Japan Society for the Promotion of Science (JSPS) through its ‘Funding Program for Next Generation World-Leading Researchers’; Grant-in-aid for Scientific Research from the Ministry of Health, Labour and Welfare. Funding for open access charge: Grant-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Agris P.F. Decoding the genome: A modified view. Nucleic Acids Res. 2004;32:223–238. doi: 10.1093/nar/gkh185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Machnicka M.A., Milanowska K., Osman Oglou, O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., et al. MODOMICS: A database of RNA modification pathways–2013 update. Nucleic Acids Res. 2013;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arragain S., Handelman S.K., Forouhar F., Wei F.Y., Tomizawa K., Hunt J.F., Douki T., Fontecave M., Mulliez E., Atta M. Identification of eukaryotic and prokaryotic methylthiotransferase for biosynthesis of 2-methylthio-N6-threonylcarbamoyladenosine in tRNA. J. Biol. Chem. 2010;285:28425–28433. doi: 10.1074/jbc.M110.106831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei F.Y., Zhou B., Suzuki T., Miyata K., Ujihara Y., Horiguchi H., Takahashi N., Xie P., Michiue H., Fujimura A., et al. Cdk5rap1-mediated 2-methylthio modification of mitochondrial tRNAs governs protein translation and contributes to myopathy in mice and humans. Cell Metab. 2015;21:428–442. doi: 10.1016/j.cmet.2015.01.019. [DOI] [PubMed] [Google Scholar]
- 5.Wei F.Y., Suzuki T., Watanabe S., Kimura S., Kaitsuka T., Fujimura A., Matsui H., Atta M., Michiue H., Fontecave M., et al. Deficit of tRNA(Lys) modification by Cdkal1 causes the development of type 2 diabetes in mice. J. Clin. Invest. 2011;121:3598–3608. doi: 10.1172/JCI58056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jenner L.B., Demeshkina N., Yusupova G., Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol. 2010;17:555–560. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
- 7.Atta M., Mulliez E., Arragain S., Forouhar F., Hunt J.F., Fontecave M. S-Adenosylmethionine-dependent radical-based modification of biological macromolecules. Curr. Opin. Struct. Biol. 2010;20:684–692. doi: 10.1016/j.sbi.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Forouhar F., Arragain S., Atta M., Gambarelli S., Mouesca J.M., Hussain M., Xiao R., Kieffer-Jaquinod S., Seetharaman J., Acton T.B., et al. Two Fe-S clusters catalyze sulfur insertion by radical-SAM methylthiotransferases. Nat. Chem. Biol. 2013;9:333–338. doi: 10.1038/nchembio.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toohey J.I. Sulfur signaling: is the agent sulfide or sulfane? Anal. Biochem. 2011;413:1–7. doi: 10.1016/j.ab.2011.01.044. [DOI] [PubMed] [Google Scholar]
- 10.Nishida M., Sawa T., Kitajima N., Ono K., Inoue H., Ihara H., Motohashi H., Yamamoto M., Suematsu M., Kurose H., et al. Hydrogen sulfide anion regulates redox signaling via electrophile sulfhydration. Nat. Chem. Biol. 2012;8:714–724. doi: 10.1038/nchembio.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iciek M., Włodek L. Biosynthesis and biological properties of compounds containing highly reactive, reduced sulfane sulfur. Pol. J. Pharmacol. 2001;53:215–225. [PubMed] [Google Scholar]
- 12.Ida T., Sawa T., Ihara H., Tsuchiya Y., Watanabe Y., Kumagai Y., Suematsu M., Motohashi H., Fujii S., Matsunaga T., et al. Reactive cysteine persulfides and Polythiolation regulate oxidative stress and redox signaling. Proc. Natl. Acad. Sci. U.S.A. 2014;111:7606–7611. doi: 10.1073/pnas.1321232111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuevasanta E., Lange M., Bonanata J., Coitino E.L., Ferrer-Sueta G., Filipovic M.R., Alvarez B. Reaction of hydrogen sulfide with disulfide and sulfenic acid to form the strongly nucleophilic persulfide. J. Biol. Chem. 2015;290:26866–26880. doi: 10.1074/jbc.M115.672816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen W., Liu C., Peng B., Zhao Y., Pacheco A., Xian M. New fluorescent probes for sulfane sulfurs and the application in bioimaging. Chem. Sci. 2013;9:2892–2896. doi: 10.1039/C3SC50754H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishii I., Akahoshi N., Yamada H., Nakano S., Izumi T., Suematsu M. Cystathionine gamma-Lyase-deficient mice require dietary cysteine to protect against acute lethal myopathy and oxidative injury. J. Biol. Chem. 2010;285:26358–26368. doi: 10.1074/jbc.M110.147439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyazaki J., Araki K., Yamato E., Ikegami H., Shibasaki Y., Oka Y., Yamamura K. Establishment of a pancreatic beta cell line that retains glucose-inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology. 1990;127:126–132. doi: 10.1210/endo-127-1-126. [DOI] [PubMed] [Google Scholar]
- 17.Xie P., Wei F.Y., Hirata S., Suzuki T., Tomizawa K. Quantitative PCR measurement of tRNA 2-methylthio modification for assessing type 2 diabetes risk. Clin. Chem. 2013;59:1604–1612. doi: 10.1373/clinchem.2013.210401. [DOI] [PubMed] [Google Scholar]
- 18.Pfeffer M., Ressler C. Beta-cyanoalanine, an inhibitor of rat liver cystathionase. Biochem. Pharmacol. 1967;16:2299–2308. doi: 10.1016/0006-2952(67)90217-1. [DOI] [PubMed] [Google Scholar]
- 19.Yadav P.K., Martinov M., Vitvitsky V., Seravalli J., Wedmann R., Filipovic M.R., Banerjee R. Biosynthesis and reactivity of cysteine persulfides in signaling. J. Am. Chem. Soc. 2015;138:289–299. doi: 10.1021/jacs.5b10494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phizicky E.M., Hopper A.K. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okamoto M., Yamaoka M., Takei M., Ando T., Taniguchi S., Ishii I., Tohya K., Ishizaki T., Niki I., Kimura T. Endogenous hydrogen sulfide protects pancreatic beta-cells from a high-fat diet-induced glucotoxicity and prevents the development of type 2 diabetes. Biochem. Biophys. Res. Commun. 2013;442:227–233. doi: 10.1016/j.bbrc.2013.11.023. [DOI] [PubMed] [Google Scholar]
- 22.Booker S.J., Grove T.L., Cicchillo R.M. Self-sacrifice in radical S-adenosylmethionine proteins. Curr. Opin. Chem. Biol. 2007;11:543–552. doi: 10.1016/j.cbpa.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shigi N. Biosynthesis and functions of sulfur modifications in tRNA. Front. Genet. 2014;5:67. doi: 10.3389/fgene.2014.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikeuchi Y., Shigi N., Kato J., Nishimura A., Suzuki T. Mechanistic insights into sulfur relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol Cell. 2006;21:97–108. doi: 10.1016/j.molcel.2005.11.001. [DOI] [PubMed] [Google Scholar]