Abstract
Recent genomic studies suggest that novel long non-coding RNAs (lncRNAs) are specifically expressed and far outnumber annotated lncRNA sequences. To identify and characterize novel lncRNAs in RNA sequencing data from new samples, we have developed COME, a coding potential calculation tool based on multiple features. It integrates multiple sequence-derived and experiment-based features using a decompose–compose method, which makes it more accurate and robust than other well-known tools. We also showed that COME was able to substantially improve the consistency of predication results from other coding potential calculators. Moreover, COME annotates and characterizes each predicted lncRNA transcript with multiple lines of supporting evidence, which are not provided by other tools. Remarkably, we found that one subgroup of lncRNAs classified by such supporting features (i.e. conserved local RNA secondary structure) was highly enriched in a well-validated database (lncRNAdb). We further found that the conserved structural domains on lncRNAs had better chance than other RNA regions to interact with RNA binding proteins, based on the recent eCLIP-seq data in human, indicating their potential regulatory roles. Overall, we present COME as an accurate, robust and multiple-feature supported method for the identification and characterization of novel lncRNAs. The software implementation is available at https://github.com/lulab/COME.
INTRODUCTION
Many long non-coding RNAs (lncRNAs) are expressed at specific stages during development or in response to specific stimuli (1–3). Recent studies suggest that there are far more potential lncRNAs than annotated RNA sequences (4–6). For instance, more than 15 000 lncRNA genes (∼27 000 loci/transcripts) were annotated by GENCODE (6) in October 2014. Within the first few months of 2015, a study examining TCGA data reported ∼58 000 lncRNA genes, with 79% of them described as novel (5). Thus, to discover novel lncRNAs associated with specific biological purposes, more RNA sequencing (RNA-seq) data are generally included in the study samples. One common approach to identify novel lncRNAs from new RNA-seq data involves the assembly of de novo transcripts from short reads and then application of a filter, i.e. a coding potential score, to remove potential coding transcripts (1).
Many coding potential calculation tools have been developed (7–16), and most use features curated from sequence information alone. These features include but are not limited to: open reading frame (ORF) features such as ORF length and coverage (7,8,10,13,15), nucleotide composition features such as k-mer sequence motif and codon usage (7,8,11,13–15), conservation scores such as pair-wise alignment score against nucleotide or protein sequence database (7–10), evolutionary features such as substitution rate and phylogenic score (12,16) and other in silico features such as predicted RNA secondary structure and ribosome release score (RRS)(7,8,13). These sequence-derived features are associated with several limitations though. First, some features, such as the predicted ORF length, require assembly of the full-length transcript. Calculation of the RRS also relies on a well-defined ORF and 3′ untranslated region (UTR). Moreover, determination of the precise 5′ and 3′ ends of a novel transcript requires very deep sequencing reads (15) or substantial experiments, such as rapid amplification of cDNA ends (17) and cap analysis gene expression (18,19). These requirements limit the application of current coding potential calculators on transcripts newly assembled from sequencing reads. Secondly, some features, such as the conservation scores calculated from blastx or tblastx, can be biased according to the length of transcript, in that a longer transcript provides a larger search space. In addition, a k-mer sequence is better sampled and estimated in a longer transcript. Lastly, some features, such as the substitution rate and phylogenic score, require multi-species alignments. However, the lncRNA transcripts are often not conserved, and therefore, are not located in the aligned regions.
Therefore, experimental features were used and integrated to separate non-coding RNAs (ncRNAs) from protein-coding RNAs (mRNAs) (4,20–23). For instance, many canonical ncRNAs (e.g. tRNAs) are not enriched in a poly(A)+ RNA-seq library, whereas most mRNAs are enriched (4,23,24). In addition, many lncRNAs show greater expression specificity than mRNAs in different tissues or during different developmental stages (3,25). Moreover, ribosome profiling data suggest that ribosomes may have different binding patterns on mRNAs and lncRNAs (26). Our previous study demonstrated that integration of experiment-based and sequence-derived features could enable classification of ncRNAs from coding sequences with high accuracy (27). However, the model focused only on canonical ncRNAs, and thus, can only predict the local regions of ∼70% of human lncRNAs.
Here, we have developed a coding potential calculator, COME (coding potential calculator based on multiple evidences), using a supervised machine learning model trained on mRNAs and lncRNAs. We introduced a decompose–compose method in COME and avoided using features (e.g. ORF length) that rely heavily on a full-length RNA transcript. Next, we showed that COME's performance is more robust than that of many well-known tools. In addition to the calculation of a single coding potential score, COME also characterized the predicted lncRNAs with multiple supporting feature scores. From these supporting features, we found that lncRNAs containing conserved local structures were significantly enriched in a database well validated by the function experiments. These lncRNAs contained local RNA structures that were conserved with various secondary structure families in Rfam, such as lncRNA families and canonical ncRNA families (signal recognition particle (SRP), snoRNA and pre-miRNA, etc). Remarkably, we associated these structural domains with RNA binding proteins (RBPs) suggesting their potential regulatory roles. Overall, we present COME as a robust and multiple-feature supported tool for the prediction and characterization of novel lncRNAs.
MATERIALS AND METHODS
Framework of COME, a coding potential calculator using multiple features
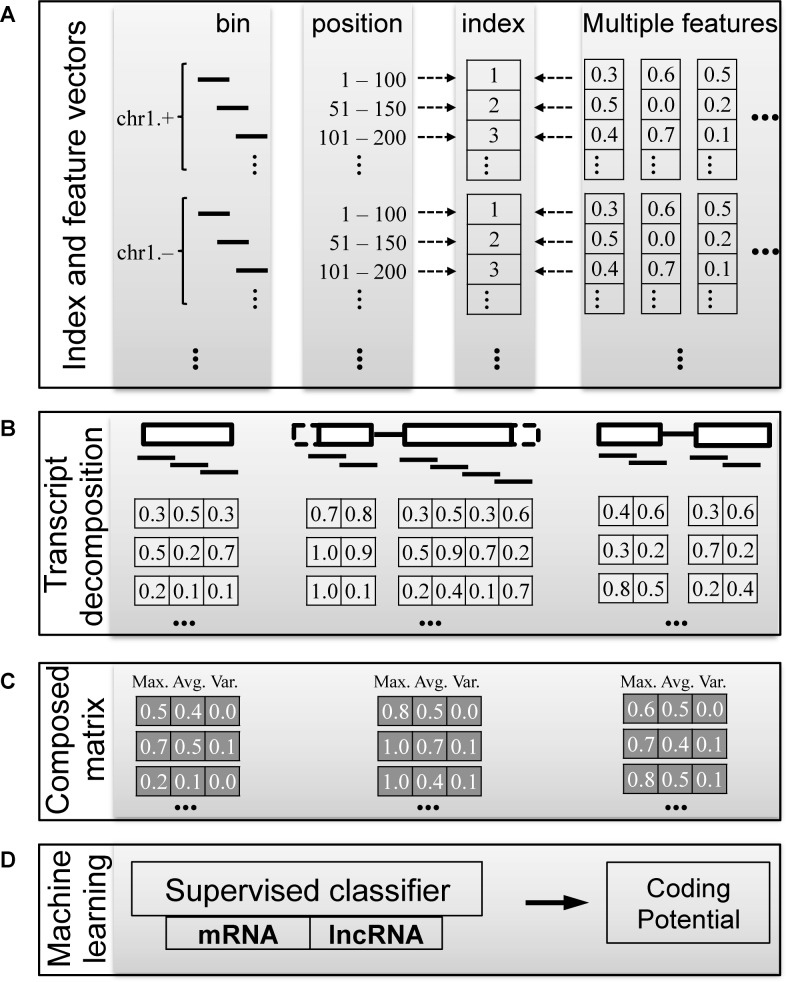
COME used machine learning models to calculate a transcript's coding potential score by integrating multiple features derived from both sequence information and experimental data. It was designed specifically for identifying and characterizing novel lncRNA transcripts assembled from RNA-seq data. Because the newly assembled transcripts could be incomplete, we developed a decompose–compose feature in the calculation procedure (Figure 1). In the decompose step, we first constructed an index for the whole genome, splitting the whole genome sequences into 100-nucleotide (nt) bins. We then calculated the input features on the indexed bins (see details in Supplementary Method). In this way, the feature values of a transcript were decomposed into multiple vectors. Subsequently, in the compose step, we used only three values (maximum, mean and variance) for each feature vector of a transcript. Thus, we composed a feature score matrix with multiple features and transcripts.
Figure 1.
COME workflow: a coding potential calculator based on multiple features. COME integrates multiple features with a supervised model to classify protein coding transcripts (mRNAs) and non-coding transcripts (lncRNAs). Multiple features (GC content, sequence conservation score, etc.) are processed by a decompose–compose procedure: feature values are initially calculated and indexed at the bin level (B). They are first indexed at the whole genome level, then mapped to each transcript (A). (C) The feature vectors of each transcript are composed at the transcript level by the maximum, mean and variance scores of the overlapping bins. (D) The probability of being mRNA predicted by the supervised model is the coding potential score for a given transcript.
Based on the composed matrix, we applied a balanced random forest (BRF) algorithm (7,27) to train on the annotated coding (mRNAs) and non-coding transcripts (lncRNAs). The predicted probability of being a coding transcript was defined as COME's coding potential score for each given transcript.
Genome index
We first indexed the human genome [Homo sapiens (hg19)] into small bins. It was first segmented into 48 segments naturally according to its chromosomes (chromosome 1–22, X and Y) and strands (forward and reverse). Each segment was then divided into 100-nt bins, with a 50-nt step size (Figure 1). We also used the same procedure to index the genomes of four other species: Mus musculus (mm10), Caenorhabditis elegans (ce10), Drosophila melanogaster (dm3) and Arabidopsis thaliana (TAIR10).
Composition of input features at the transcript level
For most features, we used the indexed bins that overlapped (>50%) with a transcript's exon(s) and assigned the feature vectors to the transcript (Supplementary Figure S1). Then, for each feature vector, we calculated three statistic scores: mean, maximum and variance. The three scores of the whole transcriptome were used to compose an input data matrix for a further machine learning procedure.
For the promoter marker (28) H3K4me3, we used the indexed bins that overlapped (>50%) with a transcript's upstream context [upstream 5000 nt for human and mouse genome, and upstream 2000 nt for fly, worm and plant genome (27)) and assigned the upstream feature vector to the transcript. When calculating the mean for H3K4me3, we calculated a CIS (context influence score) (29) to represent a weighted-mean.
Performance criteria
To quantify the classification performance, as many other methods did in the previous publications (7), we used the lncRNA transcripts as positives and mRNA transcripts as negatives to calculate the following metrics: sensitivity, specificity, accuracy, false positive rate, positive predictive value (PPV) and F-score.
Training and test sets
Training and test sets for human
The model for human data was trained and tested in two different ways. We first sampled two-thirds of Gencode annotations (v19) (6), including 15 638 lncRNA transcripts and 47 490 mRNA transcripts, as a training set. Then, we used the remaining 7819 lncRNA transcripts and 23 745 mRNA transcripts as test set 1 (T1). Test set 2 contained 2327 lncRNA transcripts selected from a human lincRNA catalog (25) and 12 327 mRNA transcripts chosen from Refseq (30). For the following performance comparisons, we used the original sets of T1 and T2 in the main figures. In addition, we also repeated the comparisons on two balanced sets, T1′ and T2′, which were up-sampled from T1 and T2 to make numbers of mRNAs and lncRNAs equal.
Independence of training and test sets
To ensure the independence of the training set and test set T1, we sampled the transcripts using a ‘block sampling’ method (27). Overlapping transcripts (≥1 nt) were assigned to the same block, and thus, transcripts sampled from any two blocks would have no overlap. In test set T2, we removed any transcripts overlapping (≥1 nt) with the training set.
Training and test sets of other four species
We also evaluated COME's performance for other four species: M. musculus, C. elegans, D. melanogaster and Arabidopsis thaliana. Their annotations were downloaded from Gencode (Version M4), Wormbase (version ws220), Flybase (version r5.45) and TAIR (version 10), respectively. The lncRNA transcripts and mRNA transcripts were divided into training and test sets using the same approach described for human data.
Optimization of the supervised machine-learning models in COME
To optimize the models in COME, we used 5-fold cross-validation on the training set (i.e. 15 638 lncRNA transcripts and 47 490 protein coding transcripts annotated by Gencode human (v19) (6)). In this imbalanced training set, we applied a BRF algorithm (7,27) that used multiple sub-training sets, each of which contained the same number of lncRNAs and mRNAs. We down-sampled the coding transcripts several times until all of them occurred in at least one sub-training set. Multiple models from multiple sub-training sets were averaged as the final model of the BRF.
Optimization of sampling methods for test
An imbalanced set would introduce bias into the test processes as well (27). By default, to make full use of all instances, we up-sampled the minority class (lncRNAs) to the number of the majority class (mRNAs) in the tests. We also tried another sampling method: down-sampling the majority class (mRNAs) to the number of minority class (lncRNAs), based on the minority class's expression level (Supplementary Figure S2a–d). We found no performance difference for the two sampling methods (Supplementary Figure S2e–h), indicating the robustness of our method.
Optimization of the bin size
In the decompose step, we indexed the genome into small bins. The smaller bin size provided more detailed descriptions but at higher computational cost. We tested four different bin sizes: 50 nt (25-nt step), 100 nt (50-nt step), 150 nt (75-nt step) and 200 nt (100-nt step). To balance the performance and computational cost (Supplementary Figure S3), we used an optimized bin size of 100 nt (50-nt step) as the default.
Optimization of the representative statistic
In the compose step, we represented a transcript's feature vector by a statistic value: mean, maximum or variance. We found that the combination of all three values achieved better performance than any individual value (Supplementary Figure S4).
Optimization of the input feature set
We used the nine features as the basic features of COME's input: GC content, DNA sequence conservation, protein sequence conservation, RNA secondary structure conservation, expression abundance calculated from various RNA-seq data (i.e. small, poly(A)+ and poly(A)- RNA-seq), and chromatin signature calculated from ChIP-seq data (i.e. H3K36me3 and H3K4me3). We adapted our previous feature process protocol (3,25,26), and listed the calculation details in Supplementary Methods as well. We used these features in our basic feature set for COME, because they were reported to be conserved for various types of ncRNAs in five species (human, mouse, worm, fly and Arabidopsis) (3,25,26). This feature set was proved to be non-redundant and better than including additional features (e.g.H3K4me1, H3K4me2, H3K4me3, H3K27ac, etc.) (27).
In addition, we further tested if adding some recently published experimental features (i.e. ribosome profiling data and expression specificity) would help to improve the performance of COME. We first tried three different scores, translation efficiency (TE) (26,31,32), RRS (26) and 3 nt periodicity score (ORFscore) (33) for ribosome profiling data. We found poor performance on distinguishing mRNAs and lncRNAs when using TE and RRS only (Supplementary Figure S5). The performance was substantially improved when the 3 nt periodicity score was included (Supplementary Figure S6). This result is consistent with previous studies showing that: (i) the TE score had little distinguish power for lncRNAs and mRNAs (26); (ii) most lncRNAs’ RRS scores were unavailable because the calculation of RRS score required high expression level of RNA-seq and Ribo-seq (26); and (iii) 3 nt periodicity better distinguished lncRNAs and mRNAs (33,34). In addition, the expression specificity score could also distinguish lncRNAs from mRNAs (Supplementary Figure S7), which was also consistent with previous studies (3,25).
However, although distinguishable between mRNAs and lncRNAs, we found trivial improvement for the final performance, by adding the new features (e.g. ribosome profiling scores and expression specificity score) to the basic feature set (including nine features) (Supplementary Figures S5–7). The performance may have already been saturated by the current feature set. Adding more features could have introduced redundancy. Moreover, the new features were usually not widely available in species other than human. Therefore, we did not use them by default. We provide the scripts of calculating them (e.g. 3 nt periodicity score with Ribo-seq data) as extra utilities in our COME software.
Evaluation of the input features
We divided the input features into three subsets: (i) sequence-derived features including GC content, DNA sequence conservation, protein conservation and RNA secondary structure conservation; (ii) expression features including expression abundance from poly(A)+, poly(A)- and small RNA sequencing; and (iii) histone features including H3K36me3 and H3K4me3 modification. The subset of sequence-derived features had the best performance among the three subsets. Still, use of all three sub-sets in combination (nine features in total) achieved the best performance (Supplementary Figure S8), and thus, we used the nine features from these three sub-sets as the default input feature set of COME.
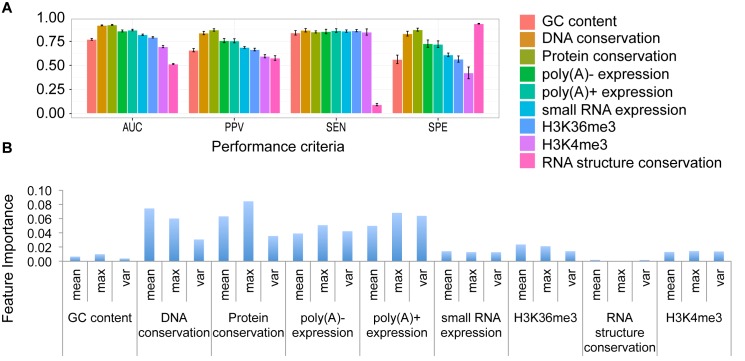
The performance of each single feature was further evaluated (Figure 2). We showed the performance with different criteria, such as PPV, sensitivity, AUC, etc. In addition, we also showed the feature importance using the decreased accuracy calculated by Random Forest. We found that, among the sequence-derived features, the protein conservation is the most powerful feature with the highest AUC score. The RNA secondary structure conservation features showed the highest specificity score, which meant most of the mRNAs had no conserved structures. We can also see this pattern later in Figure 5B.
Figure 2.
Performance of individual features of COME. We evaluate nine individual features using 5-fold cross-validation on the human training set. (A) Four criteria showing the prediction performance using individual features: area under the receiver operating characteristic curve (AUC), positive predictive value (PPV), sensitivity (SEN) and specificity (SPE). (B) Feature importance: mean decrease accuracy calculated by Random Forest.
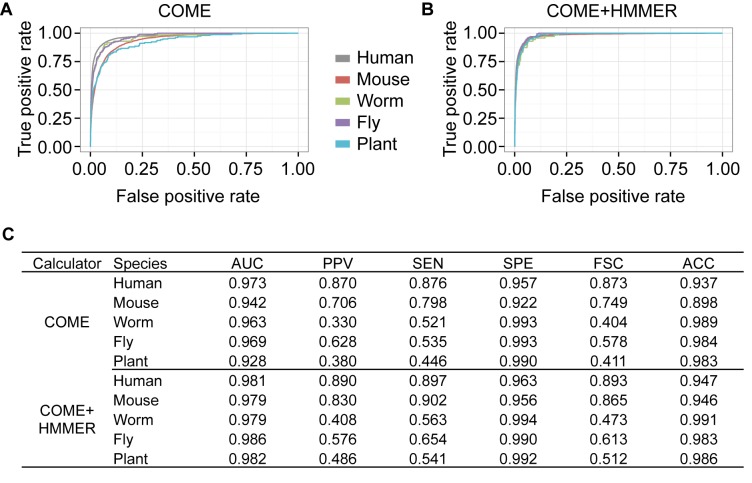
Figure 5.
COME's performances in multiple species. The performances of COME, as well as COME+HMMER, in five species: human, mouse, fly, worm and the model plant Arabidopsis. All the annotated lncRNA transcripts and mRNA transcripts from test set T1 were used for testing. We show (A and B) ROC curves and (C) six criteria: area under the receiver operating characteristic curve (AUC), PPV, sensitivity (SEN), specificity (SPE), accuracy (ACC) and F-score (FSC).
Evaluation of the experimental features’ robustness
Since COME used experimental features, including expression and histone modification profiles, which were subject to variation under different biological contexts; we evaluated the performance by replacing experimental data with different resources.
For histone modification features (i.e. H3K4me3 and H3K36me3), we recalculated the coding potential using two different datasets derived from various cell lines (35): (i) GM12878, K562, H1-hESC, HeLa-S3 and HepG21; (ii) HMEC, HSMM, HUVEC, NH-A and NHEK (Supplementary File 1). While the two histone datasets were different, the final prediction scores of COME were correlated well: the correlation of two sets varied a lot (correlation coefficient: 0.28–0.93) (Supplementary Figure S9a and b); if we only used these histone features to predict the coding potential, the results of the two datasets were also quite different (correlation coefficient: 0.36–0.51) (Supplementary Figure S9c, e and f); when combined with other features in COME, the final prediction turned out to be very robust no matter which dataset was used (correlation coefficient: 0.97–0.98) (Supplementary Figure S9d–g).
In addition to H3K36me3 and H3K4me3, we tested three extra histone makers, i.e. H3K27ac, H3K27me3 and H3K4me1, derived from same cell lines (35): GM12878, K562, H1-hESC, HeLa-S3 and HepG21 (Supplementary File 1). Compared to other three markers, H3K36me3 and H3K4me3 (the ones we used by default) showed top two AUC scores (Supplementary Figure S10). H3K36me3 and H3K4me3's combination also ranks the top one AUC score among the ten combinations. Furthermore, when combined with other features in COME, all histone combinations showed robust performance (Supplementary Figure S10).
For expression features, we also recalculated the coding potential using two different datasets derived from various cell lines (35): (i) GM12878, K562, H1-hESC, HeLa-S3 and HepG21; (ii) A549, AG04450, BJ, MCF-7 and NHEK (Supplementary File 1). The conclusion was the same as the one we got from different histone datasets: while the two expression datasets were different, the final prediction scores of COME were correlated well (correlation coefficient: 0.92–0.95) (Supplementary Figure S11).
We tested COME's performance on tissue specific lncRNAs (i.e. brain specific and testis specific lncRNAs). First of all, using seven human tissues’ expression data (36), we calculated the specificity score (3,25) for each lncRNA (at gene level). Then, top 10% specifically expressed lncRNAs were chosen, among which 2235 lncRNAs were testis specific and 204 lncRNAs were brain specific. As expected, these tissue specific lncRNAs were lowly expressed in the cell lines we used (Supplementary Figure S12a–d). Meanwhile, we sampled some coding genes that were also lowly expressed. Although the expression and histone features were not distinguishable between these lncRNAs and coding genes, the sequence features (e.g. conservation scores) showed very different patterns (Supplementary Figure S12c and d). Therefore, COME could still separate them well (AUCs: 0.993 for testis and 0.967 for brain).
In addition, since the subset of sequence features showed good performance (Supplementary Figure S8), we also provide an option in the COME software to calculate coding potential score without including experimental features, for users who do not need such supporting features.
RESULTS
COME is accurate and robust for different lncRNA annotation sets
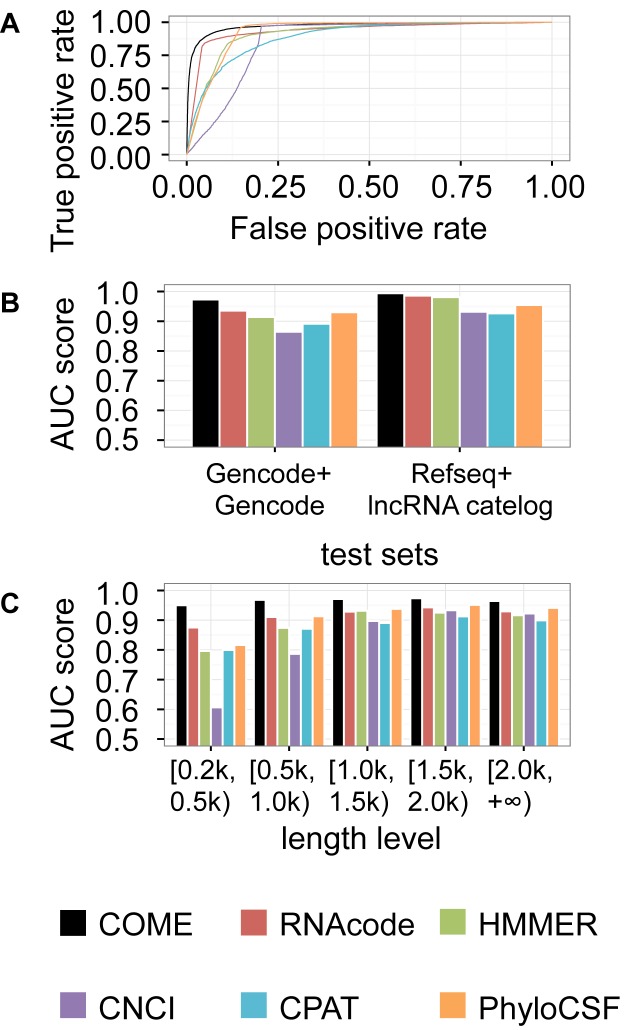
We first tested the ability of the predicted coding potential score to classify lncRNA and mRNA transcripts annotated by human Gencode (v19) (6) (described as T1 in ‘Materials and Methods’ section). We used the area under the receiver operating characteristic curve (AUC) to compare COME with five well-known coding potential calculators: CNCI (14), RNAcode (16), HMMER (37), CPAT (15) and PhyloCSF (12) (Figure 3A). Because more mRNA transcripts were annotated in Gencode, this test set (T1) was unbalanced, which would introduce bias in the performance evaluation. Therefore, we also sampled a balanced test set of T1, T1′, for the performance evaluation (Supplementary Figure S13). We found that COME performed better than the other five methods (CNCI, RNAcode, HMMER, CPAT and PhyloCSF) in both test sets.
Figure 3.
Performance comparison of COME and other coding potential calculators. We used the coding potential scores predicted by COME, CNCI, RNAcode, HMMER, CPAT or PhyloCSF to classify lncRNAs from protein coding transcripts in human data. (A) Receiver operating characteristic (ROC) curves of the six coding potential calculators for the T1 test set (mRNA annotation: Gencode; lncRNA annotation: Gencode). (B) AUC values for two test sets, T1 and T2 (mRNA annotation: Refseq; lncRNA annotation: human lincRNA catalog). (C) For the T1 test set, we also show the AUC values for transcripts of differing length.
Moreover, we used another test set (T2) annotated by a human lincRNA catalog (25) and Refseq (30), which were widely used by CNCI (14), CPAT (15) and PLEK (11). In total, 32% of the transcripts in T2 are 100% overlapped with T1's transcripts (Supplementary Figure S14). Please note that a transcript with 100% overlapping ratio does not have to be identical to the other one. One could be part of the other transcript that is longer. Actually, only 1999 transcripts are identical in both T1 (6.3%) and T2 (13.6%). Again, COME demonstrated the best performance (Figure 3B and Supplementary Figure S15). Comparing the performances between T1 and T2 sets, COME were more stable and robust than the other models. For instance, the AUC values for COME varied from 0.973 to 0.994, whereas those of the other tools showed larger performance variation (CNCI: 0.865–0.932, RNAcode: 0.936–0.986, HMMER: 0.914–0.981, CPAT: 0.891–0.926 and PhyloCSF: 0.930–0.955).
COME is accurate and robust for transcripts with different lengths
As mentioned earlier, some features (e.g. ORF length, k-mer features and blastx) used by various tools can be affected by transcript length. Therefore, we compared the performances on different sub-groups of transcripts with various lengths: 0.2–0.5 kilobases (kb), 0.5–1.0 kb, 1.0–1.5 kb, 1.5–2.0 kb and >2.0 kb (Figure 3C and Supplementary Figure S16). For RNAs of different lengths, the AUC values for CNCI, RNAcode, HMMER, CPAT and PhyloCSF varied from 0.607–0.934, 0.875–0.943, 0.796–0.925, 0.799–0.913 and 0.816–0.951, respectively. All the coding potential calculators showed better performance for RNAs with 1–2 kb bases. COME achieved the best robustness for transcripts of various lengths, with AUC values ranging from 0.950–0.974. In addition to the five methods we have compared with, we also compared COME with more tools (i.e. CPC and PLEK) using a smaller test set (Supplementary Figure S17), and still, COME showed the best accuracy (overall AUC: 0.965) as well as robustness (AUC range: 0.936–0.975).
COME improves the consistency of different coding potential calculators’ prediction results
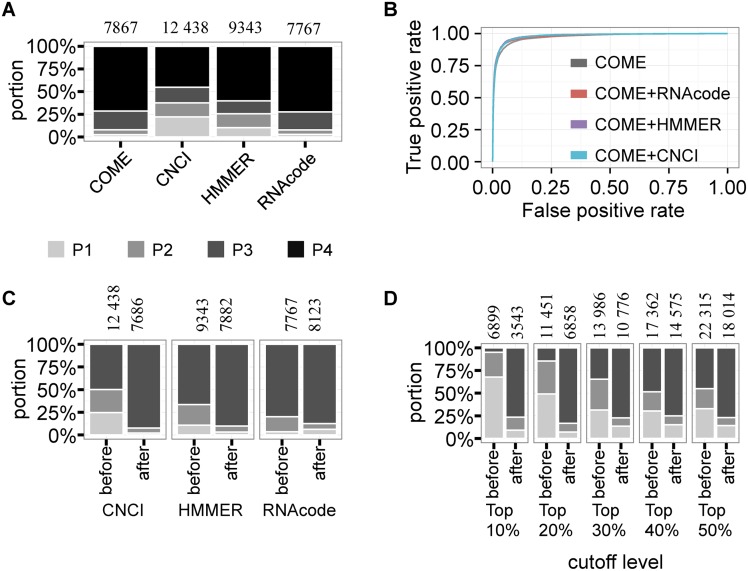
One practical issue of filtering novel lncRNA transcripts from RNA-seq data is that the filtered lncRNA sets were not consistent when using different coding potential calculators. We tested the overlapping results predicted by COME, CNCI, RNAcode and HMMER on the first test set (T1). A transcript was predicted to be a lncRNA if the coding potential score was lower than a certain cutoff, which was defined as the coding potential score that had the highest F-score in the test set. For each coding potential tool, we counted the number of predicted lncRNAs that were predicted by 1–4 tools (Figure 4A). Remarkably, COME and RNAcode had greater overlap with the other three tools (71.4 and 72.3%, respectively) and predicted relatively fewer lncRNAs (7867 and 7767 transcripts, respectively).
Figure 4.
COME enhances the consistency of other coding potential calculators. We compared the prediction overlap of four coding potential calculators: COME, CNCI, RNAcode and HMMER. (A) For T1 test set, we defined a transcript as a predicted lncRNA if its coding potential was lower than a cutoff. The cutoff was determined by the highest F-score for each method. Then, we counted the predicted lncRNA numbers according to the cutoff (shown on top of the bar). For each calculator, we also calculated the percentage of lncRNAs that were predicted by only one calculator (P1), by two calculators (P2), by three calculators (P3) and by four calculators (P4). The annotated protein coding transcripts (mRNAs) were used as negatives in this plot. (B) Receiver operating characteristic (ROC) curves of three coding potential calculators after enhancement by COME (COME+CNCI, COME+RNAcode and COME+HMMER). We also show the numbers of lncRNAs consistently predicted by multiple methods among the three coding potential calculators, before and after enhancement with COME, based on the cutoffs determined by the highest F-scores (C) and different percentiles (D).
Because COME is able to include multiple features as input, we tried to add the coding potential score of another calculator (i.e. CNCI, RNAcode, HMMER) to COME's input features. Using the same training set as above (i.e. 15 638 lncRNA transcripts and 47 490 mRNA transcripts) and the same test set T1, we trained models for the enhanced coding potential calculators: COME+CNCI, COME+RNAcode and COME+HMMER. We calculated and tested three enhanced coding potential scores using combinations of COME+CNCI, COME+RNAcode and COME+HMMER, respectively (Figure 4B and Supplementary Figure S18). A transcript was predicted to be a lncRNA if the enhanced coding potential score was lower than a certain cutoff, which was defined as the score that had the highest F-score in the test set. After the enhancement, all three enhanced coding potential scores showed higher AUC values compared to the original values: 0.936–0.978, 0.915–0.981 and 0.865–0.980 for RNAcode, HMMER and CNCI, respectively. In addition, the ROC curves and AUC values for the three enhanced coding potential scores were comparable to those of COME (0.973).
Moreover, using test set T1, we compared the prediction overlap of CNCI, RNAcode and HMMER before and after enhancement with COME. Similarly, we predicted that a transcript was a lncRNA if the coding potential score was lower than a certain cutoff, which was defined as the coding potential score that had the highest F1-score in the test set. We showed that after enhancement, all three tested coding potential scores showed greater overlap with other tools (Figure 4C). The consistency improvement remained when we used different cutoffs (i.e. top 10–50 percentile ascending ordered coding potential score; Figure 4D). Therefore, COME could not only predict coding potential with high accuracy and robustness, but also could enhance the prediction consistency of other calculators.
COME can be extended to multiple organisms
In addition to human data, we also applied COME to four other model species: M. musculus, C. elegans, D. melanogaster and Arabidopsis thaliana. We compared COME's performance for the five species on the original test set (Figure 5A) and balanced test set (Supplementary Figure S19). COME performed very well for all five species, with AUC values ranging from 0.928 (for Arabidopsis) to 0.973 (for human). We also built the enhanced models of COME+HMMER and tested them on the same test sets (Figure 5B). In summary, COME+HMMER performed even better for all five species, with AUC values ranging from 0.979 (for worm) to 0.986 (for fly). We summarized the performances of COME and COME+HMMER in the five species in Figure 5C. All of the above results demonstrate that COME can be useful for novel lncRNA identification and characterization in new samples of five model organisms.
COME characterizes lncRNAs with multiple supporting features
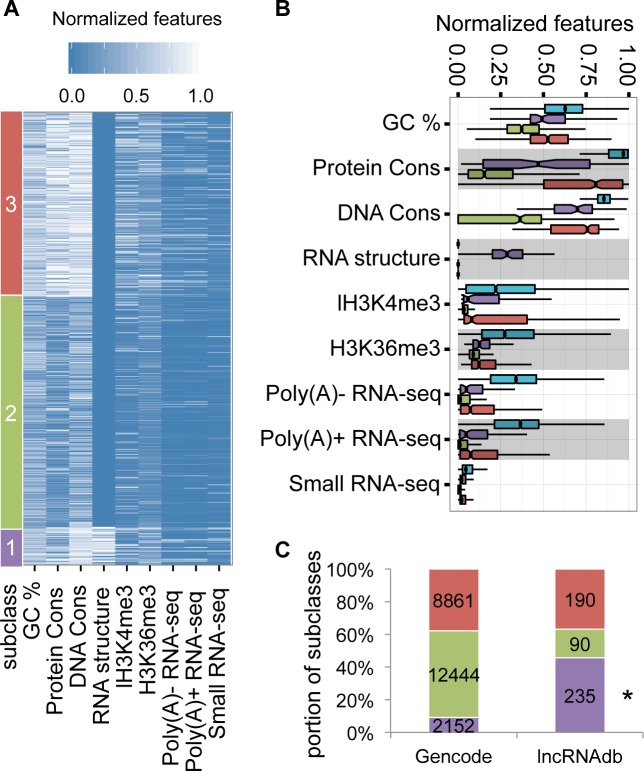
In addition to a single coding potential score for a given transcript, another advantage of COME is that it can also output multiple supporting feature scores to characterize each predicted lncRNA transcript. As an example, we clustered the human lncRNA transcripts (Gencode v19) based on COME's nine supporting features (GC content, DNA sequence conservation, protein conservation and RNA secondary structures, expression abundance from poly(A)+, poly(A)- and small RNA sequencing, H3K36me3 and H3K4me3), using a K-means algorithm (Figure 6A). The peak signal of each lncRNA transcript for each feature (maximum value of the extracted feature vector) was scaled into [0, 1]. The optimal cluster number was determined by the silhouette score (Supplementary Figure S20). Finally, we defined three lncRNA subclasses. We further used a boxplot to illustrate the feature difference among the three clusters (Figure 6B): subclass 1 showed relatively more conserved pattern in sequence features, i.e. GC content, DNA sequence conservation, protein sequence conservation and RNA secondary structures. The lncRNAs in it were also relatively highly expressed (i.e. higher values of poly(A)+, poly(A)- and small RNA sequencing data); and had relatively higher signals of H3K36me3 and H3K4me3. Subclass 2 was neither conserved in sequence features nor highly expressed. Subclass 3 is similar to subclass 1, except that the lncRNAs in it contained no conserved RNA secondary structures. As expected, when compared with mRNAs, all the three subclasses of lncRNAs showed lower values for GC content, DNA conservation, protein conservation, expression level and histone modification level (Figure 6B). As we expected, tissue specific lncRNAs were significantly enriched in subclass2 (chi-square test, P-value < 0.01), which were lowly expressed and unconserved (Supplementary Figure S12e).
Figure 6.
COME annotates lncRNAs with multiple supporting features. (A) We used the normalized features (x-axis) to cluster (k-means) the annotated human lncRNA transcripts (Gencode V19). Three subclasses were clustered according to different feature patterns. The RNA structure conservation feature was presented by the—log(E-value of Rfam hit). (B) Boxplot of feature difference among three subclasses. For better visualization, the values were normalized from 0 to 1. We included mRNAs (blue) as control. (C) Transcript numbers are shown for lncRNAs from Gencode and lncRNAdb. The small set of lncRNAs in lncRNAdb was mostly curated from experiment literatures. Star beside the subclass label indicates that the subclass is significantly enriched in lncRNAdb (chi-square test, P-value < 0.01).
Remarkably, we found that the subclasses had very different chances of being validated (Figure 6C). Among the above clustered Gencode lncRNAs, 515 transcripts (108 genes) were annotated in lncRNAdb (38), which included many lncRNAs that were supported by better function experiments. Remarkably, we found that the lncRNAs from subclass 1 were significantly enriched (chi-square test, P-value < 0.01) in the validated set of lncRNAdb. These lncRNAs were highly expressed and conserved. More importantly, almost all of them contained conserved RNA secondary structures. Because Rfam also includes many lncRNA structure families, we further excluded these lncRNA families and counted those lncRNAs only containing canonical RNA structures, such as tRNA, SRP, pre-miRNA, etc. Interestingly, lncRNAs containing local structures that were conserved with canonical ncRNA structure families were still significantly enriched in lncRNAdb (Supplementary Figure S21). These conclusions remained when evaluating the result at gene level (Supplementary Figure S21).
Overall, the supporting features of COME can improve the chances of finding functional lncRNAs. However, this enrichment could also be biased by the pre-selection criteria. For instance, some lncRNAs were selected as validation candidates by previous studies simply due to their abundant expression. Still, some interesting features, such as RNA secondary structure, can be used to inspire hypotheses for further mechanism studies.
Conserved structural domains of lncRNAs were associated with RNA binding proteins
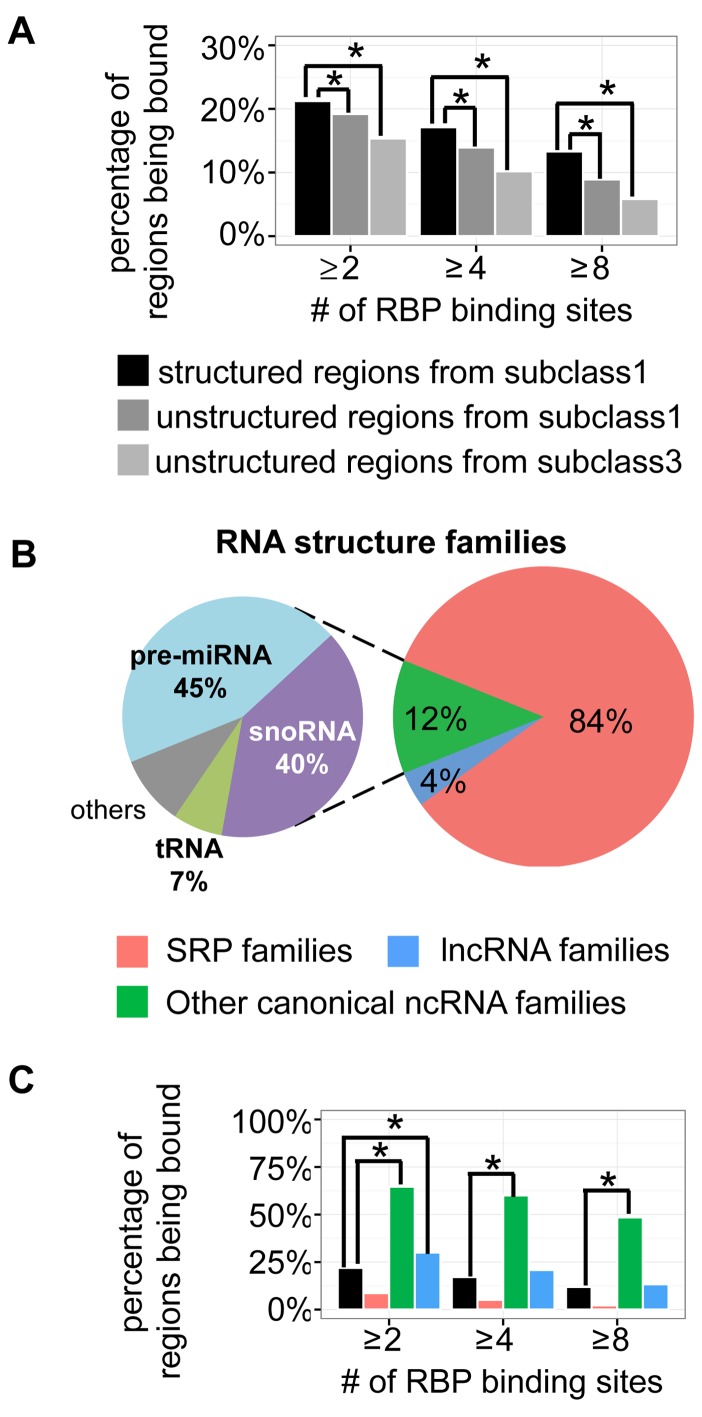
We further associated the conserved structural domains of lncRNAs with RBPs. We first collected the binding sites of 151 RBPs from ENCODE's eCLIP-seq data (39) (Supplementary File 1). Next, we overlapped the binding sites with all lncRNAs and compared the RBP binding between structured domains (bins) of lncRNAs in subclass 1 and unstructured lncRNA domains. Since the RBP binding could be biased by the expression level of investigated RNAs, we used two negative controls, (i) unstructured lncRNA domains (bins) of lncRNAs in subclass 3, which had similar expression levels to subclass 1 (Figure 6B) (P-value of Wilcox rank sum test > 0.05 using poly(A)+ RNA-seq data); (ii) unstructured domains from the same lncRNA transcripts in subclass 1. We defined a lncRNA domain as being bound by RBP if at least two binding sites were found on it. We found the percentage of being bound were significant higher for the structured lncRNA domains than controls (Figure 7A) (chi-square test, P-value < 0.01). The difference remained significant when we investigate the lncRNA domains having at least 4 and 8 RBP binding sites. Furthermore, we still observed significant enrichment when we counting RBP types (Supplementary Figure S22), instead of RBP binding sites. The association of lncRNAs’ local structures and RBPs’ binding suggested the potential regulatory roles of these RNA regions.
Figure 7.
Conserved structural domains of lncRNAs were associated with RBP binding sites. (A) The percentages of being bound by RBPs for the structured regions of lncRNAs in subclass 1, the unstructured regions of the same lncRNA transcripts in subclass 1 and the unstructured regions of lncRNAs in subclass 3. A region was counted as being bound by RBP when there are at least 2, 4 or 8 RBP binding sites on it. (B) Structure families (matched in Rfam) of the local RNA secondary structures on the lncRNAs in subclass 1. (C) RBP binding preferences of three structure families, compared with all the structured regions of lncRNAs in subclass 1.
Next, we investigated the RBP binding with different RNA structure families. By matching with Rfam families (40), we found that the conserved local structures of lncRNAs in subclass 3 mainly belonged to three structure families: SRP, other canonical ncRNA structures (e.g. snoRNA, tRNA, pre-miRNA, etc) and lncRNA family (Figure 7B). Interestingly, we found most (84%) of the local structures were SRPs. However, the RBP binding was highly enriched in the other canonical ncRNA structures (Figure 7C and Supplementary Figure S23) (chi-square test, P-value < 0.01). This enrichment might be biased by the RBP types we collected: the 151 RBPs were mainly splicing factors and UTR processing factors. This small set did not represent various regulatory roles of over 1500 RBPs in human (41), which might be associated with SRP-like structures on lncRNAs.
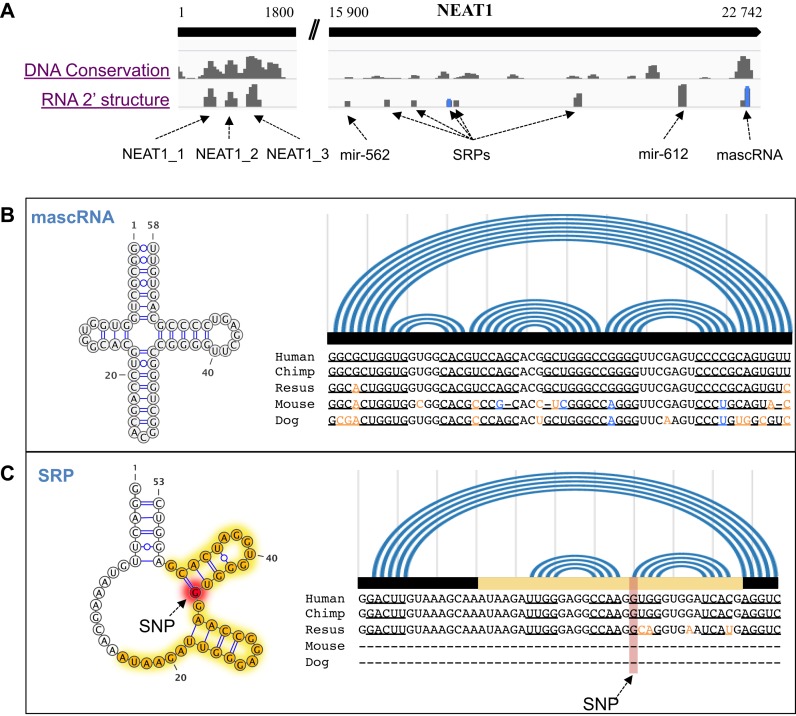
Finally, we used a well-known lncRNA, NEAT1 (nuclear enriched abundant transcript 1, i.e. MEN beta) (42–44), to illustrate the conserved local structures on a lncRNA (Figure 8A). The 5′ end of NEAT1 contains three structural domains belonged to lncRNA families (named as NEAT1 in Rfam). The 3′ end contains many conserved canonical ncRNA structures, such as SRPs, pre-miRNAs and a mascRNA. For instance, mascRNA is a conserved tRNA-like RNA (Figure 8B) processed from NEAT1 transcript, which also exists in MALAT1 (44). What's more, we observed a SRP-like structure on NEAT1 (Figure 8C). Although conserved in RNA secondary structure, the SRP-like region was less conserved at sequence level. Remarkably, we found the SRP-like structure had a G to A mutation, which was identified in melanoma (45). The G was originally paired with C in a conserved stem-loop. Moreover, based on the eCLIP-seq data, we also found this stem-loop was bound by many RBPs, such as SLBP, PTARDBP, CPSF7, CSTF2T, EIF4A3, HNRNPA1, HNRNPM, HNRNPU, NUDT21, PTBP and U2AF2. All the above evidence suggested potential regulatory roles of the structural domains of lncRNA, NEAT1.
Figure 8.
Examples of structural domains on a lncRNA, NEAT1. (A) Conserved structural domains on NEAT1. DNA conservation score is the phastCons score. RNA conservation score is the—log(E-value of Rfam hit). RNA secondary structures and sequence alignments of mascRNA (B) and a SRP-like RNA (C) are shown in detail. In the alignments, the co-variant nucleotides are shown in blue; and the structure-disrupting variances are shown in orange. The RBP binding sites are labeled as in yellow; and a SNP (G to A) site is labeled in red.
DISCUSSION
COME is a coding potential tool to utilize features derived from multiple sources and levels: it not only includes the sequence features (e.g. GC content and RNA structure conservation), but also supports the use of other experimental data. Based on multiple features and the decompose–compose method, COME is able to calculate coding potential with high accuracy, robustness and consistency. In addition, COME can annotate known and novel lncRNAs with various supporting features, which could help researchers to generate hypotheses for further functional and mechanistic studies (21).
A more practical application of a coding potential calculator is to evaluate the newly assembled transcriptome. Many of the assembled transcripts were actually transcribed fragments (transfrags), because the full-length version is difficult to assemble from RNA-seq's short reads. However, some features (e.g. ORF length) that could be affected by the transcript's assembly quality have been used by many coding potential calculators. The decompose–compose strategy and the multiple complimentary features would help COME to avoid some influences of transcript length and assembly quality. Therefore, COME has the potential to be a more practical tool. To test the performances of the various calculators for transfrags assembled de novo from RNA-seq data, we only used the re-assembled transfrags with matching levels of 50–100%, according to the reference transcripts. Transcripts with high matching levels were more likely to be known transcripts poorly assembled, rather than new isoforms. A total of 147 512 assembled transfrags were more than 50% matched in length with known transcripts [Gencode v19 (6)], and these were used as the test set (T3). The ones below 50% were more likely to be novel transcripts, and thus, we did not use them for testing. In the comparisons, we grouped the transfrags by various degrees of matching: 50–90% matched, 90–95% matched, 95–100% matched and 100% matched (Supplementary Figure S24). COME showed the best accuracy and robustness for transcripts of differing assembly quality, with AUC values varying from 0.968–0.993. However, please note that many unmatched transfrags (e.g. those had <90% similarity to the known ones) were potential novel isoforms, while others were probably poorly assembled known transcripts (e.g. those had >99% similarity to the known ones). Currently, there is no gold-standard set helping us distinguish these two cases.
We further compared the coding potential calculators' predictive power for the intergenic and genic lncRNAs. The AUC scores of all calculators were decreased to ∼80% for the genic lncRNAs, where COME still performed the best (Supplementary Figure S25).
COME's calculation is based on indexed genome and feature vectors, and therefore, it runs fast for large sets of transcripts (Supplementary Table S1). We provide downloadable data matrix and pre-trained models, along with the software implementation (https://github.com/lulab/COME).
Because COME and many other coding potential calculators require a training set for the supervised models (Supplementary Table S2), and the test set we used could be predicted by some previous coding potential calculators, the performance comparison may be unfair to other calculators and COME.
In addition, the variability in annotation quality would affect the test performance. For instance, many lncRNAs from Gencode are lacking correct transcription start site annotation. This would affect many prediction tools that require full-length transcripts. COME would be affected as well if the training set were not correctly annotated. On the other hand, as demonstrated above (Supplementary Figure S24), the decompose-compose strategy and the multiple complimentary features would help COME to avoid some influences of incorrect transcription start site annotation.
Because the current annotations are not perfect, sometimes the discrepancy between the annotation and computational prediction would provide hint to novel findings. For example, Anderson et al reported a functional small peptide encoded by an annotated lncRNA, which is conserved in both human and mouse (46). We tested this specific case and found that COME, as well as some other tools, was able to predict the ‘annotated lncRNA’ as a coding transcript (Supplementary Table S3).
Moreover, the models underlying COME include many features derived from various resources that require some pre-processing efforts. Although COME should be useful for novel lncRNA identification and characterization in new samples (e.g. cancer samples) of five model organisms, its extension to more species will require additional work. Therefore, we plan to continually update COME as annotation improves, curate more features and add more model organisms in the future.
Acknowledgments
We thank the authors of CPC and CNCI for helping us calculate the coding potential scores with their tool/server. We thank Y. Wei, T. Liu, Y. Li and B. Shi for assisting with data download, software development and some testing analyses. We thank YT. Yang for revising the manuscript and providing human eCLIP-seq data.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Plan of China [2016YFA0500803]; National High-Tech Research and Development Program of China [2014AA021103]; National Natural Science Foundation of China [31271402, 31522030]; Tsinghua University Initiative Scientific Research Program [2014z21045]; Computing Platform of the National Protein Facilities (Tsinghua University); Funding for open access charge: National Natural Science Foundation of China [31271402].
Conflict of interest statement. None declared.
REFERENCES
- 1.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G., et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di C., Yuan J., Wu Y., Li J., Lin H., Hu L., Zhang T., Qi Y., Gerstein M.B., Guo Y., et al. Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J. 2014;80:848–861. doi: 10.1111/tpj.12679. [DOI] [PubMed] [Google Scholar]
- 4.Gerstein M.B., Rozowsky J., Yan K.-K., Wang D., Cheng C., Brown J.B., Davis C.A., Hillier L., Sisu C., Li J.J., et al. Comparative analysis of the transcriptome across distant species. Nature. 2014;512:445–448. doi: 10.1038/nature13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y., Barrette T.R., Prensner J.R., Evans J.R., Zhao S., et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S., et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Achawanantakun R., Chen J., Sun Y., Zhang Y. LncRNA-ID: long non-coding RNA IDentification using balanced random forests. Bioinformatics. 2015;31:3897–3905. doi: 10.1093/bioinformatics/btv480. [DOI] [PubMed] [Google Scholar]
- 8.Biswas A.K., Zhang B., Wu X., Gao J.X. CNCTDiscriminator: coding and noncoding transcript discriminator - an excursion through hypothesis learning and ensemble learning approaches. J. Bioinform. Comput. Biol. 2013;11:1342002–1342017. doi: 10.1142/S021972001342002X. [DOI] [PubMed] [Google Scholar]
- 9.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong L., Zhang Y., Ye Z.Q., Liu X.Q., Zhao S.Q., Wei L., Gao G. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li A., Zhang J., Zhou Z. PLEK: a tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinformatics. 2014;15:311–320. doi: 10.1186/1471-2105-15-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin M.F., Jungreis I., Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275–i282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J., Gough J., Rost B. Distinguishing protein-coding from non-coding RNAs through support vector machines. PLoS Genet. 2006;2:e29. doi: 10.1371/journal.pgen.0020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun L., Luo H., Bu D., Zhao G., Yu K., Zhang C., Liu Y., Chen R., Zhao Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013;41:e166. doi: 10.1093/nar/gkt646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L., Park H.J., Dasari S., Wang S., Kocher J.P., Li W. CPAT: coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41:e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Washietl S., Findeiss S., Muller S.A., Kalkhof S., von Bergen M., Hofacker I.L., Stadler P.F., Goldman N. RNAcode: robust discrimination of coding and noncoding regions in comparative sequence data. RNA. 2011;17:578–594. doi: 10.1261/rna.2536111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frohman M.A., Dush M.K., Martin G.R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. U.S.A. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hashimoto K., Suzuki A.M., Dos Santos A., Desterke C., Collino A., Ghisletti S., Braun E., Bonetti A., Fort A., Qin X.Y., et al. CAGE profiling of ncRNAs in hepatocellular carcinoma reveals widespread activation of retroviral LTR promoters in virus-induced tumors. Genome Res. 2015;25:1812–1824. doi: 10.1101/gr.191031.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shiraki T., Kondo S., Katayama S., Waki K., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu Z.J., Yip K.Y., Wang G., Shou C., Hillier L.W., Khurana E., Agarwal A., Auerbach R., Rozowsky J., Cheng C., et al. Prediction and characterization of noncoding RNAs in C. elegans by integrating conservation, secondary structure, and high-throughput sequencing and array data. Genome Res. 2011;21:276–285. doi: 10.1101/gr.110189.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lv J., Liu H., Huang Z., Su J., He H., Xiu Y., Zhang Y., Wu Q. Long non-coding RNA identification over mouse brain development by integrative modeling of chromatin and genomic features. Nucleic Acids Res. 2013;41:10044–10061. doi: 10.1093/nar/gkt818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramos A.D., Diaz A., Nellore A., Delgado R.N., Park K.Y., Gonzales-Roybal G., Oldham M.C., Song J.S., Lim D.A. Integration of genome-wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo. Cell Stem Cell. 2013;12:616–628. doi: 10.1016/j.stem.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerstein M.B., Lu Z.J., Van Nostrand E.L., Cheng C., Arshinoff B.I., Liu T., Yip K.Y., Robilotto R., Rechtsteiner A., Ikegami K., et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu Z.J., Yip K.Y., Wang G., Shou C., Hillier L.W., Khurana E., Agarwal A., Auerbach R., Rozowsky J., Cheng C., et al. Prediction and characterization of noncoding RNAs in C. elegans by integrating conservation, secondary structure, and high-throughput sequencing and array data. Genome Res. 2011;21:276–285. doi: 10.1101/gr.110189.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., Rinn J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guttman M., Russell P., Ingolia N.T., Weissman J.S., Lander E.S. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu L., Di C., Kai M., Yang Y.C., Li Y., Qiu Y., Hu X., Yip K.Y., Zhang M.Q., Lu Z.J. A common set of distinct features that characterize noncoding RNAs across multiple species. Nucleic Acids Res. 2015;43:104–114. doi: 10.1093/nar/gku1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giannopoulou E.G., Elemento O. Inferring chromatin-bound protein complexes from genome-wide binding assays. Genome Res. 2013;23:1295–1306. doi: 10.1101/gr.149419.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pruitt K.D., Tatusova T., Maglott D.R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vasquez J.J., Hon C.C., Vanselow J.T., Schlosser A., Siegel T.N. Comparative ribosome profiling reveals extensive translational complexity in different Trypanosoma brucei life cycle stages. Nucleic Acids Res. 2014;42:3623–3637. doi: 10.1093/nar/gkt1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bazzini A.A., Johnstone T.G., Christiano R., Mackowiak S.D., Obermayer B., Fleming E.S., Vejnar C.E., Lee M.T., Rajewsky N., Walther T.C., et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014;33:981–993. doi: 10.1002/embj.201488411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calviello L., Mukherjee N., Wyler E., Zauber H., Hirsekorn A., Selbach M., Landthaler M., Obermayer B., Ohler U. Detecting actively translated open reading frames in ribosome profiling data. Nat. Methods. 2016;13:165–170. doi: 10.1038/nmeth.3688. [DOI] [PubMed] [Google Scholar]
- 35.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Necsulea A., Soumillon M., Warnefors M., Liechti A., Daish T., Zeller U., Baker J.C., Grutzner F., Kaessmann H. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505:635–640. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 37.Finn R.D., Clements J., Eddy S.R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quek X.C., Thomson D.W., Maag J.L., Bartonicek N., Signal B., Clark M.B., Gloss B.S., Dinger M.E. lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015;43:D168–D173. doi: 10.1093/nar/gku988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Nostrand E.L., Pratt G.A., Shishkin A.A., Gelboin-Burkhart C., Fang M.Y., Sundararaman B., Blue S.M., Nguyen T.B., Surka C., Elkins K., et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nat. Methods. 2016;13:508–514. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nawrocki E.P., Eddy S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerstberger S., Hafner M., Tuschl T. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guru S.C., Agarwal S.K., Manickam P., Olufemi S.E., Crabtree J.S., Weisemann J.M., Kester M.B., Kim Y.S., Wang Y., Emmert-Buck M.R., et al. A transcript map for the 2.8-Mb region containing the multiple endocrine neoplasia type 1 locus. Genome Res. 1997;7:725–735. doi: 10.1101/gr.7.7.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hutchinson J.N., Ensminger A.W., Clemson C.M., Lynch C.R., Lawrence J.B., Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39–54. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sunwoo H., Dinger M.E., Wilusz J.E., Amaral P.P., Mattick J.S., Spector D.L. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Borresen-Dale A.L., et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson D.M., Anderson K.M., Chang C.L., Makarewich C.A., Nelson B.R., McAnally J.R., Kasaragod P., Shelton J.M., Liou J., Bassel-Duby R., et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]