Abstract
We describe a Pap1–Oxs1 pathway for diamide-induced disulfide stress in Schizosaccharomyces pombe, where the nucleocytoplasmic HMG protein Oxs1 acts cooperatively with Pap1 to regulate transcription. Oxs1 and Pap1 form a complex when cells are exposed to diamide or Cd that causes disulfide stress. When examined for promoters up-regulated by diamide, effective Pap1 binding to these targets requires Oxs1, and vice versa. With some genes, each protein alone enhances transcription, but the presence of both exerts an additive positive effect. In other genes, although transcription is induced by diamide, Oxs1 or Pap1 plays a negative role with full de-repression requiring loss of both proteins. In a third class of genes, Oxs1 positively regulates expression, but in its absence, Pap1 plays a negative role. The Oxs1–Pap1 regulatory interaction appears evolutionarily conserved, as heterologous (human, mouse and Arabidopsis) Oxs1 and Pap1-homologues can bind interchangeably with each other in vitro, and at least in the fission yeast, heterologous Oxs1 and Pap1-homologues can substitute for S. pombe Oxs1 and Pap1 to enhance stress tolerance.
INTRODUCTION
In oxidative stress, different oxidants can elicit different responses and the damages caused can be distinct (1). Whereas H2O2 oxidizes a thiol to sulfenic (SOH), then to sulfinic (SO2H) or sulfonic (SO3H) acid, as well as promoting disulfide formation between a SOH and a thiol (2), the reactive electrophilic species diamide specifically triggers disulfides between thiols (3), producing a subcategory of oxidative stress that is also referred to as disulfide stress (4).
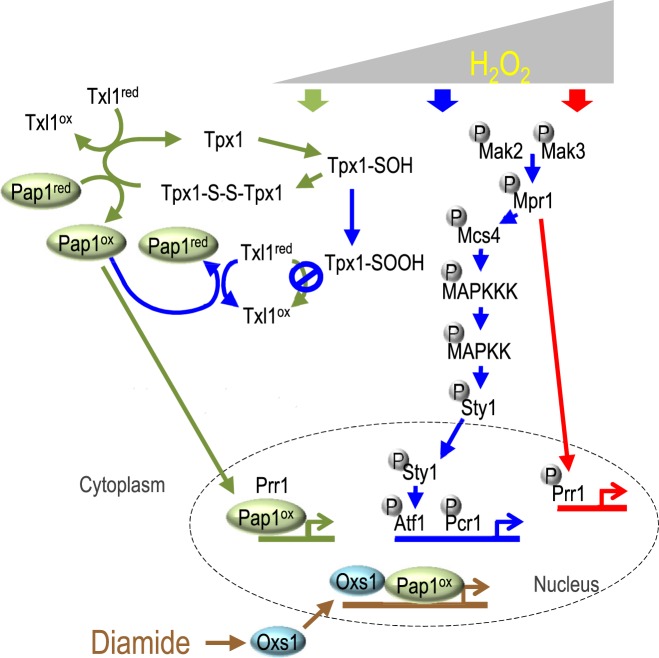
In brief, three signaling pathways respond to H2O2 in the fission yeast Schizosaccharomyces pombe (5,6) (Figure 1). At low H2O2 concentration, thioredoxin peroxidase Tpx1 is oxidized to its disulfide form, which leads to oxidization of the proteasome-associated thioredoxin-like protein Txl1, as well as Pap1 (7,8). Oxidized Pap1 relocates from the cytoplasm to the nucleus as it bypasses nuclear export by Crm1 (Exportin1), forms a heterodimer with transcription factor Prr1, and turns on the expression of target genes (9,10). At a higher H2O2 level, hyper-oxidized Tpx1 fails to oxidize Txl1, permitting reduced Txl1 to reduce Pap1 and turn off the early stress response (7,11). The Sty1–Atf1 pathway takes over with a phosphorelay system comprising of histidine kinase sensors Mak2 and Mak3 that passes the signal to the histidine-containing phosphorelay protein Mpr1, then to response regulator Mcs4, and finally through the MAPK cascade to the MAPK Sty1 (12). Sty1 then phosphorylates the basic region/leucine zipper motif (bZIP) transcription factor Atf1, which dimerizes with another bZIP protein Pcr1 to activate downstream targets (13,14). At high levels of H2O2, the phosphorylated Mpr1 is thought to activate Prr1 directly, which then activates genes responding to acute oxidative stress (15).
Figure 1.
Model of transcriptional response to H2O2 or diamide (and Cd) and the stress induced nuclear localization of Oxs1. Low, medium and high concentrations of H2O2 induce pathways shown by green, blue and red arrows, respectively. Diamide or Cd induces the Pap1–Oxs1 pathway (brown arrows). Oxidized or reduced state indicated by superscript ox or red. Thiol, sulfenic, sulfinic and disulfide shown as SH, SOH, SOOH and S-S, respectively. Circled P: phosphorylated residue.
In contrast to the wealth of knowledge on H2O2 stress, less is known of the response to diamide. Previous studies have identified Pap1, as well as its budding yeast homologue Yap1, responding to diamide or various cytotoxic drugs and heavy metals by translocating from the cytoplasm to the nucleus to activate a stress response (9,16). In this paper, we describe Oxs1, a new player that interacts with Pap1 at the target promoters in a diamide or Cd-dependent manner. Moreover, heterologous Oxs1-like proteins can also enhance diamide stress tolerance in the fission yeast. In vitro, S. pombe, human and Arabidopsis Oxs1 can interchangeably bind Pap1 or Pap1 homologues from human (cJun) and Arabidopsis (bZIP10), suggesting that Oxs1 may be a component of an evolutionarily-conserved stress response pathway.
MATERIALS AND METHODS
Genetic materials
S. pombe strains include JS23 (WT, h+ ura4.294 leu1.32); TP108-3C (h− leu1 his22 ura4 pap1::ura4+) (17); NT224 (h– leu1 ura4 sty1-1) (18); JM1066 (h+ leu1 atf1::ura4+) (14); JX26 (H90 ade6 leu1 ura4 pcr1::ura4+) (14); and the JS23-derived strains created through homologous gene disruption (Supplementary Figure S1): oxs1Δ; pap1Δ; pap1Δoxs1Δ; FLAG-oxs1; HA-pap1; FLAG-oxs1 HA-pap1; HA-pap1 oxs1Δ; and FLAG-oxs1 pap1Δ. The cDNAs were inserted into pART1 or pSLF173, expressed from the adh1 or nmt1 promoter, respectively; further details in Supplementary Materials and Methods. Genetic manipulations performed according to the Fission Yeast Handbook (http://www.biotwiki.org/foswiki/bin/view/Pombe/NurseLabManual), using YES (rich) or EMM (minimum) growth media.
RNA-Seq and qRT-PCR
Cells grown to OD600 0.3 were treated with 1.0 mM diamide for 0, 20, 60, 90 and 120 min prior to harvest of total RNA (RNeasy Mini Kit, Cat# 74104, Qiagen). RNA-seq library construction and sequencing were carried out by BGI-Tech (Shenzhen, China); expression level calculated as RPKM was compared between diamide treated samples and untreated samples. For qRT-PCR, cells grown to OD600 0.3 were treated with 1.5 mM diamide for 0–5 h before harvesting total RNA. Reverse transcription was conducted using PrimeScript™ RT reagent Kit (Cat# RR047A, TaKaRa); qRT-PCR with SYBR Premix Ex Taq™ Mix (Cat# DRR820A, TaKaRa) on LightCycler®480 II (Roche). Relative expression level normalized to act1+ (SPBC32H8.12c) of WT cells at 0 h.
In vitro GST-pull down assay
Coding regions of oxs1+ or pap1+ were inserted in-frame into pGEX-4T-1 (Cat# 28954549 GE Healthcare Lifesciences) to generate GST fusion proteins or into pET-21d (Cat# 69743-3, Novagen) to generate His fusion proteins. In vitro GST pull-down assays were as follows: Cell lysates from Escherichia coli BL21 (DE3) pLysS (Cat# BL21C-24, GE) expressing GST fusion proteins were incubated with MagneGST™ beads (Cat# V8611, Promega) at 4°C for 4 h, followed by four washes with buffer (25 mM Tris–HCl pH 7.2, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 0.5% NonidetP-40, 1 mM DTT). Beads bound with GST fusion proteins were incubated overnight at 4°C with cell lysates from E. coli BL21 expressing His fusion proteins, followed by four washes with buffer. Immunoprecipitates were subjected to western blotting (see Supplementary Materials and Methods).
Co-immunoprecipitation analysis
Cells grown to OD600 0.5 were collected and lysed in lysis buffer (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.1% NonidetP-40, 0.1% SDS, 12 mM sodium deoxycholate) with glass beads in a bead beater (Fastprep-24 MP Biomedical). Lysate was centrifuged at 12,000 rpm for 15 min at 4°C. Protein extract was incubated with 5 μg polyclonal anti-HA antibody (Cat# ab9110, Abcam) overnight at 4°C, then a 25 μl mixture (1:1) of Dynabeads® Protein A:Protein G (Cat# 10002D, Life Technologies AS) was added and incubated at 4°C for 4 h. After washing the beads 6x with buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.1% NonidetP-40, 5% glycerol pH 7.4), the immunoprecipitates were eluted by boiling in SDS-PAGE loading buffer for western blotting using Monoclonal anti-HA antibody (Cat# ab16918, Abcam) and anti-FLAG antibody (Cat# F1804, Sigma).
Chromatin immunoprecipitation
Cells were cross-linked by 1% formaldehyde for 20 min at 30°C, stopped with 125 mM glycine for 5 min, pelleted, washed twice with PBS (137 mM sodium chloride, 2.7 mM potassium chloride, and 11.9 mM phosphate buffer, pH 7.4), frozen immediately in liquid nitrogen and stored at -80°C. Nucleic acid extraction by enzymatic lysis was conducted according to the Fission Yeast Handbook. Chromatin were released and sheared to an average size of 500 bp using M220 sonicator (Covaris). The immunoprecipitation and DNA recovery procedures were as described (19) (Supplementary Materials and Methods). The immunoprecipitated DNA fragments were quantified by qPCR. Intergenic region of S. pombe chromosome I (position 465226–465326) used as negative control. Fold enrichment normalized against intergenic region of WT(HA-Pap1 knock-in) or WT strain under non-stress condition set as 1. All experiments were repeated three times.
RESULTS
A nucleocytoplasmic HMG protein mediates stress tolerance
Previously, we described using an Arabidopsis cDNA expression library to select clones that could enhance stress tolerance in S. pombe (20). Although we recovered an Arabidopsis cDNA corresponding to At1G16210, we decided to conduct the functional analysis with its S. pombe homolog SPBC29A10.12, which is predicted to encode a 207aa high mobility group (HMG) protein (Supplementary Figure S2). Expression of SPBC29A10.12 cDNA enhanced tolerance to diamide and Cd (Figure 2A), but not to H2O2, salt (NaCl or KCl), osmotic (sorbitol) or heat (42°C) stress (Supplementary Figure S3A). A deletion in SPBC29A10.12 created by homologous recombination (Supplementary Figure S1A) was indeed more sensitive to Cd and diamide but not to the other stresses (Supplementary Figure S1B). Since both diamide and Cd deplete the GSH pool; diamide promotes formation of GSSG and Cd sequesters GSH-derived (γ-Glu-Cys)n-Gly peptides in the vacuole (21), this gene may be specific for the subcategory of oxidative stress known as GSH or disulfide stress (4). Indeed, a lower GSH/GSSG ratio was found after treatment with diamide or Cd (Supplementary Figure S4) in WT and in the SPBC29A10.12Δ strain, but not when expressing the SPBC29A10.12 cDNA. Given its role in oxidative stress, we named SPBC29A10.12 oxs1+ for oxidative stress 1.
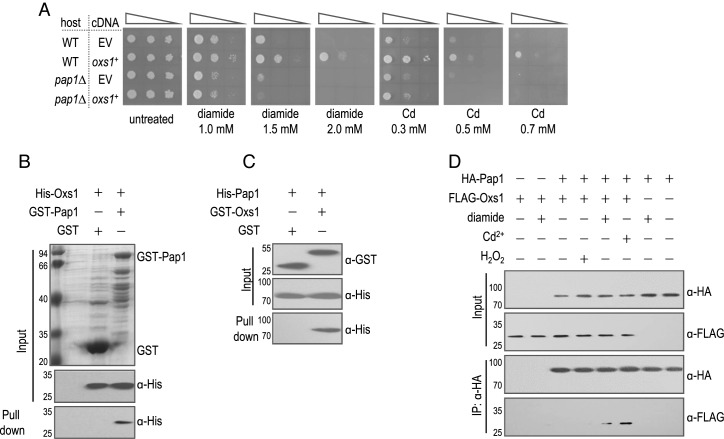
Figure 2.
Oxs1-mediated diamide tolerance requires Pap1. (A) Cells of 10-fold serially diluted containing empty vector pART1 (EV) or pART1 expressing oxs1+ spotted on EMM selective media without or with diamide or Cd. (B) GST-Pap1 or GST bound to GST affinity resin incubated with His-Oxs1. Pull-down fractions analyzed by western blotting with anti-His antibody. (C) GST-Oxs1 or GST bound to GST affinity resin incubated with His-Pap1. Pull-down fractions detected with anti-His antibody. (D) Protein extracts from strain producing HA-Pap1, FLAG-Oxs1 or both were treated without or with diamide (1 mM, 3 h), Cd (0.1 mM, 3 h) or H2O2 (0.2 mM, 5 min), immunoprecipitated with anti-HA antibody and detected by anti-HA or anti-FLAG antibody. Input fractions represent 2% of His-tagged proteins in pull-down assays. Numbers left of panels B, C and D indicate size markers in kD.
Oxs1 requires and interacts with Pap1 in a stress-dependent manner
Of the three H2O2 stress pathways in S. pombe, the Mpr1–Prr1 pathway does not appear to play a role in diamide tolerance, as prr1Δ is insensitive to diamide (22). Since sty1Δ or pap1Δ is diamide sensitive (9), we investigated whether components of these pathways could be involved. Enhanced tolerance to diamide or Cd by the oxs1+ cDNA was still observed in sty1Δ, atf1Δ or pcr1Δ strains (Supplementary Figure S3B), indicating that the Sty1–Atf1 pathway is not essential for Oxs1 function. However, enhanced tolerance to diamide or Cd was not observed in the pap1Δ mutant TP108-3C (Supplementary Figure S3B). To confirm, we generated our own pap1Δ strain (Supplementary Figure S1C) and the results were the same (Figure 2A). While this indicates that Oxs1 function requires Pap1, the converse does not hold, as overexpression of pap1+ can rescue stress sensitivity in the oxs1Δ background (Supplementary Figure S3C). This suggests that Pap1 can function without Oxs1, at least when overproduced. An additive effect was also not found, as over-expression of both pap1+ and oxs1+ failed to increase stress tolerance beyond what was achieved through pap1+ overexpression alone (Supplementary Figure S3D), while the oxs1Δ pap1Δ double mutant is as sensitive to diamide or Cd as the pap1Δ mutant (Supplementary Figure S3E). This suggests that Pap1 plays a more dominant role than Oxs1 for diamide or Cd tolerance.
To test whether Oxs1 may interact with Pap1, we first incubated His-tagged Oxs1 with GST-tagged Pap1 or the GST-only control. The resin that bound GST-Pap1 pulled down His-Oxs1 (Figure 2B). However, since the yield of GST-Pap1 from E. coli was relatively low, we switched to using GST-tagged Oxs1 to test against His-tagged Pap1. Likewise, the data show that the two proteins interact, as the affinity resin that bound GST-Oxs1 also pulled down His-Pap1 (Figure 2C). To test if this binding occurs in vivo, the native oxs1+ and pap1+ genes were replaced with versions encoding FLAG-tagged Oxs1 and HA-tagged Pap1, respectively (Supplementary Figure S1D and E). The tolerance phenotype of each single or double knock-in strain was indistinguishable from WT (Supplementary Figure S1F). Crude extracts immunoprecipitated with anti-HA antibody detected anti-FLAG cross reaction when cells were treated with diamide or Cd (Figure 2D), but not when cells were untreated or treat under conditions typically described for H2O2-induced Pap1 nuclear localization (10). This shows that the Oxs1–Pap1 interaction is specific for diamide or Cd.
Oxs1 is a co-regulator of Pap1
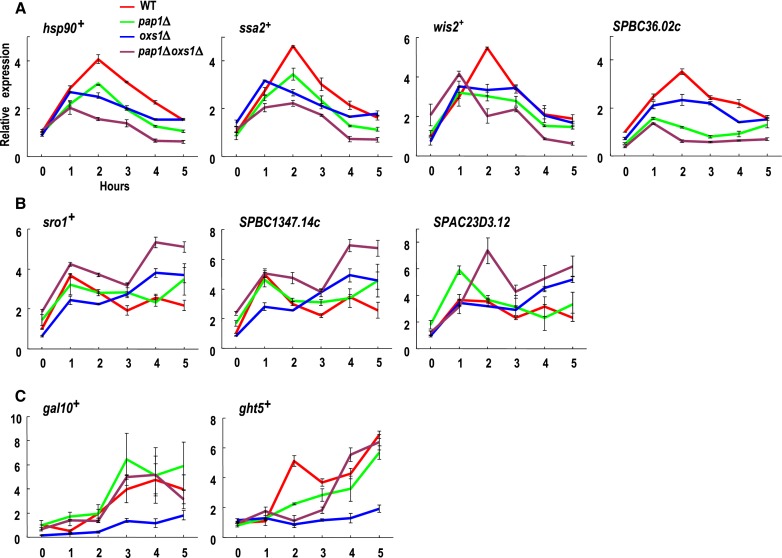
Since Pap1 is a transcription factor, and it interacts with Oxs1 in vivo, we considered the possibility that they could interact at target gene promoters. To find potential target genes for these proteins, we conducted an RNA-seq analysis. This led to finding 307 genes induced by 2-fold or more in cells grown in diamide (Cd not tested). Of these, gene annotation analysis shows that 101 of them are related to the stress response, protein folding, transport or transcription. Among the 101, 28 were induced by more than 4 fold. However, only 20 of the 28 genes showed reproducible induction by diamide in a second test by qRT-PCR (Supplementary Table S1). Hence, these 20 genes were selected for qRT-PCR in the WT, pap1Δ, oxs1Δ or pap1Δoxs1Δ strains. Their expression patterns during the 5 hours of diamide treatment place them into four classes. Class I with 4 members (hsp90+, ssa2+, wis2+, SPBC36.02c) showed reduced transcription in either a pap1Δ or oxs1Δ background and lowest expression in the pap1Δoxs1Δ double mutant (Figure 3A). This indicates that while each protein alone enhances transcription, the presence of both Pap1 and Oxs1 exerts an additive positive effect. Class II with 3 members (sro1+, SPBC1347.14c, SPAC23D3.12) were unaffected by the pap1Δ or oxs1Δ mutation, but showed elevated expression in the pap1Δoxs1Δ background (Figure 3B), suggesting that either Oxs1 or Pap1 alone is sufficient to repress transcription, and that de-repression requires loss of both proteins. Class III with two members (gal10+, ght5+) showed lower expression in the oxs1Δ strain (Figure 3C), which suggests that Oxs1 positively regulates expression. However, an expression pattern similar to that of the WT was found in the pap1Δoxs1Δ or pap1Δ genotype. This pattern may indicate that in the absence of Oxs1, the lower expression may be due to Pap1 playing a role in negative regulation. Relieve of this Pap1-mediated repression is seen in the presence of Oxs1, as in the WT, or in the absence of Pap1, as in the pap1Δ or pap1Δoxs1Δ genotype. Class IV with 11 members (Supplementary Table S1) showed expression patterns that appear unaffected by the pap1Δ, oxs1Δ, or pap1Δoxs1Δ genotype, and hence their diamide induction must be due to stress pathways unrelated to Pap1 or Oxs1.
Figure 3.
Transcription profiles of diamide-induced genes in WT and mutants. (A–C) qRT-PCR of mRNA from cells grown in EMM media treated with 1.5 mM diamide for 0–5 h; beta-actin (act1) mRNA used for normalization. Relative expression for each gene in each genotype normalized to WT at 0 h set as 1. Error bars show SEM from three independent experiments. Data from non-synchronized dividing cells.
Effective Pap1 binding to target promoters requires diamide and Oxs1
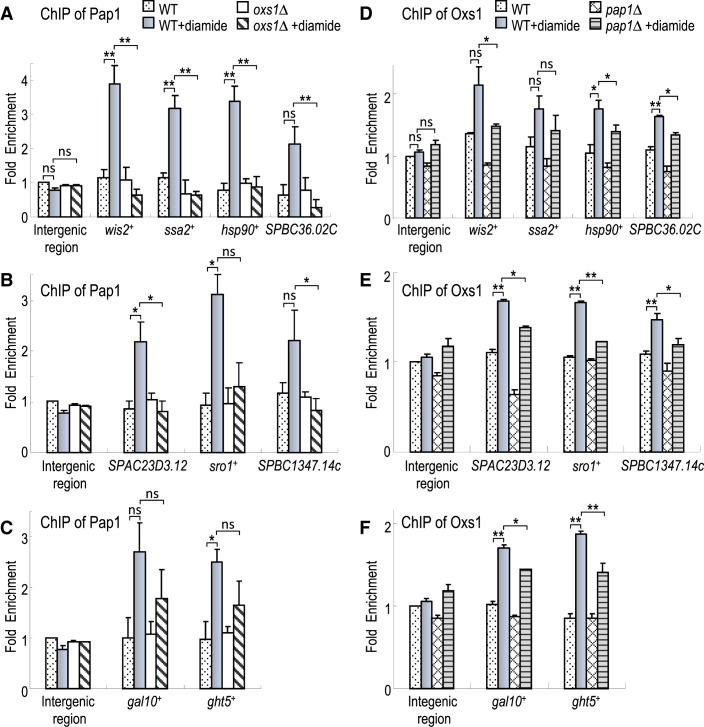
As a transcription factor, it is likely that Pap1 binds these diamide-induced promoters. To test this possibility, ChIP was conducted on the HA-Pap1 strain with anti-HA antibody followed by qPCR of the gene promoters. The data show that for all nine genes, above background amplification of their promoters was detected only when WT cells were exposed to diamide (Figure 4A–C), indicating that Pap1 is recruited to these promoters during diamide stress. When the ChIP-qPCR analysis was conducted in the oxs1Δ strain, diamide-induced Pap1 binding was abolished for the Classes I and II genes, suggesting that Oxs1 is necessary for Pap1 binding to these promoters (Figure 4A and B). For the Class III genes, Pap1 promoter binding in response to diamide was found in the oxs1Δ genotype, but at lower efficiency than in a WT background (Figure 4C). This above background level of Pap1 binding is consistent with an interpretation that in the absence of Oxs1, Pap1 exerts a repressive effect on these genes, and this repression is presumed to require promoter binding.
Figure 4.
Oxs1 and Pap1 cooperative binding to target promoters. (A–C) ChIP-qPCR of Pap1 binding to gene promoters in WT(HA-Pap1 knock-in) or oxs1Δ(HA-Pap1 knock-in) strains producing HA-Pap1 treated 5 h without or with 1.5 mM diamide. HA antibody used to immunoprecipitate HA-Pap1. (D–F) ChIP-qPCR of Oxs1 binding to gene promoters in WT or pap1Δ strain treated as in (A–C). Anti-Oxs1 polyclonal antiserum used to immunoprecipitate Oxs1; antiserum specificity shown in Supplementary Figure S5. Intergenic region of S. pombe chromosome I (position 465226 to 465326) used as negative control. Fold enrichment normalized against intergenic region of WT(HA-Pap1 knock-in) strain (A–C) or WT strain (D–F) under non-stress condition set as 1. SEM (error bars) from three independent experiments. *P < 0.05 and **P < 0.01 using unpaired Student's t tests, ns: no significant effect at P < 0.05 level.
Oxs1 binds to the same promoters induced by diamide
Since Oxs1 is necessary for Pap1 binding to target promoters, we tested if Oxs1 would also bind to the same promoters. ChIP was performed using anti-Oxs1 antibody followed by qRT-PCR. Above background amplification of these nine promoters was detected only when cells were exposed to diamide (Figure 4D–F). This effect was not significant in an oxs1Δ strain, showing that the anti-Oxs1 antibody has specificity for Oxs1 (Supplementary Figure S5). In a pap1Δ strain, Oxs1 still responds to diamide with more effective binding to nearly all of these promoters, but less effective than in the WT (pap1+) background. This suggests that as with Pap1, Oxs1 is recruited to each of these promoters during diamide stress, and that the presence of Pap1 makes Oxs1 binding to these targets more effective.
Oxs1 and Pap1 relocate to the nucleus during diamide or Cd stress
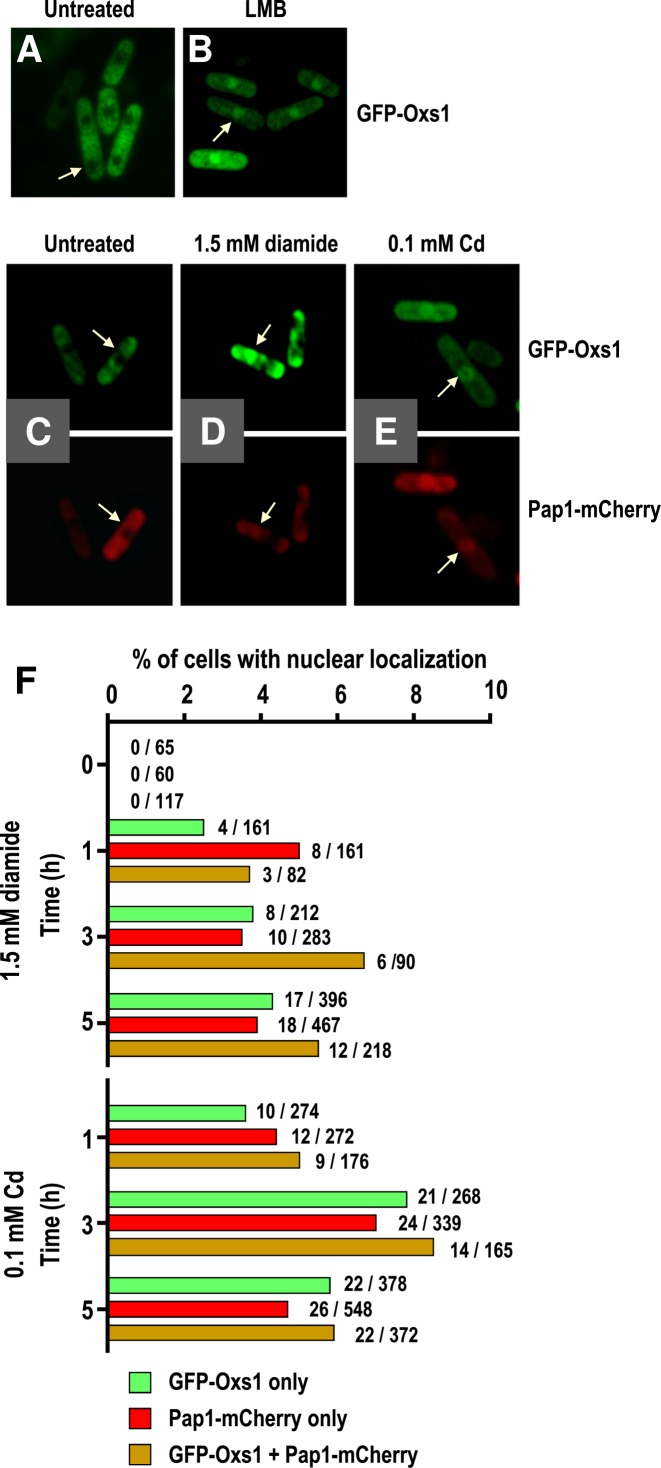
The protein subcellular localization program PSORT: http://psort.ims.u-tokyo.ac.jp/ predicts a tripartite NLS consisting of aa residues 4–10 (PKKRAEK), 23–26 (KKKK) and 129–135 (PERRFKA) as well as a nuclear export signal (NES) between aa107 to aa116 (IDDALDLLSL, conserved aa underlined; Supplementary Figure S2). This suggests that Oxs1 might be a nucleocytoplasmic shuttling protein regulated by the exportin Crm1. When Oxs1 was fused to the C-terminus of the green fluorescence protein GFP and inserted behind the adh1 promoter on a multi-copy plasmid, the GFP signal was indeed excluded from the nucleus (Figure 5A). However, when the cells were treated with leptomycin B (LMB), a Streptomyces metabolite that blocks Crm1 interaction with the NES of a cargo protein (23), the GFP-Oxs1 protein could be found in the nucleus (Figure 5B).
Figure 5.
Stress-induced nuclear localization by Oxs1 and Pap1. Representative fluorescence microscopy of GFP-tagged Oxs1 or mCherry-tagged Pap1 fusion protein. Plasmid pART1-produced GFP-Oxs1 in WT cells treated without (A) or with (B) LMB. (C–E) Plasmid pART1-produced GFP-Oxs1 or pSLF173-produced Pap1-mCherry without or with treatment of 1.5 mM diamide or 0.1 mM Cd for 1 h. (F) Percentage of cells scored as GFP only, as mCheery only, or as having both GFP and mCherry signals in the nucleus. Number of cells scored positive over total cells counted shown to the right of each bar.
As Figure 3 shows that target promoters are regulated by Oxs1 and Pap1, we examined their subcellular localization under the same treatment conditions. With plasmid-encoded GFP-Oxs1 and Pap1-mCherry fusion proteins, cells without exposure to stress showed an absence of fluorescence in the nucleus (Figure 5C). When treated with diamide or Cd, green and/or red fluorescence could be detected in the nucleus in a small percentage of the cell population (Figure 5D and E). Roughly the same percentage of cells scored positive for nuclear detection of GFP, mCherry or both (Figure 5F). This is consistent with the deduction that both proteins act in concert. However, since scoring positive for nuclear localization in this assay rests on having a sizable percentage of the signals in the nucleus, the low percentage of cells scoring as positive could be explained by the abnormally high amounts of proteins in these overexpression strains. The corollary would be that even in cells with a small percentage of total Oxs1 and Pap1 in the nucleus, and hence counted as predominantly cytoplasmic GFP-Oxs1 and Pap1-mCherry, there could be sufficient Oxs1 and Pap1 to cause the effect on target gene expression shown in Figure 3.
Overexpression of target genes for stress tolerance
For the four Class I genes up-regulated by Pap1 and Oxs1, their role in diamide tolerance was examined by expressing each individually in a multi-copy plasmid. Every gene was able to enhance tolerance to diamide in the WT background (Supplementary Figure S3F), although not as effective as overexpressing their up-stream regulator oxs1+ or pap1+. Two of them, wis2+ and hsp90+, also showed some enhanced tolerance in the oxs1Δ mutant, but none was effective in a pap1Δ background (Supplementary Figure S3G,H). Hence, the loss of Pap1, presumed to activate many other genes that respond to diamide, cannot be compensated through the simple overexpression of any one of these downstream targets.
Oxs1 stress tolerance function conserved among eukaryotic species
BLAST analysis indicates that homologs of deduced Oxs1 are present in a wide range of eukaryotes including human, mouse, rat, zebrafish, fruitfly, mosquito, nematode, Neurospora, rice, maize, and Arabidopsis, but surprisingly not in the budding yeast Saccharomyces cerevisiae (Supplementary Figure S2). Previously described as a family of unknown proteins named DUF1014 (IPR010422), there has been a recent report of a human member of this family, Ccdc124 (Coiled-Coil Domain Containing Protein 124), identified as a novel component of the centrosome during interphase and G2/M transition and is involved in cytokinesis (24).
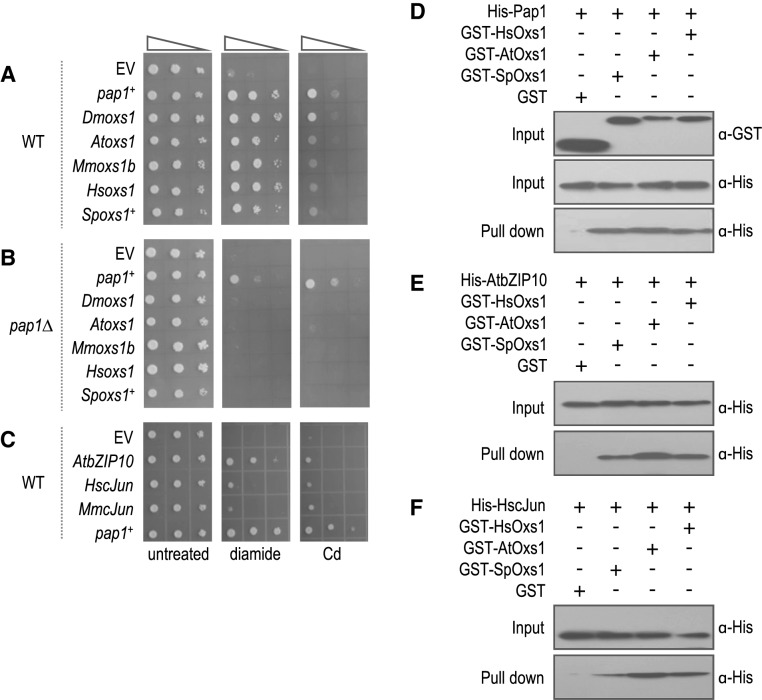
Conservation of regulatory components for oxidative stress has long been noted among diverse organisms. For instance, S. pombe Sty1 is similar to human and mouse Jun-NH2-terminal kinase (JNK) and p38 kinase (18). S. pombe Atf1 is most homologous with human and mouse ATF-2 (25) and S. pombe Pap1 with human and mouse cJun (26). In Arabidopsis, however, MAPKs AtMPK3 and AtMPK6 activate plant-specific WRKY transcription factors (27,28), but there is evidence for bZIP transcription factors involved in pathogen stress (29). To ask whether there is functional conservation among heterologous Oxs1 proteins, cDNAs encoding the Oxs1 homologues from Arabidopsis (AtOxs1, At1G16210), fruitfly (DmOxs1, Dmel_CG6013), mouse (MmOxs1b, NP_081240) and human (HsOxs1, hCG_2000823) were found to be as effective as SpOxs1 for tolerance against diamide or Cd (Figure 6A), but not to H2O2, salt, osmotic or heat (Supplementary Figure S3A). Moreover, these homologue proteins also failed to enhance diamide tolerance in the pap1Δ genotype, indicating that Oxs1 homologues also require S. pombe Pap1 for function (Figure 6B).
Figure 6.
Heterologous Oxs1 or Pap1 enhance stress tolerance and interact in vitro. (A–C) 10-fold serially diluted cells containing pART1 (EV) or pART1 expressing indicated cDNAs on EMM media without or with 2 mM diamide or 0.7 mM Cd. Photos taken after 5 days of incubation. (D–F) Western blots using anti-GST or anti-His antibodies. GST-HsOxs1, GST-AtOxs1, GST-SpOxs1 or GST control incubated with His-Pap1 (D), His-AtbZIP10 (E) or His-HscJun (F). Input control shown for GST done at the same time for (D-F).
Since S. pombe Pap1 is not present in the other eukaryotes, heterologous Oxs1 proteins might use Pap1-like proteins as co-activators, and if so, those Pap1-like proteins might behave similarly in S. pombe. When AtbZIP10 (At4g02640), along with human and mouse cJun, were tested in S. pombe, AtbZIP10 was nearly as effectively as Pap1 for diamide tolerance, and a weaker effect against Cd. The human and mouse cJun homologs also showed weak but positive effects for diamide and Cd (Figure 6C).
Physical interaction among heterologous Oxs1 and Pap1-like proteins was also detected by the in vitro binding assay. GST-tagged Oxs1, HsOxs1, AtOxs1 or the control GST protein was incubated with His-tagged S. pombe Pap1, purified through GST affinity resin, and probed with anti-His antibody. Like SpOxs1, HsOxs1 and AtOxs1 were effective in binding S. pombe Pap1 (Figure 6D). Likewise, when AtbZIP10 or HscJun was used instead of Pap1, the GST-Oxs1 protein from human, Arabidopsis or S. pombe each showed interaction (Figure 6E and F).
DISCUSSION
Of the many regulatory proteins identified in the oxidative stress response to H2O2, only loss of Sty1 and Pap1 affected diamide tolerance (9), and of the two, only Pap1 is necessary for Oxs1-mediated tolerance to diamide and Cd. Oxs1 and Pap1 can bind to each other, and moreover, this interaction is found only in cells grown with diamide or Cd, but not H2O2. We did not test further with Cd, but with diamide, the ChIP assays show that the two proteins bind the same selected set of gene promoters that are diamide induced. Moreover, this binding only occurs during diamide stress. Although it may be possible that Pap1 directs Oxs1 to the target promoters, the ChIP data suggest otherwise, that Oxs1 binds these same promoters even in the absence of Pap1. On the contrary, Pap1 binding to Class I and Class II promoters requires Oxs1, as though Oxs1 is directing the diamide stress response. The one conflicting data is that in the absence of Oxs1, Pap1 can still enhance diamide tolerance. It is possible that this effect is caused by an overproduction of Pap1 such that it either overrides the need for an Oxs1 partner, or that Pap1 also (hyper) activates other stress ameliorating genes that are not dependent on Oxs1.
Although Pap1 is well recognized to activate transcription, this role was found only with the Class I genes. With the Classes II and III genes, its role appears to be that of a negative regulator. The Classes I to III genes show that regulation by Pap1 and Oxs1 can be complex, requiring one or both proteins to be either positive or negative regulators. However, in all cases, the presence of one protein appears to help recruit the other to the same promoter. Oxs1 did not require Pap1 to bind to the target promoters, but was less effective in its absence. For the Class IV genes, in which we have not found convincing linkage to Pap1 and Oxs1, it remains possible that Sty1 may be involved in some of them, although the Sty1 downstream regulator is not likely to be Atf1 or Pcr1.
In summary, we have added a new Pap1–Oxs1 pathway in the oxidative stress response in S. pombe for diamide (Figure 1) that appears specific for disulfide or GSH stress, and this pathway also responds to Cd most likely because it also causes the GSH depletion effect of diamide (Supplementary Figure S4H). In the absence of stress, Crm1 (exportin1) keeps Oxs1 in the cytoplasm. During disulfide stress, Oxs1 moves to the nucleus. For some promoters, Oxs1 recruits Pap1 for co-activation (Class I) or co-repression (Class II). For other targets, Oxs1 neutralizes the repressive effect of an existing Pap1 (Class III). In this Pap1–Oxs1 pathway, Pap1 is presumed to be in an oxidized state in the nucleus, as has been experimentally shown with the budding yeast homologue Yap1 (30) or when S. pombe is treated with diethylmaleate (another glutathione-depleting agent) or H2O2 (31,32). Of particular interest is that since Oxs1-homologous proteins, as well as Pap1-homologous proteins are found in diverse eukaryotes from fungi, insects, worms, plants to mammals, it is likely that the Oxs1–Pap1 co-regulatory system is evolutionarily conserved. Consistent with this hypothesis, heterologous Oxs1 and Pap1-homologues can bind each other in vitro, and at least in the fission yeast, they can also pheno-copy S. pombe Oxs1 and Pap1 for stress tolerance.
Acknowledgments
We thank E. Hidalgo and J. Quinn for S. pombe strains, and Y. Li for comments on early draft manuscripts.
Footnotes
Present addresses:
Yumei He, Institute of Human Virology, Zhongshan School of Medicine, Sun Yat-Sen University, Guangzhou, China.
Wen Song, Bio-Rad Laboratories, Hercules, CA 94547, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
US DOE [EMSP 55278]; China [2010ZX08010-001, 2012M511846]; CAS Key laboratory of South China Agricultural Plant Molecular Analysis and Genetic Improvement. Funding for open access charge: Current grant funds.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wolf C., Hochgräfe F., Kusch H., Albrecht D., Hecker M., Engelmann S. Proteomic analysis of antioxidant strategies of Staphylococcus aureus: Diverse responses to different oxidants. Proteomics. 2008;8:3139–3153. doi: 10.1002/pmic.200701062. [DOI] [PubMed] [Google Scholar]
- 2.García-Santamarina S., Boronat S., Hidalgo E. Reversible cysteine oxidation in hydrogen peroxide sensing and signal transduction. Biochemistry. 2014;53:2560–2580. doi: 10.1021/bi401700f. [DOI] [PubMed] [Google Scholar]
- 3.Kosower N.S., Kosower E.M. Diamide: an oxidant probe for thiols. Methods Enzymol. 1995;251:123–133. doi: 10.1016/0076-6879(95)51116-4. [DOI] [PubMed] [Google Scholar]
- 4.Åslund F., Beckwith J. Bridge over troubled waters: sensing stress by disulfide bond formation. Cell. 1999;96:751–753. doi: 10.1016/s0092-8674(00)80584-x. [DOI] [PubMed] [Google Scholar]
- 5.Papadakis M.A., Workman C.T. Oxidative stress response pathways: fission yeast as archetype. Crit. Rev. Microbiol. 2014 doi: 10.3109/1040841X.2013.870968. doi:10.3109/1040841X.2013.870968. [DOI] [PubMed] [Google Scholar]
- 6.Veal E.A., Tomalin L.E., Morgan B.A., Day A.M. The fission yeast Schizosaccharomyces pombe as a model to understand how peroxiredoxins influence cell responses to hydrogen peroxide. Biochem. Soc. Trans. 2014;42:909–916. doi: 10.1042/BST20140059. [DOI] [PubMed] [Google Scholar]
- 7.Brown J.D., Day A.M., Taylor S.R., Tomalin L.E., Morgan B.A., Veal E.A. A peroxiredoxin promotes H2O2 signaling and oxidative stress resistance by oxidizing a thioredoxin family protein. Cell Rep. 2013;5:1425–1435. doi: 10.1016/j.celrep.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calvo I.A., Boronat S., Domènech A., García-Santamarina S., Ayté J., Hidalgo E. Dissection of a redox relay: H2O2-dependent activation of the transcription factor Pap1 through the peroxidatic Tpx1-Thioredoxin cycle. Cell Rep. 2013;5:1413–1424. doi: 10.1016/j.celrep.2013.11.027. [DOI] [PubMed] [Google Scholar]
- 9.Toone W.M., Kuge S., Samuels M., Morgan B.A., Toda T., Jones N. Regulation of the fission yeast transcription factor Pap1 by oxidative stress: requirement for the nuclear export factor Crm1 (Exportin) and the stress-activated MAP kinase Sty1/Spc1. Genes Dev. 1998;12:1453–1463. doi: 10.1101/gad.12.10.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calvo I.A., García P., Ayté J., Hidalgo E. The transcription factors Pap1 and Prr1 collaborate to activate antioxidant, but not drug tolerance, genes in response to H2O2. Nucleic Acids Res. 2012;40:4816–4824. doi: 10.1093/nar/gks141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Day A.M., Brown J.D., Taylor S.R., Rand J.D., Morgan B.A., Veal E.A. Inactivation of a peroxiredoxin by hydrogen peroxide is critical for thioredoxin-mediated repair of oxidized proteins and cell survival. Mol. Cell. 2012;45:398–408. doi: 10.1016/j.molcel.2011.11.027. [DOI] [PubMed] [Google Scholar]
- 12.Buck V., Quinn J., Pino T.S., Martin H., Saldanha J., Makino K., Morgan B.A., Millar J.B.A. Peroxide sensors for the fission yeast stress-activated mitogen-activated protein kinase pathway. Mol. Biol. Cell. 2001;12:407–419. doi: 10.1091/mbc.12.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawrence C.L., Maekawa H., Worthington J.L., Reiter W., Wilkinson C.R.M., Jones N. Regulation of Schizosaccharomyces pombe Atf1 protein levels by Sty1-mediated phosphorylation and heterodimerization with Pcr1. J. Biol. Chem. 2007;282:5160–5170. doi: 10.1074/jbc.M608526200. [DOI] [PubMed] [Google Scholar]
- 14.Sansó M., Gogol M., Ayté J., Seidel C., Hidalgo E. Transcription factors Pcr1 and Atf1 have distinct roles in stress-and Sty1-dependent gene regulation. Eukaryotic Cell. 2008;7:826–835. doi: 10.1128/EC.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quinn J., Malakasi P., Smith D.A., Cheetham J., Buck V., Millar J.B.A., Morgan B.A. Two-component mediated peroxide sensing and signal transduction in fission yeast. Antioxid. Redox Signal. 2010;15:153–165. doi: 10.1089/ars.2010.3345. [DOI] [PubMed] [Google Scholar]
- 16.Yan C., Lee L.H., Davis L.I. Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J. 1998;17:7416–7429. doi: 10.1093/emboj/17.24.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quinn J., Findlay V.J., Dawson K., Millar J.B.A., Jones N., Morgan B.A., Toone W.M. Distinct regulatory proteins control the graded transcriptional response to increasing H2O2 levels in fission yeast Schizosaccharomyces pombe. Mol. Biol. Cell. 2002;13:805–816. doi: 10.1091/mbc.01-06-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Millar J.B., Buck V., Wilkinson M.G. Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes Dev. 1995;9:2117–2130. doi: 10.1101/gad.9.17.2117. [DOI] [PubMed] [Google Scholar]
- 19.Sansó M., Vargas-Pérez I., Quintales L., Antequera F., Ayté J., Hidalgo E. Gcn5 facilitates Pol II progression, rather than recruitment to nucleosome-depleted stress promoters, in Schizosaccharomyces pombe. Nucleic Acids Res. 2011;39:6369–6379. doi: 10.1093/nar/gkr255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanvillain R., Wei S., Wei P., Kim J.H., Ow D.W. Stress tolerance to stress escape in plants: role of the OXS2 zinc-finger transcription factor family. EMBO J. 2011;30:3812–3822. doi: 10.1038/emboj.2011.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ortiz D.F., Kreppel L., Speiser D.M., Scheel G., McDonald G., Ow D.W. Heavy metal tolerance in the fission yeast requires an ATP binding cassette-type vacuolar membrane transporter. EMBO J. 1992;11:3491–3499. doi: 10.1002/j.1460-2075.1992.tb05431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohmiya R., Yamada H., Kato C., Aiba H., Mizuno T. The Prr1 response regulator is essential for transcription of ste11+ and for sexual development in fission yeast. Mol. Gen. Genet. 2000;264:441–451. doi: 10.1007/s004380000305. [DOI] [PubMed] [Google Scholar]
- 23.Kudo N., Taoka H., Toda T., Yoshida M., Horinouchi S. A novel nuclear export signal sensitive to oxidative stress in the fission yeast transcription factor Pap1. J. Biol. Chem. 1999;274:15151–15158. doi: 10.1074/jbc.274.21.15151. [DOI] [PubMed] [Google Scholar]
- 24.Telkoparan P., Erkek S., Yaman E., Alotaibi H., Bayık D., Tazebay U.H. Coiled-Coil domain containing protein 124 Is a novel centrosome and midbody protein that interacts with the Ras-guanine nucleotide exchange factor 1B and is involved in cytokinesis. PLoS One. 2013;8:e69289. doi: 10.1371/journal.pone.0069289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeda T., Toda T., Kominami K., Kohnosu A., Yanagida M., Jones N. Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 1995;14:6193–6208. doi: 10.1002/j.1460-2075.1995.tb00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toda T., Shimanuki M., Yanagida M. Fission yeast genes that confer resistance to staurosporine encode an AP-1-like transcription factor and a protein kinase related to the mammalian ERK1/MAP2 and budding yeast FUS3 and KSS1 kinases. Genes Dev. 1991;5:60–73. doi: 10.1101/gad.5.1.60. [DOI] [PubMed] [Google Scholar]
- 27.Kovtun Y., Chiu W.L., Tena G., Sheen J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. U.S.A. 2000;97:2940–2945. doi: 10.1073/pnas.97.6.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mao G., Meng X., Liu Y., Zheng Z., Chen Z., Zhang S. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell. 2011;23:1639–1653. doi: 10.1105/tpc.111.084996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaminaka H., Näke C., Epple P., Dittgen J., Schütze K., Chaban C., Holt B.F., Merkle T., Schäfer E., Harter K., Dangl J.L. bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J. 2006;25:4400–4411. doi: 10.1038/sj.emboj.7601312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gulshan K., Lee S.S., Moye-Rowley W. Differential oxidant tolerance determined by the key transcription factor Yap1 is controlled by levels of the Yap1-binding protein, Ybp1. J. Biol. Chem. 2011;286:34071–34081. doi: 10.1074/jbc.M111.251298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castillo E.A., Ayte J., Chiva C., Moldon A., Carrascal M., Abian J., Jones N., Hidalgo E. Diethylmaleate activates the transcription factor Pap1 by covalent modification of critical cysteine residues. Mol. Microbiol. 2002;45:243–254. doi: 10.1046/j.1365-2958.2002.03020.x. [DOI] [PubMed] [Google Scholar]
- 32.Vivancos A.P., Castillo E.A., Jones N., Ayte J., Hidalgo E. Activation of the redox sensor Pap1 by hydrogen peroxide requires modulation of the intracellular oxidant concentration. Mol. Microbiol. 2004;52:1427–1435. doi: 10.1111/j.1365-2958.2004.04065.x. [DOI] [PubMed] [Google Scholar]