Abstract
Neuroglobin (NGB) is predominantly expressed in the brain and retina. Studies suggest that NGB exerts protective effects to neuronal cells and is implicated in reducing the severity of stroke and Alzheimer's disease. However, little is known about the mechanisms which regulate the cell type-specific expression of the gene. In this study, we hypothesized that distal regulatory elements (DREs) are involved in optimal expression of the NGB gene. By chromosome conformation capture we identified two novel DREs located −70 kb upstream and +100 kb downstream from the NGB gene. ENCODE database showed the presence of DNaseI hypersensitive and transcription factors binding sites in these regions. Further analyses using luciferase reporters and chromatin immunoprecipitation suggested that the −70 kb region upstream of the NGB gene contained a neuronal-specific enhancer and GATA transcription factor binding sites. Knockdown of GATA-2 caused NGB expression to drop dramatically, indicating GATA-2 as an essential transcription factor for the activation of NGB expression. The crucial role of the DRE in NGB expression activation was further confirmed by the drop in NGB level after CRISPR-mediated deletion of the DRE. Taken together, we show that the NGB gene is regulated by a cell type-specific loop formed between its promoter and the novel DRE.
INTRODUCTION
Gene transcription is a complex process involving the orchestration of diverse elements such as promoters, enhancers and insulators ensuring that gene expression is under accurate control. A recent genome-wide study showed that distal enhancers play more important roles than proximal promoters in controlling cell type-specific gene expression (1). Mutations in these distal elements may cause diseases, as seen in the case of β-thalassaemia. Expression of an intact β-globin (HBB) gene may be inactivated by deletion of the locus control region (LCR) (2). The LCR is a collection of DREs that control cell type-specific and temporal expression of the genes in the HBB gene cluster (3). Identification of such gene regulatory elements is therefore important for the understanding of gene transcriptional regulation and disease pathology.
Active regulatory elements are often associated with DNaseI hypersensitive sites (DHS), which are enriched by marks of open chromatin structure and locus accessibility such as histone 3 lysine 4 mono-methylation (H3K4me1) and H3K27 acetylation (H3K27Ac), and contain clusters of transcription factor binding sites (TFBS) (4,5). These features provide clues for identifying regulatory elements, which can be located up to megabases away from the target gene (6,7), thus making their discovery challenging.
Based on the widely accepted chromatin looping model, where a loop is formed by the DNA segment between a distal enhancer and its target gene promoter thus bringing the two elements in spatial proximity (8,9), such enhancer-promoter interactions can be revealed by chromosome conformation capture (3C) (10). 3C provides a powerful tool to search for regulatory elements by identifying distal regions that interact with the gene of interest. This has been demonstrated successfully by Ghedof et al. in searching regulatory elements of the CFTR gene (11). Here, we use a similar approach to identify distal regulatory elements of the human neuroglobin (NGB) gene. To date, no regulatory element of the human NGB gene is known except for the recent characterization of the promoter (12). It is thus relevant to identify the regulatory elements because mutations of NGB have been associated with higher risk of Alzheimer's disease (AD) (13). Potentially the risk is also associated with mutations in the regulatory elements of NGB gene. Understanding NGB gene regulation may provide (i) a novel diagnostic approach for the risk of AD on individuals and (ii) a new target of therapy or preventive measures against the disease.
NGB is a neuro-protective protein which may protect neuronal cells from hypoxia, ischemia, stroke, oxidative stress, mechanical injuries and AD (14–19). It is predominantly expressed in neurons (20) and some endocrine tissues (21), but the highest expression level is found in retina (22). Besides this spatial specificity, expression of the NGB gene also shows temporal changes in human and rodent during development and aging. In the neonatal mouse brain, NGB level increases throughout development and reaches its maximum at day 14 after birth (23), whilst in the adult rat brain NGB level declines with age (24). The latter finding is in line with a postmortem study performed on adult human brain in which NGB level was found to be negatively correlated with age (13). Interestingly, the same study found that two AD risk factors, age and female sex, were associated with lower NGB levels. However, the mechanisms of spatial and temporal transcriptional regulation of the NGB gene are unknown.
We previously identified the binding sites of Sp1 and Sp3 on two functional GC-boxes in the promoter region of the NGB gene and showed that DNA methylation was associated with cell type-specific expression (12). Others subsequently reported the presence of binding sites of the early growth response protein 1 (Egr1), nuclear factor κ-light-chain-enhancer of activated B cells (Nf-κb) (25), cyclic AMP responsive element binding protein (CREB) (26) and hypoxia-inducible factor-1 (Hif-1) on the promoter of the mouse Ngb gene (27). However, these ubiquitous and stimuli-activated TFs may not explain the cell type-specific expression of NGB in full. We hypothesized that there are DREs governing the binding of these factors or cooperating with them to control NGB gene expression.
In this study, a combination of the 3C technique and in silico analysis was employed to locate potential DREs of the human NGB gene. A 300 kb region covering the NGB gene was analyzed in both neuronal and non-neuronal cell lines. Two novel regions that interact with the NGB gene were located at −70 kb upstream and at +100 kb downstream. Interestingly, the interaction frequency between the upstream region and NGB is significantly higher in a neuronal cell line whilst the downstream element showed similar interaction frequencies across cell lines. In addition, a segment of the upstream region showed cell-type specific activating function on the NGB promoter in luciferase reporter assays. CRISPR-mediated deletion further supported the notion that this segment is a bona fide distal regulatory element for the NGB gene and that it may be a key regulator for the spatial and temporal expression of the NGB gene.
MATERIALS AND METHODS
Cell culture
Human neuroblastoma SH-SY5Y cells, cervical adenocarcinoma HeLa cells, erythromyeloblastoid leukemia K562 cells and human embryonic kidney cell HEK 293T cells were purchased from American Type Culture Collection (ATCC). These cell lines were cultured in 1:1 mixture of minimum essential medium (MEM) and Ham's F12 nutrient mixture (for SH-SY5Y cells), Dulbecco's modified Eagle medium (DMEM) (for HeLa and HEK293T cells) and Roswell Park Memorial Institute (RPMI) 1640 medium (for K562 cells) respectively. All the media (Gibco) were supplemented with 10% fetal bovine serum (Hyclone) and 1% penicillin–streptomycin (Invitrogen).
qRT-PCR
RNA was extracted from the cultured cell by TRIzol (Invitrogen) following manufacturer's standard procedure. Extracted RNA was converted to cDNA using ThermoScript Reverse Transcriptase (Invitrogen) and PCR was performed with RT2 SYBR Green qPCR Master Mixes (SABiosciences). The mRNA levels were determined by two independent experiments. Sequences of the primers used are shown in Supplementary Table S1.
Chromosome conformation capture (3C) analysis
3C was performed by following a previously described protocol using EcoRI digestion (28). To generate a control library, bacterial artificial chromosomes (BAC) RP11-463C8 and RP11-493G17 clones were purchased from Children's Hospital Oakland Research Institute (CHORI). The two BAC clones carried sequences overlapping the human NGB gene region. The library was made by mixing equimolar amount of each BAC clone and used to normalize for differences in primer efficiency (29). For comparison between cell lines, each interaction frequency detected was also normalized to the interaction between exons 2 and 8 of the human α-actin (ACTA2) gene (30).
Primers were designed on the EcoRI fragment containing the NGB gene, the anchor fragment, and paired with primers in the other fragments surrounding the 300kb region of the NGB gene. Sequences of the primers used are listed in Supplementary Table S1. The PCR was performed in a touchdown manner (31).
In silico analysis for DHS and transcription factor binding sites mapping
Information on DNaseI hypersensitive sites (DHS) and ChIP-sequencing of transcription factors binding sites as well as histone modifications was obtained from the ENCODE database on the UCSC genome browser (http://genome.ucsc.edu/, Assembly February 2009 (GRCh37/hg19)) (32).
Luciferase reporter constructs
All constructs were derived from a pGL3-basic vector with the 1051 bp NGB promoter inserted between the HindIII and MluI sites (named p(−745/+306)) (12). From the 3C and in silico analysis results, two regions were selected to be inserted into the vector for further investigation. The fragments of genomic coordinates 77808992–77809573 and 77636994–77636265 on chromosome 14 (hg19) were subcloned into the p(−745/+306) promoter-only construct at the MluI site upstream of the promoter or between the SacI and BamHI sites downstream of the luciferase (LUC) gene, in either orientation. The orientation of inserts was confirmed by DNA sequencing.
Transfection and luciferase assay
SH-SY5Y and HeLa cells were seeded at densities of 3 × 104 and 1 × 104 per well respectively in 96-well plates with 100 μl corresponding medium without antibiotics. Transfection medium was prepared by mixing 60 ng of the construct, 2 ng pRL-CMV and 0.5 μl Lipofectamine (Invitrogen) in serum-free medium. After 30 min incubation at room temperature, the mixture was diluted 6-fold with serum-free medium. The medium in the 96-well plate was replaced by the diluted transfection medium and incubated for 4 h in humidified 5% CO2 incubator. Then medium with 20% FBS was added to make the final FBS content in the medium 10%. The transfected cells were further incubated for 48 h.
The Dual-Luciferase Reporter Assay System (Promega) was used to assess the luciferase activities of the transfected cells according to the manufacturer's instructions.
Chromatin immunoprecipitation (ChIP)
ChIP was performed as previously described (33). The antibodies used for the ChIP experiment were anti-GATA-2 (sc-9008X, Santa Cruz) and anti-GATA-3 (sc-268X, Santa Cruz) and non-specific rabbit IgG (sc-2027X, Santa Cruz). Primers used for the experiment are listed in Supplementary Table S1.
GATA-2 and GATA-3 knockdown cell generation
shRNA plasmids pLKO.1 control vector (SHC002), and shRNA targeting GATA-2 (pLKO-G2a and pLKO-G2b) and GATA-3 (pLKO-G3a and pLKO-G3b) were purchased from Sigma (the sequences of the shRNA inserts are shown in Supplementary Table S1). Lentiviral particles were produced by co-transfecting HEK 293T cells with the shRNA plasmids, packaging plasmid psPAX2 and envelop plasmid pMD2.G by using polyethylenimine (Sigma). DMEM was replaced by MEM:F12 mixture one day after transfection to collect the virus in the appropriate medium for infection. MEM:F12 containing the virus was then filtered and mixed with polybrene (Sigma). SH-SY5Y cells were infected with the virus for 24 h and then cultured in normal MEM:F12 for 3 days prior to selection with puromycin (1 μg/ml).
Western blotting
Protein of normal or infected SH-SY5Y cells was extracted using radio-immunoprecipitation assay (RIPA) buffer (0.15 M NaCl, 1% NP40, 0.01 M deoxycholate, 0.1% SDS, 0.05 M Tris–HCl pH 8.0) with protease inhibitor cocktail (Roche). 20 μg of the total protein was separated by SDS-PAGE (polyacrylamide gel electrophoresis) with 12% polyacrylamide gel and transferred to polyvinylidene difluoride membranes. The proteins were probed by antibodies for GATA-2 (sc-9008, Santa Cruz), GATA-3 (sc-268, Santa Cruz), β-actin (sc-69879, Santa Cruz) and NGB (Absea), and then detected by ECL Prime Western Blotting Detection Reagent (GE Healthcare).
Clustered regularly-interspaced short palindromic repeats (CRISPR)-Cas9 mediated genome editing
The CRISPR/Cas9 mediated genomic deletion procedure described by Bauer et al. (34) was modified to delete the putative regulatory Element I detected by 3C. Instead of electroporation, we performed transfection by using Lipofectamine 3000 (Thermo Fisher Scientific). Two pX330 vectors (Addgene) which express Cas9 and the desired gRNAs, together with the puromycin resistance vector were co-transfected into SH-SY5Y cells. The gRNA sequences are displayed in Supplementary Table S1. gRNA-A (chr14:77 808 950–77 808 972) and gRNA-B (chr14:77 808 916–77 808 938) were paired with gRNA-C (chr14:77 809 411–77 809 433) to make clones with different genomic deletions. Transfected cells were incubated in medium with 1 μg/ml puromycin for selection. In order to reduce heterogeneity of the culture, limiting dilution was performed on the puromycin-selected cells. Cultures showing Element I deletion were selected for analysis.
RESULTS
Differential NGB expression levels in human cell lines
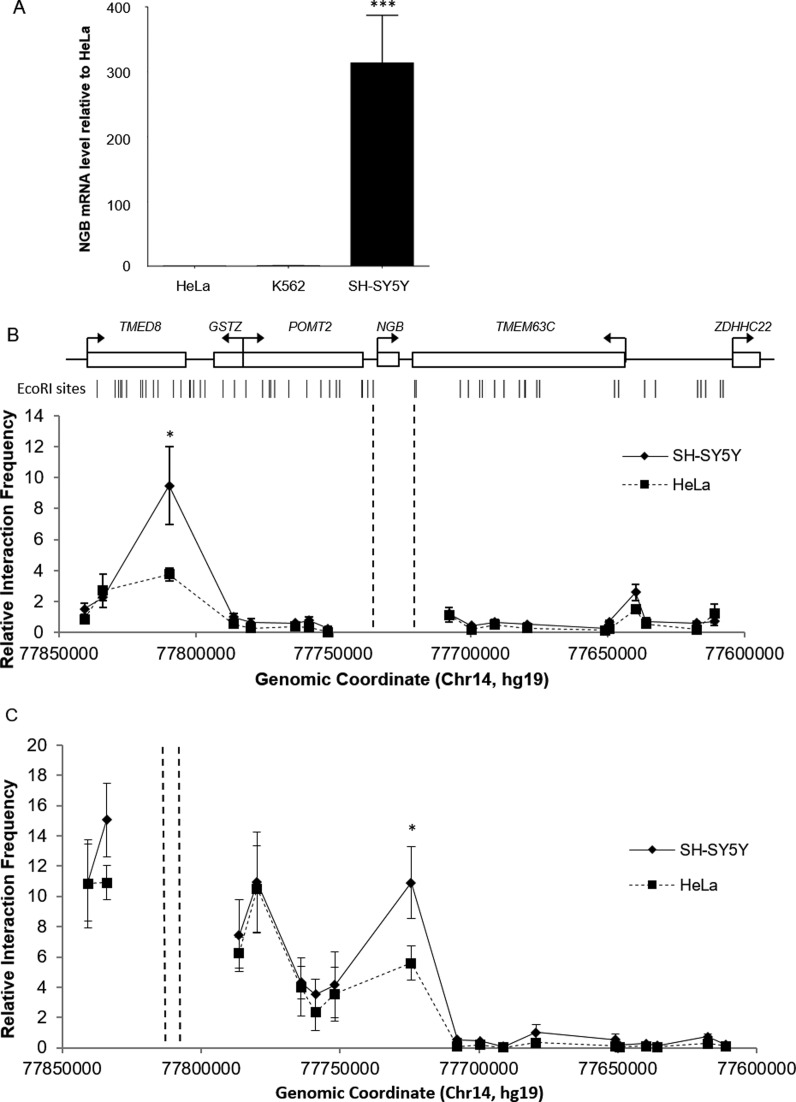
To confirm the tissue-specific expression of NGB, mRNA levels of three different human cell lines were quantified by qRT-PCR. The results showed that NGB expression in the neuroblastoma cell line SH-SY5Y was 300- and 240-fold higher than that in non-neuronal cells HeLa and K562, respectively (Figure 1A). Thus, NGB is highly expressed in SH-SY5Y neuronal cells but barely expressed in HeLa and K562 cells.
Figure 1.
(A) NGB mRNA level in three human cell lines quantified by qRT-PCR. The NGB level in SH-SY5Y cells was 300-fold of that in HeLa cells and 240-fold of that in K562 cells. ***P < 0.001 compared to HeLa (N = 3, mean ± SEM). (B and C) The 3C profiles in SH-SY5Y and HeLa cells of the 300 kb region surrounding the NGB gene. The anchor fragments are indicated by the dashed lines. Approximate positions and direction of transcription of genes, and EcoRI cutting sites within the area are displayed at the top. Significant differences between cell types were detected by Student's t test (*P < 0.05) (N = 3, mean ± SEM).
Identification of distal regions interacting with NGB gene
A 300kb region covering the NGB gene was analyzed in search for elements that physically interacted with the promoter of the NGB gene. A preliminary screening was done to select fragments within the region for 3C analysis. Eighteen fragments with active marks such as DNaseI hypersensitivity, TF binding and hyperacetylated H3 identified with the UCSC genome browser (32) (data not shown) were selected and primers were designed close to the EcoRI restriction sites of these fragments.
From the 3C results, two peaks of frequent proximity with the anchor fragment were observed in SH-SY5Y cells, while only one peak was observed in HeLa and K562 cells (Figure 1B). The peak presented in SH-SY5Y cells only was located at −70 kb upstream of the NGB gene transcription start site (TSS) and another peak was located at +100 kb downstream of the TSS. The interaction frequency between the −70 kb fragment and the anchor fragment in SH-SY5Y cells was about 2.5-fold higher than in HeLa cells. Similar interaction frequencies were observed in all the three cell lines at the +100kb downstream region of the NGB gene.
The −70 kb fragment is 3 kb in length spanning across the exons 5 and 6, intron 5 and the 3′ flanking region of transmembrane emp24 protein transport domain containing eight (TMED8) gene (chr14:77 806 771–77 809 773 (hg19)). The +100 kb fragment is a 4 kb segment located at the 5′ flanking region of transmembrane protein 63C (TMEM63C) gene (chr14:77 635 927–77 639 784 (hg19)).
3C analysis using the −70 kb upstream novel distal element as the anchor point was conducted to confirm the specific interactions occurring in SH-SY5Y cells (Figure 1C). The same set of fragments in the region was analyzed. In line with the previous results, a local peak was observed at the NGB gene-containing fragment in SH-SY5Y cells. In HeLa cells, although the interaction frequency was relatively high within the region, it was significantly lower and was about half of that in SH-SY5Y cells. There was another peak located at the promoter regions of the GSTZ and POMT2 genes. This may imply that regulatory elements contained in the −70 kb fragment could interact and regulate promoters within the region. However, we found that the interaction frequencies were similar in the two cell lines tested. In contrast, the interaction between the NGB gene containing fragment and the −70 kb upstream fragment showed cell type-specificity in SH-SY5Y cells. This suggests that the cell type-specific expression regulation of NGB is controlled by the element located at −70 kb fragment.
Mapping of potentially functional segments within the 3C fragments by in silico analysis
The potential distal regulatory elements were subjected to analysis using the UCSC genome browser (http://genome.ucsc.edu/, Assembly Feb 2009 (GRCh37/hg19)) (32). The results revealed the positions of the potentially active regulatory elements within the −70 kb and +100 kb fragments.
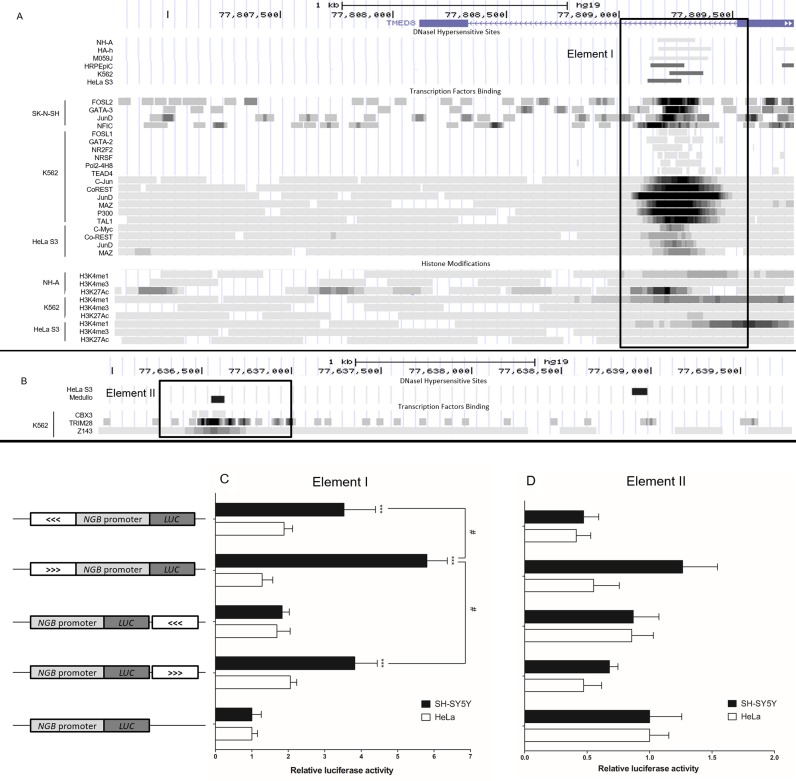
Interestingly, within the −70 kb fragment, in a ∼500 bp region within intron 5 of the TMED8 gene, we found the presence of DHS, TF binding, enrichment of H3K4me1 and H3K27Ac but the absence of H3K4me3 marks. This combination of features is characteristic of enhancer elements. Of note, the DHS was found to be present in this region (Top panel, Figure 2A) in HRPEpiC (a human retinal pigment epithelial cell line) (35), NH-A, HA-h (human astrocyte cell line), M059J (a glioblastoma cell line) (36) which all express neuroglobin, and also in HeLa and K562 cells. Apparently the locus contains DHS in multiple cell types, which is accessible for transcription factor binding.
Figure 2.
(A) Modified screen shot of UCSC genome browser page showing DHS and ChIP-seq data of the two EcoRI fragments interacting with the NGB anchor fragment. The upstream fragment (chr14:77 806 771–77 809 773 (hg19)) contains Element I used for luciferase assays. (B) The downstream fragment (chr14:77 635 927–77 639 784 (hg19)) contains Element II. The elements used for luciferase assays are outlined by rectangular boxes. (C and D) Relative luciferase activities of the constructs in SH-SY5Y and HeLa cells. On the left, structures of the constructs used for luciferase assays are shown. The white boxes represent Element I (C) and Element II (D) respectively. The arrows in the box indicate the orientation of the inserted element. All the data are normalized to the promoter-only construct. ***P < 0.001 compared to promoter-only construct, #P < 0.05 between groups (N = 6, mean ± SEM).
ChIP-seq data showed that there are a number of TFs binding at the ∼500 bp region in the −70 kb fragment. These TFs are FOSL2, GATA-3, JunD and NFIC in SK-N-SH cells (the parental cell line of SH-SY5Y), FOSL1, GATA-2, Pol2-4H8, NR2F2, NRSF, TEAD4, c-Jun, JunD, CoREST, MAZ, p300 and TAL1 in K562 cells, and c-Myc, CoREST, JunD and MAZ in HeLa S3 cells (a clone of HeLa cells) (Middle panel, Figure 2A).
Around the same region, enrichment of H3K4me1 was observed but H3K4me3 was absent in K562, HeLa and NH-A cells (Bottom panel, Figure 2A). These data suggest that the 500bp site is a potential active distal regulatory element as predicted by the presence of TF binding and histone marks. This fragment was termed Element I.
The ENCODE database revealed that at the +100 kb region downstream of the NGB gene TSS, DHS were found in HeLa and Medullo (a medulloblastoma cell line) cells at different sites (Top panel, Figure 2B), but not in K562 cells. However, ChIP-seq data showed that the presence of TFs CBX3, TRIM28 and Z143 binding sites in K562 cells approximately at the position of the DHS found in Medullo cells (lower panel, Figure 2B). The database showed no TF binding on the fragment in HeLa and Medullo cells. In all these three cell lines, neither H3K4me1, H3K4me3 nor H3K27Ac enrichment were observed (data not shown). This fragment was termed Element II.
In conclusion, analysis of these regions revealed potentially active regulatory elements within the −70 kb and +100 kb fragments. Furthermore, ChIP-seq data showed the presence of multiple TF binding sites in the −70 kb region. These elements thus served as the focus of our subsequent studies.
Activating functions of distal elements probed by luciferase assays
Luciferase reporter construct assays encompassing the two elements at −70 kb (Element I) and +100 kb (Element II) of the NGB gene were employed to delineate the functions of the two elements. These constructs harbor Element I or II inserted into p(−745/+306) (see Materials and Methods section) at different positions and orientations and were transfected into SH-SY5Y and HeLa cells.
Results displayed in Figure 2C show that Element I has an activating function which is cell type-, position- and orientation-dependent, while Element II has no significant effect on luciferase activities (in Figure 2D).
In SH-SY5Y cells, when Element I was inserted upstream of the human NGB gene promoter, luciferase activity increased about 6-fold compared to the promoter-only construct. Interestingly, when Element I was inserted downstream of the luciferase gene or flipped in the opposite orientation, the increase of luciferase activity dropped significantly to about 3.5-fold compared to the promoter-only construct. Furthermore, when Element I was inserted downstream of the luciferase gene and in the opposite orientation, there was no significant difference compared to the promoter-only construct. In HeLa cells, all the constructs showed no significant effects on the NGB promoter. Thus we conclude that Element I harbors a cell type-specific enhancer function for the NGB gene.
Transcription factor binding on Element I detected by chromatin immunoprecipitation (ChIP)
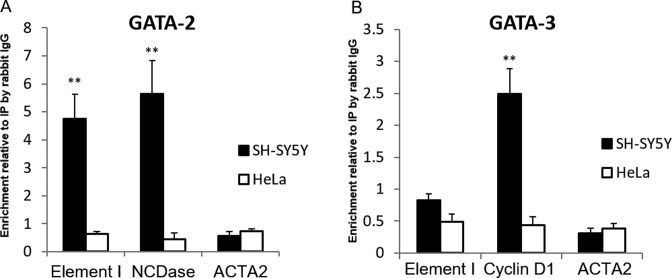
To identify potential TFs which bind to Element I in a cell type-specific manner, the sequence of Element I was analyzed using MatInspector (37). Among the potential TFs listed, there were multiple predicted sites for GATA binding proteins (GATA) (the complete list of potential TFs binding sites of Element I is shown in Supplementary Table S2). Since GATA-2 and -3 have been reported to be involved in neuronal development (38–40) and in activation of distal enhancers (41–43), ChIP was employed to investigate whether GATA-2 and/or GATA-3 were able to bind to Element I in vivo as predicted in silico.
Enrichment of GATA-2 at Element I was confirmed. The ChIP results showed that cell-type specific binding of GATA-2 at Element I was comparable to that at the promoter of neutral ceramidase (ASAH2) gene (Figure 3A). They were respectively 4.8-fold and 5.7-fold increase compared to the signal given by control immunoprecipitation (IP) with rabbit IgG. The binding of GATA-2 at the ASAH2 gene in undifferentiated SH-SY5Y cells has been demonstrated (44), thus ASAH2 served as the positive control for GATA-2 binding in this experiment. The promoter region of the ACTA2 gene where no GATA factor binding was shown, served as the negative control of both GATA factors. In HeLa cells, the binding of GATA-2 at all the three areas tested waw weak and showed no significant difference with the non-specific signal.
Figure 3.
GATA factors binding revealed by ChIP. The enrichments of the target proteins at specified sites are expressed relative to that of non-specific immunoprecipitated (IP) DNA by rabbit IgG. The binding of GATA-2 (A) but not GATA-3 (B) was observed in SH-SY5Y cells. *P < 0.05, **P < 0.01 for comparing enrichment to the signal obtained with rabbit IgG IP (N = 5, mean ± SEM).
GATA-3 binding was not detected at Element I in SH-SY5Y and HeLa cells (Figure 3B). The signals were similar to the non-specific signal. The experiment was validated by the high enrichment of GATA-3 at the cyclin D1 gene promoter in SH-SY5Y neuroblastoma cells (2.5-fold of non-specific signal). Cyclin D1 was shown to be a downstream target of GATA-3 and is abundantly expressed in neuroblastoma cells (45). Therefore, we concluded that Element I is bound by GATA-2 but not GATA-3 in SH-SY5Y cells, and not by either GATA factor in HeLa cells.
Decrease of NGB expression upon GATA-2 knockdown
To determine the role of GATA factors in NGB gene expression, shRNA was employed to reduce the expression of GATA-2 and -3. The ChIP results have shown the binding of GATA-2 to Element I in vivo. This strongly suggested the potential involvement of GATA-2 in the function of Element I as a human NGB gene enhancer. Although no binding of GATA-3 to Element I was found, GATA-3 is closely associated with GATA-2 in gene regulation, and they are able to inter-regulate. GATA-3 knockdown was therefore also performed to better elucidate the regulatory network of NGB gene expression.
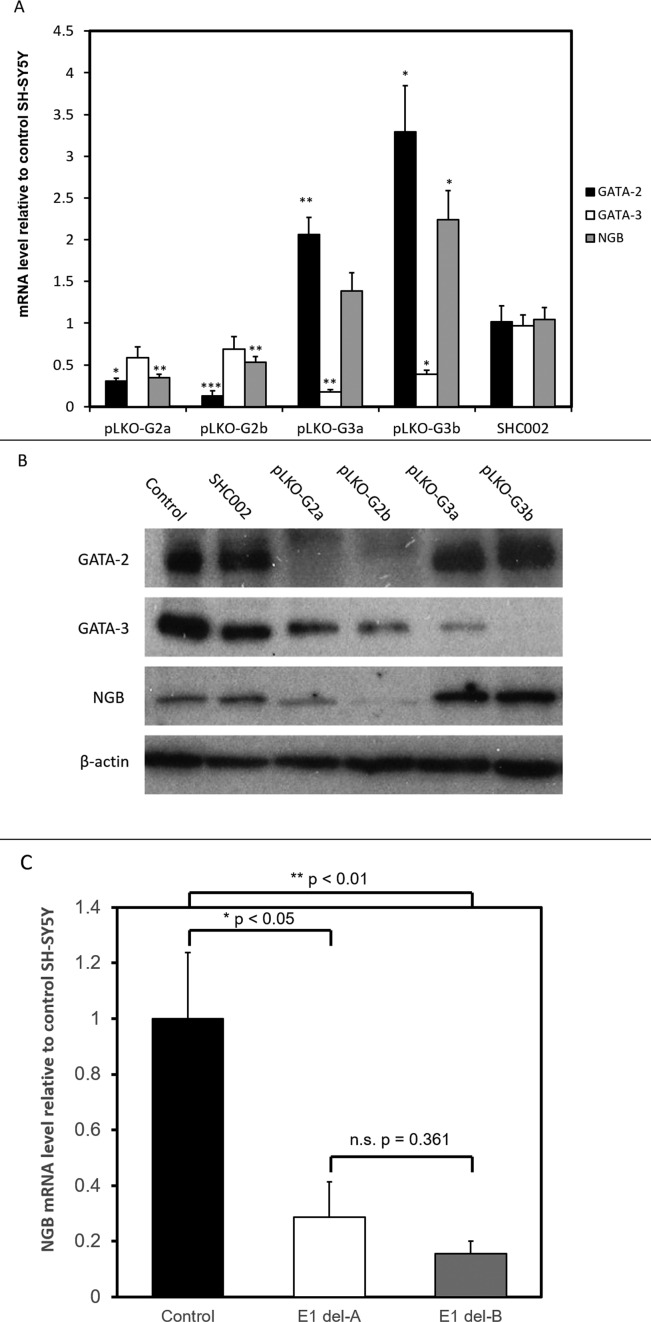
To ensure that effects on changes in gene expression of the lentivirus infected cells were independent of the infection and associated reactions, a non-targeting shRNA targeting (SHC002) was used as a control. The results showed that the GATA-2, GATA-3 and NGB levels were not changed by infection of SH-SY5Y cells with the SHC002 virus (Figure 4A).
Figure 4.
(A) The mRNA level of GATA-2, GATA-3 and NGB upon GATA factor knockdown. cDNA was synthesized from total RNA extracted from untreated SH-SY5Y cells (Control), and cells infected with control shRNA vector (SHC002) or GATA-2 (pLKO-G2a and pLKO-G2b) or GATA-3 (pLKO-G3a and pLKO-G3b) targeting shRNA constructs. The amount of mRNA is normalized to β-actin level, and expressed relative to the level in control SH-SY5Y cells. *P < 0.05, **P < 0.01, ***P < 0.001 compared to SHC002 SH-SY5Y cells (N = 3, mean ± SEM). (B) Western blot analyses of GATA-2 and -3 knockdown SH-SY5Y cells. Proteins were extracted from untreated SH-SY5Y cells (Control), and cells infected with control shRNA vector (SHC002) or GATA-2 (pLKO-G2a and pLKO-G2b) or GATA-3 (pLKO-G3a and pLKO-G3b) targeting shRNA constructs. Bands shown are from three different membranes (one for control and SHC002, one for pLKO-G2a and b, and one for pLKO-G3a and b). The amount of protein loaded and exposure times were kept constant for each membrane. (C) mRNA level of NGB in SH-SY5Y cells with and without Element I deletion. Del-A and -B are cells with Element I deleted by different gRNA pairs. The levels are normalized to β-actin level and expressed relative to the level in control SH-SY5Y cells. *P < 0.05, **P < 0.01 compared to control SH-SY5Y cells (N = 3, mean ± SEM).
The two GATA-2 targeting shRNAs (pLKO-G2a and -G2b) effectively decreased the GATA-2 mRNA levels to 30% and 13% of the control level. Interestingly, the NGB levels of the GATA-2 knockdown cells were also lowered to 35% and 53% of the control level (Figure 4A). This clearly demonstrated the correlation between GATA-2 and NGB expression in SH-SY5Y cells. GATA-3 levels in the GATA-2 knockdown cells also decreased to 58% and 69% of the control level, although the change was not statistically significant. The similar changes in gene expression pattern observed with the two different GATA-2 targeting shRNAs implies that these were not caused by off-target effects of the shRNAs (Figure 5).
Figure 5.
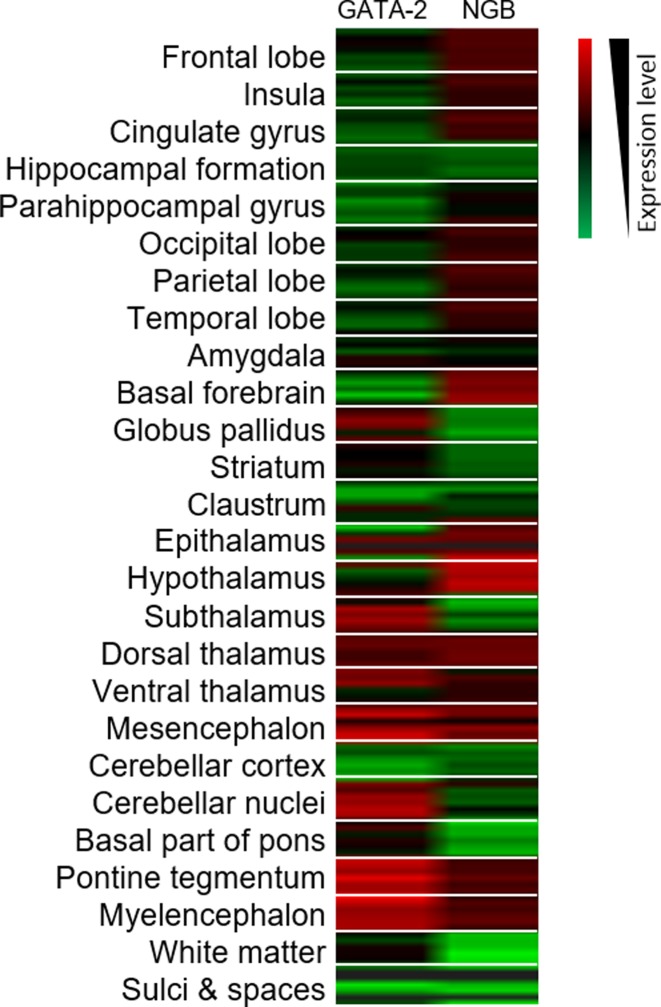
Microarray analysis of GATA-2 and NGB expression level in different parts of the brain (Image credit: Allen Institute (53)). Associated expression of the two genes is shown in dorsal and ventral thalamus, mesencephalon, pontine tegmentum and myelencephalon. Red and green on the heat map represent high and low expression respectively. For each structure, six samples are displayed.
Similarly, the two GATA-3 targeting shRNAs (pLKO-G3a and -G3b) significantly lowered GATA-3 mRNA levels to about 17% and 39% of the control level. Surprisingly, the drop in GATA-3 level was associated with an increase of GATA-2 and NGB expression. GATA-2 expression increased to 206% and 329% of the control level. NGB expression increased to 139% in pLKO-G3a infected cells and about 224% in pLKO-G3b infected cells. These associations are likely caused by the cross-regulation of GATA-3 and GATA-2, but the exact mechanism is still unknown.
In Figure 4B, the protein levels of GATA-2, GATA-3, NGB and β-actin are shown. GATA-2 expression dropped dramatically in SH-SY5Y cells infected with pLKO-G2a and pLKO-G2b. We noted that the two shRNAs also mildly decreased the expression of GATA-3. For pLKO-G3a and pLKO-G3b infected cells, GATA-3 expression was suppressed while the GATA-2 level was mildly raised. The control shRNA SHC002 showed no effect on all the proteins tested. Interestingly, the NGB protein level was decreased by GATA-2 knockdown, whilst increased by GATA-3 knockdown. The results imply the importance of GATA-2 in activating NGB expression.
Deletion of element I reduces NGB gene expression
The regulatory roles of Element I and GATA-2 were demonstrated by the luciferase reporter and knockdown experiments respectively. To further illustrate the importance of Element I in inducing expression of the NGB gene, Element I was deleted from SH-SY5Y cells by the CRISPR/Cas9 system. Element I was targeted by two different pairs of gRNAs; both gRNA pairs efficiently deleted the region overlapping Element I (Supplementary Figure S3A). We found that deletion of Element I significantly decreased NGB expression levels to 13–25% of those observed in control SH-SY5Y cells (Figure 4C). The expression levels of the intervening genes TMED8 and GSTZ1 were also decreased but to a lesser extent than the reduction observed for the NGB gene (Supplementary Figure S4). Collectively, these results provide strong evidence for the role of Element I in regulating NGB gene expression.
DISCUSSION
Extensive studies have revealed the structure, evolution, ligand binding properties and diverse functions of NGB. To date, little is known about the control of transcription of the NGB gene and its possible long-range regulatory elements. The Locus Control Region (LCR) of the HBB gene cluster comprises distal regulatory elements that activate the linked β-like globin genes by looping and interacting with their promoters to form an Active Chromatin Hub (ACH) (10,46,47). Recently, Dekker et al. (11) showed that a 3C-based approach could identify novel distal regulatory elements of the cystic fibrosis transmembrane conductance regulator gene (CFTR).
Here, with a similar approach we identified two novel elements within the 300 kb region around the locus of the NGB gene. The results of the 3C experiments demonstrated that these elements are located at around −70 kb upstream and +100 kb downstream of the NGB gene TSS. Thus, we identified the presence of potential long-range regulatory elements of the human NGB gene.
The DRE located −70 kb upstream of the NGB gene (Element I) specifically interacts with and transactivates the NGB gene promoter in neuronal cells. An active chromatin loop is likely formed between Element I and the NGB promoter upon neuronal differentiation which drives the expression of NGB gene. The interaction frequency of such looping is higher in SH-SY5Y cells than in HeLa cells (Figure 1B and C). It has been suggested that the interaction frequency may represent the ‘strength’ of the looping (48), implying Element I is fully functional in activating NGB transcription with the ‘strong’ looping observed in SH-SY5Y cells.
The GATA-2 transcription factor was shown to bind to Element I. GATA factors are known to be important in establishing and maintaining chromatin looping (48–51). Such looping can be transcriptionally activating or repressive, as demonstrated by the interplay of GATA-1 and -2 in the regulation of the KIT gene. The binding and function of the GATA factors are cell differentiation status dependent (49) and locus dependent (48). Our results demonstrated that GATA-2 knockdown was associated with suppression of NGB gene expression. Developmental studies suggest that both GATA-2 (40) and NGB (52) are expressed early in the course of neurogenesis. Searches in the Allen Brain Atlas (53) (http://human.brain-map.org) revealed that NGB and GATA-2 co-express in parts of the brain including the dorsal and ventral thalamus, mesencephalon, pontine tegmentum and myelencephalon. The expression level of the two genes are not always associated. This implies additional factors are responsible for the control of NGB gene expression. In retina, GATA-2 is expressed and governs VEGF receptor 2 expression (54). VEGF has been found to stimulate NGB expression (55). Thus, the expression of GATA-2 may have an additive effect on upregulating NGB expression by (a) stabilizing chromatin looping between Element I and the NGB promoter and (b) enhancing the effects of VEGF by increasing VEGF receptors on the cells. This may explain why the highest level of NGB is found in the retina.
Knockdown of GATA-2 caused a reduction in in NGB and GATA-3 expression. In contrast, GATA-3 knockdown led to an increase in NGB and GATA-2 expression. This phenomenon could be attributed to the genetic interactions between GATA-2 and GATA-3 during central nervous system development, as GATA-3 expression is GATA-2-dependent (40). In trophoblast cells, GATA-3 acts as a negative regulator of the GATA-2 gene by occupying distal regulatory sites. Upon trophoblast differentiation, GATA-3 is displaced by GATA-2 and GATA-2 gene transcription is activated (56). In SH-SY5Y cells with GATA-3 knockdown, the suppression was probably relieved thus GATA-2 expression increased. Therefore, there may be two possible ways by which GATA-3 regulates NGB transcription: a) GATA-3 may compete with GATA-2 for the GATA sites within Element I to affect the function of the chromatin loop; b) Expression of GATA-3 limits the level of GATA-2 which is the trans activator for NGB gene transcription.
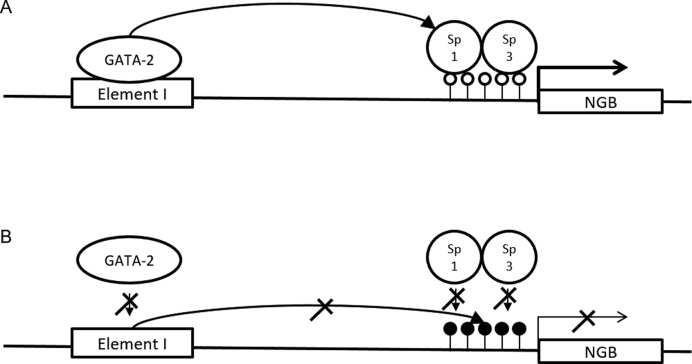
Interestingly, GATA-2 has been shown to cooperate with Sp1 to activate gene expression (57–59). Zhang et al. (12) previously demonstrated that Sp1 and Sp3 bind to and regulate the promoter of human NGB gene. Overexpression of Sp1 and/or Sp3 in SH-SY5Y cells significantly increased NGB promoter activity. The study also identified the role of DNA methylation in controlling the expression of NGB. Taken together, we propose that GATA-2 bound to the distal Element I may be brought to close proximity of the NGB promoter through interaction with Sp1 bound on the promoter to transactivate the transcriptional activation of NGB (Figure 6A). In cells that do not express NGB, the promoter is hypermethylated (data not shown). This may block the binding of Sp factors and prevent the GATA factors from the distal element from interacting productively with the promoter of the NGB gene (Figure 6B).
Figure 6.
Schematic diagrams illustrating the function of Element I in NGB gene transcription regulation. (A) In NGB-expressing cells, a chromatin loop is formed between the NGB promoter and Element I. The loop brings GATA-2 on Element I and Sp1 on the hypomethylated NGB promoter (open circle) in close proximity and activates transcription. (B) In cells not expressing NGB, the promoter of NGB gene is hypermethylated (filled circles) and binding of Sp1 and Sp3 is inhibited. This impairs the interaction between the promoter and Element I. The formation of looping may also be affected by competition between GATA-2 and -3 for the binding sites within Element I.
In addition to GATA factors, other TFs shown in the ENCODE database may contribute to the differences of the interaction frequencies between Element I and NGB gene. These include Corepressor for element-1-silencing transcription factor (CoREST) present in Element I in K562 and HeLa S3 and Neuron-restrictive silencer factor (NRSF) detected in K562. NRSF and CoREST are only expressed in non-neuronal cells and are involved in chromatin remodeling, in particular with respect to silencing of neuronal gene expression (60,61). These TFs might modify the chromatin structure around Element I in HeLa and other non-NGB expressing cells to a state which hinders the formation of an activating chromatin loop, thus decreasing the interaction frequency between the distal Element I and the NGB gene promoter. This provides an explanation for the suppression of NGB expression in K562 and Hela cells.
The luciferase reporter assays revealed the cell type-specific enhancer function of Element I in SH-SY5Y cells. However, the enhancer function was surprisingly position- and orientation-dependent, which deviates from the classical definition of enhancers which are elements activating transcription in a position- and orientation-independent manner. Although orientation-dependent enhancers are rare, they have been previously reported such as the enhancers of CCL2 (62), PROP1 (63), Proα1(I) collagen (64) and Shsp/αB-crytallin (65). The orientation-dependence phenomenon was also observed in the HBB LCR. It was shown that the expression of the HBB gene was reduced when the orientation of the LCR was changed or the order of the genes in the gene cluster was altered (66). Thus to achieve the copy number- and position-independent expression of the HBB gene, the intact gene and the LCR must be integrated into the genome. The mechanism behind the orientation dependence is unclear. It was suggested that the orientation independence of enhancers would only be achieved when all the necessary transcription factor binding sites are present in the right order and location so that the transcription factors can be assembled to confer the activating function in full (5,67). Thus the orientation- and position-independence are only valid when viewing the enhancer on the whole. The enhancers found to be orientation- and position-dependent may lack some of the transcription factor binding sites. A more extensive analysis which covers larger regions surrounding Elements I and II will be required to address this issue.
The role of Element I in activating NGB expression was supported by the CRISPR-mediated deletion. However, we found that the expression of the intervening genes TMED8, GSTZ1 and POMT2 were also affected. Since our 3C data showed that Element I also interacts with the promoter regions of the intervening genes, it is reasonable to assume that Element I controls more than one gene. The effect of Element I deletion on expression of the intervening genes was the greatest on TMED8, of which expression was decreased to 35–66% of the wild type level. Since the region deleted was within an intron of this gene, deletion of Element I may affect pre-mRNA stability and/or splicing of TMED8 RNA. Remarkably, expression of the NGB gene was more strongly reduced upon deletion of Element I. It has been observed that enhancers are most likely to activate the nearest gene (1). Despite the closer proximity of the TMED8, GSTZ1 and POMT2 promoters to Element I, our data indicate that the interactions between Element I and the NGB promoter are favored in neuronal SH-SY5Y cells. We surmise that these favorable interactions are dependent on the particular combination of transcription factors bound at Element I and the NGB promoter in SH-SY5Y cells.
In conclusion, our data show that the -70kb Element I interacts via a looping mechanism with the NGB promoter to activate and confer cell-specific NGB gene expression. GATA-2 is one of the transcription factors governing the activation of NGB gene expression. Such information will contribute to understanding the physiological and pathological roles of NGB, and to further explore its potential therapeutic and disease-preventing properties in neurological conditions such as Alzheimer's disease.
Acknowledgments
K.C.T.U. would like to acknowledge Professor R.S.S. Wu (The University of Hong Kong) for his support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
University of Hong Kong (to K.T.T., W.Z., P.P.L., Z.P.T., G.C.F.C., K.C.T.U.); Imperial College London and Medical Research Council, UK (to P.P.L., P.K.C., R.F.); School of Professional and Continuing Education, The University of Hong Kong (to Z.P.T., K.C.T.U.); Landsteiner Foundation for Blood Transfusion Research [LSBR 1040]; Netherlands Organization for Scientific Research [NWO/ZonMw TOP 40-00812-98-12128]; EU fp7 Specific Cooperation Research Project THALAMOSS [306201 to SP]. Funding for open access charge: HKU School of Professional and Continuing Education, The University of Hong Kong.
Conflict of interest statement. None declared.
REFERENCES
- 1.Heintzman N.D., Hon G.C., Hawkins R.D., Kheradpour P., Stark A., Harp L.F., Ye Z., Lee L.K., Stuart R.K., Ching C.W. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forrester W., Epner E., Driscoll M., Enver T., Brice M., Papayannopoulou T., Groudine M. A deletion of the human beta-globin locus activation region causes a major alteration in chromatin structure and replication across the entire beta-globin locus. Genes Dev. 1990;4:1637–1649. doi: 10.1101/gad.4.10.1637. [DOI] [PubMed] [Google Scholar]
- 3.Driscoll M.C., Dobkin C.S., Alter B.P. Gamma delta beta-thalassemia due to a de novo mutation deleting the 5′beta-globin gene activation-region hypersensitive sites. Proc. Natl. Acad. Sci. U.S.A. 1989;86:7470–7474. doi: 10.1073/pnas.86.19.7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berman B.P., Nibu Y., Pfeiffer B.D., Tomancak P., Celniker S.E., Levine M., Rubin G.M., Eisen M.B. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. U.S.A. 2002;99:757–762. doi: 10.1073/pnas.231608898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maston G.A., Evans S.K., Green M.R. Transcriptional regulatory elements in the human genome. Annu. Rev. Genomics Hum. Genet. 2006;7:29–59. doi: 10.1146/annurev.genom.7.080505.115623. [DOI] [PubMed] [Google Scholar]
- 6.Kleinjan D.A., Van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dekker J. Gene regulation in the third dimension. Sci. Signal. 2008;319:1793. doi: 10.1126/science.1152850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miele A., Dekker J. Long-range chromosomal interactions and gene regulation. Mol. bioSyst. 2008;4:1046–1057. doi: 10.1039/b803580f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vilar J.M.G., Saiz L. DNA looping in gene regulation: from the assembly of macromolecular complexes to the control of transcriptional noise. Curr. Opin. Genet. Dev. 2005;15:136–144. doi: 10.1016/j.gde.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 11.Gheldof N., Smith E.M., Tabuchi T.M., Koch C.M., Dunham I., Stamatoyannopoulos J.A., Dekker J. Cell-type-specific long-range looping interactions identify distant regulatory elements of the CFTR gene. Nucleic Acids Res. 2010;38:4325–4336. doi: 10.1093/nar/gkq175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang W., Tian Z., Sha S., Cheng L.Y.L., Philipsen S., Tan-Un K.C. Functional and sequence analysis of human neuroglobin gene promoter region. Biochim. Biophys. Acta. 2011;1809:236–244. doi: 10.1016/j.bbagrm.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Szymanski M., Wang R., Fallin M.D., Bassett S.S., Avramopoulos D. Neuroglobin and Alzheimer's dementia: Genetic association and gene expression changes. Neurobiol. Aging. 2010;31:1835–1842. doi: 10.1016/j.neurobiolaging.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pesce A., Bolognesi M., Bocedi A., Ascenzi P., Dewilde S., Moens L., Hankeln T., Burmester T. Neuroglobin and cytoglobin. EMBO Rep. 2002;3:1146–1151. doi: 10.1093/embo-reports/kvf248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burmester T., Hankeln T. Neuroglobin: a respiratory protein of the nervous system. Physiology. 2004;19:110–113. doi: 10.1152/nips.01513.2003. [DOI] [PubMed] [Google Scholar]
- 16.Hankeln T., Ebner B., Fuchs C., Gerlach F., Haberkamp M., Laufs T.L., Roesner A., Schmidt M., Weich B., Wystub S., et al. Neuroglobin and cytoglobin in search of their role in the vertebrate globin family. J. Inorg. Biochem. 2005;99:110–119. doi: 10.1016/j.jinorgbio.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 17.Greenberg D.A., Jin K., Khan A.A. Neuroglobin: an endogenous neuroprotectant. Curr. Opin. Pharmacol. 2008;8:20–24. doi: 10.1016/j.coph.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burmester T., Hankeln T. What is the function of neuroglobin. J. Exp. Biol. 2009;212:1423–1428. doi: 10.1242/jeb.000729. [DOI] [PubMed] [Google Scholar]
- 19.Brittain T., Skommer J., Raychaudhuri S., Birch N. An antiapoptotic neuroprotective role for neuroglobin. Int. J. Mol. Sci. 2010;11:2306–2321. doi: 10.3390/ijms11062306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burmester T., Weich B., Reinhardt S., Hankeln T. A vertebrate globin expressed in the brain. Nature. 2000;407:520–523. doi: 10.1038/35035093. [DOI] [PubMed] [Google Scholar]
- 21.Reuss S., Saaler-Reinhardt S., Weich B., Wystub S., Reuss M.H., Burmester T., Hankeln T. Expression analysis of neuroglobin mRNA in rodent tissues. Neuroscience. 2002;115:645–656. doi: 10.1016/s0306-4522(02)00536-5. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt M., Giessl A., Laufs T., Hankeln T., Wolfrum U., Burmester T. How Does the Eye Breathe? J. Biol. Chem. 2003;278:1932–1935. doi: 10.1074/jbc.M209909200. [DOI] [PubMed] [Google Scholar]
- 23.Hümmler N., Schneider C., Giessl A., Bauer R., Walkinshaw G., Gassmann M., Rascher W., Trollmann R. Acute hypoxia modifies regulation of neuroglobin in the neonatal mouse brain. Exp. Neurol. 2012;236:112–121. doi: 10.1016/j.expneurol.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 24.Sun Y., Jin K., Mao X.O., Xie L., Peel A., Childs J.T., Logvinova A., Wang X., Greenberg D.A. Effect of aging on neuroglobin expression in rodent brain. Neurobiol. Aging. 2005;26:275–278. doi: 10.1016/j.neurobiolaging.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Liu N., Yu Z., Xiang S., Zhao S., Tjärnlund-Wolf A., Xing C., Zhang J., Wang X. Transcriptional regulation mechanisms of hypoxia-induced neuroglobin gene expression. Biochem. J. 2012;443:153–164. doi: 10.1042/BJ20111856. [DOI] [PubMed] [Google Scholar]
- 26.Liu N., Yu Z., Li Y., Yuan J., Zhang J., Xiang S., Wang X. Transcriptional regulation of mouse Neuroglobin gene by cyclic AMP responsive element binding protein (CREB) in N2a cells. Neurosci. Lett. 2012;534:333–337. doi: 10.1016/j.neulet.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haines B., Demaria M., Mao X., Xie L., Campisi J., Jin K., Greenberg D.A. Hypoxia-inducible factor-1 and neuroglobin expression. Neurosci. Lett. 2012;514:137–140. doi: 10.1016/j.neulet.2012.01.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hagège H., Klous P., Braem C., Splinter E., Dekker J., Cathala G., De Laat W., Forné T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat. Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 29.Dekker J. The three'C's of chromosome conformation capture: controls, controls, controls. Nat. Methods. 2005;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- 30.Hadjur S., Williams L.M., Ryan N.K., Cobb B.S., Sexton T., Fraser P., Fisher A.G., Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Don R.H., Cox P.T., Wainwright B.J., Baker K., Mattick J.S. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson J.D., Denisenko O., Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 34.Bauer D.E., Canver M.C., Orkin S.H. Generation of genomic deletions in mammalian cell lines via CRISPR/Cas9. J. Visual. Exp. 2015;95:e52118. doi: 10.3791/52118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ostojić J., Sakaguchi D.S., de Lathouder Y., Hargrove M.S., Trent J.T., Kwon Y.H., Kardon R.H., Kuehn M.H., Betts D.M., Grozdanić S. Neuroglobin and cytoglobin: oxygen-binding proteins in retinal neurons. Invest. Ophthalmol. Vis. Sci. 2006;47:1016–1023. doi: 10.1167/iovs.05-0465. [DOI] [PubMed] [Google Scholar]
- 36.Chen X.Q., Qin L.Y., Zhang C.G., Yang L.T., Gao Z., Liu S., Lau L.T., Fung Y.W.W., Greenberg D.A., Yu A.C.H. Presence of neuroglobin in cultured astrocytes. Glia. 2005;50:182–186. doi: 10.1002/glia.20147. [DOI] [PubMed] [Google Scholar]
- 37.Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 38.Zhou Y., Yamamoto M., Engel J.D. GATA2 is required for the generation of V2 interneurons. Development. 2000;127:3829–3838. doi: 10.1242/dev.127.17.3829. [DOI] [PubMed] [Google Scholar]
- 39.Sinclair A., Göttgens B., Barton L., Stanley M., Pardanaud L., Klaine M., Gering M., Bahn S., Sanchez M.-J., Bench A. Distinct 5′ SCL enhancers direct transcription to developing brain, spinal cord, and endothelium: neural expression is mediated by GATA factor binding sites. Dev. Biol. 1999;209:128–142. doi: 10.1006/dbio.1999.9236. [DOI] [PubMed] [Google Scholar]
- 40.Nardelli J., Thiesson D., Fujiwara Y., Tsai F.-Y., Orkin S.H. Expression and genetic interaction of transcription factors GATA-2 and GATA-3 during development of the mouse central nervous system. Dev. Biol. 1999;210:305–321. doi: 10.1006/dbio.1999.9278. [DOI] [PubMed] [Google Scholar]
- 41.Lawson M.A., Whyte D.B., Mellon P.L. GATA factors are essential for activity of the neuron-specific enhancer of the gonadotropin-releasing hormone gene. Mol. Cell. Biol. 1996;16:3596–3605. doi: 10.1128/mcb.16.7.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perez-Stable C.M., Pozas A.d.l., Roos B.A. A role for GATA transcription factors in the androgen regulation of the prostate-specific antigen gene enhancer. Mol. Cell. Endocrinol. 2000;167:43–53. doi: 10.1016/s0303-7207(00)00300-2. [DOI] [PubMed] [Google Scholar]
- 43.Göttgens B., Nastos A., Kinston S., Piltz S., Delabesse E.C., Stanley M., Sanchez M.-J., Ciau-Uitz A., Patient R., Green A.R. Establishing the transcriptional programme for blood: the SCL stem cell enhancer is regulated by a multiprotein complex containing Ets and GATA factors. EMBO J. 2002;21:3039–3050. doi: 10.1093/emboj/cdf286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka K., Tamiya-Koizumi K., Hagiwara K., Ito H., Takagi A., Kojima T., Suzuki M., Iwaki S., Fujii S., Nakamura M. Role of down-regulated neutral ceramidase during all-trans retinoic acid-induced neuronal differentiation in SH-SY5Y neuroblastoma cells. J. Biochem. 2012;151:611–620. doi: 10.1093/jb/mvs033. [DOI] [PubMed] [Google Scholar]
- 45.Molenaar J.J., Ebus M.E., Koster J., Santo E., Geerts D., Versteeg R., Caron H.N. Cyclin D1 is a direct transcriptional target of GATA3 in neuroblastoma tumor cells. Oncogene. 2010;29:2739–2745. doi: 10.1038/onc.2010.21. [DOI] [PubMed] [Google Scholar]
- 46.Palstra R.-J., Tolhuis B., Splinter E., Nijmeijer R., Grosveld F., de Laat W. The β-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- 47.de Laat W., Grosveld F. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 2003;11:447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- 48.Sexton T., Bantignies F., Cavalli G. Seminars in Cell & Developmental Biology. Vol. 20. Elsevier; 2009. pp. 849–855. [DOI] [PubMed] [Google Scholar]
- 49.Jing H., Vakoc C.R., Ying L., Mandat S., Wang H., Zheng X., Blobel G.A. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol. Cell. 2008;29:232–242. doi: 10.1016/j.molcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spilianakis C.G., Flavell R.A. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat. Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- 51.Vakoc C.R., Letting D.L., Gheldof N., Sawado T., Bender M., Groudine M., Weiss M.J., Dekker J., Blobel G.A. Proximity among distant regulatory elements at the β-globin locus requires GATA-1 and FOG-1. Mol. Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 52.Haines B., Mao X., Xie L., Spusta S., Zeng X., Jin K., Greenberg D.A. Neuroglobin expression in neurogenesis. Neurosci. Lett. 2013;549:3–6. doi: 10.1016/j.neulet.2013.04.039. [DOI] [PubMed] [Google Scholar]
- 53.Hawrylycz M.J., Lein E.S., Guillozet-Bongaarts A.L., Shen E.H., Ng L., Miller J.A., van de Lagemaat L.N., Smith K.A., Ebbert A., Riley Z.L. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mammoto A., Connor K.M., Mammoto T., Yung C.W., Huh D., Aderman C.M., Mostoslavsky G., Smith L.E., Ingber D.E. A mechanosensitive transcriptional mechanism that controls angiogenesis. Nature. 2009;457:1103–1108. doi: 10.1038/nature07765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin K., Mao X., Xie L., Greenberg D.A. Interactions between vascular endothelial growth factor and neuroglobin. Neurosci. Lett. 2012;519:47–50. doi: 10.1016/j.neulet.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ray S., Dutta D., Rumi M.K., Kent L.N., Soares M.J., Paul S. Context-dependent function of regulatory elements and a switch in chromatin occupancy between GATA3 and GATA2 regulate Gata2 transcription during trophoblast differentiation. J. Biol. Chem. 2009;284:4978–4988. doi: 10.1074/jbc.M807329200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maeda K., Nishiyama C., Ogawa H., Okumura K. GATA2 and Sp1 positively regulate the c-kit promoter in mast cells. J. Immunol. 2010;185:4252–4260. doi: 10.4049/jimmunol.1001228. [DOI] [PubMed] [Google Scholar]
- 58.Cowan P.J., Tsang D., Pedic C.M., Abbott L.R., Shinkel T.A., d'Apice A.J., Pearse M.J. The human ICAM-2 promoter is endothelial cell-specific in vitro and in vivo and contains critical Sp1 and GATA binding sites. J. Biol. Chem. 1998;273:11737–11744. doi: 10.1074/jbc.273.19.11737. [DOI] [PubMed] [Google Scholar]
- 59.Zhang R., Min W., Sessa W.C. Functional analysis of the human endothelial nitric oxide synthase promoter Sp1 and GATA factors are necessary for basal transcription in endothelial cells. J. Biol. Chem. 1995;270:15320–15326. doi: 10.1074/jbc.270.25.15320. [DOI] [PubMed] [Google Scholar]
- 60.Lee M.G., Wynder C., Cooch N., Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 61.Juliandi B., Abematsu M., Nakashima K. Chromatin remodeling in neural stem cell differentiation. Curr. Opin. Neurobiol. 2010;20:408–415. doi: 10.1016/j.conb.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 62.Bonello G.B., Pham M.-H., Begum K., Sigala J., Sataranatarajan K., Mummidi S. An Evolutionarily Conserved TNF-α–Responsive Enhancer in the Far Upstream Region of Human CCL2 Locus Influences Its Gene Expression. J. Immunol. 2011;186:7025–7038. doi: 10.4049/jimmunol.0900643. [DOI] [PubMed] [Google Scholar]
- 63.Ward R.D., Davis S.W., Cho M., Esposito C., Lyons R.H., Cheng J.-F., Rubin E.M., Rhodes S.J., Raetzman L.T., Smith T.P. Comparative genomics reveals functional transcriptional control sequences in the Prop1 gene. Mamm. Genome. 2007;18:521–537. doi: 10.1007/s00335-007-9008-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Katai H., Stephenson J.D., Simkevich C.P., Thompson J.P., Raghow R. An AP-1-like motif in the first intron of human proα1 (I) collagen gene is a critical determinant of its transcriptional activity. Mol. Cell. Biochem. 1992;116:119–129. doi: 10.1007/BF00299391. [DOI] [PubMed] [Google Scholar]
- 65.Swamynathan S.K., Piatigorsky J. Orientation-dependent influence of an intergenic enhancer on the promoter activity of the divergently transcribed mouse Shsp/αB-crystallin andMkbp/HspB2 genes. J. Biol. Chem. 2002;277:49700–49706. doi: 10.1074/jbc.M209700200. [DOI] [PubMed] [Google Scholar]
- 66.Tanimoto K., Liu Q., Bungert J., Engel J.D. Effects of altered gene order or orientation of the locus control region on human β-globin gene expression in mice. Nature. 1999;398:344–348. doi: 10.1038/18698. [DOI] [PubMed] [Google Scholar]
- 67.Reményi A., Schöler H.R., Wilmanns M. Combinatorial control of gene expression. Nat. Struct. Mol. Biol. 2004;11:812–815. doi: 10.1038/nsmb820. [DOI] [PubMed] [Google Scholar]