Abstract
DNA replication is tightly regulated to occur once and only once per cell cycle. How chromatin, the physiological substrate of DNA replication machinery, regulates DNA replication remains largely unknown. Here we show that histone H3 lysine 9 demethylase Kdm4d regulates DNA replication in eukaryotic cells. Depletion of Kdm4d results in defects in DNA replication, which can be rescued by the expression of H3K9M, a histone H3 mutant transgene that reverses the effect of Kdm4d on H3K9 methylation. Kdm4d interacts with replication proteins, and its recruitment to DNA replication origins depends on the two pre-replicative complex components (origin recognition complex [ORC] and minichromosome maintenance [MCM] complex). Depletion of Kdm4d impairs the recruitment of Cdc45, proliferating cell nuclear antigen (PCNA), and polymerase δ, but not ORC and MCM proteins. These results demonstrate a novel mechanism by which Kdm4d regulates DNA replication by reducing the H3K9me3 level to facilitate formation of pre-initiative complex.
INTRODUCTION
DNA replication is tightly regulated to achieve precise duplication of genetic information once and only once per cell cycle. At the molecular level, the origin recognition complex (ORC) associates with replication origin to recruit Cdc6, Cdt1 and minichromosome maintenance (MCM) complex to form the pre-replicative complex (pre-RC) at the M-to-G1 phase transition. The pre-RC is activated by phosphorylation of S phase cyclin-dependent kinase, recruitment of Cdc45 and GINS complex and other accessory factors in a stepwise manner to form the pre-licensing complex, which will initiate DNA replication (1).
It is widely accepted that chromatin, the physiological substrate of DNA replication machinery, is one of the major obstacles for DNA replication (2,3). Several lines of evidence support the idea that the chromatin landscape plays an important role in the spatial and temporal regulation of DNA replication. For instance, the lightly packed euchromatin replicates at early S phase, while the more compact heterochromatin domains replicate at late S phase (4–6). Moreover, chromatin regulators such as histone modifiers play important roles for cells to overcome chromatin challenge during replication. For instance, acetylation of histone H4 surrounding the replication origins by HBO1 facilitates the assembly of pre-RC (7,8). In contrast, hypoacetylation and hypermethylation are required for maintaining the delayed replication timing of heterochromatin, which generally replicates in late S phase (9). Methylation on H3K27 that reduces the chromatin accessibility is required to inhibit over-replication of heterochromatin in Arabidopsis (10,11). On the other hand, it is noteworthy that mono-methylation of H4K20 by PR-Set7 was shown to associate with DNA replication origin (12–14). Degradation of PR-Set7 by CUL4(Cdt2) prevents re-replication (13), while knocking down of PR-Set7 significantly reduced S phase progression (15,16). Therefore, modifications on histones are important for the regulation of DNA replication.
Trimethylation of H3K9 (H3K9me3) is a histone mark associated with transcriptionally silenced chromatin (17,18). The euchromatin with a low level of H3K9me3 replicates at early S phase, while the heterochromatin domains enriched with H3K9me3 replicate at late S phase (4–6). Reducing the levels of methyltransferases for H3K9me3 or H3K9me3-binding protein heterochromatin protein 1 accelerates the replication timing of heterochromatin (19). Moreover, over-expression of KDM4A, one of the H3K9me3 demethylases, advances S phase entry (20). However, all the previous studies were focused on the relationship between H3K9me3 and replication timing. It was not known whether other members of the KDM4 family (KDM4B-D), which are known to play distinct roles in DNA damage response (21–24), have any roles in DNA replication. Here we present a previously unrecognized role of Kdm4d in DNA replication. We show that Kdm4d interacts with DNA replication proteins and is recruited to DNA replication origins at G1 and S phases in an ORC- and MCM-dependent manner. Furthermore, Kdm4d is required for the chromatin association of Cdc45, proliferating cell nuclear antigen (PCNA) and polymerases during S phase of the cell cycle, revealing a mechanism whereby DNA replication is regulated by the H3K9me3 demethylase.
MATERIALS AND METHODS
Tissue culture, primers and shRNAs
HeLa, HEK293T, HCT116, U2OS and NIH-3T3 cells were obtained from ATCC and cultured in standard conditions according to ATCC's instructions.
For cell synchronizations, HeLa cells were blocked in 1 mM mimosine (G1) or 2 mM thymidine (G1/S) for 24 h, or 50 ng/ml nocodazole (G2/M) for 18 h. To release synchronized cells from G1 arrest, cells were arrested in mimosine for 20 h before washing and releasing in fresh medium containing 50 ng/ml nocodazole. To release cells from G1/S arrest, cells were treated with 2 mM thymidine for 20 h before washing and releasing into fresh medium containing 50 ng/ml nocodazole.
All shRNAs were obtained from Sigma-Aldrich. sh-h1 (GCCAGAGAGACCTATGATAAT) and sh-h2 (CAGATTATCCACCCGTCAAAT) target human KDM4D. sh-m1 (CCACGGTAAGTAACGTTCCTT) and sh-m2 (ACACAGAGACTATGGTGTCTA) target mouse KDM4D. shORC2 (GCCGAACTTAAACAACTAAAT) and shMCM3 (CGGCAGGTATGACCAGTATAA) target human ORC2 and MCM3, respectively.
Primers used this study are listed in Supplementary Table S2.
Antibodies
Histone H3 (WB 1:5000), H3K9me1 (WB 1:1000), H3K9me3 (WB 1:1000) and Mcm3 (WB 1:2000) were generated in the laboratory and were used previously (25,26). Anti-GFP (ab6556, WB 1:10 000), Polδ (ab186407, WB 1:1000), Polϵ (ab74308, WB 1:1000), KDM4D (ab93694, WB 1:2000, IF 1:200, ChIP 4 μg per IP), Mcm6 (ab201683, ChIP 4 μg per IP), Orc2 (ab68348, WB 1:2000), anti-SLD5 (ab139683, WB 1:1000) and anti-CldU (ab6326) were purchased from Abcam; Orc5 (sc-20635, WB 1:1000), Cdc45 (sc-20685, WB 1:1000), PCNA (sc-56, WB 1:1000) and Tubulin (sc-9104, WB 1:5000) were purchased from Santa Cruz Biotechnology; H3K27me3 (C36B11, WB 1:1000) and H3S10ph (D2C8, WB 1:1000) were purchased from Cell Signaling Technology.
Fluorescein isothiocyanate (FITC)-conjugated anti-BrdU (347583) and anti-IdU (347580) antibodies were purchased from BD Biosciences; FITC goat anti-mouse, FITC goat anti-rabbit and Cy3 goat anti-rat antibodies were purchased from Jackson ImmunoResearch.
Flow cytometry and immunofluorescence microscopy
BrdU-PI flow cytometry was performed as described previously, with minor modification (27). In brief, cells were incubated with 10 μM BrdU for 20 min before being fixed with 70% ethanol, denatured in 2 N HCl for 30 min and stained with FITC-conjugated anti-BrdU antibody according to the manufacturer's instructions. Cells were further treated with 40 μg/ml RNase A and 200 μg/ml PI at room temperature for 30 min and analyzed by flow cytometry. The quantification was done by FlowJo single-cell analysis software.
Immunofluorescence microscopy was performed as described previously, with minor modification in which cells were extracted by extraction buffer (10 mM PIPES [pH 6.8], 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 50 mM NaF, 1 mM PMSF, protease inhibitor cocktail) with 0.5% Trion X-100 for 10 min on ice before paraformaldehyde fixation (26).
For 5-ethynyl-2′-deoxyuridine (EdU) staining, cells were pulsed with 10 μM EdU for 20 min before being fixed in 3% paraformaldehyde. Fixed cells were permeabilized by CSK buffer with 0.5% Trion X-100. Cells were stained according to the manufacturer's instructions. ImageJ software was used for fluorescence intensity quantification.
DNA fiber assay was performed as described previously (28). Statistics were generated by GraphPad Prism 6. The Student t-test was used for statistical comparisons.
Subcellular fractionation, chromatin immunoprecipitation and co-immunoprecipitation
For subcellular fractionation, 3–5 × 105 HeLa cells were extracted by extraction buffer with 0.5% Trion X-100 for 10 min on ice before being centrifuged at 13k rpm for 10 min. The insoluble material was washed once with extraction buffer and then subjected to western blot analysis.
Chromatin immunoprecipitation assay was performed as described previously (26,29). Co-immunoprecipitation assay was performed as described previously (30), in which cells were lysed using the buffer (50 mM HEPES–KOH, pH 7.4, 200 mM NaCl, 0.5% NP40, 10% glycerol, 1 mM ethylenediaminetetraacetic acid (EDTA) and proteinase inhibitors) and homogenizated for 30 times by dounce homogenizer. After clarification by centrifugation, 7.5 μl ethidium-bromide (10 mg/ml) was added to the lysates. After incubation for 30 min at 4°C, the lysates were cleared by centrifugation and incubated with 20 μl anti-GFP beads at 4°C for 4 h. The beads were washed using washing buffer (50 mM HEPES–KOH, pH 7.4, 100 mM NaCl, 0.01% NP40, 10% glycerol, 1 mM EDTA and proteinase inhibitors) five times for 5 min. Proteins bound to beads were eluted with 1× sodium dodecyl sulphate sample buffer and analyzed by western blot.
RESULTS
Kdm4d is a critical factor for DNA replication
Using an shRNA screen, we previously identified genes involved in deposition of newly synthesized histone variant H3.3 (Zhang et al., in preparation). Therefore, we decided to identify the chromatin factors involved in DNA replication by screening the same shRNA library, targeting 246 chromatin regulators and a few proteins such as Replication Protein A3 (Rpa3) known to be involved in DNA replication as controls (31). We utilized the efficiency of HeLa cells to incorporate thymidine analog EdU to monitor the replication process. Briefly, HeLa cells were first infected with lentivirus expressing a pool of five shRNAs targeting each gene. The infected cells were pulse-labeled with EdU. Cells were immediately fixed and stained by ‘click’ chemistry reaction, and incorporated EdU was measured by fluorescence microscopy. Depletion of RPA3, a subunit of the single-stranded DNA binding protein RPA involved in DNA replication, greatly impaired EdU incorporation (Supplementary Figure S1A). Fifteen genes were identified during the first round of screening (data not shown). After validation with individual shRNAs, 10 candidate genes were identified to reduce EdU incorporation by at least 40% in two independent shRNAs (Supplementary Table S1). Some of these genes, such as DNMT1, are known to affect DNA replication after depletion (32). KDM4D, the gene coding for one of the histone H3K9me3 demethylases Kdm4d, showed most significant reduction in percentage of EdU-positive cells as well as EdU fluorescence intensity after depletion (data not shown). Thus, Kdm4d was chosen for further study to understand how it is involved in DNA replication.
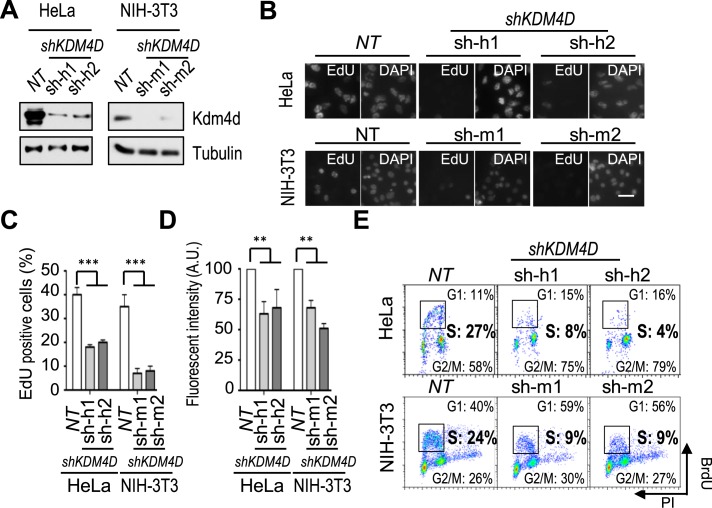
To confirm that Kdm4d has a role in DNA replication, we depleted human KDM4D from HeLa cells using 2 shRNAs (Figure 1A). Quantitative polymerase chain reaction (qPCR) on RNA isolated from Kdm4d-depleted cells showed that the expression of other members of the KDM4 family was not affected by Kdm4d depletion (Supplementary Figure S1B). As shown in Figure 1B and C, the number of EdU-positive cells, which represent the S phase cell population, was significantly reduced in KDM4D-depleted cells compared to non-target shRNA controls (NT). In addition, the EdU fluorescence intensity, which measures the replication efficiency, was also decreased in KDM4D knockdown cells (Figure 1 B and D). Similar results were observed when human KDM4D was depleted from U2OS and HCT116 cells (Supplementary Figure S1D–F), and when mouse KDM4D was depleted in NIH-3T3 cells (Figure 1A–D). As another independent assay to measure S phase cells, we performed 5-bromo-2′-deoxyuridine–propidium iodide (BrdU-PI) flow cytometry and observed that the S phase population in KDM4D-depleted cells was significantly reduced in HeLa, NIH-3T3, U2OS and HCT116 cells compared to their corresponding NT controls (Figure 1E and Supplementary Figure S1G). Taken together, these results demonstrate that Kdm4d is required for the S phase progression of both human and mouse cells.
Figure 1.
Kdm4d is required for DNA replication. (A) Analysis of Kdm4d depletion in HeLa cells and NIH-3T3 cells by western blotting. shRNAs targeting human KDM4D (sh-h1 and sh-h2), shRNAs targeting mouse KDM4D (sh-m1 and sh-m2) and non-target shRNA (NT) were used for depletion of human and mouse Kdm4d, and proteins in whole cell extracts were analyzed. (B–D) Depletion of Kdm4d in HeLa and mouse NIH-3T3 cells results in reduced 5-ethynyl-2′-deoxyuridine (EdU)-positive cells and EdU intensity. Representative images of EdU staining in HeLa and NIH-3T3 cells with and without Kdm4d are shown in B. Scale bar: 50 μm. Quantification of EdU-positive cell numbers (mean ± SD) from three independent experiments is shown in C. At least 300 cells were counted. Quantification of the EdU fluorescence intensity (mean ± SD) from three independent experiments is shown in D. At least 300 cells were counted. (E) Depletion of Kdm4d results in reduced S phase cells. HeLa and NIH-3T3 cells with and without Kdm4d depletion were stained with 5-bromo-2′-deoxyuridine (BrdU) and propidium iodide (PI) and analyzed by flow cytometry. **P < 0.01; ***P < 0.001.
Kdm4d depletion impairs both DNA replication initiation and elongation
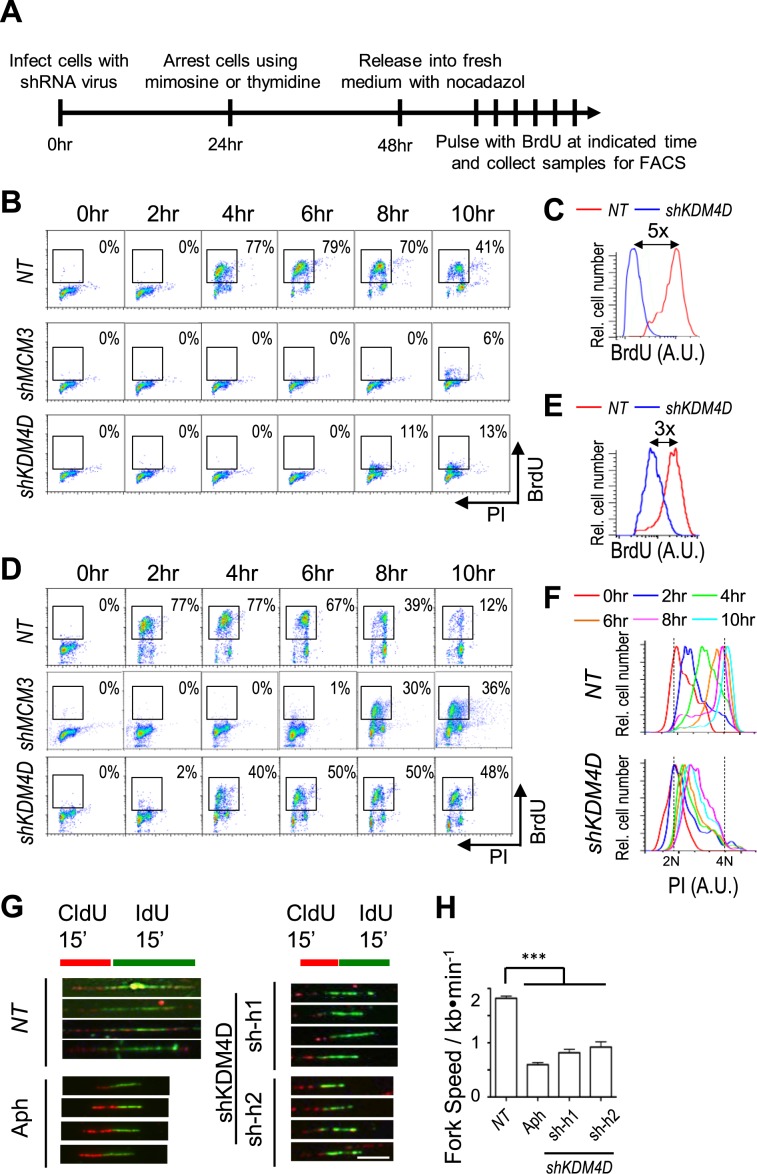
To further examine how Kdm4d depletion affects S phase cells, we synchronized cells at G1 by treating cells with mimosine or at G1/S using thymidine and analyzed how Kdm4d depletion affects cell cycle progression. As a control, we also depleted Mcm3, a subunit of the replicative helicase MCM that is involved in both initiation and elongation of DNA replication. Briefly, cells were infected with lentivirus expressing shRNAs targeting KDM4D or MCM3 for 24 h after mimosine (G1) or thymidine (G1/S) treatment for 24 h. The arrested cells were then released into fresh medium, pulsed with BrdU for 20 min at indicated time points and harvested for flow cytometry analysis (Figure 2A). Mcm3 depletion dramatically reduced S phase cells compared to NT controls in cells released from mimosine (Figure 2B). Similarly, KDM4D depletion also significantly impaired cells from entering S phase after release from mimosine block (Figure 2B), with five times less BrdU incorporation even more than 8 h after release compared to control cells (Figure 2C).
Figure 2.
Depletion of Kdm4d impairs DNA replication initiation and elongation. (A) A schematic representation for analysis of G1-to-S phase transition and S phase progression after cells were treated with mimosine or thymidine. (B) Analysis of cell cycle progression of Mcm3- or Kdm4d-depleted cells synchronized at G1 using mimosine as described in A by bromo-2′-deoxyuridine (BrdU) and propidium iodide (PI) flow cytometry. (C) Quantification of S phase cells BrdU signal intensity of non-target (NT) and Kdm4d-depleted cells at 10-hour time point in B. (D) Analysis of cell cycle progression of Mcm3- or Kdm4d-depleted cells synchronized at G1/S using thymidine by BrdU-PI flow cytometry. (E) Quantification of S phase cells BrdU signal intensity of NT and Kdm4d-depleted cells at 6-h time point in D. (F) Quantification of S phase cells PI signal intensity of NT and Kdm4d-depleted cells in D. (G and H) Depletion of Kdm4d affects elongation of DNA replication. Cells transduced with shRNA virus targeting KDM4D or treated with aphidicolin (Aph) were pulsed with 5-chloro-2′-deoxyuridine (CldU) for 15 min, followed by 5-iodo-2′-deoxyuridine (IdU) for 15 min. Then the DNA fiber was generated on a glass slide and stained with anti-CldU and anti-IdU. The representative images are shown in G. Scale bar: 2 μm. Quantification of the length of IdU tracks that follow CldU tracks (mean ± SD) from three independent experiments is shown in H. At least 500 fibers were counted. ***P < 0.001.
Compared to mimosine, thymidine synchronizes cells at G1/S transition or early S phase after replication machinery is already assembled (33). Therefore, the thymidine-arrested wild-type cells started DNA replication 2 h earlier than mimosine-arrested cells after drug removal (compare Figure 2B and D for NT). Consistent with its helicase function, depletion of Mcm3 significantly reduced replicating cells (Figure 2D). KDM4D-depleted cells released from thymidine also showed a slight delay in S phase entry (Figure 2D) and a moderate reduction in BrdU incorporation compared to NT cells (Figure 2E). However, thymidine-arrested KDM4D-depleted cells started replication 4 h earlier than mimosine-arrested cells (compare Figure 2B and D for the S phase cell number) and more efficiently (compare Figure 2C and E for BrdU intensity). These results indicate that KDM4D has a role in initiation of DNA replication.
We noted that although Kdm4d-depleted cells were able to start replication after releasing from thymidine, the PI peak of S phase population was hardly shifted (Figure 2F), suggesting that DNA replication fork progression was also slowed in Kdm4d-depleted cells. To test this idea, we measured the DNA replication fork speed in Kdm4d-depleted cells using the DNA fiber assay. The replication elongation speed of Kdm4d-depleted cells was approximately 50% of control cells (Figure 2G and H), indicating that Kdm4d also has a role in DNA replication elongation. Collectively, these results indicate that Kdm4d has a role in both initiation and elongation of DNA replication.
The effect of Kdm4d depletion on DNA replication is likely caused by an increase in H3K9me3
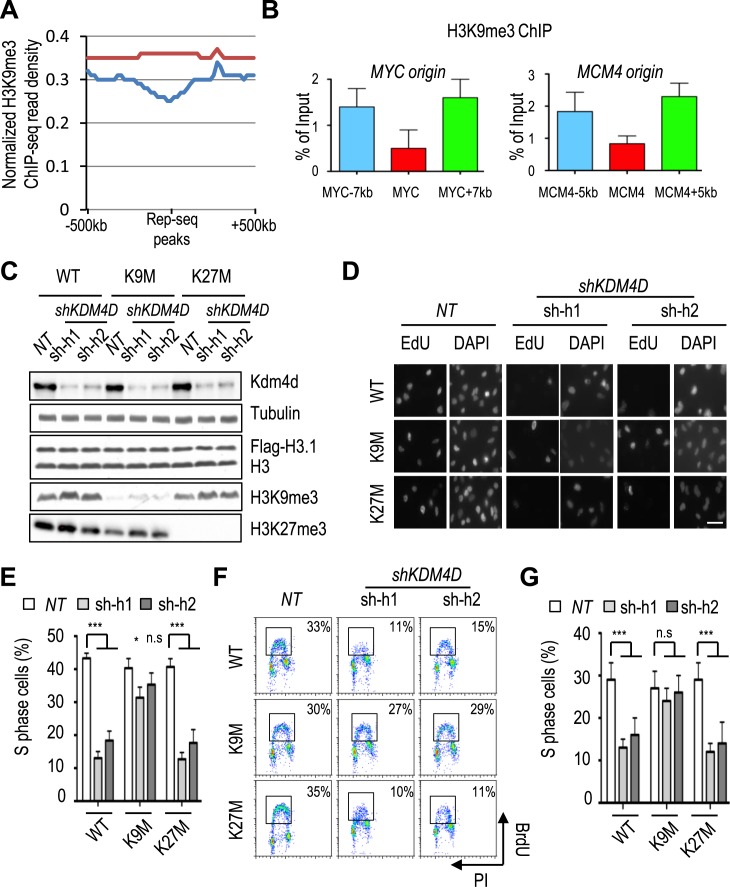
While Kdm4a-c also demethylate H3K36me3, Kdm4d specifically targets trimethylated H3K9 (34,35). We therefore tested whether the replication defect in Kdm4d-depleted cells was due to elevated levels of H3K9me3. To do this, we first analyzed H3K9me3 levels surrounding replication start sites using H3K9me3 ChIP-seq and Repli-seq datasets in HeLa cells from the Encyclopedia of DNA Elements (ENCODE) Project (36,37). The H3K9me3 levels were low at replication start sites compared to adjacent sites (Figure 3A). H3K9me3 ChIP and qPCR confirmed that H3K9me3 levels were low at the MYC and MCM4 replication origins compared to their corresponding distal sites (Figure 3B). These results indicate that H3K9me3 levels are low at DNA replication origins.
Figure 3.
Reduced H3K9me3 levels rescue the replication defect caused by Kdm4d depletion. (A and B) H3K9me3 is depleted from DNA replication origins. (A) Genome-wide scanning of H3K9me3 ChIP-seq across DNA replication start sites. (B) Chromatin immunoprecipitation (ChIP) and quantitative polymerase chain reaction (qPCR) analysis of H3K9me3 at two replication origins (MYC and MCM4). H3K9me3 ChIP was performed in HeLa cells, and ChIP DNA was analyzed by real-time PCR using PCR primers amplifying indicated origins and flanking regions. The ChIP enrichments from three independent experiments were represented by mean ± SD. (C–G) Expression of H3.1 K9M antagonizes the S phase defects caused by Kdm4d depletion. HeLa cells stably expressing wild-type (WT) H3.1, H.31 K9M or H3.1 K27M mutant were transduced for shRNAs for Kdm4d depletion. (C) Analysis of proteins in whole cell lysate by Western blotting using indicated antibodies. (D and E) Analysis of S phase cells using EdU staining. Representative EdU staining images of cells from C are shown in D. Scale bar: 50 μm. Quantification of EdU-positive cell numbers (mean ± SD) from three independent experiments is shown in D. At least 300 cells were counted. (F) Analysis of S phase cells in C by bromo-2′-deoxyuridine (BrdU) and propidium iodide (PI) flow cytometry. (G) Quantification of BrdU-positive cell numbers (mean ± SD) from two independent experiments. n.s P > 0.05; *P < 0.05; ***P < 0.001.
Next we asked whether reducing the H3K9me3 level could antagonize the S phase defects caused by KDM4D depletion by expressing histone lysine to methionine mutants, as we and others have shown recently that cells expressing the lysine to methionine mutation of histone H3, such as H3.1 K9M and H3.1 K27M, dramatically reduce methylation of corresponding lysine residues on wild-type and endogenous histones (25,38). Briefly, we generated HeLa cell lines expressing H3.1 K9M and H3.1 K27M (control), which lead to reduction in H3K9me3 and H3K27me3 levels, respectively (Figure 3C and Supplementary Figure S2A). Depletion of KDM4D resulted in an increase in H3K9me3, but not H3K27me3, in all the stable cell lines compared to NT controls (Figure 3C), but H3K9me3 levels in Kdm4d-depleted H3.1 K9M cells were still lower than those in cells expressing wild-type H3 and Kdm4d (Figure 3C). In contrast, cells expressing H3.1 K27M mutant affected H3K27me3, but not H3K9me3. Importantly, the S phase defects, as determined by immunofluorescence (Figure 3D and E) and BrdU-PI staining (Figure 3F and G), caused by KDM4D depletion were largely rescued by expressing H3.1 K9M, but not H3.1 K27M (Figure 3D and E). Similar rescue effects were observed in cells expressing H3.3 K9M mutant, which also leads to reduction of H3K9me3 (Supplementary Figure S2). Taken together, these data indicate that the replication defects induced by Kdm4d depletion are most likely caused by increased H3K9me3 levels in the Kdm4d-depleted cells.
Kdm4d associates with replication proteins
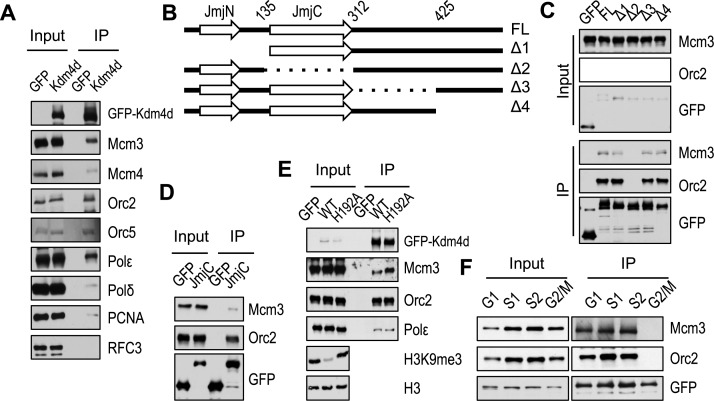
To determine if Kdm4d interacts with replication proteins, we expressed the green fluorescent protein (GFP)-tagged Kdm4d in 293T cells, precipitated GFP-Kdm4d proteins and analyzed co-immunoprecipitated proteins by western blotting using antibodies against several proteins involved in DNA replication (Figure 4A). Kdm4d associated with ORC (represented by Orc2 and Orc5) and MMC complex (represented by Mcm3 and Mcm4). In addition, Kdm4d interacted with PCNA and DNA polymerases (polymerase ϵ and polymerase δ), but Rfc3, a subunit of the clamp loader replication factor C, was not detectable in Kdm4d IP. We noted that Polδ and PCNA were pulled down in with a lower efficiency compared to other proteins, suggesting that they may interact with Kdm4d indirectly. These results indicate that Kdm4d associates with proteins involved in initiation and elongation of DNA replication.
Figure 4.
Kdm4d interacts with proteins involved in DNA replication. (A) Full-length (FL) green fluorescent protein (GFP)-tagged Kdm4d or GFP alone was expressed in 293T and immunoprecipitated (IP) by anti-GFP antibody. Proteins in whole cell extracts (Input) and IP were analyzed by western blotting using antibodies against indicated proteins. (B) Schematic diagram shows GFP-tagged Kdm4d and truncation mutants used for IP. (C) Full-length or truncated GFP-tagged Kdm4d was expressed in 293T and IP by anti-GFP antibody. Proteins in whole cell extracts (Input) and IP were analyzed by western blotting using antibodies against Mcm3 or Orc2. (D) GFP-tagged JmjC domain of Kdm4d was expressed in 293T and IP by anti-GFP antibody. (E) Wild-type (WT) and H192A mutant of KDM4D were expressed in 293T and IP by anti-GFP antibody. (F) 293T cells expressing GFP-tagged Kdm4d were synchronized by mimosine (G1), thymidine arrested and released for 2 h (S1) or 4 h (S2) or nocodazole (G2/M). The lysates were IP with anti-GFP beads.
Kdm4d contains JmjN domain, JmjC domain and a C-terminal domain (Figure 4B). The JmjN and JmjC domains are required for its demethylase activity, while the C-terminal domain is essential for its DNA damage response activity (39). We therefore analyzed which domain is involved in mediating interactions with replication proteins. Kdm4d was truncated according to the secondary structure (Figure 4B). Cells expressing truncation mutants or full-length Kdm4d were subjected to co-immunoprecipitation. Kdm4d Δ2 mutant (Δaa135-aa312), in which JmjC domain was truncated, failed to interact with ORC and MCM (Figure 4C), whereas truncations at Kdm4d C-terminus (Δ3 and Δ4) did not affect the interaction of Kdm4d with MCM and ORC. In addition, the JmjC domain alone interacted with Orc2 and Mcm3 in vivo (Figure 4D). These results indicate that JmjC domain is responsible for mediating the interaction between Kdm4d and replication proteins.
The JmjC domain is essential for the demethylase activity of Kdm4d (39). Next we tested whether the interactions between KDM4D and replication proteins depend on Kdm4d's enzyme activity. An enzymatic-dead mutant (KDM4DH192A) was generated. H3K9me3 level was reduced dramatically in 293T cells over-expressing wild-type Kdm4d, but not Kdm4dH192A mutant (Figure 4E). In contrast, Kdm4dH192A mutant exhibited no obvious effect on interaction with DNA replication proteins compared to wild-type Kdm4d (Figure 4E). These results indicated that the interactions between Kdm4d and replication proteins are dependent on JmjC domain but not on Kdm4d demethylase activity.
Because most of the replication proteins associate with chromatin from G1 to S phase (30), we asked whether the association of Kdm4d and replication proteins is regulated during the cell cycle. To test this possibility, we examined the interaction between Kdm4d and replication proteins in cells synchronized at different phases of the cell cycle. Orc2 and Mcm3 were co-precipitated with GFP-Kdm4d from G1 and S phase (early S phase, S1; late S phase, S2) cells, but not G2/M phase cells (Figure 4F), suggesting that the interaction between Orc2/Mcm3 and Kdm4d is regulated during the progression of the cell cycle.
Given the cell cycle-dependent association between Kdm4d and replication proteins, we speculated that the chromatin association of Kdm4d might be regulated in a similar way. To test this hypothesis, we first tested the localization of Kdm4d using immunofluorescence and Kdm4d-specific antibody (Supplementary Figure S3A and B). Kdm4d appeared not co-localize with replication foci in both human and mouse cell lines (Supplementary Figure S3B). Kdm4d was excluded from chromosomes in all mitotic cells (Supplementary Figure S3C). Second, we analyzed the chromatin association of Kdm4d during the cell cycle using a chromatin fractionation assay. HeLa cells were synchronized by mimosine at G1 phase and nocodazole at G2/M phase. In addition, thymidine-arrested cells were released into fresh medium for 2 h (S1, early S phase) or 4 hours (S2, late S phase; Supplementary Figure S3D). The synchronized cells were used for the chromatin fractionation assays by separating proteins into soluble fraction and chromatin fraction. As reported previously, Mcm3 was dissociated from chromatin at G2/M phase (30,40) and H3K10ph level increased at G2/M phase (Supplementary Figure S3E). Kdm4d associated with chromatin at G1 and S phases (lanes 7–9), but not at G2/M phase (lane 10, Supplementary Figure S3E). Taken together, these data indicate that Kdm4d associates with chromatin and interacts with DNA replication proteins during G1 and S phases but not G2/M phase of the cell cycle.
Kdm4d associates with replication origins at G1 phase depending on ORC and MCM
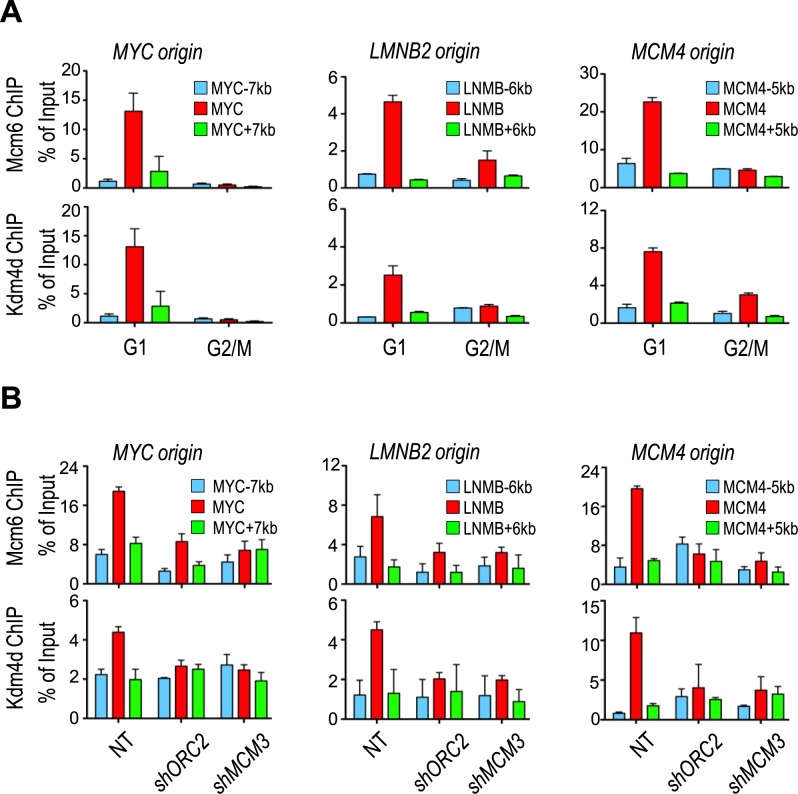
Because our results indicate that Kdm4d has a role in initiation of DNA replication and associates with chromatin at G1 and S phases of the cell cycle, we tested whether Kdm4d associated with DNA replication origins at G1 phase using chromatin immunoprecipitation (ChIP) assay. HeLa cells were synchronized at G1 phase or G2/M phase of the cell cycle. Kdm4d and Mcm6 ChIP assays were performed, and ChIP DNAs were analyzed using primers amplifying three well-studied replication origins (MYC, LMNB2 and MCM4) by qPCR. Similarly to Mcm6, Kdm4d bound replication origins only in mimosine-arrested cells (G1) but not in nocodazole-arrested cells (G2/M; Figure 5A), indicating that Kdm4d, like MCM proteins, associates with origins during G1 phase of the cell cycle (Figure 4F and Supplementary Figure S3C and D).
Figure 5.
Kdm4d associates with replication origins depending on components of pre-replicative complex. (A) Kdm4d and Mcm6 bound replication origins at G1 phase. HeLa cells were synchronized by mimosine (G1) or nocodazole (G2/M). Chromatin immunoprecipitation (ChIP) and quantitative polymerase chain reaction (qPCR) analysis using antibodies against Kdm4d or Mcm6 was performed. ChIP DNA was analyzed by real-time PCR using PCR primers amplifying indicated origins and flanking regions. The ChIP enrichments from two independent experiments were represented by mean ± SD. (B) Depletion of Orc2 or Mcm3 abolished replication origin binding of Kdm4d. ChIP-qPCR analysis using antibodies against Kdm4d or Mcm6 was performed on Orc2- or Mcm3-depleted HeLa cells. ChIP DNA was analyzed by real-time PCR using PCR primers amplifying indicated origins and flanking regions. The ChIP enrichments from two independent experiments were represented by mean ± SD.
Replication machinery is assembled at replication origins in an ordered manner, with the association of ORC with origins first and subsequent loading of MCM replicative helicase (41). Therefore, we investigated whether Kdm4d was recruited to origins by ORC and MCM. The origin association of Mcm6 was greatly reduced by depletion of Orc2, a subunit of ORC (Figure 5B), as well as in cells depleted of Mcm3. This result is consistent with the fact that MCM loading depends on ORC. Importantly, depletion of Orc2 and Mcm3 resulted in a marked reduction in the association of Kdm4d with replication origin (Figure 5B). Together, these results suggested that Kdm4d bound DNA replication origins at G1 phase depending on ORC and MCM.
Kdm4d is required for pre-initiative complex formation
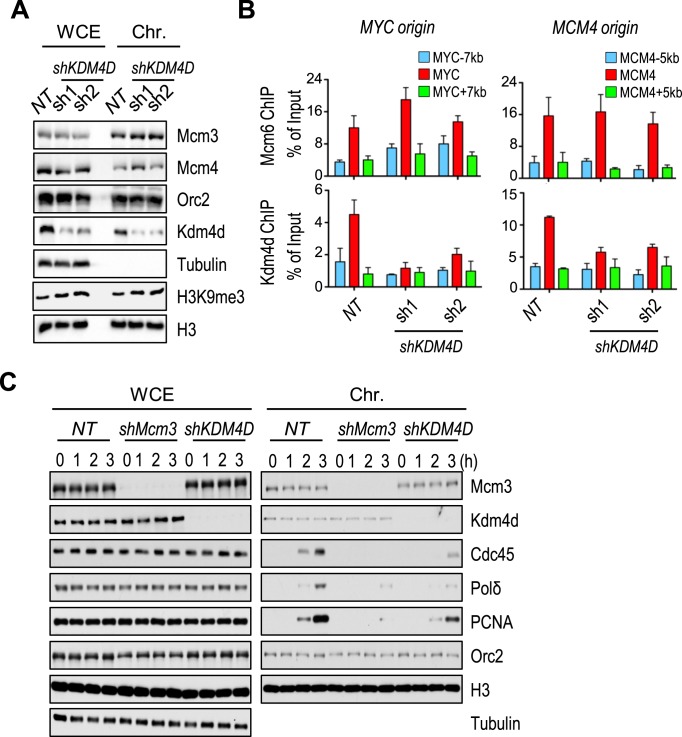
Because the association of Kdm4d with DNA replication origins depends on two pre-RC components (ORC and MCM), we examined whether the chromatin association of ORC and MCM depends on Kdm4d. The association of Orc2 and two subunits of MCM complex (Mcm3 and Mcm4) with chromatin was not affected in Kdm4d-depleted cells (Figure 6A). Mcm6 ChIP assay showed that Mcm6 was enriched at replication origins in both control and KDM4D-depleted cells (Figure 6B). These results suggest that the chromatin association of ORC and MCM, 2 key components of pre-RC, is not affected by Kdm4d depletion.
Figure 6.
Kdm4d is important for the formation of pre-initiative complex. (A) The whole cell lysates (WCE) and the chromatin fractions (Chr.) of HeLa cells infected with shRNA targeting KMD4D were analyzed by western blotting with antibodies against indicated proteins. (B) Chromatin immunoprecipitation (ChIP) and quantitative polymerase chain reaction (qPCR) assays with antibodies targeting Kdm4d or Mcm6 were performed in the Kdm4d-depleted cells. ChIP DNA was analyzed by real-time PCR using PCR primers amplifying indicated origins and flanking regions. The ChIP enrichment from two independent experiments was represented by mean ± SD. (C) The whole cell lysate and chromatin fractions from mimosine-released MCM3 or KDM4D knockdown cells were analyzed by western blotting with antibodies against indicated proteins.
After formation of pre-RC, other proteins, including Cdc45, GINS and PCNA, are loaded to form pre-initiative complex (pre-IC) (42,43). We next asked whether the recruitment of these pre-IC proteins was impaired by Kdm4d depletion. Cells expressing shRNA targeting KDM4D or MCM3 were arrested in mimosine-containing medium (G1), released into fresh medium for indicated time points and collected for chromatin fractionation assays (Figure 6C). As expected, PCNA, Cdc45 and polymerase δ failed to load on chromatin in Mcm3-depleted cells. However, Kdm4d bound chromatin normally in MCM3 knockdown cells (Figure 6C), indicating that while depletion of MCM3 affects Kdm4d binding to origins, it does not affect the overall Kdm4d chromatin association. It is known that Kdm4d also binds to heterochromatin, which account for over 90% chromatin in mammalian cells. In agreement with previous observation (Figure 5B), KDM4D knockdown did not affect chromatin association of Orc2 and Mcm3 (Figure 6C). Surprisingly, Kdm4d depletion greatly reduced or delayed the chromatin binding of PCNA, Cdc45 and polymerase δ, while having no apparent effect on the expression of these proteins (Figure 6C). These results provide an explanation for the S phase delay after Kdm4d depletion and suggest that Kdm4d regulates DNA replication after the formation of pre-RC, possibly regulating the formation of pre-IC.
DISCUSSION
Here we present the following lines of evidence supporting the idea that chromatin regulator Kdm4d plays an important role in DNA replication. First, we show that depletion of Kdm4d in four different human and mouse cell lines results in defects in S phase. Second, this S phase defect is likely due to inability of Kdm4d depleted cells to initiate and elongate DNA synthesis and is linked to increased H3K9me3. Third, we show that Kdm4d interacts with proteins involved in both initiation and elongation. Fourth, Kdm4d binds to chromatin and replication origins during G1 and S phases, and the ability of Kdm4d to bind chromatin depends on ORC and MCM, two components of pre-RC involved in initiation of DNA replication. Finally, we show that Kdm4d is required for the chromatin association of Cdc45, a component of active DNA helicase complex CMG and PCNA. These results support a model whereby Kdm4d removes H3K9me3 and provides a favorable chromatin environment for the assembly of pre-IC involved in initiation and elongation of DNA replication.
Chromatin, the physiological substrate of DNA replication machinery, in principle can affect multiple steps of DNA replication, including formation of pre-RC containing ORC, Cdc6, Ctd1 and inactive MCM2–7 helicase; formation of pre-IC involved in initiation of DNA replication; and regulation of elongation of DNA replication. Previously, various studies have documented different chromatin regulators in DNA replication. For instance, HBO1, targeting the histone H4 Lys5, Lys8 and Lys12, promotes licensing by increasing chromatin accessibility (7,8). ORC and MCM replication origin recruitment is also regulated by epigenetic factors such as heterochromatin protein 1 (HP1), high-mobility group AT-hook protein 1 (HMGA1) and histone H4K20me1 methlytransferase PR-SET7 (13,44,45).
We have shown that Kdm4d interacts with several protein components of pre-RC and pre-IC. Moreover, Kdm4d is loaded on replication origins in an ORC- and MCM-dependent manner. In addition, Kdm4d is required for the chromatin association of Cdc45, a key component of pre-IC. These results indicate that Kdm4d likely functions after the formation of pre-RC to facilitate the assembly of pre-IC during DNA replication.
Using candidate origin approaches, we found that Kdm4d bound to early replication origins, which are closed to promoters. Since Kdm4d origins binding depends on ORC and MCM, it is unlikely that Kdm4d origin binding is due to promoter binding of Kdm4d. However, we cannot exclude the possibility that promoter binding of Kdm4d may also contribute to its binding to early replication origins. H3K9me3 are marks for heterochromatin, it is possible that Kdm4d also regulates late replication origins. Future studies are needed to test this idea. Several recent studies reveal a complex relationship among ORC binding sites, promoter and early replication origins. For instance, Martin et al. reports that replication ignition events are depleted in highly transcribed regions in two independent cell lines (46). Using Okazaki fragment mapping, it was found that replication initiates primarily at non-transcribed regions (47). Interesting, about 52 000 Orc2 binding sites are largely located at open chromatin regions. It was proposed that activator proteins at promoters and enhancers shape local chromatin structure, which in turn facilitates ORC binding and DNA replication initiations (48).
A growing body of evidence has shown that proper levels of H3K9me3 are important for DNA replication. For instance, the replication timing of heterochromatin is advanced in mouse embryonic fibroblast cells lacking H3K9 methyltransferases Suv39h1/h2 (6,19,49). Studies by Black et al. elegantly demonstrated that H3K9me3 demethylase Kdm4a is amplified in ovarian cancer, and Kdm4a over-expression induces site-specific copy gain and re-replication of regions amplified in tumors (20,50). We show that Kdm4d required for efficient DNA replication globally. Collectively, these reports imply that high levels of H3K9me3 act as a barrier to DNA replication. Interestingly, Black et al. shows that Kdm4a is not required for normal DNA replication. Kdm4a was not identified in our screen. Interestingly, both Kdm4a and Kdm4d interact with proteins involved in DNA replication. We have shown that the JmjC domain is responsible for Kdm4d to bind proteins involved in DNA replication. This domain is highly conserved among members of Kdm4 family demethylases. We speculate that Kdm4d regulates DNA replication in normal cell cycle, while Kdm4a, when over-expressed in cancer cell, might over-ride Kdm4d's regulatory function (15,40).
In addition to DNA replication, KDM4 family proteins also actively regulate DNA damage response. For instance, degradation of Kdm4a triggers 53BP1 recruitment to DNA damage sites (24). Additionally, recruitment of Kdm4b and Kdm4d to DNA damage sites is required for DNA double-strand break response (21,22). However, we present lines of evidence indicating that the role of Kdm4d in DNA replication is unlikely induced by defective DNA damage response for the following reasons. First, we show that Kdm4d binds proteins involved in DNA replication and associates with DNA replication origins in G1 and S phases (Figure 4). Second, we assessed DNA replication defects immediately after Kdm4d depletion to avoid accumulation of excessive DNA damage (Figure 2). Third, Kdm4d is required for activation of G1 DNA damage checkpoints ATM and Chk2 (22), which are essential for DNA damage response, suggesting that the G1 arrest in Kdm4d-depleted cells after release from G1 block is not due to activation of the DNA damage checkpoint (Figure 2B and D). Therefore, Kdm4d likely regulates DNA replication and DNA repair separately.
Both transcription and DNA replication are performed by molecular machinery traveling along the same DNA molecule; thus, conflicts would arise when the two types of machinery encounter each other head to head. Therefore, these two processes need to be tightly coordinated. Recently, Rondinelli et al. reported that H3K4me3 demethylase Kdm5c is also critical for the assembly of pre-IC in DNA replication (51). H3K4me3 is an active marker for transcription start site (52,53). It was proposed that demethylation of H3K4 may coordinate transcription and replication initiation to prevent collision between transcription machinery and DNA replication machinery (54,55). It is also possible that H3K4me3 is an active histone mark regulating early replication origins initiation, while H3K9me3 is an inactive mark regulating late or dormant replication origins initiation. Alternatively, the crosstalk between H3K4me3 and H3K9me3 may be critical for DNA replication initiation regulation. When DNA replication starts, H3K4me3 around replication origins needs to be removed to suspend transcription, while trimethylation of H3K9 at the same chromatin locus is prevented by Kdm4d to maintain a favorable chromatin environment of pre-IC formation and origin firing.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH [GM118015]. Funding for open access charge: NIH [GM118015].
Conflict of interest statement. None declared.
REFERENCES
- 1.O'Donnell M., Langston L., Stillman B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb. Perspect. Biol. 2013;5:a010108. doi: 10.1101/cshperspect.a010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farkash-Amar S., Simon I. Genome-wide analysis of the replication program in mammals. Chromosome Res. 2010;18:115–125. doi: 10.1007/s10577-009-9091-5. [DOI] [PubMed] [Google Scholar]
- 3.Groth A., Rocha W., Verreault A., Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 4.Black J.C., Whetstine J.R. Chromatin landscape: methylation beyond transcription. Epigenetics. 2011;6:9–15. doi: 10.4161/epi.6.1.13331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Julienne H., Zoufir A., Audit B., Arneodo A. Human genome replication proceeds through four chromatin states. PLoS Comput. Biol. 2013;9:e1003233. doi: 10.1371/journal.pcbi.1003233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu R., Terry A.V., Singh P.B., Gilbert D.M. Differential subnuclear localization and replication timing of histone H3 lysine 9 methylation states. Mol. Biol. Cell. 2005;16:2872–2881. doi: 10.1091/mbc.E04-11-0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miotto B., Struhl K. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol. Cell. 2010;37:57–66. doi: 10.1016/j.molcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miotto B., Struhl K. HBO1 histone acetylase is a coactivator of the replication licensing factor Cdt1. Genes Dev. 2008;22:2633–2638. doi: 10.1101/gad.1674108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casas-Delucchi C.S., van Bemmel J.G., Haase S., Herce H.D., Nowak D., Meilinger D., Stear J.H., Leonhardt H., Cardoso M.C. Histone hypoacetylation is required to maintain late replication timing of constitutive heterochromatin. Nucleic Acids Res. 2012;40:159–169. doi: 10.1093/nar/gkr723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacob Y., Bergamin E., Donoghue M.T., Mongeon V., LeBlanc C., Voigt P., Underwood C.J., Brunzelle J.S., Michaels S.D., Reinberg D., et al. Selective methylation of histone H3 variant H3.1 regulates heterochromatin replication. Science. 2014;343:1249–1253. doi: 10.1126/science.1248357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacob Y., Stroud H., Leblanc C., Feng S., Zhuo L., Caro E., Hassel C., Gutierrez C., Michaels S.D., Jacobsen S.E. Regulation of heterochromatic DNA replication by histone H3 lysine 27 methyltransferases. Nature. 2010;466:987–991. doi: 10.1038/nature09290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Picard F., Cadoret J.C., Audit B., Arneodo A., Alberti A., Battail C., Duret L., Prioleau M.N. The spatiotemporal program of DNA replication is associated with specific combinations of chromatin marks in human cells. PLoS Genet. 2014;10:e1004282. doi: 10.1371/journal.pgen.1004282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tardat M., Brustel J., Kirsh O., Lefevbre C., Callanan M., Sardet C., Julien E. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat. Cell Biol. 2010;12:1086–1093. doi: 10.1038/ncb2113. [DOI] [PubMed] [Google Scholar]
- 14.Kuo A.J., Song J., Cheung P., Ishibe-Murakami S., Yamazoe S., Chen J.K., Patel D.J., Gozani O. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature. 2012;484:115–119. doi: 10.1038/nature10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huen M.S., Sy S.M., van Deursen J.M., Chen J. Direct interaction between SET8 and proliferating cell nuclear antigen couples H4-K20 methylation with DNA replication. J. Biol. Chem. 2008;283:11073–11077. doi: 10.1074/jbc.C700242200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beck D.B., Burton A., Oda H., Ziegler-Birling C., Torres-Padilla M.E., Reinberg D. The role of PR-Set7 in replication licensing depends on Suv4-20h. Genes Dev. 2012;26:2580–2589. doi: 10.1101/gad.195636.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bulut-Karslioglu A., De La Rosa-Velazquez I.A., Ramirez F., Barenboim M., Onishi-Seebacher M., Arand J., Galan C., Winter G.E., Engist B., Gerle B., et al. Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol. Cell. 2014;55:277–290. doi: 10.1016/j.molcel.2014.05.029. [DOI] [PubMed] [Google Scholar]
- 18.Al-Sady B., Madhani H.D., Narlikar G.J. Division of labor between the chromodomains of HP1 and Suv39 methylase enables coordination of heterochromatin spread. Mol. Cell. 2013;51:80–91. doi: 10.1016/j.molcel.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu R., Singh P.B., Gilbert D.M. Uncoupling global and fine-tuning replication timing determinants for mouse pericentric heterochromatin. J. Cell Biol. 2006;174:185–194. doi: 10.1083/jcb.200601113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Black J.C., Allen A., Van Rechem C., Forbes E., Longworth M., Tschop K., Rinehart C., Quiton J., Walsh R., Smallwood A., et al. Conserved antagonism between JMJD2A/KDM4A and HP1gamma during cell cycle progression. Mol. Cell. 2010;40:736–748. doi: 10.1016/j.molcel.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 21.Young L.C., McDonald D.W., Hendzel M.J. Kdm4b histone demethylase is a DNA damage response protein and confers a survival advantage following gamma-irradiation. J. Biol. Chem. 2013;288:21376–21388. doi: 10.1074/jbc.M113.491514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khoury-Haddad H., Guttmann-Raviv N., Ipenberg I., Huggins D., Jeyasekharan A.D., Ayoub N. PARP1-dependent recruitment of KDM4D histone demethylase to DNA damage sites promotes double-strand break repair. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E728–E737. doi: 10.1073/pnas.1317585111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khoury-Haddad H., Nadar-Ponniah P.T., Awwad S., Ayoub N. The emerging role of lysine demethylases in DNA damage response: dissecting the recruitment mode of KDM4D/JMJD2D to DNA damage sites. Cell Cycle. 2015;14:950–958. doi: 10.1080/15384101.2015.1014147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mallette F.A., Mattiroli F., Cui G., Young L.C., Hendzel M.J., Mer G., Sixma T.K., Richard S. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 2012;31:1865–1878. doi: 10.1038/emboj.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan K.M., Fang D., Gan H., Hashizume R., Yu C., Schroeder M., Gupta N., Mueller S., James C.D., Jenkins R., et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 2013;27:985–990. doi: 10.1101/gad.217778.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan K.M., Zhang H., Malureanu L., van Deursen J., Zhang Z. Diverse factors are involved in maintaining X chromosome inactivation. Proc. Natl. Acad. Sci. U.S.A. 2011;108:16699–16704. doi: 10.1073/pnas.1107616108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boos D., Yekezare M., Diffley J.F. Identification of a heteromeric complex that promotes DNA replication origin firing in human cells. Science. 2013;340:981–984. doi: 10.1126/science.1237448. [DOI] [PubMed] [Google Scholar]
- 28.Merrick C.J., Jackson D., Diffley J.F. Visualization of altered replication dynamics after DNA damage in human cells. J. Biol. Chem. 2004;279:20067–20075. doi: 10.1074/jbc.M400022200. [DOI] [PubMed] [Google Scholar]
- 29.Chan K.M., Zhang Z. Leucine-rich repeat and WD repeat-containing protein 1 is recruited to pericentric heterochromatin by trimethylated lysine 9 of histone H3 and maintains heterochromatin silencing. J. Biol. Chem. 2012;287:15024–15033. doi: 10.1074/jbc.M111.337980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huo L., Wu R., Yu Z., Zhai Y., Yang X., Chan T.C., Yeung J.T., Kan J., Liang C. The Rix1 (Ipi1p-2p-3p) complex is a critical determinant of DNA replication licensing independent of their roles in ribosome biogenesis. Cell Cycle. 2012;11:1325–1339. doi: 10.4161/cc.19709. [DOI] [PubMed] [Google Scholar]
- 31.Zuber J., Shi J., Wang E., Rappaport A.R., Herrmann H., Sison E.A., Magoon D., Qi J., Blatt K., Wunderlich M., et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vertino P.M., Sekowski J.A., Coll J.M., Applegren N., Han S., Hickey R.J., Malkas L.H. DNMT1 is a component of a multiprotein DNA replication complex. Cell Cycle. 2002;1:416–423. doi: 10.4161/cc.1.6.270. [DOI] [PubMed] [Google Scholar]
- 33.Rosner M., Schipany K., Hengstschlager M. Merging high-quality biochemical fractionation with a refined flow cytometry approach to monitor nucleocytoplasmic protein expression throughout the unperturbed mammalian cell cycle. Nat. Protoc. 2013;8:602–626. doi: 10.1038/nprot.2013.011. [DOI] [PubMed] [Google Scholar]
- 34.Krishnan S., Trievel R.C. Structural and functional analysis of JMJD2D reveals molecular basis for site-specific demethylation among JMJD2 demethylases. Structure. 2013;21:98–108. doi: 10.1016/j.str.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 35.Hillringhaus L., Yue W.W., Rose N.R., Ng S.S., Gileadi C., Loenarz C., Bello S.H., Bray J.E., Schofield C.J., Oppermann U. Structural and evolutionary basis for the dual substrate selectivity of human KDM4 histone demethylase family. J. Biol. Chem. 2011;286:41616–41625. doi: 10.1074/jbc.M111.283689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ram O., Goren A., Amit I., Shoresh N., Yosef N., Ernst J., Kellis M., Gymrek M., Issner R., Coyne M., et al. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell. 2011;147:1628–1639. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryba T., Battaglia D., Pope B.D., Hiratani I., Gilbert D.M. Genome-scale analysis of replication timing: from bench to bioinformatics. Nat. Protoc. 2011;6:870–895. doi: 10.1038/nprot.2011.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis P.W., Muller M.M., Koletsky M.S., Cordero F., Lin S., Banaszynski L.A., Garcia B.A., Muir T.W., Becher O.J., Allis C.D. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berry W.L., Janknecht R. KDM4/JMJD2 histone demethylases: epigenetic regulators in cancer cells. Cancer Res. 2013;73:2936–2942. doi: 10.1158/0008-5472.CAN-12-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu C., Wu R., Zhou B., Wang J., Wei Z., Tye B.K., Liang C., Zhu G. Structural insights into the Cdt1-mediated MCM2-7 chromatin loading. Nucleic Acids Res. 2012;40:3208–3217. doi: 10.1093/nar/gkr1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Costa A., Hood I.V., Berger J.M. Mechanisms for initiating cellular DNA replication. Annu. Rev. Biochem. 2013;82:25–54. doi: 10.1146/annurev-biochem-052610-094414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muramatsu S., Hirai K., Tak Y.S., Kamimura Y., Araki H. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol (epsilon}, and GINS in budding yeast. Genes Dev. 2010;24:602–612. doi: 10.1101/gad.1883410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gambus A., Jones R.C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R.D., Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 44.Thomae A.W., Pich D., Brocher J., Spindler M.P., Berens C., Hock R., Hammerschmidt W., Schepers A. Interaction between HMGA1a and the origin recognition complex creates site-specific replication origins. Proc. Natl. Acad. Sci. U.S.A. 2008;105:1692–1697. doi: 10.1073/pnas.0707260105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwaiger M., Kohler H., Oakeley E.J., Stadler M.B., Schubeler D. Heterochromatin protein 1 (HP1) modulates replication timing of the Drosophila genome. Genome Res. 2010;20:771–780. doi: 10.1101/gr.101790.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin M.M., Ryan M., Kim R., Zakas A.L., Fu H., Lin C.M., Reinhold W.C., Davis S.R., Bilke S., Liu H., et al. Genome-wide depletion of replication initiation events in highly transcribed regions. Genome Res. 2011;21:1822–1832. doi: 10.1101/gr.124644.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petryk N., Kahli M., d'Aubenton-Carafa Y., Jaszczyszyn Y., Shen Y., Silvain M., Thermes C., Chen C.L., Hyrien O. Replication landscape of the human genome. Nat. Commun. 2016;7:10208–10221. doi: 10.1038/ncomms10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miotto B., Ji Z., Struhl K. Selectivity of ORC binding sites and the relation to replication timing, fragile sites, and deletions in cancers. Proc. Natl. Acad. Sci. U.S.A. 2016;113:E4810–E4819. doi: 10.1073/pnas.1609060113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Panning M.M., Gilbert D.M. Spatio-temporal organization of DNA replication in murine embryonic stem, primary, and immortalized cells. J. Cell Biochem. 2005;95:74–82. doi: 10.1002/jcb.20395. [DOI] [PubMed] [Google Scholar]
- 50.Black J.C., Manning A.L., Van Rechem C., Kim J., Ladd B., Cho J., Pineda C.M., Murphy N., Daniels D.L., Montagna C., et al. KDM4A lysine demethylase induces site-specific copy gain and rereplication of regions amplified in tumors. Cell. 2013;154:541–555. doi: 10.1016/j.cell.2013.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rondinelli B., Schwerer H., Antonini E., Gaviraghi M., Lupi A., Frenquelli M., Cittaro D., Segalla S., Lemaitre J.M., Tonon G. H3K4me3 demethylation by the histone demethylase KDM5C/JARID1C promotes DNA replication origin firing. Nucleic Acids Res. 2015;43:2560–2574. doi: 10.1093/nar/gkv090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hon G.C., Hawkins R.D., Ren B. Predictive chromatin signatures in the mammalian genome. Hum. Mol. Genet. 2009;18:R195–R201. doi: 10.1093/hmg/ddp409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shlyueva D., Stampfel G., Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 54.Helmrich A., Ballarino M., Nudler E., Tora L. Transcription-replication encounters, consequences and genomic instability. Nat. Struct. Mol. Biol. 2013;20:412–418. doi: 10.1038/nsmb.2543. [DOI] [PubMed] [Google Scholar]
- 55.Helmrich A., Ballarino M., Tora L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell. 2011;44:966–977. doi: 10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.