Abstract
Acridine dyes, including proflavine and acriflavine, were commonly used as antiseptics before the advent of penicillins in the mid-1940s. While their mode of action on pathogens was originally attributed to their DNA intercalating activity, work in the early 1970s suggested involvement of the host immune responses, characterized by induction of interferon (IFN)-like activities through an unknown mechanism. We demonstrate here that sub-toxic concentrations of a mixture of acriflavine and proflavine instigate a cyclic-GMP-AMP (cGAMP) synthase (cGAS)-dependent type-I IFN antiviral response. This pertains to the capacity of these compounds to induce low level DNA damage and cytoplasmic DNA leakage, resulting in cGAS-dependent cGAMP-like activity. Critically, acriflavine:proflavine pre-treatment of human primary bronchial epithelial cells significantly reduced rhinovirus infection. Collectively, our findings constitute the first evidence that non-toxic DNA binding agents have the capacity to act as indirect agonists of cGAS, to exert potent antiviral effects in mammalian cells.
INTRODUCTION
Acriflavine (also known as trypaflavine) and its precursor proflavine are trypanocidal and antibacterial dyes derived from acridine that were first used in 1917 as topical antibacterials in wound therapy (1,2). While their use subsided after introduction of β-lactam antibiotics in the 1940s, their application remains common in parts of the world today (3). Similar to acridine, acriflavine intercalates the DNA helix in a non-covalent fashion without significant disturbance of base-pairing (4), resulting in DNA specific fluorescence commonly used in cell-based experiments. This DNA intercalating activity of acridines is directly mutagenic in bacteria and viruses, explaining some of the healing effects seen in wound therapy (2,4,5). Nonetheless, in vivo studies in 1973 established that intraperitoneal injection of acriflavine resulted in the induction of an interferon (IFN)-like activity in mice sera (although IFN was not directly detected) (6). Given that acriflavine was also found to mediate the recruitment of inflammatory cells when topically used on skin (3,7), it is most likely that some of the therapeutic properties described in wound therapy also involve recruitment of the host immune response, through an unknown mechanism.
It has recently been discovered that two other acridine derivatives, namely 10-carboxymethyl-9-acridanone (CMA) and 5,6-Dimethylxanthenone-4-acetic Acid (DMXAA), engage type-I IFN signaling through direct recruitment of murine stimulator of interferon genes (STING) (8,9). STING is an intracellular adaptor molecule involved in the detection of cytosolic DNA (10). Its activation recruits the TBK1-IRF3 axis (11) to promote the selective induction of antiviral genes, including IFN-β, triggering the expression of hundreds of interferon stimulated genes (ISGs). Critically, CMA and DMXAA were found to specifically bind to the murine variant of Sting, but not its human homolog (8,12). Crystal structure definition of the binding of CMA and DMXAA with STING helped define the molecular discrepancies between human STING and mouse Sting (8,13), leading to the concept that other compounds could be designed to bind to human STING directly (14). Critically, these findings suggest that reports of IFN-like activities of other acridines such as acriflavine, may also pertain to an engagement of the STING pathway.
Cyclic-GMP-AMP (cGAMP) synthase (cGAS) was recently found to operate upstream of STING (15). cGAS is a cytoplasmic sensor of double-stranded DNA and RNA:DNA hybrids (15,16) and catalyzes the production of cGAMP(2′-5′), the second messenger acting as a STING ligand (17). Here we demonstrate that unlike CMA and DMXAA, a mixture of acriflavine and proflavine (referred to as flavine hereafter), raises basal antiviral immunity in mouse and human cells through the indirect activation of STING, via cGAS recruitment.
MATERIALS AND METHODS
Cell culture
All mouse embryonic fibroblast (MEF) lines used in this study were immortalized with the SV40 large T antigen using pSG5-SV40-LT-Ag (gift from D. Huang, Walter and Eliza Hall Institute of Medical Research, Melbourne, Australia) (18,19). Previously described cGasCRISPR−/− MEFs (referred to as cGas-deficient MEFs) were generated by CRISPR and were derived from a parental cGaswt/wt immortalized MEF line (used as a matched control) (19). Primary Sting-deficient and matched wild-type (WT) control MEFs (18), were SV40T immortalized as previously described (20). WT MEFs used in Figure 1 were generated previously from C57BL/6 mice (18). Three different WT MEF cell lines from three different embryos were used throughout the studies, with similar responses to flavine. MEFs and Vero cells (ATCC® CCL81™) were grown in Dulbecco's modified Eagle's medium (Life Technologies) supplemented with 10% sterile fetal bovine serum (FBS) (Life Technologies) and 1× antibiotic/antimycotic (Life Technologies) (referred to as complete Dulbecco's modified Eagle's medium (DMEM)). Human fibroblasts (hTERT-BJ1 cells) were grown in complete DMEM supplemented with sodium pyruvate (Life technologies). Human embryonic kidney (HEK) cGASlow expressing low levels of murine cGAS, HEK Sting and HEK Sting CX43/45DKO cells stably expressing the murine Sting fused to an N-terminal mCherry-tag were all previously described (21). CX43/45DKO (double knock-out) lack connexin 43 and 45 (21). When needed, cells were treated with filter-sterilized flavine (Sigma A8126: acriflavine mixture: 3,6-Diamino-10-methylacridinium chloride mixed with 3,6-diaminoacridine (proflavine) resuspended in Ultrapure Distilled water (Life Technologies) to a concentration of 10 mM), DMXAA 15 μg/ml (resuspended in DMSO, Sigma D5817) or recombinant mouse IFN-β at 1000 IU/ml (22) (gift from N.A. de Weerd, A. Matthews and P.J. Hertzog, Hudson Institute). Cytotoxicity assays were performed according to the manufacturer's protocol (Lonza, ViaLight™ Plus Cell Proliferation and Cytotoxicity BioAssay Kit).
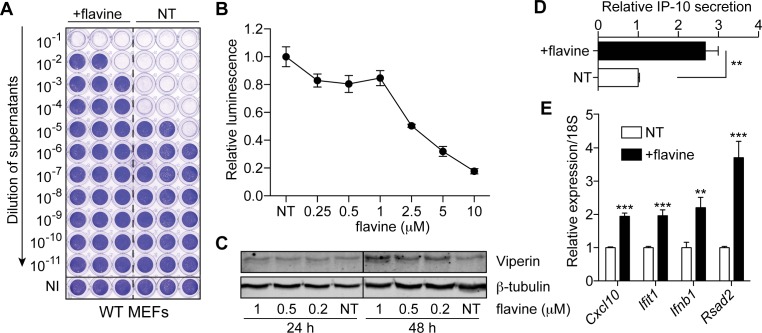
Figure 1.
Flavine treatment activates a type-I IFN antiviral response in mouse cells. (A) Wild-type (WT) MEFs were treated (in triplicate) for 72 h with 1 μM flavine prior to 24 h infection with Semliki forest virus (SFV) (MOI 2). Viral titers were assayed with log10 fold dilutions on confluent Vero cells as shown. NI: not infected (uninfected cells stain with crystal violet); NT: non-treated. (B) WT MEFs were treated with indicated doses of flavine for 24 h before being collected. NT: non-treated. Cytotoxicity assay was performed according to the manufacturer's protocol (the more cytotoxic, the lower the luminescence). Luminescence is shown relative to the NT condition (data are averaged from two independent experiments in biological triplicate ± s.e.m.). (C) Time-course and dose-response showing the activity of 24–48 h flavine treatment of WT MEFs on the levels of Viperin. (A and C) Data shown are representative of a minimum of two independent experiments. (D) IP-10 levels were measured by ELISA in supernatants of WT MEFs treated for 48 h with 1 μM flavine. The data shown are relative to NT condition (data are presented as mean of three independent experiments in biological duplicate ± s.e.m. with unpaired Mann–Whitney U test). (E) RT-qPCR analyses of selected ISGs in WT MEFs treated with 1 μM of flavine for 72 h. Data shown are averaged from three independent experiments in biological triplicate, relative to non-treated cells (± s.e.m. with unpaired Mann–Whitney U tests). **P ≤ 0.01 and ***P ≤ 0.001.
Western blotting
MEFs were seeded in 24-well plates at a density of ∼25 000 cells per well for 48 h and 50 000 cells per well for 24 h. Analyses were carried out as previously described (18). Cell lysates were separated at 80 V on 10% acrylamide gels for 2 h. Following transfer at 20 V for 1.5 h using Bolt Mini Blot Modules (Life Technologies) to Immobilon-FL (Millipore) membranes, membranes were blocked 30 min in Odyssey blocking buffer (LI-COR). The membranes were subsequently incubated overnight with 1:1000 mouse monoclonal anti-Viperin (MaP.VIP | MABF106, Millipore), rabbit anti-mouse p56 (gift from G. Sen, Cleveland Clinic, Cleveland, OH, USA), rabbit anti-mouse Mda5 (D74E4 | 5321, Cell Signaling Technology) or mouse monoclonal anti-β-tubulin (TU-06 | ab7792, Abcam). Finally, conjugated secondary antibodies with Alexa Fluor® 680 dye (Life Technologies) or IRdye800 (Rockland) were used to image the proteins at 700 or 800 nm with an Odyssey scanner (LI-COR).
Semliki forest virus (SFV) infection
A total of 120 000 MEFs or 80 000 hTERT-BJ1 cells were seeded in 24-well plates three days after 1 μM flavine treatment, and left to adhere for several hours, prior to infection with Semliki forest virus (SFV) in complete DMEM (multiplicity of infection (MOI) of 2—as determined by plaque forming units in Vero cells) (each condition was carried out in biological triplicate), as previously described (18). The cells were rinsed 2 h after infection with fresh medium complemented with 2.5% FBS, and further incubated for 22 h. Virus-containing supernatants were collected at 24 h and series-diluted (10-fold dilutions) on 80% confluent Vero cells. After 48 h, surviving Vero cells were fixed with 10% formalin and stained with 0.1% crystal violet (w/v) in 20% ethanol, before several thorough H2O washes.
Rhinovirus infection
Primary human bronchial epithelial cells (PBEC) were obtained and cultured as described previously (23). Studies were approved by the Monash Health and Monash Medical Centre Human Research Ethics Committee; consent was obtained from all subjects, and studies were conducted in accordance with the approved guidelines. Briefly, PBEC were obtained from bronchial brushings during routine bronchoscopy and cultured under submerged conditions on collagen-coated flasks (MP Biomedicals) in supplemented bronchial epithelial growth medium (BEGM; Lonza). When PBEC reached 80% confluency they were treated with flavine for 3 days. Cells were then washed and infected with rhinovirus 16 in BEGM without hydrocortisone at a MOI of 1 for 1 h, then washed and incubated in BEGM without hydrocortisone for 24 h. Supernatants and cell lysates were collected for analysis. Viral titres in supernatants were determined by titration on Ohio HeLa cells, as described previously (24).
Reverse transcription quantitative real-time PCR (RT-qPCR)
Total RNA was purified from cells using the innuPREP Micro RNA Kit (Analytik Jena) or the ISOLATE II RNA Mini Kit (Bioline). For mRNA quantification, cDNA was synthesized from isolated RNA using the High-Capacity cDNA Archive kit (Life Technologies) according to the manufacturer's instructions. RT-qPCR was carried out with the SensiFAST™ SYBR® Hi-ROX Kit (Bioline) on the HT7900 RT-PCR system (Life Technologies). Each polymerase chain reaction (PCR) was carried out in technical duplicate. Mouse and human 18S were used as reference genes. Each amplicon was sequence-verified and used to generate a standard curve for the quantification of gene expression (used in each run). Melting curves were used in each run to confirm specificity of amplification. The primers used were the following: Mouse 18S: mRn18s-FWD GTAACCCGTTGAACCCCATT; mRn18s-REV CCATCCAATCGGTAGTAGCG; Mouse Ifit1: mIfit1-RT-FWD GAGAGTCAAGGCAGGTTTCT; mIfit1-RT-REV TCTCACTTCCAAATCAGGTATGT; Mouse Rsad2: mRsad2-FWD CTGTGCGCTGGAAGGTTT; mRsad2-REV ATTCAGGCACCAAACAGGAC; Mouse Ifnb1: mIfnb1-FWD CCCTATGGAGATGACGGAGA; mIfnb1-REV CCCAGTGCTGGAGAAATTGT; Mouse Cxcl10: mCxcl10-FWD GCTGCCGTCATTTTCTGC; mCxcl10-REV CACTGGGTAAAGGGGAGTGA; Human RSAD2: hRSAD2-RT-FWD TGGTGAGGTTCTGCAAAGTAG; hRSAD2-RT-REV GTCACAGGAGATAGCGAGAATG; Human IFIT1: hIFIT1-FWD TCACCAGATAGGGCTTTGCT; hIFIT1-REV CACCTCAAATGTGGGCTTTT; Human IFIT2: hIFIT2-RT-FWD TTATTGGTGGCAGAAGAGGAAG; hIFIT2-RT-REV CCTCCATCAAGTTCCAGGTG; Human IFIT3: hIFIT3-FWD CATAAAAGCACAGACCTAACAGC; hIFIT3-REV CAGGGAATTCTTGGTGACCTC; Human 18S: h18S-FWD CGGCTACCACATCCAAGGAA; h18S-REV GCTGGAATTACCGCGGCT.
Immunofluorescence
Cells plated on coverslips were fixed in 10% formalin following flavine treatment for 48 h. Detection of γ-H2A.X and cytoplasmic DNA was carried out as previously described (18). Briefly, following cell permeabilization and blocking, DNA staining was performed using 1/50 dilution of anti-DNA (#AC-30-10 Novus Biological) for 1 h. Following phosphate buffered saline washes, A 1/250 dilution of Alexa Fluor® 647 conjugated rabbit monoclonal anti-PhosphoHistone-H2A.X (Ser139) (9720, Cell Signaling) was incubated for 1 h, together with 1/1000 dilution of goat anti-mouse AlexaFluor 568 IgM antibody (used as a secondary antibody for anti-DNA detection). Coverslips were mounted on slides using Prolong Diamond Antifade Mountant with DAPI (Life Technologies, P36971). Confocal imaging was performed using API DeltaVision Widefield and a 20X and 40X objective. Percentages of cytoplasmic positive phospho-γ-H2A.X cells (Figure 3B) were determined by counting cells with cytoplasmic bright foci (using 20X-40X objective), reported to the total number of cells (determined with DAPI staining). Ratios of co-localized cytoplasmic anti-DNA and phospho-γ-H2A.X foci per cell (Figure 3C) were determined by counting bright cytoplasmic foci (using 20X-40X objective), reported to the total number of cells. Percentages of nuclear phospho-γ-H2A.X-positive cells (Figure 4G) were determined by counting cells with nuclear staining above background signal (using 20X-40X objective), reported to the total number of cells (determined with DAPI staining). Data are from two independent experiments with two coverslips per condition per experiment. Overall, at least 170 cells have been counted per condition per independent experiment. Image analyses were performed using ImageJ v1.49.
Figure 3.
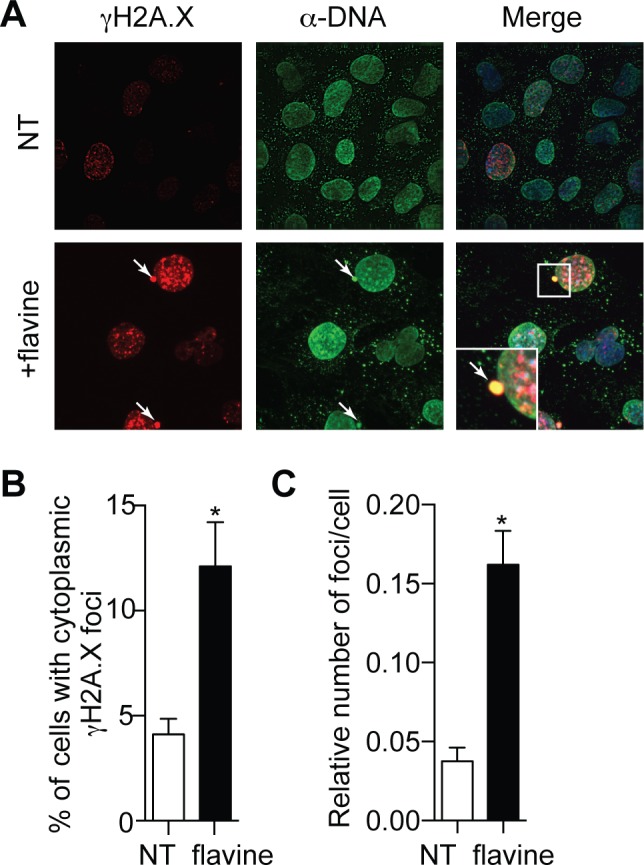
Flavine treatment promotes DNA damage and cytoplasmic DNA leaking. (A) Immunofluorescence of γ-H2A.X staining (red), and anti-dsDNA (green) and DAPI (blue—in Merge) of WT MEFs incubated with 1 μM flavine for 48 h. NT: not-treated. White arrows point to cytoplasmic DNA/phospho-γH2A.X-positive foci. (B) Percentages of cells with cytoplasmic phospho-γ-H2A.X foci and (C) ratio of DNA and phospho-γH2A.X-positive foci per cell; data are averaged from two independent experiments in biological duplicate, with greater than 170 cells counted per condition in each independent experiment (± s.e.m. and unpaired Mann–Whitney U tests are shown). *P ≤ 0.05.
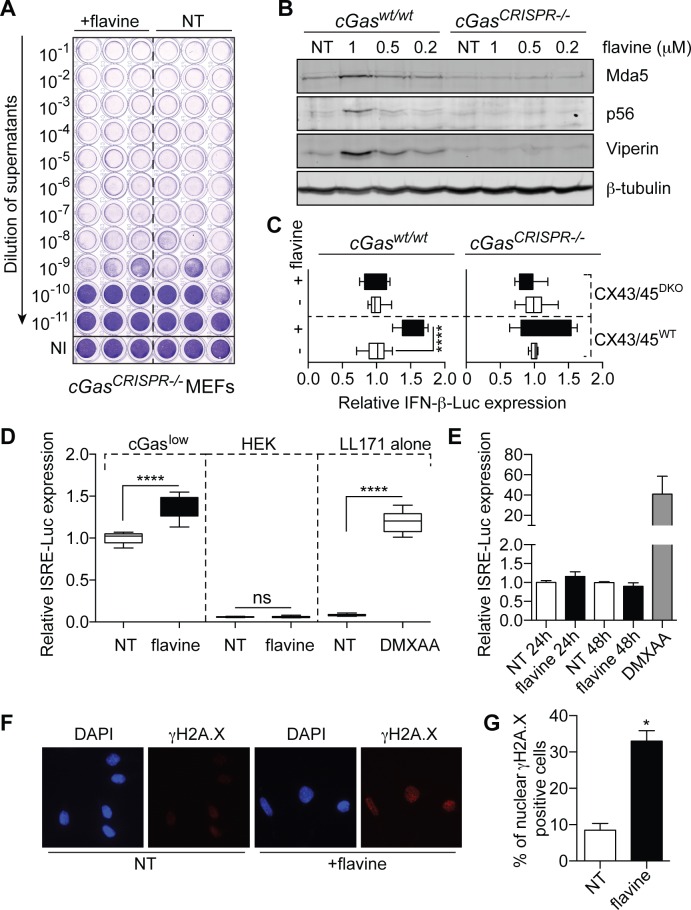
Figure 4.
cGAS-dependent antiviral activity of flavine. (A) Viral titers of cGas-deficient MEFs (19) treated for 72 h with 1 μM flavine and infected for 24 h in biological triplicate with SFV (MOI 2). Viral titers were assayed as in Figure 1. Data shown are representative of three independent experiments in three different clones of cGas-deficient MEFs (19). (B) Dose-response effect of 72 h flavine treatment on MEFs (matched wild-type or cGas-deficient) on the levels of Viperin, Mda5 and p56 by western blot. Data shown are representative of a minimum of two independent experiments. (C) HEK-Sting CX43/45WT and Sting CX43/45DKO cells expressing an IFN-β-Luciferase reporter were co-cultured with MEFs (matched wild-type or cGas-deficient) pre-treated or not with flavine for 24 h. IFN-β-Luciferase expression was reported to the NT condition for each cell line (data presented are averaged from a minimum of two independent experiments in biological triplicate ± s.e.m. and unpaired Mann–Whitney U test is shown). (D) Murine LL171 cells (L929 cells expressing an ISRE-Luciferase reporter) were cultured for 18 h, in the absence (‘LL171 alone’) or presence of HEK cells (wild-type or cGaslow expressing) pre-treated with 1 μM flavine for 24 h. ISRE-Luciferase expression is shown relative to the NT condition for the cGaslow expressing co-culture (data presented are averaged from three independent experiments in biological triplicate ± s.e.m. and unpaired Mann–Whitney U tests are shown). (E) LL171 reporter cells were treated with 1 μM flavine for 24 or 48 h before lysis. NT: not-treated. ISRE-Luciferase activity is shown relative to the NT condition. Data shown are averaged from two independent experiments in biological triplicate (± s.e.m.). (D and E) DMXAA (15 μg/ml) was used as a known agonist of mouse Sting. (F) Immunofluorescence of γ-H2A.X staining (red), and DAPI (blue) in LL171 reporter cells incubated with 1 μM flavine for 48 h. NT: not-treated. (G) Percentages of nuclear phospho-γ-H2A.X-positive cells; data are averaged from two independent experiments in biological duplicate, with >200 cells counted per condition in each independent experiment (± s.e.m. and unpaired Mann–Whitney U tests are shown). ns: not significant, *P ≤ 0.05 and ****P ≤ 0.0001.
Co-culture studies
HEK-Sting were transfected with IFN-β-Luc reporter plasmid (pLuc-IFN-β, a kind gift from K. Fitzgerald, University of Massachusetts). Briefly 0.4 μg of DNA was reverse-transfected in 500 000 HEK-Sting (CX43/45WT or CX43/45DKO) cells using Lipofectamine 2000. After 4 h incubation, the cells were co-cultured with MEF (WT or cGasCRISPR−/− MEFs) that had been previously treated with 1 μM flavine for 24 h (with a ratio of around 85 000 HEKs for 50 000 MEFs). Luciferase activity was analyzed 18 h later as previously reported (18). For co-culture studies with LL171 reporter cells (L929 expressing an IFN stimulated response element (ISRE)-Luciferase) (25), HEK cGASlow were previously treated with 1 μM flavine for 24 h prior to co-culture with LL171 cells for 18 h (ratio of 85 000 HEK for 50 000 LL171).
Statistical analyses
Statistical analyses were carried out using Prism 6 (GraphPad Software Inc.). Two-tailed unpaired non-parametric Mann–Whitney U tests were used to compare pairs of conditions. Symbols used: ns: not significant, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
RESULTS
Flavine promotes host-mediated antiviral effects through interferon induction in mouse embryonic fibroblasts
With the original intention of confirming the host-mediated antiviral effects of acriflavine in vitro, we infected immortalized MEFs with SFV, three days after pre-treatment with a non-toxic dose of flavine (Figure 1A and B). This resulted in a strong antiviral effect, with a >100-fold decrease in viral titers (Figure 1A). Flavine pre-treatment of MEFs raised the basal level of the key antiviral proteins Viperin (encoded by Rsad2), and IP-10 (encoded by Cxcl10) (Figure 1C and D), suggesting a direct contribution of the host innate immune activation (independent of a mutagenic activity of flavine on the virus), to the antiviral effect observed with flavine. In accord with involvement of type-I IFNs, flavine pre-treatment promoted the transcriptional induction of a type I ISG signature—including the antiviral genes Cxcl10, Ifit1, Ifnb1 and Rsad2 (Figure 1E).
Host-mediated antiviral effects of flavine are conserved in human cells
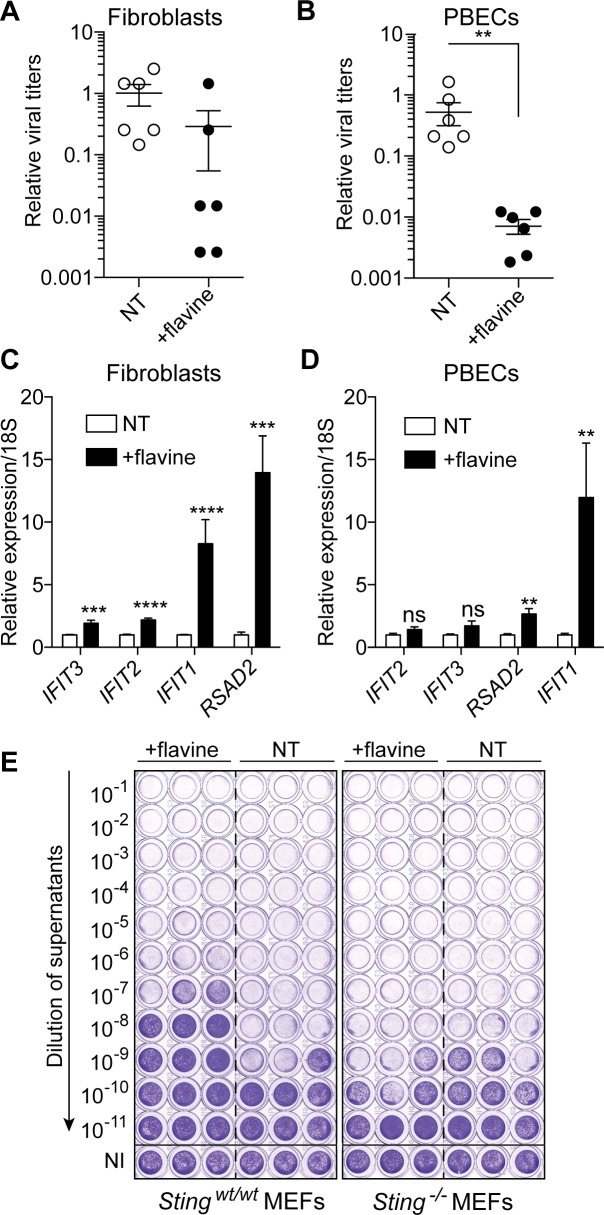
As mentioned above, while CMA and DMXAA selectively activate murine Sting, they do not activate human STING (8,13). To determine whether the effect of flavine was also species-specific, we investigated its ability to activate an antiviral response in human cells. Contrary to the species restriction exhibited by CMA and DMXAA, pre-treatment of immortalized human fibroblasts or primary bronchial epithelial cells with flavine partially protected the cells against SFV or rhinovirus infections, respectively (Figure 2A and B). This protection was concurrent with the transcriptional induction of an ISG signature in both cell models (Figure 2C and D). Given its effect in human cells, we posited that flavine operated differently from CMA and DMXAA, and possibly independently of STING. To rule out STING involvement, we next examined Sting-deficient immortalized MEFs. Unexpectedly, pre-treatment of Sting-deficient immortalized MEFs with flavine thwarted its antiviral effect against SFV (Figure 2E), establishing the Sting-dependent activity of flavine in these cells.
Figure 2.
Flavine treatment activates a type-I IFN antiviral response in human cells. (A) hTERT-BJ1 fibroblasts pre-treated for 72 h with 1 μM flavine were infected for 24 h with SFV (MOI 2) and viral titers were measured on confluent Vero cells as described in Figure 1A. (B) Primary human bronchial epithelial cells (PBECs) were treated with flavine for 72 h prior to infection with rhinovirus 16 (MOI 1) for an additional 24 h and viral titers were determined by titration on HeLa cells. (A and B) Titers are shown relative to NT condition and averaged from two independent experiments in biological triplicate (± s.e.m. with unpaired Mann–Whitney U test shown in B). (C and D) RT-qPCR analyses of selected ISGs in human hTERT-BJ1 fibroblasts (C) and PBECs (D) treated with 1 μM flavine for 72 h alone. Data shown is averaged from three (PBECs) or four (fibroblasts) independent experiments in biological duplicate, relative to non-treated cells (± s.e.m. and unpaired Mann–Whitney U tests are shown). (E) Matched wild-type and Sting-deficient SV40T MEFs were treated for 72 h with 1 μM flavine prior to 24 h infection in biological triplicate with SFV (MOI 2). Viral titers were assayed as in Figure 1. Data shown are representative of a minimum of two independent experiments. ns: not significant, **P ≤ 0.01, ***P ≤ 0.001 and ****P ≤ 0.0001.
Flavine treatment promotes DNA damage and cytoplasmic DNA leaking
The species-independent, STING-dependent effect of flavine observed here led us to propose that flavine engaged the STING pathway differently than CMA and DMXAA. This is supported in part by its delayed induction of anti-viral genes (>24 h) (Figure 1C) (8,9). We and others have recently demonstrated that DNA damage could be associated with cytoplasmic sensing of endogenous DNA products by the STING innate immune pathway (18,26,27). Given the DNA intercalating property of flavine and prior reports of its inhibitory effects on topoisomerase I and II (28), we analyzed the induction of DNA damage in MEFs following flavine treatment. First, the proportion of cells staining positively for cytoplasmic γ-H2A.X was significantly greater in flavine-treated cells (12.1% with flavine treatment versus 4.1% in non-treated cells), indicating an active recruitment of the DNA repair machinery following acriflavine intercalation with DNA and cytoplasmic leakage (Figure 3A and B), in accord with our previous observations with DNA recombinase-toxicity (18). Critically, analyses of cytoplasmic γ-H2A.X foci demonstrated that such cytoplasmic foci contained DNA (using an anti-DNA antibody) (Figure 3A and C). Collectively, these results show that DNA products (possibly in the form of complexes with histones (26)) are leaked into the cytoplasm upon flavine treatment of MEFs.
cGAS-dependent activation of STING by flavine
To determine whether such cytoplasmic DNA could be detected by cGAS, upstream of STING, cGas-deficient MEFs (19) were infected with SFV after flavine pre-treatment. Similarly to Sting-deficiency, cGas-deficiency ablated the antiviral effect of flavine on SFV (Figure 4A) and blunted the induction of antiviral proteins Viperin, p56 (encoded by Ifit1) and Mda5 (encoded by Ifih1) (Figure 4B). Others have previously noted a decrease in the basal levels of several antiviral proteins in cGas-deficient cells (29), possibly a factor also at play in our cGas-deficient MEFs. To clearly establish that flavine engaged cGAS activity, we investigated the production of cGAMP(2′-5′), the product of cGAS, in MEFs treated with flavine. Because cGAMP is transferred between adjacent cells through connexins forming gap junctions, cGAMP activity can be measured in co-cultures of mouse cGAS expressing cells and human STING-reporter recipient cells (21) (see ‘Materials and Methods’ section). Co-culture of flavine-pre-treated wild-type MEFs (i.e. cGAMP ‘donor’ cells) with recipient HEK cells expressing the murine Sting and an IFN-β-Luciferase reporter, exhibited significantly greater IFN-β expression than cells not treated with flavine (Figure 4C). This robust bystander antiviral effect was not seen in cGas-deficient cells, and required expression of connexin 43 and 45 in adjacent HEK-Sting recipient cells, thereby recapitulating the activity of cGAMP (21) (Figure 4C). In line with these findings, HEK cells stably expressing low levels of cGAS (cGASlow) (but not the parental HEK line), pre-treated with flavine resulted in the significant activation of co-cultured Sting competent mouse L929 ISRE-reporter cells (Figure 4D). Noteworthy, direct flavine treatment of L929 ISRE-reporter cells, which express cGas and Sting (15,21), failed to show induction of their reporter (Figure 4E). Further confocal analyses showed that there was no detectable cytoplasmic γ-H2A.X/DNA foci in these cells upon flavine treatment, despite DNA damage being induced (as revealed by increased nuclear γ-H2A.X staining), indicating that DNA leaking was prevented in these cells (Figure 4F and G). This observation rules out the possibility of direct binding of flavine to cGAS, recently suggested for quinacrine (a closely related acridine compound) (30). Conversely, DMXAA stimulation significantly induced expression of the L929 ISRE-reporter cells, in line with direct activation of mouse Sting (Figure 4D and E). Taken together, these results establish the engagement of the cGAS-cGAMP-STING axis by flavine in human and mouse cells, through cytoplasmic leakage of DNA products.
DISCUSSION
Flavine binding to DNA is directly responsible for an important part of its antibacterial activity, which was the main rationale for its use in wound treatment a century ago (1,2). However, if flavine binding to the host's epithelial cell DNA contributes to its antibacterial effects has not been investigated to date. In line with previous case reports of local inflammatory cell recruitment upon treatment with flavine solutions (3,7), our study indicates that flavine can have a direct activity on the host immune system, through its low-DNA damage activity. We note that acriflavine and proflavine were not found to have any carcinogenic effects when used on the skin of mice for long periods of time, or as applied to wounds in humans for over 50 years of worldwide clinical use (5). Given that our observations with flavine were carried out at below toxic ranges, our findings reposition flavine as a potential therapeutic agent in the control of virus infections. Skin infections would be an obvious target, with previous evidence that flavine can show antiviral activity against Herpes Simplex viruses (31). Further, our finding of antiviral activity against rhinovirus in human primary bronchial epithelial cells points to a potential prophylactic use of flavine to prevent upper respiratory tract infections (for instance in the form of intranasal puffer), in populations at increased risk of infections during viral outbreaks.
Mechanistically, our data demonstrate that cytoplasmic DNA accumulates upon flavine treatment, resulting in cGAS recruitment, cGAMP-like activity and STING activation. Our capacity to show cGAMP-like activities in human HEK expressing cGas, confirms that the effects reported here are directly dependent on the expression of cGAS. Importantly, the demonstration of cGAMP-like activity by flavine argues against a possible bias of cGAS deletion on the antiviral effects reported here (29) (as for instance seen with Mda5, basally lower in cGas-deficient cells—Figure 4). To our knowledge, this is the first direct evidence of cGAMP production upon DNA damage—previous reports, including our own, were limited to RNA interference-based downregulation to implicate cGAS sensing (18,26,27).
The response to sub-toxic concentrations of flavine described here is likely to be cell-type dependent, and was, for instance, absent in mouse L929 cells, which express functional levels of cGas and Sting (15,21). Although ruling out a direct activity of flavine on cGAS itself (through physical interaction—as seen with quinacrine (30)), this observation points to the existence of a specific machinery leading to the leaking of cytoplasmic DNA and subsequent cGAS engagement. Indeed, the lack of cytoplasmic DNA foci in flavine treated-L929 indicates that although responding to DNA damage, as seen with increased nuclear γ-H2A.X staining, these cells do not leak DNA into their cytoplasm. Similar to a previous report (26), we find that cytoplasmic DNA co-localizes with histones (as revealed by γ-H2A.X staining). Whether such DNA/histone complex is required for cGAS activation is not presently defined, but it is possible that residual nuclear membranes are involved in packaging this cellular structure, facilitating cGAS/STING sensing (32).
Beyond its antiviral effects, our study suggests that the recently discovered anti-cancer activities of flavine may also benefit from cGAS recruitment (28,33). Indeed, recruitment of the adaptive immune response against tumors is frequently observed in patients, in the form of infiltrating CD8+T cells and was recently proposed to be dependent on STING signaling (34,35). Critically, intratumoral STING activation through the use of cyclic dinucleotide derivatives (mimicking cGAMP activity), promoted tumor regression in animal models (36,37). The recent finding that DNA damage, through γ-irradiation and chemotherapy agents (cisplatin and etoposide) can also result in STING activation (26,27), indicates that chemo- and radio-therapies used in cancer treatment, naturally engage STING. Although this possibility relies on the functionality of the often altered cGAS-STING axis in tumor cells (38), it suggests that several existing anti-cancer treatments already have partial immunotherapy activity. Our discovery that flavine has the capacity to recruit the cGAS-STING signaling pathway, through low level DNA damage, implies that some of its previously described anti-tumoral activities (28,33), could be potentiated by immune recruitment.
Finally, and beyond the demonstration of a direct role for the host immune response to the activity of acriflavine and proflavine, this work argues that several other non-covalently binding DNA chemicals could have the capacity to act as indirect cGAS agonists. Many natural chemicals such as flavonoids are known to inhibit topoisomerase activities (39,40), whilst exhibiting antiviral effects (e.g. quercetin or the green tea catechin (-)-epigallocatechin-3-O-gallate (41)). Further studies are warranted to define whether our findings can be generalized to other non-covalent DNA binding agents, and their known activities on infections or cancers.
Acknowledgments
We thank H.J. Stunden for the SV40T immortalization of the Sting-deficient MEFs; V. Hornung for providing primary Sting-deficient MEFs, hTERT-BJ1 cells, HEK cGASlow, HEK Sting and HEK Sting CX43/45DKO; B. Kile for providing cGasCRISPR−/− MEFs; M. Pelegrin for LL171 cells; and R.E. Smith for help with the redaction of this paper.
FUNDING
Australian NHMRC [1022144 to M.P.G., 1062683 to M.P.G., B.R.G.W., in part]; Australian Research Council Future Fellowship [140100594 to M.P.G.]; Victorian Government's Operational Infrastructure Support Program. Funding for open access charge: Hudson Institute of Medical Research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wainwright M. Acridine-a neglected antibacterial chromophore. J. Antimicrob. Chemother. 2001;47:1–13. doi: 10.1093/jac/47.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Browning C.H. Aminoacridine Compounds as Surface Antiseptics. Br. Med. J. 1943;1:341–343. doi: 10.1136/bmj.1.4289.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leelavathi M., Le Y., Tohid H., Hasliza A. Contact dermatitis presenting as non-healing wound: case report. Asia Pac. Fam. Med. 2011;10:6. doi: 10.1186/1447-056X-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nasim A., Brychcy T. Genetic effects of acridine compounds. Mutat. Res. 1979;65:261–288. doi: 10.1016/0165-1110(79)90005-8. [DOI] [PubMed] [Google Scholar]
- 5.Browning C.H. Proflavine and acriflavine. Br. Med. J. 1967;2:111. doi: 10.1136/bmj.2.5544.111-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diederich J., Lodemann E., Wacker A. Basic dyes as inducers of interferon-like activity in mice. Arch. Gesamte Virusforsch. 1973;40:82–86. doi: 10.1007/BF01242639. [DOI] [PubMed] [Google Scholar]
- 7.Lim J., Goh C.L., Lee C.T. Perioral and mucosal oedema due to contact allergy to proflavine. Contact Dermatitis. 1991;25:195–196. doi: 10.1111/j.1600-0536.1991.tb01833.x. [DOI] [PubMed] [Google Scholar]
- 8.Cavlar T., Deimling T., Ablasser A., Hopfner K.P., Hornung V. Species-specific detection of the antiviral small-molecule compound CMA by STING. EMBO J. 2013;32:1440–1450. doi: 10.1038/emboj.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prantner D., Perkins D.J., Lai W., Williams M.S., Sharma S., Fitzgerald K.A., Vogel S.N. 5,6-Dimethylxanthenone-4-acetic acid (DMXAA) activates stimulator of interferon gene (STING)-dependent innate immune pathways and is regulated by mitochondrial membrane potential. J. Biol. Chem. 2012;287:39776–39788. doi: 10.1074/jbc.M112.382986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishikawa H., Barber G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu S., Cai X., Wu J., Cong Q., Chen X., Li T., Du F., Ren J., Wu Y.T., Grishin N.V., et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 12.Kim S., Li L., Maliga Z., Yin Q., Wu H., Mitchison T.J. Anticancer flavonoids are mouse-selective STING agonists. ACS Chem. Biol. 2013;8:1396–1401. doi: 10.1021/cb400264n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao P., Ascano M., Zillinger T., Wang W., Dai P., Serganov A.A., Gaffney B.L., Shuman S., Jones R.A., Deng L., et al. Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell. 2013;154:748–762. doi: 10.1016/j.cell.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao P., Zillinger T., Wang W., Ascano M., Dai P., Hartmann G., Tuschl T., Deng L., Barchet W., Patel D.J. Binding-pocket and lid-region substitutions render human STING sensitive to the species-specific drug DMXAA. Cell Rep. 2014;8:1668–1676. doi: 10.1016/j.celrep.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun L., Wu J., Du F., Chen X., Chen Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mankan A.K., Schmidt T., Chauhan D., Goldeck M., Honing K., Gaidt M., Kubarenko A.V., Andreeva L., Hopfner K.P., Hornung V. Cytosolic RNA:DNA hybrids activate the cGAS-STING axis. EMBO J. 2014;33:2937–2946. doi: 10.15252/embj.201488726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ablasser A., Goldeck M., Cavlar T., Deimling T., Witte G., Rohl I., Hopfner K.P., Ludwig J., Hornung V. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pepin G., Ferrand J., Honing K., Jayasekara W.S., Cain J.E., Behlke M.A., Gough D.J., Williams B.R.G., Hornung V., Gantier M.P. Cre-dependent DNA recombination activates a STING-dependent innate immune response. Nucleic Acids Res. 2016;44:5356–5364. doi: 10.1093/nar/gkw405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White M.J., McArthur K., Metcalf D., Lane R.M., Cambier J.C., Herold M.J., van Delft M.F., Bedoui S., Lessene G., Ritchie M.E., et al. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell. 2014;159:1549–1562. doi: 10.1016/j.cell.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gantier M.P., McCoy C.E., Rusinova I., Saulep D., Wang D., Xu D., Irving A.T., Behlke M.A., Hertzog P.J., Mackay F., et al. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. 2011;39:5692–5703. doi: 10.1093/nar/gkr148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ablasser A., Schmid-Burgk J.L., Hemmerling I., Horvath G.L., Schmidt T., Latz E., Hornung V. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature. 2013;503:530–534. doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stifter S.A., Gould J.A., Mangan N.E., Reid H.H., Rossjohn J., Hertzog P.J., de Weerd N.A. Purification and biological characterization of soluble, recombinant mouse IFNbeta expressed in insect cells. Protein Expr. Purif. 2014;94:7–14. doi: 10.1016/j.pep.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 23.Thomas B.J., Porritt R.A., Hertzog P.J., Bardin P.G., Tate M.D. Glucocorticosteroids enhance replication of respiratory viruses: effect of adjuvant interferon. Sci. Rep. 2014;4:7176. doi: 10.1038/srep07176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas B.J., Lindsay M., Dagher H., Freezer N.J., Li D., Ghildyal R., Bardin P.G. Transforming growth factor-beta enhances rhinovirus infection by diminishing early innate responses. Am. J. Respir. Cell Mol. Biol. 2009;41:339–347. doi: 10.1165/rcmb.2008-0316OC. [DOI] [PubMed] [Google Scholar]
- 25.Uze G., Di Marco S., Mouchel-Vielh E., Monneron D., Bandu M.T., Horisberger M.A., Dorques A., Lutfalla G., Mogensen K.E. Domains of interaction between alpha interferon and its receptor components. J. Mol. Biol. 1994;243:245–257. doi: 10.1006/jmbi.1994.1651. [DOI] [PubMed] [Google Scholar]
- 26.Ahn J., Xia T., Konno H., Konno K., Ruiz P., Barber G.N. Inflammation-driven carcinogenesis is mediated through STING. Nat. Commun. 2014;5:5166. doi: 10.1038/ncomms6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartlova A., Erttmann S.F., Raffi F.A., Schmalz A.M., Resch U., Anugula S., Lienenklaus S., Nilsson L.M., Kroger A., Nilsson J.A., et al. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity. 2015;42:332–343. doi: 10.1016/j.immuni.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 28.Hassan S., Laryea D., Mahteme H., Felth J., Fryknas M., Fayad W., Linder S., Rickardson L., Gullbo J., Graf W., et al. Novel activity of acriflavine against colorectal cancer tumor cells. Cancer Sci. 2011;102:2206–2213. doi: 10.1111/j.1349-7006.2011.02097.x. [DOI] [PubMed] [Google Scholar]
- 29.Schoggins J.W., MacDuff D.A., Imanaka N., Gainey M.D., Shrestha B., Eitson J.L., Mar K.B., Richardson R.B., Ratushny A.V., Litvak V., et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.An J., Woodward J.J., Sasaki T., Minie M., Elkon K.B. Cutting edge: antimalarial drugs inhibit IFN-beta production through blockade of cyclic GMP-AMP synthase-DNA interaction. J. Immunol. 2015;194:4089–4093. doi: 10.4049/jimmunol.1402793. [DOI] [PubMed] [Google Scholar]
- 31.Froissant-Richard S., Richard M. Pharmaceutical compositions based on acriflavine useful for the treatment of herpes and zona. 2001. EP 0649656 B1. Google Patents.
- 32.Motani K., Ito S., Nagata S. DNA-mediated cyclic GMP-AMP synthase-dependent and -independent regulation of innate immune responses. J. Immunol. 2015;194:4914–4923. doi: 10.4049/jimmunol.1402705. [DOI] [PubMed] [Google Scholar]
- 33.Lee K., Zhang H., Qian D.Z., Rey S., Liu J.O., Semenza G.L. Acriflavine inhibits HIF-1 dimerization, tumor growth, and vascularization. Proc. Natl. Acad. Sci. U.S.A. 2009;106:17910–17915. doi: 10.1073/pnas.0909353106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Woo S.R., Fuertes M.B., Corrales L., Spranger S., Furdyna M.J., Leung M.Y., Duggan R., Wang Y., Barber G.N., Fitzgerald K.A., et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41:830–842. doi: 10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corrales L., McWhirter S.M., Dubensky T.W., Jr, Gajewski T.F. The host STING pathway at the interface of cancer and immunity. J. Clin. Invest. 2016;126:2404–2411. doi: 10.1172/JCI86892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corrales L., Glickman L.H., McWhirter S.M., Kanne D.B., Sivick K.E., Katibah G.E., Woo S.R., Lemmens E., Banda T., Leong J.J., et al. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 2015;11:1018–1030. doi: 10.1016/j.celrep.2015.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chandra D., Quispe-Tintaya W., Jahangir A., Asafu-Adjei D., Ramos I., Sintim H.O., Zhou J., Hayakawa Y., Karaolis D.K., Gravekamp C. STING ligand c-di-GMP improves cancer vaccination against metastatic breast cancer. Cancer Immunol. Res. 2014;2:901–910. doi: 10.1158/2326-6066.CIR-13-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia T., Konno H., Ahn J., Barber G.N. Deregulation of STING signaling in colorectal carcinoma constrains DNA damage responses and correlates with tumorigenesis. Cell Rep. 2016;14:282–297. doi: 10.1016/j.celrep.2015.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boege F., Straub T., Kehr A., Boesenberg C., Christiansen K., Andersen A., Jakob F., Kohrle J. Selected novel flavones inhibit the DNA binding or the DNA religation step of eukaryotic topoisomerase I. J. Biol. Chem. 1996;271:2262–2270. doi: 10.1074/jbc.271.4.2262. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki K., Yahara S., Hashimoto F., Uyeda M. Inhibitory activities of (-)-epigallocatechin-3-O-gallate against topoisomerases I and II. Biol. Pharm. Bull. 2001;24:1088–1090. doi: 10.1248/bpb.24.1088. [DOI] [PubMed] [Google Scholar]
- 41.Kim Y., Narayanan S., Chang K.O. Inhibition of influenza virus replication by plant-derived isoquercetin. Antiviral Res. 2010;88:227–235. doi: 10.1016/j.antiviral.2010.08.016. [DOI] [PubMed] [Google Scholar]