Abstract
The DEAH-box ATP-dependent RHAU helicases specifically unfold RNA and DNA G-quadruplexes (G4s). However, it remains unclear how the RHAU's G4 unfolding activity is coupled to different stages of the ATPase cycle. Here, using a single-molecule manipulation approach, we show that binding of Drosophila RHAU stabilizes an intramolecularly folded parallel DNA G4 against mechanical unfolding in its nucleotide-free and in its AMP-PNP or ADP bound states, while it destabilizes the G4 when coupled to ATP hydrolysis. Importantly, our results show that the ADP·AlF -bound RHAU does not stabilize the G4. We also found that both a single-stranded 3′ DNA tail and the RSM domain of RHAU that binds specifically to the G4 structure, are dispensable for the stabilization of the G4, but both are required for G4 destabilization. Our study provides the first evidence that the unfolding kinetics of a G-quadruplex can be modulated by different nucleotide-bound states of the helicase.
-bound RHAU does not stabilize the G4. We also found that both a single-stranded 3′ DNA tail and the RSM domain of RHAU that binds specifically to the G4 structure, are dispensable for the stabilization of the G4, but both are required for G4 destabilization. Our study provides the first evidence that the unfolding kinetics of a G-quadruplex can be modulated by different nucleotide-bound states of the helicase.
INTRODUCTION
G-quadruplexes, or G4 structures, are four stranded nucleic acid structures formed by the circular Hoogsteen hydrogen bonding of four guanines forming a G-tetrad (1). DNA or RNA sequences containing four or more G-tracts (three or more guanines) fold intramolecularly to form G4 structures at physiological salt concentrations (1,2). Studies have shown that G4 structures exist in vivo (3,4) where they are involved in multiple biological processes including DNA replication, transcription, translation and telomere maintenance (2). Due to their high thermal and kinetic stability, G4 structures often present a physical barrier to various processes taking place on DNA and RNA (5,6). In vivo, the G4 barrier is overcome by a set of helicases that can unfold G4 structures (7–10). Functionally defective mutants of some of these helicases have been reported to cause genome instability and are associated with various diseases (10).
RHAU helicase, also named G4 resolvase 1 (DHX36 gene product, or G4R1, from human) is a superfamily 2 DEAH-box helicase that plays a role in heart development (11), hematopoiesis (12) and spermatogenesis (13). RHAU was originally identified as a RNA helicase that binds RNA G4 with sub-nanomolar affinity (14–17). Human RHAU is the major source of G4 resolving activity in Hela cell lysates (15,16). Unlike many other G4 helicases such as Pif1 and BLM that can also unfold DNA duplexes (18,19), RHAU exhibits a strong preference of unfolding G4 structures (15,16). It has been reported that RHAU regulates the metabolism of various RNAs containing G4-forming sequences, including telomerase RNA hTR (20,21), several mRNAs such as YY1 (22), PITX1 (23), Nkx2-5 (11). RHAU has also been shown to bind DNA G4 with similar affinity to RNA and can unfold DNA G4 (24), which also has crucial biological functions. In addition, a recent study revealed that RHAU directly binds to the DNA G4 structures formed on the promoter region of c-Kit, facilitating its expression and regulating spermatogonia differentiation (13). Sequence analysis showed that almost all metazoan species have RHAU orthologues (25). Among these sequences, the Drosophila melanogaster protein CG9323 (hereafter referred as DmRHAU) was shown to have a specific G4 unfolding activity similar to that of the human RHAU (25).
The primary structure of RHAU consists of two RecA-like domains, which form the helicase core region (14) (Supplementary Figure S1). Like all superfamily 2 helicases, the adenosine triphosphate (ATP) binding and hydrolysis site of RHAU is predicted to be located at the interface cleft of the two RecA-like domains. A large part of the C-terminal domain of RHAU is conserved among all DEAH-box helicases and has an unknown function. RHAU helicase has a conserved G4 specific binding motif, the RHAU specific motif (RSM), located at the N-terminal extremity (25). A solution structure of the peptide from the N-terminal region of RHAU bound to a parallel DNA G4 showed the RSM motif to fold into an α-helix that recognizes the G4 by stacking on the surface of the outer G-tetrad of the G-quadruplex (26).
Previous biochemical studies showed that G4-unfolding by RHAU requires ATP hydrolysis (16,24). It has been suggested that the ATPase cycle controls the conformation of a helicase and its interaction with nucleic acids substrate by switching between different nucleotide bound states (27–29). For example, in its nucleotide-free state the hepatitis C virus (HCV) NS3 helicase binds to single-strand DNA (ssDNA) substrates with nano-molar affinity, switching to a 100-fold weaker binding in the presence of an ATP analog (27). Such a change in affinity is crucial for the translocation of NS3 along the single-stranded nucleic acid (27). Another example is that several DEAD-box helicases show different unwinding activity in the presence of different ATP analogues that mimic different states in ATPase cycle and the results suggest that ATP binding but not the hydrolysis is necessary for the unwinding activity of these helicases (28). A recent study of human RHAU showed the G4 unfolding rate, but not the ATPase rate to be inversely correlated with the stability of the G4 substrate (30). However, it remains unclear how RHAU's G4 unfolding activity is coupled to the different stages along an ATPase cycle.
It has been experimentally demonstrated that the G4 unfolding activity of RHAU requires the recognition of the G4 structure by the N-terminal RSM domain (25). However, the isolated RSM peptide binds to G4 with micromolar affinity, much weaker than the sub-nanomolar G4 binding affinity observed for the full length human RHAU (26). This observation raises the question whether another domain in RHAU participates in G4 binding. In addition, RHAU likely interacts with ssDNA/ssRNA through the nucleic acid-binding cavity formed by the two RecA domains and the conserved C-terminal domain, as suggested by a recent X-ray crystallographic structure of MLE bound to a ssRNA, a closely related DEAH-box helicase (29). However, whether such ssDNA/ssRNA binding is needed for stable binding to G4 is unknown.
The lack of knowledge of how RHAU's G4 unfolding activity is coupled to different stages in one ATPase cycle and how it is affected by its nucleic acids substrate binding has hindered our understanding of the mechanism by which RHAU unfolds G4 structures. To address these questions, we have investigated different nucleotide bound states of DmRHAU, the specific contribution of the RSM domain as well as the effect of a ssDNA tail preceding the G4 on G4 binding and unfolding. Contrary to the unwinding activity of helicases measured in previous bulk assay mainly based on analyzing unwound products (25), our studies are based on the mechanical manipulation of a single G4 molecules using magnetic tweezers. This approach is able to detect the dynamics of G4 unfolding with high spatial and temporal resolutions (6,31). Our results suggest that the ability of RHAU to destabilize G4s is regulated by different nucleotide bound states in an ATPase cycle, and is crucially dependent on the interaction with ssDNA as well as with the G4 specifically through the RSM domain of RHAU.
MATERIALS AND METHODS
DNA and protein preparations
All oligonucleotides were purchased from Integrated DNA Technology. The G4 forming ssDNA sequences were ligated with 5′-thiol labeled 489 bp and 5′-biotin labeled 601 bp dsDNA prepared by polymerase chain reaction as described previously (6,32). The ligated products were purified by gel extraction with PureLink kit (Invitrogen). More details of the DNA constructs are included in Supplementary Data. All recombinant Drosophila melanogaster and human RHAU proteins were expressed in Escherichia coli and purified to homogeneity as described previously (24). The resulting proteins were judged to be >95% pure by SDS-PAGE analysis. The concentration of the proteins was determined spectrophotometrically. The G4 unwinding activity of each batch of purified recombinant DmRHAU was assayed on a tetramolecular G4 DNA substrate and by analyzing the reaction products by native PAGE (data not shown).
Single-molecule magnetic tweezers
A flow chamber was constructed on a (3-Aminopropyl) triethoxy silane (APTES; Sigma-Aldrich) functionalized coverslip. The thiol-end of DNA was covalently attached to the amine group of APTES via a sulfo-SMCC crosslinker (Thermo Scientific). The chamber was then blocked with a bovine serum albumin (BSA) solution (10 mg/ml BSA, 1 mM 2-mercaptoethanol, 1 × phosphate buffered saline pH 7.4 buffer) before introducing 2.8 μm-diameter streptavidin-coated paramagnetic beads (Dynal M-280, Life technologies). Unwinding assays were carried out at 23°C in a buffer composed of 10 mM Tris-HCl (pH8.0), 100 mM KCl, 2 mM MgCl2. The home-built magnetic tweezers were controlled by in-house-written LabVIEW program (National Instruments). The extension change of the construct was determined from the diffraction pattern of the bead recorded using a CCD camera that acquired ∼200 frames per second. Due to off-center attachment that often happens in magnetic tweezers experiments, which can cause bead rotation at different forces, the bead height difference between two different forces is not equal to the extension difference of the molecule (31). However, such rotation does not occur at the same force; therefore the height change at a constant force is equivalent to the extension change. Force was controlled by changing the distance between the permanent magnets and the flow chamber. The magnetic tweezers have a spatial resolution for bead stuck on surface of ∼2 nm, and the force calibration has a relative error of <10% (31). More details of the magnetic tweezers design and force calibration were described in our previous publications (31).
ATPase assay
ATPase activity was measured using an ATP-regenerating system as previously reported (33). The ATPase assay were carried out at 23°C in a buffer contained 10 mM Tris-HCl (pH8.0), 100 mM KCl, 2 mM MgCl2, 0.2 mM NADH Roche 10107735001), 200 μg/ml pyruvate kinase from rabbit muscle (Roche 10128155001), 2.5 mM Phosho(enol) pyruvic acid (Sigma P7252). First, 1 mM ATP was added to the solution, followed by different concentrations of DNA and finally 100 nM RHAU helicase to start the ATPase assay. The absorbance at 340 nm was measured using a Tecan Infinite F200 plate reader.
Nucleotides
ATP (A2383), AMP-PNP (A2647), ADP (A2754) were purchased from Sigma-Aldrich. The ATP regenerating system [200 μg/ml pyruvate kinase from rabbit muscle (Roche 10128155001), 2.5 mM Phosho(enol) pyruvic acid (Sigma P7252)] was mixed with ATP before use of ATP. The ADP (Sigma A2754) solution was treated with hexokinase (Sigma H4502) and D-glucose (1st BASE) to remove the contaminating ATP as described previously (34). The ADP solution (80 mM) was incubated with 200 mM D-glucose, 2 mM MgCl2, 0.04 U/μl hexokinase (Sigma H4502-500UN) in room temperature for 2 h. After incubation, the enzyme was eliminated from these solution using a Amicon Ultra-0.5 column. The ADP·AlF was prepared by incubating 10 mM ATP-depleted ADP, 50 mM NaF (Sigma 67414) and 10 mM AlCl3 (Sigma 563919) as described previously (35).
was prepared by incubating 10 mM ATP-depleted ADP, 50 mM NaF (Sigma 67414) and 10 mM AlCl3 (Sigma 563919) as described previously (35).
RESULTS
DmRHAU binding stabilizes the G4 structure and destabilizes it by hydrolyzing ATP
In the current study, DmRHAU was investigated based on its effects on the mechanical stability of a DNA G4 structure using single-molecule magnetic tweezers. To this end, we used the previously characterized intramolecularly folded parallel DNA G4 structure (Myc2345) formed on the c-MYC G4 forming sequence (6). A parallel-stranded G4 structure was used because previous study suggested that RHAU has a strong preferential binding to parallel-stranded DNA and RNA G4s (26). The DNA used in the magnetic tweezers experiments consisted of a single-stranded Myc2345 DNA sequence, d(GGGT GGGGA GGGT GGGG), spanned between two dsDNA handles and tethered between a coverslip surface and a streptavidin-coated paramagnetic bead (Figure 1A). Force was applied to the molecule through the bead by a pair of magnets, with the force level controlled by adjusting the distance of the magnets from the bead. Since previous studies had suggested that a 3′ ssDNA/ssRNA tail can usually facilitate DEAH-box helicase unwinding activity (36,37), we introduced a 15-nt single-stranded thymine tail (T-tail) at the 3′ end of the G4 forming sequence to facilitate DmRHAU's activity. Hereafter, we denote the construct of Myc2345 with the 15 nt T-tail by G4-15T. A 3-nt stretch was included at the 5′ side as a spacer between the G4 and the dsDNA handle.
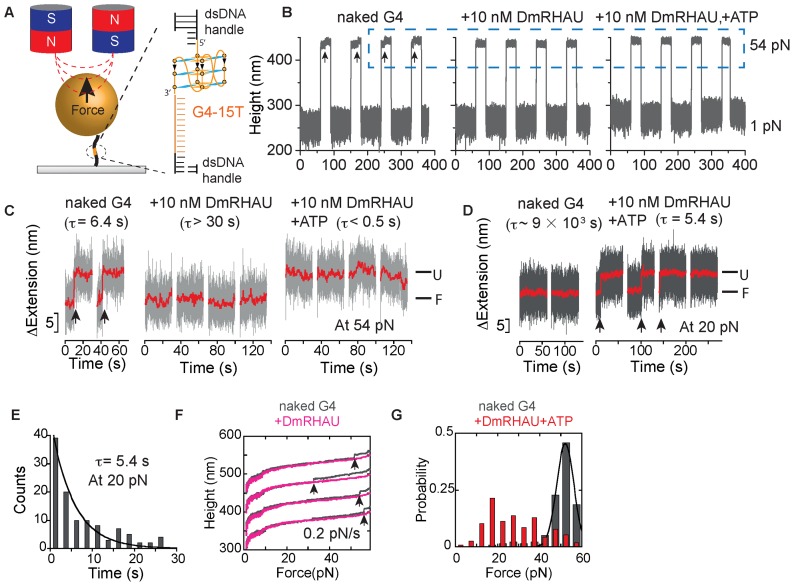
Figure 1.
(A) Schematic of the magnetic tweezers setup and the G4-15T DNA construct. A Myc2345 ssDNA sequence (G4-15T, orange) is flanked by two dsDNA handles (489 bp and 601 bp, black) and is tethered between a paramagnetic bead and a coverslip. The magnified area shows the local region containing the G4-15T sequence folded into a G4 structure. (B) Representative time trace of the bead height change during force jump cycles between 1 pN and 54 pN measured without DmRHAU, with 10 nM DmRHAU but without ATP, and with both 10 nM DmRHAU and 1 mM ATP, as indicated in the figure panel. (C) Representative time trace of extension change of a G4 molecule (grey data) after jumping to 54 pN in several force-jump cycles taken from data in Figure 1B. The extensions corresponding to folded (F) and unfolded (U) G4 are also indicated; G4 unfolding events are indicated by arrows. (D) Representative time trace of extension change of a G4 molecule after jumping to 20 pN in several force-jump cycles of 1 pN (60 s) to 20 pN (60 s) to 54 pN (30 s), in the absence (left) and presence (right) of DmRHAU and ATP. Unfolding events are indicated by arrows. In (C and D), red traces are smoothed data using adjacent averages with a 0.5-s sliding window. (E) Histogram of the lifetime of G4-15T at 20 pN (110 data obtained from 10 independent molecules) in the presence of 10 nM DmRHAU and 1 mM ATP. (F) Force-extension curve of G4-15T in the absence (grey) and presence (pink) of DmRHAU. (G) Unfolding force distribution of G4-15T in the absence (grey) and presence (red) of DmRHAU and ATP.
In order to characterize the unfolding kinetics of the intramolecularly folded G4-15T in a physiological KCl concentration, a force-jump assay was used (6). As shown in a representative trace of the bead height during four force-jump cycles (Figure 1B left panel), the force applied to the molecule was cycled between 54 pN for 30 s under which a folded G4-15T could be unfolded, and 1 pN for 60 s, under which an unfolded G4-15T could refold. The unfolding of G4-15T at 54 pN is indicated by a sudden increase in bead height of 8 ± 2 nm (n = 323, mean ± s.d.) from the coverslip surface (arrows). The probability that unfolding at 54 pN occurred during the 30 s holding time was calculated to be ∼94% from 345 cycles obtained in more than 20 independent experiments. The average lifetime of folded G4-15T at 54 pN was calculated to be τ = 6.4 ± 0.4 s from measured life time distribution (Supplementary Figure S2).
The effects of the DmRHAU on the unfolding kinetics of the G4-15T were probed using the same force-jump assay (Figure 1B middle and right panels). Figure 1C shows the zoom-in of Figure 1B (data in the dashed box) to highlight the extension change at 54 pN. Data corresponding to the first two cycles (Figure 1C, left two cycles) were obtained before DmRHAU was introduced, which shows unfolding steps of G4 alone within several seconds after jumping to 54 pN. After flowing in a solution of DmRHAU without ATP, the extension remained at the folded level throughout the 30 s holding time at 54 pN, indicating that DmRHAU strongly stabilizes the G4-15T structure against mechanical unfolding (Figure 1C, middle four cycles, also see Supplementary Figures S3 and S4 for the accuracy in assigning folded or unfolded states based on the bead height during the force-jump assay). The DmRHAU induced stabilizing effect is strong – in five independent experiments, we never observed unfolding of the G4-15T at 54 pN within the longest holding time of 300 s.
After flowing in DmRHAU and ATP, the extension of the same molecule right after jumping from 1 pN to 54 pN corresponded to the level of unfolded G4-15T (Figure 1B and C, right four cycles). This result is in sharp contrast to the stabilized folded G4-15T by DmRHAU in the absence of ATP or to the naked folded G4-15T, which has a lifetime of a few seconds at 54 pN. It indicates that the G4-15T was in an unfolded state before the force was increased to 54 pN. Force cycle experiments in the presence of DmRHAU and ATP were repeated on 34 different molecules, each for an average of 10 cycles. In the majority of the experiments (n = 26), the G4-15T molecules were in an unfolded state prior to the jump to 54 pN in all cycles. However, it should be stressed that in five experiments, the G4-15T molecules remained in a stably folded conformation at 54 pN in all cycles. In the remaining three experiments, the G4-15T molecules were in an unfolded state prior to jumping to 54 pN in the initial a few cycles but then folded and remained folded in the rest of cycles (Supplementary Figure S5). The reason why in these eight experiments DmRHAU could not unfold the G4-15T molecules remains unclear. We reason that it may indicate the presence of some DmRHAU with impaired G4 unfolding activity, which binds and stabilizes the G4. The inactive DmRHAU has a low exchange rate with free DmRHAU in solution, thereby locking the G4 in a folded state for the whole holding time. However, overall, the results from the majority (76%) of these experiments suggest that DmRHAU was able to unfold G4-15T at low forces in the presence of ATP.
Under the employed experimental conditions, it is not possible to discern whether the G4-15T was unfolded during the holding of the molecule at 1 pN for 60 s, or during the force-jump to 54 pN. Our instrumental setup typically takes less than 0.5 s for the force to increase from 1 to 54 pN. However, this time-lapse is still sufficient for G4-15T to be unfolded during the force-increase procedure (before reaching 54 pN). To test these possibilities, we carried out new force-jump cycles between 1 pN for 60 s, 20 pN for 60 s, and finally 54 pN for 30 s. The highest force of 54 pN was used to ensure complete G4 unfolding at the end of each cycle. The extension change at 20 pN during the new force-jump cycles showed that in the absence of DmRHAU and ATP, the G4-15T remained in a folded state throughout the 60s holding time (Figure 1D left two cycles). After DmRHAU and ATP were added, the extension revealed G4 unfolding signals at 20 pN in more than half of the cycles, indicating that G4-15T was in the folded state at 1 pN before the force was increased to 20 pN (Figure 1D, right four cycles. See also Supplementary Figure S6 for more time traces). Unfolding of G4-15T after increasing to 20 pN was observed in 71 ± 7% (mean ± s.e.) of all force cycles obtained from multiple tethers (129 cycles, from 6 molecules). In the remaining ∼29% of cycles, G4 was in the unfolded state right after jumping to 20 pN. The lifetime histogram of G4-15T in the presence of DmRHAU and ATP at 20 pN showed single-exponential decay with an average lifetime of τ = 5.4 ± 0.7 s (mean ± s.e.)(Figure 1E). This value is one order of magnitude higher than the time needed to jump from 1 to 20 pN. Therefore, in the remaining 29% cycles, G4-15T should be in the unfolded state at 1 pN prior to force jump. The lifetime at 20 pN is also contrasts with the ∼9 × 103 s lifetime of G4 alone (i.e. in the absence of DmRHAU) estimated at the same force (Supplementary information: ‘Force-dependent unfolding of G4-15T’) (6). Taken together, these results suggest that DmRHAU accelerates the unfolding rate of the G4 structure by three orders of magnitude at 20 pN.
To further confirm the stabilization and destabilization effects of DmRHAU on G4 structures, we also measured the unfolding force distribution of G4-15T using force-ramp measurements. Figure 1F shows representative force-extension curves for the same G4-15T before and after introduction of DmRHAU at a loading rate of 0.2 pN/s. The unfolding force distribution of G4-15T obtained from such loading rate study shows a single peak at ∼52 pN (Figure 1G, data in grey), similar to that obtained from the tail-less Myc2345 G4 (∼50 pN) measured in our previous study (6). In contrast, in all cases in the presence 10 nM DmRHAU, G4-15T remained folded during the force-increase scan up to 60 pN, indicating G4 stabilization by DmRHAU binding (Figure 1F, data in pink). Of note, the force was capped below 60 pN to avoid overstretching transition of the two dsDNA handles (38). In the presence of DmRHAU and ATP, the unfolding force distribution was significantly shifted to lower forces (Figure 1G, data in red), indicative of an ATP-dependent destabilization of G4 by DmRHAU (See also Supplementary Figure S7).
Based on these results, we conclude that DmRHAU stabilizes G4-15T in the absence of ATP, but destabilizes G4-15T in the presence of ATP. Similar results were obtained when these experiments were repeated using human RHAU (Supplementary Figure S8), suggesting that G4 stabilization and destabilization effects are conserved among RHAU helicases.
Effects of different nucleotide bound states on the destabilization of G4-15T by DmRHAU
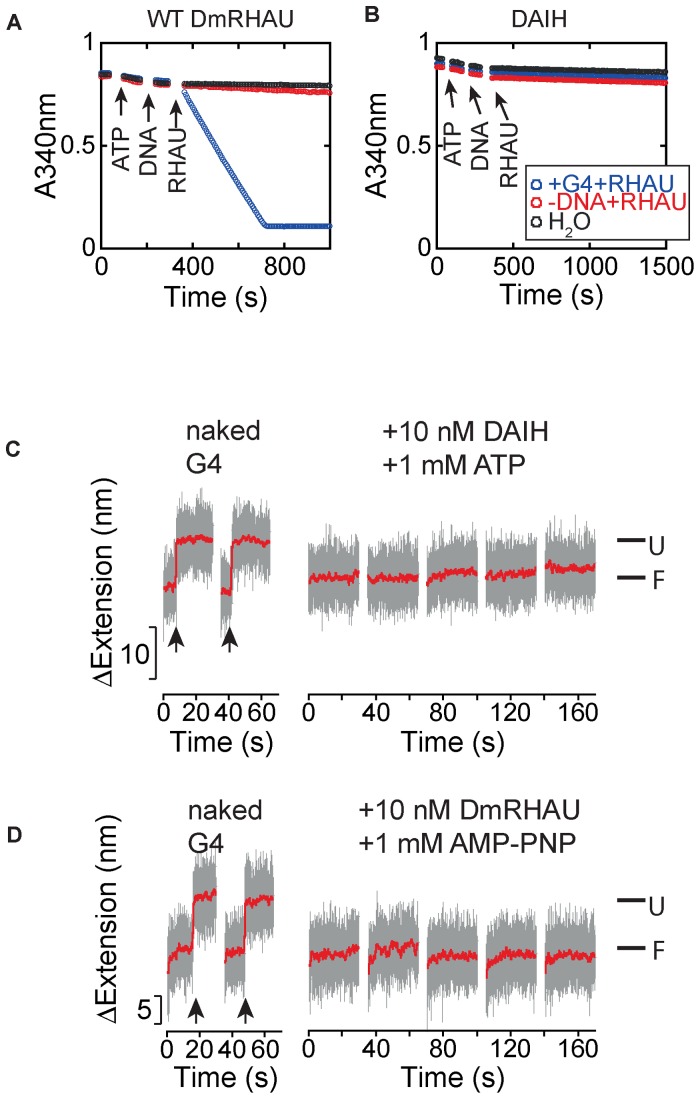
We next sought to investigate how the DmRHAU-dependent destabilization of the G4 structure is affected by different nucleotide bound states in a single ATPase cycle. To assess whether ATP binding causes destabilization, we used the force-jump assay to examine the stability of G4-15T in the presence of an ATPase-deficient mutant (DAIH) of DmRHAU. In this mutant, the Glu-279 residue within the Walker B motif (DEIH) was substituted with an alanine to abolish its ATPase activity. Using an NADH-coupled ATPase assay, we confirmed that the DAIH mutant was defective in ATPase activity while the wild-type DmRHAU showed pronounced a G4 stimulated ATPase activity (Figure 2A and B).
Figure 2.
ATP hydrolysis is required for G4 unwinding by DmRHAU. (A and B) NADH-coupled ATPase assay for (A) wild-type DmRHAU and (B) ATPase-deficient DAIH mutant in the presence and absence of 400 nM G4-15T (blue and red, respectively). Controls without DmRHAU are indicated by black data points. The steady-state ATP hydrolysis rates were determined from the slope of absorbance decrease at 340 nm. (C) Representative time trace of extension change of a G4-15T tether at 54 pN obtained in force-jump cycles of 1 pN (60 s) to 54 pN (30 s), before (left) and after (right) introduction of DAIH and ATP. (D) Representative time trace of extension change of a G4-15T tether at 54 pN obtained in force-jump cycles of 1 pN (60 s) to 54 pN (30 s), before (left) and after (right) introduction of DmRHAU and AMP-PNP.
The force-jump assay showed that G4-15T remained folded throughout the 30 s holding time at 54 pN after introduction of the DAIH mutant and ATP (Figure 2C). This result indicates that the DAIH mutant could bind and stabilize G4-15T. However, in the presence of ATP, the DAIH mutant was unable to unfold the G4 (Figure 2C), similarly to the effect of DmRHAU binding without ATP. The same results were also obtained for wild-type DmRHAU using the non-hydrolyzable ATP analogue adenosine 5′-[β, γ-imino]-triphosphate (AMP-PNP), which mimics the ATP-bound state (Figure 2D). Our results are generally consistent with previous biochemical analyses reporting that the human DAIH mutant with ATP or wild-type RHAU with AMP-PNP were all able to bind G4 but had lost the ability to unfold G4 (25).
Previously, it was reported that various superfamily 2 helicases such as HCV NS3 helicase, assume different conformations between their nucleotide-free state and when bound to ADP·AlF , an ATP-analog mimicking the ATP hydrolysis intermediate state before the ATP cleavage step (39). We reasoned that ADP·AlF
, an ATP-analog mimicking the ATP hydrolysis intermediate state before the ATP cleavage step (39). We reasoned that ADP·AlF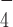 could induce a similar conformational change of DmRHAU. Using the aforementioned force-jump assay in the presence of ADP·AlF
could induce a similar conformational change of DmRHAU. Using the aforementioned force-jump assay in the presence of ADP·AlF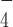 and DmRHAU, we found that G4-15T unfolded in less than 30 s after jumping to 54 pN (Figure 3). In addition, we estimated the lifetime of G4-15T to be 4 ± 0.7 s (mean ± s.e., n = 67, from 5 molecules). This unfolding rate is similar to that obtained from the naked G4, suggesting that in ADP·AlF
and DmRHAU, we found that G4-15T unfolded in less than 30 s after jumping to 54 pN (Figure 3). In addition, we estimated the lifetime of G4-15T to be 4 ± 0.7 s (mean ± s.e., n = 67, from 5 molecules). This unfolding rate is similar to that obtained from the naked G4, suggesting that in ADP·AlF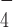 bound state RHAU cannot stabilize G4-15T. In addition, in the presence of ADP·AlF
bound state RHAU cannot stabilize G4-15T. In addition, in the presence of ADP·AlF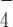 , DmRHAU bound to G4-15T with similar binding affinity as compared to in the absence of nucleotides (Supplementary Figure S9). These results indicate that ADP·AlF
, DmRHAU bound to G4-15T with similar binding affinity as compared to in the absence of nucleotides (Supplementary Figure S9). These results indicate that ADP·AlF can indeed induce a different binding mode of DmRHAU on G4, which abolishes the stabilization effect on G4-15T. We also examined the effect of ADP-bound DmRHAU and found that DmRHAU bound and stabilized G4-15T similarly to the nucleotide free DmRHAU (Supplementary Figure S10). Taken together, these results suggest that the interaction between DmRHAU and G4 is tightly regulated by different nucleotide-bound conformations. In addition, all the non-hydrolyzable nucleotides bound states analyzed in our study did not show significant destabilization effects on G4-15T suggesting that the destabilization of G4 by DmRHAU likely requires concerted conformational changes of DmRHAU driven by ATPase cycle.
can indeed induce a different binding mode of DmRHAU on G4, which abolishes the stabilization effect on G4-15T. We also examined the effect of ADP-bound DmRHAU and found that DmRHAU bound and stabilized G4-15T similarly to the nucleotide free DmRHAU (Supplementary Figure S10). Taken together, these results suggest that the interaction between DmRHAU and G4 is tightly regulated by different nucleotide-bound conformations. In addition, all the non-hydrolyzable nucleotides bound states analyzed in our study did not show significant destabilization effects on G4-15T suggesting that the destabilization of G4 by DmRHAU likely requires concerted conformational changes of DmRHAU driven by ATPase cycle.
Figure 3.
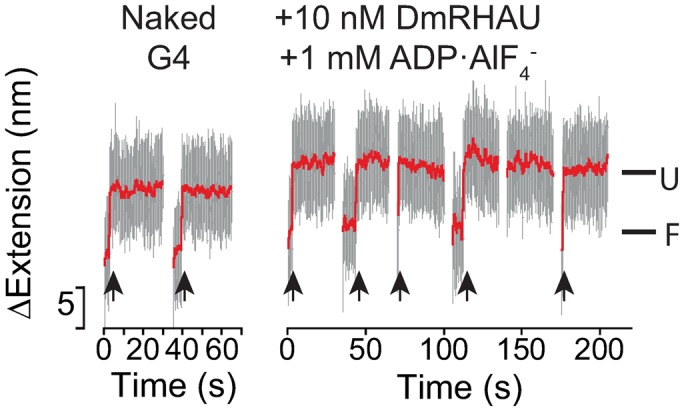
Effects of DmRHAU on G4-15T in the presence of ADP·AlF . Representative time trace of extension change of a G4-15T tether at 54 pN obtained in force-jump cycles of 1 pN (60 s) to 54 pN (30 s) before (left) and after (right) introduction of DmRHAU and ADP·AlF
. Representative time trace of extension change of a G4-15T tether at 54 pN obtained in force-jump cycles of 1 pN (60 s) to 54 pN (30 s) before (left) and after (right) introduction of DmRHAU and ADP·AlF . Unfolding events are indicated by arrows.
. Unfolding events are indicated by arrows.
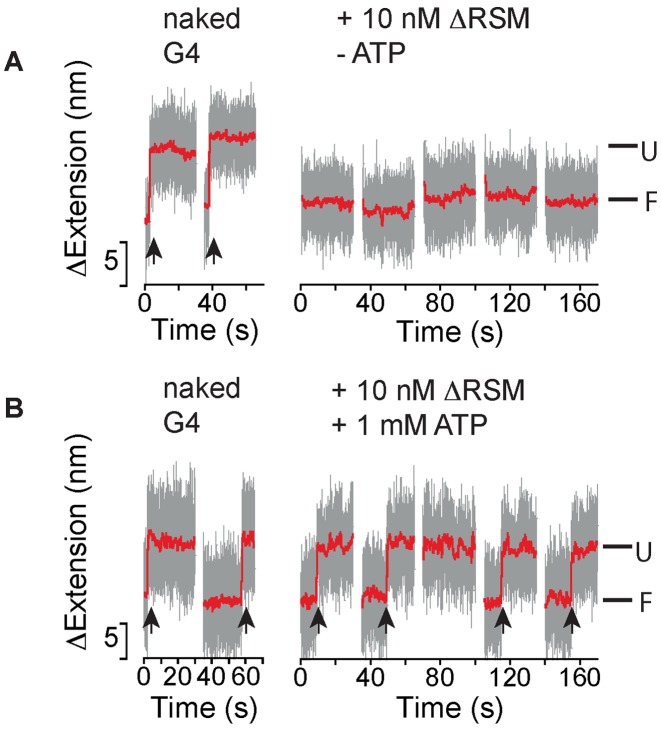
The ΔRSM mutant binds but cannot destabilize G4-15T
The 13-amino acids RSM at the N-terminus of RHAU has been shown specifically to interact with G4 (25,26). Thus, we next studied the requirements of G4 binding by the RSM in G4 stabilization and destabilization using a spliced recombinant form of DmRHAU lacking the RSM motif (ΔRSM; Supplementary Figures S1 and S11). To this end, we employed the force-jump assay using the G4-15T for ΔRSM both in the absence and presence of ATP. Surprisingly, we observed that in the absence of ATP the ΔRSM mutant was still able to bind and stabilize the G4 similarly to that observed when wild-type DmRHAU was used (Figure 4A). This result suggests that there is at least one extra yet-to-be identified G4 binding domain in DmRHAU which is crucial for G4 stabilization in the absence of ATP. The presence of such an extra G4 binding domain is also consistent with prior data that the RSM alone has a lower affinity for G4 than the full-length protein (25,26). In the presence of ATP, the average lifetime of the folded G4 at 54 pN was 11 ± 1 s (mean ± s.e., n = 39, from 3 molecules) similarly to that observed for naked G4. Compared to the sub-second lifetime at 54 pN observed for the wild-type DmRHAU in the presence of ATP, this result also indicates that the deletion of the RSM domain significantly impaired the G4 destabilizing ability of DmRHAU, which is consistent with the observation in a previous biochemical assay (25). Overall, these results suggest that the RSM domain is not dispensable for G4 binding but necessary for G4 destabilization.
Figure 4.
Effects of DmRHAU ΔRSM mutant on G4-15T. Representative time trace of extension changes of a G4-15T tether at 54 pN obtained in force-jump cycles of 1 pN (60 s) to 54 pN (30 s) using ΔRSM mutant in the (A) absence or in the (B) presence of ATP.
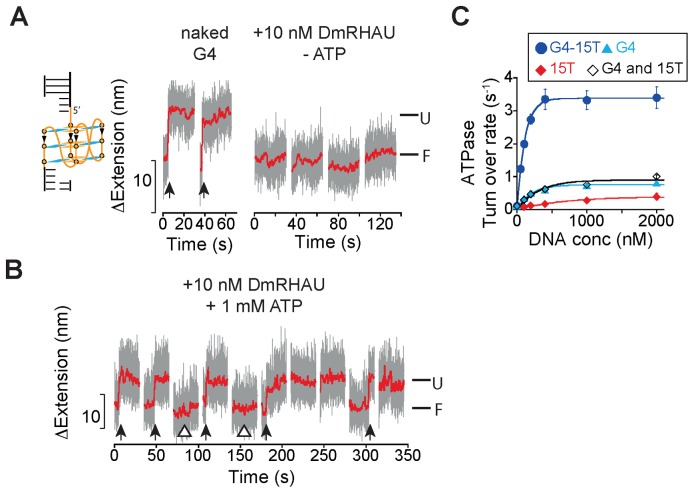
Removal of the 3′ ssDNA loading tail does not substantially reduce DmRHAU binding to G4, but results in decreased ATP-dependent G4 destabilization
A 3′ ssDNA loading tail can usually facilitate DEAH-box helicase unwinding activity (36,37). In the current study, we developed a RNase T1-coupled G4 unfolding assay and found that a 3′ loading tail of a minimum length of 8 nucleotides is also required for unfolding an intramolecularly folded G4 by DmRHAU (Supplementary Supplementary Figure S12). To further investigate the role of the ssDNA loading tail in substrate binding and in the subsequent G4 unwinding, we tested whether the DmRHAU can recognize, bind and unfold G4 devoid of a 3′ loading tail (hereafter referred as ‘tail-less G4’), using the force-jump assay. We kept 3 nucleotides at the 3′ end as a spacer between the G4 and the dsDNA handle. Our results showed that in the absence of ATP, DmRHAU bound and stabilized the tail-less G4, similarly to that observed for the 3′-tailed G4-15T (Figure 5A and Supplementary Figure S13).
Figure 5.
Effects of the 3′ ssDNA loading tail on G4 binding and destabilization. (A and B) Representative time trace of extension changes of a tail-less G4 tether at 54 pN obtained in force-jump cycles of 1 pN (60 s) to 54 pN (30 s) using DmRHAU in the (A) absence or in the (B) presence of ATP. (C) NADH-coupled ATPase activity assay for DmRHAU in the presence of various DNA substrates including G4-15T, tail-less G4, a 15-nt long oligo dT(15T), a 1:1 ratio mixture of G4 and 15T, and a control without any DNA substrate.
After introduction of DmRHAU and ATP, the tail-less G4 was not unfolded within 30 s in about 28% of the cycles, in sharp contrast to the result obtained for G4-15T under the same concentration of DmRHAU and ATP where G4 was already unfolded before jumping to 54 pN. In addition, compared to the ∼4.8 s lifetime of naked tail-less G4 at 54 pN (Supplementary Figure S2), this result also indicates that the tail-less G4 was bound and stabilized by DmRHAU even in the presence of ATP in 28% of the cases (Figure 5B, hollow triangle).
In ∼38% of the cycles (n = 99, from 3 molecules), the average lifetime was 8 ± 1 s (mean ± s. e., n = 39) (Figure 5B, arrows), similarly to that observed for naked G4. In the remaining ∼33% of the cycles, the DNA was in the extended state right after jumping to 54 pN, indicating that the G4 was already unfolded prior to the force-jump. In addition, we found that the removal of the loading tail caused nearly a 4-fold decrease in the ATPase activity of DmRHAU at saturating DNA concentrations (Figure 5C).
Overall, these results suggest that under our experimental conditions, G4 binding and stabilization by DmRHAU in the absence of ATP does not require a long ssDNA tail. An important finding is that a minimal 8-nt long 3′ ssDNA tail is required for both a robust stimulation of ATPase activity and ATP-dependent G4 destabilization activity of DmRHAU.
DISCUSSION
Our single-molecule studies show that binding of DmRHAU stabilizes an intramolecularly folded G4 in the absence of ATP or in the presence of AMP-PNP or ADP. In contrast, DmRHAU destabilizes G4-15T when it is coupled to ATP hydrolysis. Similar G4 stabilization (in the absence of ATP) and destabilization (in the presence of ATP) was also observed for human RHAU suggesting that the G4 binding mode is conserved among various RHAU orthologues. Further, we show that the G4 binding RSM domain of DmRHAU and the 3′ loading tail of the DNA substrate are dispensable for DmRHAU's stabilization of G4, whilst they are strictly needed for G4 destabilization.
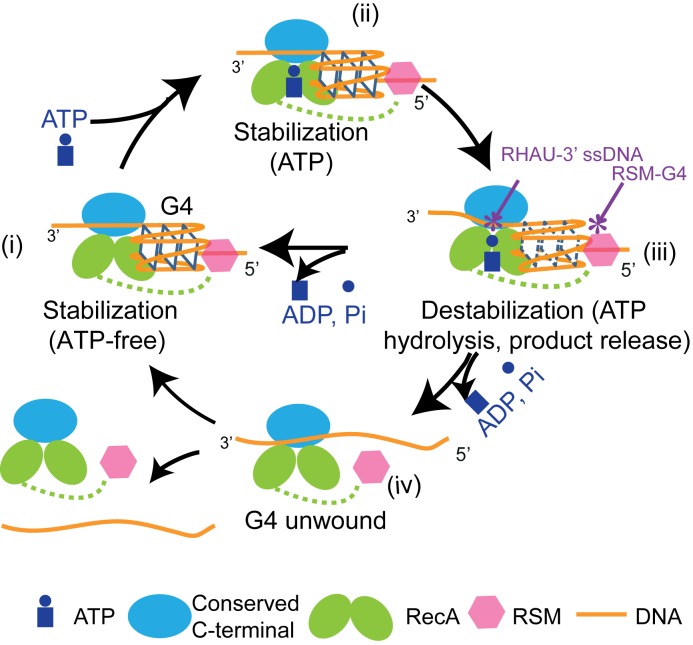
Based on the observed effects of different nucleotide-bound states of DmRHAU on G4 stability, we propose a model to account for the ATP-dependent stabilization and destabilization activities of RHAU (Figure 6). In this model, RHAU binds to a G4 structure in (i) nucleotide-free or (ii) ATP-bound form with high affinity, resulting in stabilization of G4. When RHAU enters an ATP hydrolysis (iii) intermediate state, RHAU likely undergoes conformational changes that result in loss of G4 stabilization. We speculate that subsequent conformational changes of RHAU, probably driven by product release, are involved in (iv) the final unfolding of the G4 structure. Regarding refolding of the G4, it might occur when bound to RHAU or after release. This will need further investigations.
Figure 6.
Model for ATP-dependent G4 destabilization by RHAU. RHAU (represented by its two RecA-like domains, the conserved C-terminal domain and the N-terminal RSM domain) binds to the G4 substrate in both (i) nucleotide-free and (ii) ATP-bound states. G4 destabilization occurs during (iii and iv) ATP hydrolysis and the following ADP or phosphate release steps. The RHAU–ssDNA binding interface is drawn based on the structure of a closely related DEAH-box helicase (29). The binding interface of RSM domain with G4 is drawn based on a recent NMR-solved structure (26).
We found that when bound to ADP·AlF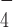 (which supposedly mimics an ATP hydrolysis intermediate state) DmRHAU loses its stabilization effect on the G4. One possible explanation is that the helicase core undergoes conformational changes that weaken the interaction between RHAU and the G4. A previous study showed that the superfamily 2 HCV NS3 co-crystalized with ADP·AlF
(which supposedly mimics an ATP hydrolysis intermediate state) DmRHAU loses its stabilization effect on the G4. One possible explanation is that the helicase core undergoes conformational changes that weaken the interaction between RHAU and the G4. A previous study showed that the superfamily 2 HCV NS3 co-crystalized with ADP·AlF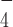 has a different conformation from its nucleotide-free form (39). In addition, two other studies of DEAH-box helicases, Prp43 co-crystalized with ADP (40) and MLE co-crystalized with ADP·AlF
has a different conformation from its nucleotide-free form (39). In addition, two other studies of DEAH-box helicases, Prp43 co-crystalized with ADP (40) and MLE co-crystalized with ADP·AlF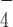 and ssRNA (29), also suggested a rotational conformational change at the RecA2 domain between ADP·AlF
and ssRNA (29), also suggested a rotational conformational change at the RecA2 domain between ADP·AlF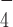 -bound state and ADP bound state. These earlier studies of some closely-related helicases suggest that the ATP hydrolysis intermediate state has a different conformation compared to the nucleotide-free or the ADP-bound states. This is likely related to the observed loss of the G4 stabilization by ADP·AlF
-bound state and ADP bound state. These earlier studies of some closely-related helicases suggest that the ATP hydrolysis intermediate state has a different conformation compared to the nucleotide-free or the ADP-bound states. This is likely related to the observed loss of the G4 stabilization by ADP·AlF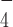 -bound DmRHAU.
-bound DmRHAU.
Our results show that the DmRHAU can tightly bind to a G4 devoid of any ssDNA loading tail, indicating a direct and attractive interaction between DmRHAU and its substrate that provides energy to stabilize the G4. In addition, we find that the 3′ single-stranded loading tail of the G4 is dispensable for the tight binding of DmRHAU, but it is required to stimulate the ATPase activity and G4 unfolding. This result suggests that the sensing of G4 by DmRHAU does not require directional translocation of the helicase along a single-stranded nucleic acid. This is consistent with a recent study reporting that human RHAU uses a local and non-processive mechanism to unwind G4 structures (30). Previous studies on RHAU identified the N-terminal RSM domain as a G4 binding domain (25,26). Our observation that DmRHAU lacking the RSM domain was able to bind the G4 indicates that RHAU must contain one or more additional G4 binding domains. Apart from the 3′ tail requirement, our results suggest that to initiate G4 unwinding, RHAU needs to be locked into a highly specific conformation through the collaborative binding of various domains with the G4 substrate.
During the review process of this work, a fluorescence resonance energy transfer (FRET)-based study of G4 destabilization by human RHAU was published (41). That work was based on the change of FRET efficiency between a Cy3 dye at the end of a 3′ ssDNA loading tail and a Cy5 dye at the 5′ side of G4. The authors observed a decreased FRET efficiency upon binding of RHAU in the absence of ATP, implying an increased distance between Cy3 and Cy5, which was interpreted as a result of G4 unwinding. However, an increased distance between the Cy3 and Cy5 dyes can alternatively be explained by a steric effect upon binding of RHAU. Therefore, we do not think there is a conflict in the experimental observations between that work and ours.
CONCLUSION
Our experiments provide the first evidence that the unfolding rates of a G4 structure can be modulated by different nucleotide-bound states of a helicase. Taking insights from previous structural studies of similar helicases crystallized with different nucleotides, our finding suggests that the conformational changes of RHAU driven by ATPase cycle play an important role in RHAU-mediated G4 unfolding. We furthermore demonstrate that besides the previously identified specific G4 binding RSM domain, there is at least another G4 binding domain that stabilizes the G4 in the absence of ATP. We show that RHAU stabilizes a G4 substrate devoid of 3′ ssDNA loading tail in the absence of ATP, suggesting that the stabilization occurs through a direct interaction between RHAU and the G4. Taken together, these findings provide new insights into the mechanism of the RHAU mediated regulation of G4 stability.
Acknowledgments
The authors thank Ahn Tuân Phan and Ryota Iino for stimulating discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Singapore Ministry of Education Academic Research Fund Tier 3 [MOE2012-T3-1-001 to J.Y. and D.R.]; National Research Foundation through the Mechanobiology Institute Singapore [to J.Y.]. Funding for open access charge: Singapore Ministry of Education Academic Research Fund Tier 3 [MOE2012-T3-1-001].
Conflict of interest statement. None declared.
REFERENCES
- 1.Sen D., Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 2.Rhodes D., Lipps H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43:8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paeschke K., Juranek S., Simonsson T., Hempel A., Rhodes D., Lipps H.J. Telomerase recruitment by the telomere end binding protein-beta facilitates G-quadruplex DNA unfolding in ciliates. Nat. Struct. Mol. Biol. 2008;15:598–604. doi: 10.1038/nsmb.1422. [DOI] [PubMed] [Google Scholar]
- 4.Biffi G., Tannahill D., McCafferty J., Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piazza A., Adrian M., Samazan F., Heddi B., Hamon F., Serero A., Lopes J., Teulade-Fichou M.-P., Phan A.T., Nicolas A. Short loop length and high thermal stability determine genomic instability induced by G-quadruplex-forming minisatellites. EMBO J. 2015;34:1718–1734. doi: 10.15252/embj.201490702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.You H., Wu J., Shao F., Yan J. Stability and kinetics of c-MYC promoter G-quadruplexes studied by single-molecule manipulation. J. Am. Chem. Soc. 2015;137:2424–2427. doi: 10.1021/ja511680u. [DOI] [PubMed] [Google Scholar]
- 7.Sun H., Karow J.K., Hickson I.D., Maizels N. The Bloom's syndrome helicase unwinds G4 DNA. J. Biol. Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 8.Fry M., Loeb L.A. Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 9.Wu Y., Shin-ya K., Brosh R.M., Jr FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paeschke K., Bochman M.L., Garcia P.D., Cejka P., Friedman K.L., Kowalczykowski S.C., Zakian V.A. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497:458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nie J., Jiang M., Zhang X., Tang H., Jin H., Huang X., Yuan B., Zhang C., Lai J.C., Nagamine Y., et al. Post-transcriptional regulation of Nkx2-5 by RHAU in heart development. Cell Rep. 2015;13:723–732. doi: 10.1016/j.celrep.2015.09.043. [DOI] [PubMed] [Google Scholar]
- 12.Lai J.C., Ponti S., Pan D., Kohler H., Skoda R.C., Matthias P., Nagamine Y. The DEAH-box helicase RHAU is an essential gene and critical for mouse hematopoiesis. Blood. 2012;119:4291–4300. doi: 10.1182/blood-2011-08-362954. [DOI] [PubMed] [Google Scholar]
- 13.Gao X., Ma W., Nie J., Zhang C., Zhang J., Yao G., Han J., Xu J., Hu B., Du Y., et al. A G-quadruplex DNA structure resolvase, RHAU, is essential for spermatogonia differentiation. Cell Death Dis. 2015;6:e1610. doi: 10.1038/cddis.2014.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tran H., Schilling M., Wirbelauer C., Hess D., Nagamine Y. Facilitation of mRNA deadenylation and decay by the exosome-bound, DExH protein RHAU. Mol. Cell. 2004;13:101–111. doi: 10.1016/s1097-2765(03)00481-7. [DOI] [PubMed] [Google Scholar]
- 15.Vaughn J.P., Creacy S.D., Routh E.D., Joyner-Butt C., Jenkins G.S., Pauli S., Nagamine Y., Akman S.A. The DEXH protein product of the DHX36 gene is the major source of tetramolecular quadruplex G4-DNA resolving activity in HeLa cell lysates. J. Biol. Chem. 2005;280:38117–38120. doi: 10.1074/jbc.C500348200. [DOI] [PubMed] [Google Scholar]
- 16.Creacy S.D., Routh E.D., Iwamoto F., Nagamine Y., Akman S.A., Vaughn J.P. G4 resolvase 1 binds both DNA and RNA tetramolecular quadruplex with high affinity and is the major source of tetramolecular quadruplex G4-DNA and G4-RNA resolving activity in HeLa cell lysates. J. Biol. Chem. 2008;283:34626–34634. doi: 10.1074/jbc.M806277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chalupníková K., Lattmann S., Selak N., Iwamoto F., Fujiki Y., Nagamine Y. Recruitment of the RNA helicase RHAU to stress granules via a unique RNA-binding domain. J. Biol. Chem. 2008;283:35186–35198. doi: 10.1074/jbc.M804857200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou R., Zhang J., Bochman M.L., Zakian V.A., Ha T. Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. Elife. 2014;3:e02190. doi: 10.7554/eLife.02190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu W.-Q., Hou X.-M., Li M., Dou S.-X., Xi X.-G. BLM unfolds G-quadruplexes in different structural environments through different mechanisms. Nucleic Acids Res. 2015;43:4614–4626. doi: 10.1093/nar/gkv361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lattmann S., Stadler M.B., Vaughn J.P., Akman S.A., Nagamine Y. The DEAH-box RNA helicase RHAU binds an intramolecular RNA G-quadruplex in TERC and associates with telomerase holoenzyme. Nucleic Acids Res. 2011;39:9390–9404. doi: 10.1093/nar/gkr630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Booy E.P., Meier M., Okun N., Novakowski S.K., Xiong S., Stetefeld J., McKenna S.A. The RNA helicase RHAU (DHX36) unwinds a G4-quadruplex in human telomerase RNA and promotes the formation of the P1 helix template boundary. Nucleic Acids Res. 2012;40:4110–4124. doi: 10.1093/nar/gkr1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang W., Smaldino P.J., Zhang Q., Miller L.D., Cao P., Stadelman K., Wan M., Giri B., Lei M., Nagamine Y., et al. Yin Yang 1 contains G-quadruplex structures in its promoter and 5′-UTR and its expression is modulated by G4 resolvase 1. Nucleic Acids Res. 2012;40:1033–1049. doi: 10.1093/nar/gkr849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Booy E.P., Howard R., Marushchak O., Ariyo E.O., Meier M., Novakowski S.K., Deo S.R., Dzananovic E., Stetefeld J., McKenna S.A. The RNA helicase RHAU (DHX36) suppresses expression of the transcription factor PITX1. Nucleic Acids Res. 2014;42:3346–3361. doi: 10.1093/nar/gkt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giri B., Smaldino P.J., Thys R.G., Creacy S.D., Routh E.D., Hantgan R.R., Lattmann S., Nagamine Y., Akman S.A., Vaughn J.P. G4 resolvase 1 tightly binds and unwinds unimolecular G4-DNA. Nucleic Acids Res. 2011;39:7161–7178. doi: 10.1093/nar/gkr234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lattmann S., Giri B., Vaughn J.P., Akman S.A., Nagamine Y. Role of the amino terminal RHAU-specific motif in the recognition and resolution of guanine quadruplex-RNA by the DEAH-box RNA helicase RHAU. Nucleic Acids Res. 2010;38:6219–6233. doi: 10.1093/nar/gkq372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heddi B., Cheong V.V., Martadinata H., Phan A.T. Insights into G-quadruplex specific recognition by the DEAH-box helicase RHAU: Solution structure of a peptide-quadruplex complex. Proc. Natl. Acad. Sci. U.S.A. 2015;112:9608–9613. doi: 10.1073/pnas.1422605112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levin M.K., Gurjar M., Patel S.S. A Brownian motor mechanism of translocation and strand separation by hepatitis C virus helicase. Nat. Struct. Mol. Biol. 2005;12:429–435. doi: 10.1038/nsmb920. [DOI] [PubMed] [Google Scholar]
- 28.Liu F., Putnam A., Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc. Natl. Acad. Sci. U.S.A. 2008;105:20209–20214. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prabu J.R., Müller M., Thomae A.W., Schüssler S., Bonneau F., Becker P.B., Conti E. Structure of the RNA helicase MLE reveals the molecular mechanisms for uridine specificity and RNA-ATP coupling. Mol Cell. 2015;60:487–499. doi: 10.1016/j.molcel.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 30.Chen M.C., Murat P., Abecassis K., Ferré-D'Amaré A.R., Balasubramanian S. Insights into the mechanism of a G-quadruplex-unwinding DEAH-box helicase. Nucleic Acids Res. 2015;43:2223–2231. doi: 10.1093/nar/gkv051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen H., Fu H., Zhu X., Cong P., Nakamura F., Yan J. Improved high-force magnetic tweezers for stretching and refolding of proteins and short DNA. Biophys J. 2011;100:517–523. doi: 10.1016/j.bpj.2010.12.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.You H., Zeng X., Xu Y., Lim C.J., Efremov A.K., Phan A.T., Yan J. Dynamics and stability of polymorphic human telomeric G-quadruplex under tension. Nucleic Acids Res. 2014;42:8789–8795. doi: 10.1093/nar/gku581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato Y., Sasayama T., Muneyuki E., Yoshida M. Analysis of time-dependent change of Escherichia coli F1-ATPase activity and its relationship with apparent negative cooperativity. Biochim. Biophys. Acta. 1995;1231:275–281. doi: 10.1016/0005-2728(95)00087-y. [DOI] [PubMed] [Google Scholar]
- 34.Jeon S.-J., Ishikawa K. A novel ADP-dependent DNA ligase from Aeropyrum pernix K1. FEBS Lett. 2003;550:69–73. doi: 10.1016/s0014-5793(03)00821-4. [DOI] [PubMed] [Google Scholar]
- 35.Bruns A.M., Pollpeter D., Hadizadeh N., Myong S., Marko J.F., Horvath C.M. ATP hydrolysis enhances RNA recognition and antiviral signal transduction by the innate immune sensor, laboratory of genetics and physiology 2 (LGP2) J. Biol. Chem. 2013;288:938–946. doi: 10.1074/jbc.M112.424416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee C.G., Chang K.A., Kuroda M.I., Hurwitz J. The NTPase/helicase activities of Drosophila maleless, an essential factor in dosage compensation. EMBO J. 1997;16:2671–2681. doi: 10.1093/emboj/16.10.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka N., Schwer B. Characterization of the NTPase, RNA-binding, and RNA helicase activities of the DEAH-box splicing factor Prp22. Biochemistry. 2005;44:9795–9803. doi: 10.1021/bi050407m. [DOI] [PubMed] [Google Scholar]
- 38.Fu H., Chen H., Zhang X., Qu Y., Marko J.F., Yan J. Transition dynamics and selection of the distinct S-DNA and strand unpeeling modes of double helix overstretching. Nucleic Acids Res. 2011;39:3473–3481. doi: 10.1093/nar/gkq1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu M., Rice C.M. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc. Natl. Acad. Sci. U.S.A. 2010;107:521–528. doi: 10.1073/pnas.0913380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He Y., Andersen G.R., Nielsen K.H. Structural basis for the function of DEAH helicases. EMBO Rep. 2010;11:180–186. doi: 10.1038/embor.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tippana R., Hwang H., Opresko P.L., Bohr V.A., Myong S. Single-molecule imaging reveals a common mechanism shared by G-quadruplex-resolving helicases. Proc. Natl. Acad. Sci. U.S.A. 2016;113:8448–8453. doi: 10.1073/pnas.1603724113. [DOI] [PMC free article] [PubMed] [Google Scholar]