Abstract
All three B cell-specific activities of the immunoglobulin (Ig) gene re-modeling system—gene conversion, somatic hypermutation and class switch recombination—require activation-induced deaminase (AID). AID-induced DNA lesions must be further processed and dissected into different DNA recombination pathways. In order to characterize potential intermediates for Ig gene conversion, we inserted an I-SceI recognition site into the complementarity determining region 1 (CDR1) of the Ig light chain locus of the AID knockout DT40 cell line, and conditionally expressed I-SceI endonuclease. Here, we show that a double-strand break (DSB) in CDR1 is sufficient to trigger Ig gene conversion in the absence of AID. The pattern and pseudogene usage of DSB-induced gene conversion were comparable to those of AID-induced gene conversion; surprisingly, sometimes a single DSB induced multiple gene conversion events. These constitute direct evidence that a DSB in the V region can be an intermediate for gene conversion. The fate of the DNA lesion downstream of a DSB had more flexibility than that of AID, suggesting two alternative models: (i) DSBs during the physiological gene conversion are in the minority compared to single-strand breaks (SSBs), which are frequently generated following DNA deamination, or (ii) the physiological gene conversion is mediated by a tightly regulated DSB that is locally protected from non-homologous end joining (NHEJ) or other non-homologous DNA recombination machineries.
INTRODUCTION
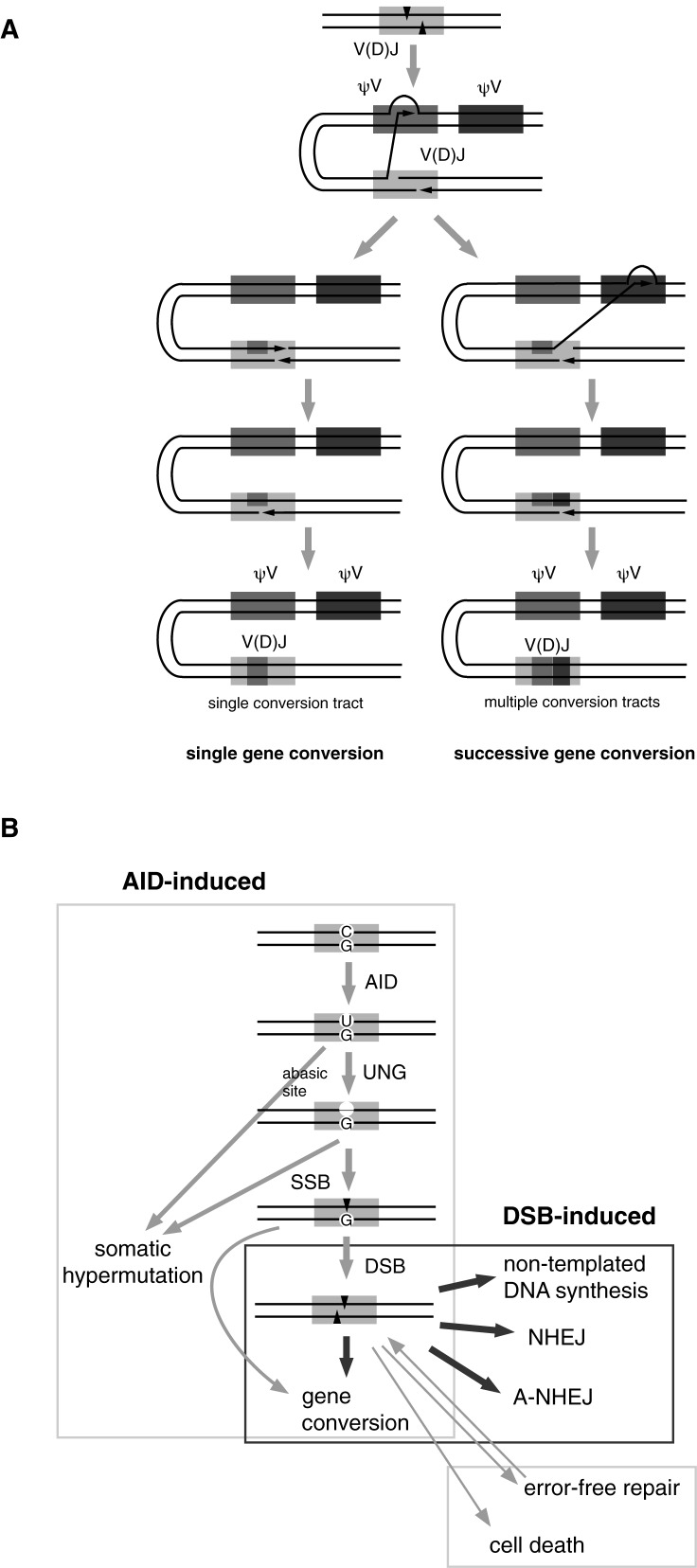
In humans and mice, a large antigen receptor repertoire is assembled from a collection of a huge number of V, D and J segments by site-specific V (D) J recombination; however, this strategy is not common to all vertebrates and antigen receptor repertoire formation systems vary by species. Chickens have only one pair of functional V and J segments in both Ig light and heavy chain gene loci (1,2); therefore, the diversity generated by V (D) J recombination is limited. Chickens have 25 pseudo V (ψV) genes in the upstream of the functional V gene in the Ig light chain locus (1) and more than 100 ψV genes in the Ig heavy chain locus (2). These ψV genes are used as templates to diversify the single functional V gene by gene conversion (1,2).
During gene conversion, genetic information is unidirectionally transferred from the ψV gene to the rearranged V (D) J gene (1). The length of the copied ψV DNA fragments range from a few bp to more than 200 bp (3). Because gene conversion is a ‘copy and paste’ genetic reaction, the ψV templates are preserved during gene conversion (1). Gene conversion occurs between ψV genes and the rearranged V (D) J gene on the same chromosome (4,5). ψV genes have been under strong selective pressure over the course of evolution, and are even more conserved than the single functional V gene (6). Since gene conversion is based on homologous recombination, ψV genes that are more homologous to the acceptor V gene sequence are more frequently used, and contribute to the stepwise editing of the acceptor V gene (7). Although the number of ψV genes is limited, gene conversion is predicted to create more potential diversity of the Ig gene than V (D) J recombination due to the flexibility of ψV gene assembly in the gene conversion system (8).
Immunoglobulin gene conversion was first identified in chickens and subsequently proposed as a mechanism acting also in other birds, such as quail, mallard duck, pigeon, turkey, cormorant, hawk (9), duck (10) and goose (11). Since humans and mice use V (D) J recombination for primary antibody gene diversification, one may wonder whether the difference between gene conversion and V (D) J recombination may be due to differences between the evolutionary strategies of birds and mammals. However, rabbits use gene conversion as a basic mechanism of antibody gene diversification (12). Cattle, sheep, swine and horses also appear to use gene conversion in addition to somatic hypermutation for B cell repertoire development (13). Recently, the guinea pig (14), Tasmanian devil (15) and prairie vole (16) have also been added to the list of mammalian species that use gene conversion for B cell repertoire formation. Thus, gene conversion has been adopted as the primary B cell repertoire formation mechanism in most mammals as well as in avian species. Most likely gene conversion evolved a long time ago in a common ancestor of avians and mammals, and was lost in the evolutionary branches to humans and mice (8).
Gene conversion mainly occurs in the gut-associated lymphoid tissues: the bursa of Fabricius in chicken (1), the appendix in rabbits (12) and the ileal Peyer's patches in cattle, sheep, swine and horses (13). These tissues are involuted several months after birth and lose their primary B cell repertoire formation function. This is in contrast to humans and mice, where B cell repertoire formation by V (D) J recombination continues throughout life in the bone marrow. In the adult chicken, gene conversion, as well as a high frequency of somatic hypermutation, is re-activated in splenic germinal centers (7,17). As gene conversion is accompanied by a low frequency of somatic hypermutation in the bursa of Fabricius (1,18), as well as in the chicken B cell line DT40 (19), gene conversion is coexistent with somatic hypermutation, both in vivo and in the cell line. Nevertheless, these two pathways can be regulated separately because the frequency of these two pathways changes depending on different tissues or cell lines, and some species even lack a gene conversion pathway.
AICDA gene was identified as a gene that is essential for class switch recombination and somatic hypermutation (20,21), which are processed by NHEJ and non-templated point mutation, respectively. In addition to these two activities, AID is also essential for gene conversion (22). In the DNA editing model, AID catalyzes cytosine deamination in the Ig gene loci (23,24); the resulting uracil is removed by the uracil glycosylase, UNG-2 (23,25) or acted upon by mismatch repair factors (26), leading to mutations at C/G. The AID-induced DNA lesion processing is highly regulated, since only differences in the location or processing of the AID-induced DNA lesion can explain the selective occurrences of the three different pathways: class switch recombination, somatic hypermutation and gene conversion.
AID-induced lesions are supposedly converted to either a single-strand break (SSB) or a double-strand break (DSB) in order to induce gene conversion, since DNA polymerases require a 3′ end of a nucleic acid in order to initiate DNA synthesis. The mega endonuclease I-SceI has been widely used to induce DSBs at genomic regions of interest (27,28). A DSB induced by I-SceI can activate switch recombination in mouse B cells (29) and enhance the gene conversion of variant surface glycoprotein coat genes in Trypanosoma (30). The crystal structures of I-SceI with uncleaved and bottom-nicked substrates suggested a sequential cleavage mechanism (31), enabling the design of I-SceI nickase variants. The K122I mutation suppresses top-strand cleavage whereas the K223I mutation suppresses bottom-strand cleavage, resulting in a bottom-strand nicking and a top-strand nicking variant, respectively (32). These mutants showed strand-specific nicking activities in vitro (32,33), although they generated DSB products over extended reaction times (32,33). I-SceI K223I was tested in vivo and promoted homology-directed gene correction in yeast up to 9-fold and in human cells up to 12-fold (33).
To our knowledge, there has been no report describing Ig gene conversion events in the absence of AID. In this study, we artificially introduced an I-SceI-induced DNA break at a fixed position of the Ig light chain gene. We introduced this break to determine whether a targeted single I-SceI-induced DSB or SSB in a rearranged V gene could replace AID-induced lesions to trigger gene conversion and somatic hypermutation. We also asked whether a single DSB could generate sufficient Ig V gene diversity, and whether usage and length of ψV genes involved in DSB-initiated gene conversion were comparable to those initiated by AID. Finally we addressed the nature of the repair pathways triggered by a DSB targeted to the Ig locus.
MATERIALS AND METHODS
Cell culture and reverse genetics
DT40 cells were cultured in chicken medium (DMEM/F-12 with 10% fetal bovine serum, 2% chicken serum, 2 mM L-glutamine, 0.1 mM β-mercaptoethanol and penicillin/streptomycin) at 39.5°C with 5% CO2. Cells were transfected by electroporation using the Gene Pulser II (BIO-RAD) at 25 μF and 700 V, as previously described (34). Targeted integration was screened by polymerase chain reaction (PCR) using PrimeSTAR GXL DNA Polymerase (Takara) with the following cycling parameters: 35 cycles of 98°C for 10 s and 68°C for 4 min. In order to activate the Tet-inducible promoter, cell lines were cultured in the presence of 100 ng/ml doxycycline (dox) (Sigma) for two weeks.
Vectors
The gene conversion reporter vector was designed as follows: (i) an I-SceI site was integrated into the CDR1; (ii) a hydrophobic array of 5 amino acids, LLLAV, was deleted from the leader sequence to functionally disrupt the leader sequence (ΔL); and (iii) VJ was fused to GFP, followed by internal ribosome entry site, bsr and a SV40 polyA signal. A vector containing the I-SceI coding sequence was kindly provided by Dr Holger Puchta. The K122I and K223I mutant I-SceI was generated by PCR-based site-directed mutagenesis of the I-SceI coding sequence. The I-SceI coding sequence was cloned into multiple cloning sites of pTRE3G-mCherry (Clontech) to generate pTre3gMcherrySce (Supplementary Figure S1). The I-SceI mutants with K122I or K223I were also cloned in the same manner. The Tet-On 3G expression vector, pChr2RsvProTetOn3gPuro (Supplementary Figure S1), was generated by cloning the Tet-On 3G coding sequence, which is driven by a RSV promoter, into a targeted integration vector directed to an intergenic locus on the chicken chromosome 2.
Cell lines
The I-SceI reporter cell lines IgLSce GFP and AID−/− IgLSce GFP were generated by targeted integration of the reporter into DT40Cre1 (35) and AID−/− DT40 (22) cell lines. Integration of the I-SceI reporter vector into the rearranged Ig light chain locus was screened using the PS148/VL533 primer pairs and was further verified by PCR amplification of the VJ intervening sequence of the un-rearranged locus using the VL513/VL514 primer pairs as previously described (36). AID−/− IgLSce GFP was transfected with a Tet-On 3G expression vector, pChr2RsvProTetOn3gPuro and integration of the vector into the target locus was screened using the CHR0208/RS16 primer pairs. This cell line, AID−/− IgLSce GFP Tet-On, was further co-transfected by pLoxGpt (35) with pTre3gMcherrySce to generate the AID−/− IgLSce GFP Tet-OnI-SceI cell line. Alternatively, the AID−/− IgLSce GFP Tet-On cell line was co-transfected by pLoxPuro (35) with either pTre3gMcherrySceK122i or pTre3gMcherrySceK223i to generate the AID−/− IgLSce GFP Tet-OnI-SceI K122I or AID−/− IgLSce GFP Tet-OnI-SceI K223I cell lines, respectively.
Sorting, cloning and sequencing
The analytical FACS was performed using FACS Calibur (BD Biosciences) (37), and preparative FACS sorting for the GFP (+) population was performed using FACS Aria (BD Biosciences). The statistical significance of the differences in Ig reversion ratios were analyzed by Student's t-test. The GFP (+) population was sorted from either the AID−/− IgLSce GFP Tet-OnI-SceI cell line cultured in the presence of 100 ng/ml dox for two weeks or the IgLSce GFP cell line two weeks after subcloning. The AID−/− IgLSce GFP Tet-OnI-SceI cell line without dox induction was used as a negative control for sequencing. These cells were further expanded and used for genomic DNA extraction using Easy-DNA kit (Invitrogen). The rearranged Ig light chain reporter gene was amplified via PCR, using the p1/p2 primer pair and Q5 high-fidelity DNA polymerase (NEB), with the following cycling parameters: 30 s of initial incubation at 98°C; 35 cycles consisting of 10 s at 98°C and 2 min at 72°C; and a final elongation step of 2 min at 72°C. The PCR product was purified using the QIAquick Gel Extraction Kit (Qiagen), digested with HindIII (NEB) and XbaI (NEB), and cloned into the pUC119 plasmid vector. Approximately 100 plasmid clones were sequenced using the VL154 primer. Although a quarter of sequences from the sorted GFP (+) population did not have sequence changes, they were considered to be derived from a contaminated GFP (−) population and were excluded from further analysis. In order to identify possible gene conversion donors, mutated sequences were aligned with a ψV gene table from a CB inbred strain chicken (1), since the ψV genes of the rearranged Ig light chain allele of the DT40 cell line appeared to be nearly identical to those of the CB inbred strain chicken (22).
The rearranged Ig heavy chain gene was amplified from AID−/− IgLSce GFP Tet-OnI-SceI DT40 cells cultured in the presence or absence of 100 ng/ml dox. The duration of dox induction was two weeks. PCR, cloning and sequencing were performed as described above, except that the p3/p4 primer pair was used for PCR and the UC1 primer was used for sequencing. Twenty-two and 23 plasmid clones were sequenced for the rearranged Ig heavy chain gene before and after dox treatment, respectively.
Oligonucleotides
p1 (VL242) GGGAAGCTTAAAGGGCATCGAGGTCCCCGGCACA
p2 (GF70) TTACGTCGCCGTCCAGCTCGACCAGGATGG
p3 (VH48) TCTCCGAAAGCTTCGGAGGAGCACCAGTCGGCTCCGCA
p4 (VH49) GCCGCGGTCTAGACAATTTTGGGGGGGGTTGAAGACTC
CHR0208 CCTCACTGAACAGGATGGCCAAGAGATGCT
PS148 TGCAGGCACAGGTGGCAGTGGGGGTCTCTG
RS16 TGTAGCTTAAATTTTGCTCGCGCACTACTC
UC1 AGCGGATAACAATTTCACACAGGA
VL154 GTGCGTGCGGGGCCGTCACTGATTGCCGTT
VL513 CACGGTGACACAAAGCAATGGGGAAATGAT
VL514 GTGACCGGTGCAAGTGATAGAAACTCATTG
VL533 GTACCAGCCATAATTACCCTGTTATCCCTA
Western blotting
The DT40 cell lines were grown in the presence or absence of 100 ng/ml dox for the described time. Total cell lysates were prepared in radioimmunoprecipitation assay (RIPA) lysis buffer (50 mM TRIS-HCL (pH 8.0), 1 mM MgCl2, 200 mM NaCl, 1 mM CaCl2, 10% Glycerol, 1% NP-40, complete protease inhibitors (1:100; Roche), and phosphatase inhibitor cocktail 2 (1:200; Sigma)). Cells were harvested and then washed with phosphate buffered saline buffer. The pellet was resuspended in RIPA buffer, incubated on ice for 30 min and centrifuged at maximum speed for 30 min. Laemmli buffer (1x) was added to the supernatant and the samples were boiled for 10 min at 95°C. Samples were separated on a 4–20% CRITERION SFX gradient precast gel (BIO-RAD). The membrane was incubated with anti-α-tubulin monoclonal antibody B-5-1-2 (1:10 000; SIGMA) and anti-I-SceI polyclonal antibody FL-86 (1:500; Santa Cruz) followed by horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG secondary antibodies (BIO-RAD). Proteins were visualized using SuperSignal West Dura Extended Duration Substrate (Thermo Scientific).
RESULTS
GFP reporter to assay DSB-induced Ig reversion
Recently, designer endonucleases, such as zinc finger endonuclease (38), TALEN (39) or CRISPR/Cas9 (40,41), have become popular gene editing tools since they can be targeted to any locus of interest. However, such endonucleases cannot be easily employed to introduce a single DNA break into a gene belonging to a family of highly homologous genes or into repetitive sequences; e.g. the chicken Ig light chain V region genomic locus, which consists of a rearranged V gene positioned in proximity to 25 highly homologous ψV genes (1). Moreover, a second Ig light chain locus is present in germline configuration in most of B cells (42).
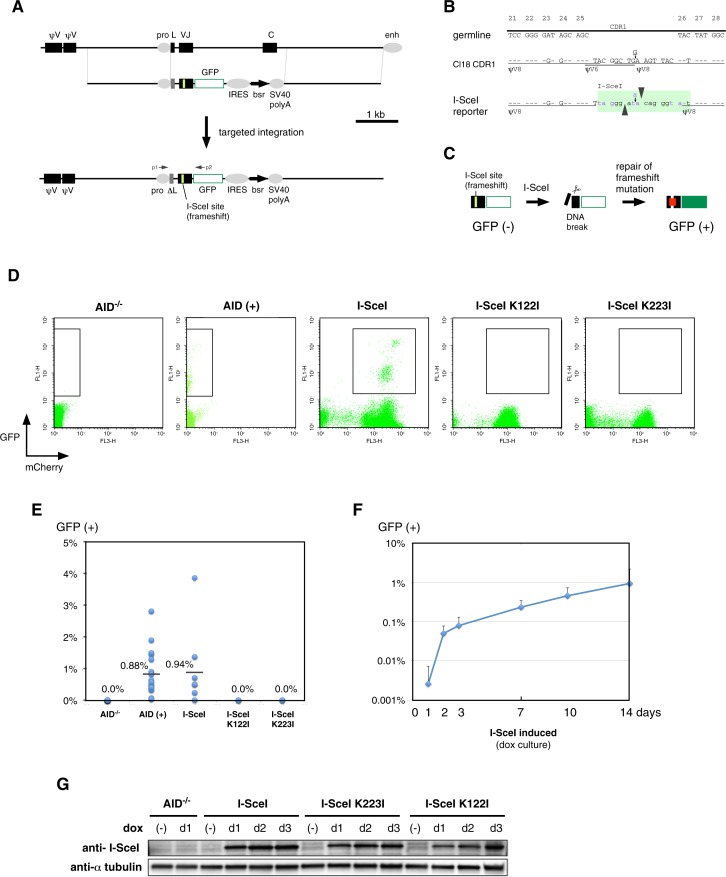
Complementarity-determining regions (CDR) 1, 2 and 3 from both the Ig light and heavy chain fold to form the antibody's antigen-binding sites. CDR1 in the Ig light chain gene is the major hotspot of Ig gene conversion (22) and somatic hypermutation (4) in the DT40 cell line. To investigate whether a DNA break can substitute for the function of AID, an I-SceI site linked to a reporter gene was knocked into the rearranged Ig light-chain locus of the DT40Cre1 cell line (35) (Figure 1A). This reporter gene contained an I-SceI site in the CDR1 (Figure 1B), and a GFP cDNA was fused in frame to the V region in order to efficiently assay frameshift reversion (Figure 1C); however, this approach failed to detect measurable GFP signals, even when the frame-shift mutation was reverted by gene conversion in the presence of AID (data not shown). Considering the possibility that the leader sequence could have caused extracellular release of the antibody-GFP fusion protein, we disrupted the leader sequence of the GFP reporter gene (ΔL in Figure 1A). This design worked as intended, as the wild-type DT40 cells carrying targeted integration of the ΔL version of the IgV-GFP reporter gene, generated 0.88% of a GFP (+) population after two weeks of continuous culture (Figure 1D and E). This result also suggests that insertion of an I-SceI recognition sequence into CDR1 of the V gene does not prevent AID from using it as substrate of gene conversion. The generated cell line was named IgLSce GFP.
Figure 1.
Ig reversion assay to analyze double-strand break (DSB) repair in the rearranged immunoglobulin (Ig) light chain gene. (A) The strategy for knocking-in the I-SceI site into complementarity determining region 1 (CDR1) of the variable (V) region in the Ig light chain gene. In this reporter, the leader sequence was functionally disrupted (ΔL) and VJ was fused to GFP to visualize frame-shift reversion. The p1/p2 primer pair was used to amplify the VJ gene. (B) I-SceI site inserted into CDR1. Since the 18-bp sequence of the I-SceI site did not contain significant homology to any of ψV genes, this site was inserted to minimize sequence changes from Cl18-type CDR1 (19). Because the I-SceI site contains three potential stop codons (purple letters), the positioning of the I-SceI site was carefully designed. While Cl18-type CDR1 contains two gene conversion tracts from ψV8 and one tract from ψV6 (19), the reporter still partially preserves the two gene conversion tracts from ψV8. (C) Ig reversion assay using GFP. (D) FACS profiles of AID−/− IgLSce GFP, IgLSce GFP, AID−/− IgLSce GFP Tet-OnI-SceI, AID−/− IgLSce GFP Tet-OnI-SceI K122I and AID−/− IgLSce GFP Tet-OnI-SceI K223I cell lines. AID-induced Ig reversion was analyzed two weeks after subcloning, and Ig reversion induced by I-SceI or its mutants was analyzed two weeks after doxycycline (dox) induction. Representative FACS profiles are shown with the GFP (+) gatings, which were also used for FACS sorting. The expression levels of I-SceI or its mutants were monitored using the intensity of the mCherry signal on the same cistron. (E) Fluctuation analysis of Ig reversion. AID-induced Ig reversion was analyzed two weeks after subcloning, where 14 and 16 subclones for the AID−/− IgLSce GFP and IgLSce GFP cell lines were analyzed, respectively. Ig reversion induced by I-SceI or its mutants was analyzed after two weeks of continuous culture in the presence of dox. Experimental replicates are eight for AID−/− IgLSce GFP Tet-OnI-SceI, and three for AID−/− IgLSce GFP Tet-OnI-SceI K122I and AID−/− IgLSce GFP Tet-OnI-SceI K223I cell lines. The differences in the Ig reversion ratios were significant between AID−/− and AID (+) (**P < 0.01), I-SceI and I-SceI K122I (*P < 0.05) and I-SceI and I-SceI K223I (*P < 0.05), but not significant between AID (+) and I-SceI (P = 0.45). Average percentages of GFP (+) cells are indicated by a bar. (F) Time course of Ig reversion of the AID−/− IgLSce GFP Tet-OnI-SceI cell line. Eight of the experimental replicates of the cell line were independently cultured for two weeks in the presence of dox. (G) Western blots using an anti-I-SceI antibody before and after dox induction. Whole cell lysates were prepared from AID−/− IgLSce GFP, AID−/− IgLSce GFP Tet-OnI-SceI, AID−/− IgLSce GFP Tet-OnI-SceI K122I and AID−/− IgLSce GFP Tet-OnI-SceI K223I cell lines cultured in the presence or absence of dox. The duration of dox induction was 1, 2 or 3 days. I-SceI and α-tubulin (loading control) were detected by Western blotting.
Ig reversion induced by a DSB
The I-SceI GFP reporter was inserted into the Ig light chain locus of AID−/− DT40 cells by targeted integration (the AID−/− IgLSce GFP cell line) followed by stable expression of the third-generation Tet-On expression vector (Supplementary Figure S1). Finally, the resulting cell line was transfected with a vector that contained the I-SceI endonuclease gene under the third-generation Tet-inducible promoter (Supplementary Figure S1). Upon doxycycline (dox) administration, expression of I-SceI was monitored by FACS according to the intensity of the mCherry signal expressed from the same cistron. Thus, we successfully established a cell line, AID−/− IgLSce GFP Tet-OnI-SceI, that strongly expressed I-SceI in a dox-dependent manner. Dox induction of this cell line generated a GFP (+) population (Figure 1D and E). This GFP (+) population appeared two days after dox induction and gradually increased, reaching 0.94% after two weeks (Figure 1F), which was comparable to one induced by AID (Figure 1E).
As controls, we generated expression vectors for two independent I-SceI nickase mutants, I-SceI K122I and K223I, that can cleave the opposite sides of the I-SceI site and thus introduce a SSB, by site-directed mutagenesis of the I-SceI coding sequence (32). We tested whether expression of I-SceI nickase mutants could revert the frame-shift mutation of the Ig light chain gene locus. Although we did not measure the nickase activity of I-SceI K122I or K223I in DT40, the nicking activity of these mutants was tested in vitro (32,33), and the function of the I-SceI K223I mutant in homology-directed repair in yeast and human cell line has been reported (33). Dox-induced protein expression was confirmed by Western blots (Figure 1G); however, flow cytometric analysis failed to observe a GFP (+) population upon expression of I-SceI nickase mutants (Figure 1D and E).
In order to investigate the degree of V gene diversity that contributed to revert the frame-shift mutation, the GFP (+) population was isolated by FACS sorting. From this GFP (+) population, the VJ-GFP reporter was amplified by PCR, cloned into a plasmid vector and sequenced.
Gene conversion induced by AID
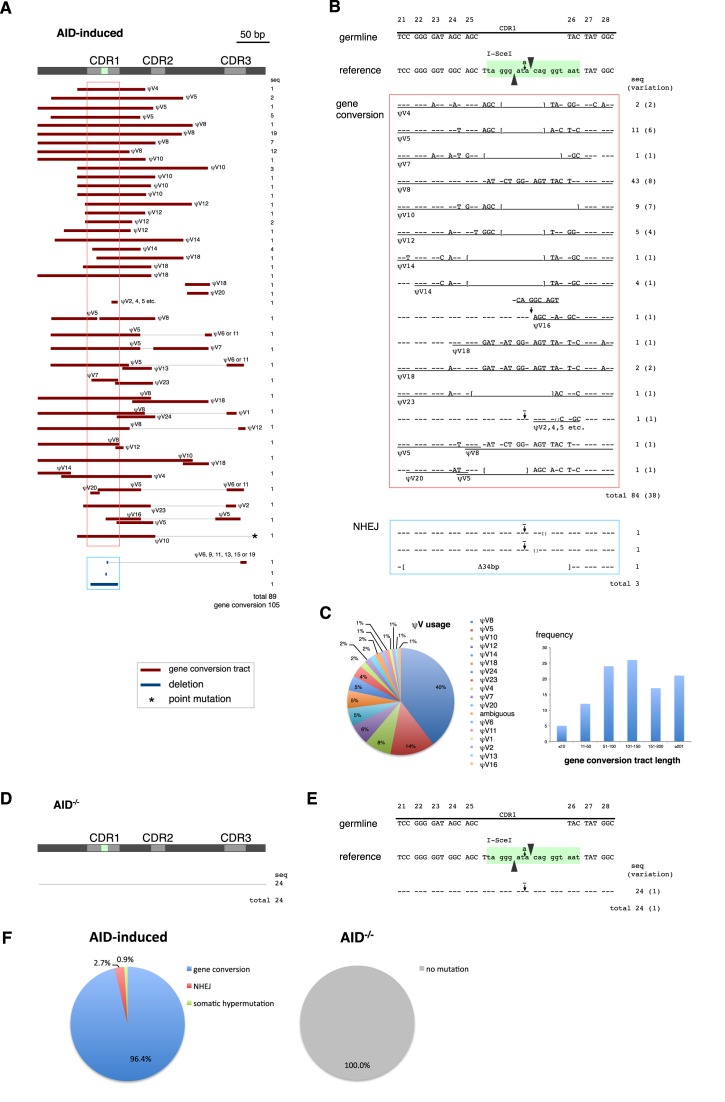
We analyzed the sequence of the VJ-GFP reporter of the IgLSce GFP cell line expressing AID. While the frame-shift mutation at the I-SceI site was reverted by either deletions or insertions in those sequences, the majority of those sequences had additional sequence changes including some resembling ψV genes (Figure 2A and B), which is a typical feature of Ig gene conversion. Thus, although the 18-bp sequence of the I-SceI site is not homologous to ψV genes, it was efficiently diversified through gene conversion.
Figure 2.
AID-induced gene conversion. (A) An overview of the AID-induced DNA sequence changes responsible for Ig reversion. Ig light chain V gene diversity was analyzed in sorted GFP (+) IgLSce GFP cell line. Sequence changes, classified into gene conversion, non-homologous end joining (NHEJ), and somatic hypermutation, are summarized on the map. The events in the same horizontal lines were derived from the same plasmid clones. The regions corresponding to CDR1 are emphasized by frames. (B) Sequences around the I-SceI site with detailed gene conversion analysis. The number of analyzed sequences and its variation are shown. Dashes indicate nucleotides identical to the sequence of the reference sequence of the mother cell line, and brackets mark the borders of deletions. Gene conversion tracts are underlined and annotated with the name of the likely pseudo V (ψV) donors. (C) ψV usage and gene conversion tract length observed in AID-induced gene conversion. (D) An overview of Ig light chain gene sequences in AID−/− DT40. (E) Sequences around the I-SceI site from AID−/− DT40. (F) Pathway choice of Ig reversion induced by AID.
Among the 89 VJ-GFP sequences obtained from the IgLSce GFP cell line, 86 showed sequence changes suggestive of gene conversion events (Figure 2A). A total of 105 gene conversion tracts from 17 of different ψV genes of various lengths were observed (Figure 2A and C). The ψV8 gene was most frequently used as a gene conversion donor, probably due to its homology with the ψV8 gene conversion tracts on the reporter (Figure 1B). In accordance with the features of gene conversion, the same ψV provided different DNA sequence tracts as substrate of gene conversion. For example, gene conversion tracts by ψV8 spanned from 118 to 212 bp. While 72 sequences had single gene conversion events, 14 sequences had two or more gene conversion events. CDR1 was the hotspot of gene conversion, as most of the gene conversion events overlapped with CDR1 (Figure 2B), and gene conversion tracts targeting CDR1 were relatively longer than gene conversion events changing CDR2 or 3 sequences (Figure 2A). While the preference for using CDR1 for gene conversion may be partly due to selective pressure to sort cells that restored GFP expression, a similar CDR1 preference was previously reported, even without specific enrichment (22). Nevertheless, gene conversion tracts were also observed in CDR2, CDR3 and framework regions. As expected, Ig light chain gene sequences remained unchanged in AID−/− DT40 cells (Figure 2D, E and F) (22).
Three events of non-templated deletion on the I-SceI site were observed and were considered deletions by NHEJ. The frequency of the NHEJ-mediated deletions appeared higher than previously described results (22), possibly because of the selection for cells that had reverted the in-frame mutation. One event of non-templated C to T point mutations was also observed, which is a common mutation signature of AID (4). Overall NHEJ mediated deletions and somatic hypermutation were rare occurrences when compared to gene conversion events (Figure 2F).
Gene conversion induced by a DSB
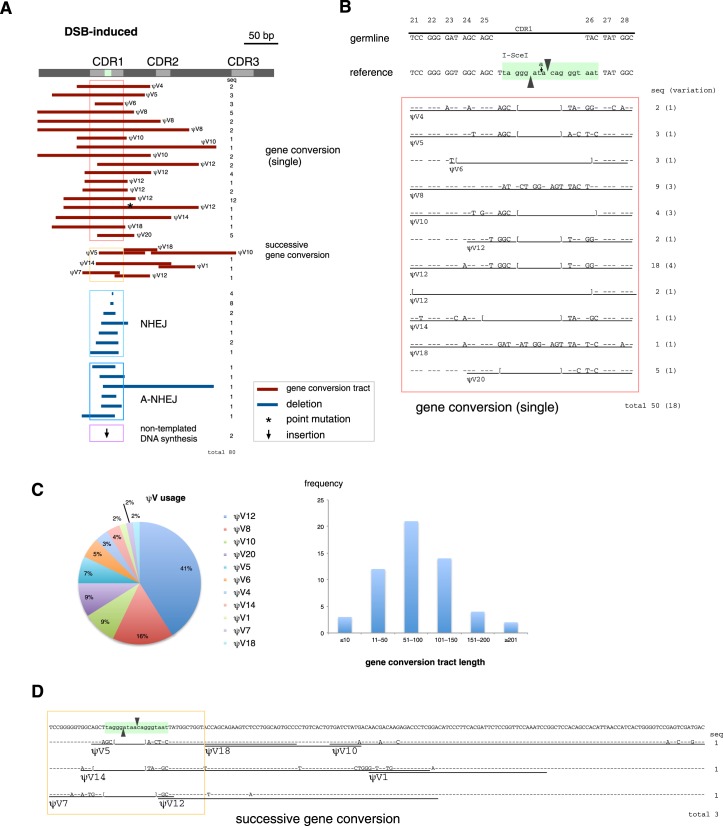
Ig light chain V gene diversity induced by a single DSB was also analyzed in sorted GFP (+) AID−/− IgLSce GFP Tet-OnI-SceI DT40 cells recovered after dox treatment. Analyzing a total of 80 VJ-GFP sequences, we found that reversion of the frame-shift mutation at the I-SceI site occurred either through deletions or insertions (Figure 3A). While some of the sequences were only repaired by deletion, other sequences had additional sequence changes that were identical to the ψV genes (Figure 3A and B), suggesting the occurrence of gene conversion events.
Figure 3.
DSB-induced gene conversion. (A) An overview of the DSB-induced DNA sequence changes responsible for Ig reversion. Ig light chain V gene diversity was analyzed in sorted GFP (+) AID−/− IgLSce GFP Tet-OnI-SceI DT40 cells recovered after dox treatment. Sequence changes, classified into gene conversion, NHEJ, alternative NHEJ (A-NHEJ), and non-templated insertion, are summarized on the map. (B) Sequences around the I-SceI site with detailed gene conversion analysis. (C) ψV usage and gene conversion tract length observed in DSB-induced gene conversion. (D) Sequence changes for successive gene conversion events. Gene conversion tracts are underlined and annotated with the name of the likely ψV donors.
The DSB-induced gene conversion pattern appeared simpler than that of AID, since most were single gene conversion events per sequence (Figure 3A), and long gene conversion tracts were relatively less frequent (Figure 3C). Single-gene conversion events were observed in 50 sequences, which had 18 variations in gene conversion tracts (Figure 3B). Among observed 11 of different ψV genes, the ψV12 gene was most frequently used as a gene conversion donor, and ψV8 was the second-most preferential pseudogene donor (Figure 3C). Thus, the preference for ψV donors may have been affected by local deletion of the sequence around the I-SceI site. Gene conversion tracts targeting CDR1 by different ψV donors, such as ψV4, 5, 8, 10, 12, 14 and 18, were the same or similar between AID- and DSB-induced gene conversion (Figures 2B and 3B). These gene conversion tracts spanned up to 212 bp, and they were sometimes extended to around 100 bp upstream and 200 bp downstream of the I-SceI site. Among the ψV donors used in a DSB-induced gene conversion, ψV20 was the most distal pseudogene donor from the functional V gene (1). Since ψV20 is located ∼20 kb upstream of the I-SceI site (1), the DNA end of the I-SceI-induced DSB was supposed to interact at a distance of at least 20 kb. Similarly to AID-induced gene conversion, even the same ψV gene produced a variety of gene conversion tracts. Thus, a DSB in the Ig V gene of DT40 cells is sufficient to trigger a diversified set of gene conversion events, comparable in frequency and nature to those induced by AID. In contrast to the Ig light chain gene, the rearranged Ig heavy chain gene sequences remained unchanged after dox induction (Supplementary Figure S2), suggesting that off-target mutation by I-SceI is limited.
To our surprise, multiple gene conversion events were observed in three sequences (Figure 3A and D). DNA synthesis initiated from the DSB within the I-SceI site is expected to destroy the I-SceI site. Indeed, all the single gene conversion tracts induced by I-SceI altered the sequence of the I-SceI site (Figure 3A and B), while AID-induced single gene conversion tracts were sometimes located outside of the I-SceI site (Figure 2A). Therefore, multiple rounds of gene conversion are unlikely to be caused by multiple rounds of I-SceI-induced DSBs. These facts raise two questions: (i) how can multiple gene conversions be achieved, and (ii) how can the gene conversion tracts be generated separately from the I-SceI site? Most likely, a single DSB can induce successive gene conversion events (Figure 3A and D). These results also indicate that gene conversion was initiated not only by the DNA end generated by I-SceI, but could also be initiated by DNA repair processes during gene conversion.
Analysis of somatic hypermutation revealed a single A to T mutation, which is rarely observed in the mutational pattern initiated by AID (4).
Non-gene conversion pathways induced by a DSB
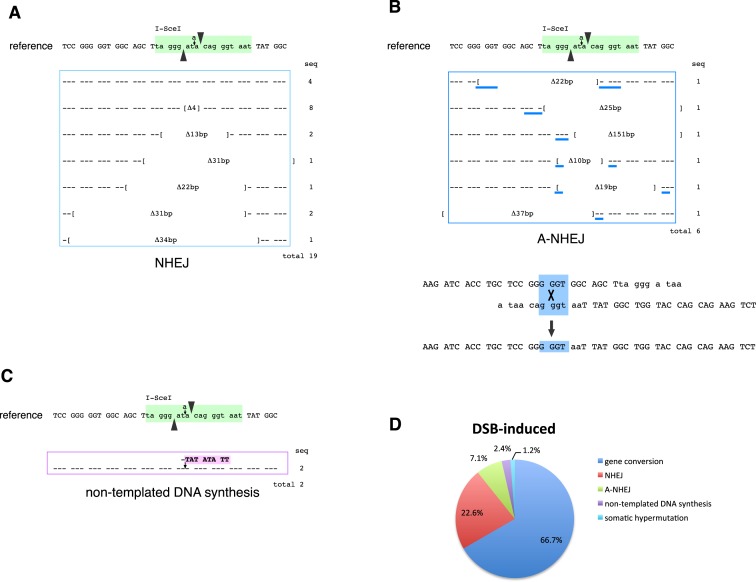
Sequencing of the VJ-GFP gene in GFP (+) AID−/− IgLSce GFP Tet-OnI-SceI DT-40 cells revealed additional DNA repair pathways responsible for reversion of the frameshift mutation (Figure 3A). We observed deletions from 1 to 34 bp around the I-SceI site in 19 sequences (Figure 4A). These deletions were sometimes extended to 16 bp upstream and 12 bp downstream of the I-SceI site. Since micro-homology was not detected around the breakpoints in those sequences, it is likely that these events are caused by NHEJ, which is another major pathway for frame-shift reversion induced by a DSB.
Figure 4.
DSB-induced DNA repair. (A) NHEJ events. Brackets mark the borders of deletions. (B) A-NHEJ events. Micro-homologies are shown by blue underlines. One example is shown to explain how deletional recombination occurs by alignment of 4 bp micro-homologies. (C) Non-templated DNA synthesis. (D) Pathway choice of Ig reversion induced by a DSB.
Micro-homology was observed around the breakpoints of deletions in six sequences (Figure 4B). These can be typical features of alternative NHEJ (A-NHEJ), which is distinguished from classical NHEJ by its use of microhomologous sequences to align the broken strands before joining. A-NHEJ is independent of Ku or DNA-PK, and the end-joining is catalyzed by DNA ligases III and I instead of DNA ligase IV (43,44). Deletions by A-NHEJ were relatively larger than those caused by NHEJ and spanned from 10 to 151 bp.
In contrast to deletion, we also observed the insertion of a ‘TATATATT’ sequence stretch (Figure 4C). Since identical or similar sequences were not found in any ψV genes, this mutation cannot be explained by gene conversion. We also exclude that the insertion resulted from duplication of the sequence at the breakpoint. Hence, this non-templated DNA synthesis event appears to be caused by an unidentified DNA polymerase.
DISCUSSION
We have shown that a DSB in CDR1 is sufficient to trigger Ig gene conversion, even in the absence of AID. A single DSB in the same position spurred a variety of gene conversions events, in a similar fashion to those initiated by AID. Taken together, our results indicate that a DSB can be an intermediate for physiological gene conversion. Our results suggest that DSB-induced repair offers higher flexibility in pathway choices than AID-induced repair, leading relatively more frequently to NHEJ or A-NHEJ.
Although the I-SceI site does not share significant homology with the ψV genes, this sequence was efficiently diversified by both AID- and DSB-induced gene conversion. This may not be surprising in light of the fact that CDRs are often diversified by the poorly homologous gene conversion tracts of different ψV genes (1,22). Paradoxically, gene conversion appears to not always require homology between the ψV genes and the target sequence; nevertheless, because ψV8 was the relatively favored gene conversion donor in both AID- and DSB-induced gene conversion (Figures 2C and 3C), gene conversion is influenced by homologous sequences flanking a non-homologous sequence. Most likely, ψV donors align not only in the short regions in CDRs, but also in rather wide regions, including conserved framework regions, based on homology. While long stretches of homologous sequences support alignment between the ψV genes and the functional V gene during gene conversion, the non-homologous regions flanked by long homologies appear to be targeted by gene conversion, contributing to the generation of diversity.
CDR1 is characterized by the highest physiological gene conversion in DT40 cells (Figure 2A) (22); this suggests that DSBs caused by AID are also predominantly located around CDR1. Interestingly, a single DSB was able to cause even two rounds of gene conversion (Figure 3D). Thus, the double-stranded DNA end can continuously access different loci of the chromosome by using microhomology during gene conversion, which can be explained by the following model: once the local homology search has been initiated by the DNA end induced by a DSB, this DNA end will copy one of the ψV gene sequences. Usually this DNA end turns back to the rearranged V (D) J locus to complete DNA synthesis, fixing a gene conversion tract (single gene conversion in Figure 5A); however, before completing DNA synthesis at the rearranged V (D) J locus, such a DNA end can be further aligned with another ψV gene to copy the second gene conversion tract. After this DNA end turns back to the rearranged V (D) J locus, the copied sequences are fixed as multiple gene conversion tracts (successive gene conversion in Figure 5A).
Figure 5.
Models of successive gene conversion and DSB-induced gene conversion. (A) A model for successive gene conversion. (B) Gene conversion and DNA repair pathways induced by either AID or a DSB.
Thus, multiple rounds of gene conversion can be initiated with a single DSB. Such events may also occur under physiological gene conversion by AID; however, it is usually impossible to distinguish whether multiple gene conversion events were initiated by a single DNA lesion in the case of AID-induced gene conversion. Hence, successive Ig gene conversion initiated by a DSB was revealed for the first time by this study. Some of the multiple gene conversion events in the presence of AID (Figure 2A) might be initiated by single AID-induced lesions.
During the successive gene conversion, template switching to the second ψV gene may use short regions of homology. This process reminded us of the ‘copy choice’ model for combinatorial assembly of different leucine-rich repeat sequences of variable lymphocyte receptor (VLR) genes in the lamprey (45). In the copy choice model for VLR assembly proposed by Nagawa et al., a replicating DNA end (i) dissociates from the original template, (ii) switches templates using short regions of identity, and (iii) copies a donor sequence elsewhere in the genome (45). Thus, the copy choice model strongly resembles the successive gene conversion model for multiple conversion tracks (Figure 5A). VLR assembly is initiated by a cytidine deaminase, a possible ancestor homologue of AID, implying a functional and evolutionary relationship between VLR assembly and Ig gene conversion.
Although a DSB induces gene conversion in the absence of AID, this does not necessarily mean that a DSB is sufficient to catalyze gene conversion in cell lines other than DT40 cells. Even in the absence of AID, components of other DNA repair, recombination or replication enzymes still remain intact in the cells. Except for AID, all the known components of the Ig gene diversification machinery are enzymes hijacked from ubiquitous DNA repair, recombination and replication machineries in the course of evolution. Indeed, gene conversion machinery commonly appears active across cell types or species, as a DSB can induce homology-directed repair in an I-SceI site-containing reporter gene in different cells. Nevertheless, AID-induced lesions rarely cause immunoglobulin gene conversion in human B cells, despite the presence of multiple pseudo-V genes in humans (46). Although it remains uncertain whether an Ig gene conversion-specific enzyme that acts downstream of AID or DSB exists, such an enzyme might contribute to the DSB-induced gene conversion. A forward genetic approach may help to screen such a hypothetical enzyme (47).
As AID-induced lesions predominantly lead to gene conversion in DT40 cells (Figure 2), DNA breaks initiated by AID are destined to a gene conversion pathway in DT40 cells. AID induces a DNA lesion as an early step of B cell-specific recombination by its cytidine deaminase activity (23,24); however, as the N-terminal of AID is responsible for Ig gene conversion and somatic hypermutation and the C-terminal domain of AID is essential for switch recombination (48,49), AID can further influence the dissected B cell-specific recombination pathways, possibly via co-factors that bind the N- or C-terminus of AID. The germinal center-associated nuclear protein is one of the candidates that affects the fate of DSB in Ig V regions in the presence of AID (50,51). An AID-induced DNA break is supposed to be protected by AID co-factors that direct the DNA lesion to the gene conversion pathway and prevent undesired DNA repair pathways.
In contrast to an AID-induced DNA break, an I-SceI-induced DSB is expected to have variation in its location or timing. Depending on the nuclear location or the cell cycle phase in which the DSB occurs, its fate may be determined by the accidental encounter with the mutator complex or a different DNA repair complex. Thus, physiological and artificial DNA breaks may be qualitatively different; artificial DSBs may have more flexibility in terms of fate decision than physiological DSBs, which are protected by the DNA mutator factory.
In the current study, surprisingly, a SSB that was induced by a nickase version of the I-SceI mutants was unable to revert frame-shift mutations at the I-SceI site (Figure 1D). These results are somehow paradoxical with previous reports that suggest the involvement of SSBs in gene conversion or homologous recombination. For instance, Nakahara et al. reported that gene conversion in DT40 is enhanced by a single-strand-specific exonuclease, E. coli Exo1 (SbcB), but not by a double-strand specific exonuclease, chicken Exo1, suggesting the role of SSBs in gene conversion (52). Nevertheless, their results that gene conversion is severely reduced in DT40 mutants deficient in key factors for DSB repair, Nbs1 or RAD54 (52), could be interpreted as that DSBs can play a role in gene conversion as well. Davis and Maizels reported that a SSB induced on the transcribed strand by an I-AniI nickase mutant can lead to homology-directed repair in a GFP reporter (53), while such homology-directed repair can be several-fold more efficiently induced by a DSB (53).
Which of the SSB or the DSB is the intermediate for physiological gene conversion? Paradoxically, another possible interpretation of the observed multiple pathway choice downstream of a DSB (Figure 4D) is that SSBs, rather than DSBs, are the dominant intermediate for physiological gene conversion (Figure 5B). Since UNG (+) DT40 in the ψV knockout-background (25) shows 7-fold less somatic hypermutation than UNG knockout DT40 (4), we previously estimated that more than 85% of AID-induced uracils are removed by error-free repair pathways through SSB formation. Considering that AID preferentially deaminates single-stranded DNA (54), SSBs are supposed to be generated much more frequently than DSBs. Although I-SceI K223I predominantly causes nicks in vitro (32,33) and promotes homology-directed repair in yeast and human cells (33), we do not exclude the possibility that I-SceI K122I or K223I was inactive due to unknown technical reasons or in a DT40-specific fashion, because we did not measure its nicking activity for ourselves. A negative proof that either a SSB or DSB is not able to induce gene conversion would be nothing less than probatio diabolica. In addition, since two SSBs occurring nearby in opposite strands can generate a DSB, some SSBs, generated physiologically or artificially, might be converted to DSBs prior to DNA repair or recombination. The answer is unlikely to be black and white, and most likely both SSBs and DSBs can cause Ig gene conversion.
The relatively frequent choice of the non-gene conversion pathways in DSB-induced Ig gene modification (Figure 4D) raises skepticism on the well-accepted hypothesis that a balance between homologous recombination and NHEJ is heavily shifted toward homologous recombination in DT40 cells. Instead, our study indicates that NHEJ is obviously active in wild-type DT40 cells. During the course of physiological gene conversion, NHEJ or A-NHEJ is more likely to be suppressed locally at the DNA lesion rather than in the whole cells. Even if homologous recombination were not up-regulated in DT40 cells, the hyper-targeted integration property of DT40 cells (55) could also be explained by assuming that the double-stranded DNA end of a linearized targeting vector is recognized as a DSB and is further processed via local control of homologous recombination.
The difference of DNA repair pathway choice downstream of AID or I-SceI-induced DSB shows that a DNA lesion in the same locus can be destined to a particular repair pathway without changing the general balance between homologous recombination and NHEJ in the cells. If B cells had to alter the general DNA repair activity to pursue B cell-specific recombination pathways, or such pathways are not restricted to Ig loci, this could cause serious genome instability as a tradeoff; however, the AID-transgenic mice suffer from carcinogenesis in T cells or the epithelium of respiratory bronchioles, but rarely from B cell lymphoma (56). Thus, the B cell genome is rather protected from AID-induced mutagenesis, possibly via specific recruitment of the AID complex to the Ig locus in a B cell-specific manner (57). In fact, the majority of germinal center B cells, which express high levels of AID (20), are not transformed to lymphoma. In order to protect the whole genome, evolution may have invented a system for locally restricting DNA repair at DNA lesions or specific loci, while the collapse of such a system may represent one of the mechanisms promoting carcinogenesis.
Acknowledgments
The authors are grateful to Holger Puchta for providing the I-SceI vector, to Takuya Abe for technical advice on tetracycline-inducible expression systems, to Stefano Casola, Svend Petersen-Mahrt, Ghadeer Shubassi and Ramveer Choudhary for critical reading of the manuscript and to Marco Foiani for his generous mentorship.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors.
Conflict of interest statement. None declared.
REFERENCES
- 1.Reynaud C.A., Anquez V., Grimal H., Weill J.C. A hyperconversion mechanism generates the chicken light chain preimmune repertoire. Cell. 1987;48:379–388. doi: 10.1016/0092-8674(87)90189-9. [DOI] [PubMed] [Google Scholar]
- 2.Reynaud C.A., Dahan A., Anquez V., Weill J.C. Somatic hyperconversion diversifies the single Vh gene of the chicken with a high incidence in the D region. Cell. 1989;59:171–183. doi: 10.1016/0092-8674(89)90879-9. [DOI] [PubMed] [Google Scholar]
- 3.McCormack W.T., Thompson C.B. Chicken IgL variable region gene conversions display pseudogene donor preference and 5′ to 3′ polarity. Genes Dev. 1990;4:548–558. doi: 10.1101/gad.4.4.548. [DOI] [PubMed] [Google Scholar]
- 4.Arakawa H., Saribasak H., Buerstedde J.M. Activation-induced cytidine deaminase initiates immunoglobulin gene conversion and hypermutation by a common intermediate. PLoS Biol. 2004;2:E179. doi: 10.1371/journal.pbio.0020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson L.M., McCormack W.T., Postema C.E., Humphries E.H., Thompson C.B. Templated insertions in the rearranged chicken IgL V gene segment arise by intrachromosomal gene conversion. Genes Dev. 1990;4:536–547. doi: 10.1101/gad.4.4.536. [DOI] [PubMed] [Google Scholar]
- 6.McCormack W.T., Hurley E.A., Thompson C.B. Germ line maintenance of the pseudogene donor pool for somatic immunoglobulin gene conversion in chickens. Mol. Cell. Biol. 1993;13:821–830. doi: 10.1128/mcb.13.2.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arakawa H., Furusawa S., Ekino S., Yamagishi H. Immunoglobulin gene hyperconversion ongoing in chicken splenic germinal centers. EMBO J. 1996;15:2540–2546. [PMC free article] [PubMed] [Google Scholar]
- 8.Arakawa H., Buerstedde J.M. Immunoglobulin gene conversion: insights from bursal B cells and the DT40 cell line. Dev. Dyn. 2004;229:458–464. doi: 10.1002/dvdy.10495. [DOI] [PubMed] [Google Scholar]
- 9.McCormack W.T., Carlson L.M., Tjoelker L.W., Thompson C.B. Evolutionary comparison of the avian IgL locus: combinatorial diversity plays a role in the generation of the antibody repertoire in some avian species. Int. Immunol. 1989;1:332–341. doi: 10.1093/intimm/1.4.332. [DOI] [PubMed] [Google Scholar]
- 10.Guan X., Wang J., Ma L., Wang X., Cheng X., Han H., Zhao Y., Ren L. Multiple germline functional VL genes contribute to the IgL repertoire in ducks. Dev. Comp. Immunol. 2016;60:167–179. doi: 10.1016/j.dci.2016.02.029. [DOI] [PubMed] [Google Scholar]
- 11.Huang T., Wu K., Yuan X., Shao S., Wang W., Wei S., Cao G. Molecular analysis of the immunoglobulin genes in goose. Dev. Comp. Immunol. 2016;60:160–166. doi: 10.1016/j.dci.2016.02.024. [DOI] [PubMed] [Google Scholar]
- 12.Becker R.S., Knight K.L. Somatic diversification of immunoglobulin heavy chain VDJ genes: evidence for somatic gene conversion in rabbits. Cell. 1990;63:987–997. doi: 10.1016/0092-8674(90)90502-6. [DOI] [PubMed] [Google Scholar]
- 13.Butler J.E. Immunoglobulin diversity, B-cell and antibody repertoire development in large farm animals. Rev. Sci. Tech. 1998;17:43–70. doi: 10.20506/rst.17.1.1096. [DOI] [PubMed] [Google Scholar]
- 14.Guo Y., Bao Y., Meng Q., Hu X., Meng Q., Ren L., Li N., Zhao Y. Immunoglobulin genomics in the guinea pig (Cavia porcellus) PloS One. 2012;7:e39298. doi: 10.1371/journal.pone.0039298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ujvari B., Belov K. Characterization of antibody V segment diversity in the Tasmanian devil (Sarcophilus harrisii) Vet. Immunol. Immunopathol. 2015;167:156–165. doi: 10.1016/j.vetimm.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 16.Qin T., Zhao H., Zhu H., Wang D., Du W., Hao H. Immunoglobulin genomics in the prairie vole (Microtus ochrogaster) Immunol. Lett. 2015;166:79–86. doi: 10.1016/j.imlet.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Arakawa H., Kuma K., Yasuda M., Furusawa S., Ekino S., Yamagishi H. Oligoclonal development of B cells bearing discrete Ig chains in chicken single germinal centers. J. Immunol. 1998;160:4232–4241. [PubMed] [Google Scholar]
- 18.Arakawa H., Kuma K., Yasuda M., Ekino S., Shimizu A., Yamagishi H. Effect of environmental antigens on the Ig diversification and the selection of productive V-J joints in the bursa. J. Immunol. 2002;169:818–828. doi: 10.4049/jimmunol.169.2.818. [DOI] [PubMed] [Google Scholar]
- 19.Buerstedde J.M., Reynaud C.A., Humphries E.H., Olson W., Ewert D.L., Weill J.C. Light chain gene conversion continues at high rate in an ALV-induced cell line. EMBO J. 1990;9:921–927. doi: 10.1002/j.1460-2075.1990.tb08190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 21.Revy P., Muto T., Levy Y., Geissmann F., Plebani A., Sanal O., Catalan N., Forveille M., Dufourcq-Labelouse R., Gennery A., et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome. Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 22.Arakawa H., Hauschild J., Buerstedde J.M. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science. 2002;295:1301–1306. doi: 10.1126/science.1067308. [DOI] [PubMed] [Google Scholar]
- 23.Di Noia J., Neuberger M.S. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419:43–48. doi: 10.1038/nature00981. [DOI] [PubMed] [Google Scholar]
- 24.Petersen-Mahrt S.K., Harris R.S., Neuberger M.S. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 25.Saribasak H., Saribasak N.N., Ipek F.M., Ellwart J.W., Arakawa H., Buerstedde J.M. Uracil DNA glycosylase disruption blocks Ig gene conversion and induces transition mutations. J. Immunol. 2006;176:365–371. doi: 10.4049/jimmunol.176.1.365. [DOI] [PubMed] [Google Scholar]
- 26.Xue K., Rada C., Neuberger M.S. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2-/- ung-/- mice. J. Exp. Med. 2006;203:2085–2094. doi: 10.1084/jem.20061067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puchta H., Dujon B., Hohn B. Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res. 1993;21:5034–5040. doi: 10.1093/nar/21.22.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abe T., Branzei D. High levels of BRC4 induced by a Tet-On 3G system suppress DNA repair and impair cell proliferation in vertebrate cells. DNA Repair (Amst) 2014;22:153–164. doi: 10.1016/j.dnarep.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zarrin A.A., Del Vecchio C., Tseng E., Gleason M., Zarin P., Tian M., Alt F.W. Antibody class switching mediated by yeast endonuclease-generated DNA breaks. Science. 2007;315:377–381. doi: 10.1126/science.1136386. [DOI] [PubMed] [Google Scholar]
- 30.Boothroyd C.E., Dreesen O., Leonova T., Ly K.I., Figueiredo L.M., Cross G.A., Papavasiliou F.N. A yeast-endonuclease-generated DNA break induces antigenic switching in Trypanosoma brucei. Nature. 2009;459:278–281. doi: 10.1038/nature07982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moure C.M., Gimble F.S., Quiocho F.A. Crystal structures of I-SceI complexed to nicked DNA substrates: Snapshots of intermediates along the DNA cleavage reaction pathway. Nucleic Acids Res. 2008;36:3287–3296. doi: 10.1093/nar/gkn178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niu Y., Tenney K., Li H., Gimble F.S. Engineering variants of the I-SceI homing endonuclease with strand-specific and site-specific DNA-nicking activity. J. Mol. Biol. 2008;382:188–202. doi: 10.1016/j.jmb.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katz S.S., Gimble F.S., Storici F. To nick or not to nick: comparison of I-SceI single- and double-strand break-induced recombination in yeast and human cells. PloS One. 2014;9:e88840. doi: 10.1371/journal.pone.0088840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saribasak H., Arakawa H. Targeted transfection of DT40 cells. Subcell. Biochem. 2006;40:419–421. doi: 10.1007/978-1-4020-4896-8_38. [DOI] [PubMed] [Google Scholar]
- 35.Arakawa H., Lodygin D., Buerstedde J.M. Mutant loxP vectors for selectable marker recycle and conditional knock-outs. BMC Biotechnol. 2001;1:7. doi: 10.1186/1472-6750-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arakawa H., Kudo H., Batrak V., Caldwell R.B., Rieger M.A., Ellwart J.W., Buerstedde J.M. Protein evolution by hypermutation and selection in the B cell line DT40. Nucleic Acids Res. 2008;36:e1. doi: 10.1093/nar/gkm616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arakawa H. Immunoglobulin gene conversion and hypermutation assay by FACS. Subcell. Biochem. 2006;40:351–352. doi: 10.1007/978-1-4020-4896-8_23. [DOI] [PubMed] [Google Scholar]
- 38.Urnov F.D., Miller J.C., Lee Y.L., Beausejour C.M., Rock J.M., Augustus S., Jamieson A.C., Porteus M.H., Gregory P.D., Holmes M.C. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 39.Bedell V.M., Wang Y., Campbell J.M., Poshusta T.L., Starker C.G., Krug R.G. 2nd, Tan W., Penheiter S.G., Ma A.C., Leung A.Y. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benatar T., Tkalec L., Ratcliffe M.J. Stochastic rearrangement of immunoglobulin variable-region genes in chicken B-cell development. Proc. Natl. Acad. Sci. U.S.A. 1992;89:7615–7619. doi: 10.1073/pnas.89.16.7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paul K., Wang M., Mladenov E., Bencsik-Theilen A., Bednar T., Wu W., Arakawa H., Iliakis G. DNA ligases I and III cooperate in alternative non-homologous end-joining in vertebrates. PloS One. 2013;8:e59505. doi: 10.1371/journal.pone.0059505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simsek D., Brunet E., Wong S.Y., Katyal S., Gao Y., McKinnon P.J., Lou J., Zhang L., Li J., Rebar E.J., et al. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011;7:e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagawa F., Kishishita N., Shimizu K., Hirose S., Miyoshi M., Nezu J., Nishimura T., Nishizumi H., Takahashi Y., Hashimoto S., et al. Antigen-receptor genes of the agnathan lamprey are assembled by a process involving copy choice. Nat. Immunol. 2007;8:206–213. doi: 10.1038/ni1419. [DOI] [PubMed] [Google Scholar]
- 46.Darlow J.M., Stott D.I. Gene conversion in human rearranged immunoglobulin genes. Immunogenetics. 2006;58:511–522. doi: 10.1007/s00251-006-0113-6. [DOI] [PubMed] [Google Scholar]
- 47.Arakawa H. A method to convert mRNA into a gRNA library for CRISPR/Cas9 editing of any organism. Sci. Adv. 2016;2:e1600699. doi: 10.1126/sciadv.1600699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barreto V., Reina-San-Martin B., Ramiro A.R., McBride K.M., Nussenzweig M.C. C-Terminal deletion of AID uncouples class switch recombination from somatic hypermutation and gene conversion. Mol. Cell. 2003;12:501–508. doi: 10.1016/s1097-2765(03)00309-5. [DOI] [PubMed] [Google Scholar]
- 49.Ta V.T., Nagaoka H., Catalan N., Durandy A., Fischer A., Imai K., Nonoyama S., Tashiro J., Ikegawa M., Ito S., et al. AID mutant analyses indicate requirement for class-switch-specific cofactors. Nat. Immunol. 2003;4:843–848. doi: 10.1038/ni964. [DOI] [PubMed] [Google Scholar]
- 50.Eid M.M., Maeda K., Almofty S.A., Singh S.K., Shimoda M., Sakaguchi N. GANP regulates the choice of DNA repair pathway by DNA-PKcs interaction in AID-dependent IgV region diversification. J. Immunol. 2014;192:5529–5539. doi: 10.4049/jimmunol.1400021. [DOI] [PubMed] [Google Scholar]
- 51.Singh S.K., Maeda K., Eid M.M., Almofty S.A., Ono M., Pham P., Goodman M.F., Sakaguchi N. GANP regulates recruitment of AID to immunoglobulin variable regions by modulating transcription and nucleosome occupancy. Nat. Commun. 2013;4:1830. doi: 10.1038/ncomms2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakahara M., Sonoda E., Nojima K., Sale J.E., Takenaka K., Kikuchi K., Taniguchi Y., Nakamura K., Sumitomo Y., Bree R.T., et al. Genetic evidence for single-strand lesions initiating Nbs1-dependent homologous recombination in diversification of Ig v in chicken B lymphocytes. PLoS Genet. 2009;5:e1000356. doi: 10.1371/journal.pgen.1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davis L., Maizels N. Homology-directed repair of DNA nicks via pathways distinct from canonical double-strand break repair. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E924–E932. doi: 10.1073/pnas.1400236111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dickerson S.K., Market E., Besmer E., Papavasiliou F.N. AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med. 2003;197:1291–1296. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buerstedde J.M., Takeda S. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell. 1991;67:179–188. doi: 10.1016/0092-8674(91)90581-i. [DOI] [PubMed] [Google Scholar]
- 56.Okazaki I.M., Hiai H., Kakazu N., Yamada S., Muramatsu M., Kinoshita K., Honjo T. Constitutive expression of AID leads to tumorigenesis. J. Exp. Med. 2003;197:1173–1181. doi: 10.1084/jem.20030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buerstedde J.M., Alinikula J., Arakawa H., McDonald J.J., Schatz D.G. Targeting of somatic hypermutation by immunoglobulin enhancer and enhancer-like sequences. PLoS Biol. 2014;12:e1001831. doi: 10.1371/journal.pbio.1001831. [DOI] [PMC free article] [PubMed] [Google Scholar]