Abstract
RecA-family recombinase-catalyzed ATP-dependent homologous joint formation is critical for homologous recombination, in which RecA or Rad51 binds first to single-stranded (ss)DNA and then interacts with double-stranded (ds)DNA. However, when RecA or Rad51 interacts with dsDNA before binding to ssDNA, the homologous joint-forming activity of RecA or Rad51 is quickly suppressed. We found that under these and adenosine diphosphate (ADP)-generating suppressive conditions for the recombinase activity, RecA or Rad51 at similar optimal concentrations enhances the DNA ligase-catalyzed dsDNA end-joining (DNA ligation) about 30- to 40-fold. The DNA ligation enhancement by RecA or Rad51 transforms most of the substrate DNA into multimers within 2–5 min, and for this enhancement, ADP is the common and best cofactor. Adenosine triphosphate (ATP) is effective for RecA, but not for Rad51. Rad51/RecA-enhanced DNA ligation depends on dsDNA-binding, as shown by a mutant, and is independent of physical interactions with the DNA ligase. These observations demonstrate the common and unique activities of RecA and Rad51 to juxtapose dsDNA-ends in preparation for covalent joining by a DNA ligase. This new in vitro function of Rad51 provides a simple explanation for our genetic observation that Rad51 plays a role in the fidelity of the end-joining of a reporter plasmid DNA, by yeast canonical non-homologous end-joining (NHEJ) in vivo.
INTRODUCTION
DNA double-strand breaks (DSBs) are generated by external and internal factors during normal metabolism, and cause various genetic disorders such as cancer (1). From bacteria (2) to humans, DSBs are repaired by either homologous recombination or non-homologous end-joining (NHEJ: (3–5)).
Homologous recombination is characterized by hybrid DNA-intermediate (homologous joint)-formation between complementary sequences of parental DNAs: one derived from a single-stranded (ss) DNA tail formed at a broken end and the other from intact double-stranded (ds) DNA that serves as a template for repair (6,7). Homologous joint formation (also called D-loop formation) is catalyzed by RecA-family recombinases in an ATP-dependent fashion (8–11). Rad51 is a eukaryotic member of the RecA-family recombinases (12), and is an essential component for homologous recombination in vivo (13,14). The Ku-heterodimer and DNA ligase IV (Lig4) are the major proteins/enzymes in canonical NHEJ, and Ku reportedly contributes to the fidelity of canonical NHEJ (15–17). We previously reported that the RAD51-deletion mutations in yeast dramatically increased errors (more than 14-fold) in the end-joining of plasmid DNA with complementary 4-base overhangs at both termini, even in the presence of active Ku-heterodimers, Lig4 and all other components of NHEJ and the observed end-joining depended on LIG4, indicating that it is canonical NHEJ. These observations suggested the role of RAD51 in the fidelity of canonical NHEJ (18: see Discussion).
Rad51 and RecA exhibit comparable molecular functions, including ssDNA-binding, dsDNA-binding, DNA-dependent ATP-hydrolysis and ATP-dependent homologous joint forming activities. The molecular structures of Rad51 and RecA share similar core-domains, with a very well conserved tertiary structure containing two flexible loops involved in DNA-binding at the corresponding positions (19–22), and form analogous right-handed helical filaments (23). Both proteins in the ATP-bound forms induce a unique extended ssDNA-structure that explains the homology-search process in solution (24,25). Since the binding to ssDNA and the unfolding of its secondary structures by filament formation precedes the binding of Rad51 or RecA to dsDNA (26–29), homologous joint formation is most effective when RecA or Rad51 is preincubated with ssDNA in the presence of adenosine triphosphate (ATP), and subsequently interacts with dsDNA (30). Notably, when RecA or Rad51 is first incubated with dsDNA in the presence of ATP, before the addition of ssDNA, homologous joint formation is suppressed (31–33). These similarities between RecA and Rad51 suggest that they also share a common mechanism to catalyze homologous joint formation.
While the helical filament formation around ssDNA, to activate ATP-bound RecA for promoting homologous joint formation, requires several minutes of incubation (30,31), the suppression by the preceding incubation of ATP-bound RecA with dsDNA is established within a minute (31), indicating that RecA can bind quickly to dsDNA (34). To find new functions that can explain their possible roles in NHEJ (18), we became interested in identifying the activity of RecA or Rad51 that is activated by the incubation with dsDNA, instead of ssDNA. We found that RecA and Rad51 each enhance the end-joining catalyzed by DNA ligases (DNA ligation), in the presence of ADP. Since far more information is available for RecA than Rad51, we examined RecA as a pilot system and subsequently verified the findings for Rad51.
MATERIALS AND METHODS
DNA, RecA, Rad51 and DNA ligases
All substrate dsDNAs were linear. The linear dsDNAs with 3′ TGCA- and 5′ AGCT four-nucleotide overhangs, and with blunt ends were prepared by digestions of closed circular dsDNAs with the restriction endonucleases PstI, HindIII and HincII, respectively, using the manufacturer's recommended conditions (Takara Bio Company, Japan and New England Biolabs). The preparation of closed circular dsDNAs from which linear dsDNA was prepared, and that of Escherichia coli RecA were cited or described previously (31). S. cerevisiae Rad51 was purified as described (33). DNA size and amount markers are Smart Ladder purchased from Nippon Gene containing linear dsDNAs of 10 000 bp (100 ng/5 μl), 8000 bp (80 ng/5 μl), 6000 bp (60 ng/5 μl), 5000 bp (50 ng/5 μl), 4000 bp (40 ng/5 μl), 3000 bp (30 ng/5 μl), 2500 bp (25 ng/5 μl), 2000 bp (20 ng/5 μl), 1500 bp (15 ng/5 μl), 1000 bp (100 ng/5 μl), 800 bp (80 ng/5 μl), 600 bp (60 ng/5 μl), 400 bp (40 ng/5 μl), 200 bp (20 ng/5 μl)).
Escherchia coli and T4 DNA ligases were purchased from Takara Bio Company, Japan.
The amounts of DNA are expressed in moles of nucleotides.
Preparation of S. cerevisiae rad51-G103E
Glycine-103 of the wild-type Rad51 (indicated as ‘Rad51’), encoded on pET3a, was replaced by glutamic acid (G103E), using a Quick Change II Site-Directed Mutagenesis Kit (Stratagene); the GGG codon was replaced by the GAG codon.
The purification of rad51-G103E was performed as previously described for the wild-type Rad51 (33), with some modifications. We monitored rad51-G103E by measuring the ssDNA-dependent ATPase activity, and by SDS-polyacrylamide gel electrophoresis (CBB staining and immunostaining with an anti-Rad51 antibody) during the purification. The detailed procedure for the purification of rad51-G103E is described in the Supplementary Methods.
It should be noted that none of the RecA, Rad51 and rad51-G103E proteins were tagged. In these studies, we avoided the use of tagged proteins, since the tags may affect the biochemical characteristics of the proteins.
Basic reaction mixture
The basic reaction mixture contained 30 mM Tris-HCl (pH 7.5), 7 mM MgCl2, 3.8% (w/v) polyethylene glycol 6000 (PEG), 0.1 mM nicotinamide adenine dinucleotide (NAD), 0.1 mM ethylenediaminetetraacetic acid (EDTA), 1.8 mM dithiothreitol (DTT) and 88 μg/ml bovine serum albumin ((BSA); New England Biolabs and Roche, molecular biology grade).
Assay for RecA- or Rad51-enhanced DNA ligation
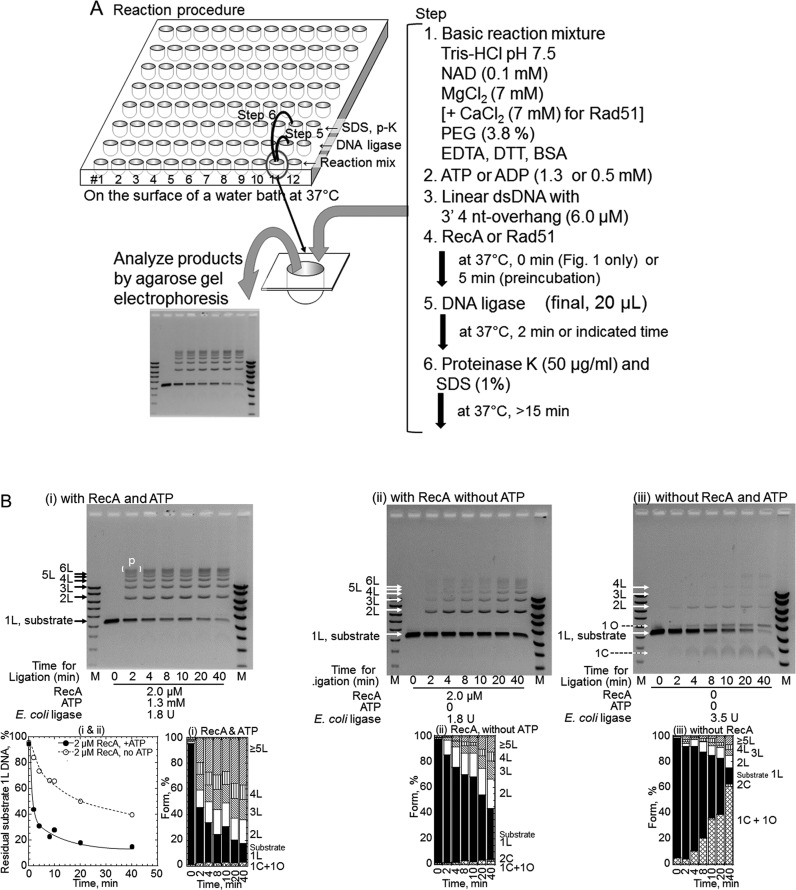
The outline of the assay is illustrated in Figure 1A. Unless otherwise stated, linear dsDNA (6.0 μM; pUC119) with 3′ four-nucleotide overhangs generated by PstI-treatment was preincubated with 2.0 μM RecA and 1.3 mM ADP or the indicated nucleotide at 37°C for 5 min in the basic reaction mixture, and ligation was initiated by the addition of the indicated amount of E. coli DNA ligase, in 20 μl of the basic reaction mixture. The reaction for each time-, each RecA amount- and each DNA ligase amount-point was initiated at an interval and performed in each well of a 96-well plate with a vent on the side (sealed with Parafilm (Bemis Co.), if necessary), which was floating (but fixed) on the surface of a water-bath at 37°C. When Rad51 was examined, 7 mM CaCl2 was added to the reaction mixture. When T4 DNA ligase was used, NAD was omitted. After the reaction, proteins were removed by SDS and proteinase K treatments, and the DNA products were fractionated by agarose gel electrophoresis in the presence of 60 nM ethidium bromide, to separate linear dsDNA species from closed circularized dsDNA species. DNA signals were detected and quantified after staining the gel with 1 μg/ml ethidium bromide. The detailed methods and conditions for the reactions, the detection of DNA signals and the quantification are described in the Supplementary Material.
Figure 1.
RecA enhances DNA ligation by Escherichia coli DNA ligase in an ATP-dependent manner. (A) Experimental system. Unless otherwise stated, the reaction mixture (20 μl each) containing the substrate linear dsDNA with 3′ four-nucleotide overhangs was prepared in a well of a 96-well plate in the indicated order. In all experiments, the reactions were performed at 37°C. The amounts of RecA, DNA ligase and nucleotide cofactors are indicated within each panel. After the reaction, the proteins were removed by SDS and proteinase K (p-K)-treatments, and the DNA products were fractionated by gel electrophoresis in the presence of 60 nM ethidium bromide, to separate linear dsDNA species from closely circularized dsDNA species. All ethidium bromide-stained gel-images were inversed to clarify signals. (B) Time course of RecA-enhanced DNA ligation. The DNA ligation of the linear dsDNA with a 3′ four-nucleotide overhang was initiated by the simultaneous addition of RecA and E. coli DNA ligase. (i) DNA ligation in the presence of RecA and ATP. (ii) DNA ligation in the presence of RecA, but without nucleotide cofactors. Note that no circular products were formed in the presence of RecA with or without ATP. (iii) The basic ligation reaction in the absence of RecA. This control contains twice the amount of DNA ligase (3.5 U) than the other reactions (1.8 U) in this figure. 1L, substrate linear dsDNA; 2L, 3L, 4L etc., linear products of dimer, trimer, tetramer and so on; [ ] with P, linear multimers larger than 7-mers; 1C and 2C, circular monomer and dimer products; 1O, nicked circular monomer products formed from substrate dsDNA with nicks introduced spontaneously during storage. Lanes M, DNA size and amount markers. Two sets of diluted marker solutions were applied to two Lanes M. The fractions of DNA species within each lane were calculated based on the signals, which were within or a slightly above the range in which the signals of the markers corresponded linearly with the amounts (see Supplementary Figure S5). These experiments were repeated twice, and the analyses at 2 min of the incubation were repeated in the following experiments. Unless otherwise stated, the cumulative bar graph below each gel profile shows the fractions of the substrates and products quantified from the gel profile above the graph.  , Substrate linear dsDNA (1L);
, Substrate linear dsDNA (1L);  , 1C+1O;
, 1C+1O;  , 2C;
, 2C;  , 2L;
, 2L;  , 3L;
, 3L;  , 4L;
, 4L;  , ≥5L. Note that the products are mostly circular monomers in the absence of RecA. The plot below gel panels (i) shows the fractions of residual substrate dsDNA (1L) against the incubation time with DNA ligase, quantified from gel panels (i) and (ii). These quantified data are only for correlation with the gel images and the fractions of signals from DNA species. • (closed circles), with RecA and ATP shown in panel (i); ∘ (open circles), with RecA without ATP shown in panel (ii).
, ≥5L. Note that the products are mostly circular monomers in the absence of RecA. The plot below gel panels (i) shows the fractions of residual substrate dsDNA (1L) against the incubation time with DNA ligase, quantified from gel panels (i) and (ii). These quantified data are only for correlation with the gel images and the fractions of signals from DNA species. • (closed circles), with RecA and ATP shown in panel (i); ∘ (open circles), with RecA without ATP shown in panel (ii).
Assay for ATP-hydrolysis by RecA
Radioactive [α-33P]ATP (1.3 mM) was incubated with RecA in the presence of 23 μM linear dsDNA (pBlueScript SK(-) or 10 μM ssDNA (phage M13mp19) at 37°C for 30 min in the basic reaction mixture (20 μl), except without NAD. The reaction was terminated and the amounts of hydrolyzed ATP were analyzed as described (31).
Immunoprecipitation experiments
A suspension of Protein A agarose beads (900 μl, Calbiochem) was mixed with the purified anti-Rad51 antibody (90 μg) in PBS-0.05% octylphenoxy poly(ethyleneoxy)ethanol, branched (IGEPAL CA630, Sigma-Aldrich Co.; total 1.2 ml) and gently shaken at 4°C for 16 h. The beads were washed with 200 mM triethanolamine (pH 8.2) and were suspended in 750 μl of the same buffer containing 20 mM dimethyl pimelimidate (purchased from Pierce Biotechnology, Inc.), followed by shaking at room temperature for 30 min to allow covalent bond formation between the protein A and the antibody. The reaction was stopped by suspending the beads in 1 ml of 50 mM Tris-HCl (pH 7.5) containing 150 mM NaCl. After washing 3 times with 1 ml TBS (20 mM Tris-HCl (pH 7.5), 150 mM NaCl and 0.05% IGEPAL CA630), the beads were resuspended in 0.5 ml of TBS.
Rad51 (1.0 μg) and E. coli ligase (1.5 μg, Takara) or Rad52 (1.0 μg) were mixed in the storage buffer (total 10 μl), and incubated on ice for 1 h. After 100 μl of TBS and 100 μl of the anti-Rad51 antibody-protein A agarose beads were added to the Rad51 and ligase mixture, the mixture was incubated at room temperature for 1 h with gentle shaking. The beads were pelleted at 1000 × g for 1 min and gently washed three times with 200 μl of TBS, and then the proteins were eluted by suspending the beads in 15 μl of sample buffer (3% SDS, 100 mM DTT, 10% glycerol, 0.02% BPB and 65 mM Tris-HCl (pH 6.8)) and heating the suspension for 1 min at 90–95°C. The supernatant was concentrated to 10 μl with an Amicon Ultra-0.5 centrifugal filter (Ultracel-30 K membrane, Millipore), and 5 μl of sample buffer was added. Aliquots (10 μl) were analyzed by SDS-polyacrylamide gel electrophoresis (10%) and Coomassie Brilliant Blue staining.
Electrophoresis mobility shift assay (EMSA) for DNA binding analyses of RecA and Rad51
Linear dsDNA with 3′ TGCA-four-nucleotide overhangs (6.0 μM; PstI digested pUC119) and RecA or Rad51 were incubated in the presence of ATP, ADP or the indicated nucleotide cofactor (1.3 mM) at 37°C for 10 min, in the basic reaction mixture (40 or 20 μl) lacking NAD and BSA. In the experiments with Rad51, we added 7 mM CaCl2 to the reaction mixtures. After the incubation, a series of the reaction mixtures was directly fractionated on an agarose gel, and to the other series of the reaction mixtures, 1 μl of 4% glutaraldehyde was added, and the mixture was further incubated for 10 min to fix the formed DNA-RecA or DNA-Rad51 complexes. As for RecA, 20 μl samples for direct analysis and for fixation were withdrawn from each of the 40 μl reaction mixtures. The DNA samples were separated by 1.0% agarose gel electrophoresis with TAE buffer (40 mM Tris-acetate, pH 8.0, 1 mM EDTA). After the electrophoresis, the DNA was stained with ethidium bromide and analyzed by a Typhoon 9410 Variable Mode Imager (GE Healthcare).
RESULTS
RecA enhances DNA ligation in an ATP-dependent manner
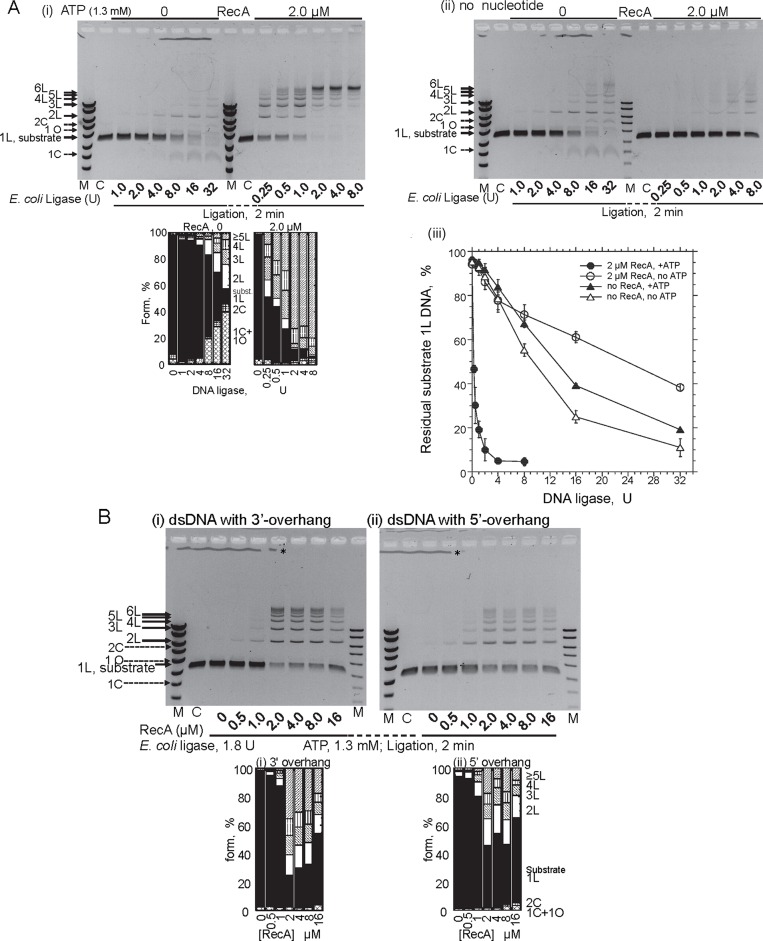
When incubated with linear dsDNA with 3′ four-nucleotide overhangs and ATP (Figure 1A), RecA greatly accelerates the end-joining to form multimers of the dsDNA catalyzed by (NAD-dependent) E. coli DNA ligase, in an ATP-dependent manner (Figure 1B, panel (i) versus panels (ii) and (iii) and plot). Linear multimers (labeled 2L, 3L, 4L, etc.) of the dsDNA substrate, including the maximum size (labeled p [ ] in Figure 1B, panel (i)), were obtained exclusively even at 2 min of the ligase-reaction, which was almost saturated within 4 min (Figure 1B, panel (i), its cumulative bar graph and plot). In the presence of ATP, RecA enhanced the overall reaction of E. coli DNA ligase catalyzed end-joining (DNA ligation) by about 40-fold, as compared to the amount of DNA ligase required for a certain decrease in the amount of residual dsDNA substrates (e.g. at 50%, 0.27 U with RecA and ATP versus 13 or 9.3 U without RecA (+ATP or without ATP; Figure 2A, panel (iii))). Without ATP, RecA inhibited the ligation somewhat (2-fold; at 50%, 24 U with RecA versus 13 or 9.3U without RecA; Figure 2A, panel (iii)).
Figure 2.
Characteristics of the ATP-dependent-DNA ligation enhancement by RecA. (A) Effects of the amounts of E. coli DNA ligase in the presence or absence of RecA, with ATP (panel (i)) or without ATP (panel (ii)). The incubation period for the ligase treatment was 2 min. Lanes C, untreated linear substrate dsDNA. Lanes M, DNA size and amount markers same as in Figure 1B. The cumulative bar graphs below the gel profile in panel (i) show the fractions of the substrates and products quantified from this gel profile. Symbols of the forms of the products and substrate are indicated in Figure 1B. (iii) Quantitative presentation of the effects of the amount of the DNA ligase. The fractions of residual substrate dsDNA (1L) at 2 min of the incubation were plotted against the amounts of DNA-ligase. Closed symbols, with 1.3 mM ATP; open symbols, without ATP; •, ∘ (circles), with RecA; ▴, Δ (triangles), without RecA. Each symbol represents an average with a standard deviation from at least three independent experiments, including those shown in panels (i) and (ii). (B) The effect of the end-structures of the linear DNA substrate. The dsDNA has a terminal 3′ TGCA-four-nucleotide overhang generated by the PstI-treatment (panel (i)), or a 5′ AGCT-four base-overhang generated by the HindIII-treatment (panel (ii)). The DNA was incubated with the ligase and the indicated amounts of RecA for 2 min. The cumulative bar graphs below these gel profiles in panels (i) and (ii) show the fractions of the substrates and products quantified from these gel profiles. Symbols of the forms of the products and substrate are indicated in Figure 1B. The differences between DNA with a 3′ overhang and DNA with a 5′ overhang were determined to be minimal, as confirmed by four additional comparisons of the gel profiles. *The irregular lines just below the wells are not DNA signals, but were caused by the irregular shapes of the gel surface formed around the comb to make the wells when the melted agarose was poured onto the glass plate.
The RecA-mediated ATP-dependent enhancement of the DNA ligation required the presence (more than 3.8%, the standard concentration in this study) of polyethylene glycol 6000 (PEG; Supplementary Figure S1). PEG is known to stimulate dsDNA end-joining by DNA ligases and RecA-catalyzed strand exchange (35) by its molecular crowding effects, which mimic the in vivo conditions. Thus, we included PEG to study the RecA-enhanced DNA ligation.
Note that without RecA, circular monomers (labeled 1C and 1O, in Figure 1B panel (iii) and Figure 2A panels (i) and (ii), and their cumulative bar graphs) are major products of the DNA ligation by E. coli ligase, and RecA suppressed the formation of circular products (Figures 1B and 2A, and their cumulative bar graphs). However, this suppression does not depend on the presence of a nucleotide cofactor (Figure 1B panels (ii) versus (iii)) and Supplementary Figure S1). Without ATP, the apparent enhancement of multimer formation by RecA is likely to reflect the compensation for the suppression of intramolecular end-joining (Figure 1B panels (ii) versus (iii), Figure 2A panel (ii) and Supplementary Figure S1).
The quick formation of the multimers required large amounts of RecA (2 μM, with 6 μM in nucleotides or 3 μM in base-pairs of dsDNA; Figure 2B), as in the case of homologous joint formation, but it was found that this was not stoichiometric requirements (see Discussion).
DNA ligation enhancement by RecA is independent of ATP hydrolysis and DNA termini-specific interactions
RecA requires ATP for various activities, but not necessarily its hydrolysis. Thus, we analyzed ATP-hydrolysis by RecA in the RecA-enhanced DNA ligation. Neither the linear dsDNA with 3′ four-nucleotide overhangs nor the dsDNA with blunt ends supported ATP-hydrolysis by RecA (Figure 3). Linear dsDNA with 5′ four-nucleotide overhangs supported RecA-catalyzed ATP-hydrolysis, to one-half of the extent obtained with ssDNA, under the current conditions (Figure 3), indicating that RecA recognizes dsDNA-ends with a 5′ four-nucleotide overhang and forms an active filament for ATP-hydrolysis (‘extended filament’: (20,36–38)). It is well known that linear dsDNA with or without the 5′ or 3′ nucleotide overhangs is a poor cofactor for the ATPase activity of RecA under the conditions for homologous joint formation (without PEG; Figure 3), and thus, the presence of PEG is responsible for the observed 5′-overhang end-recognition for ATP-hydrolysis.
Figure 3.
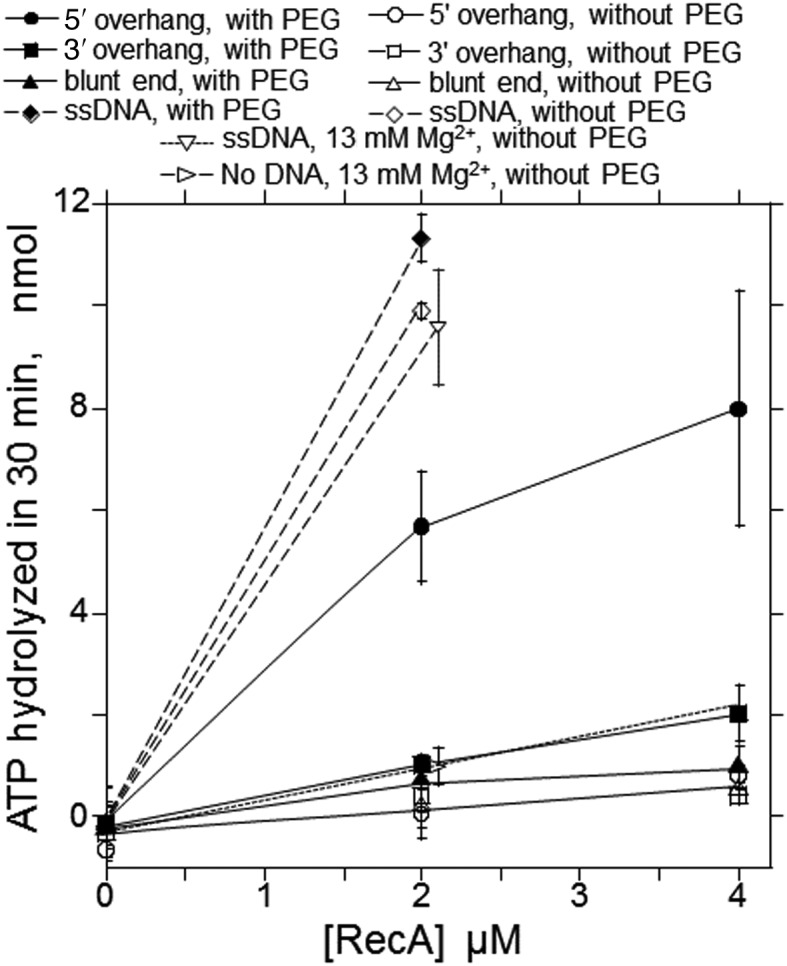
ATP-hydrolysis by RecA under the conditions for the ligation enhancement. Radioactive [α-33P]ATP (1.3 mM) was incubated with the indicated amounts of RecA for 30 min with RecA in the complete system, except for the amounts of DNA and the absence of DNA ligase and NAD. After the reaction, the amounts of hydrolyzed ATP were analyzed. The averages of more than three independent experiments were plotted with standard deviations. The small extent of ATP-hydrolysis observed in the absence of DNA was probably caused by an impurity in the RecA preparations used in these experiments, since the values varied from 0 to a few nmol among RecA preparations. Closed symbols, reactions in the basic reaction mixture (with 3.8% (w/v) PEG), open symbols, omit PEG. •, ∘ (circles), linear dsDNA with 5′ four nucleotide overhangs generated by HindIII; ▪, □ (squares), linear dsDNA with 3′ four nucleotide overhangs generated by PstI; ▴, Δ (triangles), linear dsDNA with blunt ends generated by HincII; ♦, ◊ (diamonds), ssDNA; ▿ (reversed triangles), ssDNA without PEG and in the presence of 13 mM MgCl2 (the standard reaction mixtures for homologous joint formation by RecA: (31)); ▹ (sideways triangles), without DNA (with 1.3 mM MgCl2, without PEG).
Although RecA recognizes dsDNA-ends with a 5′ four-nucleotide overhang, as indicated above, the quick formation of the multimers (observed at 2 min of incubation) was almost independent of the polarity of the terminal overhangs (3′ -TGCA made by PstI versus 5′-AGCT made by HindIII) of the dsDNA substrate (Figure 2B, panels (i) and (ii)). Thus, the ATP-dependent DNA ligation enhancement by RecA is independent of its DNA terminus-specific interactions and ATP-hydrolysis.
ADP is an efficient cofactor for DNA ligation enhancement by RecA
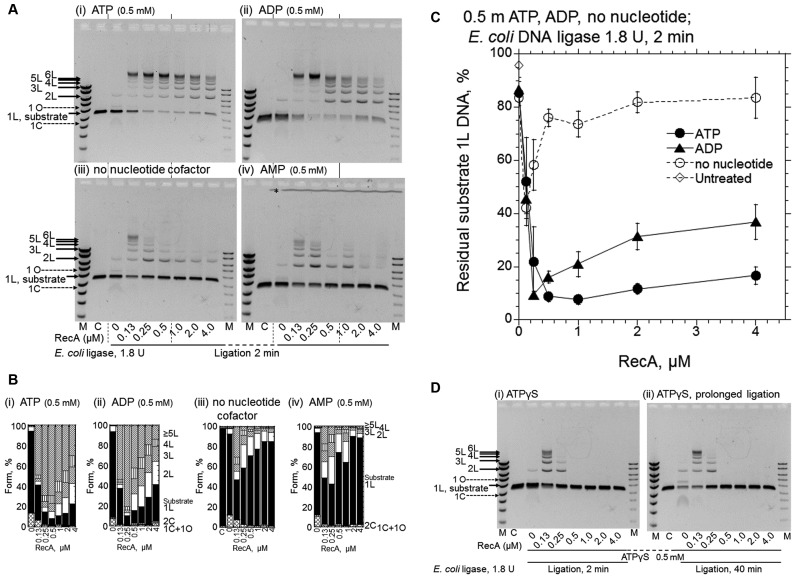
We next tested other types of adenosine derivatives instead of ATP, and found that ADP was an effective cofactor as well (Figure 4, A and B, panels (ii) versus (i) and Figure 4C). This further confirms the absence of the requirements for ATP-hydrolysis and the extended filament in the RecA-enhanced DNA ligation. Notably, the addition of ADP instantly inhibits the ATP-hydrolysis-dependent recombinase reaction catalyzed by RecA (39), and thus, even if the RecA preparation contained an ATP-bound form of RecA, it would not contribute to the observed enhancement in the presence of ADP. Unlike homologous joint formation (see (31), for an example), the RecA-enhanced DNA-ligation decreased with excess RecA, and this decrease was more significant with ADP than with ATP (Figures 2B and 4A, panels (i) versus (ii), Figure 4C and Supplementary Figure S2). Because of this decrease, there was an optimum amount of RecA for the enhancement.
Figure 4.
ADP is also an effective cofactor for RecA-enhanced DNA ligation by E. coli DNA ligase, but ATPγS causes the suppression of the DNA ligation. (A) Representative gel-profiles of the electrophoretic analysis of DNA products formed. (B) Fractions of the substrates and products quantified from the gel profiles in A shown by cumulative bar graphs. After the preincubation of the linear dsDNA with a 3′ four-nucleotide overhang with the indicated amounts of RecA and nucleotide cofactors for 5 min, the ligation was initiated by the addition of E. coli DNA ligase (1.8 U) without temperature shifts followed by a 2 min incubation. Cofactors added: panel (i), ATP; panel (ii), ADP; panel (iii), without nucleotide cofactor; panel (iv), AMP; Lanes C, untreated linear substrate dsDNA. Lanes M, DNA size and amount markers are the same as in Figure 1B. * in panel (iv) in A, see Figure 2. Symbols of the forms of the products and substrate in B are indicated in Figure 1B. (C) Quantified representation of the fractions of residual substrate dsDNA (1L) against the amounts of RecA. Each symbol represents the average and standard deviation of the results from at least three independent experiments, including those shown in the gel profiles (i–iii) shown in A. • (closed circles), with ATP; ▴ (closed triangles), with ADP; ∘ (open circles), no nucleotide cofactors; ◊ (open diamonds), untreated substrate linear dsDNA (1L) quantified from gel profile (iii), lane labeled C. (D) Effects of ATPγS. The conditions of the reactions were the same as in A, with ATPγS. The periods of the DNA ligation-reaction were 2 min in panel (i) and 40 min in panel (ii). We confirmed the reproducibility of the effects of AMP in A and ATPγS in D by repeating the experiments twice and four times, respectively.
During these tests, we noticed that the preincubation in the presence of ATP or ADP enhanced DNA ligation, especially at very low RecA concentrations (0.25 μM). As a result, the optimal concentrations of RecA for the enhancement of the ligation shifted to about a quarter to one-half (0.25–0.5 μM with ATP or ADP) of those without preincubation (1–2 μM with ATP, and 0.5 μM with ADP; Supplementary Figure S2). Thus, in the experiments shown in Figure 4 and the following experiments, we incubated the RecA with dsDNA in the presence of a nucleotide cofactor (preincubation), before the addition of E. coli DNA ligase.
The addition of AMP showed no effects (Figure 4, A and B, panels (iv) versus (iii)). A very small amount of RecA (0.13–0.25 μM) showed nucleotide cofactor-independent enhancement of DNA ligation, although to a much smaller extent than that in the presence of ATP or ADP, and was suppressed with higher amounts of RecA (>0.5 μM, Figure 4, A and B, panels (iii) and Figure 4C).
An unhydrolyzable ATP analogue, ATPγS, reportedly stabilizes RecA–DNA binding (26). The addition of ATPγS to the reaction containing more than 0.5 μM RecA resulted in the complete inhibition of DNA ligation (Figure 4D, panels (i) and (ii) versus Figure 4A, panel (iii)). At a low concentration of RecA (0.13 μM), ATPγS appeared to have no significant inhibitory effect (Figure 4D, panel (i) versus Figure 4A (iii)). This could be due to competition between the inhibitory effects of ATPγS-bound RecA and the enhancing effects of ADP-bound RecA, since ATPγS is generally contaminated with 5–10% ADP (40: Sigma Aldrich manufacturer's information).
RecA-enhanced DNA ligation requires neither species-specific interactions between RecA and a DNA-ligase nor base-pairing between complementary overhangs
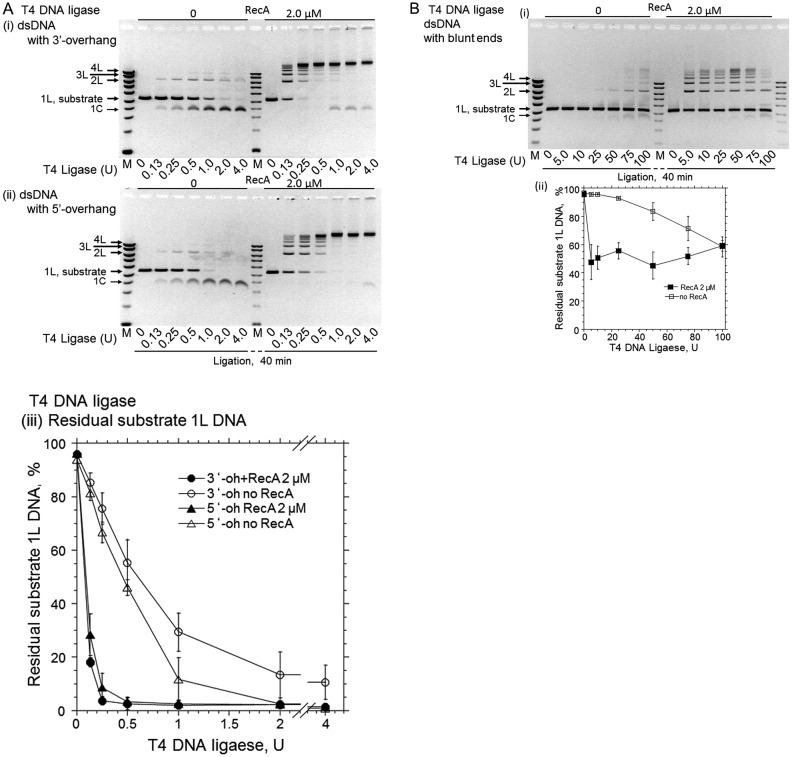
T4 phage DNA metabolism is mostly independent of the host system, and the T4 proteins involved in this function do not directly interact with E. coli proteins. RecA also enhanced the T4 DNA ligase-catalyzed DNA ligation of either 3′- or 5′-cohesive ends almost equally (Figure 5A), as compared to the amount of DNA ligase required for a certain decrease in the amount of residual dsDNA substrates (e.g. at 50%; smaller than 0.1 U with RecA versus 0.6 U without RecA after 40 min-ligation of 3′-cohesive ends; Figure 5A (iii)). This result indicated that the species-specific interactions of RecA and a DNA ligase are not required for RecA-enhanced DNA ligation, suggesting that the DNA ligation enhancement does not depend on the physical interaction between RecA and DNA ligases. This suggestion was further studied later by use of Rad51.
Figure 5.
RecA enhances T4 DNA ligase-catalyzed cohesive end- and blunt end-ligation. (A) Effects of the amounts of ligase in the presence or absence of RecA. Linear dsDNA with 3′-TGCA four nucleotide overhangs (panel (i)) or 5′-AGCT four nucleotide overhangs (panel (ii)) was incubated with the indicated amounts of T4 DNA ligase, in the presence or absence of RecA, for 40 min in the system without NAD but including ATP. In lanes M, DNA size and marker amounts are the same as in Figure 1B. The plots in panel (iii) show the fractions of residual substrate dsDNA (1L) against the amounts of T4 DNA ligase, quantified from at least three independent experiments. • (closed circles), dsDNA with 3′-four-nucleotide overhangs with RecA; ∘ (open circles), dsDNA with 3′ four-nucleotide overhangs without RecA; ▴ (closed triangles), dsDNA with 5′ four-nucleotide overhangs with RecA; Δ (open triangles), dsDNA with 5′-four-nucleotide overhangs without RecA. (B) The effects of RecA on the blunt end-ligation by T4 DNA ligase. Linear dsDNA with blunt ends was incubated with the indicated amounts of T4 DNA ligase, as in A, for 40 min, and the gel profile from a representative experiment is shown in panel (i). In lane M, DNA sizes and marker amounts are the same as in Figure 1B. The plot in panel (ii) was quantified from three independent experiments. ▪ (closed squares), dsDNA with blunt ends with RecA; □ (Open squares), dsDNA with blunt ends without RecA. It should be noted that T4 DNA ligase requires ATP as an essential cofactor, and thus the effects of nucleotide cofactors on the enhancement by RecA could not be tested.
Unlike E. coli DNA ligase, T4 DNA ligase can join blunt-ended dsDNA when added in excess (41). RecA efficiently enhanced the blunt end-ligation by T4 ligase, at concentrations similar to those for the 3′ four-nucleotide overhang-end ligation (Figure 5B). This result indicated that the base-pairing of the complementary four-nucleotide overhangs at the DNA ends is not required for the RecA-enhanced DNA ligation.
Rad51 also enhances the DNA ligation in an ADP-dependent fashion
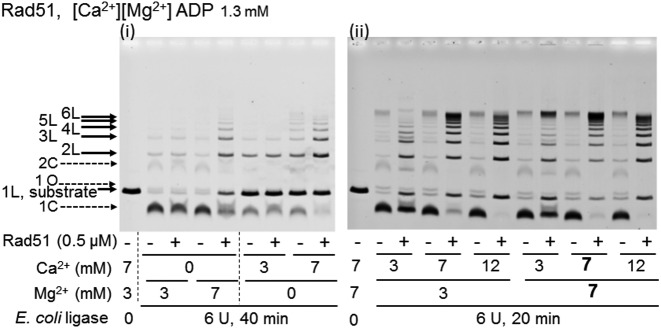
We also found that Rad51 from Saccharomyces cerevisiae enhanced E. coli ligase-catalyzed DNA ligation to produce multimers in an ADP-dependent manner, as described below. Yeast Rad51 requires Ca2+ to catalyze homologous joint molecule-formation (D-loop formation: (42,43)). Under the standard conditions for RecA (7 mM Mg2+ only), Rad51 enhanced intermolecular DNA ligation in the presence of ADP, but the circular monomer, the product of intramolecular DNA ligation, was also formed and the amount of the residual substrate dsDNA increased (Figure 6 panel (i)). The increase in the residual substrate DNA indicates the extent of the suppression of overall DNA ligation by Rad51. However, with 7 mM Ca2+ instead of Mg2+, Rad51 enhanced the DNA ligation with minimal suppression of overall ligation but strong suppression of intramolecular DNA ligation (Figure 6 panel (i)).
Figure 6.
Rad51-mediated enhancement of DNA ligation in the presence of ADP requires Ca2+. The linear dsDNA with a 3′ four-nucleotide overhang was incubated with Rad51 and E. coli DNA ligase in the presence of ADP, in the reaction mixture containing the indicted amounts of CaCl2 and MgCl2 for 40 min (panel (i)) and 20 min (panel (ii)). We repeated similar experiments twice to confirm the reproducibility of the results.
In the presence of both 7 mM Ca2+ and 7 mM Mg2+, Rad51 enhanced the intermolecular DNA ligation to the largest extent among the tested Ca2+-Mg2+ combinations and suppressed the intramolecular DNA ligation, with minimal suppression of overall ligation (i.e. with little increase in the amounts of residual dsDNA; Figure 6, panels (i) and (ii)). Therefore, we added 7 mM CaCl2 to the standard reaction mixture for RecA (7 mM MgCl2 only), as in the standard conditions for Rad51.
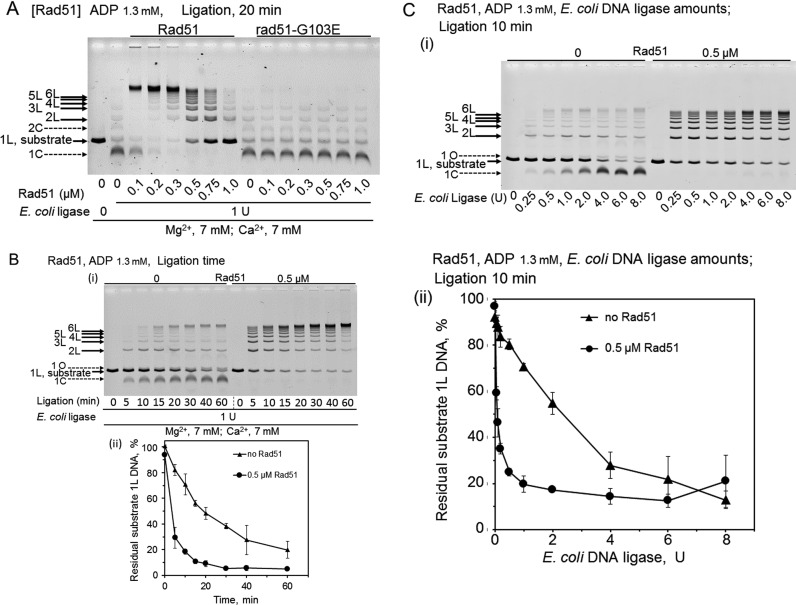
In the presence of ADP, like RecA, Rad51 enhanced DNA ligation to produce multimers and promoted the overall ligation, and prevented the formation of the circular monomer, the major product of DNA-ligase alone (Figure 7). Like RecA, a large amount of Rad51 was required, an optimal amount of Rad51 (0.2–0.3 μM Rad51 versus 6 μM dsDNA, Figure 7A; similar to that for RecA, Figure 4A panel (ii) and Figure 4C) was identified, and excess Rad51 decreased the enhancing effects on the DNA ligation (Figure 7A). The ADP-dependent Rad51-enhanced DNA ligation was also a very quick reaction with the optimal amount, and by 5 min in the reaction with E. coli DNA ligase, linear 7-mer DNAs were formed in the presence of 0.5 μM Rad51 (Figure 7B). At 0.5 μM, Rad51 enhanced DNA ligation by E. coli DNA ligase 29-fold, using the amounts of ligase required to ligate 50% of the substrate dsDNA (1L); 0.08 U with Rad51 versus 2.3 U without Rad51 (Figure 7C panel (ii)). Thus, the extents of the ligation enhancement are similar between Rad51 (29-fold) and RecA (40-fold).
Figure 7.
Features of ADP-dependent Rad51-enhanced DNA ligation and the effects of the G103E-substitution in Rad51. CaCl2 (7 mM) were added to the standard reaction mixtures (including 7 mM MgCl2). Linear dsDNA with a 3′ four-nucleotide overhang was incubated with Rad51 (wild-type) or rad51-G103E for 5 min, before the addition of E. coli DNA ligase to start the DNA ligation without temperature shifts, followed by an incubation for the indicated time. (A) Effects of the amounts of Rad51 and rad51-G103E in the presence of ADP. The linear dsDNA was treated with 1 U E. coli DNA ligase for 20 min, in the presence of the indicated amounts of Rad51 or rad51-G103E and ADP. We repeated these experiments three times to confirm the reproducibility. (B) Effects of the incubation times with or without Rad51 in the presence of ADP. The linear dsDNA was treated with 1 U E. coli DNA ligase, in the presence of ADP, and 0 or 0.5 μM Rad51 for the indicated time. The graph attached is the quantified representation of the fractions of substrate dsDNA (1L). Each symbol is the average of results obtained from two independent experiments. Error bars show the smaller and larger values, and not standard deviations only in this figure. • (closed circles), with 0.5 μM Rad51; ▴ (closed triangles), without Rad51. We repeated these experiments twice to confirm the reproducibility. (C) Effects of the amounts of E. coli DNA ligase in the presence or absence of 0.5 μM Rad51. The linear dsDNA was treated with the indicated amounts of E. coli DNA ligase in the presence of ADP and Rad51 for 10 min. (i) Representative gel-profiles of the electrophoretic analysis of DNA products. (ii) Quantified representation of the fractions of residual substrate dsDNA (1L). Each symbol is the average of results obtained from at least three independent experiments. Error bars show standard deviations. • (closed circles), with 0.5 μM Rad51; ▴ (closed triangles), without Rad51.
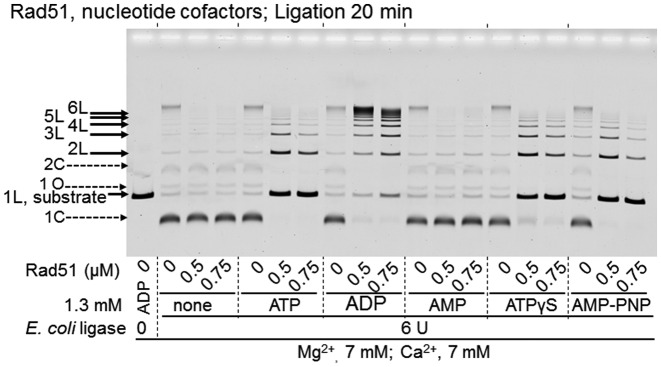
For Rad51, ADP is a suitable cofactor for DNA ligation enhancement, but ATP is not. When ADP was replaced by ATP, Rad51 appeared to enhance intermolecular DNA ligation with the inhibition of circular product formation, but the extent of overall DNA ligation was largely reduced (Figure 8). ATPγS and AMP–PNP showed similar effects to ATP at 20 min in the ligase reaction, and AMP had no effect (Figure 8). Since T4 DNA ligase itself requires ATP, ADP-dependent enhancement by Rad51 could not be tested with T4 DNA ligase.
Figure 8.
Effects of nucleotide co-factors on Rad51-enhanced DNA ligation. The linear dsDNA with a 3′ four-nucleotide overhang was treated with E. coli DNA ligase, in the presence of the indicated nucleotide cofactor and the indicated amounts of Rad51 for 20 min. Both CaCl2 and MgCl2 (7 mM) were added to the reaction mixtures. Linear dsDNA with a 3′ four-nucleotide overhang was incubated with Rad51 for 5 min, before the addition of E. coli DNA ligase to start the DNA ligation without temperature shifts, followed by an incubation for the indicated time. We repeated these experiments twice to confirm the reproducibility.
Absence of physical interaction between Rad51 and E. coli DNA ligase
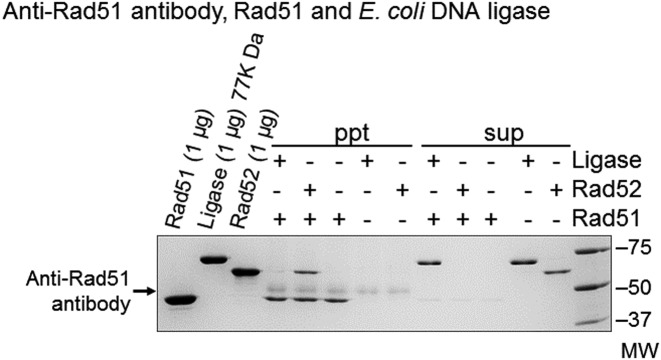
The absence of species-specific interactions between RecA and DNA ligase suggests that the DNA ligation enhancement by RecA/Rad51 does not require a physical interaction between RecA/Rad51 and DNA ligases. For further clarification, we used anti-Rad51 antibody–protein A agarose to test whether E. coli DNA ligase co-precipitated with Rad51. In a positive control, Rad52, which is known to bind Rad51 at specific sites (12,44), was precipitated with Rad51 by the anti-Rad51 antibody-protein A agarose, but E. coli DNA ligase was not precipitated at all under these conditions (Figure 9). These results clearly indicated the absence of physical interactions between Rad51 and E. coli DNA ligase. Thus, we conclude that Rad51 and likely RecA enhance DNA ligation without physical interactions with the DNA ligase that catalyzes the ligation.
Figure 9.
The absence of physical interactions between Rad51 and E. coli DNA ligase. E. coli DNA ligase (1.5 μg) or Rad52 (1 μg) was mixed with Rad51 (1 μg) and incubated in an ice-water bath for 1 h. Then, anti-Rad51 antibody-protein A agarose was added to the mixture, and incubated in the ice-water bath for another 1 h, followed by centrifugation. The proteins bound to the anti-Rad51 antibody–protein A agarose (ppt) and the proteins left in the supernatants (sup) were analyzed by SDS-polyacrylamide electrophoresis. We repeated these experiments three times to confirm the reproducibility.
DNA-binding ability of Rad51 is essential for Rad51-mediated DNA ligation enhancement
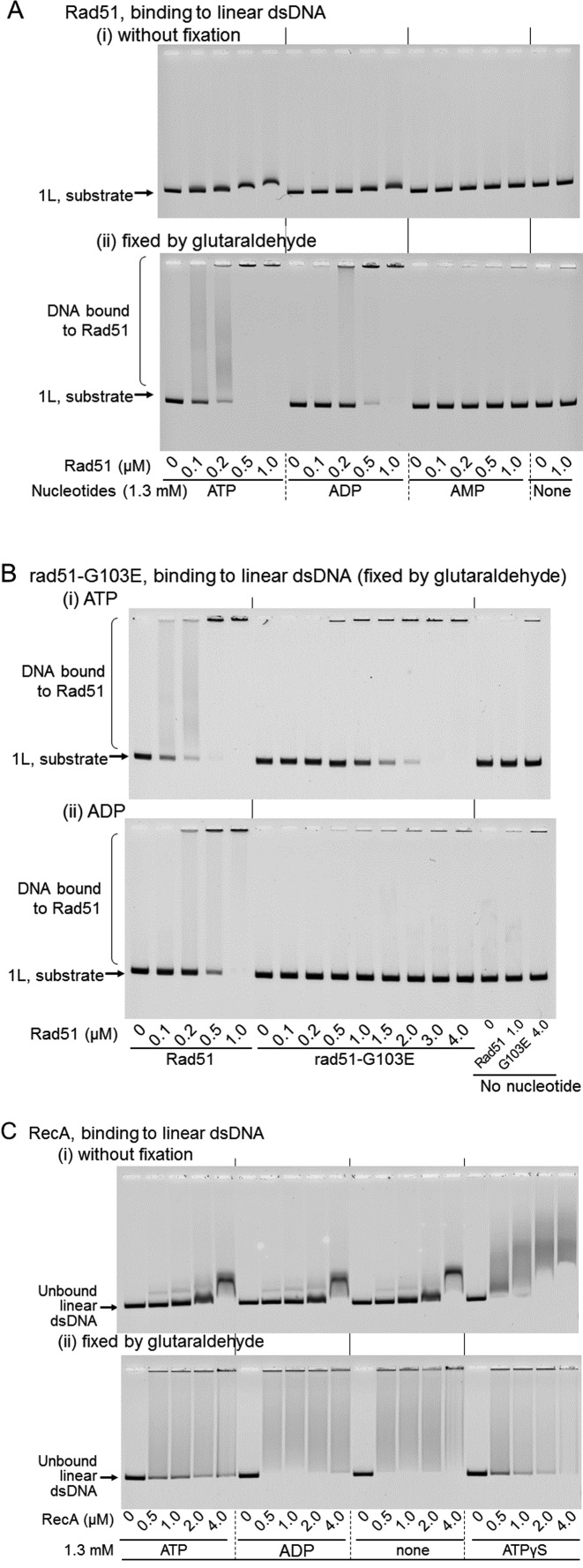
The enhancement observed in this study may result from the non-specific molecular crowding effects of Rad51/RecA, as a macromolecule. Molecular crowding does not require the physical interaction between a molecular crowding agent and the reagents involved in the observed reactions. Therefore, to exclude the possibility of non-specific molecular crowding on the observed activities, we examined the DNA-binding affinity of Rad51 and RecA by an EMSA using linear dsDNA with 3′ four-nucleotide overhangs. In the absence of nucleotide cofactors or in the presence of ADP, Rad51 and RecA might not form a stable filament with dsDNA, and thus, the binding of a small amount of Rad51 or RecA to dsDNA (3162 bp) might not yield sufficiently detectable mobility shifts. Glutaraldehyde is an efficient cross-linker, not only between proteins but also between protein and DNA (45,46), and would stabilize Rad51/RecA–dsDNA binding and promote the formation of large DNA-bound Rad51/RecA complexes exhibiting greater shifts (some of which may not enter the gel). Thus, after the dsDNA was incubated with Rad51/RecA, a series of the samples was directly loaded on the gel, and the other series of the samples was treated with glutaraldehyde before fractionation by gel-electrophoresis.
Under the conditions for the Rad51-mediated DNA ligation enhancement, which include 7 mM Mg2+, 7 mM Ca2+ and PEG, in the absence of a nucleotide cofactor or in the presence of 1.3 mM AMP, no DNA binding was detected even in the presence of 1.0 μM Rad51 and with the fixation. On the other hand, DNA-binding by Rad51 was clearly demonstrated even without fixation, and was detected at >0.1 μM and >0.2 μM in the presence of 1.3 mM ATP and ADP, respectively, with the fixation (Figure 10A). Thus, considering the results shown in Figure 8, the DNA binding activity appears essential for the Rad51-enhanced DNA ligation.
Figure 10.
DNA-binding by RecA and Rad51 in the presence of various nucleotides. (A) Binding activities of Rad51 to dsDNA. Linear dsDNA with a 3′ four-nucleotide overhang (‘1L, substrate’) was incubated in the reaction mixture contained CaCl2 for 10 min with the indicated amounts of Rad51 and the indicated nucleotide cofactor. After the incubation, portions of the reaction mixtures were directly subjected to gel electrophoresis (panel (i)), and the other portions of the samples were treated with glutaraldehyde to fix the protein–DNA complex before gel electrophoresis (panel (ii)). We repeated the experiments with ATP, ADP or no nucleotide in panel (ii) twice to confirm the reproducibility, but the experiments of panel (i) and with AMP in panel (ii) were performed once. (B) Binding activities of rad51-G103E to dsDNA. All conditions were the same as those in A, except for the use of rad51-G103E, and all samples were fixed after the incubation. Panel (i), incubated with ATP; panel (ii), incubated with ADP. We repeated these experiments at least three times to confirm the reproducibility. (C) Binding activities of RecA to dsDNA. All conditions were the same as those in A, except that RecA was used and the reaction mixture did not contain CaCl2. After the incubation, portions of the reaction mixtures were directly subjected to gel electrophoresis (panel (i)), and the other portions of the samples were treated with glutaraldehyde to fix the protein–DNA complex before gel electrophoresis (panel (ii)). The unfixed and fixed samples were analyzed by a gel-mobility shift assay. We repeated these experiments twice to confirm the reproducibility. The experiments without fixation by glutaraldehyde were performed twice in addition.
To obtain more rigorous proof for the dependence of the DNA ligation enhancement by Rad51 on the DNA-binding ability of Rad51, we tested a mutant rad51, rad51-G103E, which is known to be defective in DNA-binding (47). We purified rad51-G103E (Supplementary Figure S3A), and confirmed that the dsDNA-binding ability of this mutant rad51 was severely impaired. Specifically, under the conditions including the fixation, in the presence of ADP, no DNA binding was detected even with a 20-fold excess of the mutant (4.0 μM) over the minimum amount (0.2 μM) of Rad51 required to generate a signal for DNA-binding (Figure 10B panel (ii)), and in the presence of ATP, more than 15-fold larger amounts of the mutant rad51 (1.5 μM) were required to detect DNA-binding, as compared with Rad51 (0.1 μM; Figure 10B panel (i)). We confirmed that this mutant protein is properly folded and active, except for DNA-binding, as follows. The requirement for ssDNA in the ATPase activities of RecA/Rad51 can be replaced by a high concentration of salt (e.g. 2 M KCl). Although the activity was reduced to half of that by Rad51, the DNA-independent but strictly 2M KCl-dependent ATP hydrolysis was observed (Supplementary Figure S3B). Thus, we concluded that the activity of rad51-G103E is essentially similar to that of Rad51, except for the DNA-binding deficiency. Analyses of the ability of rad51-G103E to enhance DNA-ligase activity in the presence of ADP showed severe defects in this activity (Figure 7A).
These results clearly show that the Rad51-mediated DNA ligation enhancement depends on the dsDNA-binding ability of Rad51, and exclude the possibility that the enhancement is simply due to molecular crowding.
Optimal DNA binding is required for RecA- and Rad51-mediated DNA ligation enhancement
RecA showed mobility shifts without fixation. As judged by the minimum amounts of RecA required to detect mobility shifts and the extents of the shifts of dsDNA-signals without fixing, the highest affinity to dsDNA was found in the presence of ATPγS, then in the absence of nucleotide cofactor and with ADP, and less in the presence of ATP (Figure 10C). The DNA ligation enhancement depended on ATP or ADP. A very small enhancement of DNA ligation was detected with limited amounts of RecA in the absence of nucleotide cofactor (Figure 4A, panels (iii) and Figure 4C, and the ligation was completely inhibited in the presence of ATPγS (Figure 4D, panels (i) and (ii)). At the optimum RecA amounts for ATP/ADP-dependent DNA ligation enhancement (0.25–0.5 μM), the mobility shifts of dsDNA were not very significant without fixing, but the shifts became very clear after fixing (Figure 10C), indicating that RecA weakly binds to dsDNA at these concentrations.
In contrast, Rad51 showed higher (about 3-fold) dsDNA-binding affinity in the presence of ATP than with ADP, as compared by using the minimum amounts required for detectable dsDNA-binding (<0.1 μM and 0.2–0.5 μM, respectively), and no dsDNA-binding in the absence of nucleotide cofactors, as judged by the amounts of unbound dsDNA remaining after the fixation (Figure 10A). Rad51 required ADP to enhance the DNA ligation, and ATP suppressed the enhancement (Figure 8).
Closer examinations revealed that the suppression of the enhancement by excess RecA/Rad51 positively correlates with the DNA-binding affinity of RecA/Rad51 (Figure 4, Supplementary Figure S2 and Figure 8 versus Figure 10). These results suggest that RecA/Rad51-mediated DNA ligation enhancement depends on a balance between the DNA-binding affinity of RecA/Rad51 and that of a DNA ligase. The excessively high affinity states of RecA/Rad51 prevent the enhancement, as the bound Rad51/RecA probably interferes with the access of the DNA-ligase.
Thus, the extent of the enhancement did not simply correlate with the extents of the affinities of the proteins to dsDNA, and the forms of RecA/Rad51-dsDNA complexes would be another factor (see Discussion).
DISCUSSION
Similarities and differences between RecA and Rad51 in DNA ligation enhancement
The characteristics of DNA ligation enhancement by yeast Rad51 were very similar to those of RecA, including the extents of overall DNA ligation enhancement, the requirement for large amounts of protein, the suppression of intramolecular DNA ligation, the requirement for ADP, the presence of a similar optimal amount of protein for the enhancement with suppression at excessive protein amounts, and the quick formation of multimers (this study). These similarities support the conclusion that the enhancement of DNA ligation is a common function of RecA and Rad51, and likely of RecA-family recombinases. It is noteworthy that the conditions for the ligation enhancement by RecA and Rad51 are those that suppress the recombinase activity of these proteins to form key recombination intermediates, homologous joints (D-loops), including the presence of ADP and the preceding interactions of the protein with dsDNA before interacting with ssDNA. We reported previously that the preceding incubation of RecA with dsDNA caused the formation of a “dead-end product” for the recombinase activity (31), but this study revealed that the ‘dead-end product’ is the active form of RecA to enhance DNA ligation.
In this study, we also observed some differences in this enhancement between Rad51 and RecA. Unlike RecA, Rad51 requires Ca2+ to enhance DNA ligation (Figure 6), and ATP suppresses overall DNA ligation (Figure 8). As for its recombinase function, Rad51 required Ca2+ for the homologous joint formation, but this was completely substituted by the addition of another supporting protein (33,48), suggesting that Rad51 needs assistance from other proteins to perform functions fully equivalent to those of RecA. This would also be true for the enhancement of DNA ligation.
DNA ligation enhancement by RecA and Rad51 is not simply a consequence of molecular crowding, bundle formation and aggregation of DNA by the protein
Rad51 required their DNA binding ability to enhance DNA ligation, as revealed by the effects of nucleotide cofactors (Figure 10A versus Figure 8), and more critically by the effects of the G103E-substitution of Rad51 (Figure 7A and Figure 10B (versus Figure 10A)). This observation excludes the possibility that the DNA ligation enhancement simply occurs by molecular crowding effects, as described in the Results.
It is well known that as a recombinase to catalyze homologous joint formation, a stoichiometric amount (one RecA to 3 nucleotides of ssDNA) of RecA is required (49,50), and this amount of RecA saturates the ssDNA to form the extended RecA-filament around ssDNA (20). The enhancement of DNA ligation also required large amounts of RecA or Rad51 (Figures 2B, 4A panel (i) and (ii), Figures 4C and 7A, and Supplementary Figure S2), but the DNA ligation enhancement seems not to depend on stoichiometric interactions of RecA/Rad51 and DNA, since the optimum amounts of RecA were varied by the kinds of nucleotide cofactor (ATP and ADP; Figure 4) and the presence or absence of preincubation (Supplementary Figure S2). Thus, the large amount is required to overcome the unstable binding of Rad51/RecA to dsDNA, as shown by the difference between the results without fixing and those with fixing (Figure 10).
A trivalent cation, hexamine cobalt chloride, (without RecA) stimulates T4 DNA ligase catalyzed DNA ligation, and also effectively suppresses intramolecular DNA ligation (51). The hexamine cobalt chloride-mediated stimulation and suppression of the ligation were attributed to DNA-bundle formation, in which the trivalent cations bridge DNA-rods (51,52). In vitro, RecA forms well-ordered bundles of filaments under various conditions independently of ATP or ADP, and under crystallization conditions (19,53,54). Intramolecular DNA ligation was detected as the circularization of linear dsDNA substrates, and the suppression of intramolecular ligation of linear dsDNA substrates by RecA did not depend on ATP, ADP or other nucleotide cofactors (Figure 1B panel (ii) versus panel (iii) and Figure 2A panel (ii)). In addition, in the absence of nucleotide cofactors, Rad51 neither bound to dsDNA (Figure 10A) nor suppressed intramolecular DNA ligation (Figure 8), and the DNA-binding defective mutant rad51-G103E did not suppress the intramolecular DNA ligation (Figure 7A).
Unlike protein-free DNA bundles, the RecA- or Rad51-DNA filament bundle does not readily explain the enhancement of DNA ligation, since the DNA is located at the center of each RecA- or Rad51-filament (23,55), and thus direct and physical inter-DNA interactions cannot be formed within the bundle. In fact, RecA in the absence of nucleotide cofactors and Rad51 with ATP suppressed the intramolecular DNA ligation and allowed the intermolecular DNA ligation, but prevented overall ligation by E. coli DNA ligase (Figures 2A panel (ii) and 8).
Some preceding preliminary studies demonstrated that RecA and Rad51 enhanced DNA ligation. Rusche et al. reported the very weak stimulation of T4 DNA ligase-catalyzed intermolecular DNA ligation (up to trimer) by RecA, under conditions that stimulate the aggregation of RecA and dsDNA (56). In comparison, the DNA ligation enhancement observed under our conditions including ADP was much more robust. Baumann et al. reported that human Rad51 enhanced T4 DNA ligase-catalyzed intermolecular ligation of dsDNA with a 3′ four-nucleotide overhang or with blunt ends to a similar extent, in an overnight incubation at 16°C (11). In this and the above preceding studies, T4 DNA ligase was used, and since T4 DNA ligase requires ATP as a cofactor, the requirements for ADP or another nucleotide cofactor were not studied.
RecA forms ssDNA–RecA aggregates and aggregates consisting of ssDNA, dsDNA and RecA, under the conditions for ATP-dependent homologous joint formation (57). Aggregation of dsDNA by Rad51/RecA could be a mechanism of the DNA-ligation enhancement. However, we showed that RecA/Rad51 extensively enhanced DNA ligation in the presence of ADP (Figure 4A, panels (ii) and Figures 4C and 7C). In the presence of ADP, dsDNA is reportedly excluded from such aggregates with RecA, and even in the presence of ATP, RecA reportedly did not cause dsDNA aggregation if ssDNA was not added (57). These observations suggest that the observed enhancement by RecA in our study is unlikely to be a consequence of dsDNA aggregation.
Active forms of RecA for DNA ligation enhancement
The experiments to detect the nucleotide cofactor effects revealed the requirement for dsDNA-binding in DNA ligation enhancement by Rad51. However, these experiments did not show a simple correlation between the efficiencies of Rad51/RecA-mediated DNA ligation enhancement and their dsDNA-binding affinities. Therefore, we considered the structural differences among the Rad51/RecA–dsDNA complexes, according to the presence or absence of nucleotide cofactors. Three physically different types of RecA- and Rad51-filaments (containing DNA or without DNA) have been identified: extended filaments (active for ATP-hydrolysis) with a helical pitch of 9 nm formed in the presence of ATP or ATP-analogues, compressed (inactive as a recombinase) filaments with a helical pitch of 8.2 nm formed in the presence of ADP, and compressed (inactive) filaments with a helical pitch of 7 nm formed in the absence of nucleotide cofactor (58: see Supplementary Table S1). As a nucleotide cofactor in the enhancement, ADP is highly effective for RecA and Rad51, and ATP is good for RecA but not for Rad51, and in the absence of nucleotide cofactor, Rad51 did not show (Figure 8) and RecA did little the enhancement (Figure 4). Thus, the active forms of RecA and Rad51 for the enhancement of DNA ligation are likely to be the 8.2 nm filaments, and the 7 nm filaments are not or least active for the enhancement. It is likely that the pitch of the filament is a factor for the enhancement.
The 9 nm filaments are not active for Rad51, since the presence of ATP abrogates the ligation enhancement by Rad51. In the presence of ATP, RecA forms an extended and stable RecA–dsDNA filament that is active for ATP-hydrolysis (Supplementary Table S1). However, it was reported that in the absence of ssDNA, the establishment of the extended state with dsDNA requires tens of minutes, as shown by the long time lag until ATP-hydrolysis is detectable (34), while in the presence of ATP, RecA-mediated DNA ligation enhancement saturated within a 5 min (Figures 1 and 2) and was ATP-hydrolysis-independent (Figure 3). We previously reported that ATP-bound RecA quickly binds to dsDNA, as revealed by the rapid establishment (within a minute) of an inactive recombinase state by an incubation with dsDNA (in the absence of ssDNA, (31)). These are likely to correlate ‘a slow step in the association pathway’ of ATP-bound RecA to dsDNA (34), and it is likely that just after binding to dsDNA, the ATP-bound RecA forms compressed filaments without unwinding of the double helix (31).
RecA/Rad51 juxtaposes dsDNA-ends prepared for covalent joining by a DNA ligase
RecA/Rad51 enhanced DNA ligation is independent of species-specific interactions with DNA ligases (Figures 1 and 2 versus 5) and of physical interactions between Rad51 and a DNA ligase (Figures 7 and 9). Thus, it is likely that RecA/Rad51 acts on the substrate dsDNA, rather than directly on the DNA ligase. Electron microscopic analyses by Register and Griffith revealed the juxtaposition of linear DNA molecules along with RecA–DNA filament formation in the presence of ATP or ATPγS (59). Although the juxtaposed DNA-ends were not covalently joined by E. coli or T4 DNA-ligase in their experiments, and it was impossible to determine whether the DNA-ends were properly aligned for joining by a DNA ligase due to the resolution of electron microscopic analyses, their results suggested a function of RecA.
Considering the discussion described above, it is likely that the role of Rad51 and RecA in the ADP-dependent enhancement of DNA ligation is the juxtaposition of DNA-ends prepared for covalent joining by a DNA ligase; that is, the juxtaposition of dsDNA strands in which the ends are aligned within a distance that enables the DNA ligase to join them immediately. DsDNA termini are juxtaposed by themselves transiently and unstably, through base-stacking interactions (60) and base-pairing interactions. A possible mechanism for Rad51/RecA-enhanced DNA ligation would be that local filament formation by Rad51/RecA around the self-juxtaposed DNA-ends stabilizes the junction for ligation until the DNA ligase gains access to the junction to join it covalently, by competing with the bound RecA or Rad51. Another possibility is active juxtaposition, by the interaction between Rad51/RecA filaments locally formed at or near the ends of dsDNAs. We are testing these possible mechanisms by constructing mutant RecA and Rad51 variants.
RecA/Rad51-mediated DNA ligation enhancement as a tool for DNA technology
The robust enhancement of the tandem linear multimer formation by RecA/Rad51 in vitro can be applied to the efficient construction of genes from DNA fragments containing partial sequences, operons from DNA fragments encoding genes and cis-acting elements, and clusters of gene repeats for overproduction of the gene products, for example. In these constructs, the DNA fragments must have cohesive ends consisting of non-palindromic sequences to be aligned in a fixed orientation.
SLIC is a simultaneous cloning method of multiple DNA fragments, using RecA-catalyzed annealing (61). In this method, cohesive ends are generated by exonuclease-treatment, and after annealing, the DNA products are utilized for transformation without ligation. While this method skips the DNA-ligase treatment but requires controlled exonuclease digestion to generate about 30 n-long cohesive ends in the DNA fragments to be cloned, our RecA/Rad51-enhanced DNA ligation uses small cohesive ends, which can be generated by restriction endonuclease-treatment.
Biological roles of the RecA/Rad51-family protein-enhanced juxtaposition of DNA-ends
We previously performed transformation with a single DNA molecule to each transforming cell, using a linear single copy plasmid dsDNA that required self-circularization to replicate in the host yeast cells. These experiments showed that distinct deletion mutations of Rad51 result in a dramatic increase (more than 14-fold, from an undetectable level among 144 cases to 10%) in errors (deletions) of the rejoining of the reporter plasmid ends with a 3′ four nucleotide overhang (18). Compared with RAD51 cells, the rad51-deficient mutations did not affect the transformation efficiency of the control uncut (circular) reporter plasmid DNA (18). In contrast, in the same published study (18), the rad52 mutant showed no significant increase (1/120) in errors in NHEJ. These published results suggested the specific role of Rad51 in NHEJ, which is different from the role of Rad52.
The rejoining activities in the RAD51 and rad51-deficient cells are Lig4-dependent (62), indicating that the rejoining was mediated by a canonical NHEJ system. The only gene defective in the yeast cells used in these tests was RAD51, and the cells have all of the active components of canonical NHEJ, including Ku, which reportedly contributes to the fidelity of canonical NHEJ (15–17). However, the above observations (18) indicated that Ku is not sufficient, and Rad51 is also required for the precise end-joining of the reporter plasmids by canonical NHEJ. The Rad51-mediated juxtaposition of dsDNA-ends ready for DNA ligation shown in this study is a simple explanation of the possible function of Rad51 in promoting the fidelity of canonical NHEJ; i.e. by binding to DNA-junction regions, Rad51 quickly juxtaposes broken ends, and prevents the processing of the ends for causing error prone end-joining. Then, the canonical NHEJ machinery, including the Ku-heterodimer and Lig4, would tether the juxtaposed ends, displace the Rad51-filament and rejoin the ends covalently to complete precise NHEJ. One wonder whether Ku and Rad51 compete with each other for the binding to DNA-ends. The crystal structure of the complex of dsDNA and the Ku heterodimer (63) suggested that the Ku heterodimer is able to translocate along the dsDNA molecule, and thus, the binding of the Ku heterodimer to the terminal region and the short filament formation (not detectable as foci by microscopy) by Rad51 at the DNA ends are not mutually exclusive. In addition, the structure of the complex of dsDNA and Ku did not reveal the mechanism for the juxtaposition of DNA ends (63). Rad51 would provide a means to juxtapose DNA-ends accurately, for precise end-joining by the canonical NHEJ machinery. This study provides the first credible clue to reconsider a previously ignored possibility, a role of Rad51 in NHEJ, and further clarification is eagerly awaited.
Acknowledgments
The authors thank Yukari Iikura (Cellular & Molecular Biology Laboratory, RIKEN) for preparing the RecA variants and DNA. The authors thank Daiki Sakiyama and Shintaro Tanabe (Department of Applied Biological Science, Nihon University College of Bioresource Sciences) for technical supports of the assay with Rad51 and RecA, and for support of Rad51 preparation, respectively.
Footnotes
Present addresses:
Takehiko Shibata, Department of Chemistry, Graduate School of Science and Engineering, Tokyo Metropolitan University, Hachioji-shi, Tokyo 192-0397, Japan.
Takeshi Shinohara, Department of Life Science, College of Science, Rikkyo University, Nishi-ikebukuro, Tokyo 171-8501, Japan.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research [22247002]. Funding for open access charge:
Conflict of interest statement. None declared.
REFERENCES
- 1.Moynahan M.E., Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weller G.R., Kysela B., Roy R., Tonkin L.M., Scanlan E., Della M., Devine S.K., Day J.P., Wilkinson A., d'Adda di Fagagna F., et al. Identification of a DNA nonhomologous end-joining complex in bacteria. Science. 2002;297:1686–1689. doi: 10.1126/science.1074584. [DOI] [PubMed] [Google Scholar]
- 3.Symington L.S., Gautier J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 4.Shrivastav M., De Haro L.P., Nickoloff J.A. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 5.Paques F., Haber J.E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holliday R. A mechanism for gene conversion in fungi. Genetic Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- 7.Szostak J.W., Orr-Weaver T.L., Rothstein R.J., Stahl F.W. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 8.Shibata T., DasGupta C., Cunningham R.P., Radding C.M. Purified Escherichia coli recA protein catalyzes homologous pairing of superhelical DNA and single-stranded fragments. Proc. Natl. Acad. Sci. U.S.A. 1979;76:1638–1642. doi: 10.1073/pnas.76.4.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McEntee K., Weinstock G.M., Lehman I.R. Initiation of general recombination catalyzed in vitro by the recA protein of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1979;76:2615–2619. doi: 10.1073/pnas.76.6.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 11.Baumann P., Benson F.E., West S.C. Human rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 12.Shinohara A., Ogawa H., Ogawa T. RAD51 protein involved in repair and recombination in S. cerevisiae is a recA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 13.Nakai S., Matsumoto S. Two types of radiation-sensitive mutants in yeast. Mutat. Res. 1967;4:129–136. doi: 10.1016/0027-5107(67)90064-4. [DOI] [PubMed] [Google Scholar]
- 14.Game J.C., Mortimer R.K. A genetic study of X-ray sensitive mutants in yeast. Mutat. Res. 1974;24:281–292. doi: 10.1016/0027-5107(74)90176-6. [DOI] [PubMed] [Google Scholar]
- 15.Rathmell W.K., Chu G. Involvement of the ku autoantigen in the cellular response to DNA double-strand breaks. Proc. Natl. Acad. Sci. U.S.A. 1994;91:7623–7627. doi: 10.1073/pnas.91.16.7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boulton S.J., Jackson S.P. Saccharomyces cerevisiae ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 17.Lin W.Y., Wilson J.H., Lin Y. Repair of chromosomal double-strand breaks by precise ligation in human cells. DNA Repair (Amst) 2013;12:480–487. doi: 10.1016/j.dnarep.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miura T., Shibata T., Kusano K. Putative antirecombinase Srs2 DNA helicase promotes noncrossover homologous recombination avoiding loss of heterozygosity. Proc. Natl. Acad. Sci. U.S.A. 2013;110:16067–16072. doi: 10.1073/pnas.1303111110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Story R.M., Weber I., Steitz T.A. The structure of the E. coli recA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z., Yang H., Pavletich N.P. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–494. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 21.Conway A.B., Lynch T.W., Zhang Y., Fortin G.S., Fung C.W., Symington L.S., Rice P.A. Crystal structure of a Rad51 filament. Nat. Struct. Mol. Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- 22.Pellegrini L., Yu D.S., Lo T., Anand S., Lee M., Blundell T.L., Venkitaraman A.R. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 23.Ogawa T., Yu X., Shinohara A., Egelman E.H. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 24.Nishinaka T., Shinohara A., Ito Y., Yokoyama S., Shibata T. Base-pair switching by interconversion of sugar puckers in DNA extended by proteins of RecA-family: A model for homology search in homologous genetic recombination. Proc. Natl. Acad. Sci. U.S.A. 1998;95:11071–11076. doi: 10.1073/pnas.95.19.11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masuda T., Ito Y., Terada T., Shibata T., Mikawa T. A non-canonical DNA structure enables homologous recombination in various genetic systems. J. Biol. Chem. 2009;284:30230–30239. doi: 10.1074/jbc.M109.043810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shibata T., Cunningham R.P., DasGupta C., Radding C.M. Homologous pairing in genetic recombination: complexes of recA protein and DNA. Proc. Natl. Acad. Sci. U.S.A. 1979;76:5100–5104. doi: 10.1073/pnas.76.10.5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radding C.M. Homologous pairing and strand exchange in genetic recombination. Annu. Rev. Genet. 1982;16:405–437. doi: 10.1146/annurev.ge.16.120182.002201. [DOI] [PubMed] [Google Scholar]
- 28.Howard-Flanders P., West S.C., Stasiak A. Role of recA protein spiral filaments in genetic recombination. Nature. 1984;309:215–220. doi: 10.1038/309215a0. [DOI] [PubMed] [Google Scholar]
- 29.Kowalczykowski S.C. Mechanistic aspects of the DNA strand exchange activity of E. coli recA protein. Trends Biochem. Sci. 1987;12:141–145. [Google Scholar]
- 30.Kahn R., Radding C.M. Separation of the presynaptic and synaptic phases of homologous pairing promoted by recA protein. J. Biol. Chem. 1984;259:7495–7503. [PubMed] [Google Scholar]
- 31.Shinohara T., Ikawa S., Iwasaki W., Hiraki T., Hikima T., Mikawa T., Arai N., Kamiya N., Shibata T. Loop L1 governs the DNA-binding specificity and order for RecA-catalyzed reactions in homologous recombination and DNA repair. Nucleic Acids Res. 2015;43:973–986. doi: 10.1093/nar/gku1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sung P., Robberson D.L. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 33.Arai N., Ito D., Inoue T., Shibata T., Takahashi H. Heteroduplex joint formation by a stoichiometric complex of Rad51 and Rad52 of Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:32218–32229. doi: 10.1074/jbc.M507521200. [DOI] [PubMed] [Google Scholar]
- 34.Pugh B.F., Cox M.M. Stable binding of recA protein to duplex DNA. Unraveling a paradox. J. Biol. Chem. 1987;262:1326–1336. [PubMed] [Google Scholar]
- 35.Zimmerman S.B., Minton A.P. Macromolecular crowding: biochemical, biophysical, and physiological consequences. Annu. Rev. Biophys. Biomol. Struct. 1993;22:27–65. doi: 10.1146/annurev.bb.22.060193.000331. [DOI] [PubMed] [Google Scholar]
- 36.Dunn K., Chrysogelos S., Griffith J. Electron microscopic visualization of recA-DNA filaments: evidence for a cyclic extension of duplex DNA. Cell. 1982;28:757–765. doi: 10.1016/0092-8674(82)90055-1. [DOI] [PubMed] [Google Scholar]
- 37.DiCapua E., Engel A., Stasiak A., Koller T. Characterization of complexes between recA protein and duplex DNA by electron microscopy. J. Mol. Biol. 1982;157:87–103. doi: 10.1016/0022-2836(82)90514-9. [DOI] [PubMed] [Google Scholar]
- 38.Flory J., Tsang S.S., Muniyappa K. Isolation and visualization of active presynaptic filaments of recA protein and single-stranded DNA. Proc. Natl. Acad. Sci. U.S.A. 1984;81:7026–7030. doi: 10.1073/pnas.81.22.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shibata T., Ohtani T., Iwabuchi M., Ando T. D-loop cycle: a circular reaction sequence which comprises formation and dissociation of D-loops and inactivation and reactivation of superhelical closed circular DNA promoted by recA protein of Escherichia coli. J. Biol. Chem. 1982;257:13981–13986. [PubMed] [Google Scholar]
- 40.Lorsch J.R., Herschlag D. The DEAD box protein eIF4A. 1. A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry. 1998;37:2180–2193. doi: 10.1021/bi972430g. [DOI] [PubMed] [Google Scholar]
- 41.Sgaramella V. Enzymatic oligomerization of bacteriophage P22 DNA and of linear Simian virus 40 DNA. Proc. Natl. Acad. Sci. U.S.A. 1972;69:3389–3393. doi: 10.1073/pnas.69.11.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bugreev D.V., Mazin A.V. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl. Acad. Sci. U.S.A. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arai N., Kagawa W., Saito K., Shingu Y., Mikawa T., Kurumizaka H., Shibata T. Vital roles of the second DNA-binding site of Rad52 protein in yeast homologous recombination. J. Biol. Chem. 2011;286:17607–17617. doi: 10.1074/jbc.M110.216739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kagawa W., Arai N., Ichikawa Y., Saito K., Sugiyama S., Saotome M., Shibata T., Kurumizaka H. Functional analyses of the C terminal half of the Saccharomyces cerevisiae Rad52 protein. Nucleic Acids Res. 2014;42:941–951. doi: 10.1093/nar/gkt986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Migneault I., Dartiguenave C., Bertrand M.J., Waldron K.C. Glutaraldehyde: behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking. BioTechniques. 2004;37:790–802. doi: 10.2144/04375RV01. [DOI] [PubMed] [Google Scholar]
- 46.Kuykendall J.R., Bogdanffy M.S. Efficiency of DNA-histone crosslinking induced by saturated and unsaturated aldehydes in vitro. Mutat. Res. 1992;283:131–136. doi: 10.1016/0165-7992(92)90145-8. [DOI] [PubMed] [Google Scholar]
- 47.Zhang X.P., Lee K.I., Solinger J.A., Kiianitsa K., Heyer W.D. Gly-103 in the N-terminal domain of Saccharomyces cerevisiae Rad51 protein is critical for DNA binding. J. Biol. Chem. 2005;280:26303–26311. doi: 10.1074/jbc.M503244200. [DOI] [PubMed] [Google Scholar]
- 48.Petukhova G., Stratton S., Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 49.Shibata T., DasGupta C., Cunningham R.P., Williams J.G.K., Osber L., Radding C.M. Homologous pairing in genetic recombination. The pairing reaction catalyzed by Escherichia coli recA protein. J. Biol. Chem. 1981;256:7565–7572. [PubMed] [Google Scholar]
- 50.Tsang S.S., Muniyappa K., Azhderian E., Gonda D.K., Radding C.M., Flory J., Chase J.W. Intermediates in homologous pairing promoted by recA protein: isolation and characterization of active presynaptic complexes. J. Mol. Biol. 1985;185:295–309. doi: 10.1016/0022-2836(85)90405-x. [DOI] [PubMed] [Google Scholar]
- 51.Rusche J.R., Howard-Flanders P. Hexamine cobalt chloride promotes intermolecular ligation of blunt end DNA fragments by T4 DNA ligase. Nucleic Acids Res. 1985;13:1997–2008. doi: 10.1093/nar/13.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mandelkern M., Dattagupta N., Crothers D.M. Conversion of B DNA between solution and fiber conformations. Proc. Natl. Acad. Sci. U.S.A. 1981;78:4294–4298. doi: 10.1073/pnas.78.7.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams R.C., Spengler S.J. Fibers of RecA protein and complexes of RecA protein and single-stranded phi X174 DNA as visualized by negative-stain electron microscopy. J. Mol. Biol. 1986;187:109–118. doi: 10.1016/0022-2836(86)90410-9. [DOI] [PubMed] [Google Scholar]
- 54.Egelman E.H., Stasiak A. Structure of helical recA-DNA complexes. II. Local conformational changes visualized in bundles of recA-ATPγS filaments. J. Mol. Biol. 1988;200:329–349. doi: 10.1016/0022-2836(88)90245-8. [DOI] [PubMed] [Google Scholar]
- 55.Chen K.M., Harjes E., Gross P.J., Fahmy A., Lu Y., Shindo K., Harris R.S., Matsuo H. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature. 2008;452:116–119. doi: 10.1038/nature06638. [DOI] [PubMed] [Google Scholar]
- 56.Rusche J.R., Konigsberg W., Howard-Flanders P. Isolation of altered recA polypeptides and interaction with ATP and DNA. J. Biol. Chem. 1985;260:949–955. [PubMed] [Google Scholar]
- 57.Tsang S.S., Chow S.A., Radding C.M. Networks of DNA and recA protein are intermediates in homologous pairing. Biochemistry. 1985;24:3226–3232. doi: 10.1021/bi00334a023. [DOI] [PubMed] [Google Scholar]
- 58.Ellouze C., Takahashi M., Wittung P., Mortensen K., Schnarr M., Norden B. Evidence for elongation of the helical pitch of the RecA filament upon ATP and ADP binding using small-angle neutron scattering. Eur. J. Biochem. 1995;233:579–583. doi: 10.1111/j.1432-1033.1995.579_2.x. [DOI] [PubMed] [Google Scholar]
- 59.Register J.C., 3rd, Griffith J. RecA protein filaments can juxtapose DNA ends: an activity that may reflect a function in DNA repair. Proc. Natl. Acad. Sci. U.S.A. 1986;83:624–628. doi: 10.1073/pnas.83.3.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakata M., Zanchetta G., Chapman B.D., Jones C.D., Cross J.O., Pindak R., Bellini T., Clark N.A. End-to-end stacking and liquid crystal condensation of 6 to 20 base pair DNA duplexes. Science. 2007;318:1276–1279. doi: 10.1126/science.1143826. [DOI] [PubMed] [Google Scholar]
- 61.Li M.Z., Elledge S.J. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods. 2007;4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- 62.Miura T., Yamana Y., Usui T., Ogawa H.I., Yamamoto M.-T., Kusano K. Homologous recombination via synthesis-dependent strand annealing in yeast requires the Irc20 and Srs2 DNA helicases. Genetics. 2012;191:65–78. doi: 10.1534/genetics.112.139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walker J.R., Corpina R.A., Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]