Abstract
We report here, the design and development of an automated near real-time continuous detection system for lactate, glutamate, pyruvate and glucose using microdialysis probe. The system developed can automatically push perfusate through microdialysis probe (20, 100 and 1000 kDa MWCO cutoff probe) at low to medium flow rate of 0.5–2 μL/min with almost 100% fluid recovery. The microdialysate collected from the probe is analyzed automatically for these four metabolite biomarkers. It operates in a continuous mode with measurements of all four biomarkers once every 20 min. The dynamic range for these different markers covers the entire clinical range of traumatic brain injury. The prototype shows a low variation of ~ 7–10% across the entire clinical range for all the biomarkers with fairly good accuracy of ~95%. The instrument canrun continuously for 24 h without user intervention. With a long tubing of 1 m to and from the microdialysis probe and associated dead volume, the total lag time for actual event at the probe site versusreported concentration is roughly 1 h.
Keywords: Microdialysis, Real-time analysis, Metabolic biomarkers, Automated instrumentation, Collection efficiency, Traumatic brain injury
1. Introduction
Microdialysis is a sampling technique to monitor continuous changes in biochemical markers from extracellular fluid of tissues. Microdialysis of brain extra cellular fluids is the most widely used application though there are reports of using the technology in other organs like the liver [1], skin [2], blood [3], stomach [4] and ear [5]. This minimally invasive technique is widely used in bedside monitoring of early markers of tissue damage and ischemia for Traumatic brain injury (TBI) [6–9]. Clinical studies have indicated that neurochemical markers like glutamate [10], glucose, lactate [11], pyruvate among others are useful in prediction of development of secondary injury after TBI [12,13]. Increase in Lactate to pyruvate ratio occurs during a metabolic crisis and happens due to mitochondrial dysfunction [14]. It is a well-known marker for monitoring cell damage and ischemia after TBI and Subarachnoid Hemorrhage (SAH). Glucose, Glycerol and Glutamate are additional markers for developing ischemia. During ischemia in brain, the glutamate concentration may increase due to decrease in uptake in glial cells, whereas changes in glucose concentration can happen due to several reasons like ischemia, hyperemia, hyperglycemia or hyper/hypo metabolism [15]. Continuous monitoring of these metabolic markers will therefore be useful at identifying the risk, guide therapy and improve the outcome [6]. Currently, the only clinical bedside monitoring instrument available to analyze minute quantity of microdialysate is ISCUS [16] from MDialysis. While, the instrument is impressive in terms of its performance, it is not truly an inline system. The microdialysis probe and its collection unit are separate from the analyzer. The microdialysate is collected in separate vials which are then individually inserted in the instrument to analyze the content of the vial. It is a tedious, time consuming and error prone process that has to be performed every hour. This paper describes the first automatic inline microdialysis system that can continuously push the perfusate through microdialysis probe, analyze its content for four different biomarkers simultaneously and report the concentrations every 20 min without any intervention. There are few reports of development of inline systems for microdialysis using high performance liquid chromatography (HPLC) for analyzing the microdialysis or using electrochemical probes directly in the flow path of microdialysate. While, the HPLC systems can detect minute concentration of analytes from microdialysate [17–19], it has a large footprint and an expensive system to maintain. Moreover, the interface between microdialysis collection and LC system are not well developed [20]. Similarly electrochemical sensors placed in line with microdialysate suffer from issue of stability of the sensors and difficulty of dynamic calibration. Additionally, lack of availability of EC sensors for many important metabolites makes it difficult to develop a multiplexed detection system. Our current system is based on enzymatic reactions and optical detection in 384 well platform, which is robust, well developed and already implemented in many lab tests in different clinical applications. We report here the detailed instrument design and initial results obtained from in-vitro tests.
2. Material and methods
2.1. Materials
CMA 12 microdialysis probe (20, 100 and 1000 kDa cutoff, 4 mm length except 3 mm for 1000 KDa probe) is purchased from Harvard Apparatus. Ringer’s solution and artificial cerebrospinal fluid (ACSF) are purchased from Harvard apparatus or formulated in-house using reagent grade chemicals from Sigma and Fisher Scientific. Ringer’s salt solution powder (#M525) is from Himedia Laboratories. Glucose, glutamate and pyruvate used for in-vitro experiments are bought from Sigma. Lactate is bought from Alfa Aeser. Lactate (ECLC-100), glutamate (EGLT-100), glucose (EBGL-100) and pyruvate (EPYR-100) assay kits from BioAssay systems are used. We also have used individual reagents for further optimization of the analyte detection range, stability and sensitivity. Lactate oxidase (#L0638) and pyruvate oxidase (#P4591-100U) are bought from Sigma whereas, glucose oxidase (#0243) is from Amresco and glutamate oxidase (#YMS-80049) is from CosmoBio USA. Clear bottom low volume black 384 well plate is from Corning (#3542). Starting blocking T20 (PBS) blocking buffer from Thermo-Scientific (#37539) is used for plate preparation.
Detailed description of assay for four biomarkers can be found elsewhere [21–24]. These commercial assays are adapted for our application in following way as shown in Table 1. The reagents (substrate and enzyme mix) are mixed with the sample to start a reaction which develops a color. The total volume of reagents and sample adds up to ~40 uL of solution. This volume of fluid is enough for proper detection in a 384 well pate by absorption technique. The intensity of the product color (perceived color against white light: Purple-dark bluish) is measured at 565 nm and proportional to the concentration of the sample. The amount of sample volume and reagents required for proper detection, depends on the concentration of that particular analyte in the sample and can be changed to increase the sensitivity and dynamic range of the detection. In-vitro experiments performed with the instrument used the same reagents as described in Table 1, or a modified version of that depending on the clinical range and sensitivity requirements of some of the experiments. The reagents are drawn from a reservoir which is kept at room temperature for the whole duration of the experiment which can last up to eight hours or more. No significant deterioration of the reagents was observed over eight hours. However, care is taken to store these reagents before being used for experiments. These reagents prepared are kept at −20 °C for storage up to one month. Before the experiment, the reagents are taken out of freezer and thawed at room temperature before being transferred to the instrument.
Table 1.
Ratio of different reagents used for detection of four analytes.
| No. | Analyte | Sample−μL | Substrate–μL | Enzyme mix-μL |
|---|---|---|---|---|
| 1 | Lactate | 2 | 17.2 (27% NAD + 37.85% MTT + 35.12% buffer volume based) | 22.83 (95.93% buffer, 2.04% enzyme A, 2.04% enzyme B volume based) |
| 2 | Pyruvate | 2.5 | 4 (10% 10 × dye, 90% buffer volume based) | 36 (enzyme) |
| 3 | Glucose | 1.5 | 14.25 (1.27% dye, 98.73% buffer volume based) | 25.75 (0.71% enzyme, 99.29% buffer volume based) |
| 4 | Glutamate | 4 | 9 (26.31% NAD, 73.69% MTT volume based) | 28.9 (1.63% enzyme mix, 98.37 buffer volume based) |
A push-pull mechanism is implemented for sample collection from the microdialysis probe. This subsystem is composed of an OEM syringe pump module #702225 and two sets of 1 m tubing’s (ID 0.12 mm) from Harvard Apparatus, a single channel peristaltic pump from APT (#SP101F.013), a vacuum transducer (PX209-30VAC5 V) purchased from Omega. Flow sensors (LG16-150 and LG-16-480) are from Sensirion and solenoid valve (P/N 038T2S12-32-4) is from Biochem Fluidics. Potentiostat (Emstat3) is from Palmsens or in-house developed current sensor is used. A reagent delivery subsystem is developed in-house. XY slides (ET-150-13) from Newmark systems are used for precise plate movement. These slides have resolution of 7.5 m, accuracy of 0.0012 mm/mm of travel and repeatability of 30 m. Multichannel peristaltic pumps (Ismatec-Reglo 4 channel OEM) are from Cole Parmer. Miniature linear actuator (L12-50-100-6-p) is bought from Firgelli along with controller board. CMOS camera (DCC1545 M), Cage cube mounted turning prism mirror (CM1P01) and diffuser (DG-10-600), are bought from Thorlabs. Fujinon Lens (HF9HA-1B) is used with the camera. Multifunction data acquisition board (USB6008) and interface for I2C communication (USB8451) are bought from National Instruments. LED light source (565 nm), AC/DC converters (1470–2746-ND for 24 V, 285–1984-ND for 12 V and 102–3268-ND for 5 V) are bought from Digikey. The entire prototype is designed using Autodesk Inventor software. The control system for the prototype is written in LabVIEW.
2.2. Development of prototype
There are very few commercially available systems which can perform automatic detection of multiple biomarkers from microdialysate continuously [25]. Development of this type of system requires complex design and software control to make sure both the collection and analysis of microdialysate are performed consistently over many hours of continuous operation. Fig. 1 shows the schematic of overall operation of the prototype system. There are three major subsystems namely a) Dialysate collection unit, b) Reagent delivery subsystem and c) Detection Subsystem. The Dialysate collection unit, uses a push-pull technique to collect microdialysate in a small chamber. The flow rate and pressure inside the chamber are monitored for PID (proportional-integral-derivative) control. Once enough solution is collected, the delivery subsystem will automatically start the assay process for up to four metabolite biomarkers. The assay development will be monitored with the detection subsystem in real time. The final analysis result will be reported before the subsequent assay begins. Current prototype is capable of reporting results of four biomarkers simultaneously at a 20 min interval for up to 24 h. The overall instrument is controlled with LabVIEW running on Windows’s based computer.
Fig. 1.
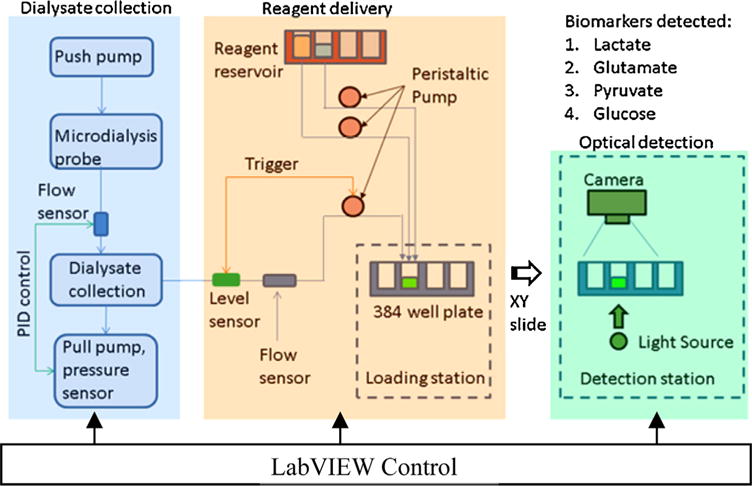
Schematic of operation for the instrument. Dialysate collection part will push and pull the microdialysate from the probe and store it intermediately. Level sensor inside the collection unit senses that certain volume of microdialysate available for analysis, which is then transferred to well plate for multiplexed optical detection of biomarkers.
2.2.1. Dialysate collection unit
Since the dialysate flow rate is usually very low (0.5–2 μL/min), it takes relatively long time to accumulate enough volume of fluid for subsequent analysis. A level detection system within the dialysate collection unit continuously monitors the volume collected and sends a trigger to the detection unit to start its operation as soon as the correct volume of dialysate becomes available. The reagent delivery subsystem will then split the microdialysate in four separate boluses and use them for detection of lactate, pyruvate, glucose and glutamate detection in a 384 well plate. Each of these subsystems is explained in details in next section.
The primary function of the dialysate collection unit is to maintain a steady sample collection efficiency by stabilizing the flow rate through the microdialysis probe and initiate the assay process once exact volume of samples are collected. As seen from Fig. 2A, a syringe pump pushes the perfusate through microdialysis probe at a predefined flow rate (0.5–2 μL/min). As the perfusate flows through the probe, it collects small metabolic molecules (by diffusion through membrane of probe) from the extracellular fluid of brain or other tissues. It is very important to maintain a constant flow through the probe, as inconsistency in the flow will increase or decrease the residence time of fluid in contact with the probe, which will further result in increase or decrease of biomarker collection efficiency and introduce outliers in the measurements.
Fig. 2.
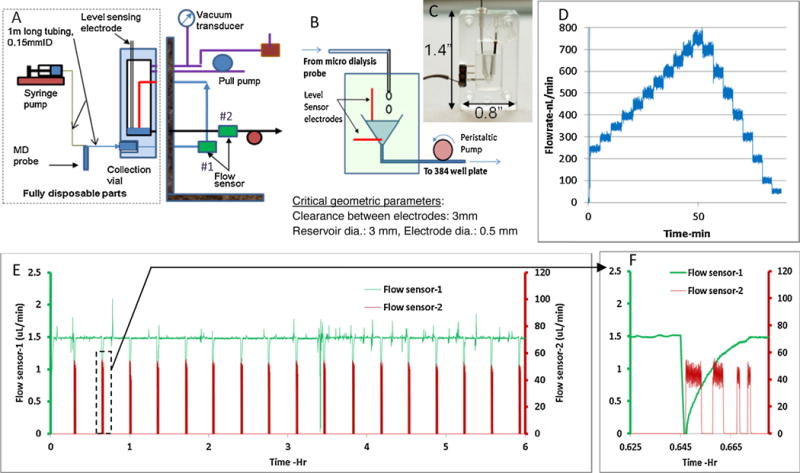
A) Schematic of push-pull system with level sensing mechanism. B,C) Detailed schematic of level sensing unit. D) Plot showing flow through the probe can be controlled in steps of 50 nL/min. E) Flow characteristics for push-pull system from a test with 100 kDa probe (1 m tubing before and after probe, 0.125 mm ID, Ringer’s solution) F) Zoomed in data for flowsensor-2, showing four distinct drawing of sample(15,10,2.5,2.5 μL) from the collection vial by the peristaltic pump after the level sensor triggering. Flow sensor-1 shows a dip due to release of vacuum before the withdrawal process.
Flow before and after the probe is mainly affected by four variables: a) backpressure to the microdialysis probe (caused by property of dialysate solution, long tubing length or very low internal diameter), b) porosity in the probe membrane, c) targeted flow rate and d) variability of probe membrane. Commercially available systems use push-pull technology, where one pump pushes the fluid to the dialysis probe and another pulls the fluid at the same rate. There is no feedback mechanism, and any fluctuation or difference in flow between the pumps will result in high pressure difference across the probe. These pressure fluctuations can affect the collection efficiency of the probe. If the pressure is high inside the probe, fluid can leak through the membrane into the extracellular space thereby reducing the collection efficiency. Similarly if the pressure is low inside the probe, the opposite effect will take place. Overall this kind of push-pull technique offers limited improvement to push only system.
This is improved by removing the pull pump from directly acting on the fluid. Rather, the pull pump will create a vacuum in the collection chamber, which then generates enough pull force to have a smooth flow inside the fluidic line connected to the probe. Fig. 2A shows the schematic of the push-pull system for the dialysate collection unit. A syringe pump pushes the fluid at constant flowrate through the microdialysis probe; where as a steady vacuum is generated at the collection chamber by a peristaltic pump. The flow is monitored by a flow sensor (#1) placed inline and before the collection vial. This flow sensor provides a feed back to the pull pump to adjust the vacuum inside the chamber, which in turn maintains the constant flow rate. The vacuum pressure generated by the pull pump is monitored with a vacuum transducer. This design is very effective and can maintain constant flow with larger MWCO (molecular weight cutoff, up to 1 million Da) probes, long tubing length (1 m to and from the probe, 0.125 mm ID) and relatively higher flowrate (up to2 μL/min). To prevent the accidental backflow from the chamber back to the microdialysis probe, there is an option of discrete droplet flow inside the chamber (Fig. 2A).
The collection unit also initiates the assay process once certain volume is collected in the chamber. A pair of platinum wires used as level sensing electrodes is placed inside the chamber to monitor the volume of microdialysate collected, as shown in Fig. 2B and C. A pulsed voltage of 0.5 VDC is applied across the electrodes at a frequency of 0.2–0.5 Hz during the sample collection. Initially the current across the electrodes is less than 1 nA since the two electrodes are in open-circuit condition. When the fluid reaches the top electrodes and closes the circuit, the current jumps to over 4 nA. This increase in current is used to trigger a solenoid valve connected to the chamber to release the vacuum so that the sample loading pump can pull the fluid out of the chamber. The system is designed to leave about 2–3 μL of fluid inside the chamber, to prevent air from entering the delivery line. Fig. 2D shows the feedback control for flow. The flow can be controlled to as low as 50 nL/min.
Fig. 2E shows an example of consistent flow data from an experiment with 100 kDa probe (1 m tubing, 0.125 mm ID) and with 1.5 μL/min target flow rate running continuously for 6 hours. Flow sensor-1 (placed before the collection unit) is used for PID control on the peristaltic pump. Flow sensor 2 is located after the collection unit and used for sample delivery confirmation when the multichannel peristaltic pump at the back of collection unit pulls the microdialysate out of the chamber for analysis. Zoomed in data for flowsensor-2 (Fig. 2F), shows four distinct withdraw of sample (15, 10, 2.5, 2.5 μL) from the collection vial by the peristaltic pump after the level sensor triggering. The inlet sensor (#1) showed a drop in flow, due to release of vacuum. The flow comes back up once the valve is closed (~ 1 min).
2.2.2. Fluidic dispensing and assay
Multiplexed dispensing of all reagents for four metabolite biomarker assays is challenging. In order to reduce cross talk between reagents and eliminate dead volume, each reagent has a dedicated pump. Each analyte is detected by reaction with only two reagents (enzyme solution and substrate solution) thereby significantly reducing the requirements of pumps. Besides these eight reagents (two each for lactate, glutamate, glucose and pyruvate), three standard solutions are used for each measurement in real time. With an additional channel for sample, total of twelve channels are used, requiring three sets of Ismatec Reglo four channel pumps (Fig. 3B). To minimize the initial dead volume in each channel and increase the reliability for low volume delivery, 0.25 mm ID tubing’s are used with peristaltic pumps. A priming procedure is used to prime all channels before each test.
Fig. 3.
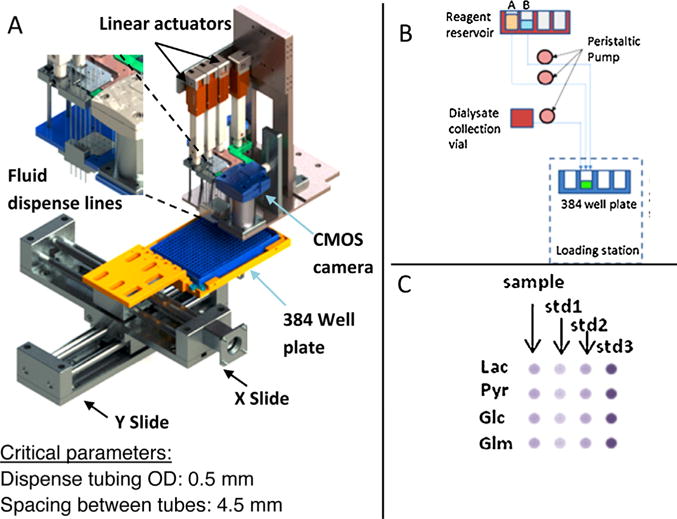
A) Integrated dispense system for 384 well plate. Fluidic dispense pins are controlled with two Firgelli linear actuators and spaced 4.5 mm apart to match the microplate. B) Schematic of reagent loading from reagent reservoir to the plate. C) Final well layout for simultaneous detection of four biomarkers for one test.
Two Firgelli linear actuators are used to control the delivery of the solutions through twelve loading pins (Fig. 3A). Additional actuators shown in the design are for integration of additional biomarkers in future. These pins are positioned to match 384 wells. Each analyte requires two reagents to complete the enzymatic reaction when mixed with sample. Each pin position corresponds to one solution and is controlled by one channel of peristaltic pump. After receiving the triggering signal from the level sensor, sample and standard solutions will be loaded to predefined wells with specific volumes, followed by the enzyme solutions and the substrate solutions. For four metabolite test, the final well layout is in 4 × 4 grid as shown in Fig. 3C. Each row is corresponding to one metabolite. For each metabolite, three wells are for internal standards and one well for sample. The movement of the microplate is controlled with X-Y slide, which has overall variation of less than 0.2 mm. The movement of 384 well plate is synchronized with dispense. After finishing reagent delivery, the 4 × 4 grid will proceed to the detection spot for real time measurement.
2.2.3. Multiplexed optical detection
Multiplexed detection of 4 small molecule markers is carried out in batch of 16 wells in 384 well plate, as shown in Fig. 3C. The 384 well plate is mounted on a movable XY slide, which is precisely controlled by protocol ensuring consistent measurement for each well. As shown in Fig. 3A, the detection system (camera light source) is placed close to the dispensing subsystem to minimize the range of plate movement and reduce the size of the overall instrument.
A dynamic calibration is also included in the system to improve the accuracy and reliability of the detection. Three standards (Low, medium and high concentration) are used, each of which contains known concentration of four markers in Ringer’s solution. During dynamic calibration standard solution is used instead of sample (microdialysate), whereas the other reagents remained the same. Fig. 4 and inset shows the dynamic calibration of analytes (plot shown for glutamate). The absorbance of glutamate (after reaction with enzymes) is shown with time for different concentrations. The color development is gradual and non-linear with time. However the rate of development is highly reproducible and accurate. The control program tracks both time and absorbance value and creates a calibration curve based on 3 standard values (low, medium and high). The sample absorbance data is then compared against the calibration curve and the concentration is calculated. A single dynamic calibration at the beginning is good for 7–8 h of experimentation. This real time calibration brings the advantages of mitigating the effects from the slow changes of the reagents, detection system and even uneven flow rates of pumps during the long time of operation (up to 24 h) (Fig. 5).
Fig. 4.
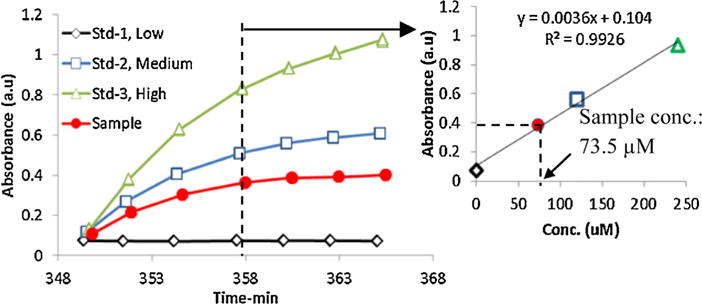
Color development (change in absorbance) over time in well as recorded by camera for single biomarker (shown for glutamate). Dynamic calibration using 3 standards is shown in inset to determine the sample concentration.
Fig. 5.

Fully functional prototype for automated multiple biomarker analysis with microdialysis probe.
3. Results and discussion
The fully functional prototype system for multiple biomarker analysis with microdialysis probe is shown in Fig. 7. The instrument measures at 24″×24″×18″. The reaction kinetics is quite different for the four biomarkers. Lactate and glucose are much faster than pyruvate and glutamate for the clinically relevant range of concentrations. The current protocol is based on 2 μL/min collection flow rate and 20 min assay interval, which requires following volume of microdialysate for each analysis:
Glucose: 3 μL, range covered: 0–4 mmol/L
Lactate: 3 μL, range covered: 0–8 mmol/L
Glutamate: 17 μL, range covered: 0–0.6 mmol/L
Pyruvate: 17 μL, range covered: 0–0.25 mmol/L
Fig. 7.

A) Schematic of experiment. In-vitro testing of the prototype with 100 kDa probe (flow 2 μL/min with Ringer’s solution). External vial concentration is changed at 180th min. B,C,D,E) Real-time concentration of 4 markers simultaneously reported by instrument in-response to step-change of concentration in external vial. F) Collection efficiency at different flow rates.
For each set of test, 40 μL volume of microdialysate collected are completely used for analysis. Volume requirement is higher for glutamate and pyruvate mainly because their clinical concentration ranges are relatively low. For flow rate of 2 μL/min, the prototype instrument can perform three tests per hour. Flow rate below 1.5 μL/min (0.5–1.5 μL/min) can be tried considering the fact that slower flow will have higher dialysis efficiency. In reality, however slower flow would not only make each test time longer (80 min for 0.5 μL/min, 26 min for 1.5 μL/min compared to 20 min for 2 μL/min), but also cause delay for the events at the probe to be detected because of the transition volume through long tubing (1 h at 2 μL/min vs. 2 h at 1 μL/min). Meanwhile, the relatively small collection efficiency increase (~6% efficiency increase, when flow is reduced from 2 μL/min to 1.5 μL/min, Fig. 7F) could not justify using slower flow rate. So for current prototype, the flow was kept at 2 μL/min. Meanwhile, it is still possible to use slower flow rate by decreasing internal dead volume, optimizing glutamate and pyruvate assay towards low concentration range and minimize the dilutions during the sample transition. Current work is focused on improving overall response time of the instrument.
Fig. 6A–D shows the comparison of calibration curves between manual assay and automated assay with the prototype. The range covered for the automated assay is relevant to clinical range for traumatic brain injury in human. Fig. 6E shows the simultaneous detection of all 4 biomarkers (glucose, glutamate, lactate and pyruvate) in the instrument for multiple runs. The biomarkers are prepared in Ringer’s solution and directly loaded into the system without the microdialysis probe to avoid introducing variations due to the probe. 23 repetitive runs (sampled from same solution with constant concentration of all markers) are completed, and the data shows the accuracy and precision between the runs. Pyruvate and glutamate have the lowest variation (~ 4%) while lactate is slightly higher (7%). Accuracy is within 95% of the target for all the biomarkers except glucose which is at 91%. The small sample volumes (3 μL) for lactate and glucose assays and error associated with dispense of that volume used would be the main reason for less accurate results than glutamate and pyruvate assays (17 μL).
Fig. 6.

A,B,C,D) Detection of 4 small molecule markers. The absorbance values obtained from prototype is compared with manual plate reader method. E) Simultaneous detection of Lactate, Glutamate, Pyruvate and Glucose in the prototype without the probe. Multiple runs show detection accuracy and precision.
Fig. 7 shows one example of continuous detection of all the four markers sampled through microdialysis probe with the prototype system. The microdialysis probe (100 kDa, 4 mm length) is dipped in an external vial (continuously stirred) containing lactate, glutamate, pyruvate and glucose in Ringer’s solution. The perfusate contained Ringer’s solution only and is pushed at constant flowrate of 2 μL/min. The pull pump and flow sensor worked in the feedback mode to make sure the flow is always maintained at targeted value. Once 40 μL of dialysate is collected, the prototype starts the detection part automatically by mixing reagents with sample and standards, followed by continuous measurement of absorptions in 4 × 4 wells. Initially the concentrations for all the biomarkers in external vials are kept at zero for 3 h to obtain the baseline data. Later the sample concentration is changed to lactate: 6 mmol/L, glutamate: 0.600 mmol/L, pyruvate: 0.25 mmol/L, glucose: 4 mmol/L at ~180th minute (from start of experiment). The solution in the external vial is switched back to buffer at ~410th min. Fig. 7B–E shows the continuous detection results. All the markers showed increase in concentration immediately after concentration increase in external vial, and finally stabilized to constant concentration after about 1 h from step change in external vial and remained same for next 2.5 h. Once the solutions are switched back to Ringer’s solution, the measured concentrations decreased to 0 in about an hour. The collection efficiency is different for different markers. The highest collection efficiencies are recorded for pyruvate at (21.2%) and lactate (25.0%), followed by glucose and glutamate at 16.5% and 10.6% respectively. Multiple test have shown that the collection efficiency tends to be low for the first use of these CMA probes, however it will slowly improve after few days of use (data not shown). The reason behind it is not clear. The low collection efficiency may be acceptable if the limit of detection for the instrument is lower than the clinical range and the variability in detection does not mask the small changes in concentration. Fig. 7F shows collection efficiency for all the four biomarkers at different flow rates (1–2 μL/min). This instrument developed so far shows good stability and variability of ~10% for all the analytes. The reagents used for detection of the markers are tested for specificity (data not shown) and cross reaction with other molecules present in extra-cellular fluid of brain.
4. Conclusion
A prototype that can perform automated continuous multiplexed detection of 4 metabolic biomarkers for assessing clinical development after traumatic brain injury is designed, developed and tested. The modular design of this prototype will allow for detection of different biomarkers from microdialysate in future by simply replacing the reagents. The challenging aspect for this system is handling minute quantity of microdialysate from the probe to instrument and simultaneously analyzing it for 4 different analytes, which is being accomplished successfully. An intermediate storage of micro-dialysate and subsequent usage of that micro-dialysate for analysis proved to be an important step in this process, as the microdialysis is a continuous process but detection is done at discrete times. Our unique collection unit design fulfills the purpose very well. The enzymatic reactions for the analysis use minute volume of reagents and sample, and corresponding accurate and reproducible metering of those volumes of fluid is important for accurate and low variability in reported concentration. The multichannel pumps used for this application is quite accurate and eliminates cross contaminations between reagents. However, regular yearly calibration may be required for these pumps to maintain high accuracy. The detection system built with simple off-the-shelf optical components proved to be quite robust for the range of absorbance used (0.04–1 a.u), combined with stable XY movement and correct reagent delivery. The accuracy for all markers is above 95% (except glucose which is at 91%) with variation of less than 11%.
Supplementary Material
Acknowledgments
Research reported in this publication is supported by the National Institute of Neurological Disorders And Stroke (NINDS) of the National Institutes of Health under Award Number R44NS076167. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We would like to thank our collaborators Dr. Claudia Robertson from Baylor College of Medicine, TX and Dr. Julie Stenken from University of Arkansas, AR for insightful discussion in developing this instrument and subsequent testing at their laboratories.
The prototypes are designed, developed and tested at SFC Fluidics, Inc. based on SBIR Phase-II grant from NINDS.
Biographies

Dr. Das received his BE in Mechanical Engineering from Jadavpur University, Calcutta, India, in 1998 followed by MS degree in Mechanical Engineering from Florida Institute of Technology, Melbourne, FL, USA, in 2003, and PhD degree in Mechanical Engineering from University of Florida, Gainesville, FL, USA, in 2007. There after he joined SFC Fluidics as a Senior Engineer and is actively involved in design and development of biomedical instruments related to detection of biomarkers from blood and microdialysate after Traumatic Brain injury. His research interests include fluid mechanics, microfluidics, optics, system integration, control system, algorithm, micro pumps and valves.

Dr. Wang received his BS in Biological Sciences and Biotechnology from Tsinghua University, Beijing, China, in 2000 followed by PhD degree in Biomedical Engineering from Brown University, Providence, RI, USA, in 2007. There after he joined SFC Fluidics as a Senior Engineer and is actively involved in design and development of diagnostic devices for point of care, especially biomarker based devices for traumatic brain injury diagnosis. His research interests include assay development, optimization and adaptation, system design, integration, testing and evaluation, microfluidics and sensor technology.

Dr. Sun received her BE in Chemistry from NanKai University, Tianjin, China, in 1986. She got her MS degree in Chemistry in 1999 and followed by PhD degree in Chemistry in 2003 from Katholieke Universiteit Leuven, Leuven, Belgium. She worked as a faculty to teach Analytical Chemistry in College of Pharmacy, Tianjin Medical University, Tianjin, China, in 1986–1997. She joined in BioDetection Instruments LLC, Fayetteville AR, as a Senior Scientist for development and evaluation of biosensors and instruments in 2004–2013. Following she joined in SFC Fluidics in 2013 to present as a Senior Scientist for development and evaluation of biomedical instruments related to detection of biomarkers from blood and micro-dialysate after Traumatic Brain injury. She was involved in more than 10 research projects funded by NIH, NSF, DOD and FDA. Her research interests includes immunoassay and bioassay development for rapid detection of protein biomarkers, foodborne pathogens, viruses, and pesticides residues.

Dr. Ledden received his BS in Physics from the University of Central Arkansas, Conway, Arkansas followed by a PhD degree in Engineering from the University of Arkansas, Fayetteville, Arkansas. After graduation he joined SFC Fluidics as Associate Scientist and is involved in design, fabrication, and testing of biomedical instruments related to drug delivery.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.snb.2016.07.097.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Richards DA, et al. Extracellular amino acid levels in the human liver during transplantation: a microdialysis study from donor to recipient. Amino Acids. 2007;33(3):429–437. doi: 10.1007/s00726-006-0480-1. [DOI] [PubMed] [Google Scholar]
- 2.Bielecka-Grzela S, Klimowicz A. Application of cutaneous microdialysis to evaluate metronidazole and its main metabolite concentrations in the skin after a single oral dose. J Clin Pharm Ther. 2003;28(6):465–469. doi: 10.1046/j.0269-4727.2003.00516.x. [DOI] [PubMed] [Google Scholar]
- 3.Huang HF, et al. Determination of baicalin in rat cerebrospinal fluid and blood using microdialysis coupled with ultra-performance liquid chromatography–tandem mass spectrometry. J Chromatogr B Anal Technol Biomed Life Sci. 2008;874(1–2):77–83. doi: 10.1016/j.jchromb.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Woo KL, Lunte CE. The development of multiple probe microdialysis sampling in the stomach. J Pharm Biomed Anal. 2008;48(1):20–26. doi: 10.1016/j.jpba.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu T, et al. Simultaneous intravenous and intramiddle-ear dosing to determine cefditoren influx and efflux clearances in middle ear fluid in freely moving chinchillas. J Pharm Sci. 2003;92(10):1947–1956. doi: 10.1002/jps.10457. [DOI] [PubMed] [Google Scholar]
- 6.Belli A, et al. Metabolic failure precedes intracranial pressure rises in traumatic brain injury: a microdialysis study. Acta Neurochir (Wien) 2008;150(5):461–469. doi: 10.1007/s00701-008-1580-3. (discussion 470.) [DOI] [PubMed] [Google Scholar]
- 7.Hutchinson PJ, et al. On-line monitoring of substrate delivery and brain metabolism in head injury. Acta Neurochir Suppl. 2000;76:431–435. doi: 10.1007/978-3-7091-6346-7_89. [DOI] [PubMed] [Google Scholar]
- 8.Johnston AJ, Gupta AK. Advanced monitoring in the neurology intensive care unit: microdialysis. Curr Opin Crit Care. 2002;8(2):121–127. doi: 10.1097/00075198-200204000-00006. [DOI] [PubMed] [Google Scholar]
- 9.Hillered L, Vespa PM, Hovda DA. Translational neurochemical research in acute human brain injury: the current status and potential future for cerebral microdialysis. J Neurotrauma. 2005;22(1):3–41. doi: 10.1089/neu.2005.22.3. [DOI] [PubMed] [Google Scholar]
- 10.Vespa P, et al. Increase in extracellular glutamate caused by reduced cerebral perfusion pressure and seizures after human traumatic brain injury: a microdialysis study. J Neurosurg. 1998;89(6):971–982. doi: 10.3171/jns.1998.89.6.0971. [DOI] [PubMed] [Google Scholar]
- 11.Goodman JC, et al. Extracellular lactate and glucose alterations in the brain after head injury measured by microdialysis. Crit Care Med. 1999;27(9):1965–1973. doi: 10.1097/00003246-199909000-00041. [DOI] [PubMed] [Google Scholar]
- 12.Marcoux J, et al. Persistent metabolic crisis as measured by elevated cerebral microdialysis lactate-pyruvate ratio predicts chronic frontal lobe brain atrophy after traumatic brain injury. Crit Care Med. 2008;36(10):2871–2877. doi: 10.1097/CCM.0b013e318186a4a0. [DOI] [PubMed] [Google Scholar]
- 13.Sarrafzadeh AS, et al. Detection of secondary insults by brain tissue pO2 and bedside microdialysis in severe head injury. Acta Neurochir Suppl. 2002;81:319–321. doi: 10.1007/978-3-7091-6738-0_81. [DOI] [PubMed] [Google Scholar]
- 14.Bellander BM, et al. Consensus meeting on microdialysis in neurointensive care. Intensive Care Med. 2004;30(12):2166–2169. doi: 10.1007/s00134-004-2461-8. [DOI] [PubMed] [Google Scholar]
- 15.Ungerstedt U, Rostami E. Microdialysis in neurointensive care. Curr Pharm Des. 2004;10(18):2145–2152. doi: 10.2174/1381612043384105. [DOI] [PubMed] [Google Scholar]
- 16.Mdialysis ISCUS Clinical Microdialysis Analyzer. 2016 Available from: http://www.mdialysis.com/analyzers/iscus-clinical-microdialysis-analyzer.
- 17.Takeuchi T. Capillary columns in liquid chromatography. Anal Bioanal Chem. 2003;375(1):26–27. doi: 10.1007/s00216-002-1619-x. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, et al. In vivo monitoring of serotonin in the striatum of freely moving rats with one minute temporal resolution by online microdialysis-capillary high-performance liquid chromatography at elevated temperature and pressure. Anal Chem. 2013;85(20):9889–9897. doi: 10.1021/ac4023605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shackman HM, et al. Microdialysis coupled on-line to capillary liquid chromatography with tandem mass spectrometry for monitoring acetylcholine in vivo. J Neurosci Methods. 2007;159(1):86–92. doi: 10.1016/j.jneumeth.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 20.Schultz KN, Kennedy RT. Time-resolved microdialysis for in vivo neurochemical measurements and other applications. Ann Rev Anal Chem. 2008;1:627–661. doi: 10.1146/annurev.anchem.1.031207.113047. [DOI] [PubMed] [Google Scholar]
- 21.BioAssaySystems. EnzyChrom Glucose Assay kit (EBGL-100) 2013 Available from: https://www.bioassaysys.com/Glucose-Assay-Kit-(EBGL-100).html#tab1.
- 22.BioAssaySystems. EnzyChrom L-Lactate Assay kit (ECLC-100) 2007 Available from: https://www.bioassaysys.com/Lactate-Assay-Kit-(ECLC-100).html#tab1.
- 23.BioAssaySystems. EnzyChrom Glutamate Assay kit (EGLT-100) 2009 Available from: https://www.bioassaysys.com/Glutamate-Assay-Kit.html.
- 24.BioAssaySystems. EnzyChrom Pyruvate Assay kit (EPYR-100) 2013 Available from: https://www.bioassaysys.com/Pyruvate-Assay-Kit.html.
- 25.Schierenbeck F, Franco-Cereceda A, Liska J. Evaluation of a continuous blood glucose monitoring system using central venous microdialysis. J Diabetes Sci Technol. 2012;6(6):1365–1371. doi: 10.1177/193229681200600615. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


