Abstract
Hyaluronan synthases (HAS) normally make large (>MDa) hyaluronan (HA) products. Smaller HA fragments (e.g. 100–400 kDa) produced in vivo are associated with inflammation and cell signaling by HA receptors that bind small, but not large, HA. Although HA fragments can arise from breakdown by hyaluronidases, HAS might also be regulated directly to synthesize small HA. Here we examined the Streptococcus equisimilis HAS (SeHAS) C-terminus, which contains a tandem B-X7-B motif (K398-X7-R406-X7-K414), by testing the effects of 27 site-specific scanning mutations and 7 C-terminal truncations on HA synthesis activity and weight-average mass. Although HAS enzymes cannot be HA-binding proteins, these motifs are highly conserved within the Class I HAS family. Fifteen Arg406 mutants made large MDa HA (86–110% wildtype size), with specific activities from 70% to 177% of wildtype. In contrast, 10 of 12 Lys398 mutants made HA that was 8–14% of wildtype size (≤250–480 kDa), with specific activities from 14% to 64% of wildtype. Four nearly inactive (2% wildtype activity) C-terminal truncation mutants made MDa HA (56–71% wildtype). The results confirm earlier findings with Cys-mutants [Weigel PH, Baggenstoss BA. 2012. Hyaluronan synthase polymerizing activity and control of product size are discrete enzyme functions that can be uncoupled by mutagenesis of conserved cysteines. Glycobiology 22:1302–1310] that HAS uses two independent activities to control HA size and HA synthesis rate; these are two separate functions. We conclude that HAS regulatory modifications that alter tandem B-X7-B motif conformation could mimic these mutagenesis-induced effects, allowing HAS in vivo to make small HA directly. The results also support a model in which the tandem-motif region is part of the intra-HAS pore and interacts directly with HA.
Keywords: enzyme function, hyaluronan binding, hyaluronan biosynthesis, hyaluronan length, regulation
Introduction
Hyaluronic acid (hyaluronan; HA) is synthesized by Class I HA synthases (HAS), which are lipid-dependent integral membrane proteins (Spicer and McDonald 1998; Itano and Kimata 2002; Weigel and DeAngelis 2007) with eight membrane domains (MDs) in vertebrates and six MDs in Streptococcus equisimilis (SeHAS) and other streptococcal species (Heldermon et al. 2001a). Mammals express three HAS isozymes encoded by HAS1, HAS2 and HAS3 genes (Spicer and McDonald 1998; Itano and Kimata 2002; Weigel and DeAngelis 2007). HA is a linear polysaccharide comprised of repeating GlcNAc(β1,4)GlcUA(β1,3) disaccharide units that are assembled at the reducing end by addition of uridine diphosphate (UDP)-sugars to a growing HA-UDP chain. We recently discovered that HAS makes novel chitin-UDP oligomers (Weigel 2015; Weigel et al. 2015b) and these activated glycans might be used as primers to initiate HA disaccharide assembly, in which case new HA chains would have a short chitin cap at their nonreducing ends. All Class I HAS enzymes use three fundamental functions (activities) during HA synthesis; (i) they synthesize disaccharide units to make HA, (ii) they create and release HA chains of particular size ranges (i.e. they control HA product length), and (iii) they translocate the growing chain, simultaneously with elongation, across the cell membrane to the cell exterior.
HA is an essential glycosaminoglycan in vertebrate extracellular matrices, where it helps maintain the physical structure and integrity of tissues, such as cartilage (Tammi et al. 2002; Knudson and Knudson 2004; Toole 2004). HA is also a major constituent of skin, vitreous humor, joint synovial fluid, and the cells surrounding oocytes prior to ovulation (Laurent et al. 1995; Tirone et al. 1997). In tissues or vitreous, HA is 4–10 MDa in molecular mass, occupies a large volume in physiological fluids and has many different functions in a range of cell types and physiologic situations (Weigel et al. 2015a). In addition to its physical functions, different cell types respond to HA (e.g. by altering gene expression profiles) in an HA size-dependent manner. Very small HA oligomers (e.g. <25 sugars) or intermediate size HA (e.g. 100–400 kDa) have different biological activities than normal large MDa HA in tissues.
First reported in 1985 (West et al. 1985), and then confirmed by other groups (Liu et al. 1996; McKee et al. 1997; Noble 2002; Slevin et al. 2007), angiogenesis is stimulated by small, not large, HA. Similarly, only smaller HA induces activated macrophages to express many genes (Noble and Jiang 2006) and induces nitric oxide synthase expression in liver Kupffer and sinusoidal endothelial cells, but not in stellate or parenchymal cells (Rockey et al. 1998). Tetrasaccharides, the smallest HA molecules, up-regulate hsp72 in synovial cells (Xu et al. 2002). HA fragments stimulate cell signaling cascades through specific cell surface receptors (Stern et al. 2006), such as CD44 (Noble 2002; Ohno et al. 2006), RHAMM (Zhang et al. 1998; Turley et al. 2002) and HARE/Stab2 (Kyosseva et al. 2008). There is a very narrow-range HA size dependence for both ERK1/2 and NF-κB signaling during HARE-mediated HA endocytosis (Pandey et al. 2013). Although all HA sizes are bound and endocytosed by HARE, only 40–400 kDa HA stimulates signaling; larger or smaller HA does not activate ERK1/2 or NF-κB.
In addition to the generation of smaller signal-competent HA by degradation of large HA, HAS itself could be regulated directly (e.g. by substrate availability, covalent modification or allosteric or cofactor modulation) to synthesize shorter HA (Jokela et al. 2011, 2014; Tammi et al. 2011; Hascall et al. 2014; Moretto et al. 2015). Our hypothesis is that the ability of HAS to modulate HA size might be mimicked, and thus demonstrated, by specific amino acid mutations that alter HAS conformation and synthase function so the enzyme makes smaller HA. Two protein structures that can bind HA with high affinity and specificity have been identified; the 93 amino acid link domain present in many matrix proteins and cell surface HA receptors (Day, 1999; Seyfried et al. 2005; Kyosseva et al. 2008) and the 9 amino acid basic B-X7-B motif (Yang et al. 1993, 1994; Jean et al. 2001; Xu et al. 2003). Mutagenesis studies and in vitro binding assays showed that two B-X7-B sequences in RHAMM bind HA and these motifs are conserved in murine and human RHAMM (Turley et al. 1993; Wang et al. 1996). Related short basic sequences (e.g. B-X-B-X2-B-X2-B) are also HA-binding motifs in other proteins (Becerra et al. 2008) or peptides (Amemiya et al. 2005).
To identify HAS regions that are important for HA size control, here we examined a tandem-motif sequence with two B-X7-B motifs at the SeHAS C-terminus. This sequence and membrane topology are highly conserved in the Class I HAS family (Figure 1A and 1B). We assessed HA synthesis activity and HA product size of site-specific scanning mutants at Lys398 and Arg406 as well as a group of C-terminal truncation (ΔCter) mutants. The results show that changes at Lys398 or within the tandem-motif greatly affect synthesis rate or the ability to make MDa HA and these two functions can be independently changed.
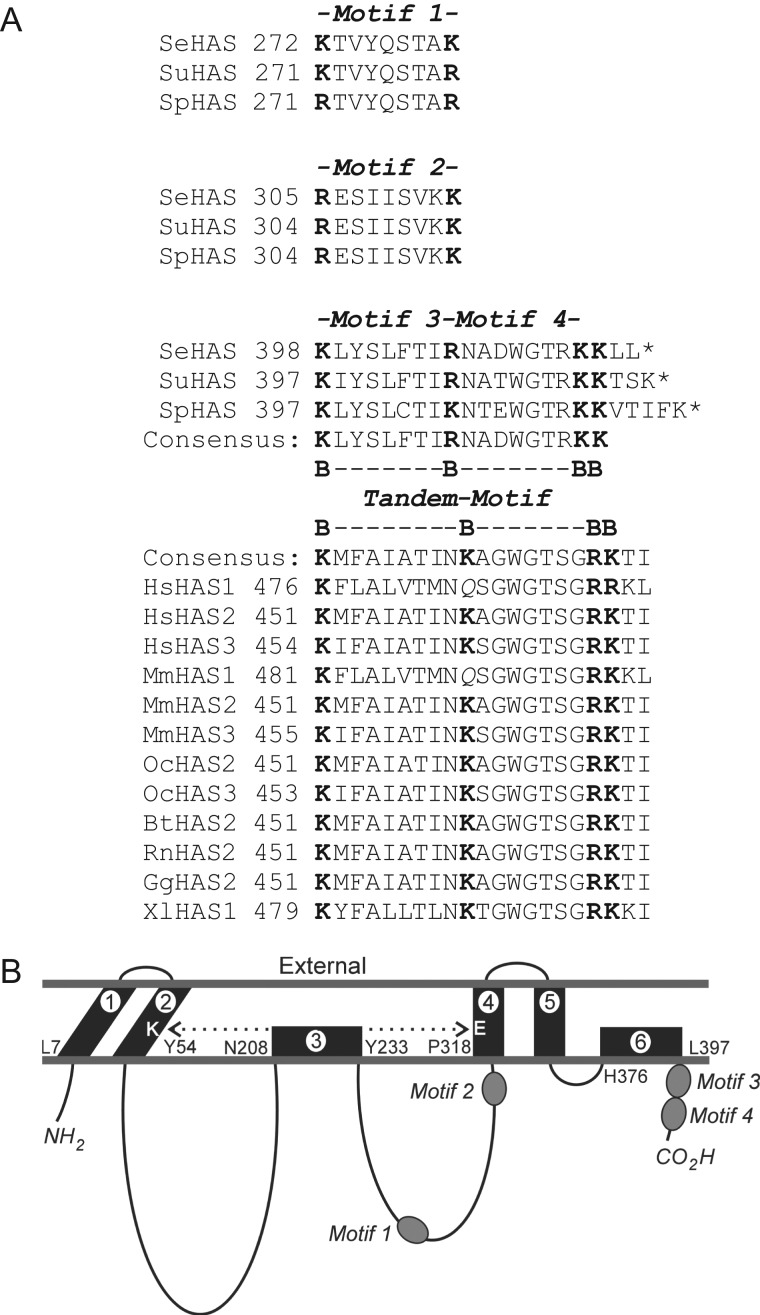
Fig. 1.
Relative positions of B-X7-B motifs in SeHAS and the Class I family. (A) Alignment of B-X7-B motifs within the Class I HAS family. Basic residues are in boldface font. Species and accession numbers for the sequences shown are: Bt, Bos taurus (HAS2 CAA06239), Gg, Gallus gallus (HAS2, 057424); Hs, Homo sapiens (HAS1, NP_001514; HAS2, NP_005319; HAS3 CAB033850); Mm, Mus musculus (HAS1 BAA11654, HAS2 AAC53309, HAS3 AAC53128); Oc, Oryctolagus cuniculus (HAS2 BAB63264, HAS3 BAB63265); Rn, Rattus norvegicus (HAS2 NP_037285); Se, Streptococcus equisimilis (AAB87874); Sp, Streptococcus pyogenes (AAA17981); Su, Streptococcus uberis (CAB46918); Xl, Xenopus laevis HAS1 (P13563). Asterisks indicate a stop codon; the C-terminus. (B) Topological organization of B-X7-B Motifs 1–4 within SeHAS. The scheme is based on previous studies that determined the topology of streptococcal HAS and identified a close interaction (dotted double arrow) between membrane domains 2 and 4 (Heldermon et al. 2001a; Kumari et al. 2006).
Results
Glycerol or low substrate-to-enzyme ratio decreases the ability of HAS to make large HA
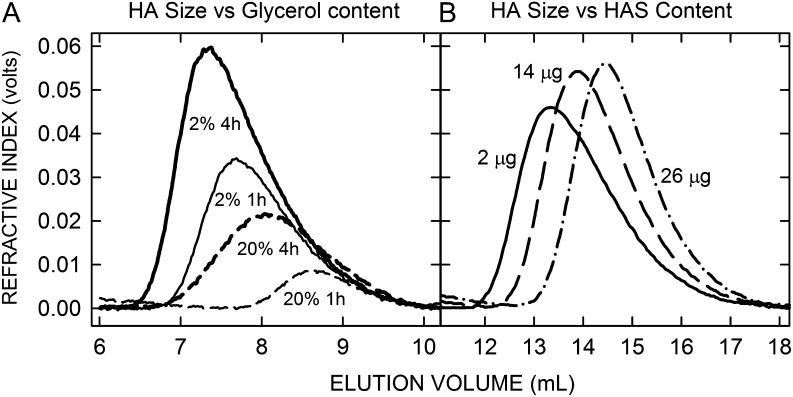
Before describing results with various HAS mutants, it is useful to know that HA product size can be decreased substantially (and inadvertently) by the assay conditions employed. For example, viscosity-increasing agents such as glycerol, a common enzyme-stabilizing reagent, inhibit purified SeHAS. HAS activity is stable up to 1 M glycerol but is then inhibited about 5%, 40% and 90% at 2 M, 3 M and 5 M glycerol, respectively (Tlapak-Simmons et al. 2004). It is not surprising that making long HA polymers would be affected by viscosity, and HAS activity also decreases with increasing concentrations of sucrose or ethylene glycol. Since glycerol is used routinely during membrane preparation, storage and HAS activity assays, we also examined the effects of glycerol on HA product size, which can be altered independently of HAS activity (Weigel and Baggenstoss 2012). Glycerol at 20% (2.2 M) inhibited enzyme activity and had a large negative effect on HA size made by wildtype (WT) SeHAS (Table I). After 1 h and 4 h of synthesis with 20% glycerol the Mw of HA made by WT was smaller (41% and 38%, respectively), than in the presence of 2% (0.22 M) glycerol. Size exclusion chromatography-multiangle laser light scattering (SEC-MALLS) analysis confirmed that the amount of HA product made (i.e. the areas under the curves) and the HA size distributions were deceased greatly at higher glycerol content (Figure 2A); after 1 h the 96% HA size range (encompassing 96% of all HA; excluding the smallest 2% and largest 2%) was 0.61–2.68 MDa with 2% glycerol but only 0.46–0.84 MDa in 20% glycerol. After 4 h, the range in 2% glycerol was 1.05–4.66 MDa and smaller, 0.36–2.54 MDa, in 20% glycerol. Based on these results, all subsequent HA assays were in the presence of only 2% glycerol.
Table I.
HAS makes smaller HA products in the presence of glycerol
| Glycerol (%) | HA MOLAR MASS (Mw) | |
|---|---|---|
| 1 h | 4 h | |
| 2 | 1.69 ± 0.15 (100) | 3.41 ± 0.06 (100) |
| 20 | 0.69 ± 0.04 ( 41)+ | 1.28 ± 0.12 ( 38)++ |
Unlabeled HA synthesized by WT SeHAS membranes at 1 and 4 h with 2% or 20% glycerol was analyzed by SEC-MALLS to determine the weight-average molar mass (Mw) of the HA products as the mean ± SEM (n = 3). Significance of differences between 2% and 20% glycerol were assessed by paired Student's t-tests: +P < 0.002; ++P < 0.00005.
Fig. 2.
Assay factors can decrease the size of HA made by HAS. (A) SEC refractive index values, reflecting HA amount in the elution volume, show that HA size made by HAS decreases with increasing glycerol concentration. Membranes containing WT SeHAS were incubated in the presence of 2% (solid lines) or 20% (dashed lines) glycerol for 1 h (thin lines) or 4 h (thick lines) at 30°C, as described in Methods. Table I summarizes the glycerol effect on Mw values. (B) HA size decreases as the ratio of HAS-to-substrates increases. SEC refractive index traces are shown for samples with identical buffer and UDP-sugar levels and 2 μg (solid line), 14 μg (dashed line) or 26 μg (dash-dot line) of SeHAS-membrane protein per assay; HAS content in WT type membranes is typically 8% of total protein (Kumari et al. 2002). Different sample volumes with approximately the same amounts of HA were loaded to emphasize changes in size distribution rather than amounts of HA made.
A second significant factor affecting HA size made by HAS is the concentration of UDP-sugars relative to enzyme (Baggenstoss and Weigel 2006). The substrate:HAS ratio directly controls HA size because as the number of HAS molecules engaged in synthesis increases, each enzyme competes for the same limiting substrate pool. As HAS-membrane protein increased from 2 to 14 and then 26 μg per assay (at constant substrate concentrations) the HA Mw values decreased, respectively, from 3.33 ± 0.02 MDa to 1.71 ± 0.16 MDa and then 1.12 ± 0.26 MDa (Figure 2B). The corresponding HA size distribution 96% ranges shifted from 2.34–4.17 MDa at 2 μg to 0.62–2.48 MDa with 14 μg and then 0.49–1.55 MDa with 26 μg/assay. To minimize this effect, we performed HAS assays using ≤2 μg membrane protein per 0.1 mL assay, unless noted otherwise.
Mutations at K398 in Motif 3 decrease HA product size by >90%
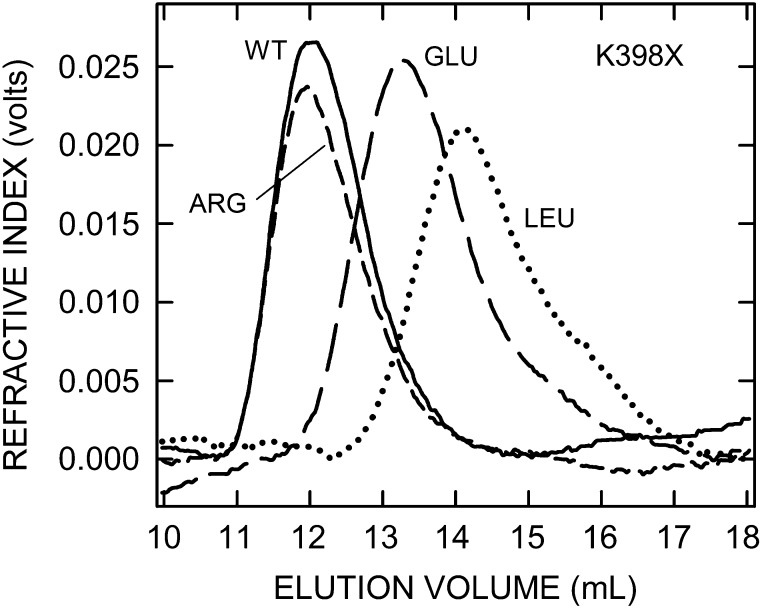
To explore the influence of the C-terminal region of SeHAS containing two tandem B-X7-B motifs (Figure 1A), we performed scanning mutagenesis at the first basic residue K398. Twelve expressed and active mutants were recovered (Table II). The most active mutant was the conservative K398R change, which produced Mw HA of 3.3 MDa (97% of WT) with nearly normal synthase activity (87% of WT). The other 11 mutants all made significantly smaller HA and had significantly less synthetic activity. SeHAS(K398E), with the opposite charge, made Mw HA of 0.7 MDa (20% WT size), but was almost inactive (1% of WT). The remaining 10 mutants had polar (Asn, Ser, Thr,), hydrophobic (Ile, Leu, Met, Val) or aromatic (Phe, Trp, Tyr) substitutions and all made smaller HA with Mw values from 250 to 480 kDa; only 8–14% of WT mass. SEC-MALLS analyses of the HA size distributions made by representative K398 mutants (Figure 3) shows the major shift to smaller sizes for HA products made by all but SeHAS(K398R), the most conservative change. The Mw and 96% HA size range for WT was 1.05–6.45 MDa and these values for K398R, K398E, and K398L were 1.15–6.75 MDa, 0.16–1.50 MDa and 78–-553 kDa, respectively. Synthase activities for these 10 mutants ranged from 27% to 64% of WT, except for K398S (14%). Their HA synthesis abilities were inhibited, but not drastically.
Table II.
Weight-average molar mass of HA synthesized by SeHAS(K398X) mutants
| K398X mutant | Mw MDa (% of WT) | Specific activity (% of WT) |
|---|---|---|
| WT | 3.38 ± 0.14 (100) | 15.4 ± 0.6 (100) |
| Arg | 3.27 ± 0.16 ( 97) | 13.4 ± 0.4 ( 87) |
| Glu | 0.68 ± 0.04 ( 20)^^ | 0.1 ± 0.0 ( 01)^^ |
| Trp | 0.48 ± 0.02 ( 13)^^ | 4.6 ± 1.4 ( 30)** |
| Val | 0.48 ± 0.02 ( 13)^^ | 8.5 ± 0.6 ( 55)** |
| Ser | 0.44 ± 0.02 ( 13)^^ | 2.1 ± 0.3 ( 14)^^ |
| Thr | 0.43 ± 0.01 ( 13)^^ | 4.4 ± 0.2 ( 29)^^ |
| Asn | 0.41 ± 0.01 ( 12)^^ | 4.5 ± 0.3 ( 29)^^ |
| Met | 0.37 ± 0.02 ( 11)^^ | 4.4 ± 0.2 ( 29)^^ |
| Phe | 0.37 ± 0.02 ( 11)^^ | 5.8 ± 0.4 ( 38)^ |
| Tyr | 0.29 ± 0.01 ( 09)^^ | 4.2 ± 0.2 ( 27)^^ |
| Ile | 0.26 ± 0.02 ( 08)^^ | 9.8 ± 0.4 ( 64)** |
| Leu | 0.25 ± 0.02 ( 08)^^ | 6.5 ± 0.3 ( 42)^ |
Unlabeled HA synthesized by membranes containing WT SeHAS or the indicated K398X mutant was analyzed, as described in Methods, to determine the weight-average molar mass (Mw) of the HA products made (ordered from largest to smallest). Values are the mean ± SEM for Mw (n = 6–11) and for specific activity (n = 4), which was determined by radio-assays (nmol GlcUA/μg HAS/h). Significant differences between mutant and WT, assessed by unpaired Student's t-tests, are indicated: **P < 0.005; ^P < 0.001; ^^P < 0.0001).
Fig. 3.
SeHAS(K398X) mutants make HA of greatly decreased size. HA was synthesized by membranes containing WT SeHAS or the indicated mutants, and samples were processed and analyzed by SEC-MALLS as described in Methods. Refractive index values reflect HA amount in the elution volume and size distributions as indicated (WT, solid line; K398R, short-dash line; K398E, long-dash line; K398L, dotted line). HA size distributions are shifted to very much smaller sizes in 11 of the 12 active K398X mutants, as shown in these examples; only the K398R mutant made normal size HA. Table II summarizes Mw values for HA made by all K398X mutants.
Most mutations at R406 in the tandem-motif do not alter HA product size
Fifteen SeHAS mutants were recovered after scanning mutagenesis at Arg406 (Table III), the middle basic residue in the tandem-motif. All mutated enzymes were active and made very large HA that was 86–110% of WT size (Mw of 3.1–4.0 MDa). The R406D mutant, with negative rather than positive charge, made the smallest HA. Significantly smaller HA was made by the R406D (86% of WT) and the R406A (91% of WT) mutants and significantly larger HA was made by R406I (110% of WT) and R406P (106% of WT). Consistent with these results, SEC profiles for the HA product size distributions indicated only slight differences among the mutants (Figure 4); e.g. 96% size ranges for R406D, R406I and R406L were 1.64–4.74 MDa, 2.73–4.63 MDa and 2.74–4.90 MDa, respectively. Unexpectedly, changes in synthase activities among all mutants were toward greater activity; only three were significantly less active (70––80% of WT), whereas eight others had significantly greater activity than WT (121–177% of WT). Most strikingly, among the 11 mutants with no significant changes in HA size, seven mutants showed significantly altered rates of HA synthesis, with five increasing and two decreasing (Table III).
Table III.
Weight-average molar mass of HA synthesized by SeHAS(R406X) mutants
| R406X mutant | Mw (MDa) | Specific activity (% WT) |
|---|---|---|
| Ile | 3.99 ± 0.11 (110)** | 13.7 ± 0.3 (121)* |
| Pro | 3.87 ± 0.01 (106)** | 8.8 ± 0.4 ( 78)* |
| Gly | 3.83 ± 0.16 (105) | 20.1 ± 0.6 (177)^ |
| Trp | 3.72 ± 0.11 (102) | 11.5 ± 0.4 (102) |
| WT | 3.64 ± 0.03 (100) | 11.3 ± 0.4 (100) |
| Phe | 3.62 ± 0.18 ( 99) | 15.1 ± 0.4 (133)** |
| Val | 3.56 ± 0.16 ( 98) | 19.0 ± 1.1 (168)** |
| Thr | 3.56 ± 0.06 ( 98) | 13.9 ± 0.9 (122) |
| Ser | 3.54 ± 0.12 ( 97) | 13.9 ± 0.8 (123)* |
| Met | 3.50 ± 0.02 ( 96) | 9.1 ± 0.1 ( 80)* |
| Leu | 3.42 ± 0.35 ( 94) | 13.5 ± 1.1 (119) |
| Lys | 3.41 ± 0.12 ( 94) | 8.0 ± 0.3 ( 70)** |
| Asn | 3.41 ± 0.31 ( 94) | 16.1 ± 0.5 (142)** |
| Cys | 3.38 ± 0.24 ( 93) | 12.3 ± 0.5 (108) |
| Ala | 3.32 ± 0.06 ( 91)^ | 16.0 ± 0.1 (141)^ |
| Asp | 3.13 ± 0.25 ( 86)* | 17.0 ± 0.4 (150)^ |
Unlabeled HA synthesized by membranes containing WT SeHAS or the indicated K406X mutant was analyzed by SEC-MALLS to determine the weight-average molar mass (Mw) of the HA products made (ordered from largest to smallest) and the specific synthase activity. Values are the mean ± SEM for Mw (n = 2–10) and for specific activity (n = 4), which was determined by radio-assays (nmol GlcUA/μg HAS/h). Significant differences between mutant and WT, assessed by unpaired Student's t-tests, are indicated: *P < 0.05; **P < 0.005; ^P < 0.001.
Fig. 4.
SeHAS(R406X) mutants make HA of near-WT size. HA was synthesized, processed and analyzed as in Figure 3. Refractive index values reflect HA amount and size distributions in the elution volume for the mutants indicated (WT, solid line; R406I, short-dash line; R406L, long-dash line; R406D, dotted line). Table III summarizes Mw values made by all R406X mutants.
SeHAS truncation mutants within the tandem-motif lose 98% HA synthesis activity, yet still make MDa HA products
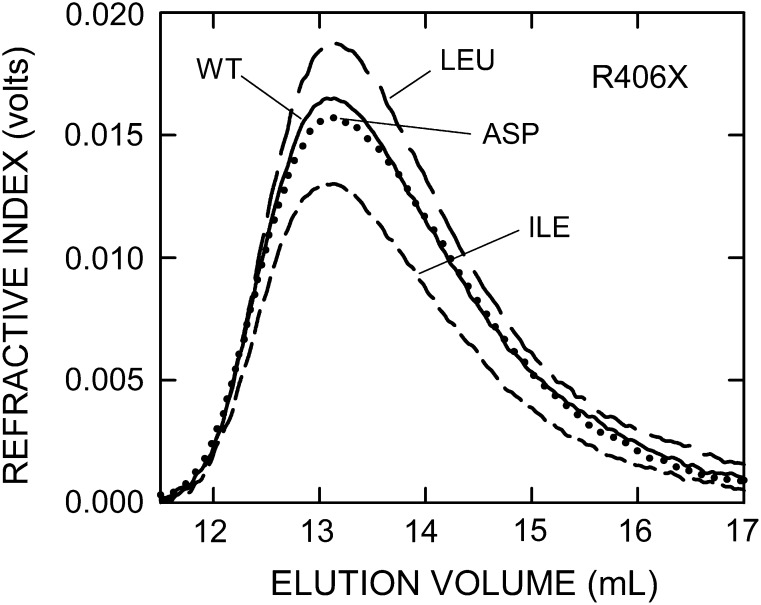
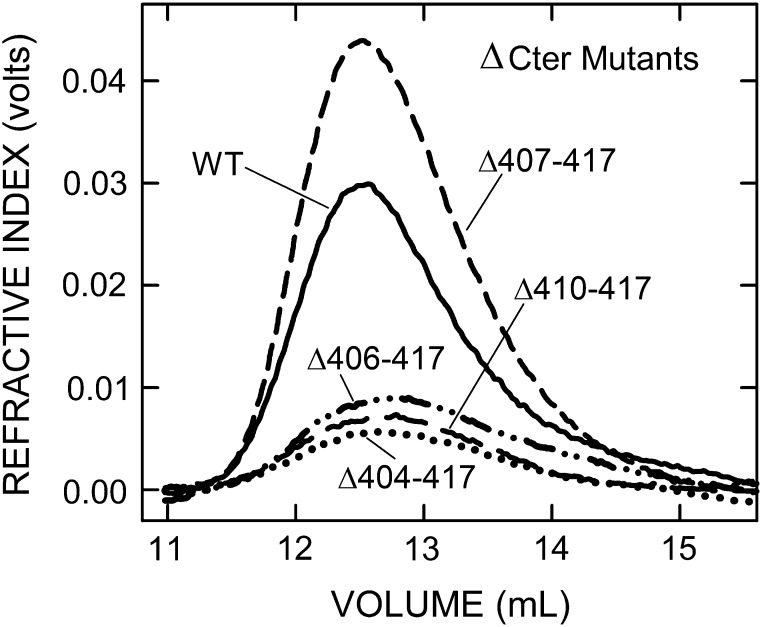
In addition to creating the above scanning mutants in the tandem-motif, we made a series of stepped truncations in the B-X7-B-X7-B region. The seven mutants either retained motif 3 and all, part or none of motif 4 or retained part or none of motif 3 and none of motif 4 (Table IV). Deletion of the last four C-terminal residues created a ∆414–417 mutant, which had good activity (58% of WT) and made large 2.8 MDa HA (81% of WT). Thus, loss of the KKLL417 C-terminus, including the last K414 of the tandem-motif, significantly decreased, but was not critical for, the control of HA size and the rate of HA synthesis. However, when the C-terminal R413 of active SeHAS(∆414–417) was deleted, the resulting SeHAS(∆413–417) was well expressed (50% of WT), but the enzyme was inactive; no HAS activity was detected. The remaining motif 4 sequence was further shortened to make SeHAS(∆410–417), which contains motif 3 and less than half of motif 4. This variant was well expressed (38% of WT) and made large HA (Mw = 2.1 MDa, 63% of WT size), but its HA synthesis activity was decreased 99.6% compared to WT. Consistent with the ∆410–417 mutant (and the others noted below) almost being inactive, much more membrane protein was needed to produce sufficient HA for analysis (Figure 5).
Table IV.
Effect of C-terminal truncations on HAS expression, specific activity, and HA size
| C-terminal sequence | Variant | HA Mass Mw (% WT) | Activity (% WT) |
|---|---|---|---|
| QPLKLYSLFTIRNADWGTRKKLL | WT | 3.39 ± 0.10 (100) | 53.0 ± 1.4 (100) |
| QPLKLYSLFTIRNADWGTR | Δ414–417 | 2.76 ± 0.12 (81)* | 30.8 ± 1.7 (58)^^ |
| QPLKLYSLFTIRNADWGT | Δ413–417 | ND | ND |
| QPLKLYSLFTIRNAD | Δ410–417 | 2.13 ± 0.12 (63)^^ | 0.20 ± 0.01 (0.4)^^ |
| QPLKLYSLFTIR | Δ407–417 | 2.40 ± 0.11 (71)^^ | 1.08 ± 0.10 (2.0)^^ |
| QPLKLYSLFTI | Δ406–417 | 1.90 ± 0.20 (56)^^ | 0.21 ± 0.02 (0.4)^^ |
| QPLKLYSLF | Δ404–417 | 2.24 ± 0.25 (66)^^ | 0.21 ± 0.03 (0.4)^^ |
| QPL | Δ398–417 | NE | NE |
Values for Mw are the mean MDa ± SEM (n = 6–11) and values for specific activity determined by MALLs are the mean nmol HA disaccharide/μg HAS/h ± SEM (n = 3–16). Boldface font denotes the basic residues comprising the tandem-motif as in Figure 1A. Significant differences between mutant and WT, assessed by unpaired Student's t-tests, are indicated (ND, not detected; NE, not expressed): *P <0.05; ^^P <0.0001.
Fig. 5.
Almost inactive C-terminal truncation mutants still make MDa HA. HA was synthesized, processed and analyzed as in Figure 3. Refractive index traces indicate HA amount and size distributions for WT (solid line) and four ∆Cter mutants: ∆410–417 (long-dash line), ∆407–417 (short-dash line), ∆406–417 (dash-dot line), ∆404–417 (dotted line). Table IV summarizes Mw values made by all the ΔCter mutants.
Further truncation to give only motif 3 at the C-terminus, SeHAS(∆407–417), also gave an enzyme that was well expressed (75% of WT), made large HA (Mw = 2.4 MDa, 71% of WT; Figure 5) but showed greatly impaired activity (98% decreased compared to WT). Removal of R406 and then TI405 gave two SeHAS mutants, similar to the (∆410–417) and (∆407–417) mutants, with similar expression levels (38% of WT), specific activities (0.4% of WT) and ability to make large HA (56–66% of WT). The final mutant, (∆398–417), lacked the entire tandem-motif and the C-terminal KLL417 sequences (Table IV). SeHAS(∆398–417) was not expressed and is presumed to be unstable, indicating that the HAS tandem-motif region is likely critical for stability. The 96% range of HA sizes, compared to WT (2.35–3.96 MDa), for the four mutants with ≤ 2% WT activity (Figure 5), respectively, were 1.14–2.94 MDa (Δ410–417), 1.29–3.35 MDa (Δ407–417), 0.56–2.74 MDa (Δ406–417) and 0.92–2.51 MDa (Δ404–417).
The pattern of HA synthesis-function and size-function retention in HAS tandem-motif mutants shows that these are two separate enzyme functions
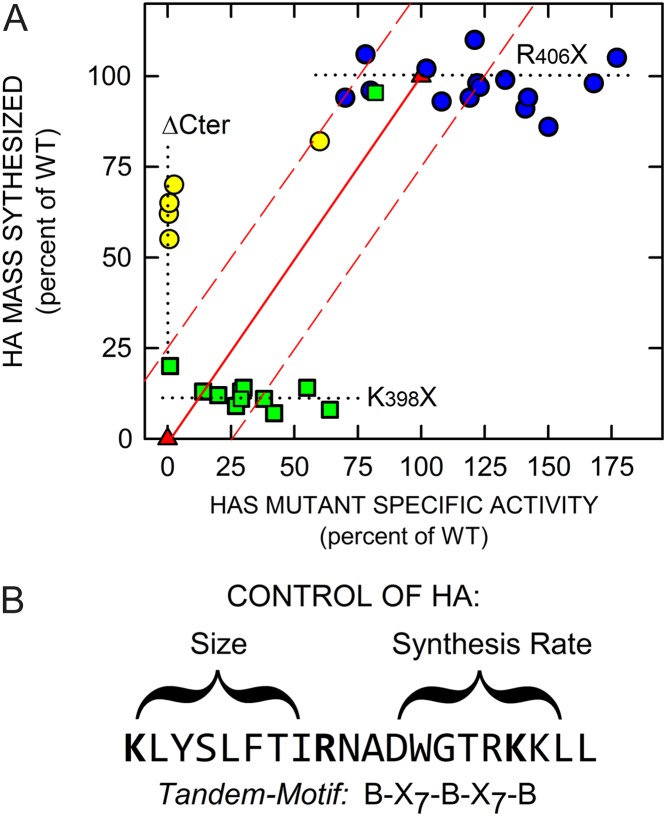
A scatter-plot of HAS enzyme activity versus HA product size highlights that the K398X, R406X and ΔCter mutants represent three different types of mutant enzyme consequences due to sequence changes in the tandem-motif region (Figure 6A). Almost all members of each mutant group were clustered along a different axis extreme: (i) 11 of 12 K398X mutants (green squares) made small HA (8–20% of WT) but of these 11 mutants, eight had 27–64% WT activity; (ii) five ΔCter mutants (yellow circles) made 56–82% WT HA size but 4 of these mutants had extremely low activity (≤ 2% of WT); and (iii) R406X mutants (blue circles) made HA of WT size (86–110%) and had high specific activity (70–177% of WT). One ΔCter mutant (Δ414–417) and the conservative K398R mutant were exceptions to this clustering. These two and 17 other R406X and K398X mutants were within ±25% of the predicted relationship for co-dependence of activity and size (red-lined box, Figure 6A). Except for the axis extremes, no mutants plotted near the line for expected co-dependence (red triangles). Thirteen other mutants were well outside this area, including: four ΔCter mutants (Δ410–417, Δ407–417, Δ406–417 and Δ404–417), two K398 (K398V and K398I) and five R406 mutants (R406G, R406V, R406N, R406A and R406D). Overall, these results indicate that the first B-X7-B motif is more involved in HA size control and the second B-X7-B motif is more involved in controlling the rate of HA synthesis (Figure 6B).
Fig. 6.
HA synthesis rate and HA size control are independent enzyme functions. (A) The specific activity and HA product size (Mw) for individual K398 (green squares), R406 (blue circles) and ΔCter (yellow circles) SeHAS mutants are shown in a scatter-plot. The three mutant groups (with only a few exceptions) cluster either (i) at large HA (55–70% of WT; yellow) with very low activity, (ii) at very small HA (8–20% of WT; green) with good activity (20–75% of WT) or (iii) at near-WT HA size with very high activity (70–177% of WT; blue). The solid red line (red triangles) indicates the expected relationship if the HAS functions that control HA synthesis and HA size are co-dependent; they should both vary in a coordinated, likely linear, way between 0% and 100%. Some mutants in each group are within (19 total) and some are outside (13 total) a region (red dashed lines), that encompasses a ±25% activity zone. The dotted lines highlight plot areas with mutants outside this zone. (B) The effects of HAS mutations in the tandem-motif are represented. The results summarized in panel A indicate that the first motif preferentially controls HA size, whereas the second motif preferentially controls the rate of HA synthesis. This figure is available in black and white in print and in color at Glycobiology online.
Discussion
The HAS mechanism of HA retention during processive synthesis is known but how HAS controls HA size is unknown
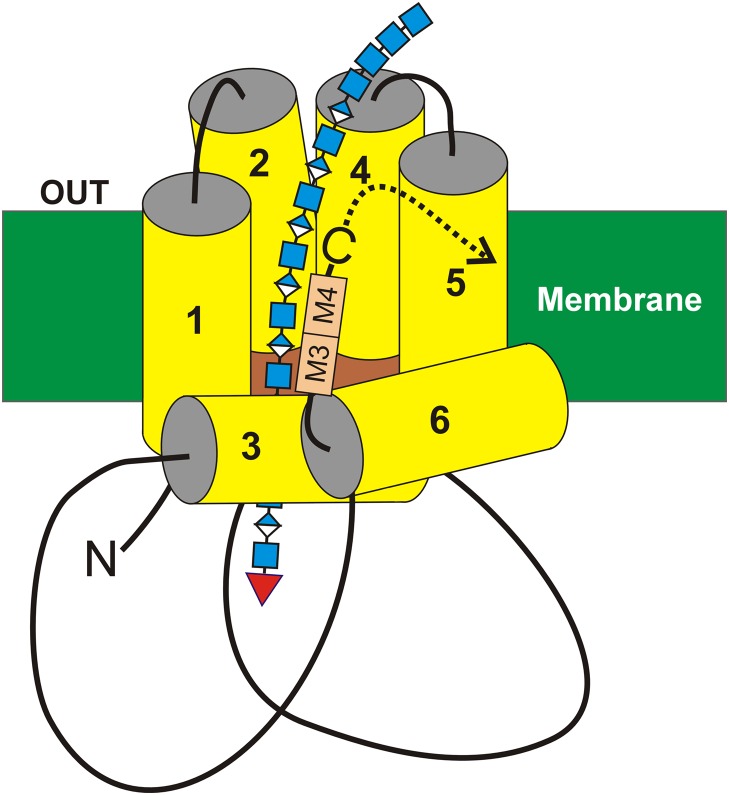
Since cloning and expressing the first HAS (DeAngelis et al. 1993), we have sought to understand how these enzymes regulate the size of assembled HA-UDP chains and how HA·HAS complexes can stay together for hours, without dissociating, during a processive synthetic process that occurs at the reducing end (Prehm 1983; Bodevin-Authelet et al. 2005; Tlapak-Simmons et al. 2005). Many RNA and DNA enzymes (e.g. polymerases and topoisomerases) have evolved to solve the dilemma of staying associated with their substrate–product, despite weak interactions between the two, by creating topological or spatial constraints so that enzyme and product cannot freely diffuse away even when they dissociate. The original proposal that HAS translocates the growing HA-UDP chain through an intra-protein pore (Tlapak-Simmons et al. 1999a) has been verified (Hubbard et al. 2012; Medina et al. 2012). We now understand that HAS enzymes function either as monomers (Tlapak-Simmons et al. 1998) or oligomers (Bart et al. 2015) and their processive mechanism utilizes an intra-HAS pore or deep protein cleft (Kumari et al. 2006) within the plasma membrane (Figure 7) that serves simultaneously to retain and to translocate the growing HA-UDP chain to the cell exterior (Hubbard et al. 2012; Medina et al. 2012; Bi et al. 2015; Weigel 2015). Many questions still remain regarding the mechanism of synthesis and what causes and controls HA release and HA product size.
Fig. 7.
A model for interaction of the HAS tandem-motif region with HA. The organization (Heldermon et al. 2001a; Kumari and Weigel 2005; Kumari et al. 2006) of the four transmembrane (MD1 & MD2 and MD4 & MD5) and two (MD3 and MD6) amphipathic MDs of SeHAS (yellow) are shown with the growing HA-UDP chain, containing alternating GlcNAcβ1,4 (blue squares) and GlcUAβ1,3 (blue-white diamonds) attached to UDP (red inverted triangle) at the reducing end. Preliminary results indicate that the nonreducing end contains a chitin oligomer cap with four GlcNAc residues, which is the primer on which HA synthesis is initiated (Weigel 2015; Weigel et al. 2015). The relative positions of the tandem B-X7-B motifs (A), M3 and M4, are indicated (tan boxes). In streptococcal HAS enzymes the C-terminus (-C) is just after the tandem-motif, whereas in vertebrate Class I HAS enzymes the additional ~140 amino acid sequence continues after the tandem-motif (dotted curved arrow). This figure is available in black and white in print and in color at Glycobiology online.
Unlike other glycosyltransferases, overall HAS synthase activity includes the control of both the rate of HA synthesis (i.e. sugar assembly; glycoside linkages/min/enzyme) and the HA size produced. These two enzyme functions are sufficiently independent that they are uncoupled (differentially rather than coordinately affected) by mutations at conserved Cys residues (Weigel and Baggenstoss 2012) or the C-terminus, as shown herein (Table IV and Figure 6). Thus, each function could be regulated and controlled by a separate enzyme sub-mechanism. The hypothesis for the present study was that mutations that alter HAS structure and function could mimic the effect of covalent, allosteric or other regulatory mechanisms, which control the two HAS functions that assemble HA and create different HA sizes. Multiple groups have created streptococcal (Heldermon et al. 2001b; Kumari et al. 2002, 2006; Kumari and Weigel, 2005; Weigel and Baggenstoss 2012) and vertebrate (Bart et al. 2015; Pummill and DeAngelis 2003; Yoshida et al. 2000; Rilla et al. 2005; Spicer et al. 2002; Spicer and Nguyen 1999) HAS mutants to examine Class I HAS function, but most studies determined synthetic activity and not HA size. Studies in which HA size was assessed found that site-specific HAS mutants produced either slightly smaller or larger HA (Kumari et al. 2002, 2006; Pummill and DeAngelis 2003; Weigel and Baggenstoss 2012). Mammalian HAS activities are regulated by localization at the plasma versus intracellular membranes (Rilla et al. 2005), covalent modification (Goentzel et al. 2006; Tammi et al. 2011; Vigetti et al. 2011; Vigetti and Passi 2014; Viola et al. 2015) and likely also by small molecule allosteric modulators and protein regulatory partners (Bart et al. 2015); these studies also assessed HA synthesis ability, but not HA size.
Biological responses to large versus small HA are very different
The cell signaling capabilities of smaller (e.g. 100–400 kDa), but not larger (MDa), HA in inflammatory diseases, tumorigenesis and metastasis (Stern et al. 2006; Simpson et al. 2015) has stimulated interest in understanding how such lower mass HA arises in vivo. Many studies infer that smaller HA arises by the degradation of extracellular large MDa HA. It is also possible, however, that one or more HAS enzymes directly synthesizes smaller (kDa) HA. The three mammalian HASs produce different HA size distributions with HAS3 producing the smallest HA, down to 100 kDa (Brinck and Heldin 1999; Itano et al. 1999). Thus, activation of HAS3 could result in the synthesis and release of signaling-competent HA without hyaluronidase action. Few studies have examined changes in both HA content and HA size distribution in tissues undergoing stress; most focus on changes in gene and protein expression of HAS1-3 and several hyaluronidases. One study of mouse lung ischemia found a 4-fold increase in 30–495 kDa HA within 4 h of ligation, which was accompanied by elevated HAS1 and HAS2 expression with no changes in hyaluronidase activity (Eldridge et al. 2011). Such results indicate that in some cases HAS activity might be more important than hyaluronidase-mediated degradation in increasing levels of smaller signaling HA in vivo. Our results indicate that different HA sizes made by HAS1-3 might be due to how the three enzymes are differentially modified and regulated, as well as to intrinsic differences in protein sequences and structure.
Mutations in the putative HA-binding motifs conserved in the HAS family can alter HA synthesis rate and HA size
The tandem-motif sequence (Figure 1B), present within the 23 amino acid C-terminus of SeHAS following MD6 (Heldermon et al. 2001a), is highly conserved among Class I HASs (Figure 1A), indicating that this region is important for HAS activity, HA translocation and/or HA size regulation. To test this, we examined scanning mutants at basic residues Lys398 and Arg406 and truncation mutants that eliminated parts or all of the tandem-motif. All 15 Arg406 mutants made large HA (86–110% of WT), whereas only one of 12 active Lys398 mutants made MDa HA (K398R, 97% of WT). This result is not surprising, since K398R maintains the positive charge in the B-X7-B motif and would be expected to have near normal function. In contrast, however, the other 11 K398X mutants made much smaller 250–680 kDa HA (7–20% of WT). A K398stop mutant was expressed but inactive, making no HA. The results indicate that Lys398, which is conserved in all Class I HASs (Figure 1A), may be a particularly important residue for control of HA size. Truncations of the tandem-motif, especially the more C-terminal B-X7-B motif, had dramatic negative effects on HA synthesis rate without altering the ability of HAS to make MDa HA (Figure 5 and Table IV). We conclude that different portions of the HAS tandem-motif region are involved in both the control of HA synthesis rate and the control of HA size and that these two discrete functions can be uncoupled by site-specific mutations at Lys398 or truncation of the K398–K414 sequence (Figure 6A). The present results confirm an earlier study (Weigel and Baggenstoss 2012) that HAS uses two discrete functions to control HA synthesis rate and HA size.
Based on the present results, the first SeHAS KLYSLFTIR406 motif may be more involved in HA size control and the following RNADWGTRK414 motif may be more involved in HA synthesis rate control (Figure 6B). Of course both functions, together with the translocation function, are necessary for HAS to synthesize HA and they can only be semi-independent, since HAS is inactive if either one of these functions is inactive. The conserved vertebrate tandem-motif consensus sequence, KMFAIATINKAGWGTSGRK, is a B-X8-B-X7-BB sequence, not a perfect B-X7-B-X7-B motif; the SeHAS tandem-motif (B-X7-B-X7-BB) with an extra K is also not canonical. The presence of conserved potential HA-binding B-X7-B motifs is suggestive, but does not mean that a motif is accessible or competent to bind HA strongly, e.g. as occurs with RHAMM (Yang et al. 1993). The level of sequence conservation across all vertebrate species, however, indicates that this is an important region for HAS function or stability, perhaps with the same functional characteristics as in SeHAS (Figure 6B).
HAS enzymes cannot be HA-binding proteins, despite the conserved putative HA-binding motifs in the enzyme family
Despite the presence of potential HA binding B-X7-B motifs, HAS enzymes cannot actually be strong HA binding proteins. Consistent with the known mechanism of processive HA synthesis (DeAngelis and Weigel 1994; Itano et al. 1999; Yoshida et al. 2000; Pummill and DeAngelis 2003; Kumari et al. 2006; Weigel 2015) and typical enzyme–substrate interactions, HAS must bind HA-UDP only in a transient and rapid on–off manner. If HAS binding to HA-UDP is of high affinity, then synthesis will be necessarily slow and highly dependent on the koff for dissociation in order for the next synthetic cycle to proceed. The in vitro rate of HA biosynthesis is 10–30 sugars/s for streptococcal HASs (DeAngelis and Weigel 1994) and 3 sugars/s for XlHAS1 (Pummill and DeAngelis 2003). If in vivo synthesis rates for mammalian HAS1-3 enzymes are ~10 sugars/s, then assembly of 12,000 sugars into a 2.3 MDa HA chain takes 20 min. Since each catalytic cycle requires that the HA-UDP product must transiently dissociate from HAS, then HA-UDP release occurs ≤ every 0.1 s and the estimated t1/2 for HAS•HA-UDP complex dissociation is 7 ms, which is very fast.
Thus, the known enzymology of HAS dictates that it cannot be, and is not, an HA-binding protein. However, it seems very feasible that these enzymes could utilize the tandem-motif domain to mediate weaker, yet specific, interactions with the nascent HA region during repetitive cycles of disaccharide assembly and translocation through an intra-HAS pore created by the six MDs (Figure 7) and associated lipids. It is also likely that the bound sugar conformations and relative orientations during synthesis may not be the same as for the free polymer, so that interactions between the tandem-motif and bound HA may be atypical for B-X7-B motifs in proteins that bind HA tightly. The conserved tandem-motif region appears to be involved in a complicated way with two core HAS functions, the assembly of sugar units at a very high rate and the retention of a growing HA-UDP chain long enough to create a >3 MDa product.
The present results demonstrate that HAS can make HA that is much smaller than normal (e.g. 10% of regular size) if modified at a single residue. Conceptually, in terms of protein conformation and function, there is no difference between the effect of a point mutation and typical enzyme regulatory mechanisms. Thus, the results support the concept that HA size could be controlled and changed in vivo as a separate regulated HAS function. Future studies of this conserved region in human HAS1-3 (e.g. K451-I471 in HsHAS2) are needed to confirm this prediction. Genomic screening of the tandem-motif region might also be productive in identifying potentially important mutations for human diseases or syndromes related to decreases in extracellular HA size (e.g. auto-inflammatory or abnormal tissue matrix syndromes).
Materials and methods
Materials and reagents
Media components were from Difco (Fisher Scientific). Uridine 5’-diphospho-[14C]glucuronic acid (UDP-[14C]GlcUA) was from Perkin-Elmer (Boston, MA). UDP-GlcUA, UDP-N-acetylglucosamine (UDP-GlcNAc) and calf intestinal alkaline phosphatase were from Sigma-Aldrich (St. Louis, MO). UltraPure grade glycerol was from Invitrogen (Carlsbad, CA). Plasmid pKK223-3 was from GE Healthcare and Escherichia coli, SURETM cells were from Stratagene (San Diego, CA). Oligonucleotide primers were obtained from Integrated DNA Technologies (Ames, IA). Other reagents were the highest grade available from Sigma-Aldrich.
Mutagenesis of SeHAS
The SeHAS gene with a fusion at the 3’-end encoding a His6 tail (SeHAS-His6) was cloned into the pKK223 vector (Tlapak-Simmons et al. 1999a). Site-directed scanning mutagenesis was carried out using the QuickChange method according to the manufacturer's instructions, as previously described (Kumari et al. 2006), using primers containing randomized nucleotides at the specific codon of interest. Primers for ΔCter mutants were designed with a stop-codon at the desired positions. The pKK223 plasmid containing the SeHAS-His6 gene was grown in E. coli SURE2 cells, purified by CsCl isopycnic centrifugation and used as the template for primer extension reactions with the appropriate primers. PCR conditions using pfu ultra DNA polymerase were 16 cycles at 95°C for 20 s, 55°C for 20 s, and 69°C for 7 min after which the PCR reaction mixture was treated with DpnI to digest methylated parental DNA. The undigested mutated DNA was transformed into SURE2 cells, and ampicillin-resistant colonies were screened in successive stages for the desired mutations by confirming expression of full-length plasmid based on agarose gel electrophoresis, then the expression of SeHAS by Western analysis, and finally the presence of the desired mutation by single-pass DNA sequencing over the site of interest. Final candidates (30% of colonies tested) were then sequenced in both directions to confirm that the complete open reading frames and promoter regions were correct.
Determination of SeHAS protein content in membranes and normalization of SeHAS activity
E. coli membranes containing either WT or mutant SeHAS were solubilized and subjected to SDS-PAGE in 10% (w/v) gels, and SeHAS protein in each membrane preparation was quantified by image analysis of Coomassie Blue-stained gels using a FluorchemTM8000 (Alpha Innotech Corp.), as described previously (Kumari et al. 2002). Integrated values for SeHAS bands in membranes were compared with standard curves using purified SeHAS (Tlapak-Simmons et al. 1999a) to estimate SeHAS protein/mg of membrane protein. These data were then used to normalize variant HAS activity in membranes compared with WT.
HA synthesis assays
Membranes containing SeHAS were prepared and assayed for HAS activity (Weigel et al. 2013) to determine specific activities either using UDP-[14C]GlcUA (Tlapak-Simmons et al. 1999b; Kumari and Weigel 2005) or by integration of refractive index values obtained from SEC-MALLS analyses of non-radiolabeled samples (Baggenstoss and Weigel 2006; Weigel and Baggenstoss 2012). Final volumes for the radioactive or MALLS assay samples were 0.1 and 0.5–1.0 mL, respectively. Membranes were suspended by brief sonication at 4°C and added to 2 mL microcentrifuge tubes to a final concentration of ≤2 μg membrane protein per 0.1 mL assay volume (except the nearly inactive ΔCter and K398X mutants, which required 5–20 times more protein to obtain sufficient HA). Assay buffer contained 18.75 mM sodium phosphate and 6.25 mM potassium phosphate, pH 7.0, 75 mM NaCl, 2% (v/v) glycerol, 0.1 mM EDTA, 1.0 mM DTT, 0.1 mM PMSF, 1.0 μM pepstatin, and 2 μM leupeptin. UDP-GlcUA and UDP-GlcNAc were added to 1 mM final concentrations, the mixture was incubated at 30°C for 10 min. For MALLS (HA size) assays, calf intestinal alkaline phosphatase was added (Baggenstoss and Weigel 2006) to a final concentration of 0.02 U/μL. For radiolabel assays, UDP-[14C]GlcUA was also added to 0.7 μM.
The HA synthesis reactions were then initiated by the addition of MgCl2 to a final concentration of 20 mM and incubated for either 1 h (for radiolabel assays) or 4 h (for MALLS assays) at 30°C in a vibrating Taitec mixer (San Jose, CA). For MALLS assays, synthesis was terminated by adding EDTA and UDP to final concentrations of 40 mM and 10 mM, respectively, chilling on ice for ~20 min and then heating in a 100°C sand bath (for 1 min/0.1 mL) to inactivate HAS. Radiolabeled assay samples were quenched by adding SDS to 2% (w/v) and then analyzed by descending paper chromatography (DeAngelis and Weigel 1994; Weigel et al. 2013). The synthesis of HA was monitored over time to ensure that the final HA size made by each mutant was not limited by the incubation time. In all cases, steady-state size ranges were reached within 2–4 h in vitro.
SEC-MALLS analyses to determine HA weight-average mass, size distribution, concentration and specific synthesis activity
Analyses were performed by chromatographic separation of samples using either one TSK-Gel G6000PWXL column (TOSOH-BIOSEP) or one or two (in series) PLaquagel-OH60 (Polymer Labs) columns at 22°C at a flow rate of 0.4–0.5 ml/min in 50 mM sodium phosphate, pH 7.0, 150 mM NaCl, 0.05% sodium azide. MALLS analysis of samples were performed continuously on the column eluate, as it passed through a DAWN DSP Laser Photometer in series with an OPTILAB DSP Interferometric Refractometer (both from Wyatt Technologies, Inc., Santa Barbara, CA). Data were analyzed, and weight-average mass (Mw) values were calculated, using Astra v4.73, a dn/dc value of 0.153, an A2 value of 0.0023, and either first-order Zimm or second-order Berry fits, depending on the HA size. First-order Zimm fits had less uncertainty in analysis of data for HA of <2 MDa, whereas second-order Berry analysis gave better fits for HA of >2 MDa. Samples (200 μL) in microcentrifuge tubes were incubated in a 100°C bath for 2 min prior to injection (to 65°C after 2 min). Column performance was monitored by regular analysis of commercial HA as a reference standard. Column washing, if needed, with 2 mL 0.1% SDS in 30% methanol removed accumulated lipid and restored normal performance and fractionation. High HA concentrations were avoided, since artificially high mass values result, likely due to chain entanglement (Baggenstoss and Weigel 2006). Since MALLS inherently provides concentration data, samples were easily monitored to be ≤0.05 mg/mL HA to minimize concentration-dependent artifacts. Specific activity was determined based on the synthesis time, the estimated membrane HAS content, as noted above, and the amount of HA disaccharides in the sample. Between the radioactive and MALLS assays, the present study might not have detected HA products that were 10–50 sugars long. However, since this is a narrower size range (5-fold) compared to the typical 20-fold HA size range observed for WT or any of the mutants, the two assays essentially cover the range of all HA sizes possibly made by HAS.
Acknowledgements
We thank Christina Baron and Amy Padgett-McCue for technical assistance with many aspects of this study.
Authors' contributions
P.H.W. conceived and coordinated the study and all co-authors (B.A.B., E.N.H., A.P.M., L.N. and J.L.W.) created or characterized mutants or performed experiments, contributed data and helped generate figures and tables. P.H.W. wrote the manuscript and all authors have approved the final version of the manuscript.
Funding
This research was supported in part by National Institutes of Health grants GM035978 (to P.H.W.) and HL130864 (to E.N.H.) and by the Ed Miller Chair in Molecular Biology (P.H.W.).
Conflict of interest
The authors declare no competing financial interests.
Abbreviations
ΔCter, C-terminal truncation; GlcNAc, N-acetyl-D-glucosamine; GlcUA, D-glucuronic acid; HA, hyaluronic acid, hyaluronate, hyaluronan; HAS, HA synthase; MD, membrane domain; SEC-MALLS, size exclusion chromatography-multiangle laser light scattering; SeHAS, Streptococcus equisimilus HAS; uridine diphosphate, UDP.
References
- Amemiya K, Nakatani T, Saito A, Suzuki A, Munakata H. 2005. Hyaluronan-binding motif identified by panning a random peptide display library. Biochim Biophys Acta. 1724:94–99. [DOI] [PubMed] [Google Scholar]
- Baggenstoss BA, Weigel PH. 2006. Size exclusion chromatography-multiangle laser light scattering analysis of hyaluronan size distributions made by membrane-bound hyaluronan synthase. Anal Biochem. 352:243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bart G, Vico NO, Hassinen A, Pujol FM, Deen AJ, Ruusala A, Tammi RH, Squire A, Heldin P, Kellokumpu S, et al. 2015. Fluorescence resonance energy transfer (FRET) and proximity ligation assays reveal functionally relevant homo- and heteromeric complexes among hyaluronan synthases HAS1, HAS2, and HAS3. J Biol Chem. 290:11479–11490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra SP, Perez-Mediavilla LA, Weldon E, Locatelli-Hoops S, Senanayake P, Notari L, Notario V, Hollyfield JG. 2008. Pigment epithelium-derived factor binds to hyaluronan. Mapping of a hyaluronan binding site. J Biol Chem. 283:33310–33320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y, Hubbard C, Purushotham P, Zimmer J. 2015. Insights into the structure and function of membrane-integrated processive glycosyltransferases. Curr Opin Struct Biol. 34:78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodevin-Authelet S, Kusche-Gullberg M, Pummill PE, DeAngelis PL, Lindahl U. 2005. Biosynthesis of hyaluronan: Direction of chain elongation. J Biol Chem. 280:8813–8818. [DOI] [PubMed] [Google Scholar]
- Brinck J, Heldin P. 1999. Expression of recombinant hyaluronan synthase (HAS) isoforms in CHO cells reduces cell migration and cell surface CD44. Exp Cell Res. 252:342–351. [DOI] [PubMed] [Google Scholar]
- Day AJ. 1999. The structure and regulation of hyaluronan-binding proteins. Biochem Soc Trans. 27:115–121. [DOI] [PubMed] [Google Scholar]
- DeAngelis PL, Papaconstantinou J, Weigel PH. 1993. Molecular cloning, identification and sequence of the hyaluronan synthase gene from group A Streptococcus pyogenes. J Biol Chem. 268:19181–19184. [PubMed] [Google Scholar]
- DeAngelis PL, Weigel PH. 1994. Immunochemical confirmation of the primary structure of streptococcal hyaluronan synthase and synthesis of high molecular weight product by the recombinant enzyme. Biochem. 33:9033–9039. [DOI] [PubMed] [Google Scholar]
- Eldridge L, Moldobaeva A, Wagner EM. 2011. Increased hyaluronan fragmentation during pulmonary ischemia. Am J Physiol Lung Cell Mol Physiol. 301:L782–L788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goentzel BJ, Weigel PH, Steinberg RA. 2006. Recombinant hyaluronan synthase 3 is phosphorylated in mammalian cells. Biochem J. 396:347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hascall VC, Wang A, Tammi M, Oikari S, Tammi R, Passi A, Vigetti D, Hanson RW, Hart GW. 2014. The dynamic metabolism of hyaluronan regulates the cytosolic concentration of UDP-GlcNAc. Matrix Biol. 35:14–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldermon C, DeAngelis PL, Weigel PH. 2001. a. Topological organization of the hyaluronan synthase from Streptococcus pyogenes. J Biol Chem. 276:2037–2046. [DOI] [PubMed] [Google Scholar]
- Heldermon CD, Tlapak-Simmons VL, Baggenstoss BA, Weigel PH. 2001. b. Site-directed mutation of conserved cysteine residues does not inactivate the Streptococcus pyogenes hyaluronan synthase. Glycobiology. 11:1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard C, McNamara JT, Azumaya C, Patel MS, Zimmer J. 2012. The hyaluronan synthase catalyzes the synthesis and membrane translocation of hyaluronan. J Mol Biol. 418:21–31. [DOI] [PubMed] [Google Scholar]
- Itano N, Kimata K. 2002. Mammalian hyaluronan synthases. IUBMB Life. 54:195–199. [DOI] [PubMed] [Google Scholar]
- Itano N, Sawai T, Yoshida M, Lenas P, Yamada Y, Imagawa M, Shinomura T, Hamaguchi M, Yoshida Y, Ohnuki Y, et al. 1999. Three isoforms of mammalian hyaluronan synthases have distinct enzymatic properties. J Biol Chem. 274:25085–25092. [DOI] [PubMed] [Google Scholar]
- Jean L, Mizon C, Larsen WJ, Mizon J, Salier JP. 2001. Unmasking a hyaluronan-binding site of the BX7B type in the H3 heavy chain of the inter-α-inhibitor family. Euro J Biochem. 268:544–553. [DOI] [PubMed] [Google Scholar]
- Jokela TA, Karna R, Makkonen KM, Laitinen JT, Tammi RH, Tammi MI. 2014. Extracellular UDP-glucose activates P2Y14 receptor and induces signal transducer and activator of transcription 3 (STAT3) Tyr705 phosphorylation and binding to hyaluronan synthase 2 (HAS2) promoter, stimulating hyaluronan synthesis of keratinocytes. J Biol Chem. 289:18569–18581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokela TA, Makkonen KM, Oikari S, Karna R, Koli E, Hart GW, Tammi RH, Carlberg C, Tammi MI. 2011. Cellular content of UDP-N-acetylhexosamines controls hyaluronan synthase 2 expression and correlates with O-linked N-acetylglucosamine modification of transcription factors YY1 and SP1. J Biol Chem. 286:33632–33640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson CB, Knudson W. 2004. Hyaluronan and CD44: Modulators of chondrocyte metabolism. Clin OrthopS. 152–S162. [PubMed] [Google Scholar]
- Kumari K, Baggenstoss BA, Parker AL, Weigel PH. 2006. Mutation of two intramembrane polar residues conserved within the hyaluronan synthase family alters hyaluronan product size. J Biol Chem. 281:11755–11760. [DOI] [PubMed] [Google Scholar]
- Kumari K, Tlapak-Simmons VL, Baggenstoss BA, Weigel PH. 2002. The streptococcal hyaluronan synthases are inhibited by sulfhydryl modifying reagents but conserved cysteine residues are not essential for enzyme function. J Biol Chem. 277:13943–13951. [DOI] [PubMed] [Google Scholar]
- Kumari K, Weigel PH. 2005. Identification of a membrane-localized cysteine cluster near the substrate binding sites of the streptococcus equisimilis hyaluronan synthase. Glycobiology. 15:529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyosseva SV, Harris EN, Weigel PH. 2008. The hyaluronan receptor for endocytosis (HARE) mediates hyaluronan-dependent signal transduction via extracellular signal-regulated kinases (ERK). J Biol Chem. 283:15047–15055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent TC, Laurent UBG, Fraser JRE. 1995. Functions of hyaluronan. Ann Rheum Dis. 54:429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DC, Pearlman E, Diaconu E, Guo K, Mori H, Haqqi T, Markowitz S, Willson J, Sy MS. 1996. Expression of hyaluronidase by tumor cells induces angiogenesis in vivo. Proc Natl Acad Sci USA. 93:7832–7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee CM, Lowenstein CJ, Horton MR, Wu J, Bao C, Chin BY, Choi AMK, Noble PW. 1997. Hyaluronan fragments induce nitric-oxide synthase in murine macrophages through a nuclear factor kappa B- dependent mechanism. J Biol Chem. 272:8013–8018. [DOI] [PubMed] [Google Scholar]
- Medina AP, Lin J, Weigel PH. 2012. Hyaluronan synthase mediates dye translocation across liposomal membranes. BMC Biochem. 13:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretto P, Karousou E, Viola M, Caon I, D'Angelo ML, De Luca G, Passi A, Vigetti D. 2015. Regulation of hyaluronan synthesis in vascular diseases and diabetes. J Diabetes Res. 2015:167283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble PW. 2002. Hyaluronan and its catabolic products in tissue injury and repair. Matrix Biol. 21:25–29. [DOI] [PubMed] [Google Scholar]
- Noble PW, Jiang D. 2006. Matrix regulation of lung injury, inflammation, and repair: The role of innate immunity. Proc Am Thorac Soc. 3:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S, Im HJ, Knudson CB, Knudson W. 2006. Hyaluronan oligosaccharides induce matrix metalloproteinase 13 via transcriptional activation of NF{kappa}B and p38 MAP kinase in articular chondrocytes. J Biol Chem. 281:17952–17960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey MS, Baggenstoss BA, Washburn J, Harris EN, Weigel PH. 2013. The hyaluronan receptor for endocytosis (HARE) activates NF-kappaB-mediated gene expression in response to 40-400-kDa, but not smaller or larger, hyaluronans. J Biol Chem. 288:14068–14079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prehm P. 1983. Synthesis of hyaluronate in differentiated teratocarcinoma cells. Mechanism of chain growth. Biochem J. 211:191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pummill PE, DeAngelis PL. 2003. Alteration of polysaccharide size distribution of a vertebrate hyaluronan synthase by mutation. J Biol Chem. 278:19808–19814. [DOI] [PubMed] [Google Scholar]
- Rilla K, Siiskonen H, Spicer AP, Hyttinen JMT, Tammi MI, Tammi RH. 2005. Plasma membrane residence of hyaluronan synthase is coupled to its enzymatic activity. J Biol Chem. 280:31890–31897. [DOI] [PubMed] [Google Scholar]
- Rockey DC, Chung JJ, McKee CM, Noble PW. 1998. Stimulation of inducible nitric oxide synthase in rat liver by hyaluronan fragments. Hepatology. 27:86–92. [DOI] [PubMed] [Google Scholar]
- Seyfried NT, McVey GF, Almond A, Mahoney DJ, Dudhia J, Day AJ. 2005. Expression and purification of functionally active hyaluronan-binding domains from human cartilage link protein, aggrecan and versican: Formation of ternary complexes with defined hyaluronan oligosaccharides. J Biol Chem. 280:5435–5448. [DOI] [PubMed] [Google Scholar]
- Simpson MA, de la Motte C, Sherman LS, Weigel PH. 2015. Advances in hyaluronan biology: Signaling, regulation, and disease mechanisms. Int J Cell Biol. 2015:690572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slevin M, Krupinski J, Gaffney J, Matou S, West D, Delisser H, Savani RC, Kumar S. 2007. Hyaluronan-mediated angiogenesis in vascular disease: Uncovering RHAMM and CD44 receptor signaling pathways. Matrix Biol. 26:58–68. [DOI] [PubMed] [Google Scholar]
- Spicer AP, McDonald JA. 1998. Characterization and molecular evolution of a vertebrate hyaluronan synthase gene family. J Biol Chem. 273:1923–1932. [DOI] [PubMed] [Google Scholar]
- Spicer AP, Nguyen TK. 1999. Mammalian hyaluronan synthases: Investigation of functional relationships in vivo. Biochem Soc Trans. 27:109–115. [DOI] [PubMed] [Google Scholar]
- Spicer AP, Tien JL, Joo A, Bowling RA Jr. 2002. Investigation of hyaluronan function in the mouse through targeted mutagenesis. Glycoconj J. 19:341–345. [DOI] [PubMed] [Google Scholar]
- Stern R, Asari AA, Sugahara KN. 2006. Hyaluronan fragments: An information-rich system. Eur J Cell Biol. 85:699–715. [DOI] [PubMed] [Google Scholar]
- Tammi MI, Day AJ, Turley EA. 2002. Hyaluronan and homeostasis: A balancing act. J Biol Chem. 277:4581–4584. [DOI] [PubMed] [Google Scholar]
- Tammi RH, Passi AG, Rilla K, Karousou E, Vigetti D, Makkonen K, Tammi MI. 2011. Transcriptional and post-translational regulation of hyaluronan synthesis. FEBS J. 278:1419–1428. [DOI] [PubMed] [Google Scholar]
- Tirone E, Dalessandris C, Hascall VC, Siracusa G, Salustri A. 1997. Hyaluronan synthesis by mouse cumulus cells is regulated by interactions between follicle-stimulating hormone (or epidermal growth factor) and a soluble oocyte factor (or transforming growth factor beta(1)). J Biol Chem. 272:4787–4794. [DOI] [PubMed] [Google Scholar]
- Tlapak-Simmons VL, Baggenstoss BA, Clyne T, Weigel PH. 1999. a. Purification and lipid dependence of the recombinant hyaluronan synthases from Streptococcus pyogenes and Streptococcus equisimilis. J Biol Chem. 274:4239–4245. [DOI] [PubMed] [Google Scholar]
- Tlapak-Simmons VL, Baggenstoss BA, Kumari K, Heldermon C, Weigel PH. 1999. b. Kinetic characterization of the recombinant hyaluronan synthases from Streptococcus pyogenes and Streptococcus equisimilis. J Biol Chem. 274:4246–4253. [DOI] [PubMed] [Google Scholar]
- Tlapak-Simmons VL, Baron CA, Gotschall R, Haque D, Canfield WM, Weigel PH. 2005. Hyaluronan biosynthesis by Class I streptococcal hyaluronan synthases occurs at the reducing end. J Biol Chem. 280:13012–13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tlapak-Simmons VL, Baron CA, Weigel PH. 2004. Characterization of the purified hyaluronan synthase from Streptococcus equisimilis. Biochem. 43:9234–9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tlapak-Simmons VL, Kempner ES, Baggenstoss BA, Weigel PH. 1998. The active streptococcal hyaluronan synthases (HASs) contain a single HAS monomer and multiple cardiolipin molecules. J Biol Chem. 273:26100–26109. [DOI] [PubMed] [Google Scholar]
- Toole BP. 2004. Hyaluronan: From extracellular glue to pericellular cue. Nat Rev Cancer. 4:528–539. [DOI] [PubMed] [Google Scholar]
- Turley EA, Belch AJ, Poppema S, Pilarski LM. 1993. Expression and function of a receptor for hyaluronan-mediated motility on normal and malignant B lymphocytes. Blood. 81:446–453. [PubMed] [Google Scholar]
- Turley EA, Noble PW, Bourguignon LYW. 2002. Signaling properties of hyaluronan receptors. J Biol Chem. 277:4589–4592. [DOI] [PubMed] [Google Scholar]
- Vigetti D, Clerici M, Deleonibus S, Karousou E, Viola M, Moretto P, Heldin P, Hascall VC, De Luca G, Passi A. 2011. Hyaluronan synthesis is inhibited by adenosine monophosphate-activated protein kinase through the regulation of HAS2 activity in human aortic smooth muscle cells. J Biol Chem. 286:7917–7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigetti D, Passi A. 2014. Hyaluronan synthases posttranslational regulation in cancer. Adv Cancer Res. 123:95–119. [DOI] [PubMed] [Google Scholar]
- Viola M, Karousou E, D'Angelo ML, Caon I, De Luca G, Passi A, Vigetti D. 2015. Regulated hyaluronan synthesis by vascular cells. Int J Cell Biol. 2015:208303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Entwistle J, Hou GP, Li QA, Turley EA. 1996. The characterization of a human RHAMM cDNA: Conservation of the hyaluronan-binding domains. Gene. 174:299–306. [DOI] [PubMed] [Google Scholar]
- Weigel PH. 2015. Hyaluronan synthase: The mechanism of initiation at the reducing end and a pendulum model for polysaccharide translocation to the cell exterior. Int J Cell Biol. 2015:367579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel PH, Baggenstoss BA. 2012. Hyaluronan synthase polymerizing activity and control of product size are discrete enzyme functions that can be uncoupled by mutagenesis of conserved cysteines. Glycobiology. 22:1302–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel PH, DeAngelis PL. 2007. Hyaluronan synthases: A decade-plus of novel glycosyltransferases. J Biol Chem. 282:36777–36781. [DOI] [PubMed] [Google Scholar]
- Weigel PH, de la Motte C, Simpson MA, Sherman LS. 2015. a. Advances in hyaluronan biology: signaling, regulation, and disease mechanisms. Int J Cell Biol. http://www.hindawi.com/journals/ijcb/si/267417/. Hindawi Pub. Corp. NY, USA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel PH, Padgett-McCue AJ, Baggenstoss BA. 2013. Methods for measuring class I membrane-bound hyaluronan synthase activity. Methods Mol Biol. 1022:229–247. [DOI] [PubMed] [Google Scholar]
- Weigel PH, West CM, Zhao P, Wells L, Baggenstoss BA, Washburn JL. 2015. b. Hyaluronan synthase assembles chitin oligomers with -GlcNAc(alpha1-->)UDP at the reducing end. Glycobiology. 25:632–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West DC, Hampson IN, Arnold F, Kumar S. 1985. Angiogenesis induced by degradation products of hyaluronic acid. Science. 228:1324–1326. [DOI] [PubMed] [Google Scholar]
- Xu HP, Ito T, Tawada A, Maeda H, Yamanokuchi H, Isahara K, Yoshida K, Uchiyama Y, Asari A. 2002. Effect of hyaluronan oligosaccharides on the expression of heat shock protein 72. J Biol Chem. 277:17308–17314. [DOI] [PubMed] [Google Scholar]
- Xu XM, Chen Y, Chen J, Yang S, Gao F, Underhill CB, Cresswell K, Zhang L. 2003. A peptide with three hyaluronan binding motifs inhibits tumor growth and induces apoptosis. Cancer Res. 63:5685–5690. [PubMed] [Google Scholar]
- Yang B, Yang BL, Savani RC, Turley EA. 1994. Identification of a common hyaluronan binding motif in the hyaluronan binding proteins RHAMM, CD44 and link protein. EMBO J. 13:286–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang BH, Zang LY, Turley EA. 1993. Identification of two hyaluronan-binding domains in the hyaluronan receptor RHAMM. J Biol Chem. 268:8617–8623. [PubMed] [Google Scholar]
- Yoshida M, Itano N, Yamada Y, Kimata K. 2000. In vitro synthesis of hyaluronan by a single protein derived from mouse HAS1 gene and characterization of amino acid residues essential for the activity. J Biol Chem. 275:497–506. [DOI] [PubMed] [Google Scholar]
- Zhang SW, Chang MCY, Zylka D, Turley S, Harrison R, Turley EA. 1998. The hyaluronan receptor RHAMM regulates extracellular-regulated kinase. J Biol Chem. 273:11342–11348. [DOI] [PubMed] [Google Scholar]