Abstract
Polypeptide GalNAc-transferases (GalNAc-Ts) constitute a family of 20 human glycosyltransferases (comprising 9 subfamilies), which initiate mucin-type O-glycosylation. The O-glycoproteome is thought to be differentially regulated via the different substrate specificities and expression patterns of each GalNAc-T isoforms. Here, we present a comprehensive in vitro analysis of the peptide substrate specificity of GalNAc-T13, showing that it essentially overlaps with the ubiquitous expressed GalNAc-T1 isoform found in the same subfamily as T13. We have also identified and partially characterized nine splice variants of GalNAc-T13, which add further complexity to the GalNAc-T family. Two variants with changes in their lectin domains were characterized by in vitro glycosylation assays, and one (Δ39Ex9) was inactive while the second one (Ex10b) had essentially unaltered activity. We used reverse transcription-polymerase chain reaction analysis of human neuroblastoma cell lines, normal brain and a small panel of neuroblastoma tumors to demonstrate that several splice variants (Ex10b, ΔEx9, ΔEx2-7 and ΔEx6/8-39bpEx9) were highly expressed in tumor cell lines compared with normal brain, although the functional implications remain to be unveiled. In summary, the GalNAc-T13 isoform is predicted to function similarly to GalNAc-T1 against peptide substrates in vivo, in contrast to a prior report, but is unique by being selectively expressed in the brain.
Keywords: GALNT/GALNT13/splicing/MALDI-TOF/neuroblastoma/cancer
Introduction
Mucin-type (GalNAc-type) O-linked glycosylation is found on Ser, Thr and some Tyr residues, and is one of the most common types of protein glycosylation found in metazoans. O-glycoproteins play important biological roles including microbial protection and clearance processes, modulation of cell–cell interactions, inflammation, immunity, tumorigenesis and metastasis (Hollingsworth and Swanson, 2004; Brockhausen, 2006; Gill et al. 2011). The GalNAc-type O-glycosylation pathway requires a complex set of enzymes localized in the Golgi apparatus. The initial step involves the transfer of α-D-GalNAc from the sugar donor (UDP-GalNAc) to Ser or Thr (and possibly Tyr) residues of acceptor protein substrates, catalyzed by members of the large polypeptide GalNAc-transferase (GalNAc-T) family (EC 2.4.1.41) (Bennett et al. 2012). Subsequent elongation of the GalNAc residue yields different “core” structures that may be further modified by the stepwise action of other Golgi glycosyltransferases, resulting in a diversity of O-glycan structures (Tarp and Clausen, 2008). It is increasingly becoming clear that most proteins trafficking the secretory pathway are O-glycosylated (Steentoft et al. 2013), and isolated O-glycosylation sites may serve highly specific roles in regulation of protein functions (Schjoldager and Clausen, 2012).
There are up to 20 distinct GalNAc-T isoforms in humans (Bennett et al. 2012). The members of the GalNAc-T family differ in both their enzymatic substrate specificity and their cell and tissue expression. The transcripts of GalNAc-T1 and T2 are most widely expressed, whereas other isoforms display more restrictive tissue expression profiles. GalNAc-T isoforms are differentially expressed spatially and temporally during development, growth and maturation (Kingsley et al. 2000; Young et al.,2003), and the expression pattern often changes with malignant transformation (Mandel et al. 1999; Kohsaki et al. 2000; Shibao et al. 2002; Gu et al. 2004; Ishikawa et al. 2004; Miyahara et al. 2004; Yamamoto et al. 2004; Landers et al. 2005; Berois et al. 2006a; Inoue et al. 2007; Wu et al. 2010).
Members of the GalNAc-T family (except GalNAc-T20) are structurally unique among metazoan glycosyltransferases because they possess a two-domain architecture consisting of a membrane-tethered catalytic domain attached via a short flexible linker to a C-terminal ricin-like lectin domain (Fritz et al. 2006; Bennett et al. 2012). The lectin domains bind GalNAc and promote secondary GalNAc addition on neighboring positions in a peptidic sequence (Wandall et al. 2007a; Pedersen et al. 2011), modulating and improving the glycosylation of glycopeptide substrates (Raman et al. 2008; Gerken et al. 2013; Lira-Navarrete et al. 2015). Although most isoforms can glycosylate both naked peptides and glycopeptides (Hagen et al. 1997; Wandall et al. 1997; Nehrke et al. 1998), GalNAc-T4 (Bennett et al. 1998) and T12 (Bennett et al. 2012) exhibit preferential, and T7 (Bennett et al. 1999a), T10 (Cheng et al. 2002, Perrine et al. 2009) and T17 (Peng et al. 2010) exclusive GalNAc-peptide substrate specificities. These activities have been further elaborated in our recent study with random glycopeptide libraries (Revoredo et al. 2016). Thus, the regulation of the O-glycoproteome and the sites and patterns of O-glycan decoration of individual glycoproteins depend on the expression of individual GalNAc-T isoforms in a given cell.
The large number of GalNAc-Ts controlling the initiation step of mucin-type O-glycosylation may be expected to provide substantial biosynthetic redundancy, and early studies of Galnt-deficient mice did not produce obvious phenotypes (Marth, 1996; Lowe and Marth, 2003). However, individual GalNAc-T isoforms serve non-redundant unique and essential cellular functions conserved through evolution (Schwientek et al. 2002; Tran et al. 2012). Nevertheless, we have only begun to understand the role of individual GalNAc-T isoforms, and most of the isoforms with more restricted expression patterns are poorly understood.
GalNAc-T13 is such an isoform with highly restricted expression in the nervous system (Zhang et al. 2003). It was recently shown that it contributes to the neuronal differentiation through glycosylation and stabilization of podoplanin (Xu et al. 2016). This isoform is highly homologous and considered to be a close paralog (subfamily member) of ubiquitous GalNAc-T1 (Bennett et al. 2012). Close paralogs of GalNAc-Ts have been found to have essentially identical peptide substrate specificities as determined by in vitro analysis (Bennett et al. 1999b; Gerken et al. 2008). However, it was previously reported that GalNAc-T13 differs in peptide substrate preferences from GalNAc-T1, and in particular with respect to important substrates such as Syndecan (Zhang et al. 2003). We are interested in GalNAc-T13 because of its potential involvement in cancer biology (Berois et al. 2006b; Matsumoto et al. 2012; Matsumoto et al. 2013; Nogimori et al. 2016). Using microarray gene expression analysis in a metastatic xenograft-derived cell model of human neuroblastoma (so-called IGR-N-91), we found that GALNT13 was the most strongly up-regulated gene (12-fold) in metastatic malignant neuroblasts compared with primary tumor xenograft (Berois et al. 2006b). Moreover, we found that GALNT13 expression in bone marrow is a strong predictor of poor clinical outcome in neuroblastoma patients (Berois et al. 2006b). However, Galnt13 was also identified as an up-regulated gene in high metastatic mouse lung cancer cell lines (Matsumoto et al. 2012). In this case, the molecular basis of aggressive behavior was explained by GalNAc-T13 leading to formation of trimeric Tn antigen on syndecan-1 (Matsumoto et al. 2013). Recently, high expression of GalNAc-T13 was also demonstrated in human lung cancer and this was correlated with poor prognosis, suggesting that this enzyme may represent a prognostic factor (Nogimori et al. 2016). In contrast, GALNT13 was also identified as one of the genes that are most down-regulated in high-grade glioma-derived tumor progenitor cells relative to their low-grade counterparts (Auvergne et al. 2013). Another interesting feature of GalNAc-T13 is the existence of alternatively spliced variants that can produce protein products. Recently two splice variants were reported, one with an incomplete (GalNAc-T13-V2) and inactive catalytic domain and the other with an alteration of the latter half of the lectin domain (GalNAc-T13-V1) (Raman et al. 2012). In this study, we report a comprehensive comparative substrate specificity analysis of GalNAc-T13 and its close paralog GalNAc-T1 using a large panel of unbiased peptide substrates and demonstrate that the two isoenzymes have essentially the same catalytic properties as evaluated by in vitro assays with peptide substrates. We further report the identification and initial characterization of nine GALNT13 splice variants.
Results
Comparative analysis of the substrate specificities of GalNAc-T13 and T1
We first probed the glycosylation capacities of the two isoforms with a small panel of peptide substrates including different syndecan-3-derived peptides (Table I). We found no difference in their in vitro activity towards these substrates using time course assays (Table II). For a more global comparison, we used a large panel of 180 peptide substrates that represent a more unbiased selection of O-glycosites identified in human cancer cell lines using the so-called SimpleCell strategy (Steentoft et al. 2013). We previously analyzed 10 GalNAc-T isoforms including GalNAc-T1 with some of these substrates, and identified both redundant and GalNAc-T isoform-specific substrates (Kong et al. 2015). The peptide assays were monitored using time course (0 h, 4 h and overnight aliquots) product development assays by matrix assisted laser desorption/ionization time-of-flight (MALDI-TOF) MS, which enables semiquantitative evaluation of the rate of reactions as well as the number of GalNAc residues incorporated (Bennett et al. 1998; Hassan et al. 2000). In contrast to previous studies (Zhang et al. 2003), we found virtually complete concordance between substrate utilization of GalNAc-T13 and GalNAc-T1 (Supplementary data, Table SI). Specifically, the same (except for a C-terminal carboxyfluorescein (FAM) tag) SDC106 peptide used previously (Zhang et al. 2003) was glycosylated by both enzymes at same velocity and with same number of sites incorporated. The same was found with two other peptides derived from the syndecan-3 molecule (SDC3a and SDC3b) (Table II).
Table I.
Partial list of the synthetic peptides used in this study
| Peptide | Sequence | MW (Da) | S+T |
|---|---|---|---|
| MUC1-b | TRPGSTAPPAC | 1057.1 | 3 |
| IgAh | VPSTPPTPSPSTPPTPSPSK | 1959.1 | 9 |
| SDC106 | AVSTTPAVLK | 986.1 | 3 |
| MUC7 | APPTPSATTPAPPSSSAPPETTAA | 2204.3 | 9 |
| Carl2 | LSESTTQLPGGGPGCA | 1474.6 | 4 |
| SDC3a | VATARATTPEAPSPPTTAAVLDTEAP | 2535.8 | 7 |
| SDC3b | APTPRLVSTATSRPRALPR | 2047.4 | 5 |
| FGF23a | IHFNTPIPRRHTRSAEDD | 2162.3 | 3 |
| FGF23b | PIPRRHTRSAEDDSERDP | 2134.2 | 3 |
| ANGPTL3 | KPRAPRTTPFLQ | 1411.7 | 2 |
| HCPitb | PRFQDSSSSKAPPPSLPSPSRLPG | 2492.7 | 7 |
| HCHIV | IRIQRGPGRAFVTIGKIGNM | 2184.6 | 1 |
| GMCSF | APARSPSPSTQPWEHV | 1746.9 | 4 |
| CD59 | WKFEHCNFNDVTTRLRENEL | 2551.8 | 2 |
aMolecular weight.
Table II.
Time course GalNAc-T assay monitored by MALDI-TOFa
| Enzyme | Assay time (h) | MUC1-b | IgAh | MUC7 | CARL2 | SDC106 | SDC3a | SDC3b | FGF23a | FGF23b | ANGPTL3 | HCPitb | HCHIV | GMCSF | CD59 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GalNAc-T1 | 1 | 0 (1) | 2 (0–1, 3) | 1 (0, 2) | 0 (1) | 0 (1) | 0 (1–2) | 0 (1) | 0 (1) | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 (1) | 3 (0–2, 4–5) | 2 (0–1, 3–4) | 0 (1) | 0 (1) | 0 (1–3) | 0 (1) | 0 (1) | 0 | 0 | 0 | 0 | 0 | 0 | |
| 24 | 1 (0) | 4 (3, 5–6) | 4 (1–3, 5) | 1 | 1 (0, 2) | 4 (0–3, 5–6) | 1 (0, 2–3) | 0 = 1 | 0 | 0 | 0 | 0 | 0 (1) | 0 | |
| GalNAc-T13 | 1 | 0 (1) | 2 (0–1, 3) | 2 (0–1, 3) | 0 (1) | 0 (1) | 0 (1–2) | 0 (1) | 0 (1) | 0 | 0 | 0 | 0 | 0 (1) | 0 |
| 4 | 0 = 1 | 2 = 3 (0–1, 4) | 2 (0–1, 3–4) | 0 (1) | 0 (1) | 0 (1–2) | 0 (1–2) | 0 (1–2) | 0 | 0 | 0 | 0 | 0 (1) | 0 | |
| 24 | 1 (0, 2) | 5 (2–4, 6–7) | 4 (1–3, 5–6) | 1 (0) | 1 (0, 2) | 4 (0–3, 5–6) | 1 (0, 2) | 0 (1–2) | 0 | 0 | 0 (1) | 0 | 0 (1) | 0 (1) | |
| GalNAc-T13-Δ39bpEx9 | 1 | 0 | 0 | 0 (1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 (1) | 0 (1) | 0 (1) | 0 | 0 | 0 (1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 24 | 0 (1) | 0 (1–2) | 1 = 2 (0, 3–4) | 0 | 0 | 0 (1–2) | 0 | 0 (1) | 0 | 0 | 0 (1) | 0 | 0 (1) | 0 | |
| GalNAc-T13-Ex10b | 1 | 0 (1) | 2 (0–1, 3) | 1 (0, 2–3) | 1 (0) | 0 (1) | 0 | 0 (1) | 0 (1) | 0 | 0 | 0 | 0 | 0 (1) | 0 |
| 4 | 0 (1) | 3 (2, 4–5) | 2 (0–1, 3–5) | 1 (0) | 1 (0) | 0 (2, 4) | 1 (0, 2) | 0 (1–3) | 0 | 0 | 0 | 0 | 0 (1) | 0 | |
| 24 | 1 (0, 2) | 5 (3–4, 6) | 5 (3–4, 6) | 1 (0) | 1 (0, 2) | 0 (1–4) | 1 (0, 2) | 1 (0, 3–4) | 0 | 0 (1) | 0 (1) | 0 | 1 (0, 2) | 0 |
aThe numbers inside each cell correspond to the number of GalNAcs incorporated in the major product. The other products, also present in the reaction mixture, are shown in brackets.
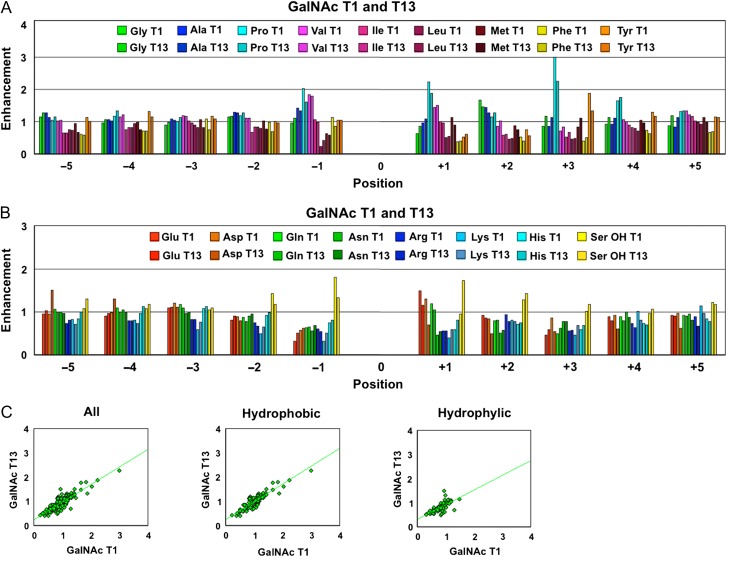
The specificity of human GalNAc-T1 (and several other human GalNAc-Ts and Drosophila PGANTs) has been previously characterized using a series of oriented random peptide substrates libraries (Gerken et al. 2006; Gerken et al. 2008; Gerken et al. 2011). To extend the comparison between GalNAc-T1 and T13, we also determined the peptide substrate specificity of GalNAc-T13 using the same procedures (see Figure 1 and Supplementary data, Figures S1 and S2). The results show, with only a few exceptions, near-identical peptide substrate preferences of the catalytic domains of these two isoforms. This was expected as the peptide binding site residues of the catalytic domains of GalNAc-T1 and T13 share 96–100% sequence identity, while their entire catalytic domains are 88% identical (Gerken et al. 2008). In contrast, we have previously reported that these two isoforms exhibit slightly different lectin domain mediated glycopeptide substrate preferences against a series of unique random glycopeptide substrates (Gerken et al. 2013). Again this difference may be consistent with the determination that GalNAc-T1 and T13 lectin domains possess a lower sequence identity of 83% (Gerken et al. 2008).
Fig. 1.
Plots of the peptide substrate preferences of GalNAc-T1 and T13 obtained from random peptide substrates, showing their near-identical catalytic domain peptide substrate specificity of these transferases. Comparison plots of the hydrophobic (A) and hydrophilic (B) amino acid (AA) residue enhancement values for GalNAc-T1 and T13. (C) The enhancement values for GalNAc-T13 plotted against those for GalNAc-T1 for all residues (left panel, r2 = 0.75), the hydrophobic residues (middle panel, r2 = 0.81) and the hydrophilic residues (right panel, r2 = 0.49). Note that in A and B position 0 represents the glycosylated Thr in the random peptide substrates as described in the Materials and Methods section. Enhancement values greater than 1 suggest that the transferase has a high preference for the residue, while values significantly less than 1 suggest that the residue exerts an inhibitory effect on the transferase. See Supplementary data (Figures S1 and S2) for the comparison of individual AA residue preferences and their standard deviations. Data for GalNAc-T1 was taken from Gerken et al. (2011). This figure is available in black and white in print and in color at Glycobiology online.
Identification of novel GalNAc-T13 splice variants
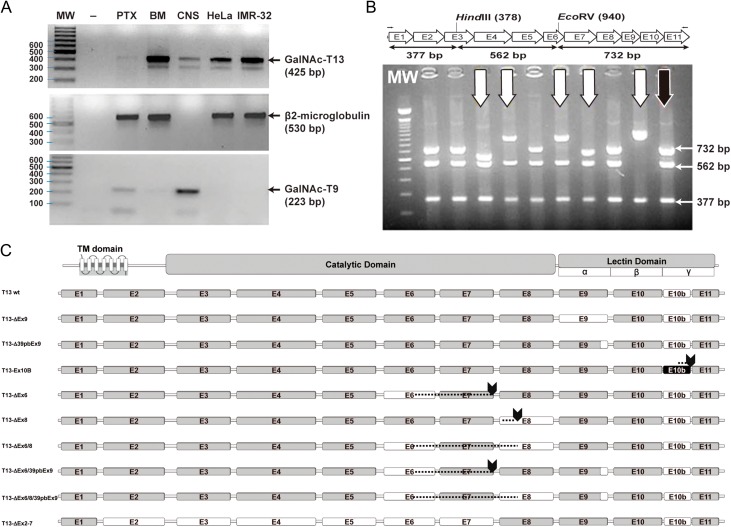
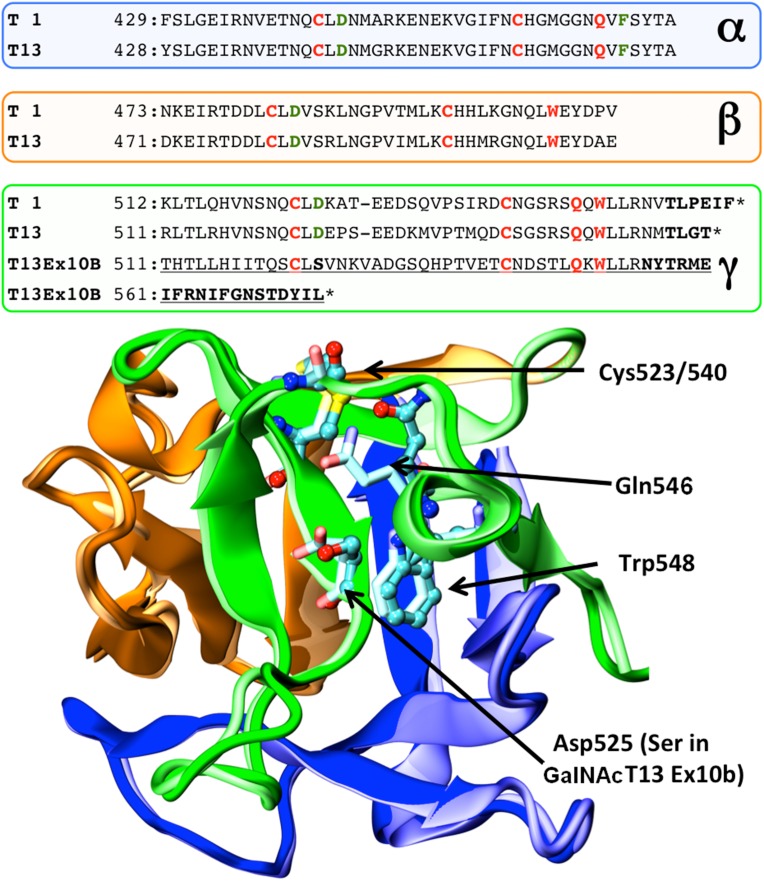
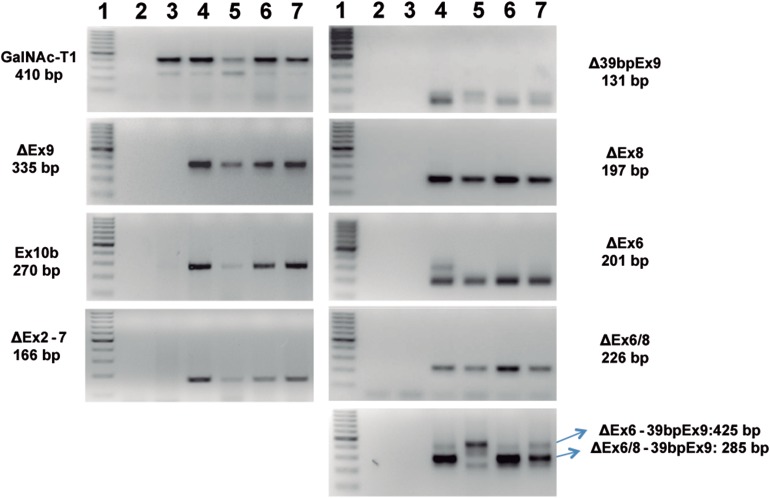
Amplification of the 3′ coding region of GALNT13 transcripts in different cell lines revealed more products than expected (Figure 2A). Cloning and sequencing identified these transcripts as alternative splice variants. Therefore, we designed a procedure for exhaustively screening new transcripts (Figure 2B). This method consisted in cloning the entire coding region, amplification by colony PCR, digestion with restriction enzymes and sequencing. We studied more than 300 clones and were able to identify 9 of the most represented GALNT13 splice variants present in the IGR-N-91 BM neuroblastoma cell line (Figure 2C). The sequences of each of the variants have been submitted to GenBank. The most abundant form (55% of the clones) with 11 exons was previously identified (Zhang et al. 2003). The second most abundant mRNA (approximately 10% of the clones) incorporates a new exon between exon 10 and 11 (named GalNAc-T13 Ex10b), and it has been previously reported and partially characterized (GalNAc-T13-V1) (Raman et al. 2012). This new exon is evolutionarily conserved, at least in higher mammals like dog (99%), chimpanzee (100%), mice (97%), rat (100%) and hamster (87%), but it is not present in human GALNT1. Exon 10b has a stop codon at its 3´end, so exon 11 is not included in the final protein. Therefore, this variant encodes an enzyme with a new gamma repeat at the lectin domain, as Ex10b conserves the disulphide-bonded cysteine residues and the QXW motif present in each carbohydrate-binding subdomain (Figure 3). In addition to GalNAc-T13 Ex10b, we found two other well-represented mRNAs that also differ in the sequence that encodes the lectin domain, one lacking the entire 9th exon (GalNAc-T13 ΔEx9) and another lacking the first 39 bp of the same exon (GalNAc-T13 Δ39bpEx9) (Figure 2C). The GalNAc-T13 ΔEx9 lacks the entire α repeat of the lectin domain. Additionally, we also identified low-frequency mRNAs encoding proteins with important deletions in the catalytic domain, probably reflecting errors in the splicing machinery, as the ones that have premature termination codons, like GalNAc-T13 ΔEx6 and GalNAc-T13 ΔEx8, both lacking the lectin domain in the definitive protein. In contrast, we identified a splice variant lacking exon 6 and 8 (GalNAc-T13 ΔEx6/8), which encodes an enzyme with a complete lectin domain (Figure 2C). We also found an mRNA that lacks all the exons between the 2th and the 7th (GalNAc-T13 ΔEx2-7), thereby missing the entire catalytic domain in the final protein. This mRNA could encode a new “lectin” like protein (Figure 2C). There are also other two variants present, GalNAc-T13 ΔEx6/39bpEx9 and GalNAc-T13 ΔEx6/8/39bpEx9. GalNAc-T13 ΔEx6/39bpEx9 encodes the same protein as the ΔEx6 variant and the ΔEx6/8/39bpEx9 encodes the same protein as the Δ6/8 variant but with a different lectin domain.
Fig. 2.
GalNAc-T13 splice variants. (A) Reverse transcription-polymerase chain reaction (RT-PCR) was performed with GalNT13_Fw and T13_XhoI_Rev primers on four different cancer cell lines and on human brain tissue. Several bands of unexpected size were observed after ethidium bromide gel staining. MW: molecular weight (100 bp DNA ladder); -: negative control (water); IGR-N-91 PTX: neuroblastoma cell line derived from a primary tumor; IGR-N-91 BM: metastatic neuroblastoma cell line; CNS: central nervous system tissue (brain); HeLa: cervical cancer cell line; IMR-32: neuroblastoma cell line. The β2M (β2-microglobulin) gene was amplified to verify cDNA quality (except for CNS, where we used GalNAc-T9 as a control). (B) Screening of GalNAc-T13 splice variants by colony PCR. Splice variants were screened by RT-PCR with T13_XhoI_Fw and T13_XhoI_Rev primers. The product was cloned into the pGEM-T Easy plasmid and analyzed by colony PCR and digestion with restriction enzymes HindIII and EcoRV. The expected product sizes were 732, 562 and 377 bp. In the illustrative gel (representative gel of 9 of more than 300 analyzed clones), the black arrow indicates digestion products of the previously characterized positive control (pGEM-T with the GalNAc-T13 coding region, confirmed by sequencing), whereas the white arrows mark clones with alternative digestion patterns. These clones were subsequently analyzed by sequencing. (C) Exon organization of the newly identified GalNAc-T13 splice variants. The transmembrane (TM), catalytic and lectin domains of GalNAc-Ts are indicated, as well as the three lectin subdomains α, β and γ. The amino acid (AA) extension of each variant is indicated on the right. Skipped exons are shown in white, Ex10b is shown in black and stop codons are marked by arrows. Dashed lines indicate regions where exon skipping introduces a change in the reading frame, resulting in a different AA sequence.
Fig. 3.
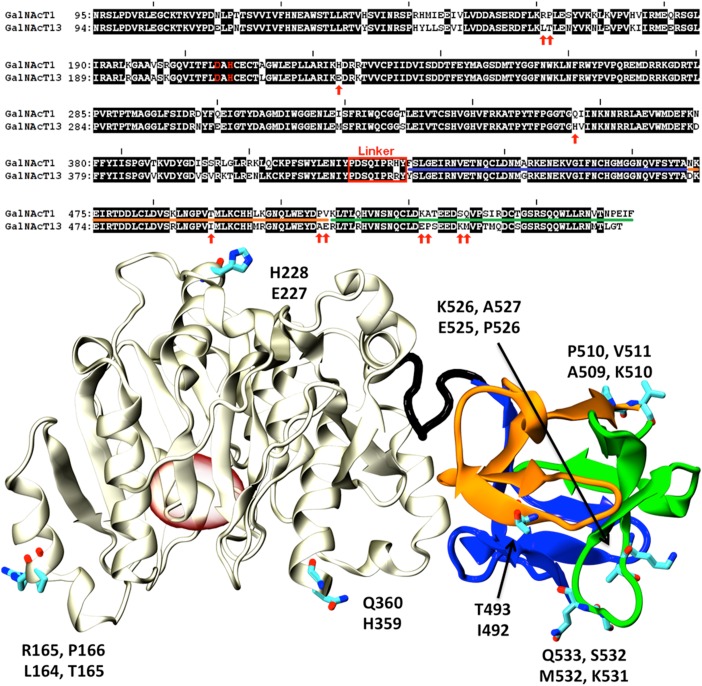
Structural model of GalNAc-T13 Ex10b. Top: sequence alignment of GalNAc-T1, GalNAc-T13 and GalNAc-T13 Ex10b in the region comprising the lectin domain. Each colored panel represents from top to bottom the three α, β, and γ-lectin subdomains. Identical residues in the GalNAc-T family are shown in red, whereas highly conserved residues are shown in green. Bold letters indicate putatively unstructured regions of the protein. Asterisks indicate the C-terminal end of the protein. Notice that the change in sequence introduced by Ex10b coincides with the beginning of the γ-subdomain. Bottom: structural superposition of the lectin domains of GalNAc-T1 (pale colors) and a homology model of Ex10b (vivid colors) looking in the direction of the γ-lectin subdomain sugar binding site. The cartoon is colored according to subdomains with the conserved residues displayed as sticks for GalNAcT-1 and balls and sticks for GalNAcT-13/Ex10b. Numbers correspond to GalNAc-T1 residues. This figure is available in black and white in print and in color at Glycobiology online.
Functional analysis of GalNAc-T13 splice variants
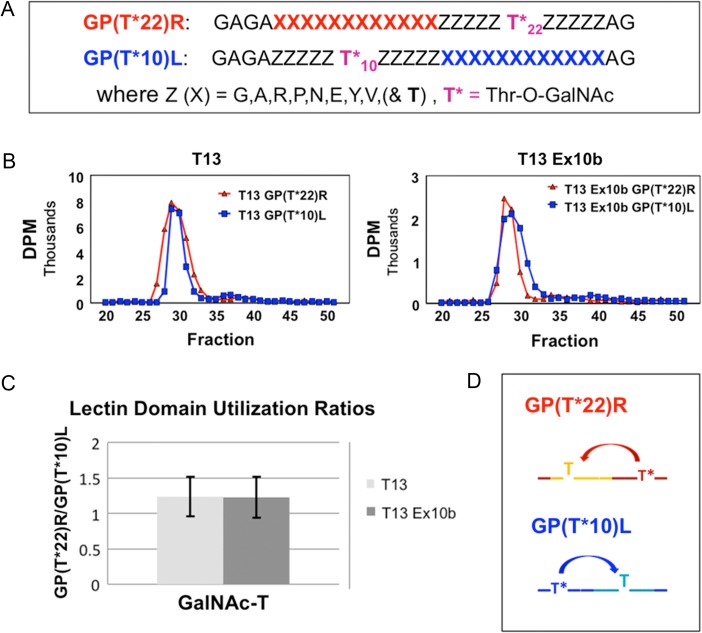
Of the nine GalNAc-T13 splice variants identified, we selected the ones differing in the lectin domain (Δ9, ∆39bpEx9 and Ex10b) for analysis. We could not obtain a pure and active ΔEx9 enzyme, as it was not effectively secreted in insect cells likely due to early degradation. Nevertheless GalNAc-T13, Ex10b and Δ39bpEx9 variants were effectively expressed. The enzymes were purified to apparent homogeneity and evaluated at equal concentrations by in vitro glycosylation assays. We found variant∆39bpEx9 largely inactive. The Ex10b variant was active similarly to wild-type (wt) GalNAc-T13 using a small panel of peptides (Table II), in agreement with a previous report (Raman et al. 2012). We further tested (Figure 4) the Ex10b variant against a series of random GalNAc-glycopeptide substrates to probe its lectin function as previously described (Gerken et al. 2013; Revoredo et al. 2016). The results indicate there is no difference in the lectin domain utilization of the Ex10b variant lectin domain compared to wt GalNAc-T13 (Figure 4B and C) with both N- and C- terminal glycopeptides being nearly equally utilized (Figure 4D). This finding is consistent with prior studies on GalNAc-T13 (Gerken et al. 2013). Note, however, that with the same glycopeptide substrates, GalNAc-T1 exhibits a several fold elevated preference for the C-terminal glycopeptide (GP(T*22)R) over the N-terminal glycopeptide (GP(T*10)L) that is significantly different from that observed for GalNAc-T13 (Gerken et al. 2013).
Fig.4.
Comparison of the lectin domain of GalNAc-T13 and its lectin domain splice variant GalNAc-T13 Ex10b. (A) Sequences of the N- and C- terminal lectin domains probing glycopeptide substrates (Gerken et al. 2013; Revoredo et al. 2016). (B) Representative gel filtration analysis of the reaction products, demonstrating highly similar substrate utilization for both GalNAc-T13 and GalNAc-T13 Ex10b. (C) Ratio of the utilization of substrate GP(T*22)R over GP(T*10)L for GalNAc-T13 (light gray) and GalNAc-T13 Ex10b (dark gray) (average values based on six and eight determinations, respectively, with error bars representing the standard deviation). (D) Schematic representation of the lectin domain probing activities of GP(T*22)R and GP(T*10)L. This figure is available in black and white in print and in color at Glycobiology online.
Structural analysis
To gain structural insights on the role of the different splice variants, we undertook a structural analysis of GalNAc-T1 and the differences with GalNAc-T13. The few non-conservative sequence variations encountered between both isoforms correspond to solvent exposed, looped regions of the GalNAc-T1 structure (Figure 5). Mapping the sequence of the splice variants characterized in this work on the structure of GalNAc-T1 indicates that ΔEx9 and Δ39bpEx9 variants result in mRNA segments coding for proteins with partial incomplete lectin domains and is very unlikely that these variants may display properly folded lectin domains.
Fig. 5.
Comparison of GalNAc-T1 and T13. Top: Sequence alignment of GalNAc-T1 and T13. Non-identical residues are presented as black letters on a white background. Non-conservative substitutions are indicated with red arrows. The linker region connecting the catalytic and lectin domains is indicated with a red rectangle. The catalytic dyad is displayed in red letters. Underlined amino acids (AAs) indicate the α, β and γ subdomains (colors according to Figure 3). Bottom: Cartoon representation of the X-ray structure of GalNAcT-1 (PDB id:1XHB (Fritz et al. 2004)). The catalytic domain is shown in gray, while the α, β and γ lectin subdomains are colored as in Figure 3. AAs indicated with red arrows in the alignment are displayed as sticks and labeled (top and bottom residues correspond to T1 and T13, respectively). The position of the active site is indicated with a red oval for reference. This figure is available in black and white in print and in color at Glycobiology online.
Variants ΔEx6, ΔEx8, ΔEx6/8, ΔEx6/39bpEx9 and ΔEx6/8/39bpEx9 result in incomplete or severely modified proteins. In particular all variants involving the skipping of exon 6 introduce a shift in the reading frame resulting in relatively large segments of modified amino acid (AA) sequence (Figure 2C). The variant ΔEx2-7 produces a sequence spanning only the last region of the catalytic domain and the entire lectin domain. Therefore, this variant would not possess any catalytic activity but is predicted to retain its capability to bind sugars groups. However, the functional significance of this is uncertain.
Perhaps the most interesting case is the Ex10b variant with a novel gamma subdomain in the lectin domain (Figure 2C). The conserved cysteines and aromatic residues required for the folding of the lectin domain remain unchanged (a partial alignment against GalNAc-T1 and T13 is shown in Figure 3). The most relevant modification in the gamma subdomain regards the substitution of a (not absolutely) conserved solvent exposed Aspartic acid mutated into Serine (Figure 3). This is in line with the identical lectin activity found for this variant and wt GalNAc-T13 (Table II and Figure 4).
Detection of GalNAc-T13 splice variants mRNA in cell lines and neuroblastoma tumors
We investigated the expression of GalNAc-T13 splice variants in IGR-N-91 BM, IGR-N-91 PTX, IMR-32 and HeLa cell lines, using a set of primer pairs designed specifically for each variant (primer sequences are available from the authors upon request). This analysis was positive for all variants in metastatic IGR-N-91 BM cell line, while no variants were detected in primary tumor IGR-N-91 PTX cell line (Figure 6). All the other assessed cell lines expressed all the splice variants. Most of the splice isoforms were detected in normal human brain (CNS), except the Δ6/8-39bpEx9 variant (Figure 6). In order to determine if splice variants are present in clinical samples, we analyzed 17 primary neuroblastoma tumors. We found GalNAc-T13 ΔEx9 in 16/17 tumors, GalNAc-T13 Δ39bpEx9 in 5/17 tumors, GalNAc-T13 Ex10b in 16/17 tumors and GalNAc-T13 ΔEx6/8 in 5/17 cases.
Fig. 6.
Reverse transcription-polymerase chain reaction of the splice variants in four different cancer cell lines and in normal brain tissue.1: molecular weight (100 bp DNA ladder), 2: negative control (water), 3: IGR-N-91 PTX (neuroblastoma cell line derived from a primary tumor), 4: IGR-N-91 BM (metastatic neuroblastoma cell line), 5: central nervous system tissue (brain), 6: HeLa and 7: IMR-32. The quality assessment of each cDNA is shown in Figure 2A. This figure is available in black and white in print and in color at Glycobiology online.
Discussion
The large human family of 20 GalNAc-T isoenzymes comprises many close paralogs that have been classified into several subfamilies based on their sequence homology and peptide/glycopeptide substrate preferences (Bennett et al. 1999b; Bennett et al. 2012). It is therefore expected that members within a subfamily would have similar enzymatic properties (Bennett et al. 1999b; Bennett et al. 2012). Here we analyzed subfamily Ia, defined by the GalNAc-T1 and T13 isoenzymes, exhibiting an overall AA sequence identity of 84.3% and an identical genomic organization with complete conservation of exons structure/architecture (Bennett et al. 2012). Testing the largest unbiased peptide substrate library studied to date, including a random peptide library and a large panel of peptides designed from a set of O-glycoproteins identified from our SimpleCell O-glycoproteome strategy (Steentoft et al. 2013), we demonstrate that the in vitro peptide substrate specificities of the two isoenzymes are essentially indistinguishable in contrast to previous reports (Zhang et al. 2003). We furthermore confirmed previous reports of an alternative coding exon in GALNT13 not found in GALNT1 that produce a functional enzyme with an alternative sequence in the gamma domain of the C-terminal lectin (Raman et al. 2012). This splice variant nevertheless possessed identical glycopeptide specificity as the wt GalNAc-T13. We also identified and further characterized an additional number of splice variants of GalNAc-T13 that did not reveal functional consequences. Thus, the two subfamily Ia isoenzymes, GalNAc-T1 and T13, appear to be functionally indistinguishable except for their different cell and tissue expression patterns. Indeed, GALNT1 and T13 genes are so similar that the first attempt to knock out the murine Galnt1 gene resulted in the knock out of T13 (Hennet et al. 1995). While the GalNAc-T isoforms in such subfamilies are predicted to share near-identical catalytic properties (Bennett et al. 1999b), it was previously reported that GalNAc-T13 exhibited unique substrate specificity compared to GalNAc-T1 against multiple acceptor peptides derived from Syndecan-3 and MUC7 (Zhang et al. 2003). In this study, we used a very large panel of peptide substrates, including the same peptide design from Syndecan-3 and a similar design from MUC7 for direct comparison, and found no evidence for differences in kinetic properties between GalNAc-T13 and T1 (Table II). The discrepancy in the activities against the Syndecan-3 and MUC7 peptides are most likely due to differences in assay conditions, as Zhang et al. (2003) utilized N- and C-terminal fluorophore-labeled peptide substrates and immobilized FLAG-tagged transferases that could affect the results. We have previously identified one significant difference in the properties of GalNAc-T1 and T13, in that the N- and C- terminal orientation of their remote glycopeptide substrates preferences differed. GalNAc-T1 preferred GalNAc-glycopeptide substrate sites with GalNAc residues positioned ~6 to ~17 residues N-terminal of the site of glycosylation, while GalNAc-T13 equally preferred N- and C-terminal GalNAc residues (Figure 4) (Gerken et al. 2013). Thus, the lectin domains of GalNAc-T1 and T13 that exhibit the highest degree of sequence dissimilarity appear to possess different glycopeptide specificities.
While in vitro enzyme analysis of this class of enzymes with peptide substrates has long been considered representative of the properties with whole protein substrates as well as in vivo (Bennett et al. 2012), we have recently discovered an example where this is not the case. Human GalNAc-T11 is the only isoform capable of glycosylating the low-density lipoprotein receptor (LDLR) in its ligand-binding region, where the short linker regions between the LDLR class A repeats contain a highly conserved O-glycosite (Steentoft et al. 2013; Pedersen et al. 2014). This unique function was verified ex vivo in an isogenic cell model system with and without expression of GalNAc-T11 as well as with in vitro glycosylation assays with a recombinant reporter protein expressed in E. coli containing the ligand-binding region of LDLR with multiple folded LDLR class A repeats (Pedersen et al. 2014). We were, however, unable to demonstrate activity with short peptides covering the LDLR linker regions (Pedersen et al. 2014; Kong et al. 2015), suggesting that for some protein substrates, proper folding may be required for substrate recognition. Thus, it appears that identification of unique substrates at least for some GalNAc-T isoforms will require analysis of actual protein substrates. For this, we have developed a quantitative O-glycoproteomics strategy with isogenic cell models with and without a single GalNAc-T suitable to identify non-redundant contributions of the isoform to the O-glycoproteome (Schjoldager et al. 2015), and we will in future use this strategy to further explore potential differences in the cellular functions of GalNAc-T1 and T13 in a relevant cell line.
For now, it appears that the biological significance of the two GalNAc-T1 and T13 isoforms may lie in the differential regulation of expression in different cells and tissues. Although GalNAc-T1 is also expressed in the nervous system, more detailed studies are needed to evaluate overlap in specific compartments and cell types of the brain. Interestingly, GALNT13 is one of the most dysregulated genes in gliomas (Auvergne et al. 2013). This scenario resembles that of the GalNAc-T3 and T6 subfamily isoforms, where T3 is widely expressed, while T6 is more restricted. Interestingly, in both these subfamilies, the isoforms with restricted expression (GalNAc-T6 and T13, respectively) have been found to be de novo expressed in cancer (Freire et al. 2006; Berois et al. 2006a; Wandall et al. 2007b; Gomes et al. 2009; Li et al. 2011; Matsumoto et al. 2012), suggesting they may serve yet unknown important functions related to cancer biology.
Studies of mouse models with deficiencies in Galnt genes have not in general provided support for major biological functions of the many GalNAc-T isoforms. In particular, Galnt13-/--deficient mice do not appear to exhibit specific phenotypic characteristics, although a somewhat surprising finding of altered expression of the truncated Tn O-glycan was reported in the brain (Zhang et al. 2003). In contrast, the Galnt1-/--deficient mouse model was found to exhibit a bleeding disorder and alterations in different functions of the immune system (Tenno et al. 2007; Block et al. 2012), and this may be explained by lack of redundancy in expression of Galnt1 and t13 in affected tissues due to the brain-restricted expression of the latter gene. However, further studies are clearly needed to address this.
Alternative splicing is one of the main sources of proteomic diversity in multicellular eukaryotes (Nilsen and Graveley, 2010). The GALNT family of genes contains members with large complex organization and up to 16 coding exons, and in addition encodes two distinct functional folded domains with the C-terminal lectin domains. Thus, these genes may be exceptional with respect to alternative splicing and functional implications, and examples of splice variants of GALNT14 and T5 have been reported (Ten Hagen et al. 1998; Wang et al. 2003). Raman et al. recently reported two splice variants of GALNT13, one of each comprising an interesting novel exon that encodes a replacement of part of the lectin domain (GalNAc-T13-V1) (Raman et al. 2012). We further probed potential functional splice variants of GALNT13 and identified a total of nine splice variants, including the GalNAc-T13-V1 variant (Raman et al. 2012), here designated GalNAc-T13-Ex10b. The GalNAc-T13 variant is particularly interesting because it originates as a result of alternative splicing with the use of a novel exon. There are other examples of glycosyltransferases variants derived through the use of different exons including human beta-galactoside alpha 2,6-sialyltransferase (ST6Gal-1) (Wang et al. 1993). In this case, three upstream exons are mutually exclusively utilized (exons Y+Z and exon X), resulting in two distinct populations of ST6Gal-1 mRNAs being synthesized. Tissue differences in ST6Gal-1 expression, at least in part, are the result of the regulation of the gene by multiple and distinct promoter regions present in the different exons.
Raman et al. originally analyzed the substrate specificity of the GalNAc-T13 Ex10b variant showing unchanged activity with a panel of peptides and a decrease in Km values with glycopeptide substrates (Raman et al. 2012). We did not find detectable differences using time course analysis that enables monitoring rate of incorporation of multiple GalNAc residues (Table II). The γ-domain of the GalNAc-T1 lectin domain was previously shown not to be important for the enzyme in vitro activity, while both the α and β domains were required for extensive glycosylation of an apomucin substrate assumed to include the GalNAc-glycopeptide activity function of the enzyme (Tenno et al. 2002). Regardless of the minor discrepancies in these results, it appears that the GalNAc-T13 Ex10b variant with its alternative γ-domain does not have major functional consequences. In contrast, the GalNAc-T13 variants ΔEx9 and Δ39Ex9, which lack 33 and 13 AA in the α-domain of their lectins, respectively, are inactive due to structural instability. This was clearly observed with the ΔEx9 variant which could not be expressed in insect cells as a secreted protein, whereas the Δ39Ex9 variant was secreted but had no detectable activity with the studied peptides. This may not be surprising, given the previous report that deletions in the γ, βγ or αβγ subdomains or point mutations that affect cysteines and disulphide bond formations in rat GalNAc-T1 give rise to non-functional enzymes (Tenno et al. 2002). Nevertheless, it is important to make it clear that in other cases, GalNAc-Ts were effectively expressed without their lectin domains and they still retained their catalytic activity, at least with peptides (Fritz et al. 2006; Raman et al. 2008).
Due to the abundance of splice variants in higher eukaryotes, it is probable that many biological processes, including those that lead to tumor development, are regulated by the relative expression of the splice isoforms of multiple genes (Relógio et al. 2005). In agreement with this, we found that the normal brain expressed alternative transcripts of GALNT13 in low levels as evaluated by our reverse transcription-polymerase chain reaction (RT-PCR) strategy, and in contrast a number of neuroblastoma cell lines studied expressed a complex and abundant set of alternative transcripts. The data presented here, however, do not suggest direct functional consequences. Recently, three GalNAc-T13 variants generated by differential exon usage were reported (Nogimori et al. 2016). None of them corresponds to the variants reported in this work, and no functional characterization was done. Nevertheless, they found that one variant has significant association with poor prognosis, while another one was related with a better prognosis in lung cancer patients. It will be important in future to evaluate if the splice variants identified here similarly may have prognostic value.
In summary, we conclude that the two close GalNAc-T paralogs of subfamily Ia, GalNAc-T13 and T1, have almost identical substrate specificities in contrast to previous reports by in vitro studies, and future in vivo studies are needed to explore potential distinct biological functions of these two isoforms. Our study confirmed that the lectin domain of GalNAc-T13 recognizes both N- and C-terminal remote prior GalNAc-glycosylation, which is not altered in the Ex10b splice variant but which differs from that of GalNAc-T1. Nevertheless, the main distinguishable feature between GalNAc-T1 and T13 is their strikingly different regulation and expression patterns. Thus, this subfamily resembles the GalNAc-T3/T6 subfamily, although clearly further studies are needed to evaluate the cellular functions as exemplified by studies of the GalNAc-T11 isoform.
Materials and methods
Cloning and expression of recombinant GalNAc-T variants
Expression of soluble secreted truncated GalNAc-T13 and splice variants in insect cells was performed based on a procedure previously reported (Bennett et al. 1999a). The pAcGP67 baculovirus expression constructs of the soluble coding region of the human GalNAc-T13 and the ∆Ex9, ∆39bpEx9 and Ex10b variants were prepared starting with the cloning in pGEM-T of the different sequences using the primers T13_EcoRI_His_Fw and T13_XhoI_Rev (primer sequences are available from the authors upon request). A DNA sequence encoding a His-tag was included in the forward primer. Then, the plasmids and the BD BaculoGold™ pAcGP67 Baculovirus Transfer Vector (BD Pharmingen, San Diego, CA) were digested with EcoRI (New England Biolabs, Beverly, MA). The inserts were purified from a 1.5% agarose electrophoresis gel using the GFX PCR DNA and gel band purification kit (Amersham Pharmacia Biotech Inc., Piscataway, NJ). After quantifying the different purified fragments in a gel, using the Low DNA Mass Ladder (Invitrogen, Carlsbad, CA) as a standard, the ligation with the vector was done using the T4 DNA ligase (Invitrogen). The cloned vectors were analyzed by digestion and sequencing to discard the presence of any important mutation.
Plasmids pAcGP67-T13, pAcGP67-T13 ∆Ex9, pAcGP67-T13 ∆39bpEx9 and pAcGP67-T13 Ex10b were co-transfected with BaculoGold™ DNA (BD Pharmingen), and recombinant baculovirus were obtained after two successive amplifications in Sf9 insect cells (Life Technologies, Gaithersburg, MD), according to the manufacturer's instructions. Amplified viruses were used for infection of High Five insect cells (Life Technologies) grown in Express Five® SFM (Gibco BRL, Rockville, MD) in upright roller bottles shaken 100 rpm at 26.5°C (cell density of approximately 2 × 105). Secreted, soluble recombinant proteins were harvested (centrifugation at 2000 × g, 4°C, 10 min) and then the His tagged enzymes were affinity purified by Ni-NTA as previously described (Vester-Christensen et al. 2013). The samples were diluted in a 10 mM MES, 10 mM NaCl pH 5.8 buffer and were further purified by cation exchange chromatography using a Mono S 5/50 GL column (GE Healthcare, Buckinghamshire, UK) and eluting with a gradient of NaCl from 0.01 to 1 M in 20 CV. The presence of the enzymes in the different fractions and the corresponding purity were assayed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis stained for proteins with Coomassie Blue R-250 and by western blot using an anti-His antibody. The fractions that contained the enzymes were pooled, and the activity was analyzed by standard GalNAc-transferase assays.
In vitro GalNAc-transferase assays
In vitro glycosylation assays were performed as product development assays in 25 µL of buffer (25 mM cacodylic acid sodium, pH 7.4, 10 mM MnCl2, 0.25% Triton X-100), 4 mM UDP-GalNAc (Sigma-Aldrich), 0.1 μg of purified enzyme and 10 μg of acceptor peptide (Schafer-N and NeoBioSci) (Table I and Supplementary data,Table SI). The reactions were left overnight at 37°C with gentle mixing. For time course evaluation, 1 μL of the reaction mixtures were taken at 1, 4 and 24 h and analyzed by MALDI-TOF MS.
MALDI-TOF MS analysis
Evaluation of incorporation of GalNAc residues into peptide substrates was performed by MALDI-TOF MS. Sampling of reactions (1 µL) were diluted 10-fold in 0.1% TFA/H2O and 1 μL mixed with 1 μL of matrix. The matrix was 2,5-dihydroxybenzoic acid (25 mg/mL, Sigma-Aldrich) dissolved in a 1:1 mixture of methanol and water. Mass spectra were acquired on a 4800 MALDI TOF/TOF Analyzer mass spectrometer (Applied Biosystems/MDS Sciex, Concord, Canada). Recorded data were processed using the Data Explorer software.
Glycosylation of random peptide substrates and determination of transferase substrate specificity
Three oriented random glycopeptide libraries in the form of GAGAXXXXXTXXXXXAGA (where X = G,A,P,V,L,E,Q,R,H (PVI), G,A,P,I,M,F,D,N,R,K (PVII) or G,A,P,V,Y,E,N,S,R,K (PVIII)) were utilized to obtain the GalNAc-T13 catalytic domain specificity as described previously (Gerken et al. 2011). Briefly, reactions were carried out with 10 mM MnCl2, 50 mM sodium cacodylate, pH 6.5, 1.3 mM 2-mercaptoethanol, 2 mM UDP-GalNAc (containing a total of 100 µCi UDP-[3H]-GalNAc), 1/100 dilution of Protease Inhibitor Cocktails P8340 and P8849 (Sigma-Aldrich), 0.003% sodium azide, 5 mg/mL random peptide substrate (~1.7 mM) and 50 µL of GalNAc-T13 (~0.1 mg/mL) to a final volume of 250 µL. Reactions were incubated overnight at 37°C and the glycopeptide products isolated as previously described (Gerken et al. 2011). The comparison of the Edman AA sequence analysis (ABI Procise 494) of the starting random peptide and the isolated random glycopeptide products revealed the so-called enhancement values (Gerken et al. 2011). Data for GalNAc-T13 were based on one to five determinations on each of the random peptide libraries and the data averaged (see Figure 1 and Supplementary data, Figures S1 and S2). Enhancement values represent the propensity of a particular AA residue type to be utilized by the transferase, where values greater than 1 suggest a high preference while values significantly less than 1 indicate a low preference, suggesting an inhibitory effect.
Cell lines and tissues
The neuroblastoma metastatic model, IGR-N-91, derived from an 8-year-old high-risk neuroblastoma patient, was obtained by in vitro culturing of malignant neuroblasts from the bone marrow and successive xenografts into nude mice, as previously described (Ferrandis et al. 1994). Sublines were established from primary tumor xenograft (IGR-N-91 PTX) and bone marrow metastasis (IGR-N-91 BM), and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 2 mMol/L L-glutamine, 1 mMol/L sodium pyruvate and 10% fetal bovine serum (FBS), at 37°C in a 5% CO2-humidified atmosphere. IMR-32 (another neuroblastoma cell line) and HeLa (cervical cancer cell line), obtained from ATCC (American Type Culture Collection, Rockville, MD), were cultured in the same conditions up to 80% confluence. Cells were washed with sterile phosphate buffered saline (PBS) and detached from monolayer culture using 0.05% trypsin in PBS at 37°C. Harvested cells in DMEM medium containing FBS were pelleted after centrifugation, washed with sterile PBS and then used for RNA extraction.
Primary neuroblastoma tumor tissues were obtained from patients of Institut Gustave Roussy (France) with the approval of the appropriate ethics committees and according to the national law of people taking part in biomedical research. Tumors were immediately snap-frozen before being stored in liquid nitrogen until nucleic acids extraction. The central nervous system sample was obtained from the surgery of a patient with brain injury.
Screening of GalNAc-T13 splice variants
Total RNA from cell lines and tissues was extracted with Tri-Reagent (Sigma-Aldrich), according to the manufacturer's instructions. First-strand cDNA was synthesized using 1 µg of RNA with MMLV reverse transcriptase (Amersham, Piscataway, NJ). The reaction mixture consisted of 200 U of enzyme, 2 µL of 10 mmol/L of each deoxynucleoside triphosphate (dNTP) and 1 µL of 250 ng of random hexamers in a 20 µL total reaction volume. After incubation at 37°C for 1 h, the mixture was heated at 96°C, snap-cooled and stored at −20°C until use. The RT-PCR was performed with a high-fidelity polymerase, Platinum pfx DNA polymerase (Invitrogen), and the T13_XhoI_Fw and T13_XhoI_Rev primers (primer sequences are available from the authors upon request). The PCR products were purified with the GFX PCR DNA and gel band purification kit (Amersham) and then cloned with the pGEM-T Easy Vector System I (Promega Corp., Madison, WI). Next, E. coli XL1 Blue were transformed and afterwards analyzed by colony PCR with the T13_XhoI_Fw and T13_XhoI_Rev primers. The PCR products where digested with HindIII and EcoRV and were evaluated by agarose electrophoresis. The clones that deviated from the theoretical pattern were sequenced.
GalNAc-T lectin domain specificity from random glycopeptide substrates
The lectin domain probing random glycopeptide substrates GP(T*22)R and GP(T*10)L (and their non-glycosylated controls GP(A22)R and GP(A10)L), whose sequences are given in Figure 4A, were utilized to compare the remote GalNAc-O-Thr glycopeptide specificity of GalNAc-T13 and GalNAc-T13 Ex10b as previously described (Gerken et al. 2013). Reactions were carried out using 68 mM sodium cacodylate, pH 6.5, 1.8 mM 2-mercaptoethanol, 10 mM MnCl2, 50 µM [3H]-UDP-GalNAc (~6 × 108 DPM/µmol), 5 mg/mL (~1.5 mM) of substrates GP(T*22)R, GP(A22)R, GP(T*10)L and GP(A10)L and 100–150 µL of GalNAc-T13 or GalNAc-T13 Ex10b (0.06–0.1 mg/ml) to a total reaction volume of 250–300 µL. Reactions were incubated at 37°C and 50–80 µL aliquots removed for analysis after incubating for 15 min, 45 min, 2 h, 4 h and overnight for wt GalNAc-T13 and due to limited transferase, 2 h, 4 h and overnight for GalNAc-T13 Ex10b. In all cases, reactions with wt GalNAc-T13 and GalNAc-T13 Ex10b were performed concurrently under identical reaction conditions. Glycopeptide workup and isolation was performed as described by Gerken et al. (2013), ending with chromatography on Sephadex G10. The lyophilized pooled glycopeptide fractions were taken up in 1 mL of H2O and transferase activity quantified by [3H]-GalNAc content normalized to the 220 and 280 nm OD values. With this approach, any losses in peptide substrate during sample processing were corrected for by normalizing to the peptide OD values. The C-terminal/N-terminal random glycopeptide utilization ratios (GP(T*22)R/ GP(T*10)L) were obtained from these normalized values as shown in Figure 4C.
RT-PCR of GalNAc-T13 and the splice variants
The β2M (β2-microglobulin) gene was amplified to verify cDNA quality (except for CNS, where we used GalNAc-T9 as a control). Different RT-PCR reactions were optimized in order to amplify fragments of GalNAc-T13 and its splice variants, by means of a single round of 35 cycles for GalNAc-T13 and Ex10b variants and 40 cycles for the ∆Ex2-7 variant, or a nested PCR for ∆Ex6, ∆Ex8, ∆Ex9, ∆39bpEx9, ∆Ex6/39bpEx9, ∆Ex6/8 and ΔEx6/8/39bpEx9 variants (primer sequences are available from the authors upon request). For wt GalNAc-T13 amplification, 1 µL of cDNA was added to a final volume of 25 µL of a PCR mixture containing 10 mM Tris-HCl (pH 8.8), 50 mM KCl, 3 mM MgCl2, 400 nM of each primer (GALNT13-Fw/T13_XhoI_Rev), 200 µM dNTPs and 1 unit of Taq DNA polymerase (Fermentas, Hunover, MD). Thirty-five cycles were performed as follows: 30 seconds at 95°C, 30 seconds at 60°C and 1 min at 72°C; the PCR product analyzed by electrophoresis in a 2% agarose gel resulted in a 425 bp band. Same conditions but 2 mM MgCl2 (and 2 μL of cDNA for ΔEx2-7) were used for Ex10b and ∆Ex2-7 amplifications using T13_Ex10b_Fw/T13_Ex11_Rev and T13_XhoI_Fw/T13_Δ2-7_Rev pair of primers, respectively. The annealing temperatures were 60°C and 62°C, whereas the obtained PCR products were 270 and 166 bp, respectively. The same first round was performed for the seven remaining variants, seeding 1 µL of cDNA in a final volume of 25 µL. The PCR mixture contained 10 mM Tris-HCl (pH 8.8), 50 mM KCl, 1.5 mM MgCl2, 400 nM of each primer (T13_784_Fw/T13_Ex11_Rev), 200 µM dNTPs and 1 unit of Taq DNA polymerase, was amplified for 20 cycles at 59°C of annealing temperature and the PCR product obtained had 820 bp. The second round was performed for 25 cycles for variant ΔEx9, 30 cycles for variants ΔEx8 and ΔEx6/8, and 35 cycles for variants ΔEx6, ∆39bpEx9, ∆Ex6/39bpEx9 and ΔEx6/8/39bpEx9. One microliter of the first round product was seeded and the second round was done in the same conditions, although 2 mM MgCl2 was used in the case of ∆8 and ∆9 variants. The pair of primers and the PCR products were T13_Δ6_Fw/T13_Ex7_Rev (201 bp); T13_Ex7_Fw/T13_Δ8_Rev (197 bp); T13_Ex7_Fw/T13_1393_Rev (336 bp); GalNT13_Fw/T13_Δ39bpEx9_Rev (131 bp); T13_Δ6_Fw/T13_Δ8_Rev (226 bp) and T13_Δ6_Fw/T13_Δ39bpEx9_Rev (425 and 285 bp) for ∆Ex6, ∆Ex8, ∆Ex9, ∆39bpEx9, ∆Ex6/8, ΔEx6/39bpEx9 and ΔEx6/8/39bpEx9 variants, respectively. PCR products (15 µL) were analyzed by electrophoresis on 2% agarose gels by direct visualization after ethidium bromide staining. The identity of each band was confirmed by sequencing.
Molecular modeling
Protein sequences were aligned with T-Coffee (Notredame et al. 2000). A molecular model of the lectin domain of GalNAc-T13 was constructed by comparative modeling using Modeller (Sali et al. 1995) (doi:10.1002/prot.340230306), using as template the X-ray structure of GalNAc-T1 (Fritz et al. 2004). The model comprises residues 429 to 554 which span the entire lectin domain. A total of 10,000 models were generated, from which the best was selected on the base of the Modeller's objective function and stereochemical properties were assessed using Procheck (Laskowski et al. 1996). The selected model contains 92% of AAs in most favored regions.
Funding
Mizutani Foundation for Glycoscience (number 70059 to E.O.); Programa Grupos de Investigación, CSIC, Universidad de la República, Uruguay (number 908 to E.O.); Fondo para la Convergencia Estructural del MERCOSUR (COF 03/11 to E.O.); Danish National Research Foundation (DNRF104 to H.C.); Scholarship from the Agencia Nacional de Investigación e Innovación (POS_2011_1_3327 to M.F.F.); Scholarship from Comisión Académica de Posgrado, Universidad de la República (to M.F.F.); The National Institutes of Health [grant U01-GM113534] (to T.A.G.).
Conflict of interest statement
None declared.
Abbreviations
AA, amino acids; bp, base pair; CV, column volume; dNTPs, deoxynucleosides triphosphate; FBS, fetal bovine serum; GalNAc, N-acetyl-D-galactosamine; GalNAc-T, UDP-GalNAc: polypeptide N-acetylgalactosaminyltransferase; GALNT, GalNAc-transferase gene; GP(A22)R and GP(A10)L, non-glycosylated controls of GP(T*22)R and GP(T*10)L; GP(T*22)R and GP(T*10)L, random glycopeptidesas defined in Figure 4A; His, histidine; MALDI-TOF, matrix assisted laser desorption/ionization time-of-flight; MES, 2-(N-morpholino)ethanesulfonic acid; MMLV, Moloney Murine Leukemia Virus; MS, mass spectrometry; MUC7, mucin 7; Ni-NTA, nickel-nitrilotriacetic acid; OD, Optical density; PBS, phosphate buffered saline; PCR, polymerase chain reaction; RT-PCR, reverse transcription-polymerase chain reaction; Ser, serine; T*, Thr-O-GalNAc; Thr, threonine; TFA, trifluoroaceticacid; Tris, tris(hydroxymethyl)aminomethane; Tyr, tyrosine; UDP, uridine diphosphate; wt, wild-type.
Supplementary data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
References
- Auvergne RM, Sim FJ, Wang S, Chandler-Militello D, Burch J, Al Fanek Y, Davis D, Benraiss A, Walter K, Achanta P et al. 2013. Transcriptional differences between normal and glioma-derived glial progenitor cells identify a core set of dysregulated genes. Cell Rep. 3:2127–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, Tabak LA. 2012. Control of mucin-type O-glycosylation. A classification of the polypeptide GalNAc-transferase gene family. Glycobiology. 22:736–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EP, Hassan H, Hollingsworth MA, Clausen H. 1999. a. A novel human UDP-N-acetyl-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase, GalNAc-T7, with specificity for partial GalNAc-glycosylated acceptor substrates. FEBS Lett. 460:226–230. [DOI] [PubMed] [Google Scholar]
- Bennett EP, Hassan H, Mandel U, Hollingsworth MA, Akisawa N, Ikematsu Y, Merkx G, van Kessel AG, Olofsson S, Clausen H. 1999. b. Cloning and characterization of a close homologue of human UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-T3, designated GalNAc-T6. Evidence for genetic but not functional redundancy. J Biol Chem. 274:25362–25370. [DOI] [PubMed] [Google Scholar]
- Bennett EP, Hassan H, Mandel U, Mirgorodskaya E, Roepstorff P, Burchell J, Taylor-Papadimitriou J, Hollingsworth MA, Merkx G, van Kessel AG et al. 1998. Cloning of a human UDP-N-acetyl-alpha-D-Galactosamine:polypeptide N-acetylgalactosaminyltransferase that complements other GalNAc-transferases in complete O-glycosylation of the MUC1 tandem repeat. J Biol Chem. 273:30472–30481. [DOI] [PubMed] [Google Scholar]
- Berois N, Mazal D, Ubillos L, Trajtenberg F, Nicolas A, Sastre-Garau X, Magdelenat H, Osinaga E. 2006. a. UDP-N-acetyl-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase-6 as a new immunohistochemical breast cancer marker. J Histochem Cytochem. 54:317–328. [DOI] [PubMed] [Google Scholar]
- Berois N, Blanc E, Ripoche H, Mergui X, Trajtenberg F, Cantais S, Barrois M, Dessen P, Kågedal B, Bénard J et al. 2006. b. ppGalNAc-T13: a new molecular marker of bone marrow involvement in neuroblastoma. Clin Chem. 52:1701–1712. [DOI] [PubMed] [Google Scholar]
- Block H, Ley K, Zarbock A. 2012. Severe impairment of leukocyte recruitment in ppGalNAcT-1-deficient mice. J Immunol. 188:5674–5681. [DOI] [PubMed] [Google Scholar]
- Brockhausen I. 2006. Mucin-type O-glycans in human colon and breast cancer. Glycodynamics and functions. EMBO Rep. 7:599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Tachibana K, Zhang Y, Guo JM, Kahori Tachibana K, Kameyama A, Wang H, Hiruma T, Iwasaki H, Togayachi A et al. 2002. Characterization of a novel human UDP-GalNAc transferase, pp-GalNAc-T10. FEBS Lett. 531:115–121. [DOI] [PubMed] [Google Scholar]
- Ferrandis E, Da Silva J, Riou G, Bénard J. 1994. Coactivation of the MDR1 and MYCN in human neuroblastoma during the metastatic process in the nude mouse. Cancer Res. 54:2256–2261. [PubMed] [Google Scholar]
- Freire T, Berois N, Sóñora C, Varangot M, Barrios E, Osinaga E. 2006. UDP-N-acetyl-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (ppGalNAc-T6) mRNA as a potential new marker for detection of bone marrow-disseminated breast cancer cells. Int J Cancer. 119:1383–1388. [DOI] [PubMed] [Google Scholar]
- Fritz TA, Raman J, Tabak LA. 2006. Dynamic association between the catalytic and lectin domains of human UDP-GalNAc:polypeptide α-N-acetylgalactosaminyltransferase-2. J Biol Chem. 281:8613–8619. [DOI] [PubMed] [Google Scholar]
- Fritz TA, Hurley JH, Trinh LB, Shiloach J, Tabak LA. 2004. The beginnings of mucin biosynthesis: the crystal structure of UDP-GalNAc:polypeptide alpha-N-acetylgalactosaminyltransferase-T1. Proc Natl Acad Sci USA. 101:15307–15312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken TA, Revoredo L, Thome JJ, Tabak LA, Vester-Christensen MB, Clausen H, Gahlay GK, Jarvis DL, Johnson RW, Moniz HA et al. 2013. The lectin domain of the polypeptide GalNAc transferase family of glycosyltransferases (ppGalNAcTs) acts as a switch directing glycopeptide substrate glycosylation in an N- or C-terminal direction, further controlling mucin type O-glycosylation. J Biol Chem. 288:19900–19914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken TA, Jamison O, Perrine CL, Collette JC, Moinova H, Ravi L, Markowitz SD, Shen W, Patel H, Tabak LA. 2011. Emerging paradigms for the initiation of mucin-type protein O-glycosylation by the polypeptide GalNAc transferase family of glycosyltransferases. J Biol Chem. 286:14493–14507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken TA, Ten Hagen KG, Jamison O. 2008. Conservation of peptide acceptor preferences between Drosophila and mammalian polypeptide-GalNAc transferase ortholog pairs. Glycobiology. 18:861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken TA, Raman J, Fritz TA, Jamison O. 2006. Identification of common and unique peptide substrate preferences for the UDP-GalNAc:polypeptide alpha-N-acetylgalactosaminyltransferases T1 and T2 derived from oriented random peptide substrates. J Biol Chem. 281:32403–32416. [DOI] [PubMed] [Google Scholar]
- Gill DJ, Clausen H, Bard F. 2011. Location, location, location. New insights into O-GalNAc protein glycosylation. Trends Cell Biol. 21:149–158. [DOI] [PubMed] [Google Scholar]
- Gomes J, Marcos NT, Berois N, Osinaga E, Magãlhaes A, Pinto-de-Sousa J, Almeida R, Gärtner F, Reis CA. 2009. Expression of UDP-N-acetyl-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase-6 in gastric mucosa, intestinal metaplasia, and gastric carcinoma. J Histochem Cytochem. 57:79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C, Oyama T, Osaki T, Li J, Takenoyama M, Izumi H, Sugio K, Kohno K, Yasumoto K. 2004. Low expression of polypeptide GalNAc N-acetylgalactosaminyl transferase-3 in lung adenocarcinoma: Impact on poor prognosis and early recurrence. Br J Cancer. 90:436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen FK, Ten Hagen KG, Beres TM, Balys MM, VanWuyckhuyse BC, Tabak LA. 1997. cDNA cloning and expression of a novel UDP-N-acetyl-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase. J Biol Chem. 272:13843–13848. [DOI] [PubMed] [Google Scholar]
- Hassan H, Reis CA, Bennett EP, Mirgorodskaya E, Roepstorff P, Hollingsworth MA, Burchell J, Taylor-Papadimitriou J, Clausen H. 2000. The lectin domain of UDP-N-acetyl-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase-T4 directs its glycopeptide specificities. J Biol Chem. 275:38197–38205. [DOI] [PubMed] [Google Scholar]
- Hennet T, Hagen FK, Tabak LA, Marth JD. 1995. T-cell-specific deletion of a polypeptide N-acetylgalactosaminyl-transferase gene by site-directed recombination. Proc Natl Acad Sci USA. 92:12070–12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth MA, Swanson BJ. 2004. Mucins in cancer. Protection and control of the cell surface. Nat Rev Cancer. 4:45–60. [DOI] [PubMed] [Google Scholar]
- Inoue T, Eguchi T, Oda Y, Nishiyama K, Fujii K, Izumi H, Kohno K, Yamaguchi K, Tanaka M, Tsuneyoshi M. 2007. Expression of GalNAc-T3 and its relationships with clinicopathological factors in 61 extrahepatic bile duct carcinomas analyzed using stepwise sections—special reference to its association with lymph node metastases. Mod Pathol. 20:267–276. [DOI] [PubMed] [Google Scholar]
- Ishikawa M, Kitayama J, Nariko H, Kohno K, Nagawa H. 2004. The expression pattern of UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyl transferase-3 in early gastric carcinoma. J Surg Oncol. 86:28–33. [DOI] [PubMed] [Google Scholar]
- Kingsley PD, Hagen KG, Maltby KM, Zara J, Tabak LA. 2000. Diverse spatial expression patterns of UDP-GalNAc:polypeptide N-acetylgalactosaminyl-transferase family member mRNAs during mouse development. Glycobiology. 10:1317–1323. [DOI] [PubMed] [Google Scholar]
- Kohsaki T, Nishimori I, Nakayama H, Miyasaki E, Enzan H, Nomoto M, Hollingsworth MA, Onishi S. 2000. Expression of UDP-GalNAc: polypeptide N-acetylgalactosaminyltransferase isozymes T1 and T2 in human colorectal cancer. J Gastroenterol. 35:840–848. [DOI] [PubMed] [Google Scholar]
- Kong Y, Joshi HJ, Schjoldager KT, Madsen TD, Gerken TA, Vester-Christensen MB, Wandall HH, Bennett EP, Levery SB, Vakhrushev SY et al. 2015. Probing polypeptide GalNAc-transferase isoform substrate specificities by in vitro analysis. Glycobiology. 25:55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landers KA, Burger MJ, Tebay MA, Purdie DM, Scells B, Samaratunga H, Lavin MF, Gardiner RA. 2005. Use of multiple biomarkers for a molecular diagnosis of prostate cancer. Int J Cancer. 114:950–956. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. 1996. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 8:477–486. [DOI] [PubMed] [Google Scholar]
- Li Z, Yamada S, Inenaga S, Imamura T, Wu Y, Wang KY, Shimajiri S, Nakano R, Izumi H, Kohno K et al. 2011. Polypeptide N-acetylgalactosaminyltransferase 6 expression in pancreatic cancer is an independent prognostic factor indicating better overall survival. Br J Cancer. 104:1882–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lira-Navarrete E, de Las Rivas M, Compañón I, Pallarés MC, Kong Y, Iglesias-Fernández J, Bernardes GJ, Peregrina JM, Rovira C, Bernadó P et al. 2015. Dynamic interplay between catalytic and lectin domains of GalNAc-transferases modulates protein O-glycosylation. Nat Commun. 6:6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe JB, Marth JD. 2003. A genetic approach to Mammalian glycan function. Annu Rev Biochem. 72:643–691. [DOI] [PubMed] [Google Scholar]
- Mandel U, Hassan H, Therkildsen MH, Rygaard J, Jakobsen MH, Juhl BR, Dabelsteen E, Clausen H. 1999. Expression of polypeptide GalNAc-transferases in stratified epithelia and squamous cell carcinomas: immunohistological evaluation using monoclonal antibodies to three members of the GalNAc-transferase family. Glycobiology. 9:43–52. [DOI] [PubMed] [Google Scholar]
- Marth JD. 1996. Complexity in O-linked oligosaccharide biosynthesis engendered by multiple polypeptide N-acetylgalactosaminyltransferases. Glycobiology. 6:701–705. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Zhang Q, Akita K, Nakada H, Hamamura K, Tsuchida A, Okajima T, Furukawa K, Urano T, Furukawa K. 2013. Trimeric Tn antigen on syndecan 1 produced by ppGalNAc-T13 enhances cancer metastasis via a complex formation with integrin α5β1 and matrix metalloproteinase 9. J Biol Chem. 288:24264–24276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Zhang Q, Akita K, Nakada H, Hamamura K, Tokuda N, Tsuchida A, Matsubara T, Hori T, Okajima T et al. 2012. pp-GalNAc-T13 induces high metastatic potential of murine Lewis lung cancer by generating trimeric Tn antigen. Biochem Biophys Res Commun. 419:7–13. [DOI] [PubMed] [Google Scholar]
- Miyahara N, Shoda J, Kawamoto T, Furukawa M, Ueda T, Todoroki T, Tanaka N, Matsuo K, Yamada Y, Kohno K et al. 2004. Expression of UDP-N-acetyl-alpha-D-galactosamine-polypeptide N-acetylgalactosaminyltransferase isozyme 3 in the subserosal layer correlates with postsurgical survival of pathological tumor stage 2 carcinoma of the gallbladder. Clin Cancer Res. 10:2090–2099. [DOI] [PubMed] [Google Scholar]
- Nehrke K, Hagen FK, Tabak LA. 1998. Isoform-specific O-glycosylation by murine UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase-T3, in vivo. Glycobiology. 8:367–371. [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature. 463:457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogimori K, Hori T, Kawaguchi K, Fikui T, Mii S, Nakada H, Matsumoto Y, Yamauchi Y, Takahashi M, Furukawa K et al. 2016. Increased expression levels of ppGalNAc-T13 in lung cancers: significance in the prognostic diagnosis. Int J Oncol. 49:1369–1376. [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 302:205–217. [DOI] [PubMed] [Google Scholar]
- Pedersen NB, Wang S, Narimatsu Y, Yang Z, Halim A, Schjoldager KT, Madsen TD, Seidah NG, Bennett EP, Levery SB et al. 2014. Low density lipoprotein receptor class A repeats are O-glycosylated in linker regions. J Biol Chem. 289:17312–17324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen JW, Bennett EP, Schjoldager KT, Meldal M, Holmér AP, Blixt O, Cló E, Levery SB, Clausen H, Wandall HH. 2011. Lectin domains of polypeptide GalNAc transferases exhibit glycopeptide binding specificity. J Biol Chem. 286:32684–32696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, Togayachi A, Kwon YD, Xie C, Wu G, Zou X, Sato T, Ito H, Tachibana K, Kubota T et al. 2010. Identification of a novel human UDP-GalNAc transferase with unique catalytic activity and expression profile. Biochem Biophys Res Commun. 402:680–686. [DOI] [PubMed] [Google Scholar]
- Perrine CL, Ganguli A, Wu P, Bertozzi CR, Fritz TA, Raman J, Tabak LA, Gerken TA. 2009. Glycopeptide-preferring polypeptide GalNAc transferase 10 (ppGalNAc T10), involved in mucin-type O-glycosylation, has a unique GalNAc-O-Ser/Thr-binding site in its catalytic domain not found in ppGalNAc T1 or T2. J Biol Chem. 284:20387–20397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman J, Guan Y, Perrine CL, Gerken TA, Tabak LA. 2012. UDP-N-acetyl-α-D-galactosamine:polypeptide N-acetylgalactosaminyltransferases: completion of the family tree. Glycobiology. 22:768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman J, Fritz TA, Gerken TA, Jamison O, Live D, Liu M, Tabak LA. 2008. The catalytic and lectin domains of UDP-GalNAc:polypeptide alpha-N-Acetylgalactosaminyltransferase function in concert to direct glycosylation site selection. J Biol Chem. 283:22942–22951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relógio A, Ben-Dov C, Baum M, Riggiu M, Gemund C, Benes V, Darnell RB, Valcárcel J. 2005. Alternative splicing microarrays reveal functional expression of neuron-specific regulators in Hodgkin lymphoma cells. J Biol Chem. 280:4779–4784. [DOI] [PubMed] [Google Scholar]
- Revoredo L, Wang S, Bennett EP, Clausen H, Moremen KW, Jarvis DL, Ten Hagen KG, Tabak LA, Gerken TA. 2016. Mucin-type O-glycosylation is controlled by short- and long-range glycopeptide substrate recognition that varies among members of the polypeptide GalNAc transferase family. Glycobiology. 26:360–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. 1995. Evaluation of comparative protein modeling by MODELLER. Proteins. 23:318–326. [DOI] [PubMed] [Google Scholar]
- Schjoldager KT, Joshi HJ, Kong Y, Goth CK, King SL, Wandall HH, Bennett EP, Vakhrushev SY, Clausen H. 2015. Deconstruction of O-glycosylation--GalNAc-T isoforms direct distinct subsets of the O-glycoproteome. EMBO Rep. 16:1713–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjoldager KT, Clausen H. 2012. Site-specific protein O-glycosylation modulates proprotein processing – deciphering specific functions of the large polypeptide GalNAc-transferase gene family. Biochim Biophys Acta. 1820:2079–2094. [DOI] [PubMed] [Google Scholar]
- Schwientek T, Bennett EP, Flores C, Thacker J, Hollmann M, Reis CA, Behrens J, Mandel U, Keck B, Schäfer MA et al. 2002. Functional conservation of subfamilies of putative UDP-N-acetylgalactosamine:polypeptide N-acetylgalactosaminyltransferases in Drosophila, Caenorhabditis elegans, and mammals. One subfamily composed of l(2)35Aa is essential in Drosophila. J Biol Chem. 277:22623–22638. [DOI] [PubMed] [Google Scholar]
- Shibao K, Izumi H, Nakayama Y, Ohta R, Nagata N, Nomoto M, Matsuo K, Yamada Y, Kitazato K, Itoh H et al. 2002. Expression of UDP-N-acetyl-alpha-D-galactosamine-polypeptide GalNAc N-acetylgalactosaminyl transferase-3 in relation to differentiation and prognosis in patients with colorectal carcinoma. Cancer. 94:1939–1946. [DOI] [PubMed] [Google Scholar]
- Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L et al. 2013. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32:1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarp MA, Clausen H. 2008. Mucin-type O-glycosylation and its potential use in drug and vaccine development. Biochim Biophys Acta. 1780:546–563. [DOI] [PubMed] [Google Scholar]
- Ten Hagen KG, Hagen FK, Balys MM, Beres TM, Van Wuyckhuyse B, Tabak LA. 1998. Cloning and expression of a novel, tissue specifically expressed member of the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family. J Biol Chem. 273:27749–27754. [DOI] [PubMed] [Google Scholar]
- Tenno M, Ohtsubo K, Hagen FK, Ditto D, Zarbock A, Schaerli P, von Andrian UH, Ley K, Le D, Tabak LA et al. 2007. Initiation of protein O-glycosylation by the polypeptide GalNAcT-1 in vascular biology and humoral immunity. Mol Cell Biol. 27:8783–8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenno M, Saeki A, Késdy FJ, Elhammer AP, Kurosaka A. 2002. The lectin domain of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase 1 is involved in O-glycosylation of a polypeptide with multiple acceptor sites. J Biol Chem. 277:47088–47096. [DOI] [PubMed] [Google Scholar]
- Tran DT, Zhang L, Zhang Y, Tian E, Earl LA, Ten Hagen KG. 2012. Multiple members of the UDP-GalNAc: polypeptide N-acetylgalactosaminyltransferase family are essential for viability in Drosophila. J Biol Chem. 287:5243–5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vester-Christensen MB, Bennett EP, Clausen H, Mandel U. 2013. Generation of monoclonal antibodies to native active human glycosyltransferases. Methods Mol Biol. 1022:403–420. [DOI] [PubMed] [Google Scholar]
- Wandall HH, Irazoqui F, Tarp MA, Bennett EP, Mandel U, Takeuchi H, Kato K, Irimura T, Suryanarayanan G, Hollingsworth MA et al. 2007. a. The lectin domains of polypeptide GalNAc-transferases exhibit carbohydrate-binding specificity for GalNAc: lectin binding to GalNAc-glycopeptide substrates is required for high density GalNAc-O-glycosylation. Glycobiology. 17:374–387. [DOI] [PubMed] [Google Scholar]
- Wandall HH, Dabelsteen S, Sørensen JA, Krogdahl A, Mandel U, Dabelsteen E. 2007. b. Molecular basis for the presence of glycosylated onco-foetal fibronectin in oral carcinomas: the production of glycosylated onco-foetal fibronectin by carcinoma cells. Oral Oncol. 43:301–309. [DOI] [PubMed] [Google Scholar]
- Wandall HH, Hassan H, Mirgorodskaya E, Kristensen AK, Roepstorff P, Bennett EP, Nielsen PA, Hollingsworth MA, Burchell J, Taylor-Papadimitriou J et al. 1997. Substrate specificities of three members of the human UDP-N-acetyl-alpha-D-galactosamine:Polypeptide N-acetylgalactosaminyltransferase family, GalNAc-T1, -T2, and -T3. J Biol Chem. 272:23503–23514. [DOI] [PubMed] [Google Scholar]
- Wang H, Tachibana K, Zhang Y, Iwasaki H, Kameyama A, Cheng L, Guo JM, Hiruma T, Togayachi A, Kudo T et al. 2003. Cloning and characterization of a novel UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase, pp-GalNAc-T14. Biochem Biophys Res Commun. 300:738–744. [DOI] [PubMed] [Google Scholar]
- Wang X, Vertino A, Eddy RL, Byers MG, Jani-Sait SN, Shows TB, Lau JT. 1993. Chromosome mapping and organization of the human beta-galactoside alpha 2,6-sialyltransferase gene. Differential and cell-type specific usage of upstream exon sequences in B-lymphoblastoid cells. J Biol Chem. 268:4355–4361. [PubMed] [Google Scholar]
- Wu C, Guo X, Wang W, Wang Y, Shan Y, Zhang B, Song W, Ma S, Ge J, Deng H et al. 2010. N-Acetylgalactosaminyltransferase-14 as a potential biomarker for breast cancer by immunohistochemistry. BMC Cancer. 10:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Pang W, Lu J, Shan A, Zhang Y. 2016. Polypeptide N-acetylgalactosaminyltransferase 13 contributes to neurogenesis via stabilizing the mucin-type O-glycoprotein podoplanin. J Biol Chem. 291:23477–23488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Nakamori S, Tsujie M, Takahashi Y, Nagano H, Dono K, Umeshita K, Sakon M, Tomita Y, Hoshida Y et al. 2004. Expression of uridine diphosphate N-acetyl-α-D-galactosamine:polypeptide N-acetylgalactosaminyl transferase 3 in adenocarcinoma of the pancreas. Pathobiology. 71:12–18. [DOI] [PubMed] [Google Scholar]
- Young WW Jr, Holcomb DR, Ten Hagen KG, Tabak LA. 2003. Expression of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase isoforms in murine tissues determined by real-time PCR: a new view of a large family. Glycobiology. 13:549–557. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Iwasaki H, Wang H, Kudo T, Kalka TB, Hennet T, Kubota T, Cheng L, Inaba N, Gotoh M et al. 2003. Cloning and characterization of a new human UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase, designated pp-GalNAc-T13, that is specifically expressed in neurons and synthesizes GalNAc alpha-serine/threonine antigen. J Biol Chem. 278:573–584. [DOI] [PubMed] [Google Scholar]