Abstract
Computer simulation of genomic data has become increasingly popular for assessing and validating biological models or to gain understanding about specific datasets. Multiple computational tools for the simulation of next-generation sequencing (NGS) data have been developed in recent years, which could be used to compare existing and new NGS analytical pipelines. Here we review 23 of these tools, highlighting their distinct functionality, requirements and potential applications. We also provide a decision tree for the informed selection of an appropriate NGS simulation tool for the specific question at hand.
Introduction
Next-generation sequencing (NGS) techniques are the standard nowadays for the generation of genomic data, producing ever-increasing amounts of information rapidly and at a low cost. These techniques allow us to sequence DNA and RNA very quickly, facilitating the acquisition of massive genomic, transcriptomic, DNA-protein interaction and epigenomic datasets, and are radically changing the way we look at genomes1–3. Given their higher parallelism and smaller reaction volumes compared to conventional Sanger sequencing, NGS methods offer larger amounts of data, shorter sequencing time and reduced costs, albeit at the cost of increased error rates and shorter reads4. NGS clearly facilitates the accumulation of large data sets, but the downstream processing of these data is still an important bottleneck5. Not surprisingly, NGS data result in numerous bioinformatics challenges, including storage, transmission, manipulation and analysis. Better computational methods and more efficient software tools are constantly being developed in order to provide faster processing and more accurate inferences. However, it is essential that these methods are benchmarked against existing tools with similar functionality, in order to show their superiority at least in some aspect. In general, computational methods can be benchmarked using empirical and/or simulated data. Although validation with empirical data is essential as it represents real scenarios, the true process underlying it is usually unknown, complicating its use for the assessment of accuracy (that is, how close the estimated value is to the ‘true’ value). On the other hand, in silico data allow us to generate as much data as desired and under controlled scenarios with predefined parameters for which the ‘true’ values are known, nicely complementing the validation with real data6,7. Thus, computer simulation of genetic and genomic data has become increasingly popular for assessing and validating biological models or to gain understanding about specific datasets. Simulations alone can be used as guidance for the development of new computational tools8, for debugging and to evaluate software performance9,10. Computer simulations also allow us to generate new hypotheses11, help in the design of sequencing projects12,13, and are absolutely essential to verify distinct inferences such as the correctness of an assembly14, the accuracy of gene prediction15 or the power to reconstruct accurate genotypes and haplotypes2,16. Several computational tools for the simulation of NGS data have been developed in the past few years. These tools have very diverse input requirements and functionalities, which makes it quite difficult to choose the most appropriate one for the problem at hand.
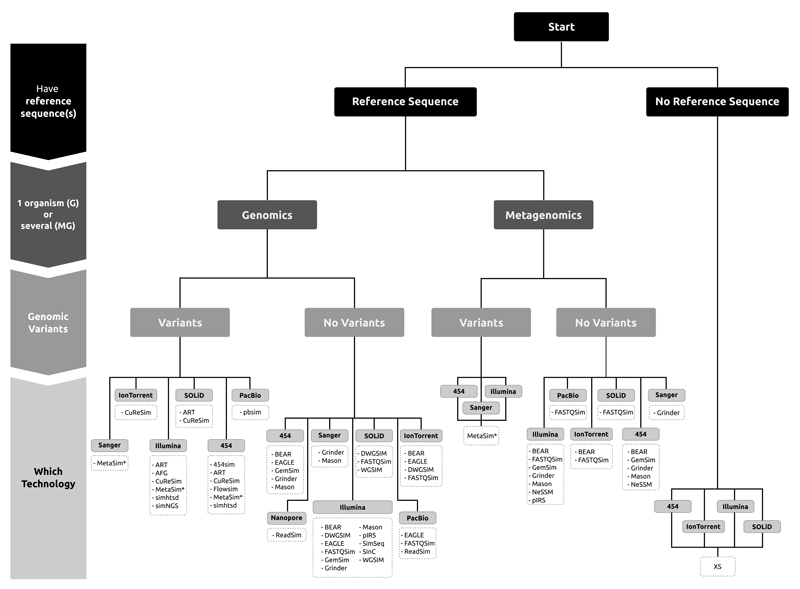
Here we present, to our knowledge, the first review of available software tools for the simulation of genomic NGS data. Note that we focus on the simulation of DNA sequences and do not discuss RNA sequencing (RNAseq) simulation, which has its own characteristics. We review 23NGS simulation tools that were either recently published or developed, that were — in most cases — still maintained and that were freely available. We discuss their various features, such as the required input, the interaction with the user, the sequencing platforms, the type of reads, the error models, the possibility of introducing coverage bias, the simulation of genomic variants and the output provided. This is done within the framework of potential applications, providing readers with guidelines for the identification of the NGS simulators that are best suited for their purposes (Fig. 1).
Figure 1. NGS genomic simulators decision tree.
Selection of a NGS simulator requires a set of sequential decisions. First we should reason whether we have a reference sequence or not. Then we need to decide whether we want to simulate reads from one or several organisms. Next we will specify whether we want to introduce genomic variants (in addition to those already existing in the reference sequence(s)). Finally we need to determine the sequencing technology of interest.
An overview of current NGS technologies
The most popular NGS technologies on the market are Illumina’s sequencing by synthesis, which is probably the most widely used platform at present17, Roche’s 454 pyrosequencing (454), SOLiD sequencing-by-ligation (SOLiD), IonTorrent semiconductor sequencing18 (IonTorrent), Pacific Biosciences’s (PacBio) single molecule real-time sequencing19, and Oxford Nanopore Technologies (Nanopore) single-cell DNA template strand sequencing. These strategies can differ, for example, regarding the type of reads they produce or the kind of sequencing errors they introduce (Table 1). Only two of the current technologies (Illumina and SOLiD) are capable of producing all three sequencing read types —SINGLE END, PAIRED END and MATE PAIR. Read length is also dependent on the machine and the kit used; in platforms like Illumina, SOLiD, or IonTorrent it is possible to specify the number of desired base pairs per read. According to the sequencing run type selected it is possible to obtain reads with maximum lengths of 75 bp (SOLiD), 300 bp (Illumina) or 400bp (IonTorrent). On the other hand, in platforms like 454, Nanopore or PacBio, information is only given about the mean and maximum read length that can be obtained, with average lengths of 700 bp, 10 kb and 15 kb and maximum lengths of 1 kb, 10 kb and 15 kb, respectively. Error rates vary depending on the platform from <=1% in Illumina to ~30% in Nanopore. Further overviews and comparisons of NGS strategies can be found in 5,20–22.
Table 1. Main characteristics of current NGS technologies.
| Technology | Run Type | Maximum Read Length | Quality Scores | Error Rates | References | ||
|---|---|---|---|---|---|---|---|
| Single-read | Paired-end | Mate-pair | |||||
| Illumina | X | X | X | 300 bp | > Q30 | 0.0034 – 1% | 65 |
| SOLiD | X | X | X | 75 bp | > Q30 | 0.01 – 1% | 66 |
| IonTorrent | X | X | 400 bp | ~ Q20 | 1.78% | 22 | |
| 454 | X | X | ~700 bp (up to 1 Kb) | > Q20 | 1.07 – 1.7% | 59,67 | |
| Nanopore | X | 5.4 – 10 Kb | NAY | 10 – 40% | 68–72 | ||
| PacBio | X | ~15 Kb (up to 40 Kb) | < Q10 | 5 – 10% | 22,73–75 | ||
Simulation parameters
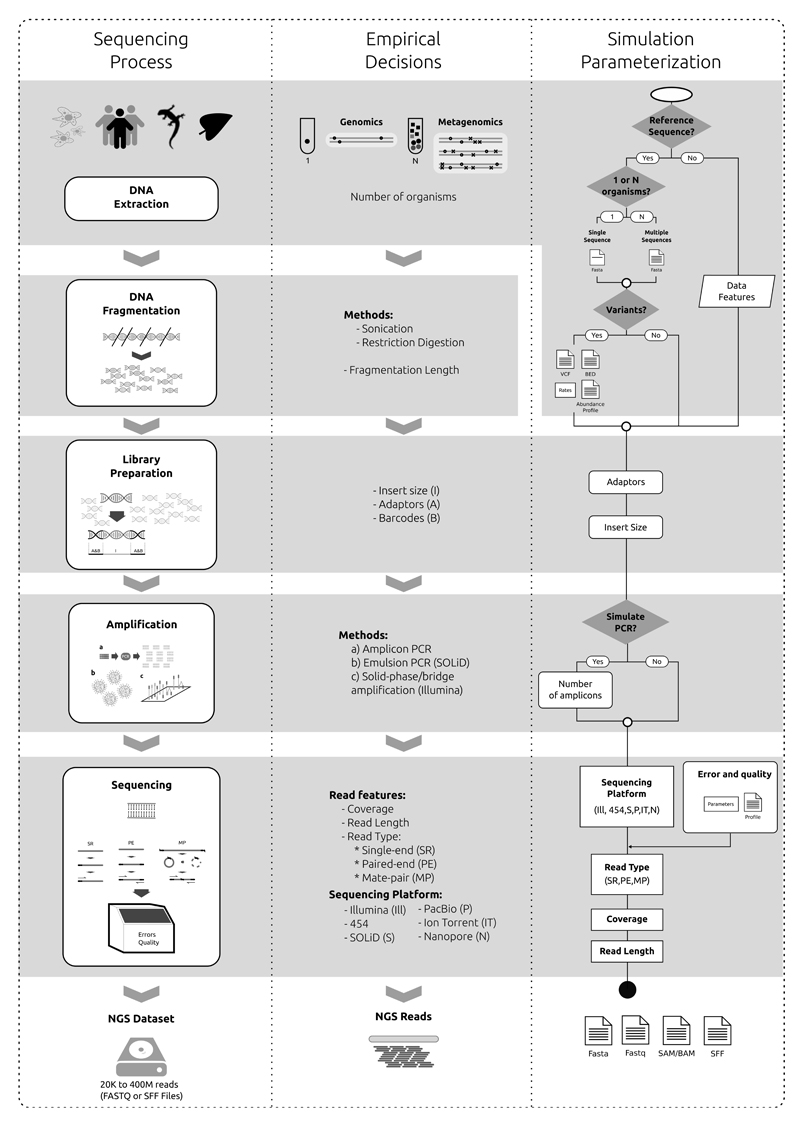
The existing sequencing platforms use distinct protocols that result in datasets with different characteristics1. Many of these attributes can be taken into account by the simulators (Fig. 2), although there is not a single tool that incorporates all possible variations. The main characteristics of the 23 simulators considered here are summarized in Tables 2 and 3. These tools differ in multiple aspects, such as sequencing technology, input requirements or output format, but maintain several common aspects. With some exceptions, all programs need a REFERENCE SEQUENCE, multiple parameter values indicating the characteristics of the sequencing experiment to be simulated (read length, error distribution, type of variation to be generated, if any, etc.) and/or a PROFILE (a set of parameter values, conditions and/or data used for controlling the simulation), which can be provided by the simulator or estimated de novo from empirical data. The outcome will be aligned or unaligned reads in different standard file formats, such as FASTQ, FASTA or BAM. An overview of the NGS data simulation process is represented in Fig. 3. In the following sections we delve into the different steps involved.
Figure 2. General overview of the sequencing process and steps that can be parameterized in the simulations.
NGS simulators try to imitate the real sequencing process as closely as possible by considering all the steps that could influence the characteristics of the reads. a | NGS simulators do not take into account the effect of the different DNA extraction protocols in the resulting data. However, they can consider whether the sample we want to sequence includes one or more individuals, from the same or different organisms (e.g., pool-sequencing, metagenomics). Pools of related genomes can be simulated by replicating the reference sequence and introducing variants on the resulting genomes. Some tools can also simulate metagenomes with distinct taxa abundance. b | Simulators can try to mimic the length range of DNA fragmentation (empirically obtained by sonication or digestion protocols) or assume a fixed amplicon length. c | Library preparation involves ligating sequencing–platform dependent adaptors and/or barcodes to the selected DNA fragments (inserts). Some simulators can control the insert size, and produce reads with adaptors/barcodes. d | | Most NGS techniques include an amplification step for the preparation of libraries. Several simulators can take this step into account (for example, by introducing errors and/or chimaeras), with the possibility of specifying the number of reads per amplicons. e | Sequencing runs imply a decision about coverage, read length, read type (single-end, paired-end, mate-pair) and a given platform (with their specific errors and biases). Simulators exist for the different platforms, and they can use particular parameter profiles, often estimated from real data.
Table 2. General information about 23 NGS genomic simulators.
| Simulator | Technology | G vs M | Run types | REF | Characterization | Processes | Outputs | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Input | Profile Process | PCR | GV | QS | |||||||||||||
| PA | RE | PR | DF | PA | GU | SW | RE | AL | FO | ||||||||
| 454Sim | 454 | G | SR | x | x | x | x | x | SFF | ||||||||
| ART | 454, ILL, SOL | G | SR, PE, MP | x | x | x | x | x | x | x | x | x | x | SFF/FQ | |||
| ArtificialFastqGenerator | ILL | G | PE | x | x | x | x | x | x | FQ | |||||||
| BEAR | 454, ILL, ITO | G,M | SR, PE | x | x | x | x | x | x | FQ | |||||||
| CuReSim | 454, ILL, SOL, ITO | G | SR | x | x | x | x | FQ | |||||||||
| DWGSIM (dnaa) | ILL, SOL, ITO | G | SR, PE, MP | x | x | x | x | x | x | x | FQ | ||||||
| EAGLE | 454, ILL, PCB, ITO | G | SR, PE | x | x | x | x | x | x | x | FQ | ||||||
| FASTQSim | ILL, SOL, PCB, ITO | G,M | SR | x | x | x | x | x | x | x | FQ | ||||||
| Flowsim | 454 | G | SR, PE | x | x | x | x | x | x | x | x | SFF | |||||
| GemSim | 454, ILL | G,M | SR, PE | x | x | x | x | x | x | x | FQ | ||||||
| Grinder | 454, ILL, SNG | G,M | SR, PE, MP | x | x | x | x | x | x | x | x | FQ | |||||
| Mason | 454, ILL, SNG | G | SR, PE, MP | x | x | x | x | x | x | x | x | FA/FQ | |||||
| MetaSim | 454,ILL, SNG | G,M | SR, PE, MP | x | x | x | x | FA | |||||||||
| NeSSM | 454, ILL | M | SR, PE | x | x | x | x | x | x | FQ | |||||||
| pbsim | PCB | G | CLR/CCS | x | x | x | x | x | x | FQ | |||||||
| pIRS | ILL | G,M | PE | x | x | x | x | x | x | x | x | FQ | |||||
| ReadSim | PCB, ONT | G | SR | x | x | x | x | x | x | FQ | |||||||
| simhtsd | 454, ILL | G | SR, PE | x | x | x | x | FQ | |||||||||
| simNGS | ILL | G | SR, PE | x | x | x | x | x | x | FQ | |||||||
| SimSeq | ILL | G | SR, PE, MP | x | x | x | x | x | x | x | x | SAM/BAM | |||||
| SInC | ILL | G | PE | x | x | x | x | x | x | x | FQ | ||||||
| wgsim | ILL, SOL | G | SR | x | x | x | x | x | x | FQ | |||||||
| xs | 454, ILL, SOL, ITO | G | SR,PE | x | x | x | x | FQ | |||||||||
454: Roche’s 454. ILL: Illumina. SOL: SOLiD. ITO: Ion Torrent. PCB: Pacific Biosciences. ONT: Oxford Nanopore Technologies. SNG: Sanger. G: Genomics. M: Metagenomics. PA: Parameters. RE: Reads. PR: Profile. DF: Default Profile. GU: Guide to generate profiles. SW: Specific software to generate profile. PCR: Polymerase Chain Reaction. GV: Genomic variants. QS: Quality scores. FO: Format. AL: Alignments. FA: Fasta. FQ: Fastq. SFF: Standard Flowgram Format. SAM Sequence Alignment Map. BAM: Compressed SAM File. Also accessible in http://darwin.uvigo.es/ngs-simulators/
Table 3. Technical information about 23 NGS genomic simulators.
| Simulators | Programming Language | Operative System | Interface | Processing | License | Open Source |
|---|---|---|---|---|---|---|
| 454Sim | C++/Perl | Win, Lnx, MOS | CLI | NP,P | GNU GPL v1 | Y |
| ART | C++/Perl | Win, Lnx, MOS | CLI | P | GNU GPL | Y |
| ArtificialFastqGenerator | Java | Win, Lnx, MOS | CLI | P | GNU GPL v3 | Y |
| BEAR | Python/Perl | Lnx | CLI | P | AU | Y |
| CuReSim | Java | Win, Lnx, MOS | CLI | P | * | N |
| DWGSIM (dnaa) | C/Perl/Python | Lnx | CLI | P | GNU GPL v2 | Y |
| EAGLE | C++ | Lnx | CLI | NP,P | BSD | Y |
| FASTQSim | Bash/Python | Lnx | CLI | NP,P | GNU GPL v3 | Y |
| Flowsim | Haskell | Lnx | CLI | P | GNU | Y |
| GemSim | Python | Win, Lnx, MOS | CLI | P | GNU GPL v3 | Y |
| Grinder | Perl | Win, Lnx, MOS | CLI, GUI, API | P | GPL | Y |
| Mason | C++ | Win, Lnx, MOS | CLI | P | GPL/LGPL. | Y |
| MetaSim | Java | Win, Lnx, MOS | CLI, GUI | P | PRO / AU | N |
| NeSSM | C/Cuda/Perl | Lnx | CLI | NP,P | AU | Y |
| pbsim | C++ | Lnx | CLI | P | GNU GPL v2 | Y |
| pIRS | C++/Perl | Lnx | CLI | NP,P | GNU GPL v2 | Y |
| ReadSim | Python | Win, Lnx, MOS | CLI | P | * | Y |
| simhtsd | Perl | Lnx | CLI | P | GNU GPL v3 | Y |
| simNGS | C | Lnx, MOS | CLI | P | GNU GPL v3 | Y |
| SimSeq | Java | Lnx | CLI | P | MIT | Y |
| SInC | C++ | Lnx | CLI | NP,P | CCANCL V2.0 | N |
| wgsim | C | Lnx | CLI | P | MIT | Y |
| xs | C++ | Lnx | CLI | P | GNU GPL v3 | Y |
Information related to this topic is not available. Win: Windows. Lnx: Linux. MOS: MacOS. CLI: Command line interface. GUI: Graphical User Interface. API: Application Programming Interface. NP: No parallel processing. P: Parallel processing (accepts multi-threading). GNU GPL: GNU General Public License. PRO: Proprietary software. AU: Academic use only. BSD: Berkeley Software Distribution. CCANCL: Creative Commons Attribution Non-Commercial License.
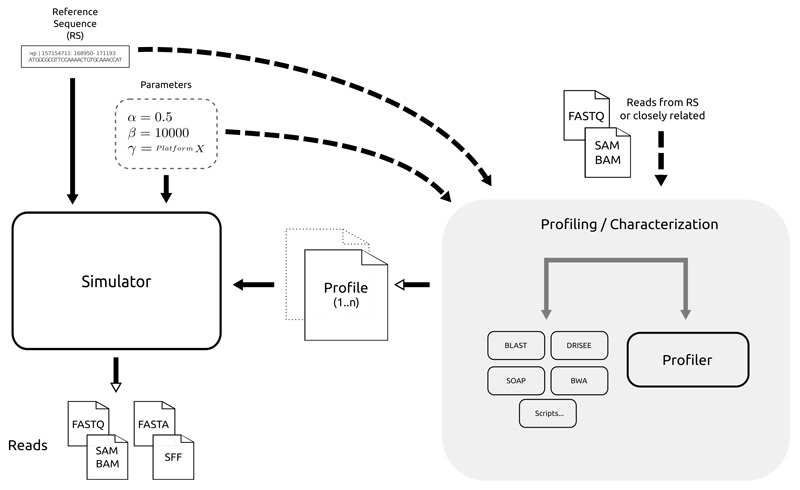
Figure 3. General overview of NGS simulation.
The simulation process begins with the input of a reference sequence (most cases) and simulation parameters. Some of the parameters can be given via a profile, that is estimated (by the simulator or other tools) from other reads or alignments. The outcome of this process may be reads (with or without quality information) or genome alignments in different formats.
Reference Sequence
Most NGS simulators require a reference sequence from which they will generate the simulated reads. This reference sequence can be a particular genomic region, multiple genomic regions concatenated, a chromosome, or a complete genome. The only exception in this regard is the simulator XS23, which just requires the read length, sequencing technology and nucleotide composition to generate completely de novo reads. Most of the current NGS simulators use a haploid genome as the reference sequence. Some tools such as EAGLE24, pIRS9, ReadSim25,26 and SimSeq27 simulate reads from different ploidies. While in EAGLE and ReadSim one can specify any ploidy (or even a specific chromosome for EAGLE), pIRS and SimSeq simulate reads from diploid genomes given a haploid reference. Furthermore, several tools are able to generate pools of reads from multiple reference sequences, in some cases using an ABUNDANCE PROFILE that defines the proportion of reads that are generated from each sequence.
Profiles
Most simulators require the setting of many parameters. This can be done in the command line and/or using a profile. Profiles can specify parameter distributions or discrete values for different biological features (e.g. GC-content, indel and substitution rates) and/or technological features (e.g. insert sizes, read lengths, error rates and QUALITY SCORES). Note that there are not standard formats for profiles and the information they include can change for the different tools. Because for many users it might be difficult to decide on particular parameter values or to construct their own profile, some simulators provide default profiles. Alternatively, many tools offer a way to estimate de novo profiles from empirical data. Several simulators are able to generate new profiles from alignments of reads mapped to a reference genome (SAM/BAM files) or from real sequencing data from a previous sequencing run (FASTQ files). Thus, BEAR28, NeSSM29 and pIRS provide guidelines for the use of alignment and mapping tools such as BWA30, BLAST31, SOAP32 or SOAP233, and for error estimation programs such as DRISEE34, together with other scripts for parsing the data or for other tasks. ART8, , FASTQsim13, GemSim16, SimSeq27 and SInC17 packages provide their own standalone tools for the generation of error, quality and/or abundance profiles. ART and SInC generate quality profiles based on specific error models and/or the quality score distribution extracted from empirical data. NeSSM generates quality and error profiles. The quality profiles define the quality score given to each base along the read and are estimated based on an existing set of reads. The error profiles define the proportion of the different error types (substitutions and indels) and are estimated with specific scripts. pIRS generates quality profiles using mapped reads and known variations from re-sequencing data. The program BEAR, focused on metagenomics, generates error, quality and abundance profiles. For the generation of the error profile it uses a modified version of DRISEE to infer error rates by clustering artefactual duplicate reads in the supplied dataset. For the quality profile it uses the output of the error model to determine the average quality score assigned to erroneous nucleotides per position per read28. In addition, it generates an abundance profile from the relative frequency of the different taxa in a metagenomic dataset.
Finally, other simulation programs such as ArtificialFastqGenerator35 and CuReSim10 do not use a profile, their simulation parameter values are specified directly via the command-line.
Accounting for PCR Amplification
DNA amplification with polymerase chain reaction (PCR) is currently a necessary step in the preparation of libraries for the Illumina, 454, IonTorrent and SOLiD36,37 sequencing platforms. One may be interested therefore in modeling the bias introduced by PCR1,38,39, as done by ART, Flowsim40,41 and Grinder7.
ART, which simulates reads for Illumina, 454 and SOLiD, can mimic PCR bias by specifying the number of reads (SR or PE) generated per AMPLICON8. Flowsim is a suite of executables that simulate the entire 454 pyrosequencing process; using its module “kitsim” one can simulate the attachment of adapters to the end of each amplicon, which later on serve as primers for their PCR amplification simulated by “duplicator”40,41.
Grinder was specifically developed to simulate amplicon sequencing from user supplied PCR-primer collections, introducing known experimental artifacts like chimeras39 and spurious copy number variants. Grinder can generate chimeras in two ways: 1) by appending consecutive segments at given breakpoints, where both amplicon sequences and breakpoints are randomly selected; and 2) by concatenating fragments at breakpoints determined by specific K-MERS that must be shared by the amplicons. In addition, the presence of several gene copies in a genome may affect the composition of the amplicon library, contributing with extra amplicon reads. Grinder models this bias by sampling species proportionally to their relative abundance and to the number of copies of the amplicon in their genome7.
Read Features
In an NGS experiment, the number, length and type of reads are determined by the specific sequencing machine and the library preparation. It is possible to simulate a specific amount of reads with different lengths and types according to the sequencing technology assumed. The number of reads can be specified or estimated according to the desired COVERAGE. Also, it is possible to select a fixed length, the length of the longest read or a length distribution. The read type can be specified directly or indirectly by defining particular insert sizes. By default, most simulators assume single-end reads.
Base call errors
NGS technologies rely on a complex interplay between chemistry, hardware and optical sensors. Adding to this complexity is the software that analyzes the sensor data and predicts the individual bases. This last step is usually referred to as BASE CALLING42. The base calling converts the signals into actual sequence data with quality scores (known as Phred Q Scores43,44). The different sequencing platforms usually assume an explicit error model in order to assign a measure of uncertainty to each base call45.
Error-rate models determine the probability of erroneous substitutions, insertions or deletions at a given position within a read28,46. For the generation of realistic reads, it is necessary to understand and incorporate as much as possible the different sources of sequencing error. Each sequencing platform has a specific error rate (Table 1), which can also vary within the same technology and among reads16. The importance of taking this into account and simulating sequencing data based on specific error models should not be underestimated.
Simulators may generate sequence errors in different ways: based on the quality scores (ArtificialFastqGenerator); by introducing particular errors at specific positions (SimSeq); by using specific error parameters for each platform/technology, which can be user-defined (ART, Mason6, pIRS) or fixed by the program (DWGSIM47, FASTQsim); using vriable error rates within reads (simhtsd48, wgsim49); using error distributions (Grinder); or generating specific errors along with some noise (simNGS50). In the following subsections we describe in more detail the different errors that are modeled and their occurrence in sequencing platforms, as well as how the different simulators implement them.
Indel errors
It has been reported that Illumina platforms rarely contain indel errors9, whereas for 454 and IonTorrent insertions and deletions (indels) are actually the main source of error, although they occur at very low rates51. However, in 454, assessing the correct number of polynucleotide sites (HOMOPOLYMERS) is often quite difficult because light signal changes among homopolymers with similar lengths can be undetectable 5,52–56. PacBio yields long single-molecule reads that are prone to false indels from non-fluorescing nucleotides52,54, which are stochastically modeled by the PacBio read simulator pbsim57. With Nanopore it is also possible to have indel errors; insertions occur when the strand slips back and forth so that a given position is read more than once, and deletions occur when the rate of strand displacement in the pore sensor exceeds the rate of data acquisition57. ReadSim, which is so far the only simulator available for Nanopore, assumes fixed error rates for indels and substitutions. Indel rates can be specified via the command line, or using a configuration profile in the cases of ART, CuReSim, Grinder, Mason, MetaSim58, NeSSM, pbsim, ReadSim, SInC and XS. Some programs like BEAR, EAGLE and GemSim include utilities or use external tools like DRISEE for the estimation of indel rates from FASTQ or SAM files. On the other hand, 454 and IonTorrent homopolymer specific errors59 may be extracted from a profile determining the position and corresponding error rate (as in ART), or introduced under the form of homopolymeric stretches using a specified empirical model (as in MetaSim, Flowsim or Grinder).
Substitution errors
Substitution errors are dominant in Illumina and SOLiD platforms. These may occur when incorrect bases are introduced during clonal amplification of templates (by PCR)9,54,60 or when the optical signals are translated into bases. In the latter process the green laser is used to detect G and T at the same time, using afterwards a filter to distinguish between G and T. A and C are detected in a similar way but by using a red laser. Thus, base call errors may arise because of insufficient discrimination of the respective base emission spectra51. It is also known that SOLiD sequencers are unable to read through palindromic regions, presumably due to the formation of hairpin structures, therefore interpreting such regions as miscellaneous random sequences. ART simulates this kind of error. As with indels, substitutions errors rates have to be defined in the command line or within a profile.
Some NGS platforms can make position-specific substitution errors, with reads having significantly lower quality in the later cycles. In Illumina these type of errors possibly arise from either single-strand DNA folding or sequence-specific alterations in enzyme preference1,52,54,60 and can be modeled by GemSim and pIRS. There is a similar case for 454 platforms59. Flowsim, 454sim and MetaSim can simulate two kinds of sequencing flows with a degradation model. The positive flow, interpreted as the occurrence of one or more bases, is modelled as a Normal distribution; the negative flow –no base or noise–, is modelled as a Log Normal distribution. The degradation model is introduced as a standard deviation that gradually increases the chance of error along the sequence.
Quality Scores
The quality score is a prediction of the probability of an error in a base call43,44,46,61. The distribution of base quality scores is position dependent, and the mean quality score decreases as a function of increasing base position for most of the available technologies8. Some NGS read simulators separate the quality score from sequencing error, even though they are correlated measurements. Several strategies can be used to simulate the quality scores, in most cases using empirical information. 454sim, EAGLE, Flowsim and simNGS use fixed quality scores profiles that are based on previous studies. ART, ArtificialFastqGenerator, BEAR, FASTQsim, GemSim, NeSSM, SimSeq and SInC also include utilities that allow the user to derive quality profiles from FASTQ files. On the other hand, pIRS determines both the base and quality score in relation to the cycle number and to the base position on the simulated read, using empirical parameters. Alternatively, the distribution of the quality scores can be controlled by the user. Some programs use a simple parameter that determines a fixed quality score for every read (ArtificalFastqGenerator, CuReSim, DWGSIM, ReadSim, simhtsd, wgsim and XS). Grinder assigns two quality scores, depending on whether the simulated base call is correct or not. More complex, realistic simulators use a Gaussian distribution (XS) or a Position Specific Normal Distribution (Mason) with mean, standard deviation and quality standard deviation for the first and last base. For PacBio the distribution of errors is considered to be constant along the chromosomes22 and programs like pbsim use a Uniform distribution to assign the quality scores. In Illumina, each PE read can have equal or different quality scores. Simulators that explicitly allow two different quality distributions for PE reads are ArtificialFastqGenerator, DWGSIM, EAGLE, SimSeq and SInC.
Sequencing depth
Sequencing depth or coverage is not continuous along genomes. This can be due to chance62 but also to the GC bias introduced during DNA amplification by PCR63,64, as sequencing depth increases in regions with elevated GC content38,51.This coverage bias is taken into account by ArtificialFastqGenerator, BEAR, EAGLE, NeSSM and pIRS. ArtificialFastqGenerator calculates the GC content of different genomic regions from the reference sequence and then samples coverage levels for these regions from a Normal distribution. BEAR, EAGLE NeSSM and pIRS use data from previous studies to determine the variation of the GC content along the reference sequence, resulting in the simulation of variable regional coverage.
Simulating genomic variants
Apart from sequencing error (Fig. 4), many tools can also introduce different types of genomic variants in the simulated reads17 like single nucleotide polymorphisms (SNPs), indels, inversions, translocations, copy number variants (CNVs) and short tandem repeats (STRs) (Table 4).
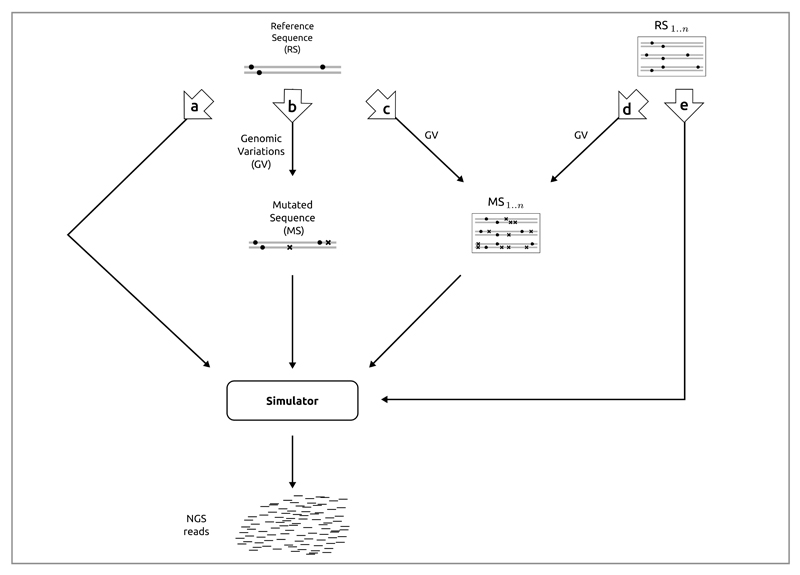
Figure 4. Flows available to generate reads with and without genomic variation.
Dots represent variants present in the reference sequence(s), and crosses represent the newly introduced variants (mutated sequences). a | Simulation of reads from a single reference sequence without adding new genomic variants. b | Generation of reads from a single mutated sequence generated from a single reference sequence. c | Reads are generated from a set of mutated sequences that were generated from a single reference sequence. d | Generation of reads from a set of mutated sequences obtained from a set of reference sequences. e | Reads are obtained directly from a set of reference sequences without introducing additional genomic variants.
Table 4. Genomic variants.
| Simulators | Genomic Variants | |||||||
|---|---|---|---|---|---|---|---|---|
| MGC | PLO | SNPs | Indels | INVs | TRA | CNVs | STRs | |
| BEAR | x | |||||||
| DWGSIM (dnaa) | x | x | x | x | x | |||
| EAGLE | x | x | x | x | x | x | ||
| FASTQSim | x | x | x | |||||
| GemSim | x | x | x | |||||
| Grinder | x | x | x | |||||
| Mason | x | x | ||||||
| NeSSM | x | |||||||
| pIRS | x | x | x | x | ||||
| ReadSim | x | x | x | x | ||||
| SimSeq | x | |||||||
| SInC | x | x | x | |||||
| wgsim | x | x | x | |||||
Variation that can be introduced in the reference sequences: MGC, Metagenomic community; PLO, Ploidy; SNPs, Single Nucleotide Polymorphisms; Indels, Insertions and/or deletions; INVs, Inversions; TRA, Translocations; CNVs, Copy Number Variants; STRs, Short Tandem Repeats.
The general strategy is to create a mutated sequence by introducing genomic variants in the reference sequence before the generation of reads (Fig. 4). In most cases, these variants are simulated using a given mutation rate, so the mutated sequence differs by a given percentage from the reference sequence. Programs like DWGSIM and EAGLE require instead a file with known mutations (in plain text, VCF or BED-like format). FASTQsim includes a separate tool that builds a mutation file from real data, using a NGS dataset (FASTQ files) and a reference genome, being best suited for re-sequencing.
Some programs are capable of generating population-level diversity by creating several mutated sequences from a single reference sequence (Fig. 4). Programs like GemSim and Mason can generate sets of related haplotypes differing by at least one SNP from the reference sequence. In GemSim users may also create their own tab-delimited haplotype file providing the specific position and mutation introduced.
Tools like GemSim, BEAR, Grinder and NeSSM can introduce genomic variants in a given set of reference sequences belonging to different taxa to create a set of mutated genomes that will resemble a metagenomic community (Fig. 4). As mentioned before, these programs use an abundance profile so the reads are generated from these sequences with a probability proportional to “taxa” abundance.
Output
The generated NGS reads may be stored in different file formats. According to the specific NGS technology simulated, one can get SFF files (standard flowgram format) from 454 platforms (454sim and Flowsim), and FASTA or FASTQ files for IonTorrent, Illumina, PacBio, SOLiD and Nanopore. Other possible output files include alignment files, either in MAF (Multiple Alignment Format) or SAM/BAM formats. These can be outputted by default (as in Mason, pbsim and SimSeq), or as an option, complementary to the simulated reads (as in ART).
Conclusions
NGS is having a big impact in a broad range of areas that benefit from genetic information, from medical genomics, phylogenetic and population genomics, to the reconstruction of ancient genomes, epigenomics and environmental barcoding. These applications include approaches such as de novo sequencing, resequencing, target sequencing or genome reduction methods. In all cases, caution is necessary in choosing a proper sequencing design and/or a reliable analytical approach for the specific biological question of interest. The simulation of NGS data can be extremely useful for planning experiments, testing hypotheses, benchmarking tools and evaluating particular results. Given a reference genome or dataset, for instance, one can play with an array of sequencing technologies to choose the best-suited technology and parameters for the particular goal, possibly optimizing time and costs. Yet, this is still not the standard practice and researchers often base their choices on practical considerations like technology and money availability. As shown throughout this Review, simulation of NGS data from known genomes or transcriptomes can be extremely useful when evaluating assembly, mapping, phasing or genotyping algorithms e.g. 2,7,10,13,64 exposing their advantages and drawbacks under different circumstances.
Altogether, current NGS simulators consider most, if not all, of the important features regarding the generation of NGS data. However, they are not problem-free. The different simulators are largely redundant, implementing the same or very similar procedures. In our opinion, many are poorly documented and can be difficult to use for non-experts, and some of them are no longer maintained. Most importantly, for the most part they have not been benchmarked or validated. Remarkably, among the 23 tools considered here, only 13 have been described in dedicated application notes, 3 have been mentioned as add-ons in the methods section of bigger articles, and 5 have never been referenced in a journal. Indeed, peer-reviewed publication of these tools in dedicated articles would be highly desirable. While this would not definitively guarantee quality, at least it would encourage authors to reach minimum standards in terms of validation, benchmarking, and documentation. Collaborative efforts like the Assemblathon e.g. 27 or iEvo (http://www.ievobio.org/) might be also a source of inspiration. Meanwhile, we hope that the decision tree presented in Fig. 1 helps users making appropriate choices.
Online Summary.
There is a large number of tools for the simulation of genomic data for all currently available NGS platforms, with partially overlapped functionality. Here we review 23 of these tools, highlighting their distinct functionalities, requirements and potential applications.
The parameterization of these simulators is often complex. The user may decide between using existing sets of parameters values called profiles or re-estimating them from its own data.
Parameters than can be modulated in these simulations include the effects of the PCR amplification of the libraries, read features and quality scores, base call errors, variation of sequencing depth across the genomes and the introduction of genomic variants.
Several types of genomic variants can be introduced in the simulated reads, such as SNPs, indels, inversions, translocations, copy-number variants and short-tandem repeats.
Reads can be generated from single or multiple genomes, and with distinct ploidy levels. NGS data from metagenomic communities can be simulated given an “abundance profile” that reflects the proportion of taxa in a given sample.
Many of the simulators have not been formally described and/or tested in dedicated publications. We encourage the formal publication of these tools and the realization of comprehensive, comparative benchmarkings.
Choosing among the different genomic NGS simulators is not easy. Here we provide a guidance tree to help users choosing a suitable tool for their specific interests.
Acknowledgements
This work was supported by the European Research Council (ERC-617457- PHYLOCANCER to D.P.) and the Spanish Government (Research grants BFU2012-33038 and BFU2015-63774-P to D.P.; FPI graduate fellowship BES-2013-067181 to M.E. and Juan de la Cierva postdoctoral fellowship FPDI-2013-17503 to S.R.). We thank two anonymous reviewers and members of the Phylogenomics lab (http://darwin.uvigo.es) for the critical comments.
Glossary
- Coverage Bias
A bias in the amount of reads for a particular region. For example, sequencing depth increases in regions of elevated GC content.
- Single Read
Single-read sequencing involves sequencing DNA fragment from only one end.
- Paired-End Reads
In paired-end sequencing, a single fragment is sequenced from both 5' and 3' ends, giving rise to reads in both forward and reverse (FR) orientation, where read 1 is the forward read and read 2 is the reverse. The sequenced fragments may be separated by a certain number of bases (depending on insert size and read length) or overlapping.
- Mate-Pair Reads
Mate-pair sequencing means generating long-insert paired-end DNA libraries. The inserts are circularized and fragmented and the labeled fragments (corresponding to the ends of the original DNA ligated together) are purified, ligated to another set of adapters and finally PE sequenced. The resulting inserts include two DNA segments that were originally separated by 2-5 kb, facilitating mapping and assembly.
- Reference Sequence
A particular genomic region, multiple genomic regions concatenated, a chromosome, or a complete genome from which NGS reads will be generated.
- Profile
A set of biological (GC-content, indel and substitution rates) and/or technological (insert sizes, read lengths, error rates and quality scores) parameter distributions or values that will be used in a specific simulation.
- Abundance Profile
Set of probabilities that represent the proportion of taxa within a community (and dataset).
- Quality Score
A prediction of the probability of an error in a base call.
- Amplicon
A piece of DNA or RNA resulting from an amplification event (for example as in PCR), either natural or artificial.
- K-mer
A k-mer refers to all the possible subsequences of length k that can be obtained from a given sequence.
- Coverage (or Sequencing Depth)
Number of times a certain nucleotide has been sequenced.
- Base Calling
The analysis of the information obtained from the machine sensors during NGS and posterior prediction of the individual bases. This converts the signal into actual sequence data with quality scores.
- Homopolymer
A sequence of multiple identical nucleotides.
Biographies
Merly Escalona
Merly Escalona is a PhD student at the University of Vigo, Spain. She is a computer scientist with a MSc in Adaptive and Intelligent Software Systems who is currently working with NGS simulations, genome reduction data analysis methods and phylogenetic inference.
Sara Rocha
Sara Rocha received her PhD from the University of Porto in 2011, where she focused in colonization and diversification patterns of reptiles from Western Indian Ocean islands. She is broadly interested in phylogenetic inference and its use on understanding the processes that lead to genes, populations and species divergence. Currently she is a postdoctoral fellow at the University of Vigo, participating in projects related to phylogenetic and species-tree inference from closely related genomes using next-generation sequencing (NGS) data.
David Posada
David Posada is Professor of Genetics at the University of Vigo in Spain. He is a computational evolutionary biologist interested in phylogenetic methodology and its application for the study of different organisms. He has developed popular tools for the statistical selection of nucleotide substitution models, network reconstruction or phylogeographic analysis. He has recently become interested in the study of tumor evolution using next-generation sequencing data, and in particular in the application of population genetics and phylogenetic concepts for the analysis of intratumoral heterogeneity.
Footnotes
Competing interests
The authors declare no competing interests.
Online links
- ArtificialFastqGenerator: http://sourceforge.net/projects/artfastqgen/
- DWGSIM: https://github.com/nh13/DWGSIM
- simNGS and simLibrary: http://www.ebi.ac.uk/goldman-srv/simNGS/
- Wgsim: http://github.com/lh3/wgsim
Further Information
Link 1: [http://darwin.uvigo.es/ngs-simulators/]
Access to this interactive links box is free online.
References
- 1.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen R, Paul JS, Albrechtsen A, Song YS. Genotype and SNP calling from next-generation sequencing data. Nat Rev Genet. 2011;12:443–451. doi: 10.1038/nrg2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koboldt DC, Steinberg KM, Larson DE, Wilson RK, Mardis ER. The Next-Generation Sequencing Revolution and Its Impact on Genomics. Cell. 2013;155:27–38. doi: 10.1016/j.cell.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang XV, Blades N, Ding J, Sultana R, Parmigiani G. Estimation of sequencing error rates in short reads. BMC Bioinformatics. 2012;13:185. doi: 10.1186/1471-2105-13-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu L, et al. Comparison of Next-Generation Sequencing Systems. J Biomed Biotechnol. 2012;2012:1–11. doi: 10.1155/2012/251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holtgrewe M. Technical Report. Institut für Mathematik und Informatik, Freie Universität Berlin; 2010. Mason - A Read Simulator for Second Generation Sequencing Data. [Google Scholar]
- 7.Angly FE, Willner D, Rohwer F, Hugenholtz P, Tyson GW. Grinder: A versatile amplicon and shotgun sequence simulator. Nucleic Acids Res. 2012;40:e94. doi: 10.1093/nar/gks251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang W, Li L, Myers JR, Marth GT. ART: a next-generation sequencing read simulator. Bioinformatics. 2012;28:593–594. doi: 10.1093/bioinformatics/btr708. [This paper describes probably the most popular NGS simulator nowadays, with well-supported and detailed documentation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu X, et al. pIRS: Profile-based Illumina pair-end reads simulator. Bioinformatics. 2012;28:1533–1535. doi: 10.1093/bioinformatics/bts187. [DOI] [PubMed] [Google Scholar]
- 10.Caboche S, Audebert C, Lemoine Y, Hot D. Comparison of mapping algorithms used in high-throughput sequencing: application to Ion Torrent data. BMC Genomics. 2014;15:264. doi: 10.1186/1471-2164-15-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoban S, Bertorelle G, Gaggiotti OE. Computer simulations: tools for population and evolutionary genetics. Nat Rev Genet. 2012;13:110–122. doi: 10.1038/nrg3130. [DOI] [PubMed] [Google Scholar]
- 12.Shendure J, Aiden EL. The expanding scope of DNA sequencing. Nat Biotechnol. 2012;30:1084–1094. doi: 10.1038/nbt.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shcherbina A. FASTQSim: platform-independent data characterization and in silico read generation for NGS datasets. BMC Res Notes. 2014;7:533. doi: 10.1186/1756-0500-7-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knudsen B, Forsberg R, Miyamoto MM. A computer simulator for assessing different challenges and strategies of de Novo sequence assembly. Genes. 2010;1:263–282. doi: 10.3390/genes1020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mavromatis K, et al. Use of simulated data sets to evaluate the fidelity of metagenomic processing methods. Nat Methods. 2007;4:495–500. doi: 10.1038/nmeth1043. [This paper describes the use of NGS simulations for benchmarking NGS analytical methods.] [DOI] [PubMed] [Google Scholar]
- 16.McElroy KE, Luciani F, Thomas T. GemSIM: general, error-model based simulator of next-generation sequencing data. BMC Genomics. 2012;13:74. doi: 10.1186/1471-2164-13-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pattnaik S, Gupta S, Rao AA, Panda B. SInC: an accurate and fast error-model based simulator for SNPs, Indels and CNVs coupled with a read generator for short-read sequence data. BMC Bioinformatics. 2014;15:40. doi: 10.1186/1471-2105-15-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rothberg JM, et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 19.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 20.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 21.Shendure J, Mitra RD, Varma C, Church GM. Advanced sequencing technologies: methods and goals. Nat Rev Genet. 2004;5:335–344. doi: 10.1038/nrg1325. [DOI] [PubMed] [Google Scholar]
- 22.Quail M, et al. A tale of three next generation sequencing platforms: comparison of Ion torrent, pacific biosciences and illumina MiSeq sequencers. BMC Genomics. 2012;13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pratas D, Pinho AJ, OS Rodrigues JM. XS: a FASTQ read simulator. BMC Res Notes. 2014;7:40. doi: 10.1186/1756-0500-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janin L. EAGLE - Enhanced Artificial Genome Engine. Github Repository. 2014 [online] https://github.com/sequencing/EAGLE. [Google Scholar]
- 25.Lee H. ReadSim: Simple reads simulator for pacbio & nanopore. Sourceforge - Repository. 2012 [online] http://sourceforge.net/projects/readsim/ [This paper describes the only NGS simulator, at least among those considered in this review, capable of producing Oxford Nanopore data.] [Google Scholar]
- 26.Lee H, et al. Error correction and assembly complexity of single molecule sequencing reads. bioRxiv. 2014:6395. doi: 10.1101/006395. [DOI] [Google Scholar]
- 27.Earl D, et al. Assemblathon 1: A competitive assessment of de novo short read assembly methods. Genome Res. 2011;21:2224–2241. doi: 10.1101/gr.126599.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson S, Trost B, Long JR, Pittet V, Kusalik A. A better sequence-read simulator program for metagenomics. BMC Bioinformatics. 2014;15:S14. doi: 10.1186/1471-2105-15-S9-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia B, et al. NeSSM: A Next-Generation Sequencing Simulator for Metagenomics. PLoS ONE. 2013;8:e75448. doi: 10.1371/journal.pone.0075448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 32.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 33.Li R, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 34.Keegan KP, et al. A Platform-Independent Method for Detecting Errors in Metagenomic Sequencing Data: DRISEE. PLoS Comput Biol. 2012;8:e1002541. doi: 10.1371/journal.pcbi.1002541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frampton M, Houlston R. Generation of Artificial FASTQ Files to Evaluate the Performance of Next-Generation Sequencing Pipelines. PLoS ONE. 2012;7:e49110. doi: 10.1371/journal.pone.0049110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 37.Morozova O, Marra Ma. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92:255–264. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Aird D, et al. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 2011;12:R18. doi: 10.1186/gb-2011-12-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haas BJ, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balzer S, Malde K, Lanzén A, Sharma A, Jonassen I. Characteristics of 454 pyrosequencing data-enabling realistic simulation with flowsim. Bioinformatics. 2011;27:i420–i425. doi: 10.1093/bioinformatics/btq365. [This paper presents one of the most popular simulators for 454 pyrosequencing long reads.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balzer S, Malde K, Jonassen I. Systematic exploration of error sources in pyrosequencing flowgram data. Bioinformatics. 2011;27:304–309. doi: 10.1093/bioinformatics/btr251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ledergerber C, Dessimoz C. Base-calling for next-generation sequencing platforms. Briefings in Bioinformatics. 2011;12:489–497. doi: 10.1093/bib/bbq077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ewing B, et al. Base-Calling of Automated Sequencer Traces Using Phred I. Accuracy Assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 44.Ewing B, et al. Base-Calling of Automated Sequencer Traces Using Phred II. Error Probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 45.Kao W-C, Stevens K, Song YS. BayesCall: A model-based base-calling algorithm for high-throughput short-read sequencing. Genome Res. 2009;19:1884–1895. doi: 10.1101/gr.095299.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Illumina Inc. Quality Scores for Next-Generation Sequencing.Illumina Technotes on Sequencing. 2011 [online] http://www.illumina.com/documents/products/technotes/technote_Q-Scores.pdf.
- 47.Homer N. DWGSIM: Whole Genome Simulator for Next-Generation Sequencing. Github Repository. 2010 [online] https://github.com/nh13/DWGSIM. [Google Scholar]
- 48.Bodi K. simhtsd: Simulate High-thoughput Sequencing Data. Sourceforge - Repository. 2009 [online] http://sourceforge.net/projects/simhtsd/ [Google Scholar]
- 49.Li H. wgsim - Read simulator for next generation sequencing. Github Repository. 2011 [online] http://github.com/lh3/wgsim. [Google Scholar]
- 50.Massingham T, Goldman N. simNGS and simLibrary: Software for simulating Next-Gen sequencing data. European Molecular Biology Laboratory - European Bioinformatics Institute. Goldman Group. 2010 [online] http://www.ebi.ac.uk/goldman-srv/simNGS/ [Google Scholar]
- 51.Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [This paper describes the most relevant biases affecting the generation of NGS data.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kircher M, Kelse J. High-throughput DNA sequencing - concepts and limitations. BioEssays. 2010;32:524–536. doi: 10.1002/bies.200900181. [DOI] [PubMed] [Google Scholar]
- 53.Loman NJ, et al. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol. 2012;30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- 54.Robasky K, Lewis NE, Church GM. The role of replicates for error mitigation in next-generation sequencing. Nat Rev Genet. 2013;15:56–62. doi: 10.1038/nrg3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang X, Chockalingam SP, Aluru S. A survey of error-correction methods for next-generation sequencing. Briefings in Bioinformatics. 2013;14:56–66. doi: 10.1093/bib/bbs015. [DOI] [PubMed] [Google Scholar]
- 56.Ekblom R, Smeds L, Ellegren H. Patterns of sequencing coverage bias revealed by ultra-deep sequencing of vertebrate mitochondria. BMC Genomics. 2014;15:467. doi: 10.1186/1471-2164-15-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ono Y, Asai K, Hamada M. PBSIM: PacBio reads simulator--toward accurate genome assembly. Bioinformatics. 2013;29:119–121. doi: 10.1093/bioinformatics/bts649. [This paper presents one of the most popular simulators for the Pacific Biosciences sequencing platform.] [DOI] [PubMed] [Google Scholar]
- 58.Richter DC, Ott F, Auch AF, Schmid R, Huson DH. MetaSim—A Sequencing Simulator for Genomics and Metagenomics. PLoS ONE. 2008;3:e3373. doi: 10.1371/journal.pone.0003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2006;441:120–120. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakamura K, et al. Sequence-specific error profile of Illumina sequencers. Nucleic Acids Res. 2011;39:e90–e90. doi: 10.1093/nar/gkr344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kwon S, Park S, Lee B, Yoon S. In-depth analysis of interrelation between quality scores and real errors in illumina reads; 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC); 2013. pp. 635–638. [DOI] [PubMed] [Google Scholar]
- 62.Lander ES, Waterman MS. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics. 1988;2:231–239. doi: 10.1016/0888-7543(88)90007-9. [DOI] [PubMed] [Google Scholar]
- 63.Sims D, Sudbery I, Ilott NE, Heger A, Ponting CP. Sequencing depth and coverage: key considerations in genomic analyses. Nat Rev Genet. 2014;15:121–132. doi: 10.1038/nrg3642. [DOI] [PubMed] [Google Scholar]
- 64.Li B, et al. Evaluation of de novo transcriptome assemblies from RNA-Seq data. Genome Biol. 2014;15:553. doi: 10.1186/s13059-014-0553-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ross MG, et al. Characterizing and measuring bias in sequence data. Genome Biol. 2013;14:R51. doi: 10.1186/gb-2013-14-5-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Glenn TC. Field guide to next-generation DNA sequencers. Mol Ecol Resour. 2011;11:759–769. doi: 10.1111/j.1755-0998.2011.03024.x. [DOI] [PubMed] [Google Scholar]
- 67.Gilles A, et al. Accuracy and quality assessment of 454 GS-FLX Titanium pyrosequencing. BMC Genomics. 2011;12:245. doi: 10.1186/1471-2164-12-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quick J, Quinlan AR, Loman NJ. A reference bacterial genome dataset generated on the MinION™ portable single-molecule nanopore sequencer. GigaScience. 2014;3:22. doi: 10.1186/2047-217X-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Loman NJ, Quick J, Simpson JT. A complete bacterial genome assembled de novo using only nanopore sequencing data. bioRxiv. 2015 doi: 10.1101/015552. [DOI] [PubMed] [Google Scholar]
- 70.Jain M, et al. Improved data analysis for the MinION nanopore sequencer. Nat Methods. 2015;12:351–356. doi: 10.1038/nmeth.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Laver T, et al. Assessing the performance of the Oxford Nanopore Technologies MinION. Biomol Detect Quantif. 2015;3:1–8. doi: 10.1016/j.bdq.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Madoui M-A, et al. Genome assembly using Nanopore-guided long and error-free DNA reads. BMC Genomics. 2015;16:327. doi: 10.1186/s12864-015-1519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carneiro MO, et al. Pacific biosciences sequencing technology for genotyping and variation discovery in human data. BMC Genomics. 2012;13:375. doi: 10.1186/1471-2164-13-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Koren S, et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol. 2012;30:693–700. doi: 10.1038/nbt.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Salmela L, Rivals E. LoRDEC: accurate and efficient long read error correction. Bioinformatics. 2014;30:3506–3514. doi: 10.1093/bioinformatics/btu538. [DOI] [PMC free article] [PubMed] [Google Scholar]