Abstract
The ribosomally synthesized and post-translationally modified peptide (RiPP), pyrroloquinoline quinone (PQQ), is a dehydrogenase cofactor synthesized by, but not exclusively used by, certain prokaryotes. RiPPs represent a rapidly expanding and diverse class of natural products—many of which have therapeutic potential—and the biosynthetic pathways for these are gaining attention. Five gene products from the pqq operon (PqqA, PqqB, PqqC, PqqD, and PqqE) are essential for PQQ biosynthesis. The substrate is the peptide PqqA, which is presented to the radical SAM enzyme PqqE by the small protein PqqD. PqqA is unstructured in solution, and only binds to PqqE when in complex with PqqD. PqqD is a member of a growing family of RiPP chaperone proteins (or domains in some cases) that present their associated peptide substrates to the initial RiPP biosynthesis enzymes. An X-ray crystal dimer structure exists for Xanthomonas campestris PqqD (PDB ID: 3G2B), but PqqD is now known to act as a monomer under physiological conditions. In this study, the PqqD truncation from naturally fused Methylobacterium extorquens (Mex) PqqCD was overexpressed in Escherichia coli and MexPqqA was chemically synthesized. Solution NMR 1H-,15N-HSQC chemical shift studies have identified the PqqD residues involved in binding PqqA, and 1H, 13C, and 15N peak assignments for PqqD alone and for PqqD bound to PqqA are reported herein.
Keywords: Pyrroloquinoline quinone biosynthesis, PQQ, PqqA, PqqD, NMR resonance assignments, RiPP
Biological context
4,5-Dihydro-4,5-dioxo-1H-pyrrolo[2,3-f] quinolone-2,7,9-tricarboxylic acid (pyrroloquinoline quinone or PQQ), is a ribosomally synthesized and post-translationally modified peptide (RiPP) that acts as a dehydrogenase cofactor for certain alcohol and aldose sugar dehydrogenases in prokaryotes (Duine 1999; Anthony 2004). Five gene products from the pqq operon are required for PQQ biosynthesis (Shen et al. 2012). Two of these five products, PqqA, a 20 to 30 residue peptide with an absolutely conserved EXXXY sequence near its C-terminal end (Goosen et al. 1989; Houck et al. 1989; Unkefer et al. 1995; van Kleef and Duine 1988), and PqqD, which tightly binds PqqA and forms a ternary complex with PqqA and PqqE (Latham et al. 2015), are the focus of the NMR studies described in this paper.
PQQ is of interest for several reasons. It is a tricyclic, redox active o-quinone that is not formed by direct post-translational modification of active site residues, but instead is synthesized by way of a RiPP pathway (Goodwin 1998; Anthony 2001; Latham et al. 2015). PQQ is a significant antioxidant, and when present supports mitochondrial biogenesis and function in a wide range of organisms (Bauerly et al. 2011; Bauerly et al. 2006; Chowanadisai et al. 2010; Harris et al. 2013; Stites et al. 2006; Singh et al. 2015; Zhang et al. 2015). Additionally, PQQ demonstrates probiotic properties in mammals; studies with rats and mice have demonstrated decreased growth, reduced immune response, and declining reproductive success when subjects were deprived of PQQ in their diets (Kasahara and Kato 2003; Killgore et al. 1989; Steinberg et al. 2003; Steinberg et al. 1994). While initially considered a cofactor for prokaryotes only, a recent publication identified a fungal enzyme for which PQQ serves as cofactor (Matsumura et al. 2014). Finally, plant studies indicate that the presence of PQQ promotes growth (Okhee et al. 2008).
Only one structural model of PqqD has been published, and this is a dimeric crystal structure of Xanthomonas campestris (Xc) PqqD (PDB ID: 3G2B). However, the physiological state of PqqD is monomeric (Latham et al., 2015), so the biological relevance of the XcPqqD structure is uncertain. In this study, the PqqD portion of the natural Methylobacterium extorquens (Mex) fusion PqqCD was expressed in Escherichia coli to give 13C and 15N isotopically labeled protein, which was purified and subjected to NMR spectroscopic analysis. The interaction of the isotopically labeled MexPqqD with unlabeled and chemically synthesized MexPqqA, which binds with a Kd of ∼ 200 nM (Latham et al. 2015), was also probed by NMR spectroscopy. Here we present the MexPqqD resonance and secondary structure assignments in the absence and presence of MexPqqA in the pursuit of the physiological structure of PqqD and the mapping of the interaction surface of PqqA on PqqD.
Methods and experiments
Recombinant protein expression and purification
Materials
The T4 DNA ligase and restriction enzymes were obtained from New England BioLabs (Ipswich, MA). Polymerase was obtained from Agilent Technologies (Santa Clara, CA). Oligonucleotides were obtained from Eurofins (Huntsville, AL). All DNA sequencing was performed by the University of California's DNA Sequencing Facility (Berkeley, CA).
Preparation of PqqA
The unlabeled peptide, MexPqqA ΔM1, C12S (derived from the wild type, UniProt # Q49148), was synthesized and purified to >80% purity by CPC Scientific (Emeryville, CA) and then used at that purity. The peptide sequence, KWAAPIVSEISVGMEVTSYESAEIDTFN, incorporated a serine in place of the cysteine at residue position 11 to eliminate spurious dimer formation.
Preparation of 15N- and 13C-labeled, recombinant PqqD
The MexpqqD gene (corresponding to amino acids 280-372 from the natural MexPqqCD fusion) was cloned into the pET28a vector (EMD Millipore) using the NdeI and XhoI restriction sites. The cloned gene, incorporating an N-terminal His6-tag, was sequence verified and used to transform E. coli BL21 (DE3) for gene expression. Transformed E. coli BL21 (DE3) cells were grown aerobically at 37°C in M9 minimal media supplemented with 1 g/L NH4Cl (99% 15N, Cambridge Isotopes, Tewksbury, MA), 4 g/L D-glucose (U-13C, Cambridge Isotope Laboratories, Tewksbury, MA) and 50 μg/mL kanamycin. Cells were induced with 1 mM IPTG when the OD600 reached 0.6. Following a 12 h induction at 20°C, the cells were harvested by centrifugation at 6,500 rpm for 10 min. The cells were suspended in five times the mass of cell paste of 50 mM PBS (pH 7.5) and 50 mM imidazole. The cells were lysed by sonication, and the lysate was centrifuged at 20,000 rpm for 15 min. The supernatant was loaded onto a 5 mL HisTrap FF column (GE Healthcare) and the column was washed at 4°C with lysis buffer to remove non-tagged protein, and then with 50 mM PBS (pH 6.5) and 300 mM imidazole to elute the tagged protein. The desired fractions were combined, concentrated, and buffer exchanged over PD-10 columns (GE Healthcare) equilibrated with 25 mM phosphate buffer (pH 6.5). Yield for His6-tagged 13C-,15N-labeled MexPqqD: 27 mg/L culture.
Experimental quantities
NMR experiments were performed using D2O matched Shigemi microtubes, 5 mm O.D. (Shigemi, Inc.). The experimental solution for PqqD alone contained 285 μL of 5.0 mg/ml (0.40 mM) 13C-,15N-labeled MexPqqD in 25 mM potassium phosphate, pH 6.5, 7 μL of 50 mM sodium azide, and 15 μL HPLC grade D2O (final concentrations: 4.6 mg/ml (0.37 mM) MexPqqD, 1.1 mM sodium azide, 4.9% D2O).
The experimental solution for PqqD bound to PqqA (in approximately 4-fold molar excess) was identical to the PqqD alone with the inclusion of 1.35 mg lyophilized, unlabeled MexPqqA (final concentrations: 4.6 mg/ml (0.37 mM) MexPqqD, 4.4 mg/ml MexPqqA (1.4 mM), 1.1 mM sodium azide, 4.9% D2O).
NMR spectroscopy
All NMR data were recorded at 25°C on Bruker AVANCE™ III 850 or 900 MHz NMR spectrometers, each with 5 mm TCI CryoProbes including shielded z-gradient. Two sets of NMR data were acquired with the two samples, 13C-,15N-labeled PqqD and 13C-,15N-labeled PqqD + unlabeled PqqA. The tight binding of PqqD and PqqA precluded a titration approach. Data were processed with nmrPipe (Delaglio, et al. 1995). Proton chemical shifts were calibrated with respect to the water signal relative to DSS ((CH3)3Si(CH2)3SO3Na); 15N and 13C chemical shifts were indirectly referenced to DSS (Live, et al. 1984). Linear predictions were applied to the 15N and 13C dimensions to double the data size and improve digital resolution. A cosine square window function and “auto” zero filling were applied to all 1H, 15N and 13C dimensions. Data were analyzed with Sparky (Goddard and Kneller).
Experiments
Sequence-specific backbone assignments were completed using AutoAssign with two 3D spectra: HNCACB and CBCA(CO)NH (Zimmerman, et al. 1997; Muhandiram, et al. 1994). The HNCACB creates both intra- and inter-residue correlations, whereas the CBCA(CO)NH creates only inter-residue correlations. Combining these two spectra, backbone chemical shifts, including 1HN, 15N, 13CA, 13CB, were assigned. 13C′ chemical shifts were assigned using a 3D HNCO and the first 2D 1H-13C plane of 3D HNCACO (Kay, et al. 1994). 1H and 13C side chain assignments were performed with HCCH-TOCSY, H(CCCO)NH and C(CCO)NH (mixing time: 16 ms) (Montelione, et al. 1992; Kay, et al. 1993). HA assignments and scalar J coupling 3JHNHA were obtained from 3D HNHA spectrum (Vuister and Bax, 1994). The 1Hδ and 1Hε resonances of aromatic residues were assigned using 2D (HB)CB(CGCD)HD and (HB)CB(CGCDCE)HE (Yamazaki, et al. 1993).
Results
Backbone and sidechain 1H, 15N, and 13C chemical shifts were assigned at 100% with the exception of 14 of 27 aromatic 13C's (52%) and 4 of 21aromatic 1H's (19%), which were not assigned. A superposition of the 2D 1H-15N HSQC spectra for PqqD (blue peaks) and PqqD + PqqA (red peaks) is shown in Figure 1. From this plot, changes in the chemical shifts of 1H and 15N can clearly be identified.
Figure 1.
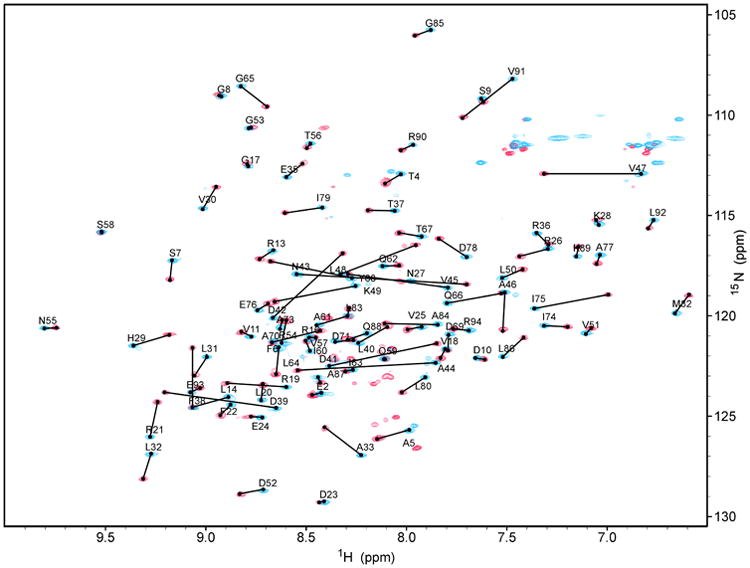
Overlay of 2D 1H-15N HSQC spectra for 13C-,15N-labeled MexPqqD (blue) and 13C-,15N-labeled MexPqqD + unlabeled MexPqqA (red) showing peak shifts.
Secondary structure information
The chemical shifts of 1Hα, 13Cα, 13Cβ and 13CO resonances were used as an input to TALOS+ and CSI2.0 to predict the secondary structures of the two samples (Shen, et al. 2009; Wishart and Sykes, 1994). As shown in Figure 2, the secondary structures predicted from the two methods for PqqD in each sample are very similar. The main difference observed between the two samples, PqqD and PqqD + PqqA, is that one β-strand from the sequence fragment RTFDL of PqqD is significantly longer in the presence of PqqA. In addition, the order parameters predicted by TALOS+ and the flexibility predicted by CSI2.0 are consistent between each sample. Besides the N-terminus of PqqD (residues 1 and 8), the region between residues 51 and 56 in both samples indicates high mobility and disorder.
Figure 2.
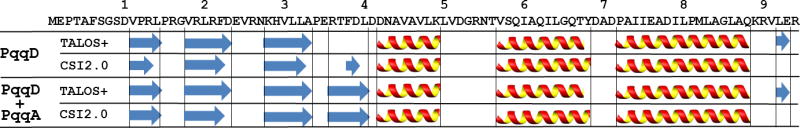
Secondary structures predicted by TALOS+ and CSI2.0 for MexPqqD (the first two rows) and MexPqqD + MexPqqA (the last two rows). The MexPqqD sequence is shown above the secondary structure predictions with sequence decades indicated. Arrows and helices represent β-strand and α-helix, respectively.
Assignments and data deposition
The complete backbone and side chain chemical shift assignments have been deposited in the BioMagResBank database (www.bmrb.wisc.edu) with accession numbers 26634 and 26690 for samples PqqD and PqqD + PqqA, respectively.
Acknowledgments
Financial support for this work came from the National Institutes of Health grants GM-066569 (CMW) and GM-039296 (JPK). Special thanks to the Minnesota NMR Center for both professional expertise and access to spectrometers, and to the Minnesota Supercomputing Institute for their assistance in installing and configuring the annealing software to run on Mesabi (reducing 20 hour anneals to 30 minutes).
References
- Anthony C. Pyrroloquinoline quinone (PQQ) and quinoprotein enzymes. Antioxidant Redox Signal. 2001;3:757–774. doi: 10.1089/15230860152664966. [DOI] [PubMed] [Google Scholar]
- Anthony C. The quinoprotein dehydrogenases for methanol and glucose. Arch Biochem Biophys. 2004;428(1):2–9. doi: 10.1016/j.abb.2004.03.038. [DOI] [PubMed] [Google Scholar]
- Bauerly K, Harris C, Chowanadisai W, Graham J, Havel PJ, Tchaparian E, Satre M, Karliner JS, Rucker RB. Altering pyrroloquinoline quinone nutritional status modulates mitochondrial, lipid, and energy metabolism in rats. PLoS One. 2011;6(7):e21779. doi: 10.1371/journal.pone.0021779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerly KA, Storms DH, Harris CB, Hajizadeh S, Sun MY, Cheung CP, Satre MA, Fascetti AJ, Tchaparian E, Rucker RB. Pyrroloquinoline quinone nutritional status alters lysine metabolism and modulates mitochondrial DNA content in the mouse and rat. Biochim Biophys Acta. 2006;1760(11):1741–8. doi: 10.1016/j.bbagen.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Choi O, Kim J, Kim JG, Jeong Y, Moon JS, Park CS, Hwang I. Pyrroloquinoline quinone is a plant growth promotion factor produced by Pseudomonas fluorescens B16. Plant Physiology. 2008;146:657–668. doi: 10.1104/pp.107.112748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowanadisai W, Bauerly KA, Tchaparian E, Wong A, Cortopassi GA, Rucker RB. Pyrroloquinoline quinone stimulates mitochondrial biogenesis through cAMP response element-binding protein phosphorylation and increased PGC-1alpha expression. J Biol Chem. 2010;285(1):142–52. doi: 10.1074/jbc.M109.030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Duine JA. The PQQ story. J Biosci Bioeng. 1999;88(3):231–6. doi: 10.1016/s1389-1723(00)80002-x. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG. SPARKY 3. University of California; San Francisco: [Google Scholar]
- Goodwin PM, Anthony C. The biochemistry, physiology and genetics of PQQ and PQQ-containing enzymes. Adv Microb Physiol. 1998;40:1–80. doi: 10.1016/s0065-2911(08)60129-0. [DOI] [PubMed] [Google Scholar]
- Goosen N, Horsman HP, Huinen RG, van de Putte P. Acinetobacter calcoaceticus genes involved in biosynthesis of the coenzyme pyrrolo-quinoline-quinone: nucleotide sequence and expression in Escherichia coli K-12. J Bacteriol. 1989;171(1):447–55. doi: 10.1128/jb.171.1.447-455.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris CB, Chowanadisai W, Mishchuk DO, Satre MA, Slupsky CM, Rucker RB. Dietary pyrroloquinoline quinone (PQQ) alters indicators of inflammation and mitochondrial-related metabolism in human subjects. J Nutr Biochem. 2013;24(12):2076–84. doi: 10.1016/j.jnutbio.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Houck DR, Hanners JL, Unkefer CJ, van Kleef MA, Duine JA. PQQ: biosynthetic studies in Methylobacterium AM1 and Hyphomicrobium X using specific 13C labeling and NMR. Antonie Van Leeuwenhoek. 1989;56(1):93–101. doi: 10.1007/BF00822589. [DOI] [PubMed] [Google Scholar]
- Kasahara T, Kato T. Nutritional biochemistry: A new redox-cofactor vitamin for mammals. Nature. 2003;422(6934):832. doi: 10.1038/422832a. [DOI] [PubMed] [Google Scholar]
- Kay LE, Xu GY, Yamazaki T. Enhanced-sensitivity triple-resonance spectroscopy with minimal H2O saturation. J Magn Reson. 1994;A109:129–133. [Google Scholar]
- Kay LE, Xu GY, Singer AU, Muhandiram DR, Forman-Kay JD. A gradient-enhanced HCCH-TOCSY experiment for recording side-chain 1H and 13C correlations in H2O samples of proteins. J Magn Reson. 1993;B101:333–337. [Google Scholar]
- Killgore J, Smidt C, Duich L, Romero-Chapman N, Tinker D, Reiser K, Melko M, Hyde D, Rucker RB. Nutritional importance of pyrroloquinoline quinone. Science. 1989;245(4920):850–2. doi: 10.1126/science.2549636. [DOI] [PubMed] [Google Scholar]
- Latham JA, Iavarone AT, Barr I, Juthani PV, Klinman JP. PqqD is a novel peptide chaperone that forms a ternary complex with the radical S-adenosylmethionine protein PqqE in the pyrroloquinoline quinone biosynthetic pathway. J Biol Chem. 2015;290(20):12908–18. doi: 10.1074/jbc.M115.646521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Live DH, Davis DG, Agosta WC, Cowburn D. Long Range Hydrogen Bond Mediated Effects in Peptides: 15N NMR Study of Gramicidin S in Water and Organic Solvents. J Am Chem Soc. 1984;106:1939–1941. [Google Scholar]
- Matsumura H, Umezawa K, Takeda K, Sugimoto N, Ishida T, Samejima M, Ohno H, Yoshida M, Igarashi K, Nakamura N. Discovery of a eukaryotic pyrroloquinoline quinone-dependent oxidoreductase belonging to a new auxiliary activity family in the database of carbohydrate-active enzymes. PLoS One. 2014;9(8):e104851. doi: 10.1371/journal.pone.0104851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montelione GT, Lyons BA, Emerson SD, Tashiro M. An efficient triple resonance experiment using carbon-13 isotropic mixing for determining sequence-specific resonance assignments of isotopically-enriched proteins. J Am Chem Soc. 1992;114:10974–75. [Google Scholar]
- Muhandiram DR, Kay LE. Gradient-enhanced triple-resonance three-dimensional experiments with improved sensitivity. J Magn Reson. 1994;B103:203–216. [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–22. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YQ, Bonnot F, Imsand EM, RoseFigura JM, Sjolander K, Klinman JP. Distribution and properties of the genes encoding the biosynthesis of the bacterial cofactor, pyrroloquinoline quinone. Biochemistry. 2012;51(11):2265–75. doi: 10.1021/bi201763d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AK, Pandey SK, Saha G, Gattupalli NK. Pyrroloquinoline quinone (PQQ) producing Escherichia coli Nissle 1917 (EcN) alleviates age associated oxidative stress and hyperlipidemia, and improves mitochondrial function in ageing rats. Exp Gerontol. 2015;66:1–9. doi: 10.1016/j.exger.2015.04.001. [DOI] [PubMed] [Google Scholar]
- Steinberg F, Stites TE, Anderson P, Storms D, Chan I, Eghbali S, Rucker R. Pyrroloquinoline quinone improves growth and reproductive performance in mice fed chemically defined diets. Exp Biol Med. 2003;228(2):160–6. doi: 10.1177/153537020322800205. [DOI] [PubMed] [Google Scholar]
- Steinberg FM, Gershwin ME, Rucker RB. Dietary pyrroloquinoline quinone: growth and immune response in BALB/c mice. J Nutr. 1994;124(5):744–53. doi: 10.1093/jn/124.5.744. [DOI] [PubMed] [Google Scholar]
- Stites T, Storms D, Bauerly K, Mah J, Harris C, Fascetti A, Rogers Q, Tchaparian E, Satre M, Rucker RB. Pyrroloquinoline quinone modulates mitochondrial quantity and function in mice. J Nutr. 2006;136(2):390–6. doi: 10.1093/jn/136.2.390. [DOI] [PubMed] [Google Scholar]
- Unkefer CJ, Houck DR, Britt BM, Sosnick TR, Hanners JL. Biogenesis of pyrroloquinoline quinone from 13C-labeled tyrosine. Methods Enzymol. 1995;258:227–35. doi: 10.1016/0076-6879(95)58049-2. [DOI] [PubMed] [Google Scholar]
- van Kleef MA, Duine JA. L-tyrosine is the precursor of PQQ biosynthesis in Hyphomicrobium X. FEBS Lett. 1988;237(1-2):91–7. doi: 10.1016/0014-5793(88)80178-9. [DOI] [PubMed] [Google Scholar]
- Vuister GW, Bax A. Measurement of four-bond HN-Hα J-couplings in staphylococcal nuclease. J Biomol NMR. 1994;4:193–200. doi: 10.1007/BF00175247. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD. The 13C Chemical-Shift Index: A simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Forman-Ka JD, Kay LE. Two-dimensional NMR experiments for correlating 13Cβ. and 1Hδ/ε chemical shifts of aromatic residues in 13C-labeled proteins via scalar couplings. J Am Chem Soc. 1993;115:11054–11055. [Google Scholar]
- Zhang J, Meruvu S, Bedi YS, Chau J, Arguelles A, Rucker R, Choudhury M. Pyrroloquinoline quinone increases the expression and activity of Sirt1 and -3 genes in HepG2 cells. Nutr Res. 2015;35(9):844–9. doi: 10.1016/j.nutres.2015.06.014. [DOI] [PubMed] [Google Scholar]
- Zimmerman DE, Kulikowski CA, Huang Y, Feng W, Tashiro M, Shimotakahar S, Chien C, Power R, Montelione GT. Automated analysis of protein NMR assignments using methods from artificial intelligence. J Mol Biol. 1997;269:592–610. doi: 10.1006/jmbi.1997.1052. [DOI] [PubMed] [Google Scholar]


