Abstract
The CXCR4 chemokine receptor is implicated in a number of diseases including HIV infection and cancer development and metastasis. Previous studies have demonstrated that configurationally restricted bis-tetraazamacrocyclic metal complexes are high-affinity CXCR4 antagonists. Here, we present the synthesis of Cu2+ and Zn2+ acetate complexes of six cross-bridged tetraazamacrocycles to mimic their coordination interaction with the aspartate side chains known to bind them to CXCR4. X-ray crystal structures for three new Cu2+ acetate complexes and two new Zn2+ acetate complexes, demonstrate metal-ion dependent differences in the mode of binding the acetate ligand concomitantly with the requisite cis-V configured cross-bridged tetraazamacrocyle. Concurrent density functional theory molecular modelling studies produced an energetic rationale for the unexpected [Zn(OAc)(H2O)]+ coordination motif present in all of the Zn2+ cross-bridged tetraazamacrocycle crystal structures, which differs from the chelating acetate [Zn(OAc)]+ structures of known unbridged and side-bridged tetraazamacrocyclic Zn2+ containing CXCR4 antagonists.
Keywords: CXCR4 chemokine receptor, cross-bridged tetraazamacrocyle, copper, zinc, acetate binding
Graphical Abstract
Complexes of cross-bridged tetraazamacrocycles exhibit Cu2+ bound to acetate in a monodentate fashion yielding square pyramidal geometries with the acetate occupying an equatorial position (base of the pyramid) and having a relatively short Cu—O bond (~1.95 Å), which may explain the strong binding of Cu2+ cross-bridged bis-cyclam CXCR4 antagonists. The Zn2+ complexes all locate acetate equatorially, hydrogen bonded to a cis water molecule to form distorted octahedral coordination geometries. Crystallographic and computational studies determined that the short cross-bridge opposite the acetate/water ligands forces equatorial N-Zn-N bond angles to be much less than 90°, which leaves a large equatorial space most energetically favourably filled by the acetate/water cis ligands interacting by a hydrogen bond.
Introduction
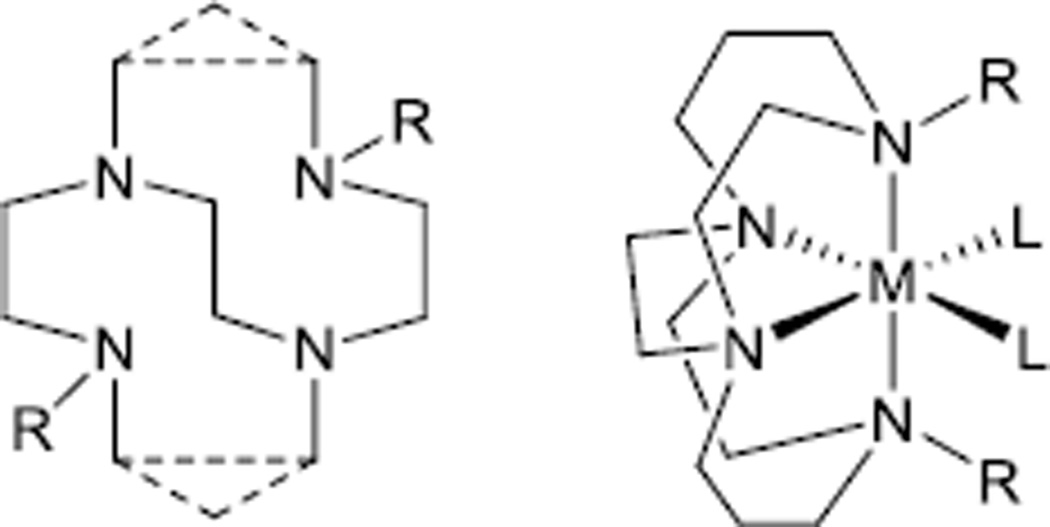
Due to the kinetic stability of their transition metal complexes the highly rigid cross-bridged tetraazamacrocycles[1] (Figure 1), have been of increasing interest in applications where complex stability is vital, such as biological imaging[2] and aqueous oxidation catalysis.[3] Octahedral,[3c] trigonal bipyramidal,[4] or square-pyramidal,[5] coordination geometries (Figure 1) are generally observed where the macrocycle takes up axial and cis-equatorial positions of metal complex structures, since the cross-bridge restricts the configuration of the complex to a folded, cis geometry. Locating the two remaining coordination positions cis to each other in octahedral complexes is important in oxidation applications[3a] and also provides an optimal arrangement for protein-binding complexes.[6]
Figure 1.
Generic cross-bridged tetraazamacrocyclic ligand and metal complex
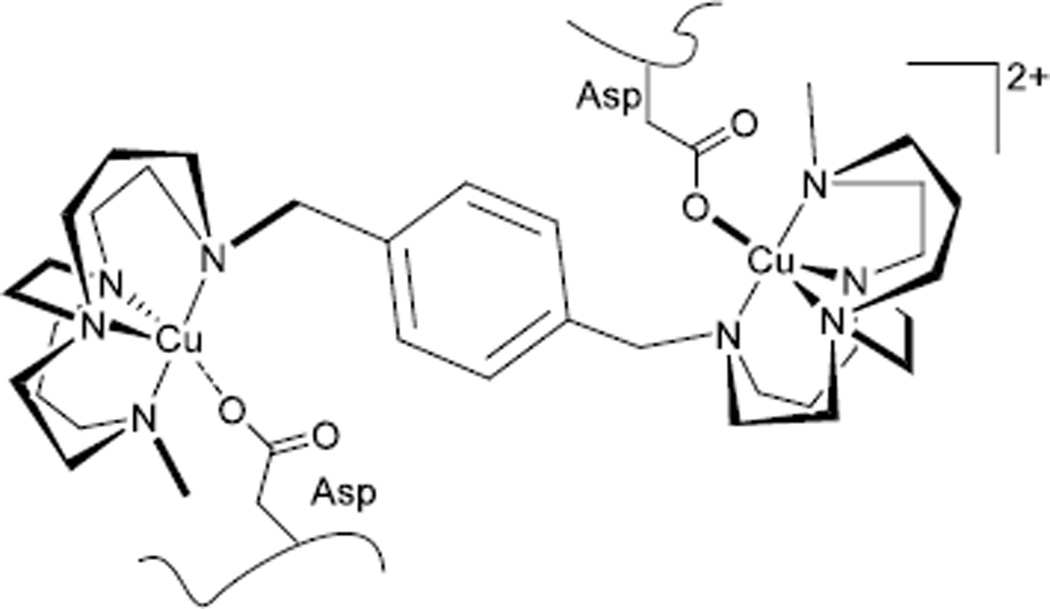
Linked bis cross-bridged tetraazamacrocyclic copper(II) and zinc(II) complexes (Figure 2) that we have recently designed take advantage of these properties and have been demonstrated to efficiently bind to the chemokine receptor CXCR4 with long residence times and high affinity.[6a] In a healthy organism CXCR4 plays an essential developmental role at the embryonic stage. It has also been shown to have a key role in the growth, survival and metastasis of cancer cells and is overexpressed on multiple tumour types.[7] CXCR4 is implicated in other disease states including HIV infection where it acts as a co-receptor for viral cell entry.[8] [9] The binding mode of xylyl bridged bis-tetraazamacrocyclic compounds with CXCR4 has been demonstrated, via site directed mutagenesis, to utilise two aspartate residues (Asp 171 and Asp 262).[10] Hydrogen bonding interactions are replaced by coordination bonds on addition of the metal centres. One aspect of our CXCR4 antagonist studies has been to investigate the aspartate-metal ion binding by synthesising acetate complexes of cross-bridged complexes. The aim was to grow X-ray quality crystals containing acetate ligands bound to the metal ion as a model for the metal ion-aspartate interaction taking place in the biological system.[9] This study characterises the geometric and electronic requirements for generating strong-binding CXCR4 antagonists by obtaining and examining these structures.
Figure 2.
CXCR4 antagonist cross-bridged complex in contact with CXCR4.
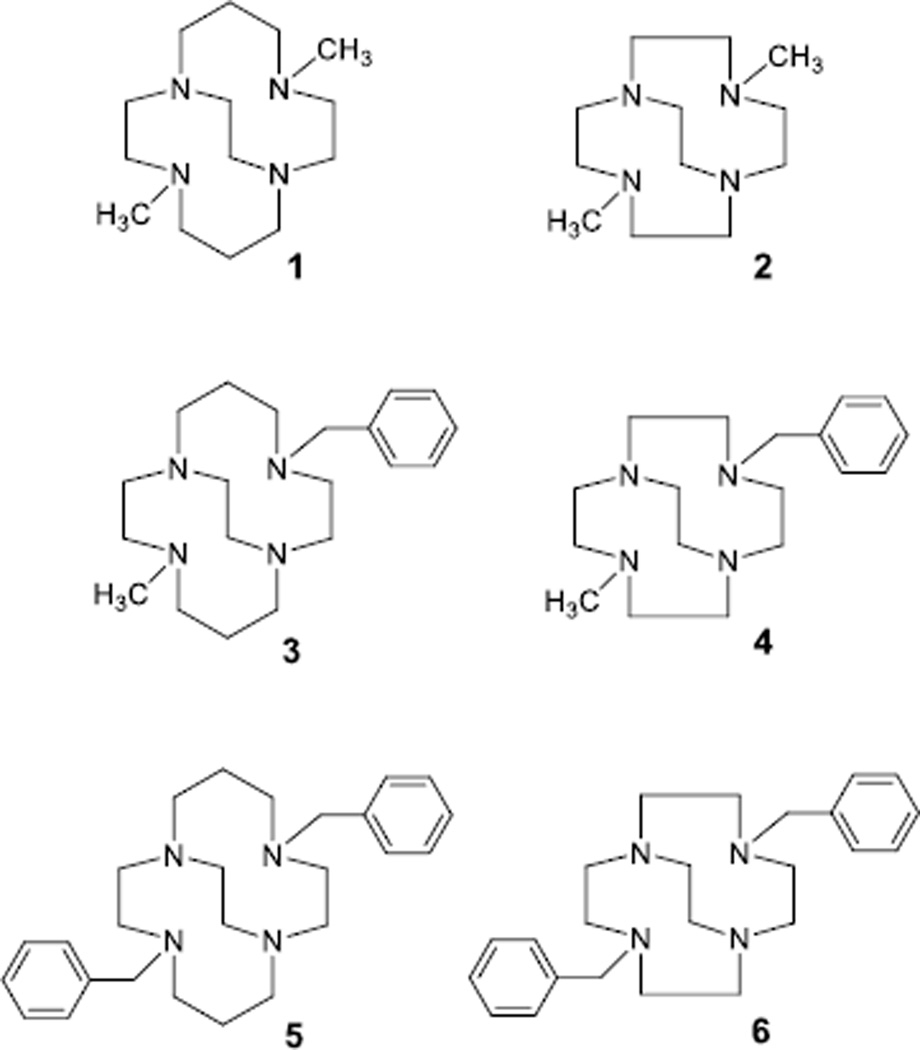
Generally, xylyl bridged bis-linked tetraazamacrocycle complexes produce very few X-ray quality crystals, with only a few examples of these structures published.[11] Our own experience with growing crystals of these complexes has been similarly unproductive. However, producing X-ray quality crystals of bridged mono-macrocycle transition metal complexes has been much more successful in our hands.[3a, 3c, 4–5, 12] We have synthesised a number of dimethyl, monobenzyl-monomethyl, and dibenzyl pendant arm containing cross-bridged tetraazamacrocycles to provide the best models for our bis-macrocycle antagonists,[3c] which are linked through a xylene moiety (Figure 3). These model ligands provide the same cross-bridged macrocycle geometric constraint around the metal ion, as well as placing zero, one, or two bulky benzyl groups on the coordinated nitrogen atoms. The xylene-linked bis-macrocycle complexes typically have one aromatic ring and one methyl group on coordinated nitrogens (Figure 2).
Figure 3.
Structures of ligands discussed in this study
Here, we describe the synthesis, characterisation, and X-ray crystal structural study of these ligands complexed to Cu2+ and Zn2+ ions which are also coordinated to acetate as a model for the aspartate side-chains of CXCR4. Their interaction with acetate sheds light on the binding modes of the highly potent bis-linked complexes with CXCR4.
Results and Discussion
Synthesis and Characterisation
Preparation of Ligands and Metal complexes
Cu2+ complexes of ligands 1–3 and 5 are known {[Cu1Cl]PF6,[4] [Cu2Cl]Cl and [Cu2(CH3CN)2][PF6]2,[13] [Cu3(OH2)][ClO4]2,[14] [Cu5(CH3CN)][PF6]2[15]} as are Zn2+ complexes of ligands 1–2 {Zn1Cl2 and Zn2Cl2}.[13] However, none of these involve acetate as a coordinated ligand. We have previously communicated [Cu3(OAc)]PF6,[6a] but fully disclose its structure and characterisation here. Since copper(II) and zinc(II) complexes of cross-bridged tetraazamacrocycles have shown an ability to interact with aspartate side chains of the biologically important CXCR4 chemokine receptor (Figure 2),[9] we sought to understand the interactions of carboxylate groups with such complexes. The ubiquitous, and commonly used in inorganic chemistry, acetate anion provides the simplest model for the interaction of these metal centres with carboxylate groups. Therefore, we began the process of synthesising and structurally characterising the Cu2+ and Zn2+ complexes of mono-macrocyclic cross-bridged analogues (ligands 1–6) of our potent bis-macrocycle cross-bridged CXCR4 antagonists[6a] (Figure 2) as simpler models for examination.
Ligands 1 and 2[16] were available in our laboratories and thought to be most likely to produce acetate complexes that would crystallise based on previous experience.[1] However, their lack of any bulky benzyl groups might make the resulting structures less representative of how the xylyl-linked bis-macrocycle compounds could interact with a carboxylate. Ligands 5 and 6 were also available from previous work.[16] They bear two pendant benzyl groups, which should more closely approximate the bis ligands. However, we had concerns that the two bulky benzyl arms might provide too much of a steric challenge to acetate binding. As a compromise, we developed the synthesis of ligands 3 and 4, which give the most accurate approximation of our bis-macrocycle ligands (Figure 2), where each metal ion has one methyl pendant and the xylyl linker in the vicinity of the metal ion. The synthesis of ligand 3[6a, 14] follows the Weisman synthesis of cross-bridged tetraazamacrocycles,[16] but utilises a stepwise alkylation of the key macrocycle-glyoxal condensate, first with benzyl bromide, then with methyl iodide, prior to the ring-opening reduction reaction that yields the ethylene cross-bridge. Ligand 4 was synthesised following the same strategy and goes through a known bis-quaternary ammonium salt.[17]
Complexation of the ligands was carried out using anhydrous metal acetate salts in anhydrous solvents (acetonitrile, DMF, or methanol depending on the solubility of the ligand) in an inert atmosphere glovebox and proceeded smoothly in all cases at room temperature with overnight stirring. Although these complexes are not air sensitive, we have found protection of the ligand from sources of water are helpful in complexation reactions as they are strongly basic and protonation can defeat complexation.[12a, 18] Once the complexation had occurred, the reaction solutions were removed from the glovebox and concentrated to dryness, which generally yielded viscous oils as products. In order to produce more easily handled solid complexes, as well as to purify the products, anion metathesis reactions with NH4PF6 in dry methanol were carried out. With only two cis coordination sites available, we believed only one acetate anion would be likely to coordinate to the metal ion, leaving an uncoordinated acetate anion that could be replaced with PF6−. Hexafluorophosphate anions precipitated the complex cations from methanol as microcrystalline powders. In some cases, excess NH4PF6 co-precipitated with the complex, as evidenced by elemental analysis.
Electronic Structure
The electronic spectra of the Cu2+ acetate complexes of ligands 1–6 in acetonitrile show the expected ligand field transitions for d9 Cu2+ (see Table S1) similar to those of other Cu2+ complexes with cross-bridged cyclam and cyclen ligands,1–4 with the presence of the acetate not causing significant differences from other bound monodentate ligands.[4, 13–15]
Use of 64Cu2+ complexes of tetraazamacrocycles, including ethylene cross-bridged examples, as radiopharmaceuticals has been an active area of research.[14, 19] Cross-bridged tetraazamacrocycles offer an advantage for this purpose in that they very slowly decomplex from the Cu2+ ion in aqueous solution due to the rigidity and topological complexity provided by the short cross-bridge.[18–19, 20] The likely major mechanism of inactivation of Cu2+ complexes in vivo is through loss of ligand resulting in free, inactive Cu2+.[20b, 20c] However, an additional proposed indicator of in vivo stability is resistance toward reduction to Cu+ followed by loss of the more labile Cu+ ion.[20a] Reversibility of the Cu2+/Cu+ reduction wave, which indicates an ability of the ligand to accommodate both Cu2+ and Cu+, has been correlated with in vivo stability.[19a, 20a] We sought to examine the reduction potentials and reversibility of the reduction processes of all of the Cu2+ acetate complexes of 1–6 by carrying out cyclic voltammetry experiments, in order to further study their potential in vivo utility.
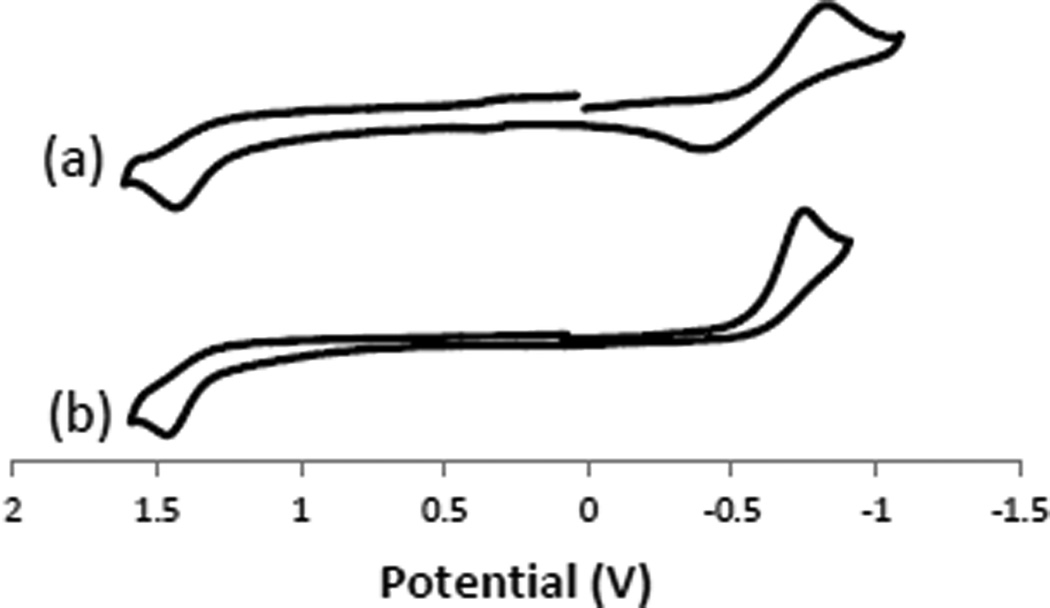
Cyclic voltammetry at a scan rate of 200 mV s−1 of 1 mM acetonitrile solutions (Figure 4, Table 1) was performed on all of the copper complexes. The three cyclam-based complexes (with ligands 1, 3, and 5) gave similarly shaped voltammograms (Figure 4a), which were importantly different than the voltammograms (Figure 4b) of the three cyclen-based complexes (with ligands 2, 4, and 6). The difference between these two sets of complexes is the return oxidation wave from Cu+ to Cu2+ for the cyclam-based complexes, which is not present for any of the cyclen-based complexes. The larger cyclam macrocycles are indeed able to accommodate the larger Cu+ ion, and the Cu+ complexes produced can be quasi-reversibly oxidised back to Cu2+. The smaller cyclen macrocycles do not provide complementary ligands for larger and more labile Cu+, and the reduced forms decompose before they can be re-oxidised. This result is similar to what has been observed[13] for [Cu1Cl]+ and [Cu2Cl]+, where the former has an observable return oxidation, but the latter does not. As noted above, reversibility of the Cu2+/Cu+ reduction wave has been used as an indicator of potential in vivo stability, along with kinetic inertness towards aqueous hydrolysis. In combination with the kinetic stability of the cyclam-based ligand 1[20b, 20c] the reversibility of the reduction of these cyclam-based ligands bodes well for their in vivo stability.
Figure 4.
Cyclic voltammograms of (a) [Cu3(OAc)]+ and (b) [Cu6(OAc)]+. These voltammograms are representative of the data for the (a) cyclam-based and (b) cyclen-based complexes studied. Complexes were 0.001 M in acetonitrile with 0.1 M TBAPF6. Voltages are vs SHE.
Table 1.
Electrochemical results for copper acetate complexes of ligands 1–6.
| Complex | Eox (Cu2+/3+) [V] | Ered (Cu2+/+) [V] | Eox (Cu+/2+) [V] |
|---|---|---|---|
| [Cu1(OAc)]+ | +1.477 | −0.877 | −0.307 |
| [Cu3(OAc)]+ | +1.434 | −0.830 | −0.390 |
| [Cu5(OAc)]+ | +1.516 | −0.641 | −0.156 |
| [Cu2(OAc)]+ | +1.417 | −0.893 | ----- |
| [Cu4(OAc)]+ | +1.731 | −0.591 | ----- |
| [Cu6(OAc)]+ | +1.465 | −0.637 | ----- |
Interestingly, reduction from Cu2+ to Cu+ does appear to be effected systematically by the change of methyl groups to benzyl groups. In the series of cyclam-based [CuL(OAc)]+ complexes where L goes from 1 (two methyl) to 3 (one benzyl and one methyl) to 5 (two benzyl), the reduction potential to Cu+ changes significantly from −0.877 V to −0.830 V, to −0.641 V, respectively. This trend is also seen in the cyclen-based series of complexes where the reduction potentials change from −0.893 V to −0.591 V to −0.637 V for the complexes of ligand 2 (two methyl), 4 (one benzyl and one methyl), and 6 (two benzyl), respectively. Addition of one or two benzyl groups can cause a shift towards a less negative reduction potential of more than 230 mV. This result indicates that the presence of the benzyl substituent favours the formation of Cu+, making it occur at a less negative potential, which may indicate less stability in vivo for the benzyl-containing complexes than the dimethyl complexes.[19a, 20a] On the basis of the structure of [Cu5]+ the benzyl group(s) can fold towards the metal ion and occupy empty coordination sites.[15] In the [Cu5]+ structure, both benzyl groups do so and occupy a gap in a highly distorted tetrahedral coordination geometry of the Cu+ ion where one N-Cu+-N bond angle is much larger (171.85°) than the ideal 109.5°. The benzyl groups clearly stabilise Cu+ in this crystal structure, and may be able to do so in solution as well, explaining the large shift towards favourability of reduction to Cu+ upon mono- or di-benzylation seen in the cyclic voltammetry.
Although less relevant for in vivo complex stability, all six complexes show irreversible oxidations to Cu3+. The irreversibility of these oxidations has been explained for similar complexes as an inability of the neutral tetraazamacrocycle to adequately stabilise reactive Cu3+ cation, which requires strong bonds to stabilise this high valent state.[13] It should be noted that the complex [Cu4(OAc)]+ has a significantly higher (~200 mV) oxidation potential (+1.731 V) than any other complex, the reason for this is not clear. It should be noted that we do not have an X-ray crystal structure of this compound, and it is possible that there is a significant structural difference, at least in the acetonitrile solution in which the electrochemistry experiment was performed, between it and the other two cyclen complexes which would explain the large Eox difference.
CXCR4 Affinity
AMD3100,[8, 21] and the high potency CXCR4 antagonists that we have developed[6a, 11a, 22] are bis-macrocyclic with an aryl (xylyl) linker. Our previously collected data indicates that monomacrocyclic compounds will also have affinity for the receptor but this will be lower than for the bismacrocyclic derivatives. The main reason for synthesising the monomacrocyclic compounds (metal complexes of 3 – 6) was to utilise them as simpler structural analogues to allow us to obtain X-ray structural data that models aspartate or glutamate coordination to the metal centre. We do not anticipate taking any of these compounds into further biological evaluation or in vivo studies as they have greater potential for off target binding. However it is still of interest to determine the receptor affinity and investigate the structure activity relationships for this subset of compounds.
Preliminary screening assays were carried out for the free chelators showing IC50 values of greater than 10 µM indicating no measurable affinity for these compounds. This is consistent with previously analysed free macrocyclic chelators where the H-bonding potential of the chelator has been disrupted by alkylation and they are only activated on inclusion of the metal centre to give the potential for coordinate bond formation.[11a]
Two assays were performed to confirm that CXCR4 binding occurred for the monomacrocylic metal complexes: a competition binding assay with fluorescently tagged CXCL12; and a chemokine-induced calcium signalling assay, see Table 1. The IC50 values were determined for the ability of the compounds to block both the binding and the signalling of CXCL12, the natural ligand of CXCR4. The inhibition of the fluorescently tagged CXCL12 generally returns higher potency IC50 determinations.
A comparison of macrocycle ring size for the complexes i.e. 1 vs. 2, 3 vs. 4 and 5 vs. 6 show some evidence for a preference for the cyclen ring size for zinc(II) and the cyclam ring for copper(II). This could relate to coordinational flexibility of the zinc(II) d10 metal ion. For chelators 1–3 the copper(II) complexes are more active than the zinc(II) complexes indicating that these chelators offer an optimal arrangement for both coordination to the copper(II) ion and secondary interactions with the protein structure.
The most potent compounds are [Cu3(OAc)]+ and [Zn4(OAc)]+ with [Zn6(OAc)]+ / [Zn5(OAc)]+ also highly active. The IC50 values are ca. 10 nM in the CXCL12 binding inhibition assay and 50 nM in the signaling assay, showing that, in these assays, they are of similar potency to AMD3100 and approach the activity of the high affinity bismacrocyclic metal complexes that we have developed (e.g, with chelator 8).[6a, 11a, 23] However they are likely to have off target binding and shorter residence times at the receptor.
Modelling aspartate binding
Our previously published configurationally restricted tetraazamacrocylic transition metal complexes have a stronger binding interaction with CXCR4 than non-restricted analogues. To understand the strong binding of these compounds we have modelled analogues with acetate using single-crystal x-ray crystallography and density functional theory.
Crystallography
Macrocycle/Metal Interactions
For the purposes of this discussion, two closely related crystal structures from recently published work will be included for relevant comparisons: [Zn9(OAc)(H2O)]+ and [Cu3(OAc)]+.[6a, 20b] Crystallographic details for the seven new crystal structures in this work, along with selected bond lengths and angles, are presented in Tables S2–3. Several general observations can be made about this collection of crystals structures prior to the detailed description of acetate binding. Below, each point is outlined in text and illustrated with a figure.
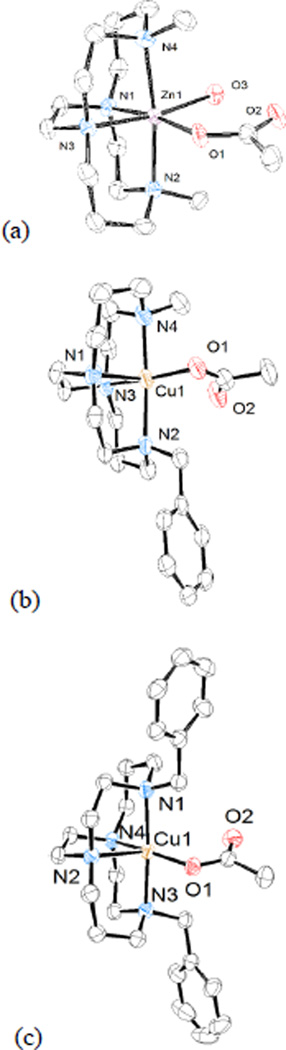
First, the cis-V configuration that is dictated by the ligand cross-bridge is observed as expected for all of the complexes structurally characterised here. Figure 5 illustrates this observation for the cyclam family of ligands. Structure (a) is of [Zn1(OAc)(H2O)]+, the dimethyl bridged cyclam complex of Zn2+, (b) is of [Cu3(OAc)]+, the monobenzyl-monomethyl-bridged cyclam complex of Cu2+, and (c) is of [Cu5(OAc)]+, the dibenzyl bridged cyclam complex of Cu2+. Neither the identity of the metal ion, nor that of the alkyl substituents affects this configuration. This same configuration has been seen in all known metal complexes with ethylene cross-bridged cyclams and cyclens.
Figure 5.
Structures of (a) [Zn1(OAc)(H2O)]+ (b)[6a] [Cu3(OAc)]+ and (c) [Cu5(OAc)]+ (from PF6− salt) demonstrating cis-V configuration for all three cyclam based ligands.
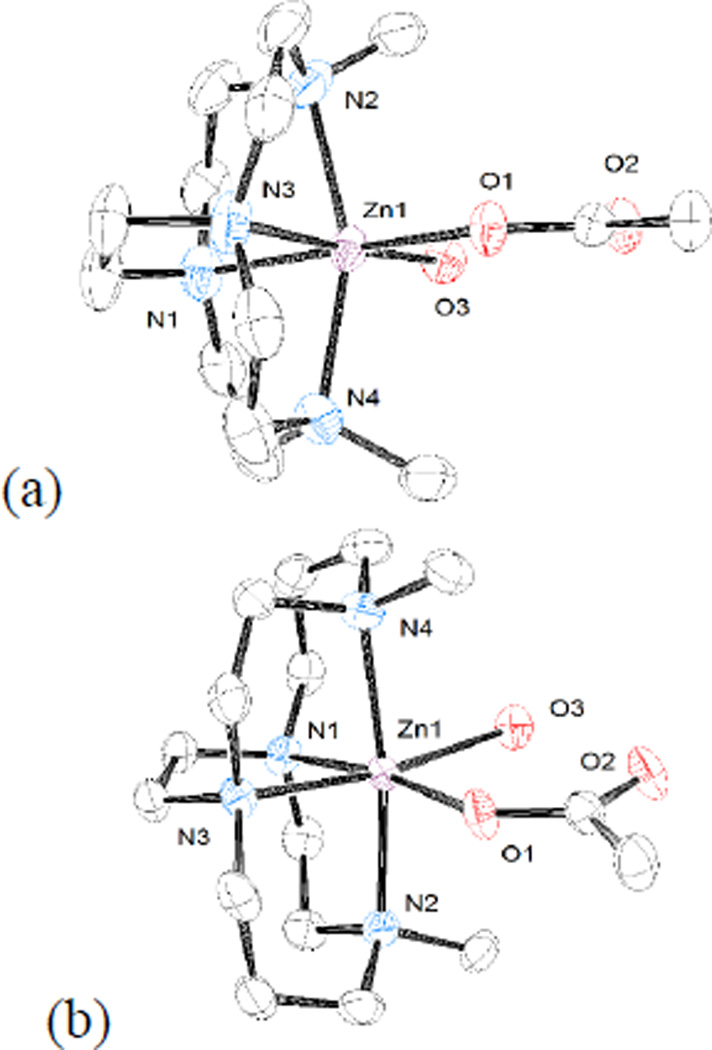
Second, the ring size of the parent macrocycle influences how fully the metal ion is engulfed by the bridged macrocycle. Figure 6 illustrates this trend using (a) the dimethyl bridged cyclen complex of Zn2+, [Zn2(OAc)(H2O)]+, and (b) dimethyl bridged cyclam complex of Zn2+, [Zn1(OAc)(H2O)]+. The easiest to use parameter to discuss this trend is the Nax-Zn-Nax bond angle, where Nax is an axially coordinated nitrogen. This bond angle is 157.59(10)° in the smaller cyclen case of [Zn2(OAc)(H2O)]+, while it is 171.89(12)° for the cyclam case of [Zn1(OAc)(H2O)]+. The larger the bond angle, the closer to linearity and thus the better fit, or complementarity, between the ligand and an idealised octahedral geometry for the Zn2+ ion. The same trend is observed in all of the cyclen vs. cyclam cases and is well known for cross-bridged complexes.[1]
Figure 6.
Structures of (a) [Zn2(OAc)(H2O)]+ and (b) [Zn1(OAc)(H2O)]+ demonstrating that the larger cyclam ligands more fully engulf the metal ion and have a more linear Nax-M-Nax angle.
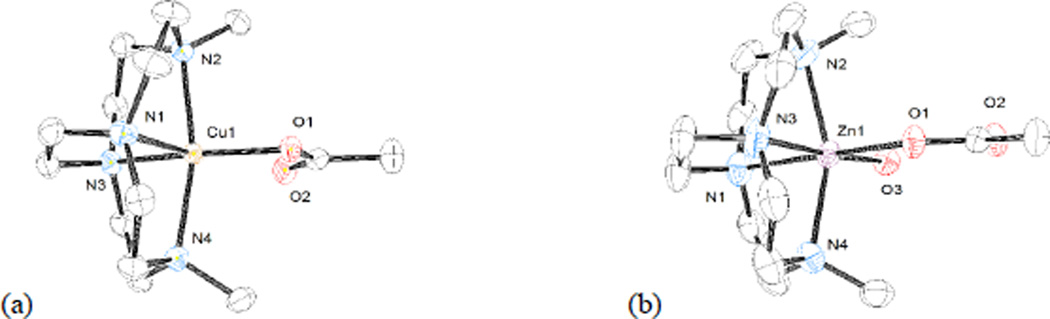
Third, the size of the metal ion also influences the complementarity of ligand ring size to metal ion coordination geometry. The Cu2+ ion has a radius of 79 pm in five-coordinate complexes, while Zn2+ has a radius of 88 pm in six-coordinate complexes.[24] The larger the metal ion, the more difficulty the ligand has in providing it with its preferred geometry. This trend is illustrated in Figure 7, where the copper complex of the dimethyl bridged cyclen ligand, [Cu2(OAc)]+, is shown in (a) and the zinc complex of the same ligand, [Zn2(OAc)(H2O)]+, is shown in (b). The Nax-Zn-Nax bond angle for the smaller copper is 164.04(8)°, while this angle is only 157.59(10)° for the larger zinc ion. The same trend is observed (though not pictured) for the dibenzyl bridged cyclam complex of Cu2+, [Cu5(OAc)][PF6], where this angle is 176.74(8)°; while for [Zn1(OAc)(H2O)]+, the Zn2+ dimethyl bridged cyclam complex, the angle is 171.89(12)°.
Figure 7.
Structures of (a) [Cu2(OAc)]+ and (b) [Zn2(OAc)(H2O)]+ demonstrating the effect of metal ion radius on Nax-M-Nax bond angle.
Acetate Binding
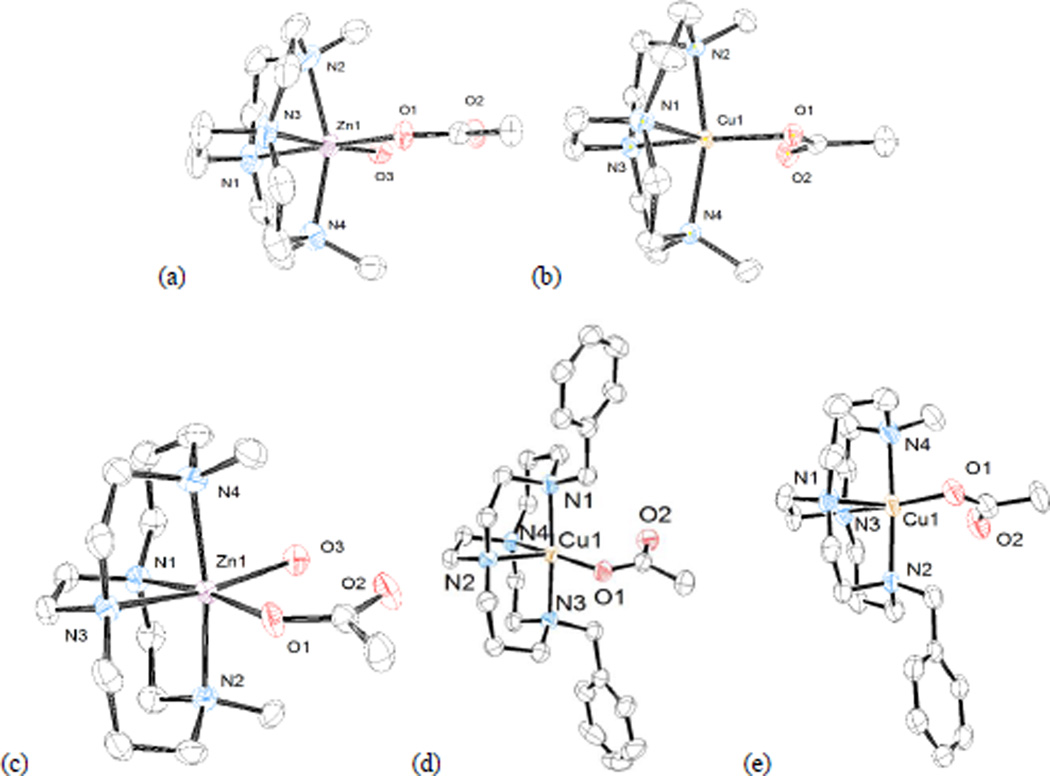
Since acetate is acting as a model for aspartate residues in CXCR4, its mode of coordination to the most biologically active metal ions Cu2+ and Zn2+ is the most interesting feature about this set of structures.[6a, 11a] A detailed description of the structure of each complex in which acetate is bound to either Cu2+ or Zn2+ will be presented first, followed by a discussion of the information that these structures offer us about binding of acetate. Figure 8 contains a representation of each of these structures:
Figure 8.
Structures of (a) [Zn2(OAc)(H2O)]+ (b) [Cu2(OAc)]+ (c) [Zn1(OAc)(H2O)]+ (d) [Cu5(OAc)]+ (from PF6- salt) (e) [Cu3(OAc)]+
[Zn2(OAc)(H2O)]+ is shown in Figure 8a. The zinc ion is in a distorted octahedral geometry, where both axial and two adjacent equatorial site are occupied by the bridged macrocycle ligand in a cis-V configuration. The zinc ion protrudes markedly from the small ligand cavity, with a Nax-Zn-Nax bond angle of 157.59(10)°. The remaining equatorial sites are occupied by the monodentate acetate and a water molecule. A hydrogen bond exists between the water molecule and the unbound oxygen of the acetate ion (O-O distance, 2.620(3) Å).
[Zn1(OAc)(H2O)]+ is shown in Figure 8c. The zinc ion is similarly in a distorted octahedral geometry, where both axial and two adjacent equatorial site are occupied by the bridged macrocycle ligand in a cis-V configuration. The zinc ion is better engulfed by the larger ligand cavity, with a Nax-Zn-Nax bond angle of 171.89(12)°. The remaining equatorial sites are occupied by the monodentate acetate and a water molecule. A hydrogen bond between the water molecule and the unbound oxygen of the acetate ion has an O-O distance of 2.653(4) Å=[Cu2(OAc)]+ is shown in Figure 8b. The copper ion is in a distorted square pyramidal geometry with one of the bridge-head nitrogens in the axial position. The other three nitrogens and the bound acetate oxygen occupy the base of the pyramid. The bond length to the bound oxygen, Cu(1)-O(1), is 1.9565(17) Å. The distance to the second oxygen is much greater, Cu(1)-O(2), at 2.576 Å. The Addison Parameter (τ) quantifies trigonal bipyramidal/square pyramidal geometry for five-coordinate complexes,[25] with a value of 1.0 a pure trigonal bipyramid and a value of 0.0 for a pure square pyramid. Axial donors of tetraazamacrocycle Cu2+ complexes above 2.5 Å are not thought to interact with the metal centre and are not considered as bonds.[26] In this case, if the long 2.576 Å Cu(1)-O(2) interaction is ignored and the complex treated as five-coordinate, then τ = 0.007 indicating a geometry closest to square pyramidal. However, including the long Cu(1)-O(2) interaction would make a distorted octahedral six-coordinate geometry. For the purposes of discussion of this and all similarly structured Cu2+-acetate complexes below, we will not tally the long Cu(1)-O(2) interaction as a bond and continue to describe the structures as a five-coordinate square pyramidal examples.
We were able to crystallise [Cu5(OAc)]+ as two separate salts, one having an uncoordinated acetate anion and the other a PF6− anion. The acetate salt has a nearly identical, yet crystallographically distinct, structure to the PF6− salt. Both of these cations can be described as having distorted square pyramidal geometries. Figure 8d contains the cation of [Cu5(OAc)]+ from the PF6− salt, and is used to represent both nearly identical structures. The copper ion is in a slightly distorted square pyramidal geometry (τ = 0.19) with one of the bridge-head nitrogens in the axial position. The other three nitrogens and the bound acetate oxygen occupy the base of the pyramid. The bond length to the bound oxygen, Cu(1)-O(1), is 1.9469(18) Å. The distance to the second oxygen is much greater, Cu(1)-O(2), at 2.887 Å. A PF6− anion balances the charge. Both benzyl groups of the ligand are held away from the coordinated acetate. The benzyl groups are approximately perpendicular to the plane of the acetate ligand, and are also approximately perpendicular to one another.
[Cu3(OAc)]+ is shown in Figure 8e. This complex and its crystal structure were communicated previously,[6a] but full discussion of its structure is relevant and appears for the first time here. The copper ion is in a distorted square pyramidal geometry (τ = 0.22) with one of the bridge-head nitrogens in the axial position. The other three nitrogens and the bound acetate oxygen occupy the base of the pyramid. The bond length to the bound oxygen, Cu(1)-O(1), is 1.946(2) Å. The distance to the second oxygen is much greater, Cu(1)-O(2), at 2.657 Å. The benzyl group is held up away from the bound acetate, and is approximately perpendicular to the plane of the acetate ion.
In analysing these acetate-bound structures, it is clear that steric interactions with the benzyl groups does not interfere with the ability of acetate to bind. Both monobenzyl and dibenzyl ligands support the coordination of acetate. The bulky benzyl groups appear to have some flexibility as to their positioning with respect to the open binding sites not occupied by the macrocyclic ligands. If favourable acetate binding can occur, the benzyl groups can be rotated out of the way. This is an important observation, because the linked macrocycle dimers contain a xylyl group between them. Confirming what is apparent from CXCR4 binding and antagonism studies, these structures show us that steric bulk is not a major issue for binding carboxylate ligands in the open coordination sites.
The identity of the metal ion is more important than ring size or alkyl substitution in determining the mode of acetate binding. In all cases, the copper(II) complexes are bound in an asymmetric way to the acetate. One of the acetate oxygen atoms is always bound strongly to copper, with a bond length near 1.95 Å, while the other oxygen is much further away, typically around 2.70 Å. The resulting geometry can best be described as square pyramidal. For five-coordinate complexes, a square pyramidal geometry seems to be an arrangement favoured by Cu2+ in these cross-bridged macrocycles, as demonstrated in numerous Cu2+ complexes already in the literature.[4, 13–15] Only rarely[13] is the Cu2+ ion found in an octahedral geometry
An important observation, and one communicated previously,[6a] is that the cross-bridge forces a preorganised, cis-V optimised geometry with a prescribed equatorial position (pyrimadal base) for the acetate to bind. Due to the typical Jahn-Teller distortions found in d9 Cu2+ complexes, the axial position is most often elongated, and thus coordination to this site is weakened. Similarly, the equatorial positions are shortened, and thus coordination to them is strengthened. Unbridged cyclam-Cu2+ complexes typically locate a fifth ligand in the axial (weaker bound) position and the four macrocycle nitrogen donors in all four of the equatorial (stronger bound) sites.[27] However, in all of the cross-bridged complexes of Cu2+ presented here, the axial position is forced to be occupied by a macrocycle nitrogen, simply because the bridge prevents the folded macrocycle from reaching all four equatorial positions. Instead, one equatorial position is unoccupied by the macrocycle and is the site of acetate binding. This shorter, stronger binding equatorial position for acetate (or aspartate) binding may explain why a cross-bridged bis-cyclam CXCR4 antagonist has a higher affinity and longer residence time at CXCR4 than the unbridged Cu-AMD3100 analogue.[6a] The multiple crystal structures presented here containing a cross-bridged macrocycle, Cu2+, and a bound acetate all exhibit the same equatorial acetate binding and reaffirm the advantageous structural properties of cross-bridged CXCR4 antagonists.
In contrast, the Zn2+ ion in these cross-bridged macrocycles binds acetate in a monodentate fashion only, but fills its sixth coordination site with an additional water molecule in the three structures presented here. Zn2+, being a spherical, d10 ion, has a greater preference for an octahedral coordination geometry than the d9 Cu2+. One option to obtain this more symmetric geometry is for the Zn2+ to bind both oxygen atoms of acetate in a bidentate coordination mode. Apparently, this is not the most stable arrangement in these complexes in the solid state however, because in each case, the sixth coordination site around zinc is occupied not by the second acetate oxygen, but rather by a water molecule. The bound water molecule then acts as a hydrogen bond donor to the uncoordinated oxygen atom of the bound acetate, with the distance between the non-bonded acetate oxygen and the bound water oxygen between 2.619 Å and 2.653 Å in each case. Seemingly, the binding of water to Zn2+ in addition to the hydrogen bonding interaction is more favourable than the chelation of the second acetate oxygen to zinc. Since water is readily available around CXCR4, this same binding mode is possible between our zinc containing antagonist compounds and the aspartate residues of CXCR4. The water molecule could be involved in further hydrogen bonding interactions with surrounding amino acid residues when the complex is bound to the protein.
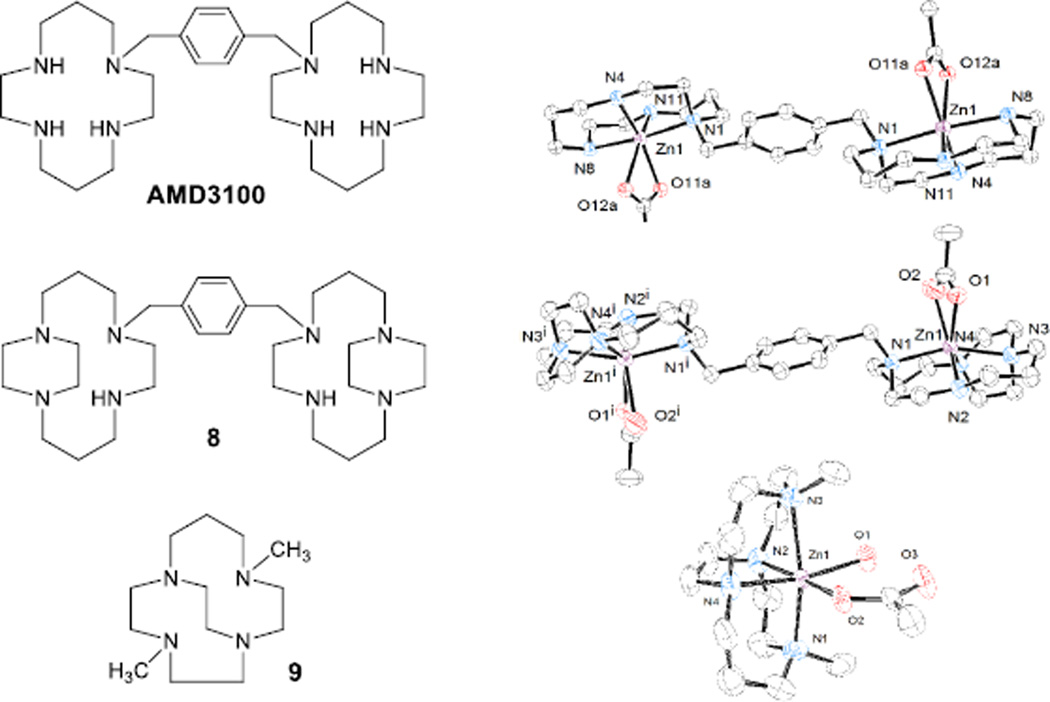
Three interesting previously published tetraazamacrocycle Zn2+-acetate complexes, two of which are known efficient CXCR4 antagonists, deserve comparison. Sadler’s group published the crystal structure of the di-Zn2+ complex of AMD3100 (Figure 9) where the zinc ion is bound to acetate.[28] Like the two crystal structures of our mono-ring compounds discussed above, the unbridged cyclam rings in AMD3100 are found in the cis-V configuration in this structure. However, the acetate bound to the zinc ion is bound in an aniso-bidentate fashion (Zn-O bond distances of 2.09 Å and 2.41 Å) rather than in a monodentate fashion with a water ligand at the sixth position as in our cross-bridged complexes. In one of our previous studies of symmetric ethylene side-bridged bis-cyclam (Figure 9, structure 8),[11a] a crystal structure of the [Zn28(OAc)2]2+ cation shows a similar aniso-bidentate interaction with acetate (Zn-O bond distances of 2.11 Å and 2.41 Å) and no water molecule bound to the zinc. Because of the ethylene side-bridges, each macrocycle in this complex is in a trans-II configuration, however. Finally, we recently published a series of complexes of the intermediate ring-size cross-bridged homocyclen (Figure 9, structure 9) analogue of 1 and 2, including the Zn2+-acetate complex.[20b] Like the Zn2+ complexes of other cross-bridged ligands presented here, this complex features an octahedral Zn2+ ion coordinating one monodentate acetate hydrogen bonded to a coordinated water molecule. Clearly, the cross-bridged architecture leads to this Zn(OAc)(H2O) structural feature despite ring size and substitution pattern.
Figure 9.
Ligand structures (left) and X-ray crystal structures (right) of known Zn2+-acetate complexes for structural comparison
Additional examples of the cross-bridged architecture leading to the M(OAc)(H2O) structural unit can be found in the crystal structures of [Ni1(OAc)(H2O)]+ and [Ni5(OAc)(H2O)]+, which we have recently published.[22] In these structures, very similar six-coordinate geometries to the cross-bridged Zn2+ structures discussed above are present, with monodentate acetate ligands hydrogen bonded to aqua ligands that complete pseudo-octahedral structures.
Examining these five zinc-acetate structures, it appears that the presence of the ethylene cross-bridge is the deciding factor as to whether the bound acetate will interact in an aniso-bidentate versus a monodentate fashion with a hydrogen bond to a water molecule which is providing the sixth ligand to Zn2+. Table 3 lists the structural parameters used in this analysis. The Nax-Zn-Nax bond angle has often been useful in determining how well a cross-bridged tetraazamacrocycle engulfs its bound metal ion.[1, 12a] However, it appears not to be correlated to the acetate binding mode in these structures. [Zn2(AMD3100)(OAc)2]2+ and [Zn1(OAc)(H2O)]+ both have very similar Nax-Zn-Nax bond angles (174.67° and 171.70(11)°, respectively). However, one binds the acetate in an aniso-bidentate fashion while the other has a monodentate acetate hydrogen bonded to a water molecule in the sixth coordination site.
Table 3.
X-ray structural parameters determining acetate binding mode in Zn2+ complexes.
| Complex | Nax-Zn-Nax Angle (°) | Neq-Zn-Neq Angle (°) | O-Zn-O Angle (°) | OAc Binding Mode |
|---|---|---|---|---|
| [Zn2(AMD3100)(OAc)2]2+ | 174.67 | 105.32 | 58.34 | aniso-bidentate |
| [Zn28(OAc)2]2+ | 158.40 | 117.91 | 56.31 | aniso-bidentate |
| [Zn1(OAc)(H2O)]+ | 171.89 | 83.44 | 88.70 | monodentate/H2O |
| [Zn9(OAc)(H2O)]+ | 169.88 | 83.52 | 89.52 | monodentate/H2O |
| [Zn2(OAc)(H2O)]+ | 157.59 | 81.80 | 90.83 | monodentate/H2O |
The [Zn28(OAc)2]2+ (158.40°), and [Zn2(OAc)(H2O)]+ (157.59°) complexes have similar Nax-Zn-Nax bond angles, but different acetate binding modes. In the case of [Zn2(OAc)(H2O)]+, this small angle distortion from the preferred linear angle of an octahedral complex is due to the small cyclen macrocycle size, while in [Zn28(OAc)2]2+ this small angle has been explained as a movement of Zn2+ out of the cyclam macrocycle plane in order to better interact with acetate.[11a] Finally, intermediate ring size complex [Zn9(OAc)(H2O)]+ has an intermediate Nax-Zn-Nax bond angle of 169.88°.[20b] Again, no correlation between this bond angle and acetate binding mode is apparent as the planar acetate in all five complexes is nearly parallel to the Neq-Zn-Neq plane and is not large enough to interact with the axial nitrogens or their substituents. In contrast, the Neq-Zn-Neq and O-Zn-O bond angles clearly correlate to the acetate binding mode. In the unbridged AMD3100 complex and the side-bridged ligand 8 complex, the Neq-Zn-Neq bond angles are much larger (105.32° and 117.91°, respectively) than the ideal 90° bond angles of an octahedral complex. These large angles are in contrast with the small O-Zn-O angles of the aniso-bidentate acetate ligand on the opposite side of the complex (58.34° and 56.31°, respectively).
Having only the small-angle acetate chelated on one side is favoured, when the more flexible unbridged or side-bridged macrocycle can occupy more of the equatorial space. In clear contrast to these two complexes are the three cross-bridged macrocycle complexes of ligands 1, 2, and 9. Their Neq-Zn-Neq bond angles (83.44°, 81.80°, and 83.52°) are constrained by the ethylene cross-bridge tying the two equatorial nitrogen donors together. With these angles restricted to less than the ideal 90° of an octahedron, there is much more equatorial space available on the opposite side of the complex where the acetate is bound. Rather than binding acetate in an aniso-bidentate fashion with small O-Zn-O bond angles, which would not effectively fill the equatorial space around the Zn2+ cation, these complexes bind acetate in a monodentate mode and bind an additional H2O ligand in the sixth coordination site to better fill the equatorial space. Energy lost due to the absence of the chelating interaction is apparently made up for by coordination of the water molecule and the presence of the hydrogen bond between acetate and water. Additional preference for these structures is likely to be gained by the near-ideal O-Zn-O bond angles of the ligand 1, 2, and 9 complexes (88.70°, 90.83°, and 89.52°). Computational studies of unbridged, side-bridged, and cross-bridged Zn2+-acetate complexes were carried out to determine the energetics of the acetate binding modes observed.
Computational Studies of Zn2+-Acetate-Water Coordination
DFT calculations have previously been reported on copper(II) cross- and side-bridged derivatives, showing a geometry optimised five-coordinate distorted square based pyramidal complex for both, with cross-bridged exhibiting shorter Cu-O acetate bond distances and increased stability compared to cross-bridged derivatives.[23, 29] Because of the systematic differences in Zn2+ coordination of acetate dependent on the presence or absence of the macrocyclic cross-bridge in the structures discussed above, we wanted to further examine this phenomenon. We used density functional theory to probe the energy differences between acetate binding modes for unbridged, side-bridged, and cross-bridged cyclam-based complexes containing Zn2+ and acetate/water. We hoped that the combined structural and computational studies might produce useful conclusions about the general binding of carboxylate groups to Zn2+ macrocyclic complexes, as well as specific information about the best ligand design practices for M2+ containing CXCR4 antagonists.
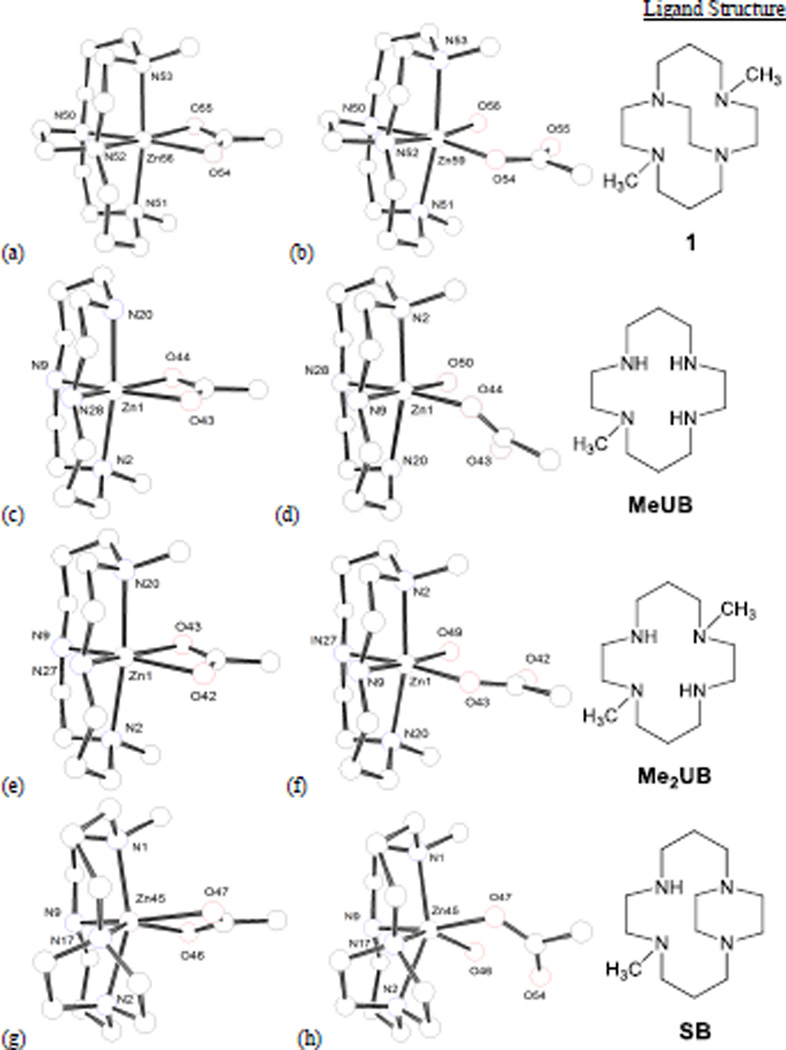
Figure 10 shows the four ligands studied and the M06/6-311+G(d,p) geometry optimised [Zn(OAc)]+ and [Zn(OAc)(OH2)]+ structures for the eight complexes generated. In order, these complexes are named after the ligand structures as: dimethyl-cross-bridged (1), monomethyl-unbridged (MeUB), dimethyl-unbridged (Me2UB), and monomethyl-side-bridged (SB), respectively. MeUB is the closest mono-macrocycle analogue of the two cyclams in AMD3100. Me2UB is the simplest unbridged analogue of 1. SB is the simplest mono-macrocycle analogue of 8. To analyse the relative stability of the single acetate binding versus the acetate/water binding the energy change for the reaction: [(L)Zn(OAc)]+ + H2O → [(L)Zn(OAc)(OH2)]+ in the gas phase was calculated using the M06/6-311+G(d.p) results. These results are listed in Table 4.
Figure 10.
Minimised structures from computational study. (a) [Zn1(OAc)]+, (b) [Zn1(OAc)(H2O)]+, (c) [Zn(MeUB)(OAc)]+, (d) [Zn(MeUB)(OAc)(H2O)]+, (e) [Zn(Me2UB)(OAc)]+, (f) [Zn(Me2UB)(OAc)(H2O)]+, (g) [Zn(SB)(OAc)]+, (h) [Zn(SB)(OAc)(H2O)]+
Table 4.
Energy Changes (ΔE, ΔH, and ΔG) for the [Zn-OAc]1+ + H2O → [Zn-(OAc/OH2)]1+ reaction from M06/6-311+G(d,p) calculations. ΔE is the electronic energy change for the reaction, not including zero-point energy, while ΔH and ΔG include the zero-point energy and thermal corrections within the harmonic oscillator approximation. All energy differences are in kJ/mol.
| Ligand (L) | ∆rxnEo0K (electronic) | ∆rxnHo298K | ∆rxnGo298K |
|---|---|---|---|
| dimethyl-cross-bridged (1) | −57.3 | −51.8 | −7.5 |
| monomethyl-unbridged (MeUB) | −52.7 | −46.3 | −0.9 |
| dimethyl-unbridged (Me2UB) | −51.0 | −45.9 | −0.5 |
| monomethyl-side-bridged (SB) | −24.9 | −18.7 | 26.5 |
From the energetic results, it is clear that the dimethyl-cross-bridged (Ligand 1) complex prefers to be 6-coordinate with binding from both an acetate and water, much more so than any of the unbridged or side-bridged complexes. However, the ΔGo298 value suggests that the single acetate binding complex is preferred for the monomethyl-side-bridged (SB) complex. These calculated energies agree with the crystallographically observed coordination modes for cross-bridged and side-bridged Zn-acetate complexes presented above. There is little difference between the energies of the two calculated coordination modes, as quantified in ΔGo298 for both unbridged ligands, MeUB and Me2UB. The justification for pursuing side- and cross-bridged analogues of AMD3100 was to produce topologically constrained complexes that were preorganised to bind CXCR4 in specific configurations.[6a, 11a] Unbridged cyclams are configurationally flexible,[27] and thus could potentially need to reorganise in order to bind CXCR4 in an optimal fashion. This set of calculations is congruent with these ideas, as the two bridged ligand complexes clearly select for a single coordination mode, while the unbridged ligand complexes do not.
Geometric parameters for the optimised structures in Figure 10 are listed in Table 5 and may indicate the structural reasons for the calculated energies. The most unique set of parameters shows that the (SB) complex is a significantly distorted octahedral structure, with a trans-II configuration (Figure 10 g and h). Both SB structures contain the smallest Nax-Zn-Nax, and largest Neq-Zn-Neq angles of their coordination mode, which is testament to the distortion from octahedral the side-bridged trans-II configuration produces. The 2-carbon side bridge essentially functions as a bulky group forcing the equatorial nitrogen atoms apart, which causes the Zn2+ ion to move out of the macrocycle plane in order to bind acetate and/or water.[11a]
Table 5.
Geometric parameters for the M06/6-311+G(d,p) minimised structures.
| [Zn(OAc)]+ structure | [Zn(OAc)(H2O)]+ structure | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Bond Angles (°) | Distance (Å) | Bond Angles (°) | Distance (Å) | |||||||
| Ligand | Nax-Zn-Nax | Neq-Zn-Neq | O-Zn-O | Zn-O | Nax-Zn-Nax | Neq-Zn-Neq | O-Zn-O | Zn-OOAc | Zn-OH2O | O-OH-Bond |
| CB | 172.4 | 84.4 | 62.2 | 2.116, 2.120 | 169.8 | 81.9 | 92.6 | 2.022 | 2.180 | 2.553 |
| MeUB | 172.1 | 102.9 | 61.5 | 2.136, 2.152 | 169.2 | 98.1 | 88.9 | 2.034 | 2.265 | 2.610 |
| Me2UB | 171.9 | 100.4 | 61.9 | 2.128, 2.134 | 168.8 | 95.4 | 91.3 | 2.025 | 2.201 | 2.556 |
| SB | 155.9 | 116.7 | 60.7 | 2.076, 2.245 | 149.4 | 109.8 | 85.1 | 2.058 | 2.196 | 2.565 |
From Figure 10h, significant distortions of the bound acetate and water out of the equatorial plane are required to accommodate the [Zn(OAc)(H2O)]+ hydrogen bonded unit, no doubt contributing to the positive value of ΔGo298. The smaller chelated acetate in Figure 10g is more easily accommodated by the reduced equatorial space around Zn2+ caused by the large Neq-Zn-Neq angle of the SB ligand. In addition to the distorted octahedral structure, the Zn–O bond lengths in the chelated acetate (SB) complex are quite different in length, 2.08 Å and 2.25 Å. These lengths are clearly aniso-bidentate and similar to the bond lengths from the crystal structure of [Zn28(OAc)2]2+ (Figure 10) where the Zn–O distances are 2.112 Å and 2.407 Å.
In contrast to the SB structures, the geometry about Zn2+ in the [Zn1(OAc)(H2O)]+ complex (Figure 10b) is much less distorted. The smallest Neq-Zn-Neq angles in both coordination modes are forced by the 2-carbon cross-bridge, only 81.9° in the aqua complex. This minimised equatorial bulk allows for the largest O-Zn-O bond angle in the same complex (92.6°), as the [Zn(OAc)(H2O)]+ unit can occupy a larger equatorial space. In contrast, the [Zn1(OAc)]+ structure has small Neq-Zn-Neq (84.4°) and O-Zn-O (62.2°) equatorial angles forced by the cross-bridge and the chelation of acetate, respectively, that do not as effectively “fill” the equatorial space around Zn2+. Overall, the distortion from octahedral is less in the [Zn1(OAc)(H2O)]+ structure than any of the other seven structures, leading to the most favourable ΔGo298.
The MeUB and Me2UB structures primarily demonstrate the configurational flexibility suggested by the energetic calculations. In the [Zn(L)(OAc)]+ structures, both unbridged ligands give much larger Neq-Zn-Neq angles (102.9° and 100.4°) than the 84.4° angle of the CB complex, but much smaller than the 116.7° angle of the SB complex. These intermediate (and topologically unconstrained) angles allow the ligand to expand around the equator to fill space not occupied by the small chelated acetate (O-Zn-O angle only 61.4° and 61.9°, respectively). After water binds these unbridged complexes, the Neq-Zn-Neq angles (98.1° and 95.4°) contract to allow the [Zn(OAc)(H2O)]+ motif to form with nearly ideal octahedral O-Zn-O bond angles of 88.9° and 91.3°, respectively. Although the aqua complexes are slightly energetically favoured (Table 4), the configurational flexibility of these unbridged ligands leads to reasonable geometries for both coordination modes. From Figure 10d, the asymmetric MeUB ligand does cause distortion of the acetate out of the equatorial plane as it appears to flex away from the lone methyl group. This distortion leads to the longest H-bonding O-O distance (2.610 Å) of any of the optimised aqua complexes. The acetate is returned to the equatorial plane in the Me2UB complex, as the steric requirements of both methyl groups exert balanced effects on the equatorial plane. Consequently, the H-bonding O-O distance is shortened to 2.556 Å, intermediate between the CB and SB complex H-bonding O-O distances.
These computational results reinforce the conclusions derived from the crystal structures. In the unbridged and the side-bridged complexes, the Neq-Zn-Neq bond angles are much larger, and these large angles favour the small O-Zn-O angles of the aniso-bidentate acetate ligand on the opposite side of the complex. In contrast, the cross-bridged macrocycle complexes are constrained to small, < 90° Neq-Zn-Neq bond angles ensuring there is much more equatorial space available on the opposite side of the complex where the acetate is bound. These complexes fill this space by binding acetate in a monodentate mode and binding an additional H2O ligand in the sixth coordination site to produce near-ideal 90° O-Zn-O bond angles and produce stabilised 6-coordinate pseudo-octahedral structures.
Conclusions
A novel benzyl methyl cross-bridged cyclen ligand has been prepared to complement the five other known dimethyl, dibenzyl, and benzyl methyl cyclam and cyclen derivatives. These ligands have been complexed to Cu2+ and Zn2+ concurrently with an acetate anion, which serves as a model carboxylate ligand for the aspartate side chains shown to bind xylyl bridged bis-cyclam CXCR4 antagonists.
X-ray crystal structures of three such Cu2+ and three Zn2+ complexes were obtained, to complement recently published analogues. All of these structures were examined to learn about preferences for Cu2+ and Zn2+ macrocycle complexes in binding carboxylate ligands, which could possibly be applied to CXCR4 antagonist design.
The cross-bridged Cu2+ complexes exhibited Cu2+ bound to acetate in a monodentate fashion yielding square pyramidal geometries with the acetate occupying an equatorial position (base of the pyramid) and having a relatively short Cu—O bond (~1.95 Å). This topologically constrained position forced on the acetate by the cross-bridge may explain the relatively strong binding of Cu2+ cross-bridged bis-cyclam CXCR4 antagonists relative to side- and unbridged analogues, where longer axial-type binding of acetate is possible. Cyclic voltammetry of the Cu2+ complexes showed that the cyclam-based complexes are quasi-reversibly reduced to Cu+, indicating high stability for in vivo studies which will be particularly important to maintain low toxicity and also for radiolabelling with copper isotopes for nuclear imaging. However, the cyclen-based complexes gave only irreversible reduction to Cu+ indicating that they are less well suited to these applications.
The Zn2+ complexes of cross-bridged tetraazamacrocycles all located the acetate ligand equatorially, hydrogen bonded to a cis water molecule to form distorted octahedral coordination geometries. Crystallographic and computational studies determined that the short cross-bridge opposite the acetate/water ligands forces equatorial N-Zn-N bond angles to be much less than 90°, which leaves a large equatorial space most energetically favourably filled by the acetate/water cis ligands interacting by a hydrogen bond. A side-bridged tetraazamacrocycle Zn2+-acetate structure from the literature has an equatorial N-Zn-N bond angles much greater than 90°, which appears to favour anisobidentate acetate binding as there is insufficient space available for an additional water ligand. An unbridged AMD3100 Zn-acetate structure, as well as computational models of two other unbridged macrocycles, have intermediate Neq-Zn-Neq bond angles near 100°, which can change substantially based on acetate chelation vs. acetate-water coordination. Energetically, these unbridged complexes don’t appear to strongly favour either coordination mode due to an evident configurational flexibility.
The biological properties of the monomacrocyclic complexes were evaluated and all were found to be active in targeting the CXCR4 receptor in vitro. Four of the single ring cross bridged complexes have high affinity for the receptor target, in some cases matching the potency of AMD3100 which was included as a control. On considering these results in the context of the computational and crystallographic results, the key point of variability is the range of potencies for coordinationally flexible zinc(II) ion. This could be explained by the variation in substituents influencing the position in the binding pocket, as well as the impact on secondary interactions with both the bound water and the aryl substituents. These data are not necessarily predictive of the activity of analogous bismacrocyclic compounds as the factors influencing their potency may not be consistent.
The crystal structures and computational analysis presented suggest strong structural preferences applicable to CXCR4 binding. Cu2+ cross-bridged complexes are constrained by the cross-bridge to bind carboxylates in equatorial positions, leading to short, strong interactions compared to unconstrained macrocycle complexes. Zn2+ cross-bridged complexes are constrained to bind carboxylates in a monodentate fashion, cis to an aqua ligand that hydrogen bonds to the unbound carboxylate oxygen. We have characterised the key features that optimise the binding interactions for tetraazamacrocyclic copper(II) and zinc(II) complexes with aspartate or glutamate amino acid side chains. This will allow future design of novel antagonist constructs for the CXCR4 chemokine receptor but can also be more generally applied to any other proteins with accessible surface carboxylate containing residues. The chemokine receptor family of proteins are all rich in aspartates and glutamates indicating the potential for the positioning of optimised binding units to recognise other receptors relevant to human diseases that can be exploited for therapy or diagnostic imaging.
Experimental Section
General
Elemental analyses were performed by Quantitative Technologies Inc. Electron impact mass spectra were collected on a Shimadzu QP2010 GCMS instrument equipped with a direct insertion probe (DIP). Solid samples of the metal complexes were inserted directly into the mass spectrometer and heated until ionization occurred. Electrospray Mass spectra were collected at the Oklahoma University Health Sciences Center Laboratory for Molecular Biology and Cytometry Research on a Bruker-Daltonics HCT Ultra ion trap mass spectrometer. NMR spectra were obtained on a Varian Bruker AVANCE II 300 MHz NMR Spectrometer. Electronic spectra were recorded using a Shimadzu UV-240 UV-Vis Spectrometer. Electrochemical experiments were performed on a BAS100B Electrochemical Analyser. A button Pt electrode was used as the working electrode with a Pt-wire counter electrode and a Ag-wire pseudo-reference electrode. Scans were taken at 200 mV/s. Acetonitrile solutions of the complexes (1 mM) with tetrabutylammonium hexafluorophosphate (0.1 M) as a supporting electrolyte were used. The measured potentials were referenced to SHE using ferrocene (+0.400 V versus SHE) as an internal standard. All electrochemical measurements were carried out under N2.
Synthesis
Anhydrous solvents, and starting materials, including anhydrous Cu(OAc)2 and Zn(OAc)2, were purchased from Aldrich and used as received. Ligands 4,11-dimethyl-1,4,8,11-tetraazabicyclo[6.6.2]hexadecane (1), 4,11-dibenzyl-1,4,8,11-tetraazabicyclo[6.6.2]hexadecane (5), and 4,10-dibenzyl-1,4,7,10-tetraazabicyclo[5.5.2]tetradecane (6) were synthesised according to literature procedures.[16] Ligand 4,10-dimethyl-1,4,7,10-tetraazabicyclo[5.5.2]tetradecane (2) was prepared according to a recent synthesis.[20b] 4-benzyl-11-methyl-1,4,8,11-tetraazabicyclo[6.6.2]hexadecane (3), which has recently been published by some of us[6a] and 4-benzyl-10-methyl-1,4,7,10-tetraazabicyclo[5.5.2]tetradecane (4), were made by mono-benzylating the appropriate macrocycle-glyoxal condensate, which was then methylated at the non-adjacent nitrogen with iodomethane. While 4 has not been published, the immediate mono-benzyl mono-methyl glyoxal condensate precursor has.[17]
Ligand 4
Mono-benzyl-mono-methyl cyclen glyoxal was synthesised according to a literature procedure.[17] 14.326 g of bis-quaternary ammonium salt was dissolved in 1170 mL of 95% EtOH under N2 and ~15 equivalents (14.0 g) of NaBH4 were slowly added, then left to stir under N2 for 5 days at room temperature. Portions of 6 M HCl were then added to the flask to decompose the NaBH4 until a pH of ~1–2 was reached. The EtOH was then evaporated and the remaining aqueous solution was made basic to a pH of ~14 by addition of 30% by mass KOH, after which an additional 10 g of KOH was added. The solution was then extracted with 5 × 100 mL portions of benzene and set to dry over Na2SO4 overnight. After gravity filtration, solvent evaporation removed the benzene and the product 4 (yellow oil) was dried under vacuum. Yield: 6.992 g (93.6%). 1H NMR (300 MHz, C6D6) 2.22 (m, 1H N-α-CH2), 2.36 (s, 3H, CH3), 2.45–3.00 (m, 16H, N-α-CH2), 3.20 (m, 5H, N-α-CH2), 7.28 (m, 5H, CHaromatic). 13C NMR (100 MHz, C6D6) 42.77 (N-α-CH2), 55.60 (N-α-CH3), 55.91 (N-α-CH2), 56.51 (N-α-CH2), 57.35 (N-α-CH2), 59.10 (N-α-CH2), 60.08 (N-α-CH2), 126.58 (CHaromatic), 127.04 (CHaromatic), 127.80 (CHaromatic), 139.59 (Caromatic). MS (EI) m/z 303.3 [LH]+. Elemental analysis(%) calcd. C18H30N4: C 71.48, H 10.00, N 18.52; Found C 71.29, H 9.89, N 18.58.
General Complexation Procedure for Acetate Complexes
(1.00 mmol) of the ligand (1–6) and (1.00 mmol) of the anhydrous metal(II) acetate salt (Cu or Zn) were added to 25 ml of either dry acetonitrile (ligands 2 and 4), dry DMF (ligands 5 and 6), or dry methanol (ligand 1 and 3) in an inert atmosphere glovebox. The reaction was stirred at room temperature for 18 h. For ligand 2, 4, 5, and 6 complexes, the crude [M(L)(OAc)][(OAc)] solution was removed from the glovebox, filtered to remove any trace solids, and evaporated to dryness, typically giving oils. In the ligand 1 and 3 complex cases, the methanol reaction solution were removed from the glovebox and filtered, but not evaporated, before proceeding to the next step. These crude products were dissolved in 10 mL of methanol, to which was added over the course of a few minutes a 5 mL methanol solution of 5 equivalents (0.815 g, 5.00 mmol) of NH4PF6. Powders of the [M(L)(OAc)]PF6 salts precipitated overnight in a freezer at −5 °C, were collected on a fine glass frit, washed with cold methanol and ether, and dried under vacuum.
Complexation reactions
[Cu1(OAc)]PF6: Blue powder. Yield: 0.136 g (26%). X-ray quality crystals were obtained from evaporation of a methanol solution. Elemental analysis(%) calcd. [CuC14H30N4(C2H3O2)]PF6 • 0.5 H2O (530.982 g/mol): C 36.19.70, H 6.45, N 10.55; Found C 36.08, H 6.48, N 10.52. MS (ES) m/z 376 [CuL(OAc)]+. [Zn1(OAc)]PF6: White powder. Yield: 0.110 g (21%). X-ray quality crystals were obtained from ether diffusion into an acetone solution. Elemental analysis (%) calcd. [ZnC14H30N4(C2H3O2)]PF6 • 1.5 H2O (550.831 g/mol): C 34.89, H 6.59, N 10.17; Found C 34.74, H 6.51, N 10.12. MS (ES) m/z 379 [ZnL(OAc)]+. [Cu2(OAc)]PF6: Blue powder. Yield: 0.430 g (87%). X-ray quality crystals were obtained from ether diffusion into a methanol solution. Elemental analysis(%) calcd. [CuC12H26N4(C2H3O2)]PF6 (493.920 g/mol): C 34.04, H 5.92, N 11.34; Found C 33.90, H 6.02, N 11.27. MS (ES) m/z 350 [CuL(OAc)]+. [Zn2(OAc)]PF6: White powder. Yield: 0.416 g (84%). X-ray quality crystals were obtained from evaporation of a 1,2-dichloroethane solution. Elemental analysis(%) calcd. [ZnC12H26N4(C2H3O2)]PF6 (495.754 g/mol): C 33.92, H 5.90, N 11.30; Found C 33.68, H 5.86, N 11.30. MS (ES) m/z 349 and 351 [ZnL(OAc)]+. [Cu3(OAc)]PF6: This compound has been published.[6a] [Zn3(OAc)]PF6: White powder. Yield: 0.438 g (63%). Elemental analysis(%) calcd. [ZnC20H34N4(C2H3O2)]PF6 • 0.6 NH4PF6 (697.707 g/mol): C 37.87, H 5.69, N 9.24; Found C 37.83, H 5.71, N 9.23. MS (EI) m/z 396 [ZnL]+. [Cu4(OAc)]PF6: Dark blue powder. Yield: 0.134 g (20%). Elemental analysis(%) calcd. [CuC18H30N4(C2H3O2)]PF6 • 0.6 NH4PF6 (667.819 g/mol): C 35.97, H 5.34, N 9.65; Found C 35.84, H 5.04, N 9.76. MS (EI) m/z 424 [CuL(OAc)]+. [Zn4(OAc)]PF6: White powder. Yield: 0.212 g (36%). Elemental analysis(%) calcd. [ZnC18HN(C2H3O2)]PF6 • 0.5 H2O (580.860 g/mol): C 41.36, H 5.90, N 9.65; Found C 41.27, H 5.74, N 9.64. MS (ES) m/z 425 [ZnL(OAc)]+. [Cu5(OAc)]PF6: Blue-green powder. Yield: 0.573 g (83%). X-ray quality crystals of the crude acetate salt were obtained from ether diffusion into an acetonitrile solution. X-ray quality crystals of the purified hexafluorophosphate salt were obtained from ether diffusion into an acetone solution. Elemental analysis(%) calcd. [CuC26H38N4(C2H3O2)]PF6 • H2O (692.185 g/mol): C 48.59, H 6.26, N 8.09; Found C 48.65, H 6.18, N 8.21. MS (ES) m/z 528.3 and 530.3 [CuL(OAc)]+. [Zn5(OAc)]PF6: White powder. Yield: 0.558 g (80%). Elemental analysis(%) calcd. [ZnC26H38N4(C2H3O2)]PF6 • H2O (694.019 g/mol): C 48.46, H 6.25, N 8.07; Found C 48.57, H 6.30, N 8.16. MS (ES) m/z 529.3 [ZnL(OAc)]+. [Cu6(OAc)]PF6: Light blue powder. Yield: 0.571 g (83%). Elemental analysis(%) calcd. [CuC24H34N4(C2H3O2)]PF6 • 0.2 NH4PF6 • 0.4 H2O (685.922 g/mol): C 45.53, H 5.67, N 8.57; Found C 45.89, H 5.39, N 8.18. MS (ES) m/z 500 [CuL(OAc)]+. [Zn6(OAc)]PF6: Off-white powder. Yield: 0.598 g (92%). Elemental analysis(%) calcd. [ZnC24H34N4(C2H3O2)]PF6 (647.950 g/mol): C 48.20, H 5.76, N 8.65; Found C 48.20, H 5.73, N 8.68. MS (ES) m/z 499 [ZnL(OAc)]+.
Chemokine (CXCL12-AF647) binding inhibition assay
Human peripheral blood lymphocytes (PBL) were washed once with assay buffer (Hanks’ balanced salt solution with 20 mM HEPES buffer and 0.2% bovine serum albumin, pH 7.4) and then incubated for 15 min at room temperature with the sample diluted in assay buffer at the indicated concentrations. Subsequently, CXCL12-AF647 (25 ng/ml) was added to the compound-incubated cells. The cells were incubated for 30 min at room temperature. Thereafter, the cells were washed twice in assay buffer, fixed in 1% paraformaldehyde in PBS, and analysed on the FL4 channel of a FACSCalibur flow cytometer equipped with a 635-nm red diode laser (Becton Dickinson, San Jose, CA, USA). The percentages of inhibition of CXCL12-AF647 binding were calculated according to the formula: [1 – ((MFI – MFINC) / (MFIPC – MFINC))] × 100 where MFI is the mean fluorescence intensity of the cells incubated with CXCL12-AF647 in the presence of the inhibitor. Experiments were carried out in triplicate and presented as an average.
Chemokine-induced calcium signalling assay
Ca2+ mobilization assays were performed by the use of a fluorometric imaging plate reader (FLIPR) (Molecular Devices, Sunnyvale, USA) as described previously.[30] Briefly, CXCR4-positive U87 cells were loaded with the fluorescent calcium indicator Fluo-3 acetoxymethyl (Molecular Probes, Leiden, The Netherlands) in the appropriate culture medium for 45 min at 37°C, after which the cells were washed three times in Hanks balanced salt solution buffer containing 20 mM HEPES and 0.2% bovine serum albumin (pH 7.4). The cells were then incubated in the dark at 37 °C for 15 min with the compounds. Changes in intracellular calcium concentration upon addition of CXCL12 (SDF-1), the specific ligand for CXCR4, was simultaneously measured in all 96 wells of a black-wall microtiter plate and in real time with the FLIPR device. The data were expressed as fluorescence units versus time and were analysed using the program Softmax PRO 4.0 (Molecular Devices), and IC50 values were calculated using GraphPad Prism 4.0 software (San Diego, CA). Experiments were carried out in triplicate and presented as an average.
Computational Methods
Density functional theory calculations were performed utilizing the M06 functional with the 6-311+G(d,p) basis set, as implemented in Gaussian 09.[31] Full geometry optimizations and vibrational frequency calculations were performed with this method using an ultrafine integration grid. All calculations were run in the singlet electronic state with a charge of 1+, with the exception of the charge neutral H2O.
X-ray Crystallography
Single crystal X-ray diffraction data were collected in series of ω-scans using a Stoe IPSD2 image plate diffractometer utilizing monochromated Mo radiation (λ = 0.71073 Å). Standard procedures were employed for the integration and processing of the data using X-RED.[32] Samples were coated in a thin film of perfluoropolyether oil and mounted at the tip of a glass fibre located on a goniometer. Data were collected from crystals held at 150 K in an Oxford Instruments nitrogen gas cryostream.
Crystal structures were solved using routine automatic direct methods implemented within SHELXS-97.[33] Completion of structures was achieved by performing least squares refinement against all unique F2 values using SHELXL-97.[33] All non-H atoms were refined with anisotropic displacement parameters. Hydrogen atoms were placed using a riding model. Where the location of hydrogen atoms was obvious from difference Fourier maps, C H bond lengths were refined subject to chemically sensible restraints.
CCDC 1470920-1470926 contain the supplementary crystallographic data for this paper. These data can be obtained free of charge from The Cambridge Crystallographic Data Centre via www.ccdc.cam.ac.uk/data%5Frequest/cif.
Supplementary Material
Table 2.
CXCR4 IC50 values (nM) of the evaluated compounds
| Complex | CXCL12 inhibition (nM) |
Ca2+ flux IC50 CXCR4a (nM) |
|---|---|---|
| [Cu1(OAc)]+ | 28.0 | 186.0 |
| [Zn1(OAc)]+ | 153.5 | 2236.0 |
| [Cu2(OAc)]+ | 16.8 | 298.1 |
| [Zn2(OAc)]+ | 117.7 | 642.5 |
| [Cu3(OAc)]+ | 12.4 | 53.5 |
| [Zn3(OAc)]+ | 195.0 | 2793.8 |
| [Cu4(OAc)]+ | 26.4 | 203.5 |
| [Zn4(OAc)]+ | 21.0 | 40.0 |
| [Cu5(OAc)]+ | 29.8 | 155.8 |
| [Zn5(OAc)]+ | 14.8 | 68.5 |
| [Cu6(OAc)]+ | 44.5 | 231.6 |
| [Zn6(OAc)]+ | 15.0 | 71.0 |
| [Cu8(OAc)]+ | 9.5 | 52.8 |
| [Zn8(OAc)]+ | 0.6 | 1.5 |
| [Cu9(OAc)]+ | 68.8 | 884.1 |
| [Zn9(OAc)]+ | 129.0 | 1163.4 |
| AMD3100 | 11.9 | 87.6 |
CXCR4-positive U87 cell line. IC50 is the concentration of the compound required to inhibit 50% of binding of AF647-labeled CXCL12 (CXCL12 inhibition) the CXCL12 (SDF-1) induced Ca2+ signalling.
Acknowledgments
TJH acknowledges the Health Research award for project number HR13-157, from the Oklahoma Center for the Advancement of Science and Technology. This project was supported by the National Center for Research Resources and the National Institute of General Medical Sciences of the National Institutes of Health through Grant Number 8P20GM103447. TJH acknowledges the Research Corporation (CC6505) for funding. TJH also acknowledges the Henry Dreyfus Teacher-Scholar Awards Program for support of this work. DS acknowledges the financial support of the KU Leuven grants GOA 15/19 TBA and PF10/18.
References
- 1.Hubin TJ. Coord. Chem. Rev. 2003;241:27–46. [Google Scholar]
- 2.a Sprague JE, Peng Y, Fiamengo AL, Woodin KS, Southwick EA, Weisman GR, Wong EH, Golen JA, Rheingold AL, Anderson CJ. J. Med. Chem. 2007;50:2527–2535. doi: 10.1021/jm070204r. [DOI] [PubMed] [Google Scholar]; b Weiss ID, Jacobson O. Theranostics. 2013;3:76–84. doi: 10.7150/thno.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a Hubin TJ, McCormick JM, Collinson SR, Buchalova M, Perkins CM, Alcock NW, Kahol PK, Raghunathan A, Busch DH. J. Am. Chem. Soc. 2000;122:2512–2522. [Google Scholar]; b Collinson SR, Alcock NW, Hubin TJ, Busch DH. J. Coord. Chem. 2001;52:317–331. [Google Scholar]; c Hubin TJ, McCormick JM, Collinson SR, Alcock NW, Clase HJ, Busch DH. Inorg. Chim. Acta. 2003;346:76–86. [Google Scholar]; d Feng Y, England J, Que L., Jr Acs Catalysis. 2011;1:1035–1042. [Google Scholar]
- 4.Hubin TJ, McCormick JM, Alcock NW, Clase HJ, Busch DH. Inorg. Chem. 1999;38:4435–4446. doi: 10.1021/ic990491s. [DOI] [PubMed] [Google Scholar]
- 5.Hubin TJ, Alcock NW, Busch DH. Acta Crystallogr. Sect. C: Cryst. Struct. Commun. 1999;55:1404–1406. [Google Scholar]
- 6.a Khan A, Nicholson G, Greenman J, Madden L, McRobbie G, Pannecouque C, De Clercq E, Ullom R, Maples DL, Maples RD, Silversides JD, Hubin TJ, Archibald SJ. J. Am. Chem. Soc. 2009;131:3416. doi: 10.1021/ja807921k. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Archibald SJ, Smith R. Comprehensive Inorganic Chemistry II. Second. Amsterdam: Elsevier; 2013. pp. 661–682. [Google Scholar]
- 7.a Fulton A. Chemokine Receptors in Cancer. Humana Press; 2009. [Google Scholar]; b Teicher BA, Fricker SP. Clin. Cancer. Res. 2010;16:2927–2931. doi: 10.1158/1078-0432.CCR-09-2329. [DOI] [PubMed] [Google Scholar]
- 8.De Clercq E. Nat. Rev. Drug Discov. 2003;2:581–587. doi: 10.1038/nrd1134. [DOI] [PubMed] [Google Scholar]
- 9.Khan A, Greenman J, Archibald SJ. Curr. Med. Chem. 2007;14:2257–2277. doi: 10.2174/092986707781696618. [DOI] [PubMed] [Google Scholar]
- 10.a Gerlach LO, Skerlj RT, Bridger GJ, Schwartz TW. J. Biol. Chem. 2001;276:14153–14160. doi: 10.1074/jbc.M010429200. [DOI] [PubMed] [Google Scholar]; b Vinader V, Ahmet DS, Ahmed MS, Patterson LH, Afarinkia K. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0078744. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Wong RSY, Bodart V, Metz M, Labrecque J, Bridger G, Fricker SP. Mol. Pharmacol. 2008;74:1485–1495. doi: 10.1124/mol.108.049775. [DOI] [PubMed] [Google Scholar]
- 11.a Valks GC, McRobbie G, Lewis EA, Hubin TJ, Hunter TM, Sadler PJ, Pannecouque C, De Clercq E, Archibald SJ. J. Med. Chem. 2006;49:6162–6165. doi: 10.1021/jm0607810. [DOI] [PubMed] [Google Scholar]; b Soibinet M, Dechamps-Olivier I, Guillon E, Barbier JP, Aplincourt M, Chuburu F, Le Baccon M, Handel H. Eur. J. Inorg. Chem. 2003:1984–1994. [Google Scholar]; c El Ghachtouli S, Cadiou C, Dechamps-Olivier I, Chuburu F, Aplincourt M, Roisnel T, Turcry V, Patinec V, Le Baccon M, Handel H. Eur. J. Inorg. Chem. 2008:4735–4744. [Google Scholar]
- 12.a Hubin TJ, Alcock NW, Clase HJ, Busch DH. Supramol. Chem. 2001;13:261–276. [Google Scholar]; b Hubin TJ, Alcock NW, Clase HJ, Seib LL, Busch DH. Inorg. Chim. Acta. 2002;337:91–102. [Google Scholar]
- 13.Hubin TJ, Alcock NW, Morton MD, Busch DH. Inorg. Chim. Acta. 2003;348:33–40. [Google Scholar]
- 14.Silversides JD, Smith R, Archibald SJ. Dalton Trans. 2011 doi: 10.1039/c0dt01395a. [DOI] [PubMed] [Google Scholar]
- 15.Hubin TJ, Alcock NW, Busch DH. Acta Crystallogr. Sect. C: Cryst. Struct. Commun. 2000;56:37–39. doi: 10.1107/s010827019901255x. [DOI] [PubMed] [Google Scholar]
- 16.Weisman GR, Wong EH, Hill DC, Rogers ME, Reed DP, Calabrese JC. Chem. Commun. 1996:947–948. [Google Scholar]
- 17.Rohovec J, Gyepes R, Cisarova I, Rudovsky J, Lukes I. Tetrahedron Lett. 2000;41:1249–1253. [Google Scholar]
- 18.Hubin TJ, McCormick JM, Collinson SR, Alcock NW, Busch DH. Chem. Commun. 1998:1675–1676. [Google Scholar]
- 19.a Wadas TJ, Wong EH, Weisman GR, Anderson CJ. Curr. Pharm. Des. 2007;13:3–16. doi: 10.2174/138161207779313768. [DOI] [PubMed] [Google Scholar]; b Anderson CJ, Wadas TJ, Wong EH, Weisman GR. Q. J. Nucl. Med. Mol. Imaging. 2008;52:185–192. [PMC free article] [PubMed] [Google Scholar]; c Zeng D, Ouyang Q, Cai Z, Xie X-Q, Anderson CJ. Chem. Commun. 2014;50:43–45. doi: 10.1039/c3cc45928d. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Silversides JD, Burke BP, Archibald SJ. Comptes Rendus Chimie. 2013;16:524–530. [Google Scholar]
- 20.a Woodin KS, Heroux KJ, Boswell CA, Wong EH, Weisman GR, Niu WJ, Tomellini SA, Anderson CJ, Zakharov LN, Rheingold AL. Eur. J. Inorg. Chem. 2005:4829–4833. [Google Scholar]; b Matz DL, Jones DG, Roewe KD, Gorbet M-J, Zhang Z, Chen Z, Prior TJ, Archibald SJ, Yin G, Hubin TJ. Dalton Trans. 2015;44:12210–12224. doi: 10.1039/c5dt00742a. [DOI] [PubMed] [Google Scholar]; c Jones DG, Wilson KR, Cannon-Smith DJ, Shircliff AD, Zhang Z, Chen Z, Prior TJ, Yin G, Hubin TJ. Inorg. Chem. 2015;54:2221–2234. doi: 10.1021/ic502699m. [DOI] [PubMed] [Google Scholar]
- 21.a Bridger GJ, Skerlj RT, Thornton D, Padmanabhan S, Martellucci SA, Henson GW, Abrams MJ, Yamamoto N, Devreese K, Pauwels R, Declercq E. J. Med. Chem. 1995;38:366–378. doi: 10.1021/jm00002a019. [DOI] [PubMed] [Google Scholar]; b Gerlach LO, Jakobsen JS, Jensen KP, Rosenkilde MR, Skerlj RT, Ryde U, Bridger GJ, Schwartz TW. Biochemistry (Mosc.) 2003;42:710–717. doi: 10.1021/bi0264770. [DOI] [PubMed] [Google Scholar]; c Este JA, Cabrera C, De Clercq E, Struyf S, Van Damme J, Bridger G, Skerlj RT, Abrams MJ, Henson G, Gutierrez A, Clotet B, Schols D. Mol. Pharmacol. 1999;55:67–73. doi: 10.1124/mol.55.1.67. [DOI] [PubMed] [Google Scholar]
- 22.Smith R, Huskens D, Daelemans D, Mewis RE, Garcia CD, Cain AN, Freeman TNC, Pannecouque C, De Clercq E, Schols D, Hubin TJ, Archibald SJ. Dalton Trans. 2012;41:11369–11377. doi: 10.1039/c2dt31137b. [DOI] [PubMed] [Google Scholar]
- 23.McRobbie G, Valks GC, Empson CJ, Khan A, Silversides JD, Pannecouque C, De Clercq E, Fiddy SG, Bridgeman AJ, Young NA, Archibald SJ. Dalton Trans. 2007:5008–5018. doi: 10.1039/b705800d. [DOI] [PubMed] [Google Scholar]
- 24.Shannon RD. Acta Crystallogr. Sect. A: Found. Crystallogr. 1976;32:751–767. [Google Scholar]
- 25.Addison AW, Rao TN, Reedijk J, Vanrijn J, Verschoor GC. J. Chem. Soc., Dalton Trans. 1984:1349–1356. [Google Scholar]
- 26.Comba P, Jurisic P, Lampeka YD, Peters A, Prikhod'ko AI, Pritzkow H. Inorg. Chim. Acta. 2001;324:99–107. [Google Scholar]
- 27.Bosnich B, Poon CK, Tobe ML. Inorg. Chem. 1965;4:1102–1108. [Google Scholar]
- 28.Liang XY, Parkinson JA, Weishaupl M, Gould RO, Paisey SJ, Park HS, Hunter TM, Blindauer CA, Parsons S, Sadler PJ. J. Am. Chem. Soc. 2002;124:9105–9112. doi: 10.1021/ja0260723. [DOI] [PubMed] [Google Scholar]
- 29.Unpublished Data - Manuscript revisions submitted [Google Scholar]
- 30.Princen K, Hatse S, Vermeire K, De Clercq E, Schols D. Cytometry Part A. 2003;51A:35–45. doi: 10.1002/cyto.a.10008. [DOI] [PubMed] [Google Scholar]
- 31.a Zhao Y, Truhlar DG. Theor. Chem. Acc. 2008;120:215–241. [Google Scholar]; b Gaussian 09. Wallingford CT, USA: Gaussian, Inc; 2009. [Google Scholar]
- 32.X-AREA v 1.64. Darmstadt, Germany: STOE & Cie GmbH; 2012. [Google Scholar]
- 33.Sheldrick GM. Acta Crystallogr. Sect. A: Found. Crystallogr. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.