Abstract
Bone Morphogenetic Proteins (BMPs) pattern the dorsal-ventral axis of bilaterian embryos; however, their roles in the evolution of body plan are largely unknown. We examined their functional evolution in fly embryos. BMP signaling specifies two extraembryonic tissues, the serosa and amnion, in basal-branching flies such as Megaselia abdita, but only one, the amnioserosa, in Drosophila melanogaster. The BMP signaling dynamics are similar in both species until the beginning of gastrulation, when BMP signaling broadens and intensifies at the edge of the germ rudiment in Megaselia, while remaining static in Drosophila. Here we show that the differences in gradient dynamics and tissue specification result from evolutionary changes in the gene regulatory network that controls the activity of a positive feedback circuit on BMP signaling, involving the tumor necrosis factor alpha homolog eiger. These data illustrate an evolutionary mechanism by which spatiotemporal changes in morphogen gradients can guide tissue complexity.
DOI: http://dx.doi.org/10.7554/eLife.20894.001
Research Organism: D. melanogaster, Other
Introduction
The specification of different cell fates by morphogen gradients has been a longstanding focus within developmental biology. While it is well established that gradients of diffusible morphogens produce complex pattern during development, their role as drivers of morphological evolution has mostly been inferred from theoretical studies, due to the challenge of quantifying and functionally assessing their activities in species outside of select genetic model organisms (Turing, 1952; Kondo and Miura, 2010; Green and Sharpe, 2015; Marcon et al., 2016). Bone Morphogenetic Proteins (BMPs) pattern the embryonic dorsal-ventral axis of bilaterian embryos, raising the question of the role of the BMP gradient in the evolution of body plans (Bier and De Robertis, 2015). To address this question, we compared the functions of embryonic BMP gradients in two fly species that differ in tissue complexity downstream of BMP signaling.
In Drosophila melanogaster embryos, the BMP gradient forms through directed extracellular BMP movement and initiates a positive feedback circuit leading to a bistable pattern of BMP signaling by the end of the blastoderm stage (reviewed in O'Connor et al., 2006; Shilo et al., 2013; Wharton and Serpe, 2013). High levels of BMP signaling, centered on the dorsal midline, specify a single extraembryonic tissue, the amnioserosa. However, in basal-branching flies, including Megaselia abdita (Phoridae), BMP signaling specifies two extraembryonic tissues, the serosa and the amnion (Schmidt-Ott and Kwan, 2016). Previously, we showed that the dynamics of BMP signaling in the blastoderm are similar between Megaselia and Drosophila, but differ in the early gastrula when the Megaselia gradient broadens while the Drosophila gradient remains static (Rafiqi et al., 2012). Here, we show that differences in the control of a positive feedback circuit involving eiger (egr) (Gavin-Smyth et al., 2013) are responsible for the altered dynamics of BMP signaling and amnion specification in Megaselia. We hereby reveal an evolutionary mechanism by which morphogen gradients can alter the complexity of tissue types between species.
Results and discussion
BMP signaling during gastrulation is necessary and sufficient for amnion specification
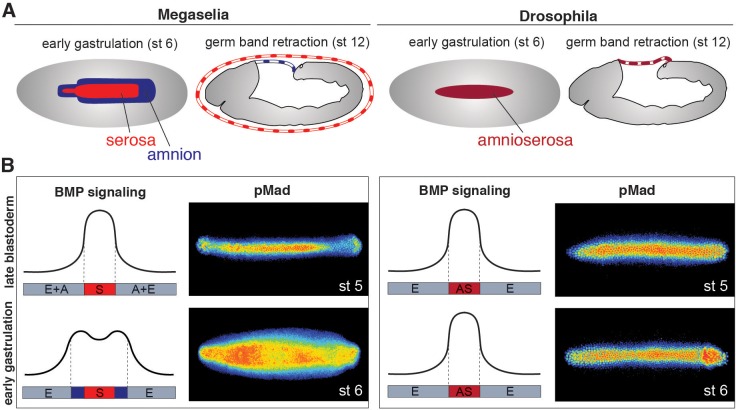
In Megaselia and Drosophila, BMP signaling specifies extraembryonic membranes (Figure 1A) and can be quantified by staining with an antibody specific to the activated phosphorylated form of Mad (pMad), an essential transcriptional effector of the BMP pathway (Dorfman and Shilo, 2001). During early blastoderm stages in both species BMP signaling is initially low and broadly distributed over the dorsal regions of the embryo but refines into a narrow dorsal stripe of high activity by the onset of gastrulation. However, during early gastrulation in Megaselia, the BMP signaling domain broadens to encompass the edge of the germ rudiment comprising the presumptive amnion, while the domain in Drosophila remains static (Figure 1B).
Figure 1. Extraembryonic tissue and BMP signaling differ between Megaselia and Drosophila.
(A) Schematics of Megaselia embryos with serosa and amnion and of Drosophila embryos with amnioserosa at the beginning of gastrulation (stage 6, left) and during early germ band retraction (stage 12, right), modified from Rafiqi et al. (2012). Here, as in all subsequent figures, blastoderm and gastrula stages are shown in dorsal view while later stages are shown in lateral view with the dorsal side up unless specified otherwise. Anterior is always left. (B) Schematic pMad intensity profiles at the dorsal midline relative to prospective serosa (S, red), amnion (A, blue), amnioserosa (AS, maroon), and embryonic tissues (E, grey) in Megaselia and Drosophila. Representative embryos stained for pMad on right.
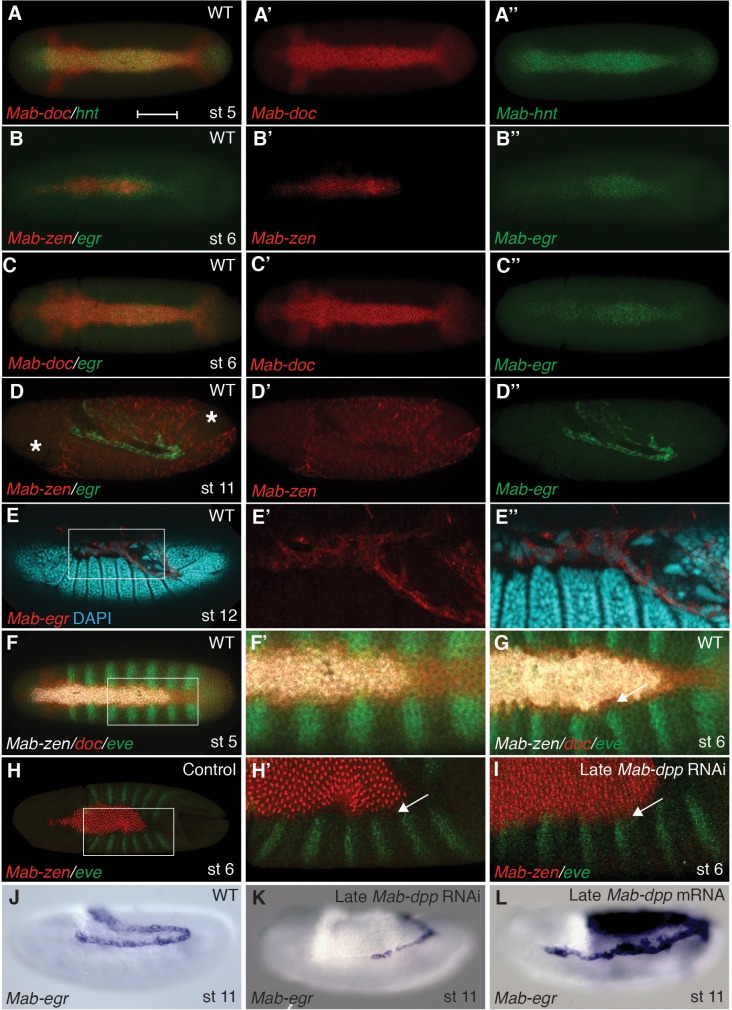
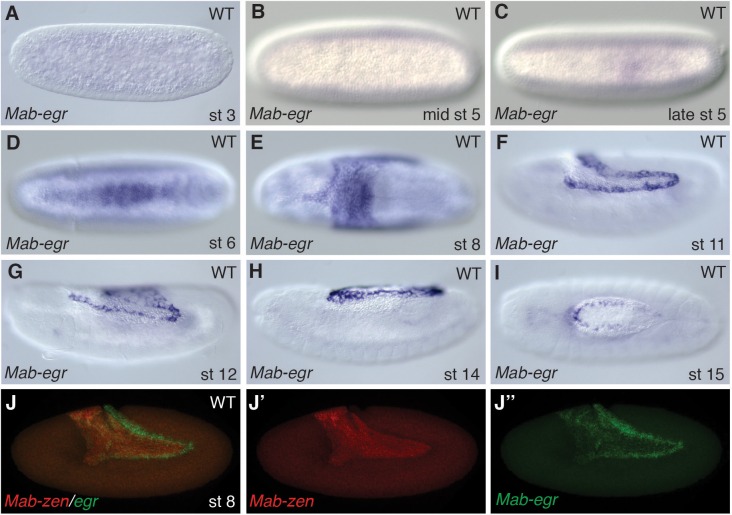
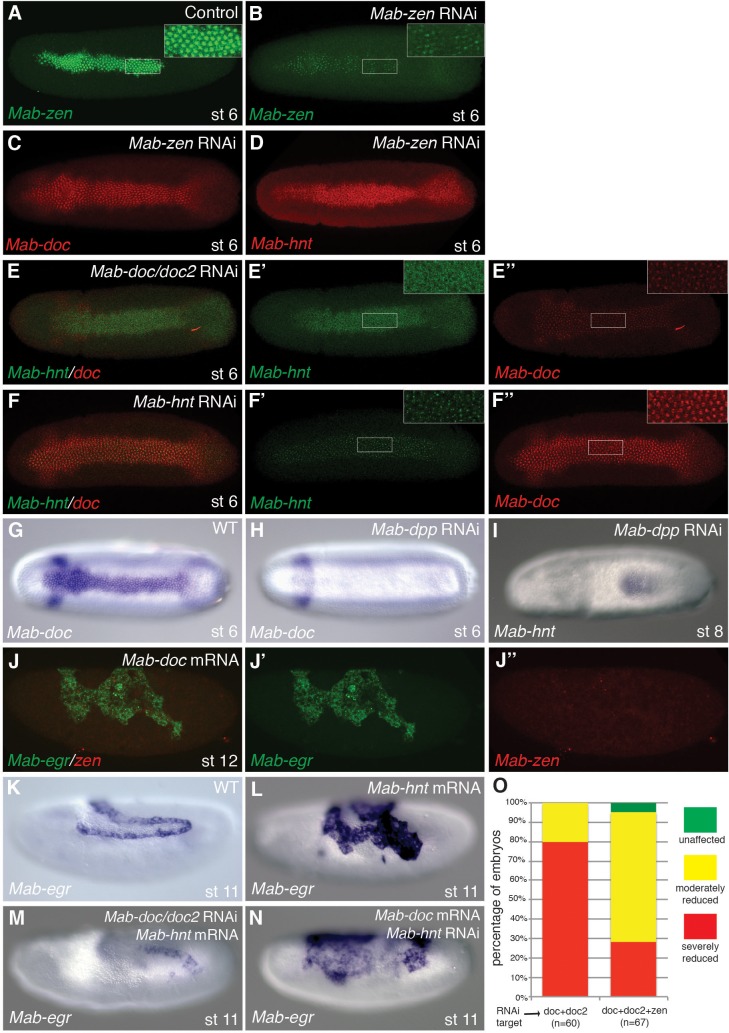
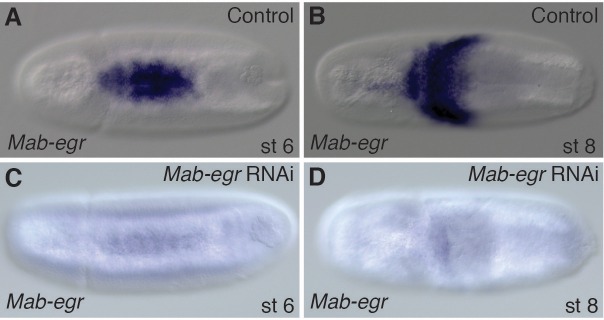
In Megaselia, serosa and amnion specification can be visualized with a combination of genetic markers. A homolog of zerknüllt (Mab-zen), which encodes a homeodomain protein, marks and specifies serosa cells in blastoderm (stage 5) and gastrula embryos (stage 6) (Rafiqi et al., 2008). Homologs of hindsight (Mab-hnt) and dorsocross (Mab-doc, Mab-doc2) (Figure 2A and Figure 2—figure supplement 1), which encode zinc-finger and T-box proteins respectively, are also expressed in stage 5 and 6 embryos in a slightly wider domain than Mab-zen (Figure 2—figure supplement 2) (Rafiqi et al., 2012), encompassing both the prospective serosa and amnion. Lastly, a homolog of the TNF alpha gene eiger (Mab-egr) is expressed in the serosa and amnion of gastrulating Megaselia embryos (Figure 2B–C). Mab-egr expression continues until dorsal closure, but from germ band extension (stage 11) onwards it is expressed only in the amnion cells, which at this stage are polyploid and much larger than the adjacent embryonic cells (Figure 2D–E and Figure 2—figure supplement 3). The time course of serosa and amnion specification is suggested by the dorsal repression of an embryonic marker, Megaselia even-skipped (Mab-eve) (Rafiqi et al., 2012). In wild-type blastoderm embryos, the repression of Mab-eve extends laterally from the dorsal midline to the boundary of the Mab-zen domain, but after gastrulation begins Mab-eve expression withdraws further to abut the Mab-doc/hnt domain (Figure 2F–G and Figure 2—figure supplement 2), likely as a result of repression by BMP signaling. Conversely, overexpression of Mab-eve suppresses Mab-zen expression (Figure 2—figure supplement 4). Thus, amnion specification might not be completed before the onset of gastrulation. At stage 6, the amnion anlage of Megaselia is defined as the thin band of cells expressing Mab-doc/doc2, Mab-hnt, and Mab-egr but not Mab-zen while, after germ band extension, mature amnion cells are defined by their large size and Mab-egr expression.
Figure 2. Specification of amnion by BMP signaling in Megaselia.
(A) Mab-hnt and Mab-doc expression at the late blastoderm stage. Scale bar = 100 µm. (B, C) Mab-egr and Mab-zen (B) and Mab-egr and Mab-doc (C) expression at early gastrulation. (D) Mab-egr and Mab-zen expression after germ band extension. Asterisks denote tears in the serosa during sample preparation. (E) Mab-egr expression during germ band retraction. The serosa has been removed and nuclei have been labeled with DAPI. Boxed region enlarged (E’–E’’). (F, G) Mab-doc, Mab-zen and Mab-eve expression at late blastoderm stage (stage 5) (F, enlargement F’) and early gastrulation (stage 6) (G) with arrow pointing to abutting Mab-eve and Mab-doc expression domains. (H, I) Mab-zen and Mab-eve expression in early gastrula control embryo (H, enlargement H’) and following Mab-dpp knockdown after 50% blastoderm cellularization (I). Arrows, gap between the Mab-eve and Mab-zen domains (H’) that is suppressed in the knockdown embryo (I). (J–L) Mab-egr expression at germ band extension in wild-type embryo (J), and after Mab-dpp knockdown (K) or Mab-dpp overexpression (L) after 50% blastoderm cellularization.
Figure 2—figure supplement 1. Expression of Mab-doc2.
Figure 2—figure supplement 2. Time course of Mab-zen, Mab-doc and Mab-eve expression.
Figure 2—figure supplement 3. .Expression of Mab-egr.
Figure 2—figure supplement 4. Overexpression of Mab-eve represses Mab-zen expression.
To test the hypothesis that temporally distinct BMP signaling sequentially specifies the two extraembryonic membranes, we decreased BMP signaling during gastrulation by Mab-dpp knockdown after 50% blastoderm cellularization and monitored the expression of Mab-eve. Knockdown of Mab-dpp by injection of double stranded RNA (dsRNA) at the early blastoderm stage completely suppresses both serosa and amnion specification and results in the circumferential expression of Mab-eve (Rafiqi et al., 2012). In contrast, in all 13 late Mab-dpp knockdown embryos fixed during early gastrulation, the expression of Mab-zen was not affected; however, in five embryos the repression of Mab-eve in the amnion anlage was incomplete (Figure 2H–I). Late Mab-dpp knockdown reduced Mab-egr expression in a majority of stage 11/12 embryos (55%, n = 40) whereas late ectopic Mab-dpp expression caused an expansion of the Mab-egr domain in at least 35% of the embryos (n = 57) (Figure 2J–L). Taken together, these data provide evidence that BMP signaling during gastrulation is necessary and sufficient for amnion specification.
Mab-doc promotes amnion development by elevating BMP signaling in the amnion anlage at the onset of gastrulation
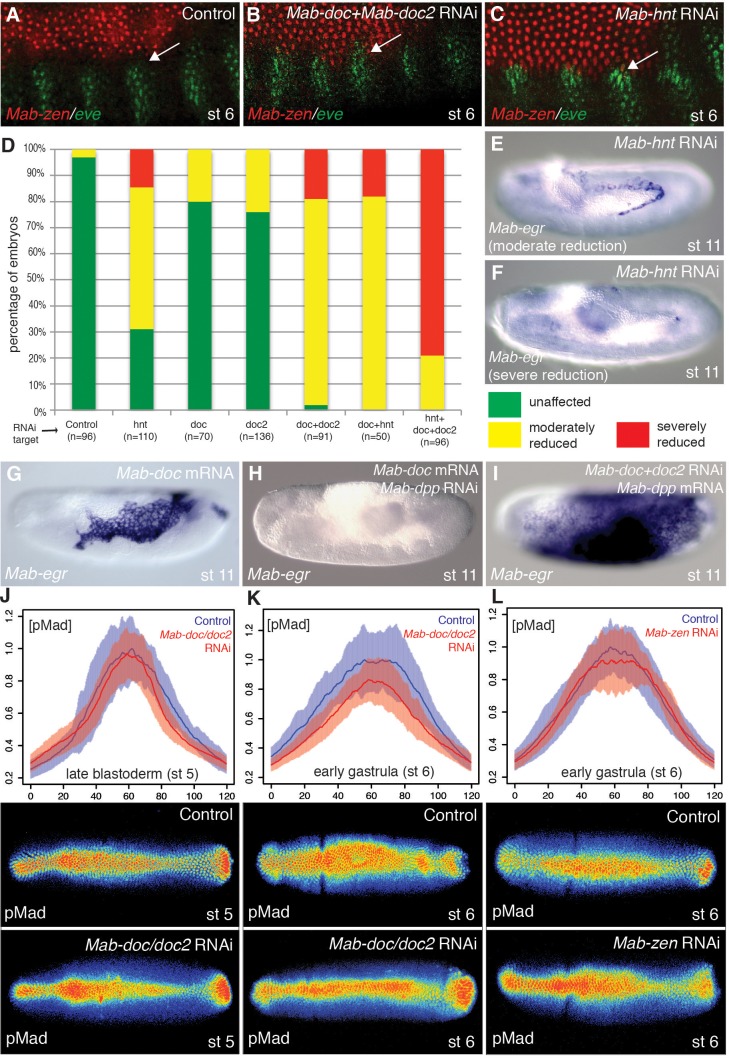
In Drosophila, both BMP signaling and zen are necessary at the blastoderm stage for the expression of the three doc paralogs and hnt in the amnioserosa anlage, although the essential function of these genes in amnioserosa maintenance becomes apparent only after gastrulation (Yip et al., 1997; Reim et al., 2003). However, in Megaselia, BMP signaling activates Mab-doc and Mab-hnt independently from Mab-zen (Figure 3—figure supplement 1A–I), suggesting these genes could play a role in amnion specification. Following knockdown of Mab-doc/doc2 or Mab-hnt activity by RNAi, we observed confluent expression domains of Mab-zen and Mab-eve during early gastrulation (4/9 and 5/11 embryos, respectively; Figure 3A–C), and a decrease in Mab-egr expression at stage 11/12 (Figure 3D–F). Knockdown of the activities of all three genes eliminated Mab-egr expression in germ-band extended embryos (Figure 3D), indicating that Mab-doc/doc2 and Mab-hnt together are essential for amnion specification in Megaselia. Conversely, overexpression of Mab-doc or Mab-hnt induced ectopic amnion, as evidenced by an enlargement of the Mab-egr domain at stage 11/12 (Figure 3G and Figure 3—figure supplement 1J–L). Overexpression of Mab-doc could bypass the requirement for Mab-hnt, while overexpression of Mab-hnt could not bypass the requirement for Mab-doc (Figure 3—figure supplement 1M–N), consistent with the hypothesis that Mab-doc and Mab-hnt share a common target necessary for amnion formation that is primarily dependent upon doc activity.
Figure 3. Mab-doc and Mab-hnt elevate BMP signaling to specify amnion in Megaselia.
(A–C) Mab-zen and Mab-eve expression in early gastrula control embryo (A) and after Mab-doc/doc2 knockdown (B) or Mab-hnt knockdown (C). Arrows, gap between the Mab-eve and Mab-zen domains (A) that is suppressed in the knockdown embryos (B, C). (D–F) Bar chart (D) quantifying the reduction of Mab-egr expression at stage 11/12 after Mab-hnt and/or Mab-doc/doc2 knockdown, and representative embryos of moderately reduced (yellow, E), or severely reduced (red, F) phenotypes. (G–I) Mab-egr expression at germ band extension following Mab-doc overexpression (G), Mab-doc overexpression and Mab-dpp knockdown (H), and Mab-dpp overexpression and Mab-doc/doc2 knockdown (I). (J–L) Mean and shaded standard deviation of pMad intensities in control injected embryos (blue) and in Mab-doc/doc2 knockdown embryos (red) at the cellular blastoderm stage (n = 10, control n = 10) (J), at early gastrulation (n = 11, control n = 11) (K), and in Mab-zen knockdown embryos (red) at early gastrulation (n = 10, control n = 17) (L) with representative embryos stained for pMad underneath each plot.
Figure 3—figure supplement 1. Function and regulation of Mab-doc and Mab-hnt.
The activities of Mab-doc and Mab-hnt could promote the amnion formation in an instructive manner, by activating the amnion gene network of Megaselia, or they might promote the amnion formation in a permissive manner, e.g., by elevating BMP signaling. Overexpression of Mab-doc in Mab-dpp knockdown embryos did not induce any expression of Mab-egr in stage 11/12 embryos (n = 44) (Figure 3H). Conversely, overexpression of Mab-dpp in Mab-doc/doc2 knockdown embryos resulted in ectopic expression of Mab-egr at stage 11/12 (36%, n = 47) (Figure 3I). Thus, BMP signaling is sufficient to direct the expression of amnion specific genes in the absence of Mab-doc/doc2 activity. To confirm that this result was not due to an excessive non-physiological level of Mab-Dpp produced by the injected mRNA, we asked whether the endogenous level of BMP signaling at the dorsal midline in the blastoderm embryo would be sufficient to specify amnion in the absence of both Mab-doc/doc2 and the serosal determinant Mab-zen. Knockdown of Mab-zen partially restored amnion in Mab-doc/doc2 knockdown embryos (Figure 3—figure supplement 1O). Taken together, these data suggest Mab-doc promotes amnion formation permissively.
To directly test whether Mab-doc activity elevates BMP signaling, we quantified the intensity of pMad staining in embryos after Mab-doc/doc2 knockdown. While Mab-doc/doc2 knockdown had little effect on pMad levels during the late blastoderm stage compared to control embryos (Wilcoxon rank sum test, p=0.3697; Figure 3J), in early gastrula stage embryos, knockdown of Mab-doc/doc2 resulted in significantly reduced pMad levels compared to control embryos (Wilcoxon rank sum test, p=0.01165; Figure 3K). In contrast, knockdown of Mab-zen did not alter the average level of pMad at the beginning of gastrulation (Wilcoxon rank sum test, p=0.2367; Figure 3L). The observation that Mab-doc/doc2 is dispensable for amnion cell fate specification but necessary for wild-type levels of BMP signaling at the early gastrula stage strongly support the model that amnion formation is driven by a Mab-doc-dependent elevation of BMP signaling in the amnion anlage at the onset of gastrulation.
Mab-doc-dependent control of Mab-egr expression contributes to a positive feedback circuit that promotes BMP signaling during gastrulation
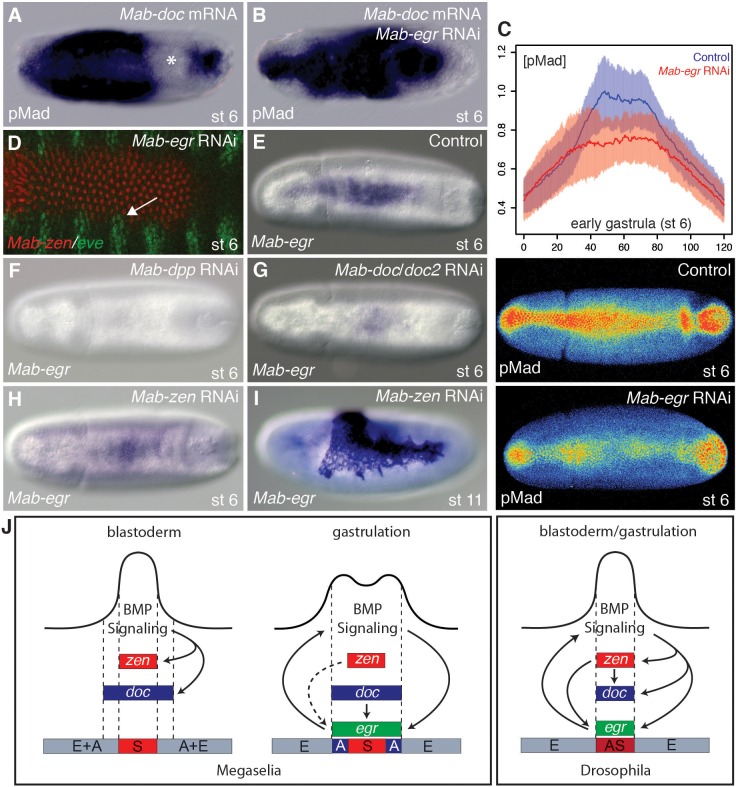
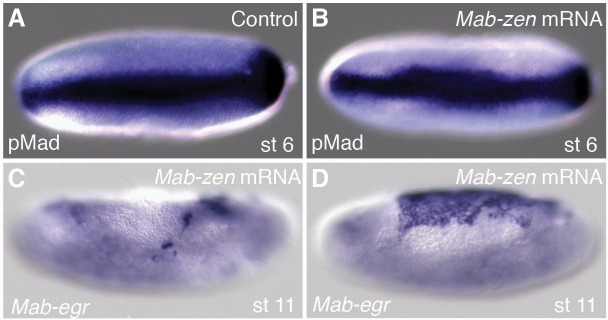
We then explored the mechanism by which Mab-doc promotes BMP signaling at the gastrula stage. Embryos injected centrally with Mab-doc mRNA displayed a local expansion of the pMad domain during gastrulation (15/15) coupled with the frequent depletion of endogenous pMad in adjacent regions (12/15) (Figure 4A). This result parallels a phenotype observed in Drosophila where injection of mRNA encoding activated BMP receptors into the blastoderm embryo causes an increase in BMP ligand-receptor interactions, coupled with a decrease in BMP ligand-receptor binding in nearby regions (Wang and Ferguson, 2005). These data indicate that a positive feedback circuit downstream of BMP signaling increases local receptor-ligand interactions, and that, due to a limiting amount of BMP ligand, ligand-receptor interactions decrease in nearby regions. Conversely, Megaselia embryos injected with Mab-zen mRNA (n = 11) had a similar pMad domain to injected control embryos (n = 12) and developed a reduced or abnormal amnion (44/51) (Figure 4—figure supplement 1). These results suggest Mab-doc, but not Mab-zen, locally activates a positive feedback circuit in the Megaselia embryo, where BMP ligands are limiting.
Figure 4. Mab-egr is downstream of Mab-doc and promotes BMP signaling.
(A, B) pMad staining following Mab-doc overexpression (A) or Mab-doc overexpression and Mab-egr knockdown (B). The asterisk marks site of endogenous pMad depletion. (C) Mean intensity and standard deviation of pMad staining in control injected embryos (blue, n = 10) and Mab-egr knockdown embryos (red, n = 9) at early gastrulation with representative embryos stained for pMad underneath the plot. (D) Mab-zen and Mab-eve expression at early gastrulation after Mab-egr knockdown with arrow indicating suppressed gap between the Mab-eve and Mab-zen domains. (E–H) Mab-egr expression in wild type (E), Mab-dpp knockdown (F), Mab-doc/doc2 knockdown (G), and Mab-zen knockdown (H) embryos at early gastrulation. (I) Mab-egr expression in a Mab-zen knockdown embryo after germ band extension. (J) BMP gene regulatory networks in Megaselia abdita and Drosophila melanogaster.
Figure 4—figure supplement 1. Effect of Mab-zen overexpression on BMP signaling.
Figure 4—figure supplement 2. Efficiency of Mab-egr RNAi.
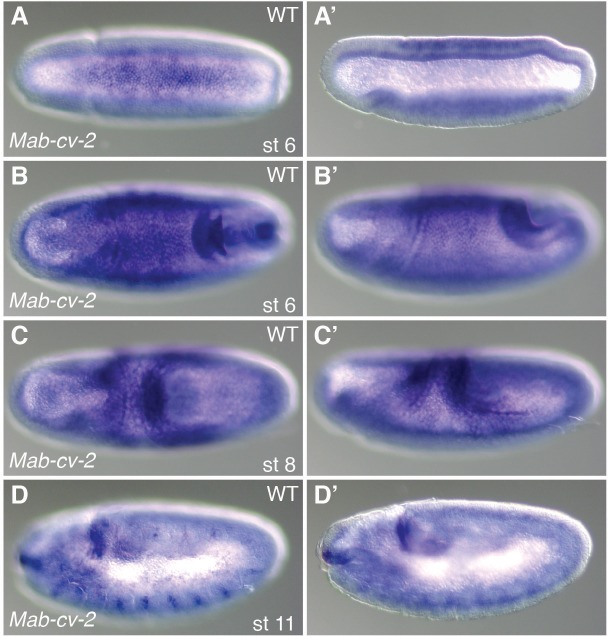
Figure 4—figure supplement 3. Expression profile of Mab-cv-2.
Figure 4—figure supplement 4. Mab-cv-2 promotes amnion specification.
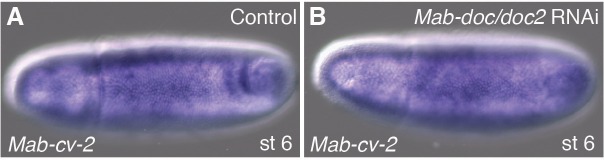
Figure 4—figure supplement 5. Regulation of Mab-cv-2 is largely independent of Mab-doc/doc2.
Recent experiments in Drosophila identified egr activity as a component of the positive feedback circuit (Gavin-Smyth et al., 2013). To determine whether Mab-egr could be a component of a positive feedback circuit in Megaselia, we asked whether knockdown of Mab-egr could modify the phenotype caused by injection of Mab-doc mRNA. While the pMad domain in all Mab-doc injected embryos was locally expanded (14/14) (Figure 4B), only a few embryos (2/14) displayed a depletion of endogenous pMad in adjacent regions. These data indicate that Mab-egr increases the ability of cells overexpressing Mab-doc to compete for BMP ligands during early gastrulation.
In Drosophila, loss of egr reduces intensity of pMad staining by 50% (Gavin-Smyth et al., 2013). Similarly, we found that, at the onset of gastrulation, pMad levels in Mab-egr knockdown embryos were reduced by about 50% on average (Wilcoxon rank sum test, p=0.00381) (Figure 4C and Figure 4—figure supplement 2). Moreover, confluent expression domains of Mab-eve and Mab-zen could be observed in such embryos (3/10) (Figure 4D). As Mab-egr expression extends to the edge of the gastrulating germ rudiment, these observations suggest that Mab-egr promotes amnion specification downstream of Mab-doc/doc2 by elevating BMP signaling during gastrulation in prospective amnion cells.
The pMad gradients of Mab-egr RNAi embryos were on average slightly broader than in Mab-doc/doc2 RNAi embryos (Figures 3K and 4C), suggesting that Mab-doc/doc2 might control more than one gene with a role in shaping the BMP gradient, similar to Drosophila where the BMP-dependent feedback circuit appears to involve more genes than just egr (Gavin-Smyth et al., 2013). As a potential second Mab-doc/doc2 target we examined the Megaselia ortholog of cv-2 (Figure 4—figure supplement 3), which encodes an extracellular, context- and concentration-dependent modulator of BMP signaling (Conley et al., 2000; Serpe et al., 2008). However, although Mab-cv-2 likely also promotes BMP signaling in Megaselia (Figure 4—figure supplement 4), it appears to function independently of the Mab-doc/doc2-dependent feedback loop (Figure 4—figure supplement 5).
Lastly, we examined the regulation of Mab-egr expression. In Drosophila, egr expression begins at the syncytial blastoderm stage under the control of both BMP signaling and zen, whereas in Megaselia, Mab-egr expression begins at the onset of gastrulation. In Mab-dpp knockdown embryos, Mab-egr expression was completely absent (Figure 4E–F). In Mab-doc/doc2 knockdown embryos, Mab-egr expression was greatly reduced (Figure 4G). Mab-doc/doc2/hnt triple knockdown did not further reduce Mab-egr expression during gastrulation (not shown). Mab-zen knockdown embryos displayed only a slight reduction in Mab-egr expression during gastrulation; however, at germ band extension, Mab-zen knockdown embryos displayed an increase in the number of Mab-egr expressing cells due to the transformation of serosa into amnion (Figure 4H–I). Thus, in Megaselia, Mab-egr is primarily under the control of Mab-doc, not Mab-zen.
Conclusions
While previous work has demonstrated that BMP gradients can form and be stabilized through molecular feedback circuits (Bier and De Robertis, 2015), we have shown here that positive feedback circuits can also be an important target in the evolution of body plans. Specifically, the distinct BMP gradients of Megaselia and Drosophila, which result in the specification of distinct extraembryonic tissue complements, are the result of spatiotemporal changes in an egr-dependent positive feedback circuit (Figure 4J).
Given that Megaselia has retained distinct serosa and amnion tissues, the function of the BMP gradient in this species might be similar to the ancestral condition in higher flies. What changes to the underlying genetic network during evolution would have been necessary to transform the shape of the Megaselia BMP gradient into that seen in Drosophila? In blastoderm embryos of Megaselia, the BMP gradient establishes the expression of Mab-zen in prospective serosa tissue, and Mab-doc/doc2 and Mab-hnt in prospective serosa and amnion tissues. While this patterning phase is not sufficient to differentiate between serosa and amnion tissue, it sets the stage for Mab-doc/doc2-dependent Mab-egr expression during gastrulation. The Drosophila gene network that regulates egr expression differs at least in two ways. First, doc (along with hnt) is expressed downstream of zen. Conceptually, this regulatory difference is sufficient to explain the difference of egr expression between the two species during gastrulation, and hence also the difference in BMP signaling and tissue specification. We therefore propose that this change led the evolutionary transition. Once doc was downstream of zen, the latter might have gradually gained control over egr. This scenario is consistent with our observation that even in Megaselia, Mab-zen slightly promotes Mab-egr expression. Second, Drosophila acquired a BMP-independent zen expression domain in the syncytial blastoderm, which is not observed in other dipterans (Goltsev et al., 2007; Rafiqi et al., 2008). The acquisition of this early zen domain could have promoted egr expression in the blastoderm of Drosophila, where egr is part of a zen-dependent network that confers robustness to the BMP gradient (Gavin-Smyth et al., 2013).
Our data suggest that the ancestral function of the positive feedback circuit was to promote amnion specification through BMP signaling. While the identity of regulatory factors of the positive feedback circuit may be evolutionarily labile (in Tribolium doc and hnt appear to be dispensable for amnion specification [Horn and Panfilio, 2016]), we propose that the mechanism of amnion specification through feedback-driven BMP signaling dynamics applies to a wide range of insects, because in Tribolium the pMad domain also gradually shifts to become elevated in the presumptive amnion during early gastrula stages (van der Zee et al., 2006; Nunes da Fonseca et al., 2008). The principle of evolving tissue complexity through changes in positive feedback circuits of morphogen gradients has not yet been documented in other developmental contexts, but it may also apply to unrelated traits, such as eyespots on butterfly wings (Monteiro, 2015).
Materials and methods
dsRNA and mRNA synthesis, injections and fixation
RNAi was performed as described (Rafiqi et al., 2008, 2010). In negative controls, embryos were injected with dsRNA against the huckebein homolog from another fly species, Eba-hkb (Lemke et al., 2010). dsRNAs for Eba-hkb, Mab-hnt, Mab-zen, and Mab-dpp were prepared as described (Rafiqi et al., 2008; Lemke et al., 2010; Rafiqi et al., 2010). The following primers were used to synthesize dsRNA against Mab-doc: (5’-CCAAGCCTTCTAATACGACTCACTATAGGGAGAGACGAGGATGGCGAGTACTG-3’ and 5’-CAGAGATGCATAATACGACTCACTATAGGGAGAGTTCCCACCAATGGTTGTGC-3’), Mab-doc2: (5’-CCAAGCCTTCTAATACGACTCACTATAGGGAGATGAGTGGTGTGGATATCGCG-3’ and 5’-CAGAGATGCATAATACGACTCACTATAGGGAGAAGCAAGGACAGTGTGACCAT-3’), Mab-cv-2: (5’-CAGAGATGCATAATACGACTCACTATAGGGAGAACGGCGCAAATCCGACTGTTGT-3’ and 5’-CCAAGCCTTCTAATACGACTCACTATAGGGAGAAACGCAGAGTGGAGCCGCTT-3’), and Mab-egr: (5’-CGCCGCGGTCTACATCACTGTT-3’ and 5’-CGCCGCGGTCTACATCACTGTT-3’). T7 promoters are underlined. To create the template for capped Mab-doc, Mab-hnt, Mab-dpp, and Mab-eve mRNAs, complete ORFs were PCR-amplified from embryonic cDNA using primers with attB recombination sites attached at the 5’ ends. The following primers were used: Mab-hnt (5’-GGGGACAAGTTTGTACAAAAAAGCAGGCTACCATGCTTCATGCAACCAACC-3’ and 5’-GGGGACCACTTTGTACAAGAAAGCTGGGTCTACTTCTCAACACCCAAGAACTTG-3’), Mab-doc (5’-GGGGACAAGTTTGTACAAAAAAGCAGGCTAAAATGATTACCATGAATGAATTAGTG-3’ and 5’-GGGGACCACTTTGTACAAGAAAGCTGGGTCTAACATTGCGCAACACCCAAAA-3’), Mab-dpp (5’-GGGGACAAGTTTGTACAAAAAAGCAGGCTAAAATGCGCGCATGGCTT-3’ and 5’-GGGGACCACTTTGTACAAGAAAGCTGGGTTCATCGACATCCACATCCAAC-3’), and Mab-eve (5’-GGGGACAAGTTTGTACAAAAAAGCAGGCTAAAATGCAAGGATACAGAAACTACA-3’ and 5’-GGGGACCACTTTGTACAAGAAAGCTGGGTTTAGGCCTCACTCTCTGTCTT-3’). The attB recombination sites are underlined. The ORFs were cloned into a destination vector which was modified from the pSP35T (Amaya et al., 1991) using Gateway Cloning (Life Technology). Capped mRNAs were prepared using SP6 polymerase with the mMessage mMachine Kit (Ambion). Sequence information of Mab-doc2 (KY302676), Mab-egr (KY302677), and Mab-cv-2 (KY302678) is provided in Genbank. For microinjection, embryos were collected and aligned on a glass slide along a 0.2 mm glass capillary, briefly desiccated, and covered with halocarbon oil (Sigma H773) at room temperature as described (Rafiqi et al., 2011). Stages of the embryos were defined according to (Wotton et al., 2014). Embryos were injected before the syncytial blastoderm stage (~1:30–2:30 hr at 18°C after egg deposition) unless otherwise specified. Injected embryos were then heat fixed and manually devitellinized as described (Rafiqi et al., 2011) before in situ hybridization and immunostaining.
RNA in situ hybridization, immunohistochemistry and image analysis
RNA probes were labeled with digoxigenin (Mab-egr, Mab-dpp), fluorescein (Mab-doc, Mab-doc2, Mab-zen and Mab-hnt) and biotin (Mab-eve) as described (Tautz and Pfeifle, 1989; Kosman, 2004). Probe templates for Mab-eve, Mab-zen, and Mab-hnt were synthesized as described (Bullock et al., 2004; Rafiqi et al., 2008, 2010). Other probes were synthesized from PCR templates obtained from an embryonic cDNA library using the following primers: Mab-egr5’ (5’-CCAAGCCTTCTAATACGACTCACTATAGGGAGATGAGCTGCTGCCAGAGCGTT-3’ and 5’-CAGAGATGCAATTAACCCTCACTAAAGGGAGATGTGCATTTTGTGATTATTGAAAGT-3’), Mab-egr3’ (5’-CCAAGCCTTCTAATACGACTCACTATAGGGAGAAACTATGAGACAAATACTTAACGGA-3’ and 5’-CAGAGATGCAATTAACCCTCACTAAAGGGAGATCGAGCGATTGACGTCTCAGT-3’), Mab-doc (5’-CCAAGCCTTCTAATACGACTCACTATAGGGAGAGACGAGGATGGCGAGTACTG-3’ and 5’-CAGAGATGCAATTAACCCTCACTAAAGGGAGAGTTCCCACCAATGGTTGTGC-3’), Mab-doc2 (5’-CCAAGCCTTCATTAACCCTCACTAAAGGGAGATGAGTGGTGTGGATATCGCG-3’ and 5’-CCAAGCCTTCTAATACGACTCACTATAGGGAGATGAGTGGTGTGGATATCGCG-3’), Mab-cv-2 (5’-CAGAGATGCAATTAACCCTCACTAAAGGGAGAACGGCGCAAATCCGACTGTTGT-3’ and 5’-CCAAGCCTTCTAATACGACTCACTATAGGGAGAAACGCAGAGTGGAGCCGCTT-3’) and the synthesis of other probes were previously described (Rafiqi et al., 2008, 2010). T3 and T7 promoters are underlined in the forward and reverse primers, respectively. The following procedures for RNA in situ hybridization and immunostaining were done as described (Rafiqi et al., 2012). For RNA in situ hybridization, Megaselia embryos were heat fixed, while for immunostaining, they were fixed by formaldehyde except for quantification (see below). pMad was detected with a rabbit monoclonal antibody against Smad3 phosphorylated on Serine 423 and Serine 425 (Epitomics, Cat# 1880–1) at 1:250 dilution. For two-color fluorescent in situ hybridization, confocal scans were done with a Zeiss LSM510 laser-scanning microscope. All subsequent image quantification and analysis of confocal micrographs were done in ImageJ (Schneider et al., 2012). To quantify pMad staining intensity, embryos were stained with pMad as described (Rafiqi et al., 2012) after a modified fixation protocol. To preserve better morphology for quantification, heat fix was used instead of formaldehyde. The embryos were treated with a boiling solution of 0.7% NaCl and 0.05% Triton X-100 followed by a heptane and methanol devitellinization step. Postfixation was done with 5% formaldehyde in a 3:1 mixture of phosphate buffered saline (PBT; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, 0.1% Triton X-100 pH 7.4) and methanol. This was followed by a second heptane and methanol devitellinization step. Embryos at early gastrulation were staged after the initiation of cephalic furrow and before the dorsal-most point of the proctodeum reached 20% of total egg length. The quantification of pMad staining in injected Megaselia embryos followed the Drosophila protocol (Gavin-Smyth et al., 2013). To compare whether there was a significant reduction of pMad intensity in Mab-egr RNAi compared to wild type, a Wilcoxon rank sum test was performed in R [R Core Team (2012). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org].
Gene trees
Amino acid sequences of Dorsocross homologs in different species with the following reference numbers were retrieved (Aedes aegypti; Aae-docA XP_001648597.1 and Aae-docB XP_001663692.1), (Anopheles gambiae; Aga-docA XP_315924.3 and Aga-docB EAA11871.5), (Drosophila melanogaster; Doc1 AAF50328.2, Doc2 AAF50329.1 and Doc3 AAF50331.1), (Drosophila pseudoobscura; Dps-Doc1 EAL31211.1, Dps-Doc2 EAL31212.2, and Dps-Doc3 EAL31210.1), (Drosophila grimshawi; Dgr-Doc1 EDV96918.1, Dgr-Doc2 EDV96917.1 and Dgr-Doc3 EDV96915.1). Full-length protein alignments were created using the MUSCLE program with default parameters (Edgar, 2004). The best amino acid substitution model was estimated using AIC in ProtTest 3 (Darriba et al., 2011) and the LG model was chosen. Maximum likelihood trees were calculated using PhyML 3 (Criscuolo, 2011). Bootstrap values were based on 1000 replicas.
Acknowledgements
We thank Jeff Klomp (University of Chicago) and Steffen Lemke (University of Heidelberg) for technical advice and discussions, and Sally Horne-Badovinac (University of Chicago) and Robert Ho (University of Chicago) for comments on the manuscript. This work was supported by the National Science Foundation Grant IOS-1121211 to US-O and an award of the University of Chicago Hinds Fund to CWK.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Funding Information
This paper was supported by the following grants:
National Science Foundation IOS-1121211 to Urs Schmidt-Ott.
University of Chicago Hinds Fund graduate student award to Chun Wai Kwan.
Additional information
Competing interests
The authors declare that no competing interests exist.
Author contributions
CWK, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article.
JG-S, Analysis and interpretation of data, Drafting or revising the article.
ELF, Analysis and interpretation of data, Drafting or revising the article.
US-O, Conception and design, Analysis and interpretation of data, Drafting or revising the article.
References
- Amaya E, Musci TJ, Kirschner MW. Expression of a dominant negative mutant of the FGF receptor disrupts mesoderm formation in Xenopus embryos. Cell. 1991;66:257–270. doi: 10.1016/0092-8674(91)90616-7. [DOI] [PubMed] [Google Scholar]
- Bier E, De Robertis EM. Embryo development. BMP gradients: A paradigm for morphogen-mediated developmental patterning. Science. 2015;348:aaa5838. doi: 10.1126/science.aaa5838. [DOI] [PubMed] [Google Scholar]
- Bullock SL, Stauber M, Prell A, Hughes JR, Ish-Horowicz D, Schmidt-Ott U. Differential cytoplasmic mRNA localisation adjusts pair-rule transcription factor activity to cytoarchitecture in dipteran evolution. Development. 2004;131:4251–4261. doi: 10.1242/dev.01289. [DOI] [PubMed] [Google Scholar]
- Conley CA, Silburn R, Singer MA, Ralston A, Rohwer-Nutter D, Olson DJ, Gelbart W, Blair SS. Crossveinless 2 contains cysteine-rich domains and is required for high levels of BMP-like activity during the formation of the cross veins in Drosophila. Development. 2000;127:3947–3959. doi: 10.1242/dev.127.18.3947. [DOI] [PubMed] [Google Scholar]
- Criscuolo A. morePhyML: improving the phylogenetic tree space exploration with PhyML 3. Molecular Phylogenetics and Evolution. 2011;61:944–948. doi: 10.1016/j.ympev.2011.08.029. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman R, Shilo BZ. Biphasic activation of the BMP pathway patterns the Drosophila embryonic dorsal region. Development. 2001;128:965–972. doi: 10.1242/dev.128.6.965. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin-Smyth J, Wang YC, Butler I, Ferguson EL. A genetic network conferring canalization to a bistable patterning system in Drosophila. Current Biology. 2013;23:2296–2302. doi: 10.1016/j.cub.2013.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goltsev Y, Fuse N, Frasch M, Zinzen RP, Lanzaro G, Levine M. Evolution of the dorsal-ventral patterning network in the mosquito, Anopheles gambiae. Development. 2007;134:2415–2424. doi: 10.1242/dev.02863. [DOI] [PubMed] [Google Scholar]
- Green JB, Sharpe J. Positional information and reaction-diffusion: two big ideas in developmental biology combine. Development. 2015;142:1203–1211. doi: 10.1242/dev.114991. [DOI] [PubMed] [Google Scholar]
- Horn T, Panfilio KA. Novel functions for Dorsocross in epithelial morphogenesis in the beetle Tribolium castaneum. Development. 2016;143:3002–3011. doi: 10.1242/dev.133280. [DOI] [PubMed] [Google Scholar]
- Kondo S, Miura T. Reaction-diffusion model as a framework for understanding biological pattern formation. Science. 2010;329:1616–1620. doi: 10.1126/science.1179047. [DOI] [PubMed] [Google Scholar]
- Kosman D, Mizutani CM, Lemons D, Cox WG, McGinnis W, Bier E. Multiplex detection of RNA expression in Drosophila embryos. Science. 2004;305:846. doi: 10.1126/science.1099247. [DOI] [PubMed] [Google Scholar]
- Lemke S, Busch SE, Antonopoulos DA, Meyer F, Domanus MH, Schmidt-Ott U. Maternal activation of gap genes in the hover fly Episyrphus. Development. 2010;137:1709–1719. doi: 10.1242/dev.046649. [DOI] [PubMed] [Google Scholar]
- Marcon L, Diego X, Sharpe J, Müller P. High-throughput mathematical analysis identifies turing networks for patterning with equally diffusing signals. eLife. 2016;5:e14022. doi: 10.7554/eLife.14022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro A. Origin, development, and evolution of butterfly eyespots. Annual Review of Entomology. 2015;60:253–271. doi: 10.1146/annurev-ento-010814-020942. [DOI] [PubMed] [Google Scholar]
- Nunes da Fonseca R, von Levetzow C, Kalscheuer P, Basal A, van der Zee M, Roth S. Self-regulatory circuits in dorsoventral axis formation of the short-germ beetle Tribolium castaneum. Developmental Cell. 2008;14:605–615. doi: 10.1016/j.devcel.2008.02.011. [DOI] [PubMed] [Google Scholar]
- O'Connor MB, Umulis D, Othmer HG, Blair SS. Shaping BMP morphogen gradients in the Drosophila embryo and pupal wing. Development. 2006;133:183–193. doi: 10.1242/dev.02214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiqi AM, Lemke S, Ferguson S, Stauber M, Schmidt-Ott U. Evolutionary origin of the amnioserosa in cyclorrhaphan flies correlates with spatial and temporal expression changes of zen. PNAS. 2008;105:234–239. doi: 10.1073/pnas.0709145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiqi AM, Lemke S, Schmidt-Ott U. Postgastrular zen expression is required to develop distinct amniotic and serosal epithelia in the scuttle fly Megaselia. Developmental Biology. 2010;341:282–290. doi: 10.1016/j.ydbio.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Rafiqi AM, Lemke S, Schmidt-Ott U. The scuttle fly Megaselia abdita (Phoridae): a link between Drosophila and Mosquito development. Cold Spring Harbor Protocols. 2011;2011:pdb.emo143. doi: 10.1101/pdb.emo143. [DOI] [PubMed] [Google Scholar]
- Rafiqi AM, Park CH, Kwan CW, Lemke S, Schmidt-Ott U. BMP-dependent serosa and amnion specification in the scuttle fly Megaselia abdita. Development. 2012;139:3373–3382. doi: 10.1242/dev.083873. [DOI] [PubMed] [Google Scholar]
- Reim I, Lee HH, Frasch M. The T-box-encoding dorsocross genes function in amnioserosa development and the patterning of the dorsolateral germ band downstream of Dpp. Development. 2003;130:3187–3204. doi: 10.1242/dev.00548. [DOI] [PubMed] [Google Scholar]
- Schmidt-Ott U, Kwan CW. Morphogenetic functions of extraembryonic membranes in insects. Current Opinion in Insect Science. 2016;13:86–92. doi: 10.1016/j.cois.2016.01.009. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serpe M, Umulis D, Ralston A, Chen J, Olson DJ, Avanesov A, Othmer H, O'Connor MB, Blair SS. The BMP-binding protein crossveinless 2 is a short-range, concentration-dependent, biphasic modulator of BMP signaling in Drosophila. Developmental Cell. 2008;14:940–953. doi: 10.1016/j.devcel.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilo BZ, Haskel-Ittah M, Ben-Zvi D, Schejter ED, Barkai N. Creating gradients by morphogen shuttling. Trends in Genetics. 2013;29:339–347. doi: 10.1016/j.tig.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Turing AM. The chemical basis of morphogenesis. Philosophical Transactions of the Royal Society B: Biological Sciences. 1952;237:37–72. doi: 10.1098/rstb.1952.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zee M, Stockhammer O, von Levetzow C, Nunes da Fonseca R, Roth S. Sog/Chordin is required for ventral-to-dorsal Dpp/BMP transport and head formation in a short germ insect. PNAS. 2006;103:16307–16312. doi: 10.1073/pnas.0605154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YC, Ferguson EL. Spatial bistability of Dpp-receptor interactions during Drosophila dorsal-ventral patterning. Nature. 2005;434:229–234. doi: 10.1038/nature03318. [DOI] [PubMed] [Google Scholar]
- Wharton KA, Serpe M. Fine-tuned shuttles for bone morphogenetic proteins. Current Opinion in Genetics & Development. 2013;23:374–384. doi: 10.1016/j.gde.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotton KR, Jiménez-Guri E, García Matheu B, Jaeger J. A staging scheme for the development of the scuttle fly megaselia abdita. PLoS One. 2014;9:e84421. doi: 10.1371/journal.pone.0084421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip ML, Lamka ML, Lipshitz HD. Control of germ-band retraction in Drosophila by the zinc-finger protein Hindsight. Development. 1997;124:2129–2141. doi: 10.1242/dev.124.11.2129. [DOI] [PubMed] [Google Scholar]