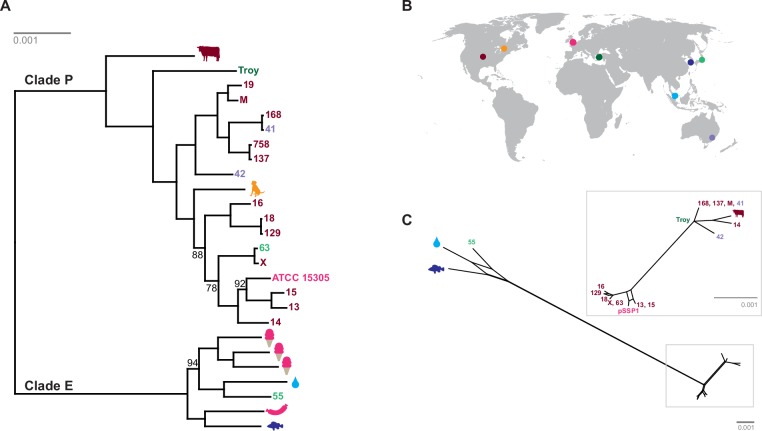
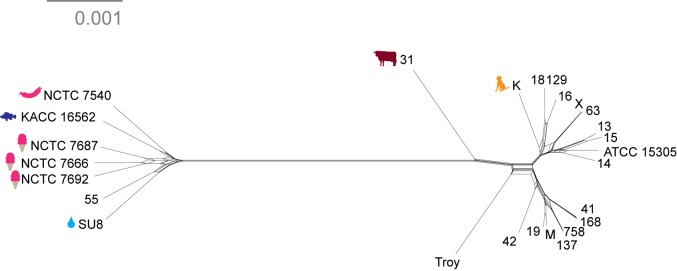
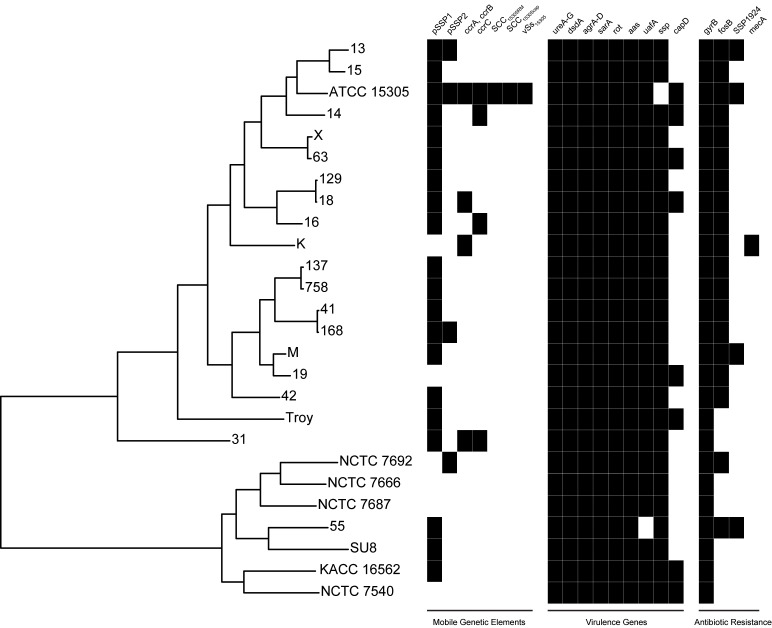
Figure 4. Phylogenetic analysis of Staphylococcus saprophyticus.
(A) Maximum likelihood tree estimated using RAxML (Stamatakis, 2014) (Figure 4—source data 3) from an alignment of S. saprophyticus genomes (Figure 4—source data 1, Figure 4—source data 2). Bootstrap values less than 100 are labeled. Silhouettes indicate bacterial sample source. Isolates without silhouettes are from human clinical samples isolated from urine. Color corresponds to country of isolation as seen on the map. Full sample descriptions are in Supplementary file 1H. (B) Source countries of bacterial samples. (C) Neighbor-net network of S. saprophyticus plasmid sequences (Figure 4—source data 4) related to pSST1 created in SplitsTree4 (Huson and Bryant, 2006). The boxed inset is an enlarged version of the portion of the network from Clade P isolates. Some S. saprophyticus isolates do not encode pSST1-like plasmids, and therefore, they are not included in the network. Starts and stops of recombinant regions of the alignment can be found in Figure 4—source data 5.
DOI: http://dx.doi.org/10.7554/eLife.20983.025