Abstract
Paget’s disease of bone (PDB) is a common metabolic bone disorder with a significant genetic component. To date, only one gene associated with PDB has been identified, the p62-Sequestosome1 gene (SQSTM1), and more than 20 mutations of this gene have been reported in PDB, the most common being the P392L substitution. In order to search for differentially expressed genes in PDB, we investigated the relative gene expression profile of candidate genes in osteoclast (OCL) cultures from 12 PDB patients and six unmatched healthy controls with known genetic status regarding p62, including healthy carriers of the P392L mutation. We selected 48 OCL-expressed candidate genes that may be involved in relevant pathways of PDB pathogenesis, such as OCL signaling, survival, bone resorption activity, or adhesion. In OCL cultures derived from peripheral blood mononuclear cells, total RNA extraction was performed, followed by real-time PCR experiments. Relative quantification analysis utilized the qBase method where relative expression levels were normalized with respect to a set of reference primer pairs for three housekeeping genes. When compared to non-mutated healthy controls, OCL cultures from PDB patients displayed a significant down-regulation in genes involved in apoptosis (CASP3 and TNFRSF10A), in cell signaling (TNFRSF11A), in the OCL bone resorbing function (ACP5 and CTSK) and in the gene coding for Tau protein (MAPT) (all comparisons, p<0.0001). Comparison of relative gene expression in PDB patients with P392L mutation versus PDB patients without SQSTM1 mutation did not provide significant differential gene expression. However, we observed a non-significant decrease in the expression of several genes such as IL6ST, HIF1A, OSTM1, TNFRSF10B and -10D, PDK1, MAPT and CASP3 in healthy carriers of the P392L mutation. These results provide important information about the mis-regulated activities of pagetic OCL, and highlight the role of altered apoptosis pathways in these cells. They also suggest that the SQSTM1 P392L mutation plays a role in PDB pathogenesis, even at early preclinical stages in healthy carriers of the P392L mutation.
Keywords: Osteoclast, Apoptosis, Paget’s disease of bone, Gene expression profile, SQSTM1 gene
Introduction
Paget’s disease of bone (PDB) is the second most common skeletal disorder, next to osteoporosis, affecting up to 3% of the population over 55 years of age. PDB is characterized by focal, disorganized increases in bone turnover [1]. Osteoclasts (OCLs) have been described as the cells primarily affected in PDB with the initial phase of the disease typified by excessive bone resorption [2]. Pagetic OCLs are both larger and more numerous than OCLs in a healthy adult, are both hyperactive and hypersensitive to osteoclastogenic factors such as 1,25-(OH)2D3 or the receptor activator of NF-κB Ligand (RANKL) [3,4], and are resistant to apoptosis [5]. Although a viral etiology has been suggested for PDB, several studies have revealed a strong genetic component. In approximately one third of cases, the disease has an autosomal dominant pattern of inheritance [6,7]. Among the seven loci reported, the 5q35-qter region is the only one for which a gene has been identified, namely the Sequestosome1 gene (SQSTM1) that encodes the p62 protein [8]. More than twenty missense or truncating mutations of the SQSTM1 gene have been reported, although the P392L substitution is the most frequent [9,10]. All of these mutations are clustered either within or near the C-terminal region of the p62 protein that embodies the ubiquitin-associated (UBA) domain.
The presence of p62-P392L mutation may contribute to the overactive state of OCLs in PDB [5]. Normal OCL precursors transfected with the p62MUT gene (P392L) display markedly increased responsivity to RANKL and TNF-α, but not to 1, 25-(OH)2 D3, unlike OCL precursors from PDB patients, and when mature have an increased ability to resorb bone [11]. In addition, transgenic mice with targeted expression of the human p62P392L gene in the OCL lineage develop increased OCL numbers and display progressive bone loss, although no Paget-related bone lesions are observed [11]. Thus, SQSTM1 mutations do not appear sufficient to induce pagetic lesions, but may predispose to PDB by increasing the susceptibility of OCL precursors to osteoclastogenic cytokines and/or the osteoclastogenic potential of the bone microenvironment [11,12].
Differential gene expression profile in PDB has been recently investigated in two case-control studies. The first study, using cDNA microarrays and real-time PCR, investigated differential gene expression profiles in osteoblasts and bone marrow stromal cells of PDB patients or healthy controls. An overexpression of Dickkopf homolog1 (Dkk1), Interleukin-1 (IL-1), Interleukin-6 (IL-6) and alkaline phosphatase, and an underexpression of bone sialoprotein and osteocalcin mRNA were observed in pagetic osteoblasts [13]. The second study investigated candidate genes involved in interferon-mediated signaling in peripheral monocytes and lymphocytes in 23 PDB patients versus 23 controls; a significant overexpression of genes of the interferon pathway was reported, along with a down-regulation of the tumor necrosis factor alpha (TNF-α) gene [14]. While clearly indicating that a number of genes are differentially expressed in bone cells from PDB patients compared to healthy donors, these two studies did not investigate whether the donors were carriers of mutations in the SQSTM1 gene or not.
Our aim was to study the relative gene expression profile of 48 candidate genes involved in relevant pathways of PDB pathogenesis (OCL signaling, survival, function and cell adhesion) in OCL cultures from PDB patients, all of whom had been genotyped for the presence of mutations in the p62 UBA domain, and to compare these genes expression to those from OCLs of healthy donors with no SQSTM1 mutation, or healthy SQSTM1 P392L-mutation carriers.
Patients and methods
Patients
All research performed for this trial was approved by the Centre Hospitalier de l’Université Laval (CHUL) Ethics Committee and all individuals signed an informed consent document before entering into the study. Twelve PDB patients and six unmatched healthy controls were selected for this study. Phenotype assessments comprised of complete skeletal evaluation, including total serum alkaline phosphatase, total body bone scan and skull and pelvis x-rays. Criteria for diagnosis of PDB included: abnormal skeletal x-rays and abnormal bone scan, with or without elevated total serum alkaline phosphatase levels. Individuals aged 40 years or more were considered healthy when they possessed normal alkaline phosphatase levels with a normal bone scan.
All individuals used in this study were previously genotyped for the SQSTM1 gene, as reported [9]. The P392L mutation was the only SQSTM1 mutation identified in individuals participating in this study. We selected patients for our analysis according to the presence or the absence of the p62 P392L mutation, with about half of the PDB patients (7 out of 12) and half of healthy controls (3 out of 6) carrying this mutation. The healthy carriers were relatives of PDB patients. All clinical characteristics and genotypes for SQSTM1 are summarized in Table 1.
Table 1.
Clinical characteristics and SQSTM1 genotypes of patients with Paget’s disease of bone and healthy individuals.
| Individual | Age | Sexa | Phenotypeb | P392L mutation |
ALP (xULN)c |
Nb bonesd |
Last BPe
(months) |
|---|---|---|---|---|---|---|---|
| 1 | 71 | F | PDB | Yes | 0.78 | 1 | 33 |
| 2 | 70 | F | PDB | Yes | 0.82 | 4 | 27 |
| 3 | 67 | M | PDB | Yes | 0.62 | 5 | 34 |
| 4 | 80 | M | PDB | Yes | 1.3 | 7 | 25 |
| 5 | 68 | F | PDB | Yes | 0.41 | 2 | 0f |
| 6 | 65 | M | PDB | Yes | 0.73 | 10 | 110 |
| 7 | 79 | F | PDB | Yes | 2.32 | 8 | 92 |
| 8 | 72 | F | PDB | No | 0.71 | 2 | 19 |
| 9 | 78 | M | PDB | No | 0.74 | 3 | 48 |
| 10 | 75 | F | PDB | No | 0.65 | 2 | 126 |
| 11 | 73 | F | PDB | No | 0.73 | 3 | 0g |
| 12 | 83 | F | PDB | No | 0.58 | 1 | 9 days |
| 13 | 57 | F | Healthy | Yes | 0.74 | 0 | – |
| 14 | 64 | F | Healthy | Yes | 0.46 | 0 | – |
| 15 | 43 | F | Healthy | Yes | 0.49 | 0 | – |
| 16 | 65 | F | Healthy | No | 0.41 | 0 | – |
| 17 | 49 | F | Healthy | No | 0.67 | 0 | – |
| 18 | 44 | M | Healthy | No | 0.66 | 0 | – |
F=female, M=male.
PDB=Paget’s disease of bone.
ALP=Total serum alkaline phosphatase x ULN=upper limit of normal range, as determined by each clinical laboratory.
Number of affected bones.
Time since last bisphosphonate (BP) treatment.
Currently treated by alendronate 70 mg weekly.
Currently treated by risedronate 35 mg weekly.
Osteoclast cultures
Peripheral blood mononuclear cells (PBMCs) were isolated from heparinized blood by density-gradient centrifugation, washed, and suspended in Opti-MEM with the antibiotics, glutamine, and 2% FCS. They were plated at a density of 3×106/ml on eight-well chamber/slides (Lab-Tek, Biosciences, Bedford, MA). After incubating overnight, the cells were washed to remove any non-adherent cells. The selected PBMCs were cultured for 3 weeks in medium supplemented with GM-CSF (100 pg/ml) for the first 3 days, with the remainder of the time in the same medium supplemented with M-CSF (25 ng/ml) and RANKL (75 ng/ml). The medium was changed every 2–3 days. These culture conditions yielded multinucleated cells (MNCs) that expressed OCL markers, calcitonin receptor and RANK, and possessed the ability to resorb bone, as previously described [5,15].
Quantitative real-time PCR
For the quantitative real-time RT-PCR analysis, total RNA was extracted from fully matured OCL using the RNeasy plus kit (Qiagen, Mississauga, Canada), which excludes genomic DNA. By utilizing this method, at least 50 μg of RNA was harvested from each cell sample. After being assayed and checked for quality, 1 μg of total RNA was used to perform cDNA synthesis. cDNA synthesis was performed using the high-efficiency QuantiTect Primer Assay kit (Qiagen, Mississauga, Canada), and sent for quantitative PCR amplification (RNomics Platform, Laboratoire de Génomique Fonctionnelle, University of Sherbrooke, QC). Human primers of candidate genes were generated and validated, and the real-time PCR reaction was conducted with 200 nM of primers. A further 100 ng of cDNA and 5 μl of Power SYBR-green Master Mix were added to the reaction mix to give a total volume of 10 μl. Amplification and detection of the candidate genes and of three reference housekeeping genes (hydroxymethylbilane synthase (HMBS), proteasome 26S subunit ATPase 4 (PSMC4), succinate dehydrogenase complex subunit A (SDHA)) were conducted with a Realplex 2 Master Cycler (Eppendorf, Mississauga, Canada).
The quantification and normalization of results were based on the computation of target threshold cycle (Ct) values and reference gene Ct values in qBase software. All samples were run in triplicate for the target and reference genes. Relative expression levels were normalized with respect to a set of reference primer pairs (i.e. the mean of the three housekeeping genes), and to technical and experimental errors. Relative expression quantification analysis relied on the qBase method [16]. Briefly, this method constitutes an improvement over the classical delta-delta-Ct method, which was extended to take into account multiple stably expressed reference genes for improved normalization.
We selected 48 OCL-related genes and investigated their relative gene expression in OCL cultures from our 4 groups of subjects. This selection was based on pagetic OCL characteristics, focusing on OCL signaling pathways, particularly RANKL signaling and apoptotic pathways. Candidate genes (encoded proteins): ACP5 (Tartrate-resistant acid phosphatase-TRAP), AKT1 (Protein kinase Akt), CALCR (Calcitonin receptor), CASP3-CASP8-CASP9 (Caspase -3, -8, -9), CDH1 (E-cadherin), CREM (cAMP Response Element Modulator), CTSK (Cathepsin K), CYLD (cylindromatosis deubiquitinase), DDR1-DDR2 (Discoidin domain receptor tyrosine kinase -1, -2), DKK1 (Dickkopf homolog 1), FCGR3A (Immunoglobulin G Fc receptor III), HIF1A (Hypoxia-inducible factor 1α), IL6ST (IL-6 signal transducer-gp130), IRAK1 (IL-1 receptor-associated kinase-1), ITGAV-ITGB3 (Integrin αv, Integrin β3), KIAA1432 (Connexin43-interacting protein 150), MAP2K5 (mitogen activated protein kinase kinase 5), MAPT (Microtubule-associated protein Tau), NFATC1 (nuclear factor of activated T-cells, cytoplasmic), NFKBIA-NFKBIB (NFκB inhibitor α and β), OSCAR (Osteoclast-associated receptor), OSTM1 (osteopetrosis-associated TM protein 1), PDK1 (pyruvate dehydrogenase kinase isoenzyme 1), PRKCZ (PKCζ), PTK2 (protein-tyrosine kinase 2), SQSTM1, SRC (C-src tyrosine kinase), TAF2 (TATA box binding protein-associated factor 2), TANK (TRAF family member-associated NFkB activator), TGFBR1-TGFBR2 (TGFβ receptor -1, -2), TNFRSF10 -A, -B, -C, -D (TRAIL receptor -1, -2, -3, -4), TNFRSF11A (RANK), TNFSF10 (TNF-related apoptosis-inducing ligand (TRAIL)), TRAF6 (TNF receptor-associated factor 6), TRIP6 (thyroid receptor-interacting protein 6), TRPV5 (vanilloid transient receptor potential 5), TYROBP (TYRO protein tyrosine kinase-binding protein (DAP12)), VDR (vitamin D receptor), WDFY3 (WD repeat and FYVE domain containing 3 (ALPHY)).
All the primers were designed based on sequences reported in Ace view database. Three primer pairs were designed for each gene, in order to choose the best primer pair after validation. All primers pairs were validate by qPCR amplification of a total XPressRef Universal human mRNA (Cedarlane, Burlington, ON). Amplification products were submitted to capillary electrophoresis in order to validate the size of the expected product. In addition, a melting curve was generated and only those primers giving one product were selected. If all the three primer pairs gave such results, the pair giving the smallest Ct was selected. Finally, the primer pair efficiency was obtained from a standard curve experiment where a series of dilution of the same sample was correlated to the Ct values. qPCR amplification was performed separately for each sample.
Statistical analysis
To study the differential gene expression pattern in the four groups (PDB patients without any SQSTM1 mutation, PDB patients carrying the P392L SQSTM1 mutation, healthy donors without any SQSTM1 mutation, and healthy donors carrying the P392L SQSTM1 mutation), we compared the mean normalized relative expression in 3 intergroup analyses (all PDB patients versus healthy donors with no SQSTM1 mutation; PDB patients without any SQSTM1 mutation versus PDB patients carrying the P392L mutation; and healthy donors with no SQSTM1 mutation versus healthy donors carrying the P392L mutation). Statistical analysis was performed using analysis of variance (ANOVA) after adjusting for age. Analyses were corrected for multiple testing using the Bonferroni correction. A multiple-test corrected p-value<0.00035 was considered statistically significant. The fold change was calculated as the ratio of the mean relative expression for each group. The natural logarithm (Ln) of the fold change was then calculated. Results are presented in order of increasing p-value, whatever the Ln fold change (Ln fold <1 or >−1).
Results
Gene expression profile in PDB patients versus healthy controls
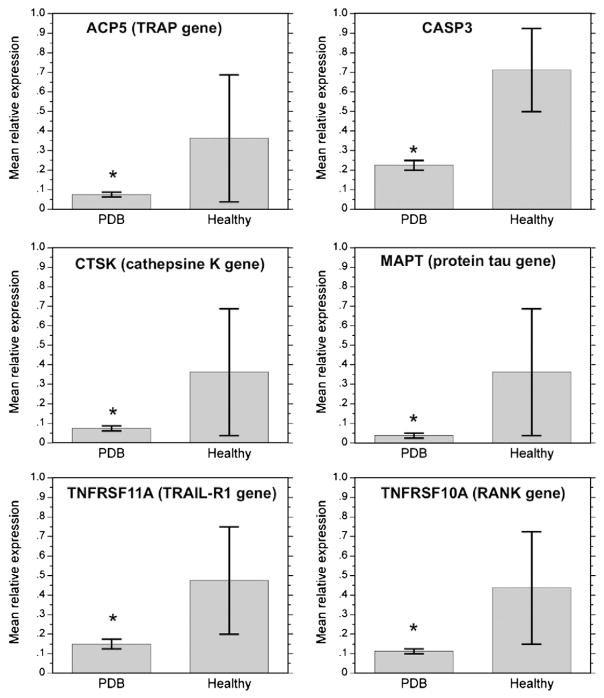
The OCL gene expression profile comparison from all PDB patients regardless of whether the SQSTM1 mutation was present or not versus non-mutated healthy controls is shown in Table 2. In PDB patients, there was a significant down-regulation in genes involved in apoptosis [CASP3 (0.22±0.02 versus 0.71±0.22, p<0.0001) and TNFRSF10A (0.11±0.01 versus 0.44±0.29, p<0.0001)], in cell signaling [TNFRSF11A (0.15±0.03 versus 0.48±0.28, p=0.0001)], in the OCL bone resorbing function [ACP5 (0.07±0.01 versus 0.36± 0.32, p<0.0001) and CTSK (0.08±0.01 versus 0.36±0.32, p<0.0001)], and in the gene coding for Tau protein (MAPT) (0.04± 0.01 versus 0.36±0.32, p<0.0001). For all these genes, the multiple-test corrected p-value was statistically significant, i.e.<0.00035 (Table 2, Fig. 1). Changes in the relative expression of other genes were also observed, reflected by a Ln Fold change <−1 or >1. In this way, some genes were down-regulated in PDB patients, although non-significantly, such as TNFRSF10B, DDR2, TGFBR1, HIF1A, ITGAV, ITGB3, CDH1 and CYLD. Finally, a non-significant up-regulation of FCGR3A was observed (Table 2).
Table 2.
Relative expression levels normalized to 3 internal control genes in OCLs from all patients with PDB and healthy controls without any SQSTM1 mutation.
| Gene symbol | Mean relative expression±SE in PDB patients | Mean relative expression±SE in healthy controls | Ln of fold changea | Uncorrected p-value |
|---|---|---|---|---|
| Down-regulated genes | ||||
| ACP5 | 0.07±0.01 | 0.36±0.32 | −1.66 | <0.0001# |
| CASP3 | 0.22±0.02 | 0.71±0.22 | −1.20 | <0.0001# |
| CTSK | 0.08±0.01 | 0.36±0.32 | −1.53 | <0.0001# |
| MAPT | 0.04±0.01 | 0.36±0.32 | −2.19 | <0.0001# |
| TNFRSF10A | 0.11±0.01 | 0.44±0.29 | −1.37 | <0.0001# |
| TNFRSF11A | 0.15±0.03 | 0.48±0.28 | −1.15 | 0.0001# |
| TNFRSF10B | 0.12±0.04 | 0.46±0.28 | −1.37 | 0.001 |
| DDR2 | 0.08±0.03 | 0.37±0.32 | −1.53 | 0.004 |
| TGFBR1 | 0.20±0.05 | 0.62±0.24 | −1.14 | 0.005 |
| HIF1A | 0.12±0.04 | 0.54±0.46 | −1.50 | 0.007 |
| ITGAV | 0.10±0.04 | 0.38±0.31 | −1.34 | 0.010 |
| ITGB3 | 0.13±0.04 | 0.37±0.32 | −1.01 | 0.040 |
| CDH1 | 0.16±0.06 | 0.44±0.29 | −0.98 | 0.047 |
| CYLD | 0.31±0.08 | 0.74±0.22 | −0.88 | 0.048 |
| TNFRSF10D | 0.18±0.05 | 0.39±0.30 | −0.81 | 0.095 |
| WDFY3 | 0.21±0.10 | 0.55±0.24 | −0.95 | 0.12 |
| TRPV5 | 0.16±0.08 | 0.50±0.50 | −1.16 | 0.13 |
| TYROBP | 0.18±0.04 | 0.29±0.19 | −0.47 | 0.25 |
| Up-regulated genes | ||||
| FCGR3A | 0.32±0.09 | 0.07±0.03 | 1.54 | 0.23 |
Gene relative expression levels are reported as mean±standard error (SE). p-values were calculated from an ANOVA test using the reported means± SE, (#) p-values<0.00035 were considered statistically significant. Up-regulated and down-regulated genes are presented by increasing level of p-value, whatever the Ln fold change (<1 or >−1).
The fold changes are the ratios of the mean relative expression levels in PDB patients over the mean relative expression levels in healthy donors.
Fig. 1.
Mean relative expression levels normalized to internal control gene in osteoclasts from all patients with PDB regardless the presence of SQSTM1 mutation and healthy controls without any SQSTM1 mutation. Graphs represent the mean relative expression±standard error of the 6 genes that were significantly down-regulated in PDB patients compared to healthy non-mutated controls (*multiple-test corrected p-value<0.00035).
Impact of the p62 P392L mutation on the gene expression profile of PDB patients and healthy individuals
When comparing the relative gene expression between OCLs from PDB patients carrying the p62 P392L mutation and PDB patients with no mutation in the SQSTM1 gene, no difference in SQSTM1 gene expression was observed (0.35±0.10 in PDB patients with the mutation versus 0.25±0.04 in PDB patients without the mutation, p=0.45). However, changes in the relative expression of some genes were observed with a Ln Fold change <−1 or >1. In PDB patients carrying the SQSTM1 mutation, a non-significant down-regulation in CALCR (0.08±0.04 versus 0.27±0.18, p=0.26) and a non-significant up-regulation of genes such as NFKBIB, OSCAR, and DKK1 were observed in the mutated group as compared to the non-mutated group (Table 3).
Table 3.
Relative expression levels normalized to 3 internal control genes in osteoclasts from patients with Paget’s disease of bone (PDB) carrying the P392L mutation versus PDB patients without any SQSTM1 mutation.
| Gene symbol | Mean relative expression±SE in PDB patients with P392L mutation | Mean relative expression±SE in PDB patients without any SQSTM1 mutation | Ln of fold changea | Uncorrected p-value |
|---|---|---|---|---|
| Down-regulated genes | ||||
| CALCR | 0.08±0.04 | 0.27±0.18 | −1.26 | 0.26 |
| Up-regulated genes | ||||
| CTSK | 0.10±0.02 | 0.05±0.01 | 0.57 | 0.15 |
| NFKBIB | 0.26±0.13 | 0.06±0.01 | 1.47 | 0.27 |
| ACP5 | 0.08±0.02 | 0.05±0.01 | 0.46 | 0.28 |
| OSCAR | 0.26±0.14 | 0.06±0.01 | 1.50 | 0.29 |
| PDK1 | 0.33±0.14 | 0.13±0.02 | 0.94 | 0.31 |
| TNFRSF10B | 0.15±0.06 | 0.07±0.00 | 0.84 | 0.32 |
| NFATC1 | 0.35±0.13 | 0.16±0.04 | 0.80 | 0.32 |
| TNFRSF10D | 0.23±0.09 | 0.10±0.02 | 0.80 | 0.33 |
| DKK1 | 0.31±0.16 | 0.09±0.09 | 1.25 | 0.33 |
Gene relative expression levels are reported as mean±standard error (SE). p-values were calculated from an ANOVA test using the reported means±SE. Up-regulated and down-regulated genes are presented by increasing level of p-value, whatever the Ln fold change (<1 or >−1).
The fold changes are the ratios of the mean relative expression levels in PDB patients carrying the P392L mutation over the mean relative expression levels in PDB patients without any SQSTM1 mutation.
When comparing the normalized relative gene expression between OCL cultures from healthy individuals carrier of a P392L mutation to healthy controls without any SQSTM1 mutation, we failed to identify any difference in SQSTM1 gene expression (0.34±0.30 in healthy carriers of P392L mutation versus 0.45±0.28 in healthy non-mutated individuals, p=0.19). Nevertheless, we observed a non-significant decreased expression in several genes such as IL6ST, HIF1A, OSTM1, TNFRSF10-B and -D, PDK1, MAPT and TAF2 (Table 4). Interestingly, some of these genes (HIF1A, TNFRSF10-B, ITGAV, MAPT, RANK, TGFBR1, CASP3, ACP5 and CTSK) were also down-regulated in PDB patients as compared to healthy controls (Tables 2 and 4).
Table 4.
Relative expression levels normalized to 3 internal control genes in OCLs from healthy individuals carrying the P392L mutation versus healthy controls without any SQSTM1 mutation.
| Gene symbol | Mean relative expression±SE in healthy subjects with P392L mutation | Mean relative expression±SE in healthy subjects without any SQSTM1 mutation | Ln of fold changea | Uncorrected p-value |
|---|---|---|---|---|
| Down-regulated genes | ||||
| IL6ST | 0.05±0.00 | 0.24±0.11 | −1.52 | 0.003 |
| HIF1A | 0.07±0.00 | 0.54±0.46 | −2.04 | 0.004 |
| OSTM1 | 0.09±0.03 | 0.47±0.27 | −1.67 | 0.006 |
| CDH1 | 0.19±0.09 | 0.44±0.29 | −0.82 | 0.012 |
| TNFRSF10B | 0.08±0.04 | 0.46±0.28 | −1.82 | 0.013 |
| PDK1 | 0.07±0.02 | 0.22±0.11 | −1.09 | 0.021 |
| TNFRSF10D | 0.06±0.02 | 0.39±0.30 | −1.88 | 0.028 |
| MAP2K5 | 0.21±0.08 | 0.41±0.17 | −0.68 | 0.029 |
| TAF2 | 0.10±0.02 | 0.44±0.19 | −1.47 | 0.041 |
| ITGAV | 0.07±0.01 | 0.38±0.31 | −1.68 | 0.058 |
| MAPT | 0.03±0.02 | 0.36±0.32 | −2.66 | 0.062 |
| TNFRSF11A | 0.20±0.07 | 0.48±0.28 | −0.90 | 0.062 |
| TGFBR1 | 0.15±0.05 | 0.62±0.24 | −1.45 | 0.082 |
| PTK2 | 0.13±0.07 | 0.35±0.18 | −1.02 | 0.10 |
| CASP3 | 0.17±0.07 | 0.71±0.22 | −1.43 | 0.12 |
| TNFSF10 | 0.01±0.00 | 0.03±0.02 | −1.58 | 0.12 |
| ITGB3 | 0.12±0.04 | 0.37±0.32 | −1.13 | 0.13 |
| ACP5 | 0.11±0.06 | 0.36±0.32 | −1.19 | 0.15 |
| CTSK | 0.10±0.07 | 0.36±0.32 | −1.26 | 0.17 |
| CASP8 | 0.06±0.02 | 0.14±0.05 | −0.81 | 0.17 |
| KIAA1432 | 0.06±0.02 | 0.15±0.05 | −0.91 | 0.17 |
| TANK | 0.11±0.04 | 0.19±0.08 | −0.54 | 0.19 |
| CYLD | 0.20±0.09 | 0.74±0.22 | −1.29 | 0.19 |
Gene relative expression levels are reported as mean±standard error (SE). p-values were calculated from an ANOVA test using the reported means±SE. Down-regulated genes are presented by increasing level of p-value, whatever the Ln fold change (<1 or >−1).
The fold changes are the ratios of the mean relative expression levels in healthy individuals with P392L mutation over the mean relative expression in healthy individuals without any SQSTM1 mutation.
Discussion
PDB is characterized by the presence of a large number of OCLs which display large degrees of misregulated activity. Pagetic OCLs are larger, possess a higher number of nuclei per OCL, and are more active as compared to OCLs from healthy adults. In addition, OCL precursors exhibit increased sensitivity to osteoclastogenic factors, such as RANKL [2]. Although mutations of RANK, RANKL or OPG genes have been reported in some rare heritable bone disorders that share a number of characteristics with PDB, they have not as yet been reported in PDB [17]. Therefore, the sensitivity of PDB OCLs towards these regulating signals is hindered, implicating the post-stimulatory signaling pathways as important players in PDB OCL dysfunction. An imbalance between OCL formation and apoptosis may also affect the PDB phenotype, suggesting that their control mechanisms could additionally be defective [5]. In both precursors and mature OCLs, the interaction between RANKL and RANK results in signaling cascades that require the recruitment of TNFR-associated factor 6 (TRAF6), and ultimately activate transcription factors, particularly NF-κB and NFATc1 [18]. The cytosolic p62 protein, encoded by the SQSTM1 gene, is a scaffolding protein that interacts with the RANK signaling complex, and is one of the functional links reported between RANKL and TRAF6-mediated NF-κB activation [19].
We studied the differential expression of genes in OCLs of PDB patients and healthy controls, as well as the impact of the p62 P392L mutation, the most frequent SQSTM1 mutation reported in PDB. We selected 48 genes on the basis of pagetic OCL characteristics, focusing on OCL signaling pathways, particularly the RANKL signaling and apoptotic pathways. Comparing OCLs from all the PDB patients and unmutated healthy controls, we observed a significant down-regulation of the genes involved in apoptosis (CASP3 encoding Caspase 3 and TNFRSF10-A encoding TRAIL-R1), in cell signaling (TNFRSF11A encoding RANK), in the OCL bone resorbing function (ACP5 encoding TRAP and CTSK encoding cathepsin K), and of the gene coding for the Tau protein (MAPT). Comparing the gene expression profile in OCLs from healthy individuals with and without the SQSTM1 mutation, we observed a non-significant down-regulation of IL6ST, HIF1A, OSTM1, TNFRSF10-B and -D, PDK1, MAPT and TAF2 in the presence of the P392L mutation. However, comparison of relative gene expression in PDB patients with the P392L mutation versus PDB patients without any SQSTM1 mutation did not reveal any significant change in the expression of these genes. We believe that the apparent lack of impact of the p62 P392L mutation could be related to other mutations or to environmental factors that are probably involved in the pagetic phenotype, and may supercede and obscure the specific effects of the p62 mutation in pagetic OCLs. The small size of our groups may explain why, although we did observe changes in the gene expression profile, defined by an Ln Fold change <−1 or >1, most of these changes were not significant, particularly when we studied the impact of the P392L mutation and compared the 2 groups of PDB patients or the 2 groups of healthy subjects on the basis of the presence or absence of the mutation. However, although this analysis is only descriptive, we believe that it does provide important information about the misregulated activities of pagetic OCL, and the role of the SQSTM1 P392L mutation in PDB pathogenesis.
OCL apoptosis and its regulation are of critical importance to our understanding of OCL behavior during bone resorption and remodeling [20]. In a previous study, we showed that pagetic OCLs were resistant to apoptosis regardless of whether the p62 mutation was present or not [5]. Our results confirmed a role of apoptosis pathways in PDB pathogenesis, as the genes encoding Caspase 3, the central-effector caspase, as well as TRAIL-R1, which have been implicated in human OCL apoptosis [15] were significantly down-regulated in PDB patients compared to healthy controls. The p62 P392L mutation could have an impact on apoptosis pathways as a down-regulation of CASP3 and of other genes involved in OCL apoptosis such as those of TRAIL-R2 and TGFβ-R1 [21] was also observed in healthy carriers of P392L mutation compared to unmutated healthy subjects, although these changes were non significant. Other apoptotic factors could be involved as an up-regulation of several anti-apoptotic genes, notably the Bcl-2 gene, was previously reported [22]. Whereas apoptosis appears to be hindered in pagetic OCLs, the impact of PDB mutations in the p62 gene and the mechanisms by which apoptotic pathways are altered are still largely unknown.
The decrease of RANK gene expression, along with the decrease in the expression of genes involved in OCL activity such as ACP5 encoding TRAP or CTSK encoding cathepsin K, in PDB patients compared to unmutated controls, were surprising as OCLs are characteristically overactive in PDB. However other pathways could be concomitantly up-regulated, such as the co-stimulating ITAM pathway [18]; supporting this hypothesis was our observation of an increase in FcRgamma gene expression in pagetic OCLs, although non-significant.
The discovery of mutations in the SQSTM1 gene in numerous patients has identified protein p62 as an important modulator of bone turnover. P62 is a scaffold protein which mediates various cell functions, including the control of NF-κB signaling, protein trafficking and gene transcription [23]. The p62-P392L mutation, as well as the other mutations that have been described in PDB, affect the interactions of the UBA domain with multiubiquitin chains, suggesting that a loss of ubiquitin-chain binding by p62 is probably important in the development of PDB [24,25]. Both p62 and TRAF6 are up-regulated during RANKL-induced osteoclastogenesis, and genetic inactivation of p62 in mice leads to impaired osteoclastogenesis, due to defective NF-κB activation [26]. Here we were unable to identify any differential gene expression of SQSTM1 and TRAF6 genes in PDB patients in comparison with healthy controls. Although an increase in p62 expression was reported at a protein level [5,27], SQSTM1 gene overexpression was previously reported in PDB in only one study where RNA was extracted from Epstein–Barr virus immortalized B-cell lymphocytes cultures [27]. Other studies have also observed a similar expression of the SQSTM1 gene in peripheral monocytes, lymphocytes and osteoblasts from PDB patients and healthy controls [13,14]. Finally, we observed a significant down-regulation of the gene encoding the microtubule-associated Tau protein (MAPT) in PDB patients compared to unmutated controls. Tau is a protein associated with the formation of tangled protein filaments, and accumulation of p62-Tau aggregates have been involved in neurodegenerative diseases. After K63-polyubiquitination, Tau is shuttled for degradation to the proteasome by p62, and any changes in p62 expression could lead to disturbances in Tau degradation, accumulation and aggregation [28]. Although the role of Tau protein in PDB disease has not been defined, our results suggest a down-regulation of the MAPT gene in PDB patients compared to healthy controls. This could be a consequence of p62 disregulation, although the significance of this observation remains to be determined.
The small size of the samples of PDB patients, and of their unmatched healthy controls is certainly the major weakness of our study. This is why the findings reported here must be considered to be purely descriptive. In addition, our analysis was restricted to selected candidate genes involved in cell signaling and survival/apoptosis pathways, and our cultured cell population was heterogeneous although consisting mainly of OCL-like cells. Nevertheless, our analysis does provide results that are promising for further studies investigating the molecular basis of pagetic OCL behavior. A larger cohort of PDB patients and a more systematic approach of genome-wide transcriptome will be warranted to further elucidate the early events in PDB development and to improve our understanding of the role of p62 protein and SQSTM1 mutations in PDB pathogenesis.
Acknowledgments
This work was supported by the Canadian Institutes of Health Research. Dr Michou and Dr Roux are supported by career awards from the Fonds de la Recherche en Santé du Québec. The authors thank Danielle Poulin for her assistance in recruiting patients and healthy controls, Edith Gagnon for her assistance in the SQSTM1 gene sequencing, and Dr. K. Shawn Davison for the careful revision of the manuscript.
References
- 1.Seitz S, Priemel M, Zustin J, Beil FT, Semler J, Minne H, et al. Paget’s disease of bone: histologic analysis of 754 patients. J Bone Miner Res. 2009;24:62–9. doi: 10.1359/jbmr.080907. [DOI] [PubMed] [Google Scholar]
- 2.Roodman GD, Windle JJ. Paget disease of bone. J Clin Invest. 2005;115:200–8. doi: 10.1172/JCI24281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Menaa C, Barsony J, Reddy SV, Cornish J, Cundy T, Roodman GD. 1,25-Dihydroxyvitamin D3 hypersensitivity of osteoclast precursors from patients with Paget’s disease. J Bone Miner Res. 2000;15:228–36. doi: 10.1359/jbmr.2000.15.2.228. [DOI] [PubMed] [Google Scholar]
- 4.Menaa C, Reddy SV, Kurihara N, Maeda H, Anderson D, Cundy T, et al. Enhanced RANK ligand expression and responsivity of bone marrow cells in Paget’s disease of bone. J Clin Invest. 2000;105:1833–8. doi: 10.1172/JCI9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chamoux E, Couture J, Bisson M, Morissette J, Brown JP, Roux S. The p62 P392L mutation linked to Paget’s disease induces activation of human osteoclasts. Mol Endocrinol. 2009;23:1668–80. doi: 10.1210/me.2009-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hocking L, Slee F, Haslam SI, Cundy T, Nicholson G, van Hul W, et al. Familial Paget’s disease of bone: patterns of inheritance and frequency of linkage to chromosome 18q. Bone. 2000;26:577–80. doi: 10.1016/s8756-3282(00)00278-7. [DOI] [PubMed] [Google Scholar]
- 7.Laurin N, Brown JP, Lemainque A, Duchesne A, Huot D, Lacourciere Y, et al. Paget disease of bone: mapping of two loci at 5q35-qter and 5q31. Am J Hum Genet. 2001;69:528–43. doi: 10.1086/322975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laurin N, Brown JP, Morissette J, Raymond V. Recurrent mutation of the gene encoding sequestosome 1 (SQSTM1/p62) in Paget disease of bone. Am J Hum Genet. 2002;70:1582–8. doi: 10.1086/340731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morissette J, Laurin N, Brown JP. Sequestosome 1: mutation frequencies, haplotypes, and phenotypes in familial Paget’s disease of bone. J Bone Miner Res. 2006;21(Suppl 2):P38–44. doi: 10.1359/jbmr.06s207. [DOI] [PubMed] [Google Scholar]
- 10.Cundy T, Bolland M. Paget disease of bone. Trends Endocrinol Metab. 2008;19:246–53. doi: 10.1016/j.tem.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 11.Kurihara N, Hiruma Y, Zhou H, Subler MA, Dempster DW, Singer FR, et al. Mutation of the sequestosome 1 (p62) gene increases osteoclastogenesis but does not induce Paget disease. J Clin Invest. 2007;117:133–42. doi: 10.1172/JCI28267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiruma Y, Kurihara N, Subler MA, Zhou H, Boykin CS, Zhang H, et al. A SQSTM1/p62 mutation linked to Paget’s disease increases the osteoclastogenic potential of the bone microenvironment. Hum Mol Genet. 2008;17:3708–19. doi: 10.1093/hmg/ddn266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naot D, Bava U, Matthews B, Callon KE, Gamble GD, Black M, et al. Differential gene expression in cultured osteoblasts and bone marrow stromal cells from patients with Paget’s disease of bone. J Bone Miner Res. 2007;22:298–309. doi: 10.1359/jbmr.061108. [DOI] [PubMed] [Google Scholar]
- 14.Nagy ZB, Gergely P, Donath J, Borgulya G, Csanad M, Poor G. Gene expression profiling in Paget’s disease of bone: upregulation of interferon signaling pathways in pagetic monocytes and lymphocytes. J Bone Miner Res. 2008;23:253–9. doi: 10.1359/jbmr.071021. [DOI] [PubMed] [Google Scholar]
- 15.Roux S, Lambert-Comeau P, Saint-Pierre C, Lepine M, Sawan B, Parent JL. Death receptors, Fas and TRAIL receptors, are involved in human osteoclast apoptosis. Biochem Biophys Res Commun. 2005;333:42–50. doi: 10.1016/j.bbrc.2005.05.092. [DOI] [PubMed] [Google Scholar]
- 16.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whyte MP. Paget’s disease of bone and genetic disorders of RANKL/OPG/RANK/NF-kappaB signaling. Ann N Y Acad Sci. 2006;1068:143–64. doi: 10.1196/annals.1346.016. [DOI] [PubMed] [Google Scholar]
- 18.Shinohara M, Takayanagi H. Novel osteoclast signaling mechanisms. Curr Osteoporos Rep. 2007;5:67–72. doi: 10.1007/s11914-007-0005-1. [DOI] [PubMed] [Google Scholar]
- 19.Moscat J, Diaz-Meco MT, Wooten MW. Signal integration and diversification through the p62 scaffold protein. Trends Biochem Sci. 2007;32:95–100. doi: 10.1016/j.tibs.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Roux S, Brown J. Osteoclast apoptosis in rheumatic diseases characterized by a high level of bone resorption (osteoporosis, rheumatoid arthritis, myeloma and Paget’s disease of bone) Curr Rev Rheumatol. 2009;5:98–110. [Google Scholar]
- 21.Houde N, Chamoux E, Bisson M, Roux S. Transforming growth factor-beta1 (TGF-beta1) induces human osteoclast apoptosis by up-regulating Bim. J Biol Chem. 2009;284:23397–404. doi: 10.1074/jbc.M109.019372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brandwood CP, Hoyland JA, Hillarby MC, Berry JL, Davies M, Selby PL, et al. Apoptotic gene expression in Paget’s disease: a possible role for Bcl-2. J Pathol. 2003;201:504–12. doi: 10.1002/path.1463. [DOI] [PubMed] [Google Scholar]
- 23.Geetha T, Wooten MW. Structure and functional properties of the ubiquitin binding protein p62. FEBS Lett. 2002;512:19–24. doi: 10.1016/s0014-5793(02)02286-x. [DOI] [PubMed] [Google Scholar]
- 24.Cavey JR, Ralston SH, Hocking LJ, Sheppard PW, Ciani B, Searle MS, et al. Loss of ubiquitin-binding associated with Paget’s disease of bone p62 (SQSTM1) mutations. J Bone Miner Res. 2005;20:619–24. doi: 10.1359/JBMR.041205. [DOI] [PubMed] [Google Scholar]
- 25.Najat D, Garner T, Hagen T, Shaw B, Sheppard PW, Falchetti A, et al. Characterization of a non-UBA domain missense mutation of sequestosome 1 (SQSTM1) in Paget’s disease of bone. J Bone Miner Res. 2009;24:632–42. doi: 10.1359/jbmr.081204. [DOI] [PubMed] [Google Scholar]
- 26.Duran A, Serrano M, Leitges M, Flores JM, Picard S, Brown JP, et al. The atypical PKC-interacting protein p62 is an important mediator of RANK-activated osteoclastogenesis. Dev Cell. 2004;6:303–9. doi: 10.1016/s1534-5807(03)00403-9. [DOI] [PubMed] [Google Scholar]
- 27.Collet C, Michou L, Audran M, Chasseigneaux S, Hilliquin P, Bardin T, et al. Paget’s disease of bone in the French population: novel SQSTM1 mutations, functional analysis, and genotype–phenotype correlations. J Bone Miner Res. 2007;22:310–7. doi: 10.1359/jbmr.061106. [DOI] [PubMed] [Google Scholar]
- 28.Babu JR, Geetha T, Wooten MW. Sequestosome 1/p62 shuttles polyubiquitinated tau for proteasomal degradation. J Neurochem. 2005;94:192–203. doi: 10.1111/j.1471-4159.2005.03181.x. [DOI] [PubMed] [Google Scholar]