Abstract
FGF-16 has been reported to be preferentially expressed in the adult rat heart. We have investigated the expression of FGF-16 in the perinatal and postnatal heart and its functional significance in neonatal rat cardiac myocytes. FGF-16 mRNA accumulation was observed by quantitative RT-PCR between neonatal days 1 and 7, with this increased expression persisting into adulthood. FGF-2 has been shown to increase neonatal rat cardiac myocyte proliferative potential via PKC activation. Gene array analysis revealed that FGF-16 inhibited the upregulation by FGF-2 of cell cycle promoting genes including cyclin F and Ki67. Furthermore, the CDK4/6 inhibitor gene Arf/INK4A was upregulated with the combination of FGF-16 and FGF-2 but not with either factor on its own. The effect on Ki67 was validated by protein immunodetection, which also showed that FGF-16 significantly decreased FGF-2-induced Ki67 labeling of cardiac myocytes, although it alone had no effect on Ki67 labeling. Inhibition of p38 MAPK potentiated cardiac myocyte proliferation induced by FGF-2 but did not alter the inhibitory action of FGF-16. Receptor binding assay showed that FGF-16 can compete with FGF-2 for binding sites including FGF receptor 1. FGF-16 had no effect on activated p38, ERK1/2, or JNK/SAPK after FGF-2 treatment. However, FGF-16 inhibited PKC-α and PKC-ε activation induced by FGF-2 and, importantly, IGF-1. Collectively, these data suggest that expression and release of FGF-16 in the neonatal myocardium interfere with cardiac myocyte proliferative potential by altering the local signaling environment via modulation of PKC activation and cell cycle-related gene expression.
Keywords: fibroblast growth factor, protein kinase C, Ki67
FGF-16, A member of the fibroblast growth factor family, is a 207 amino acid protein containing a core region of 120 amino acids that binds heparin and the FGF receptor (FGFR; Ref. 33). It shares 75 and 62% amino acid sequence similarity with its closest relatives FGF-9 and FGF-20, respectively (26, 32). FGF-16 is highly conserved between species and has been cloned from human, rat, mouse, chicken, and zebrafish (30). FGF-16 was originally characterized in the rat and shown to be expressed preferentially in brown adipose tissue in the embryo (26) and linked subsequently with proliferation of brown adipocytes (18) and limb bud development in zebrafish (30). Interestingly, FGF-16 induced migration, but not proliferation, of primary human endothelial cells in culture (1). Even so, recent studies (20, 21) have implicated FGF-16 in cardiac myocyte proliferation and the development of coronary vasculature in the embryonic mouse. Evidence for the role of FGF-16 in cardiovascular development is so far indirect, as the work reported explored the phenotype of mice lacking FGF-9 or FGFR-1 and/or FGFR-2; however, FGF-16 appears to be part of the FGF signaling network acting downstream of Wnt/β-catenin signaling that is required for expansion of anterior heart field progenitor cells (8). Beyond this, however, no functional studies of FGF-16, especially its role in postnatal heart, have been presented.
In the rat, FGF-16 expression in the late embryo is found predominantly in brown adipose tissue, with a faint signal in the heart. By contrast, in the adult, RNA was detected preferentially in the heart by RNA blotting with only a faint signal in brown adipose tissue (26). With the exception of this intriguing pattern of expression, there are no reports as to 1) which cardiac cell types (myocytes vs. nonmyocytes) are responsible for the production of FGF-16 in the postnatal heart, 2) when expression occurs (perinatal vs. adult), 3) whether the protein is released, and 4) what its function is in this context. Here, we have begun to address these issues and found that adult accumulation of FGF-16 RNA is associated with an induction of FGF-16 expression in myocardial cells and the release of FGF-16 from cardiac myocytes during the early postnatal period. This period is associated with significant changes in the proliferative potential of cardiac myocytes (22), and our data indicate that FGF-16 may influence this transition through interference with other growth factor activated cell signaling.
MATERIALS AND METHODS
Animals
All animals were housed and treated according to standards and guidelines set by the Canadian Council for Animal Care. The investigation conforms to the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health (NIH Publication No. 85–23, revised 1996). The protocol for primary culture of neonatal rat ventricular myocytes was approved by the Bannatyne Campus Protocol Management and Review Committee at the University of Manitoba.
Primary cell culture
Neonatal rat cardiac myocytes were isolated as described previously (9). Cells were fractionated by centrifugation on a HEPES-buffered Percoll (40.5 and 58.5% by volume) gradient. Myocytes (lower layer) were plated on collagen-coated plates at a density of 1 million cells/60-mm diameter plate or 0.65 million/ 35-mm plate in media consisting of Ham’s F-10 (Sigma-Aldrich, Oakville, ON, Canada) supplemented with 10% FBS, 10% horse serum, and 1× Penstrep antibiotic. Nonmyocytes (upper layer) were plated in defined media consisting of DMEM F-12 (GIBCO Invitrogen, Mississauga, ON, Canada), supplemented with 0.05% FBS, 0.66% by volume albumin (Sigma-Aldrich), 1× Penstrep, 115 μM ascorbic acid (Sigma-Aldrich), and 1× insulin transferrin selenium (GIBCO Invitrogen). The next day (~20 h later) the cells were conditioned with defined media for 24 h. Then, the medium was replenished with fresh defined media containing the appropriate concentration of recombinant FGF-2 (Upstate Biotechnology, Lake Placid, NY) and/or FGF-16 (Peprotech, Rocky Hill, NJ) for 24 h in Gene-Array and Ki67 Labeling Index Assay and 2–20 min for signal transduction assays.
Conditioned media from cultures
Media were allowed to condition for 48 h, harvested, and supplemented with 0.2% Tween 20 (Bio-Rad Laboratories, Missisauga, ON, Canada) and protease inhibitors (one protease inhibitor cocktail tablet from Roche Diagnostics per 50 ml). A slurry of washed heparin Sepharose beads (Pfizer-Pharmacia, Calgary, AB, Canada) was then added to the media (100 μl/50 ml) and incubated, with rocking, for 2 h at room temperature. The beads were pelleted and washed with 250 mM NaCl and then 100 mM NaCl, each buffered with 10 mM Tris · HCl pH 7.0, 1 mM EDTA, and protease inhibitors. The final pellet was subjected to 15% SDS-PAGE.
For deglycosylation after extraction, the final 100-mM NaCl wash of the beads was done in the absence of protease inhibitors. One microliter of Endo HF glycosidase enzyme (New England Biolabs, Pickering, ON, Canada) was added per 50-μl sample and then incubated for 1 h at 37°C.
Real-time reverse transcription-PCR
Rat tissue RNA (age related) was obtained from Zyagen Laboratories (San Diego, CA). Total RNA from cells was isolated using the RNeasy mini plus RNA extraction kit (Qiagen, Missassauga, ON, Canada), and cDNA was synthesized by reaction with MMLV reverse transcriptase (Invitrogen, Burlington, ON, Canada) and oligo-dT-primers according to standard protocols. Real-time quantitative RT-PCR was performed on an iCycler (Bio-Rad) in 30-μl reactions using platinum Taq DNA polymerase (Invitrogen) and gene-specific primers for FGF-16 (forward: 5′-GAGGAGAGCT-GTTTGGATCG-3′; reverse 5′-GGGTGAGCCGTCTTTATTCA-3′; 155-bp amplicon) and GAPDH (forward: 5′-ATCCCGCTAAC-ATCAAATGG-3′; reverse: 5′-GTGGTTCACACCCATCACAA-3′; 170-bp amplicon). The baseline was set automatically, and the cycle threshold value was measured during the exponential phase of the amplification. The efficiency of amplification of each pair of primers was determined by serial dilutions of templates, and both were >0.99. Plasmids containing the amplicon sequences for FGF-16 and GAPDH were used to quantitate DNA concentration and to determine the relative FGF-16 expression level in each sample [expression level = (ng FGF-16 per μg cDNA)/(ng GAPDH per μg cDNA)]. Tests were run in duplicate on three separate PCR reactions.
RNA blotting
RNA blotting was done essentially as previously described (41). Hybridization with radiolabeled murine FGF-16 cDNA (XhoI fragment, 624 bp, Genbank Accession No. AF292104) was done in NorthernMax prehybridization/hybridization buffer (Ambion) at 42°C for 24 h, and then the blot was washed and exposed to Biomax film.
Protein immunoblotting
All samples for SDS-PAGE were mixed 1:5 with 5× loading buffer (10% SDS, 1.5 M dithiothreitol, and 0.3 M Tris · HCl pH 6.8) and boiled for 5 min before loading on a SDS-PAGE gel (8–15%), followed by transfer to a polyvinylidine fluoride (PVDF) membrane (Immobilon, Millipore, Billerica, MA). Membranes were blocked for 1 h in TBS-T (10 mM Tris · HCl pH 8.0 or 7.6, 150 mM NaCl, and 0.5% vol/vol Tween-20) containing 5% skim milk powder. The blot was then incubated with primary (FGF-16) antibody or normal rabbit IgG (Sigma-Aldrich) for 1 h at room temperature, washed, and incubated with secondary antibody (horse-radish peroxidase) for 1 h. After further washing, the membrane was incubated with Pico West Supersignal and exposed to film (Amersham Biosciences, Piscataway, NJ). Rabbit FGF-16 antibodies were generated against a synthetic peptide (ALNKDGSPREGYRTKRHQK) conjugated to keyhole limpet hemocyanin (44) and purified by Quality Controlled Biochemicals (Hopkinton, MA).
Quantitative (real-time) RT-PCR-based gene array assay
To analyze cell cycle gene regulation, a commercially available targeted cDNA array of 84 cell cycle regulatory genes, five housekeeping genes, as well as RNA quality control, and RT and PCR efficiency controls (Cell Cycle PCR Array, SuperArray Bioscience, Frederick, MD) were used on a Bio-Rad iCycler according to the instructions of the manufacturer. Complementary DNA was synthesized using SuperArray RT first strand kit. The baseline and threshold values for the real-time PCR were defined manually and were kept the same across the PCR array. Only results without genomic DNA contamination and no RT inhibition were calculated. Data were generated from three independent cardiac myocyte preparations (n = 3) as described by and analyzed with the Excel-based PCR Array Data Analysis Template provided by the manufacturer. Details of the mathematical analysis are given in Table 1.
Table 1.
Cell cycle related gene expression after FGF-2 and/or FGF-16 treatment in neonatal cardiac myocytes
| Genes | Category | FGF-2 | FGF-16 | FGF-2 + FGF-16 |
|---|---|---|---|---|
| Ccnf (cyclin F) | 6 | 2.80* | 1.26 | 1.83 |
| Mki67 (Ki67) | 2 | 2.22* | 1.28 | 1.49 |
| Arf/INK4A | 5/7 | 1.25 | 1.20 | 2.38* |
Categories of functional gene grouping: 1, G1 phase and G1/S transition; 2, S phase and DNA replication; 3, G2 phase and G2/M transition; 4, M phase; 5, cell cycle checkpoint and cell cycle arrest; 6, regulation of the cell cycle; 7, negative regulation of cell cycle (www.superarray.com). Cultures of neonatal cardiomyocytes were treated with vehicle, FGF-2 (1 ng/ml), FGF-16 (100 ng/ml), or FGF-2 combined with FGF-16 for 24 h, followed by extraction of total RNA. Real-time RT-PCR was carried out on a 96-gene mini array. Data are mean fold change of mRNA expression in treatment group vs. control from 3 separate experiments. Gene expression level, expressed as inverse relationship to threshold cycle (Ct) and doubling of amount of product with every cycle, is calculated using the equation L = 2−Ct. After correction using housekeeping gene expression, relative expression level for each treatment group is given by [L(GOI)experiment/L(HKG)experiment]/[L(GOI)control/L(HKG)control], where L is gene expression level, GOI is gene of interest, and HKG is housekeeping gene average value. When calculated using L = 2−Ct, relative expression is equivalent to 2−ΔΔCt.
Numbers pass dual criteria where 1) gene expression level is >200% of control and 2) P < 0.01, when treatment is compared with control by repeated measures ANOVA and the Dunnett posttest for comparisons of multiple treatments to a common control.
Ki67 labeling index assay
Cells were fixed using 1% paraformaldehyde in PBS at 4°C for 15 min. Primary antibodies [Ki67 (Vector Laboratories, Burlington, ON), 1:1,000; α-actinin (Sigma-Aldrich), 1:500] were applied in PBS with 1% BSA overnight at 4°C, followed by secondary antibodies [FITC or Texas red, conjugated, 1:100 (Jackson Laboratory, Bar Harbor, ME)] at room temperature for 1.5 h, then counter stained with Hoechst-33342 (Calbiochem-Behring, San Diego CA), and mounted on slides. Cells were counted using a confocal microscope (12–15 fields per slide) to determine the proportion of α-actinin-positive cells that also stained for Ki67 or labeling index (LI).
Receptor binding assay
Isolated cardiac myocytes or H9c2 myoblasts (wild type or stably transfected with FGFR-1; Ref. 42) were plated onto 96-well plates at a density of 3 × 104 cells per well. After 24 h, the cells were washed twice with binding buffer (300 mM sucrose, 100 mM NaCl, 50 mM HEPES, 1.2 mM CaCl2, 1.2 mM MgCl2, and 0.5% BSA). Iodinated FGF-2 (60 pM, corresponding to 1 ng/ml) was added to each well along with cold competitor (FGF-2 or FGF-16), and the cells were incubated for 1 h at 37°C and then washed with binding buffer (2×), 2 M NaCl, and 20 mM HEPES. Cells were lysed on ice in two changes of 0.5 M NaOH and 0.1% Triton X-100 for 10 min each and were transferred to glass vials. Scintillation fluid was added, and counts were read after 1 h at room temperature. The counting efficiency (cpm/dpm) was calculated to correct for quenching (15, 48), and the results were interpreted with the assumption that there was no preferential internalization of any receptor under these conditions (i.e., that all receptors are represented in the assay).
Signal transduction
For assessment of phosphorylated MAPK, cells were washed (2×) with ice-cold PBS and flash frozen with liquid nitrogen. For processing, 0.5 ml of lysis buffer (50 mM Tris · HCl pH 6.8, 10 mM NaF, 2% SDS, 25 mM β-glycerophosphate, 1 mM Na3O4V, 1 mM EDTA, 1 mM EGTA, and protease inhibitor) was added. Cells were removed with a rubber policeman, sonicated briefly, and centrifuged to remove insoluble material. MAPK phosphorylation/activation in the supernatant was assessed by protein immunoblotting as described in Protein immunoblotting using a phosphor-specific MAPK sampler kit (Cell Signaling Technology, Boston, MA).
For assessment of PKC levels, cells were fractionated into cytosolic and membrane components after growth factor stimulation (1 ng/ml FGF-2, 100 ng/ml FGF-16, and/or 10 nM IGF-1); IGF-1 was a generous gift from S. Mishra (University of Manitoba). Cells were mechanically lysed (23-G needle) in homogenization buffer and spun to remove nuclei, and after further centrifugation (186,000 g for 2 h), the supernatant was collected as the cytosolic fraction. The pellet was resuspended in PBS with 0.5% Triton X-100 and protease inhibitors, homogenized, and sonicated briefly. After incubation on ice for 30 min and further centrifugation (154,000 g for 30 min), the supernatant was collected and assayed as the membrane fraction. PKC levels in fractions were assessed by SDS-PAGE and protein immunoblotting as described in Protein immunoblotting. Antibodies against PKC-α and PKC-ε were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Statistical analysis
Statistical analysis was performed using Instat by GraphPad. Multiple column comparisons were achieved using one-way or, in the case of the PCR gene array data, repeated measures ANOVA followed by the appropriate posttest: Tukey-Kramer for comparison of all columns, Bonferroni for the comparison of selected columns, and Dunnett for comparison of multiple columns to a common control value.
RESULTS
RNA expression profile for FGF-16 in perinatal and post-natal heart
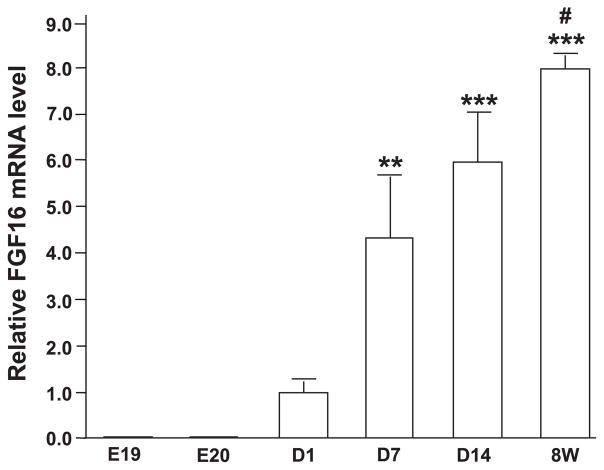
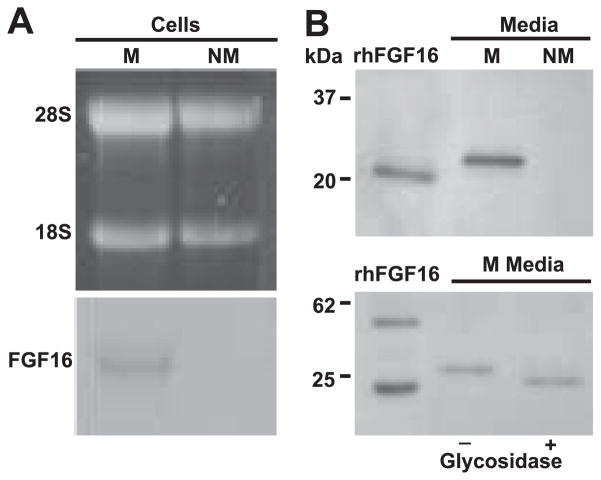
The temporal expression of FGF-16 was detected by real-time RT-PCR from total RNA of rat heart at late embryonic days 16 and 20 as well as postnatal days 1, 7, and 14, and week 8 (adult). In contrast to the very low FGF-16 expression at late embryonic stages, a marked increase in FGF-16 mRNA accumulation was observed to begin between postnatal days 1 and 7 and then continuing to increase into adulthood (Fig. 1). To further examine FGF-16 in neonatal cardiac cells, RNA isolated from neonatal rat cardiac myocytes and nonmyocyte cells was probed with the FGF-16 cDNA. A transcript of the expected size (1.8 kb) was detected in cardiac myocytes but not nonmyocyte RNA (Fig. 2A). To investigate FGF-16 protein and evidence of release, heparin-binding proteins were isolated from conditioned media collected from each culture type, fractionated by SDS-PAGE, and assessed by immunoblotting. A band of 26.5 kDa (estimated based on size markers) was seen in the cardiac myocyte but not the nonmyocyte media (Fig. 2B, top). The size of the band was larger than that predicted (23.7 kDa) based on the amino acid sequence of murine FGF-16. Treatment of conditioned medium protein from neonatal cardiac myocytes with glycosidase resulted in a reduction of product size from 26.5 to 24.1 kDa, with the same mobility as human recombinant FGF-16 (Fig. 2B, bottom).
Fig. 1.
FGF-16 mRNA expression during the perinatal and postnatal period in rat heart. Total RNA from late embryonic days (E19, E20); postnatal days (D1, D7, and D14); as well as 8-wk-old adult hearts (8W) were reverse transcribed into cDNA, followed by real-time PCR using platinum Taq DNA polymerase and gene-specific primers for FGF-16 and GAPDH; all FGF-16 values were normalized to GAPDH expression. Data are means ± SE. Values for E19 and E20 are (16.46 ± 7.189) × 10−3 and (17.90 ± 3.855) × 10−3, respectively. **P < 0.01 and ***P < 0.001, compared with E19; #P < 0.05, compared with D7 (n ≥ 3) by one-way ANOVA with the Bonferroni posttest for selected multiple comparisons.
Fig. 2.
FGF-16 is produced by neonatal cardiac myocytes in culture. A: total RNA (40 μg) isolated from cell cultures enriched for either myocytes (M) or nonmyocytes (NM) was visualized with ethidium bromide (top) and then blotted and probed for FGF-16 transcripts. A 1.8-kb FGF-16 transcript (relative to the 28S and 18S ribosomal RNA bands) was detected in myocytes but not nonmyocytes. B, top: neonatal rat cardiac cell cultures enriched for myocytes and nonmyocytes were processed and conditioned media from each cell type were enriched for heparin binding proteins. Samples were separated by SDS-PAGE and probed using custom polyclonal FGF-16 antiserum. A band of 26.5 kDa was seen in myocytes but not nonmyocytes. (B, bottom). Conditioned media were incubated with glycosidase buffer in the absence (−) and presence (+) of enzyme. Fractions were separated by SDS-PAGE and probed with commercial FGF-16 antibody. Recombinant human (rh) FGF-16 (10 ng) and mobilities of markers (B, left) are included as specificity or size controls, respectively. B, bottom, left lane: rhFGF-16 appears as both a monomer and a dimer.
FGF-16 modulates FGF-2-induced cell cycle gene expression in neonatal cardiac myocytes
The induction of FGF-16 expression in the early postnatal heart correlates with the decline of cardiac myocyte proliferative potential at early postnatal stages, raising the possibility of FGF-16 regulation of cardiac myocyte growth. To determine the influence of FGF-16 on the cardiac myocyte cell cycle, a real-time RT-PCR-based commercial gene array was used. This gene array assays a total of 84 genes in 7 functional categories that include positive and negative regulation of the cell cycle, phase transitions, checkpoints, and DNA replication (see Table 1; the online version of this article also contains supplemental data with a complete list of genes and their functional categories). Neonatal rat cardiac myocytes in culture were treated with FGF-2 (1 ng/ml) for a dual purpose: 1) to act as a positive control for cell cycle entry (36), and 2) as a means to test the effect of FGF-16 against a background of enhanced proliferative potential. Two criteria were used to define an “effect”: the average response from three independent experiments involving separate myocyte cultures must be 1) increased or decreased by at least twofold, and 2) statistically significant (treatment vs. control) after paired ANOVA (P < 0.05). As shown in Table 1, FGF-2 (1 ng/ml) treatment upregulated several genes that promote the cell cycle, including cyclin F (2, 5, 17) and Ki67 (40). FGF-16 alone, even at a high concentration (100 ng/ml), had no marked effect on any cell cycle gene compared with control. However, at this concentration, FGF-16 abrogated the FGF-2-induced increase of cyclin F and Ki67 expression. Furthermore, the CDK4/6 inhibitor gene Arf/INK4A (31, 37) was upregulated by the combined effect of FGF-2 and FGF-16.
FGF-16 interferes with FGF-2-induced Ki67 labeling in neonatal cardiac myocytes
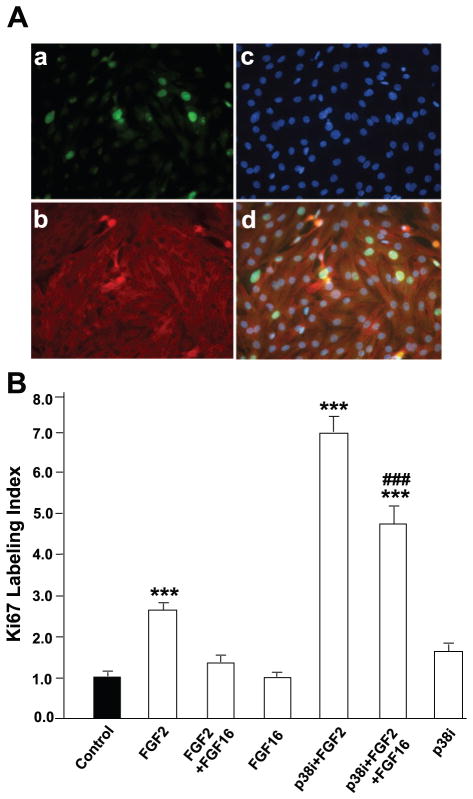
Ki67 is present during all active phases of the cell cycle but not in quiescent (G0) cells (40), making it an indicator of cell cycle entry. To validate the gene array at the protein level, Ki67 expression was assessed through immunodetection in α-actinin-positive cells (Fig. 3A). The Ki67 LI is defined as the fraction (percentage) of α-actinin-positive cells counted in a microscopic field that also stained for Ki67. The absolute value for the LI in untreated cardiac myocytes was 1.68 ± 0.24% (n = 67 fields). Consistent with the gene array assay, FGF-16 treatment alone had no effect on the Ki67 LI. However, FGF-16 decreased FGF-2-induced Ki67 LI significantly (Fig. 3B).
Fig. 3.
FGF-16 interferes with FGF-2-induced Ki67 labeling index in neonatal rat cardiomyocytes. A: cells cultured on coverslips were treated with 1 ng/ml FGF-2 and 100 ng/ml FGF-16 in the presence or absence of 10 μM p38 inhibitor (p38i, SB203580) for 24 h, fixed, and stained simultaneously with anti-Ki67 antibody (a), α-actinin (b), and Hoechst 33342 staining for detection of nuclei (c); d is merged image of a, b, and c. In this manner, the Ki67 labeling index was determined as the fraction (percentage) of Ki67-positive myocytes (identified by α-actinin staining). B: labeling index was counted in 59–67 fields for each treatment group, and value for control (untreated) cells was set to 1.0. Absolute means ± SE for control group (n = 67) are 1.68 ± 0.24%. ***P < 0.001, compared with control; ###P < 0.001, compared with FGF-2 treatment in the presence of p38 inhibitor, using one-way ANOVA and Tukey-Kramer posttest for multiple comparisons.
Inhibition of p38 MAPK was shown to potentiate or enhance proliferation of postnatal cardiac myocytes after treatment with FGF-1 (13), thereby implicating elevated p38 MAPK in the lack of proliferative potential for cardiac myocytes. To test whether 1) FGF-2 can substitute for FGF-1, and 2) p38 MAPK mediates the interference of FGF-16 with FGF-2-induced cardiac myocyte growth, cells were treated with FGF-2 with p38 inhibitor (SB203580, 10 μM; Ref. 13), with or without FGF-16. The Ki67 LI was determined as described in FGF-16 interferes with FGF-2-induced Ki67 labeling in neonatal cardiac myocytes. The combination of FGF-2 with p38 inhibition increased the Ki67 LI significantly over FGF-2 or inhibitor treatment alone (Fig. 3B). However, when FGF-16 was added to cultures treated with FGF-2 and p38 inhibitor, there was a significant (but not complete) loss of proliferative potential, indicated by an decrease in the Ki67 LI (Fig. 3B).
FGF-16 can compete with FGF-2 for binding to cell surface FGFRs
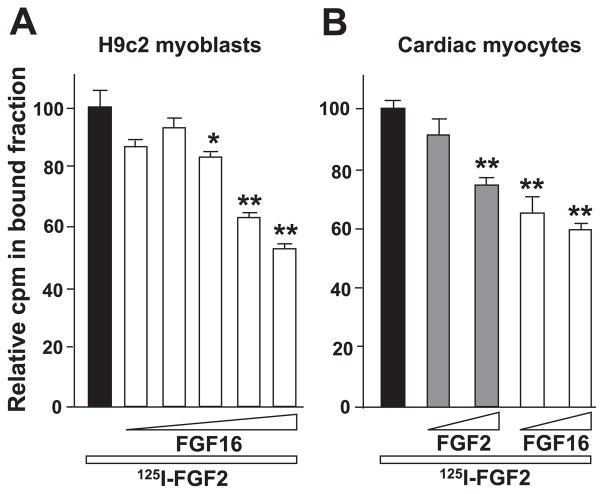
Proliferation of cardiac myocytes by FGF-2 has been linked to signaling via the cell surface receptor FGFR-1 (41, 45). We have shown previously that overexpression of FGFR-1 increased cell division both in rat myoblast H9c2 cells stably transfected with FGFR-1 (42) and in primary cardiac myocytes (41). To assess binding of FGF-16 to FGFR-1, the ability of unlabeled recombinant human (rh) FGF-16 to compete with binding of 125I-FGF-2 to H9c2 cells (wild type and transfected) was investigated. Effective binding and competition were not observed with incubation at 4°C for 2 h, so the time and temperature were adjusted to 1 h at 37°C to balance the optimization of binding with the minimization of receptor internalization. With increasing concentrations of cold FGF-16, the amount of 125I-FGF-2 binding to H9c2/FGFR-1 cells (but not wild-type H9c2 cells) was decreased significantly. Binding of iodinated FGF-2 to H9c2/FGFR-1 cells was competed with control (wild-type H9c2 cell) levels with 295-, 606-, and 1,227-fold molar excess of FGF-16 (Fig. 4A). Similar competition was observed with unlabeled rhFGF-2 (data not shown). The high concentration of cold FGF needed to achieve visible competition with 125I-FGF-2 was not unexpected, as an increased affinity of iodinated growth factor for its receptor was reported previously (23, 28). To confirm the presence of FGF-2/FGF-16 binding sites in neonatal rat cardiac myocytes, cultures were incubated with 125I-FGF-2 in the presence of unlabeled FGF-2 or FGF-16 competitor (100- and 1,000-fold molar excess). Both FGF-2 and FGF-16 competed in a specific manner (Fig. 4B). The possibility of receptor internalization is acknowledged but does not change the observation that both cold FGF-2 and cold FGF-16 competed with 125I-FGF-2 for binding sites in a dose-dependent manner.
Fig. 4.
A: FGF-16 competes with FGF-2 for binding specifically at FGF receptor (R)-1. H9c2 myoblasts stably transfected with FGFR-1 were incubated with 125I-labeled FGF-2 (1.0 ng/ml) at 37°C for 1 h in the presence of increasing amounts of unlabeled FGF-16 (representing 77-, 154-, 295-, 606-, and 1,227-fold molar excess). Absolute value for control mean ± SE was 1,164.25 ± 161.13 cpm (n = 4). *P < 0.05 and **P < 0.01, compared with control (Dunnett posttest). B: FGF-16 competes with FGF-2 for receptor binding in cardiac myocytes. Cardiac myocytes were incubated with 125I-labeled FGF-2 (1.1 ng/ml) in the presence of 100- or 1,000-fold molar excess of cold competitor (FGF-2 or FGF-16, as indicated). Relative counts per minute values are shown for each group. Absolute means ± SE for control group (no competition) are 411.5 ± 30 (n = 6). **P < 0.01, compared with control (Dunnett posttest).
FGF-16 interferes with PKC activation by FGF-2
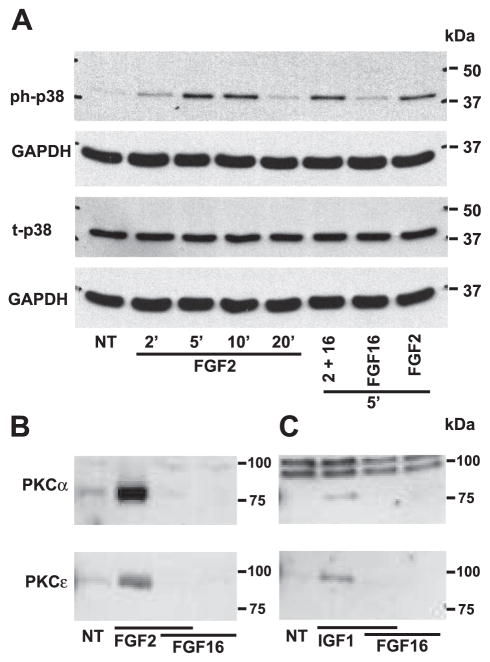
FGF-2 triggers multiple signaling pathways in cardiac myocytes including both MAPK and PKC (10). To investigate the effect of FGF-16 on MAPK activation, protein immunoblotting was performed for both total and phosphorylated p38 MAPK, ERK1/2, and JNK/SAPK expression after treatment with FGF-2 or FGF-16, alone or in combination. No change in total p38 levels was observed with any treatment (Fig. 5A). A time course of treatment with 1 ng/ml FGF-2 indicated that 5 min was a point of maximal p38 phosphorylation (Fig. 5A). At this time point, FGF-16 had a minimal but detectable effect on p38 phosphorylation compared with control, but no further effect was evident when used in combination with FGF-2, compared with FGF-2 alone (Fig. 5A). FGF-2 had a similar effect on other MAPKs, including ERK1/2, and JNK/SAPK, but FGF-16 had no effect, either on its own or on the effect of FGF-2 (data not shown).
Fig. 5.
FGF-16 modulates signal transduction in neonatal rat cardiomyocytes. A: cells were treated with 1 ng/ml FGF-2 for 2, 5, 10, and 20 min, or 1 ng/ml FGF-2 and 100 ng/ml FGF-16 for 5 min, or 100 ng/ml FGF-16 alone for 5 min. Cell extracts were analyzed by protein immunoblotting for phosphorylated p38 MAPK (ph-p38) and total p38 (t-p38), and each blot was stripped and reprobed with GAPDH as a control for gel loading. B: membrane (sarcolemmal) fractions from cells treated with FGF-2 (1 ng/ml) and/or FGF-16 (100 ng/ml) for 5 min were assessed by protein immunoblotting for PKC-α (80 kDa) or PKC-ε (90 kDa). C: same experiment as in B except for growth factor treatment with IGF-1 (10 nM) and/or FGF-16 (100 ng/ml) for 2 min. NT, no treatment. Positions of molecular mass markers (kDa) are shown at right.
FGF-2 is known to stimulate PKC isoforms, including the conventional (calcium-dependent) PKC-α (43) and the novel (calcium-independent) PKC-ε (12). The levels of PKC-α and PKC-ε in the cytosolic vs. membrane fraction were detected after FGF-2/FGF-16 treatment by SDS-PAGE and protein immunoblotting. The 80-kDa PKC-α and 90-kDa PKC-ε bands were detected in the cytosolic fraction, but, although present at relatively high levels, there was no obvious change by FGF treatments (not shown). By contrast, FGF-2 but not FGF-16 increased PKC-α and PKC-ε levels in the membrane fraction (Fig. 5B). However, when 100 ng/ml FGF-16 was added in combination with 1 ng/ml FGF-2, the observed increase was lost (Fig. 5B). In addition, we found that treatment with FGF-16 at lower concentrations (10 ng/ml and in 2 of 3 replicates using 1 ng/ml) also resulted in significant inhibition of FGF-2-induced PKC activation, suggesting a dose-dependent effect of FGF-16 (data not shown).
To test whether the ability of FGF-16 to modulate PKC activity was specific to FGF-2/FGFR and consistent with receptor competition, IGF-1 was used to replace FGF-2 for PKC activation (14). Treatment with 10 nM IGF-1 for 2 min increased membrane PKC levels (Fig. 5C). Combined treatment with FGF-16 (100 ng/ml) inhibited the increase in both membrane PKC-α and PKC-ε levels (Fig. 5C).
DISCUSSION
The preferential expression of FGF-16 in the adult heart appears to be a unique characteristic among members of the FGF family (26). In this study, we provide evidence that FGF-16 RNA expression is induced in the early postnatal myocardium and its protein is produced and released from neonatal cardiac myocytes. It is generally accepted that cardiac myocytes lose their ability to divide shortly after birth (27). The mechanism, however, is poorly understood, particularly as growth factors such as FGF-2 and IGF-1 are implicated in cardiac myocyte proliferation during embryonic development (19, 38) and they continue to be present in the postnatal period to adulthood (10, 39). The timing of FGF-16 expression in the postnatal heart corresponds with cardiac myocyte exit from the cell cycle (22) and suggests a role in postnatal heart development. Our data indicate that FGF-16 can interfere with the proliferative potential of neonatal cardiac myocytes when they are stimulated by exogenous FGF-2 treatment, even though FGF-16 alone has no visible effect on cell cycle gene expression. FGF-2 increases neonatal rat cardiac myocyte proliferative potential, and specifically DNA synthesis, via PKC-ε activation (16). Although FGF-16 could compete for FGF-2 receptor binding, our data are also consistent with FGF-16 exerting its effect further downstream on PKC signaling, as FGF-16 can also interfere with PKC activation by IGF-1. Furthermore, our gene array indicated effects on the expression of Arf/INK4A only upon the combination of FGF-16 with FGF-2, but not with either FGF-2 or FGF-16 independently, suggesting a multireceptor-mediated effect and cell signaling cross talk between FGF-2 and FGF-16. In contrast to PKC activation, we were not able to link the inhibitory effect of FGF-16 with MAPK activation in spite of evidence that p38 is directly involved in the inhibition of cardiac myocyte proliferation (13). Regardless, these data indicate that induction of FGF-16 in the mammalian myocardium at birth is expected to alter the local signaling environment and its activity on cell cycle-related gene expression is consistent with the reduction in proliferation seen in the neonatal heart.
Transfection and overexpression of FGF-16 in both insect (Sf9) and mammalian (COS-1) cells suggested that the protein is secreted by way of an uncleaved signal sequence in two parts: a hydrophobic region at the N terminus and one in a more central location in a manner similar to that described for FGF-9 (24–26). Our data, however, are the first to demonstrate release of the endogenous glycosylated FGF-16 protein from a mammalian cell (Fig. 2B). While glycosylation is an important posttranslational modification with a wide variety of functional consequences (46), experiments from our laboratory using cell survival as an endpoint suggest that recombinant FGF-16 and glycosylated FGF-16 (from conditioned media of transfected cells) do not differ in their biological activity (44). In addition, glycosylation does not appear to influence the activity of other FGFs (3, 4, 6) and recombinant (unglycosylated) FGF-16 showed biological activity in embryonic cardiac myocytes (21). The choice of recombinant FGF-16 for use in the work presented here allows for more precise control of dosage and eliminates other factors present in conditioned media that may complicate or mask experimental results.
Expression of FGF-16 in embryonic myocardium (embryonic days 10.5 and 12.5) was previously reported to be restricted to epicardial and endocardial cells (21). Release of FGF-16, however, could affect adjacent cardiac myocytes in the myocardium. Exposure of the embryonic mouse myocardium to recombinant FGF-16 (in the form of soaked beads applied to living tissue in culture) increased DNA synthesis and, thus, the proliferative potential of cardiac myocytes (21). This activity is consistent with the ability of FGF-16 to stimulate brown adipocyte proliferation during embryonic development (18). While embryonic cardiac myocytes do not appear to significantly express FGF-16 but may respond positively to FGF-16 in terms of proliferative potential, neonatal cardiac myocytes not only express and release FGF-16 but FGF-16 also appears to negatively regulate their growth. Our gene array data indicate that FGF-16 inhibits FGF-2-stimulated expression of cyclin F, a protein that promotes G2 (2) and G2/M transition in the cell cycle by regulating cyclin B1 localization to the nucleus (5, 17). FGF-16 also inhibits FGF-2-induced Ki67 expression, a marker of cell cycle activity expressed in G1, S, G2, prophase, and metaphase but not in quiescent (G0) cells (40). Furthermore, the CDK4/6 inhibitor Arf/INK4a (p16), an important negative regulator of G1 progression (31, 37), was upregulated by the combination of FGF-16 and FGF-2. These effects of FGF-16 on cell cycle gene expression indicate its interference with the growth-promoting effect of FGF-2.
Our data suggest that FGF-16 by itself may not be sufficient to activate a signal transduction pathway; however, it may be involved in shaping the influence of other stimuli or growth factors. In cardiac myocytes, PKC-α and PKC-ε have been associated with proliferation induced by growth factors, including FGF-2 (11, 16) and IGF-1 (7), as well as cardioprotection (35). In a human fibroblast cell line, PKC inhibition has been reported to arrest cells in G1 via accumulation of the cyclin-dependent kinase inhibitor p27 (29). PKC thus represents an important mediator of cell signaling in cardiac myocytes through which FGF-16 could affect changes to proliferative potential in postnatal cells.
Previously, we have shown that functional FGF-2 receptors are present in the postnatal heart (10, 23) and FGFR activity in cardiac cells is traditionally associated with PKC activation (10). The ability of FGF-16 to interfere with FGF-2-induced proliferative potential and/or membrane levels of PKCs in cardiac myocytes might be explained by ligand (FGF-16 for FGF-2) competition for a cell surface receptor (FGFR). Three sets of observations, however, indicate this explanation alone may not be sufficient. First, FGF-16 in combination with FGF-2 upregulated Arf/INK4A, but neither FGF-16 nor FGF-2 alone significantly affected Arf/INK4A expression, as indicated by our cell cycle gene array (Table 1). Secondly, while FGF-2 was able to stimulate MAPKs (ERK1/2, JNK/SAPK, and p38), competition for this effect through FGF-16 treatment was not observed, suggesting that the effect of FGF-16 is exerted downstream of ligand-receptor interaction. Finally, FGF-16 was able to interfere with membrane PKC-α and PKC-ε levels induced by IGF-1 as well, a growth factor that activates different receptors than FGF-2. It is therefore apparent that the action of FGF-16 is not solely dependent on receptor competition with FGF-2. Its own unique effect and signaling pathway independent of FGF-2 may be attributed to the considerable complexity of FGF/FGFR interactions. Both FGF-16 and FGF-2 can bind with high affinity to more than one receptor subtype (34, 47), but the relative strength of activation for each differed. This may lead to functional differences that are critical for the regulation of both physiological and pathological processes. Differentiation of receptor action is complicated by the large number of splicing variants and FGFR isoforms and the fact that truly selective inhibition of receptor isoforms is simply not feasible.
Another possible mechanism for FGF-16 inhibition of FGF-2-stimulated growth was suggested by the negative regulation of cardiac myocyte proliferation by p38 (13). Although a slight effect of FGF-16 on its own was observed (Ref. 44; Fig. 5A), we did not observe any regulation of cell cycle gene expression by FGF-16 itself and were unable to link the inhibitory regulation of FGF-16 on FGF-2-induced growth to p38 MAPK activation. We found p38 inhibition enhanced the FGF-2-induced Ki67 LI; however, it did not abolish the inhibitory effect of FGF-16 (Fig. 3). Consistent with this, immunoblot assays showed that FGF-16 did not change FGF-2-induced p38 activation (Fig. 5). The growth response of cardiac myocytes, then, appears to involve a complex array of signaling pathways the activity of which is determined not only by the presence of ligands that directly activate them but also by ligands that may also serve to potentiate (or suppress) the growth response under certain physiological (i.e., developmental) or pathological conditions. Indeed, the suggestion that FGF-16 may be a negative regulator for postnatal cardiac growth may translate to the potential to interfere with the pressure-induced hypertrophic process mediated by other growth factors (including FGF-2). The prospect of an antihypertrophic effect of FGF-16 is very intriguing, particularly from a clinical point of view. As systems are used to further study the function of FGF-16 in vivo, this concept will be explored further.
In summary, FGF-16 expression increases in the myocardium at birth where this heparin-binding protein has the capacity to be produced and released in a glycosylated form and to modify the signaling environment. The appearance of FGF-16 in the postnatal heart correlates with a change in the proliferative potential of cardiac myocytes. A role for FGF-16 in this transition is suggested by its ability to interfere with the proliferation potential of neonatal cardiac myocytes through a mechanism involving modulation of PKC activation and an effect on cell cycle-related gene expression.
Acknowledgments
GRANTS
This work was funded by a grant from the Canadian Institutes of Health Research (MOP-62742). S. Y. Lu is the recipient of a Manitoba Institute of Child Health Postdoctoral Fellowship.
References
- 1.Antoine M, Wirz W, Tag CG, Gressner AM, Wycislo M, Muller R, Kiefer P. Fibroblast growth factor 16 and 18 are expressed in human cardiovascular tissues and induce on endothelial cells migration but not proliferation. Biochem Biophys Res Commun. 2006;346:224–233. doi: 10.1016/j.bbrc.2006.05.105. [DOI] [PubMed] [Google Scholar]
- 2.Bai C, Richman R, Elledge SJ. Human cyclin F. EMBO J. 1994;13:6087–6098. doi: 10.1002/j.1460-2075.1994.tb06955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bates B, Hardin J, Zhan X, Drickamer K, Goldfarb M. Biosynthesis of human fibroblast growth factor-5. Mol Cell Biol. 1991;11:1840–1845. doi: 10.1128/mcb.11.4.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellosta P, Talarico D, Rogers D, Basilico C. Cleavage of K-FGF produces a truncated molecule with increased biological activity and receptor binding affinity. J Cell Biol. 1993;121:705–713. doi: 10.1083/jcb.121.3.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bicknell KA, Coxon CH, Brooks G. Forced expression of the cyclin B1-CDC2 complex induces proliferation in adult rat cardiomyocytes. Biochem J. 2004;382:411–416. doi: 10.1042/BJ20031481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clements DA, Wang JK, Dionne CA, Goldfarb M. Activation of fibroblast growth factor (FGF) receptors by recombinant human FGF-5. Oncogene. 1993;8:1311–1316. [PubMed] [Google Scholar]
- 7.Clerk A, Aggeli IK, Stathopoulou K, Sugden PH. Peptide growth factors signal differentially through protein kinase C to extracellular signal-regulated kinases in neonatal cardiomyocytes. Cell Signal. 2006;18:225–235. doi: 10.1016/j.cellsig.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Cohen ED, Wang Z, Lepore JJ, Lu MM, Taketo MM, Epstein DJ, Morrisey EE. Wnt/beta-catenin signaling promotes expansion of Isl-1-positive cardiac progenitor cells through regulation of FGF signaling. J Clin Invest. 2007;117:1794–1804. doi: 10.1172/JCI31731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Detillieux KA, Meij JT, Kardami E, Cattini PA. Alpha1-adrenergic stimulation of FGF-2 promoter in cardiac myocytes and in adult transgenic mouse hearts. Am J Physiol Heart Circ Physiol. 1999;276:H826–H833. doi: 10.1152/ajpheart.1999.276.3.H826. [DOI] [PubMed] [Google Scholar]
- 10.Detillieux KA, Sheikh F, Kardami E, Cattini PA. Biological activities of fibroblast growth factor-2 in the adult myocardium. Cardiovasc Res. 2003;57:8–19. doi: 10.1016/s0008-6363(02)00708-3. [DOI] [PubMed] [Google Scholar]
- 11.Doble BW, Dang X, Ping P, Fandrich RR, Nickel BE, Jin Y, Cattini PA, Kardami E. Phosphorylation of serine 262 in the gap junction protein connexin-43 regulates DNA synthesis in cell-cell contact forming cardiomyocytes. J Cell Sci. 2004;117:507–514. doi: 10.1242/jcs.00889. [DOI] [PubMed] [Google Scholar]
- 12.Doble BW, Ping P, Kardami E. The epsilon subtype of protein kinase C is required for cardiomyocyte connexin-43 phosphorylation. Circ Res. 2000;86:293–301. doi: 10.1161/01.res.86.3.293. [DOI] [PubMed] [Google Scholar]
- 13.Engel FB, Schebesta M, Duong MT, Lu G, Ren S, Madwed JB, Jiang H, Wang Y, Keating MT. p38 MAP kinase inhibition enables proliferation of adult mammalian cardiomyocytes. Genes Dev. 2005;19:1175–1187. doi: 10.1101/gad.1306705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foncea R, Galvez A, Perez V, Morales MP, Calixto A, Melendez J, Gonzalez-Jara F, Diaz-Araya G, Sapag-Hagar M, Sugden PH, LeRoith D, Lavandero S. Extracellular regulated kinase, but not protein kinase C, is an antiapoptotic signal of insulin-like growth factor-1 on cultured cardiac myocytes. Biochem Biophys Res Commun. 2000;273:736–744. doi: 10.1006/bbrc.2000.3008. [DOI] [PubMed] [Google Scholar]
- 15.Jordan WC, Spiehler V, Haendiges R, Helman EZ. Evaluation of alternative counting methods for radioimmunoassay of hepatitis-associated antigen (HB-Ag) Clin Chem. 1974;20:733–737. [PubMed] [Google Scholar]
- 16.Kardami E, Banerji S, Doble BW, Dang X, Fandrich RR, Jin Y, Cattini PA. PKC-dependent phosphorylation may regulate the ability of connexin43 to inhibit DNA synthesis. Cell Commun Adhes. 2003;10:293–297. doi: 10.1080/cac.10.4-6.293.297. [DOI] [PubMed] [Google Scholar]
- 17.Kong M, Barnes EA, Ollendorff V, Donoghue DJ. Cyclin F regulates the nuclear localization of cyclin B1 through a cyclin-cyclin interaction. EMBO J. 2000;19:1378–1388. doi: 10.1093/emboj/19.6.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konishi M, Mikami T, Yamasaki M, Miyake A, Itoh N. Fibroblast growth factor-16 is a growth factor for embryonic brown adipocytes. J Biol Chem. 2000;275:12119–12122. doi: 10.1074/jbc.275.16.12119. [DOI] [PubMed] [Google Scholar]
- 19.Laustsen PG, Russell SJ, Cui L, Entingh-Pearsall A, Holzenberger M, Liao R, Kahn CR. Essential role of insulin and insulin-like growth factor 1 receptor signaling in cardiac development and function. Mol Cell Biol. 2007;27:1649–1664. doi: 10.1128/MCB.01110-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavine KJ, White AC, Park C, Smith CS, Choi K, Long F, Hui CC, Ornitz DM. Fibroblast growth factor signals regulate a wave of Hedgehog activation that is essential for coronary vascular development. Genes Dev. 2006;20:1651–1666. doi: 10.1101/gad.1411406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J, Ornitz DM. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev Cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Li F, Wang X, Capasso JM, Gerdes AM. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J Mol Cell Cardiol. 1996;28:1737–1746. doi: 10.1006/jmcc.1996.0163. [DOI] [PubMed] [Google Scholar]
- 23.Liu L, Pasumarthi KB, Padua RR, Massaeli H, Fandrich RR, Pierce GN, Cattini PA, Kardami E. Adult cardiomyocytes express functional high-affinity receptors for basic fibroblast growth factor. Am J Physiol Heart Circ Physiol. 1995;268:H1927–H1938. doi: 10.1152/ajpheart.1995.268.5.H1927. [DOI] [PubMed] [Google Scholar]
- 24.Miyakawa K, Hatsuzawa K, Kurokawa T, Asada M, Kuroiwa T, Imamura T. A hydrophobic region locating at the center of fibroblast growth factor-9 is crucial for its secretion. J Biol Chem. 1999;274:29352–29357. doi: 10.1074/jbc.274.41.29352. [DOI] [PubMed] [Google Scholar]
- 25.Miyakawa K, Imamura T. Secretion of FGF-16 requires an uncleaved bipartite signal sequence. J Biol Chem. 2003;278:35718–35724. doi: 10.1074/jbc.M300690200. [DOI] [PubMed] [Google Scholar]
- 26.Miyake A, Konishi M, Martin FH, Hernday NA, Ozaki K, Yamamoto S, Mikami T, Arakawa T, Itoh N. Structure and expression of a novel member, FGF-16, on the fibroblast growth factor family. Biochem Biophys Res Commun. 1998;243:148–152. doi: 10.1006/bbrc.1998.8073. [DOI] [PubMed] [Google Scholar]
- 27.Nadal-Ginard B, Kajstura J, Leri A, Anversa P. Myocyte death, growth, and regeneration in cardiac hypertrophy and failure. Circ Res. 2003;92:139–150. doi: 10.1161/01.res.0000053618.86362.df. [DOI] [PubMed] [Google Scholar]
- 28.Neufeld G, Gospodarowicz D. The identification and partial characterization of the fibroblast growth factor receptor of baby hamster kidney cells. J Biol Chem. 1985;260:13860–13868. [PubMed] [Google Scholar]
- 29.Nishi K, Schnier JB, Bradbury EM. The accumulation of cyclin-dependent kinase inhibitor p27kip1 is a primary response to staurosporine and independent of G1 cell cycle arrest. Exp Cell Res. 1998;243:222–231. doi: 10.1006/excr.1998.4166. [DOI] [PubMed] [Google Scholar]
- 30.Nomura R, Kamei E, Hotta Y, Konishi M, Miyake A, Itoh N. Fgf16 is essential for pectoral fin bud formation in zebrafish. Biochem Biophys Res Commun. 2006;347:340–346. doi: 10.1016/j.bbrc.2006.06.108. [DOI] [PubMed] [Google Scholar]
- 31.Nozato T, Ito H, Watanabe M, Ono Y, Adachi S, Tanaka H, Hiroe M, Sunamori M, Marum F. Overexpression of cdk Inhibitor p16INK4a by adenovirus vector inhibits cardiac hypertrophy in vitro and in vivo: a novel strategy for the gene therapy of cardiac hypertrophy. J Mol Cell Cardiol. 2001;33:1493–1504. doi: 10.1006/jmcc.2001.1412. [DOI] [PubMed] [Google Scholar]
- 32.Ohmachi S, Watanabe Y, Mikami T, Kusu N, Ibi T, Akaike A, Itoh N. FGF-20, a novel neurotrophic factor, preferentially expressed in the substantia nigra pars compacta of rat brain. Biochem Biophys Res Commun. 2000;277:355–360. doi: 10.1006/bbrc.2000.3675. [DOI] [PubMed] [Google Scholar]
- 33.Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2:REVIEWS3005. doi: 10.1186/gb-2001-2-3-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M. Receptor specificity of the fibroblast growth factor family. J Biol Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 35.Padua RR, Merle PL, Doble BW, Yu CH, Zahradka P, Pierce GN, Panagia V, Kardami E. FGF-2-induced negative inotropism and cardioprotection are inhibited by chelerythrine: involvement of sarcolemmal calcium-independent protein kinase C. J Mol Cell Cardiol. 1998;30:2695–2709. doi: 10.1006/jmcc.1998.0832. [DOI] [PubMed] [Google Scholar]
- 36.Pasumarthi KB, Kardami E, Cattini PA. High and low molecular weight fibroblast growth factor-2 increase proliferation of neonatal rat cardiac myocytes but have differential effects on binucleation and nuclear morphology. Evidence for both paracrine and intracrine actions of fibro-blast growth factor-2. Circ Res. 1996;78:126–136. doi: 10.1161/01.res.78.1.126. [DOI] [PubMed] [Google Scholar]
- 37.Pei XH, Xiong Y. Biochemical and cellular mechanisms of mammalian CDK inhibitors: a few unresolved issues. Oncogene. 2005;24:2787–2795. doi: 10.1038/sj.onc.1208611. [DOI] [PubMed] [Google Scholar]
- 38.Pennisi DJ, Ballard VL, Mikawa T. Epicardium is required for the full rate of myocyte proliferation and levels of expression of myocyte mitogenic factors FGF2 and its receptor, FGFR-1, but not for transmural myocardial patterning in the embryonic chick heart. Dev Dyn. 2003;228:161–172. doi: 10.1002/dvdy.10360. [DOI] [PubMed] [Google Scholar]
- 39.Ren J, Samson WK, Sowers JR. Insulin-like growth factor I as a cardiac hormone: physiological and pathophysiological implications in heart disease. J Mol Cell Cardiol. 1999;31:2049–2061. doi: 10.1006/jmcc.1999.1036. [DOI] [PubMed] [Google Scholar]
- 40.Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 41.Sheikh F, Fandrich RR, Kardami E, Cattini PA. Overexpression of long or short FGFR-1 results in FGF-2-mediated proliferation in neonatal cardiac myocyte cultures. Cardiovasc Res. 1999;42:696–705. doi: 10.1016/s0008-6363(99)00008-5. [DOI] [PubMed] [Google Scholar]
- 42.Sheikh F, Jin Y, Pasumarthi KB, Kardami E, Cattini PA. Expression of fibroblast growth factor receptor-1 in rat heart H9c2 myoblasts increases cell proliferation. Mol Cell Biochem. 1997;176:89–97. [PubMed] [Google Scholar]
- 43.Sheikh F, Sontag DP, Fandrich RR, Kardami E, Cattini PA. Overexpression of FGF-2 increases cardiac myocyte viability after injury in isolated mouse hearts. Am J Physiol Heart Circ Physiol. 2001;280:H1039–H1050. doi: 10.1152/ajpheart.2001.280.3.H1039. [DOI] [PubMed] [Google Scholar]
- 44.Sontag DP. PhD thesis. Winnipeg, Canada: University of Manitoba; 2005. Characterization of Fibroblast Growth Factor-16 Expression and Biological Function in the Heart. [Google Scholar]
- 45.Speir E, Tanner V, Gonzalez AM, Farris J, Baird A, Casscells W. Acidic and basic fibroblast growth factors in adult rat heart myocytes. Localization, regulation in culture, and effects on DNA synthesis. Circ Res. 1992;71:251–259. doi: 10.1161/01.res.71.2.251. [DOI] [PubMed] [Google Scholar]
- 46.Varki A. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao LR, Navalitloha Y, Singhal S, Mehta J, Piao CS, Guo WP, Kessler JA, Groothuis DR. Hematopoietic growth factors pass through the blood-brain barrier in intact rats. Exp Neurol. 2007;204:569–573. doi: 10.1016/j.expneurol.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]