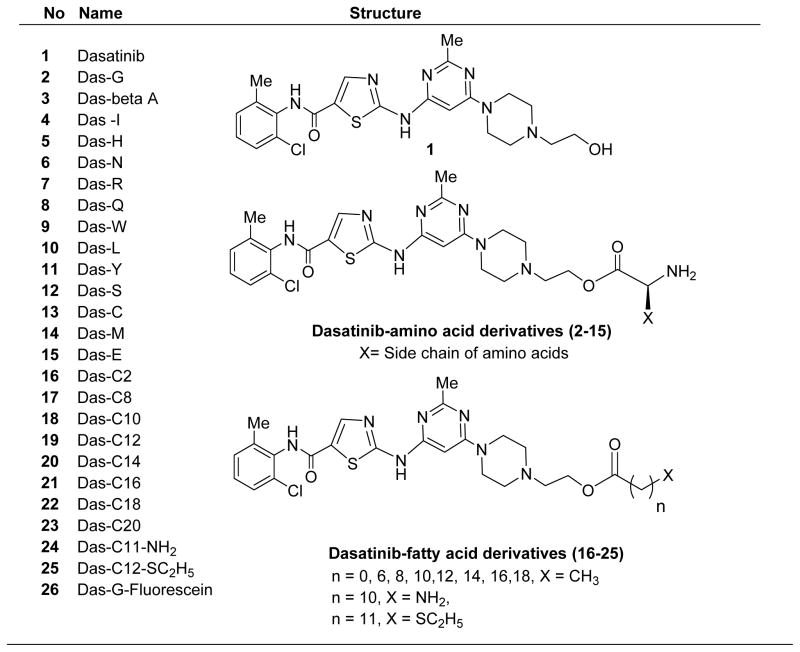
Abstract
Derivatives of dasatinib were synthesized via esterification with 25 carboxylic acids including amino acids and fatty acids by extending the inhibitor to interact with more diverse sites and to improve specificity. Dasatinib-L-arginine derivative (Das-R, 7) was the most potent of the inhibitors tested with IC50 values of 4.4 nM, <0.25 nM, and <0.45 nM against Csk, Src, and Abl kinases, respectively. The highest selectivity ratio obtained in our study, 91.4 Csk/Src belonged to compound 18 (Das-C10) with an IC50 of 3.2 μM for Csk compared to 35 nM for Src. Furthermore, many compounds displayed increased selectivity toward Src, as compared with Abl. Compounds 15 (Das-E) and 13 (Das-C) demonstrated the largest gains (10.2 and 10.3 Abl/Src IC50 ratios). Das-R (IC50 = 2.06 μM) was significantly more potent than Das (IC50 = 26.3 μM) against Panc-1 cells while they both showed an IC50 < 51.2 pM against BV-173 and K562 cells. Molecular modeling and binding free energy simulations revealed a good agreement with the experimental results and rationalized differences in selectivity of the studied compounds. Integration of experimental and computational approaches in the design and biochemical screening of dasatinib derivatives facilitated rational engineering and diversification of dasatinib scaffold, providing useful insights into mechanisms of kinase selectivity.
Keywords: amino Acid, dasatinib, fatty acid, inhibitor, tyrosine kinase
Dasatinib-Amino Acid and Dasatinib Fatty Acid Conjugates
A number of dasatinib conjugates (25 compounds) were synthesized to improve specificity towards Src, Csk, and Abl kinases. We reasoned that by extending dasatinib from the ATP binding pocket into variable region, the derivatives obtain new kinase-specific interactions.

Introduction
Protein tyrosine kinases (PTKs) are involved in numerous pathologies, including many cancers. Furthermore, PTK inhibitors have proven to be invaluable research and therapeutic tools. Ideally, such molecules would be able to target the aberrant kinases without interfering with normal physiological kinase signaling. Current PTK inhibitors as ATP-binding site inhibitors have limited specificity1, most likely due to the high degree of conservation within their targets’ ATP binding pockets. This problem stems from a lack of structural diversity in the ATP binding pocket, similar in all kinases. The potent tyrosine kinase inhibitor (TKI) dasatinib, N-(2-chloro-6-methylphenyl)-2-((6-(4-(2-hydroxyethyl)piperazin-1-yl)-2-methylpyrimidin-4-yl)amino)thiazole-5-carboxamide (BMS-354825), is a case in point. It binds to the ATP binding site of many PTKs and is a potent broad spectrum PTK inhibitor.2
The discovery of dasatinib was announced by Bristol-Myers Squibb in 2004 as an orally administered anticancer compound.3 In 2006, the company was granted FDA tentative approval for dasatinib, via the accelerated review process.4 Initially, approved indications for use applied only to adult patients diagnosed with either Philadelphia chromosome-positive (Ph+) chronic myeloid leukemia (CML) in the chronic phase, or Ph+ acute lymphoblastic leukemia (ALL), who were intolerant or resistant to standard imatinib (Gleevec, by Novartis) treatment. In 2010, full approval was granted for the treatment of imatinib-resistant Ph+ ALL/CML, once further safety and efficacy were established. This extended the approved indications to include less developed Ph+ CML, still in the accelerated or blast phases.5
The Ph+ profile of many ALL/CML cancers is readily observed via standard karyotype, as it involves a large and very specific translocation between chromosomes 9 and 22.6 This chromosomal aberration induces abnormal expression of a BCR (Breakpoint Cluster Region, 22q11.21) – Abl (9q34.12) fusion-protein. The fusion of the two proteins disrupts the normal regulatory mechanisms of each. In particular, the loss of the N-terminal domains of Abl removes key components of substrate recognition, cellular localization, and catalytic inactivation. The truncated kinase is responsible for creating a highly distorted pattern of phosphorylation, upsetting normal cell signaling dynamics, and leading to the oncogenic progression of ALL or CML. TKIs, such as imatinib and dasatinib, inhibit cancer progression by blocking the kinase activities of BCR-Abl and/or other kinases that are known to contribute oncogenic signaling, such as Src Family Kinases (SFK).
Dasatinib inhibits SFKs, BCR-Abl, c-KIT, EphA2, PDGFR-beta kinases with nanomolar or sub-nanomolar IC50.5 It has also been crystalized as a bound ligand to the Abl, BMX, BTK, EphA4, Lyn, p38alpha, p38MAP, and Src kinases.7–14 The common manner in which it binds to each of these kinases, suggests it should be similar for many other kinases, too. This may, in part, explain side-effects of the compound at clinical dosage. Moreover, the clinical dosage, itself, may be limited by toxicity due to off-target kinase inhibition. This can lead to the persistence of cancer in small, drug-resistant populations and, ultimately relapse of patients who were previously in remission. In some patients, mutations arise during the course of treatment, or before. In all of these cases, where resistance has developed, alternative therapies must be sought.
The problems associated with balancing toxicity with effective dosage are not unique to dasatinib. All currently available TKIs are expected to lose effectiveness at some point to restart disease progression. From this perspective, it seems obvious that greater kinase specificity should be a top priority in the development of novel therapeutic inhibitors. However, it seems likely that many kinases share too much similarity to reasonably expect true specificity from ATP analogs, even from drugs with the potency of a molecule like dasatinib.
Outside of the ATP-binding cleft of PTKs there are several regions of great variability amongst kinases, as demonstrated in Figure 1. We reasoned that by extending dasatinib from the ATP binding pocket into these variable regions, the derivatives may obtain new kinase-specific interactions, which would lead to inhibitory specificity toward a given kinase. This concept is further strengthened by the finding that the loop that connects α-helixes D and E in Csk harbors the key residues of a substrate-docking site that is unique to the Csk family kinases, Csk and Chk.15–16 Conveniently, dasatinib binds to the kinase in such a way that its hydroxyl group is located just outside of the ATP-binding cleft and very close to the Csk substrate-docking site. This observation led to the strategy of modifying the hydroxyl group of dasatinib with various functional groups in the hope that some of them will interact with the substrate docking site or nearby kinase-specific motifs, resulting in inhibitory specificity. It has been demonstrated previously that the hydroxyl group can be modified without affecting the interaction of dasatinib with the ATP-binding site, it can be used to facilitate attachment to chromatographic resins for affinity purification.17–19
Figure 1.
Dasatinib inhibition of kinase activity is mediated by molecular interaction with highly conserved residues from the ATP-binding cleft of the catalytic domain. (A) The MUSCLE aligned protein sequences from the catalytic domains of Abl, Csk, Src, and Lyn illustrate the level of conservation throughout various regions of the catalytic domain among these diverse PTKs. The darker shaded portions of the alignMent indicate greater conservation. The ligand-bound crystal structure of Lyn (B) demonstrates how dasatinib is docked within the catalytic domain. The inhibition of kinase activity is achieved via molecular interaction with amino acids of the ATP-binding cleft. These residues are displayed in stick form in the structure and also indicated by black arrowheads within the alignMent. The region of the catalytic domain between the end of αD-helix and the beginning of αE-helix is boxed within the alignMent and structure to highlight its variability and proximity to the binding site.
By the addition of small molecules to the dasatinib scaffold, it may be possible to extend the binding site to include regions of the kinase that are more diverse in order to impart greater potential PTK specificity. We hypothesized that specificity may be developed by extending the inhibitor out from the traditional ATP-binding pocket to interact with more diverse sites.
We sought to evaluate this strategy by linking dasatinib to a series of amino acids and fatty acids via esterification reactions. The selection of amino acids and fatty acids for conjugation with dasatinib was based on their diverse structures and physicochemical properties, and the presence of different functional groups. Following favorable molecular modeling results, we employed this technique to build a small library of 25 compounds (Figure 2 and Figures S1, S2, Supporting Information). Three non-receptor PTKs (Csk, Abl, and Src) were used for evaluating these compounds in terms of their potency and specificity. Several selected compounds were then further explored to reveal some unique properties and potential applications of these novel inhibitors.
Figure 2.
Dasatinib was used as a scaffold for the attachment of various amino acids and fatty acids.
Results
Chemistry
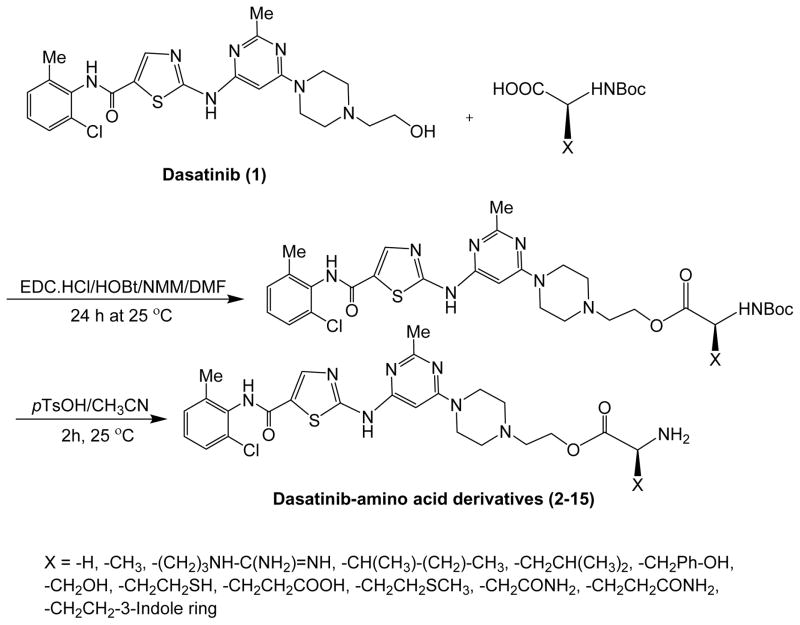
Schemes 1 and 2 depict the synthesis of amino acid and fatty acid conjugates of dasatinib by using an ester linkage. In brief, dasatinib was coupled to Boc-protected amino acids using N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC)/hydroxybenzotriazole (HOBt) and N-methylmorpholine (NMM) as coupling and activating reagents under an anhydrous condition using DMF as a solvent. The conjugation of fatty acids with dasatinib was conducted with under a similar condition using the same coupling and activating reagents. However, in the case of long-chain fatty acids, such as stearic acid and eicosanoic acid, the use of dichloromethane was needed to improve the solubility of fatty acids during the coupling reaction. Furthermore, for long chain fatty acids, more time (24–72 h) was required in the coupling reaction. The Boc protection group from the coupled amino acids derivatives was removed in the presence of p-toluenesulfonic acid (pTsOH) in acetonitrile. The crude products were purified by flash silica gel column chromatography. All synthesized compounds were characterized by using 1H and 13C nuclear magnetic resonance (NMR) spectroscopy and high-resolution time-of-flight electrospray mass spectrometry. The purity of final products (≥95%) was confirmed by reverse phase analytical HPLC. Synthesis yields ranged 80 to 91% for fatty acid conjugates and 20–84% for amino acid conjugates.
Scheme 1.
Synthesis of dasatinib-amino acid conjugates (2-15).
Scheme 2.
Synthesis of dasatinib-fatty acid conjugates (16-25).
Modeling to Establish the Theoretical Feasibility of Binding and Rule Out Steric Hindrance of Dasatinib Derivatives
In order to establish the theoretical feasibility of modeling experiments, we first conducted control docking simulations of dasatinib with the protein conformations obtained from the crystal structures of dasatinib complexes with Abl (pdb id 2GQG), Csk (pdb id 3QLG), Src (pdb id 3G5D), Lyn (pdb id 2ZVA), Btk (pdb id 3K54, 3OCT), Bmx (pdb is 3SXR), EphA4 (pdb id 2Y6O), and P38 kinase (pdb id 3KFA). The lowest energy docked poses of dasatinib complexes reproduced crystallographic inhibitor conformations within RMSD = 1.5 Å (root mean square deviation). The predicted structures of dasatinib complexes were subjected to 200 ns molecular dynamics (MD) simulations, and the obtained trajectories were subsequently used in MM-GBSA calculations of binding free energies (Figure S3, Supporting Information). For consistency of comparison with the experiment, we utilized the results of kinase inhibitors screening in in vitro competition binding assays that were followed by determination of quantitative dissociation constants Kd for high-affinity inhibitors.20–21
Molecular docking and binding free energy simulations validated the utility of the computational approach in rational design of dasatinib analogs by reproducing the experimental binding affinities of dasatinib across a spectrum of kinase targets. In particular, the computed binding affinities correctly discriminated weak Dasatinib binding to p38α kinase from significantly stronger binding with Abl, Src, and Lyn kinases.
Equipped with a good agreement with experimental results for a range of known crystal structures of dasatinib complexes, we embarked on modeling of the selected derivatives with Abl, Csk, and Src kinases to evaluate binding propensities of dasatinib derivatives and rule out a potential steric hindrance. In docking of all derivatives, we used the crystal structures of dasatinib complexes with Abl, Csk, and Src. As a control, the selected dasatinib derivatives were also docked into the crystal structure of the Lyn kinase10, which is a member of the clinically relevant SFK family. The resolution and completeness of the crystal structure of the Lyn kinase outside of the dasatinib binding pocket was important in our modeling, given that the additions to the core compound could potentially interact with a region outside the traditional binding pocket. We chose to model each of the 20 common amino acids, along with the addition of phosphotyrosine, as carboxyesters of dasatinib to establish the feasibility of our approach.
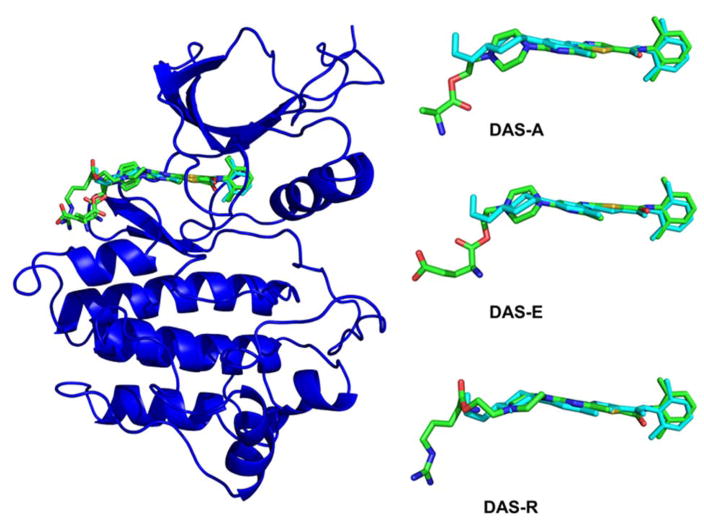
Combining accuracy and ease of use, we chose Autodock Vina to perform our in silico modeling. Autodock Vina’s performance in dockings has been relatively well established, and its algorithms require no assumptions to be made concerning the ionization of side-chains.22–23 The first feature that the models predict is the preferred orientation of the compounds that were attached to dasatinib. Despite allowance to rotate freely, almost all of the simulated dockings favor interaction with the region surrounding αD-helix, rather than the equally proximal ATP binding lobe of the kinase (Figure 3).
Figure 3.
The predicted docked poses of dasatinib derivatives DAS-A (3), DAS-E (15), and DAS-R (7) were superimposed with the crystallographic conformation of dasatinib. Crystal structure of dasatinib-bound Abl kinase (pdb id 2GQG, blue). The docked conformations of dasatinib derivatives to Abl are shown in sticks colored according to the atom type, and the crystallographic conformation of dasatinib bound to Abl is shown in cyan sticks.
All of the compounds were docked, and the top hits for each compound (Figure S4, Supporting Information) were subsequently subjected to 200 ns MD simulations. MM-GBSA calculations using the obtained trajectories provided binding free energies and compound ranking as reported in Figure S5 (Supporting Information).
The core dasatinib portion of each derivative was nearly identical and showed only small deviation from the original crystallized ligand after structural superposition. The binding free energies of dasatinib (compound 1) with Abl, Csk and Src were in good agreement with the experimental binding constants. The computed binding energetics of all derivatives were within 0.9 kcal/mol of the dasatinib biding free energy, indicating that the proposed chemical modifications could be well tolerated. The favorable binding energies of dasatinib derivatives (Figure S5, Supporting Information) suggested that these small molecules may potentially enhance dasatinib binding by maintain scaffold interactions andf making additional favorable contacts outside the ATP binding pocket.
Design and Synthesis
By utilizing computational predictions in rational design of dasatinib analogues, we explored chemical esterification with various amino acids as these adducts were predicted to extend out from the ATP binding pocket while preserving the crystallographic mode of dasatinib scaffold. Alongside these compounds, additional derivatives were synthesized by esterification with several fatty acids. These particular derivatives were chosen to evaluate their potential use as linkers to sites more distal to the ATP binding pocket. Using his approach, 25 compounds were successfully synthesized and purified including 14 amino acid and 10 fatty acid adducts of dasatinib (compounds 2-25). Compound 26 was later synthesized according to the previously reported procedure24 as a fluorescein-glycine ester to demonstrate further the utility of such chemical derivatives.
Compound Screening
Each of the amino acid and fatty acid conjugates of dasatinib was screened at a fixed concentration: 250 pM against Abl, and Src, and 8 nM against Csk. The respective concentrations were selected based on IC50 of dasatinib against each enzyme. Their percent inhibition relative to dasatinib was then determined. The results of this screening (Figure 4) offered insight as to the potency of each adduct, relative to dasatinib. Moreover, this relative profile could be compared between Abl, Csk, and Src kinases. Thus, this screening helped to highlight potential patterns of enhanced or diminished inhibition relating to the type of adduct.
Figure 4.
Results of screening of compounds at a fixed concentration against each of the panel PTKs. The activity of uninhibited enzyme was compared to dasatinib-inhibited enzyme activity to establish a baseline of inhibition. Each of the compounds were then compared to dasatinib inhibition for each enzyme. Bars above the x-axis show increased potency while those below indicate a decreased capacity to inhibit the kinase, relative to dasatinib. The error bars represent the SEM of at least three determinations.
In general, fatty acyl-derived compounds (16-25) dramatically decreased inhibitor potency. Among all of these compounds, only compound 16 (Das-C2) inhibited comparably to dasatinib. Each of the other fatty acid derivatives (17-25) exhibited less than 60% of dasatinib’s inhibition toward each kinase. Figure 4 shows that Das-C10, Das-C12, and Das-C11SC2H5 have inhibitor potencies in a similar range as compound 24. This screening profile suggests that hydrophobicity, not size, accounts for much of these inhibitors’ reduced potency. The size of compound 24 with the amino group is similar, in particular, to that of Das-C10 and Das-C12 which do not have polar groups. Furthermore, losses in potency due to long, hydrophobic chains may be recovered, at least in part, by linkage to a polar functional group (like –NH2).
The amino acid derived compounds (2-15) showed significantly greater potency over fatty acyl derivatives, with the exception of compound 13 with a cysteine residue (Das-C). Compound 15 containing a glutamic acid (Das-E) also displayed a significantly reduced potency when compared to other amino acid derivatives. Compound 7 containing an arginine residue (Das-R) appeared to be the most potent of the inhibitors tested against all three kinases (Figure 4). Interestingly, the side-chain guanidinium group of compound 7 (Das-R) was predicted to form a hydrogen bond with the backbone of the kinase, just outside the ATP-binding cleft.
The major changes in potency by our derivatives are interesting and perhaps informative but are not the main focus of our study. Since we endeavor to identify an active agent with altered kinase selectivity, the compounds’ variable potency between kinases was our greatest concern. Compounds 21 (Das-C16), 24 (Das-C11NH2), and 23 (Das-C20) showed a significantly decreased capacity to inhibit Abl, as compared to the other kinases (Figure 4). Compound 7 (Das-R), however, demonstrated its potency gains were greatest for Abl. Additionally, compound 16 (Das-C2) displayed significantly increased potency toward Csk when compared to Abl and Src. The screening assay’s sensitivity, however, limited the extraction of further selectivity results.
IC50 determination
The screening, while relatively rapid, is not incredibly sensitive and, thus, is limited in the task of identifying changes in kinase specificity. We further characterized the inhibition of selected compounds via IC50 assays toward each of the kinases. Compound 7 (Das-R) was chosen because it was one of the best compounds screened for each of the kinases in terms of potency and selectivity. We then chose compounds 3, 13, and 15 (Das-A, Das-C, Das-E) to use as a basis for comparison and to tease out potential differences in selectivity. For the fatty acid-derived compounds, we again chose to compare the most potent compound, 16 (Das-C2), one longer chained derivative (21, Das-C16), two moderately long-chained compounds (17/Das-C8, 18/Das-C10), and the potential linker compound 24 (Das-C11NH2) for further investigation.
The results of the IC50 assays, summarized in Table 1, reflect the potency of the compounds toward each of the PTKs. As predicted by the screenings, compound 7 (Das-R) displayed increased potency against all kinases with an average 37% decrease in IC50 compared to dasatinib. The greatest gain in potency was observed for Abl by compound 7 (-R) with an IC50 of <0.45 nM as compared to the <0.75 nM value for dasatinib, representing a 42% decrease. Conversely, compound 13 (Das-C) was the worst performing of the amino acid derivatives with IC50’s ranging from 1.7–5.3x higher than dasatinib toward our PTK panel. Interestingly, compound 15 (Das-E) performed about as well as compound 3 (Das-A), despite the latter showing possible signs of greater potency in the screenings.
Table 1.
Selected compounds were evaluated against each of the enzymes in our panel. Potency was measured via IC50 in standard P32 kinase assay with appropriate substrate. Selectivity is represented as the ratio of IC50 for one enzyme over another. The change in IC50, as compared to dasatinib, is calculated and shown in the right-most columns to illustrate how potency was affected.
| Compd No. | Name | IC50 [nM]#
|
Selectivity
|
Net change in IC50 (%Dasatinib)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Abl | Csk | Src | C/A | C/S | A/S | Abl | Csk | Src | Avg | ||
| 1 | Dasatinib | <0.78 | 7 | <0.37 | 9.0 | 18.9 | 2.1 | -- | -- | -- | -- |
| 7 | Das-R | <0.45 | 4.4 | <0.25 | 9.8 | 17.6 | 1.8 | −42 | −37 | −32 | −37 |
| 16 | Das-C2 | 2.1 | 5.5 | <0.33 | 2.6 | 16.7 | 6.4 | 169 | −21 | −11 | 46 |
| 3 | Das-A | 3 | 21 | <0.39 | 7.0 | 53.8 | 7.7 | 285 | 200 | 5 | 163 |
| 15 | Das-E | 4.4 | 20 | <0.43 | 4.5 | 46.5 | 10.2 | 464 | 186 | 16 | 222 |
| 24 | Das-C11-NH2 | 4.9 | 30 | 1 | 6.1 | 30.0 | 4.9 | 528 | 329 | 170 | 342 |
| 13 | Das-C | 68 | 500 | 6.6 | 7.4 | 75.8 | 10.3 | 8618 | 7043 | 1684 | 5782 |
| 17 | Das-C8 | 51 | 600 | 7.5 | 11.8 | 80.0 | 6.8 | 6438 | 8471 | 1927 | 5612 |
| 18 | Das-C10 | 220 | 3200 | 35 | 14.5 | 91.4 | 6.3 | 28105 | 45614 | 9359 | 27693 |
| 21 | Das-C16 | 680 | 5100 | 75 | 7.5 | 68.0 | 9.1 | 87079 | 72757 | 20170 | 60002 |
The standard errors were in the range of 0.034–0.095 nM for Src, 0.041–0.242 nM for Csk, and 0.041–0.109 nM for Abl. All kinase inhibition assays were performed in duplicate at least three times.
To garner more quantitative insight into potential gains or losses in enzyme selectivity, the IC50 of each compound toward a particular kinase can be compared to the IC50 of the same compound for another kinase, in the form of a ratio. The IC50 of dasatinib was 7 nM, <0.37 nM, and <0.78 nM against Csk, Src, Abl, respectively, giving a ratio of 18.9 Csk/Src and a ratio of 2.1 Abl/Src clearly indicating inhibiting both Abl and Src kinases in rather non-selective fashion. When comparing Abl and Src, however, a ratio of 2.1 A/S for dasatinib implies that the compound inhibits both kinases in rather non-selective fashion. In this manner, ratios greater than 10 or less than 0.1 reflect IC50s that differ by more than an order of magnitude, and the inhibitor is generally considered to be selective or specific for the kinase with the lower IC50. Our approach generated numerous inhibitory compounds with enhanced kinase selectivity.
Comparisons of the specificities for each of the compounds toward Csk shows that two compounds, 7 (Das-R) and 16 (Das-C2), showed little change in their preference for Src (17.6 A/S and 16.7 C/S, respectively). However, each of the remaining compounds increased their preference for Src dramatically with C/S IC50 ratios of 30.0 or higher. The highest selectivity ratio obtained in our study, 91.4 C/S, was from this group and belonged to compound 18 (Das-C10) with an IC50 for Csk = 3.2 μM compared to 35 nM for Src. This marks a difference of almost two orders of magnitude between the IC50 values. Additionally, each of these compounds displayed increased selectivity toward Src, as compared with Abl. Of particular note, compound 15 (Das-E) and compound 13 (Das-C) demonstrated the largest gains (10.2 and 10.3 A/S IC50 ratios). These two dasatinib derivatives represent the only bona-fide Src specific inhibitors in our dataset, as they possess IC50’s at least an order of magnitude higher for both Abl and Csk, and present a significant increase in kinase specificity when compared to dasatinib.
Modeling to Rationalize Differences in Specificity
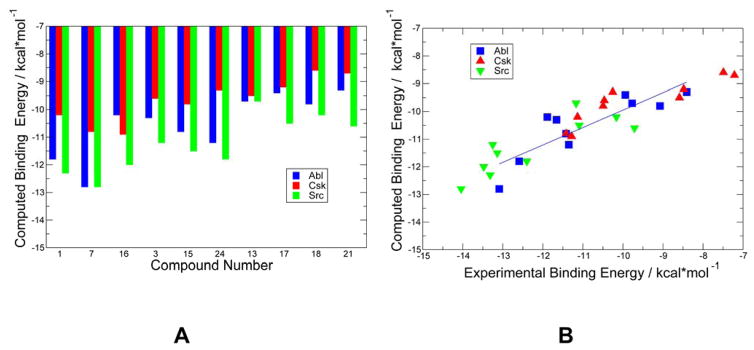
We used computational modeling to evaluate binding energetics and differences in selectivity for the panel of studied compounds (Table 1). In this analysis, 200 ns MD simulations followed by MM-GBSA binding free energy computations were performed separately for each of these 10 compounds with Abl, Csk, and Src kinases respectively. We focused our analysis on compounds 3, 7, and 15 (Das-A, Das-R, Das-E) as a basis for comparison. In agreement with the experimental data, the results indicated that compound 7 (Das-R) has a better affinity with Abl and Src kinases. Moreover, we noticed that compound 15 (Das-E) displayed a reduced potency when compared to other amino acid derivatives as was observed in the experiments, whereas compound 7 (Das-R) exhibited the strongest binding affinity against all three kinases. In addition, we also observed that the Csk/Src selectivity is markedly improved for compounds 7 and 15. This is also consistent with the experimental data, albeit the experimental Csk/Src selectivity ratio is greater for compounds 3 and 15. By extracting binding constants from the IC50 data using Cheng-Prusoff equation25–26, we correlated the experimental and computed binding free energies (Figure 5). Because the enzyme concentrations necessary for the assays were significantly higher than the true Ki’s, extracting Ki’s from IC50 values results in at best an estimate.
Figure 5.
(A) The computed binding free energies for a selected panel of compounds that were evaluated against Abl, Csk, and Src kinases. The original potencies that was measured in inhibition constants IC50 were converted into Ki values with the aid of Cheng-Prusoff equation.23,24 In these calculations, the enzyme concentration was set to 0.4 nM. (B) The correlation between experimental binding constants and computed binding free energies. The data for Abl are shown in blue squares, for Csk in red triangles-up, and for Src in green triangles-down.
A strong correspondence between computation and experiment with a correlation coefficient R ~ 0.892 provided support to modeling results and allowed for a preliminary analysis of structural basis underlying binding affinity and selectivity profiles of studied compounds. The computational analysis suggested that high-affinity dasatinib derivatives may benefit from forming favorable interactions with the αD-helix residues (Figure S6, Supporting Information). For instance, we observed that Das-A may approach the αD-helix in Abl and form stable hydrogen bonding with E329 and Y326 residues (Figure S6, Supporting Information). Similarly, Das-R appeared to maintain stable hydrogen bonding with the carbonyl oxygen of E329. Although only structural determination could provide the ultimate verification of these predictions, the binding energetics analysis based on the predicted docked poses and MD simulations reproduced experimental binding constants with an excellent accuracy (Figure 5). Particularly, computational analysis successfully predicted the Csk/Src selectivity trend seen in the studied compounds. According to our analysis, the observed differences in Csk/Src binding affinity can be partly attributed to a cumulative effect of the side-chain interactions with the αD-helix of the kinases. For instance, the differences in specific interactions with the αD-helix residues may be among important determinants underlying the observed Csk/Src selectivity of compound 7 (Das-R). Indeed, the side-chain guanidinium group of this dasatinib derivative may form strong and stable hydrogen bond interactions with E349 and E353 in Src kinase, whereas only some of these interactions may be preserved in Csk kinase (Figure S7, Supporting Information). According to our results, the αD-helix residues may contribute to the structural stability and favorable binding affinities of dasatinib derivatives with the panel of all studied kinases. The importance of the αD-helix residues in binding of dasatinib derivatives may also arise from their proximity and involvement in stabilization of the catalytic spine (C-spine) residues (V394, I392, and L346) (Figure S7, Supporting Information). The C-spine is involved in the positioning of the adenine ring of ATP that arranges the two lobes so that the catalytic residues are aligned optimally for catalysis.27–28
To explore the role of complementarity in determining the binding free energy differences we computed the average complementarity determined by the LPC program29 as complementarity function CF defined as:
Normalized complementarity is defined as CF divided by the solvent-accessible surface of the uncomplexed ligand. In this equation, S1 is the sum of all surface areas of all “desirable” atomic contacts between ligand and receptor. Si is the sum of all surface areas of all “undesirable” atomic contacts and E is the repulsion term as defined in reference.29
In general, energetically favorable contacts (e.g., hydrophobic-hydrophobic) are classified as legitimate contact, whereas energetically unfavorable contacts (e.g., hydrophobic-hydrophilic) are classified as illegitimate ones. More specifically, according to the LPC program, the “desirable” contacts include hydrophobic-hydrophobic, hydrophobic-aromatic, hydrophobic-neutral, donor-acceptor and other typical classes of favorable intermolecular contacts. A more complete description of this approach has been published28 and references therein. Normalized complementarity is defined as CF divided by the solvent-accessible surface of the uncomplexed ligand.
We observed that the complementarity of dasatinib and compound 7 (Das-R) is greater as compared to other compounds. Additionally, the Csk/Src differences in the normalized complementarity are greater than between Abl and Src kinases (Figure S8, Supporting Information).
Assaying the dynamic nature of dasatinib esters
Given the profound influence of drug metabolism on inhibitor efficacy, we sought to characterize how our synthesis strategy may impart a unique and favorable stability profile. In vivo, numerous carboxylesterases have been shown to hydrolyze ester bonds of compounds (similar to those in the dasatinib derivatives). Mouse plasma contains relatively high concentrations of carboxylesterases and frequently serve as proxies for the stability studies. Selected compounds, Das-C2 and Das-C8 were pre-treated with mouse plasma at 0, 15, 30, 60, and 120 min. If the derivative is a suitable substrate for one or more carboxylesterase, the products of the subsequent reaction should include the core dasatinib molecule. We used LC/MS/MS to monitor the formation of dasatinib. The assays relied upon the careful consideration of proper compound concentration and a significant difference in potency between the derivative and dasatinib.
As the dasatinib derivatives vary substantially in size and chemical properties, it is worth noting that stability of the derivatives likely differs between compounds, according to variations in substrate specificities of carboxylesterases. With this dynamic in mind, we chose representative compounds for the assays, namely 16 (Das-C2) and 17 (Das-C8). From the results, summarized in Supporting Information (Table S1 and Figure S9), it appears that compound 17 (Das-C8) is remarkably unaffected by carboxylesterases, even after 2 h pre-treatment. The stability of 17 (T1/2 > 480 min) was a high contrast to that of 16 (Das-C2), which showed to be less stable T1/2 < 5 min). Compound 16 (Das-C2) was completely metabolized in mouse plasma by the 15 min time point, and rapid dasatinib formation was also monitored from this compound by using HPLC and mass spectrometry. In contrast, Das-C8 (17) was observed to be stable in mouse plasma (and no dasatinib formation was seen at specified time). The difference can be due to the presence of larger fatty acyl chain in Das-C8 (17) that generated sterical hindrance. The dynamic offered by these esterified derivatives can be quite advantageously applied toward engineering a unique drug metabolic profile.
Development of a Fluorescent Dasatinib Derivative
To expand upon the vast potential offered by dasatinib derivatives, we sought to construct a fluorescent probe for in vitro characterization of kinases. Tethered to dasatinib by a glycine linker, a fluorescein-conjugated derivative was synthesized (compound 26) (Figure 2). The compound was assayed against Src, finding an IC50 of 1 nM. The affinity of the dasatinib-fluorescein compound was then measured using a fluorescence polarization binding assay.
In such an assay, polarized light of a specific wavelength is used to excite the fluorescein molecule, which in turn emits light at a higher wavelength. The degree to which the emitted light is depolarized depends on the free rotation of fluorescein, which is inversely correlated with ligand binding. Thus, the binding affinity of a fluorescein-ligand conjugate for a given receptor can be measured by recording the polarization of light in a serial dilution of the receptor and fitting the data to a standard ligand-binding curve.
We sought to measure the affinity of our ligand for a form of Src that is deficient in kinase activity (kdSrc). This mutant kinase is often able to be purified to significantly higher concentrations than active kinase, which adds to the convenience of the assay and broadens its potential application (as discussed later). The binding curve generated from these assays (shown in Figure S10A, Supporting Information) reveals an affinity of 15 nM toward kdSrc by our fluorescein-dasatinib conjugate (compound 26). This value was higher than expected and may reflect differences between active/inactive kinase forms or limitations of the assay’s lowest detection limit.
To explore the utility of this fluorescent probe, we employed a variation of this assay to determine the binding affinity of unlabeled dasatinib via a competitive binding assay. In this assay, fixed concentrations of the fluorescent probe and kinase were used to produce a high polarization signal. The reactions were then titrated with unlabeled dasatinib while recording the change in polarization. The resulting data was graphed and curve fitting analysis provided a binding constant of 3 nM for dasatinib (Figure S10B, Supporting Information).
The results of these fluorescence polarization binding assays with the kinase-deficient enzyme are fairly consistent with the IC50’s derived from the radio-ligand kinase assays. The difference between the measured affinities of the fluorescent probe and dasatinib accurately predict the relative potency of the compounds as kinase inhibitors. Combined with the ease of polarization binding assays, the utility of compound 26 (-G-Flu) as a probe for inhibitor development is clear.
Cytotoxicity cell-based assays were conducted in human B leukemia cell lines (BV- 173, Creative Bioarray # CSC-C0203) and bone marrow lymphoma (K652, ATTC # CCL-243), and pancreatic epithelioid carcinoma (Panc-1, ATCC # CRL-1469) cell lines. BV-173 cell line is an example of Philadelphia chromosome (Ph1)+ chronic myeloid leukemia derived from the peripheral blood of a 45-year-old man with CML in blast crisis in 1980. K562 is an example of bone marrow leukemia cells30. Since, dasatinib has been shown previously potency in treating pancreatic cancer31. We also tested the activity of selected compounds against Panc-1 cell lines. Thus, the IC50 values of selected compounds 1 (Das), 7 (Das-R), 13 (Das-C), 15 (Das-E), 16 (Das-C2), 21 (Das-C16), 24 (Das-C11NH2), and staurosporine (as control drug) were obtained after 72 h incubation (Table 2; Figure S11 (Supporting Information)). The IC50 values were obtained in the range of 26.3 μM to <51.2 pM. Das-R (IC50 = 2.06 μM) and Das-C16 (IC50 = 0.72 μM) were significantly more potent than Das (IC50 = 26.3 μM) against Panc-1 cells while they showed an IC50 < 51.2 pM against BV-173 and K562 cells. All Das conjugates were significantly more potent in BV-173 and K562 cells than Panc-1 cells. Indeed, Das-C was not active (IC50 >100 μM) against Panc-1 cells while it showed IC50 values of 658 nM and 1330 nM against BV-173 and K562 cells, respectively. A similar pattern was observed for Das-C2 that exhibited no activity against Panc-1 while was very potent against BV-173 and K562.
Table 2.
IC50 values of representative compounds using MTT cell viability assay in three cancer cell lines.*
| Compound | IC50 (M) | ||
|---|---|---|---|
| BV-173 | K562 | Panc-1 | |
| 1 (Das) | <51.2 pM | <51.2 pM | 2.63E-05 |
| 7 (Das-R) | <51.2 pM | <51.2 pM | 2.06E-06 |
| 13 (Das-C) | 1.33E-09 | 6.58E-10 | >100 μM |
| 15 (Das-E) | 6.07E-11 | <51.2 pM | 6.41E-05 |
| 16 (Das-C2) | <51.2 pM | <51.2 pM | N/A |
| 21 (Das-C16) | <51.2 pM | <51.2 pM | 7.16E-07 |
| 24 (Das-C11NH2) | <51.2 pM | <51.2 pM | 5.92E-06 |
| Staurosporine | 4.77E-09 | 2.35E-07 | 1.55E-08 |
N/A indicates the IC50 value is not available because the data values did not fit any IC50 curve.
All the data points are average of duplicate experiments.
Discussion
Library construction
Based upon the wealth of available structural data and past successes of dasatinib modifications for affinity-based interaction studies, we hypothesized that the exceptional potency of dasatinib would support its use as a molecular scaffold for the synthesis of novel kinase inhibitors with enhanced specificity. The solved crystal structures of dasatinib in complex with several PTK catalytic domains demonstrate the common manner in which the inhibitor binds PTK active sites. Particularly intriguing to us, is a hydroxyl group of dasatinib that extrudes from the binding pocket to the surface of the kinase. In literature, there are numerous examples of dasatinib (and other kinase inhibitors, as well) that have been modified at this position through the coupling of compounds to agarose resin or other molecules.17, 32–34 These modifications enabled efficient extraction of PTKs that are potential dasatinib targets from various cancer cells and tissues. These affinity-driven applications rely upon specific interaction or recognition of kinases, which suggests that the inhibitor portions of these compounds retain, at least in part, their ability to bind PTK active sites. We reasoned that if large resin attachment does not abolish the affinity of dasatinib, then smaller chemical adducts should be well tolerated, too.
In the present study, we sought to take advantage of this feature of dasatinib by attaching a series of small molecules in an effort to impart novel inhibitor characteristics. In the spirit of rapid library development, we designed a synthesis strategy that was efficient, convenient, and easily employed to obtain derivatives with a wide range of chemical properties. To meet these criteria, we opted for the synthesis directly from dasatinib (free base) by esterification of its hydroxyl group with carboxylic acid containing compounds (amino acids and fatty acids).
A potential drawback to any conjugation, of course, is the potential destruction of key features of the original molecule. The hydroxyl group was reported to extend out towards the solvent –exposed protein in the crystal structure of dasatinib with Abl kinase.8 Upon review of the crystallized ligand, it seems likely that the proper binding orientation of the core molecule is stabilized by energetically favoring exclusion of the hydroxyl group from the binding pocket. Docking studies of this molecule without the hydroxyl group support this idea, as calculated binding energies rise and predicted conformation deviates substantially from the known crystal structures. Additionally, this polar group’s inclusion probably enhances the solubility or bioavailability of hydrophobic dasatinib core molecule. In a recent report, dasatinib was conjugated with a number of hydrophobic compounds and showed promising angiostatic activity.35 Thus, our conjugation of dasatinib, which targets this position for modification, threatens to disturb proper binding conformation and solubility-related characteristics. The potency with which the derivatives inhibit the PTKs in our panel suggests proper orientation is likely achieved by most, if not all compounds.
Conjugation alters kinase selectivity
The structural diversity of target kinases is a central element to this study, as we endeavor to identify the different types of inhibitor interactions that confer specificity. For this reason, we chose three kinases that represent divergent non-receptor PTK families, distinct in structure and function. An ideal inhibitor would target just one of these kinases through recognition of one or more of its distinct structural features, not found in more distantly related PTKs. Therefore, our diverse panel of kinases serves to increase our chances of identifying novel compounds that more closely resemble an ideal inhibitor.
With that idea in mind, it is important to recognize that our kinase inhibitor scaffold, dasatinib, already has a built-in bias toward certain kinases, particularly the Abl and Src families, as compared to others, like Csk. This suggests two things: first, that dasatinib recognizes structural elements common to all three kinases, and second, that Csk possesses features that limit this interaction. It is, therefore, reasonable to assume that complete reversal in this pattern of specificity is unlikely to occur without a major change in binding mode. The inhibitor would not only have to overcome its previously limited interaction with Csk, but also would have to develop some new interaction that is not tolerated by Abl and Src. Therefore, when starting from a biased inhibitor, altered selectivity is probably most likely to occur as a result of either the elimination or exaggeration of an inherent bias.
From this line of reasoning, it makes sense that our library of derivatives displays no signs of dramatic reversals in kinase selectivity pattern. All of the compounds showed better inhibit Src than Abl, and Abl was always inhibited more than Csk. This apparent limitation aside, our measures of selectivity do change dramatically for a number of inhibitors, simply by enhancing a pre-existing bias of dasatinib for or against a given kinase. In one of the most dramatic examples, the IC50 of compound 15 (Das-E) increased only mildly for Src (+16%) but greatly for Csk (+186%) and even more so for Abl (+464%). While other compounds such as 13 (Das-C) and 18 (Das-C10) also possessed significantly altered kinase selectivity, compound 15 (-E) was particularly unique for its minimal loss of potency toward Src.
The changes in kinase inhibitor specificity are quite remarkable considering the limited size of the library generated. Compound 15 (Das-E) demonstrates the ease with which dasatinib derivatives can be developed to alter significantly kinase selectivity. An IC50 for Src that is more than a magnitude lower than for other kinases is certainly quite notable and, we believe, represents a major step in the development of truly specific PTK inhibitors. Furthermore, this success can be built upon by expanding this strategy to include many new dasatinib derivatives for study and comparison to compound 15 (Das-E).
Compound 17 (Das-C8) is low nanomolar inhibitor that displays enhanced specificity for Abl and Csk compared to dasatinib. The highest selectivity ratio obtained in our study, 91.4 Csk/Src belonged to compound 18 (Das-C10) with an IC50 of 3.2 μM for Csk compared to 35 nM for Src. Moreover, each of these compounds displayed increased selectivity toward Src, as compared with Abl.
Compound 7 containing an arginine residue (Das-R) appeared to be the most potent of the inhibitors tested against all three kinases (Figure 4). The results of docking and MD simulations have supported structural models in which compound 7 could indeed form additional hydrogen bonds with Abl and Src kinases, just outside the ATP-binding cleft. More specifically, compound 7 formed hydrogen bonding with the carbonyl oxygen of E329 and strengthened the inhibitor coupling to stable C-spine. Although only structural determination could provide the ultimate verification of these predictions, the binding energetics analysis based on the predicted docked poses and MD simulations reproduced experimental binding constants with an excellent accuracy (Figure 5). Particularly, computational analysis successfully predicted the Csk/Src selectivity trend seen in the studied compounds, including compound 7. In addition, the experimental data supported our predictions as compound 7 increased potency against all kinases. Dasatinib derivatives represent significant extensions of the original inhibitor and computational analysis has highlighted the fact that the increased number of strong hydrogen bonds can be often offset by unfavorable solvation and entropic factors. Our comprehensive analysis has reproduced well the changes in binding affinity and selectivity, also pointing to the important and relatively unexplored kinase areas that can be targeted to modulate selectivity in this series.
According to modeling results, we found that for the fatty acid-derived compounds, two moderately long-chained compounds (17/Das-C8, 18/Das-C10) may affect conformational dynamics and interactions with the catalytic domain, particularly with the catalytic spine (C-spine) residues. The computed binding free energies for a panel of compounds (including 17 and 18) showed a rather significant change in activity for 17 and 18 favoring Src over Csk. In molecular dynamics simulations, we found that the crystal structure of Csk kinase in complex with dasatinib (pdb id 3QLG) was fairly rigid, including D-helical region and adjacent small helix that are in proximity to C-spine residues (I392,V394, L346, L451, L455). Molecular docking and MD simulations showed that rigidity of these regions in Csk structure would not allow the formation of favorable hydrophobic contacts without substantial restrictions of conformational entropy of the ligand, as the protein could not readily adopt to the long hydrophobic tail of the inhibitor. In contrast, the crystal structures of Abl (pdb id 2GQG), and Src (pdb id 3G5D) in complexes with dasatinib are more flexible in these regions and allow the formation of multiple hydrophobic contacts with Das-C8 and Das-C10 analogs without a large conformational entropy loss. The interactions made by Das-C8 and Das-C10 with Src kinase contribute to the stabilization of the C-spine and the enhanced compound selectivity Src over Csk kinase. The importance of the αD-helix residues in the binding of dasatinib derivatives may also arise from their proximity and involvement in the stabilization of the C-spine residues.
Structural Basis of Increased Selectivity
The structural variation of PTKs outside the ATP-binding pocket is localized to a few small areas. One such region is located at the end of αD-helix of the peptide binding lobe and extends to αE-helix. Even among closely related PTKs, like the Src Family Kinases, a great deal of structural variations exists here. This region has also been implicated as a key site of substrate recognition in some kinases.15 Furthermore, αD-helix is in proximity (within a few angstroms) to the hydroxyl group of dasatinib.
One hurdle to developing dasatinib derivatives in this manner has to do with the size of the compounds being tethered to the inhibitor. If these are too short or too long, they will likely miss the targeted αD-helix. Likewise, bulkier compounds threaten to interfere via steric hindrance. One means by which we sought to alleviate such concerns was to simulate the binding of potential compounds using molecular modeling software. Retrospective analysis of these models, in conjunction with our biochemical data and the available crystal structures of Abl, Csk, and Src, offer valuable insight into the changes in specificity we observed from our compounds.
The first and, perhaps, most exciting feature that the models predict is the preferred orientation of the compounds that were attached to dasatinib. Despite allowance to rotate freely, almost all of the simulated dockings favor interaction with the region surrounding αD-helix, rather than the equally proximal ATP binding lobe of the kinase. Undoubtedly, this orientation better positions the inhibitor for possible contact with structural features that are unique to any given kinase.
Key features of αD-helix may also explain the altered selectivity we observed of the fatty acid conjugated derivatives. The extreme effects that these molecules have on potency are likely due to exclusionary interactions with the polar surface of the kinase that surrounds the exit from the inhibitor binding pocket. This can easily be seen by coloring the surfaces of the kinases according to hydrophobicity. In Src, however, the pattern of hydrophobicity at the end region of αD-helix is such that a clear pathway to αE-helix (also fairly hydrophobic) is maintained. This may explain the longer-chained fatty acid derivatives’ enhanced capacity to inhibit Src, relative to the other kinases.
The structural evidence examined thus far suggests that αD-helix offers the greatest potential avenue of interaction with the synthesized compounds. The novel specificities of the dasatinib derivatives we generated certainly imply unique interactions have been formed with the PTKs they inhibit. While more conclusive evidence remains to be collected, it appears highly likely that the modulation of kinase-specific inhibition observed in this study is a direct result of novel interactions outside the ATP binding pocket, particularly αD-helix.
Implications in Drug Metabolism
The ADME characteristics of a given kinase inhibitor are vital to its usefulness as a therapeutic tool. How well the drug gets taken into the target cells and how long it remains in the active form are direct functions of the chemical properties inherent in the molecule. Particularly, susceptibility of a drug to the hosts’ metabolic processes is a major part of this. The stability assays performed in this study highlight an important feature of our dasatinib derivatives. The implications in drug metabolism are species-dependent: humans have quite different carboxylesterases from mice. Thus the results here reported are useful to drive experiments on animal (mouse) models, not to predict bioavailability in humans.
Consider the compound 16 (Das-C2), which displays a low nanomolar IC50. While this study does not address bioavailability, we have observed that the compound is readily hydrolyzed to dasatinib by carboxylesterases as shown by the formation of dasatinib by LC/MS/MS. Therefore, if titrated properly, this compound may offer considerable value as a prodrug for the extended release of dasatinib.
The utility of other derivatives, perhaps not metabolized by carboxylesterases, also opens the door for new applications. Compound 17 (Das-C8) is another low nanomolar inhibitor that actually displays enhanced specificity for Abl and Csk compared to dasatinib. While overall potency is diminished to some degree, the added specificity may provide the proper trade-offs to enable more effective outcomes. Perhaps the most interesting feature of this compound, however, is its inability to be broken down quickly by the plasma carboxylesterases. The fact that the larger hydrophobic chain appears to disrupt the inhibitor’s metabolic break-down offers evidence that this strategy may be employed to subvert other enzymatic components of a host’s drug metabolism, to effectively increase half-life of the compound. Ester linkage was selected as a convenient approach to test the feasibility of developing specificity in vitro. Other more stable linkages can be used to generate compounds with appropriate stability for applications in vivo.
The broad applications offered by an altered drug metabolic profile are numerous and multifaceted. Any derivative, no matter how small the alteration, can potentiate major changes. The huge chemical diversity that is possible with the strategy we describe here, however, can be rapidly and conveniently applied to screen for desired characteristics, without resynthesizing the core dasatinib molecule. With these ideas in mind, further investigation of the unique ADME features of these compounds and newly synthesized dasatinib derivatives seems merited, given the potential therapeutic benefits of increased stability and extended release.
Fluorescence-Labeled Dasatinib Derivatives
Compound 26 (-G-Flu) is a dual-purpose inhibitor with broad potential applications. As an inhibitor, it can bind to a kinase tightly while enabling its visible detection. This may be particularly useful as a molecular diagnostic tool for imaging. Likewise, in vitro techniques may find this compound useful in FACS for monitoring kinase expression. Since this compound is capable of measuring the binding of unlabeled inhibitor, it will be especially adaptable to high-throughput kinase inhibitor drug discovery.
Conclusions
Dasatinib-amino acid and dasatinib-fatty acid conjugates were synthesized to generate derivatives with better PTK selectivity across a studied panel of Abl, Csk, and Src kinases. Dasatinib conjugation changed the kinase selectivity. While dasatinib inhibits both Src and Abl with IC50 values in low nM, a number of compounds were found to be more selective against Src. The highest selectivity ratio obtained in our study, 91.4 Csk/Src belonged to compound 18 (Das-C10) with an IC50 of 3.2 μM for Csk compared to 35 nM for Src. Compounds 15 (Das-E) and 13 (Das-C) also demonstrated large selectivity gains (10.2 and 10.3 Abl/Src IC50 ratios). Cytotoxicity assay reveal that Das-R (IC50 = 2.06 μM) was significantly more potent than Das (IC50 = 26.3 μM) against Panc-1 cells while they both showed an IC50 < 51.2 pM against BV-173 and K562 cells. Molecular modeling studies were consistent with the experiments and provided structural basis of the binding affinity and selectivity profiles. Moreover, several unique characteristics of these inhibitors were explored for their potential utility as prodrugs or research tools. These data provide insights for developing PTK specific inhibitors using known scaffold by using the structural features in the kinase. Together, these findings highlight the great potential for this strategy with broad applications for the design and development of kinase-specific inhibitors.
Experimental Section
General
Dasatinib (Sprycel®) was obtained from LC Laboratories, MA. N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), 1-hydroxybenzotriazole (HOBT), 4-methylmorpholine (NMM), p-toluenesulfonic acid monohydrate (pTsOH), anhydrous N,N-dimethylformamide, fatty acids, staurosporine, and other miscellaneous chemicals were purchased from Sigma-Aldrich Chemical Co. (Milwaukee, WI). Boc-protected amino acids were purchased from Chem-Impex International Inc, Wood dale, IL, USA. [γ-32P]-ATP (6,000 Ci mol−1) was purchased from PerkinElmer for kinase assays, and pre-cast protein gels were bought from BioRad. All reagents used for bacterial culture and protein expression were purchased from Fisher. EDC and HOBt were used as coupling reagents. CellTiter 96® Non-Radioactive Cell Proliferation Assay (MTT) reagent was purchased from Promega (Madison, WI). NMM was used as a base. The chemical structures of all the synthesized compounds were characterized by nuclear magnetic resonance spectra (1H NMR, 13C NMR) using Bruker Biospin (300 MHz) and Varian NMR spectrometer (500 MHz). TMS (tetramethylsilane) was used as the internal standard and chemical shifts were reported in parts per million (ppm). The High-resolution quadrupole time-of-flight QqTOF mass spectrometer (Applied Biosystems) was used to confirm the chemical structures of compounds. Melting point were measured with a Mel-Temp with fluke 51 digital thermometer and were uncorrected. All compounds were purified in column chromatography (≥95%), and the purity of compounds was confirmed by analytical HPLC.
General Procedure for the Synthesis of Dasatinib-Amino acid Conjugates (2-15)
Dasatinib (150 mg, 0.31 mmol) was dissolved in anhydrous DMF (2.0 mL) in a 15 mL glass vial fitted with rubber septum under dry nitrogen and stirred in an ice bath. The Boc-protected amino acid (3 equiv, 0.92 mmol) and EDC.HCl (180 mg, 3.05 equiv, 0.94 mmol) were added under nitrogen followed by stirring to dissolve the reagents and addition of NMM (101 μL, 3 equiv, 0.92 mmol). The mixture was stirred for 1 h. The ice bath was removed after 1 h, and the reaction was continued for additional 20 h. The analysis of small aliquot using ESI mass spec confirmed the disappearance of starting material and the formation of the product. The reaction mixture was quenched with water (20 mL) and extracted with ethyl acetate (70 mL × 3 times). The organic layer was washed with sodium bicarbonate (5%, 50 mL) followed by washing with water (50 mL) and brine (50 mL). The organic layer was separated and dried on anhydrous sodium sulfate. The crude solid product was obtained after evaporation of the solvents and was purified using flash chromatography using a gradient system of hexane/ethyl acetate or hexane/dichloromethane/methanol as eluting systems. The Boc group removal was carried out in the presence of pTsOH. The dry Boc-Amino acid-dasatinib compound (~0.31 mmol) was dissolved in acetonitrile (5 mL). The pTsOH (408 mg, 7 equiv, 2.15 mmol) was added to the reaction mixture, and the mixture was continued to stir for 1.5 h. The addition of pTsOH made the reaction clear and transparent. The solvents were removed from the reaction mixture using a rotatory evaporator under vacuum. The residue was dissolved in ethyl acetate (100 mL), water (50 mL), and sodium bicarbonate (229 mg) and transferred to a separating funnel. The water layer were washed with ethyl acetate (25 mL × 3). Combined ethyl acetate layer was washed with brine (50 mL × 2) and dried over anhydrous sodium sulfate. The solvent was removed using a rotatory evaporator to provide the crude solid compound, which was further purified by using combiflex chromatography using a gradient system of hexane/ethyl acetate as the eluting system.
General Procedure for Synthesis of Dasatinib-Fatty Acid Conjugates (16-25)
Synthesis of fatty acyl derivatives of dasatinib was accomplished according the general procedure described above except that no deprotection was needed for removal of Boc group. The Boc group for Boc-11-aminoundecanoic acid was removed as described above for amino acids. Dasatinib (150 mg, 0.307 mmol) was added to 25 mL round flask containing anhydrous DMF (3 mL). The reaction mixture was stirred to dissolve the compound followed by keeping the reaction mixture in ice. Fatty acid (3 equiv), EDC.HCl (180.0 mg, 3.05 equiv), HOBt (128 mg, 3.05 equiv), and NMM (102 μL, 3 equiv) were added, and the reaction mixture was continued to stirred in ice bath for 1 h. After 1 h, the ice bath was removed, and the reaction was continued for additional 24 h. A small aliquot was removed and tested with mass spec to confirm the product formation and disappearance of starting compound. Then, the reaction mixture was quenched with water (20 mL) and extracted with ethyl acetate (70 mL × 3 times). The organic layer was washed with sodium bicarbonate (5%, 50 mL) followed by wash with water (50 mL) and brine (50 mL). The organic layer was separated and dried on anhydrous sodium sulfate. The crude (solid) compound was obtained after evaporating the solvents. The crude compounds were purified using flash chromatography and dichloromethane/ethyl acetate as eluting solvents.
Molecular Modeling
All crystal structures of Dasatinib complexes used in molecular modeling were obtained from the Protein Data Bank (RCSB PDB www.rcsb.org). In simulations, all crystallographic water molecules were removed, and missing hydrogen atoms of the protein were added. All ionizable residues were considered in the standard ionization state at the neutral pH condition. The missing residues, unresolved structural segments, and disordered loops were modeled and evaluated with the ModLoop server36–37 and the ArchPRED server38.
In order to accurately reproduce the crystal structure in control dockings, the receptor was held rigid and the rotatable bonds (torsions) of the core dasatinib structure were frozen with the exception of the C13-N5 and N6-C20 bonds. Based on the superimposition of other solved structures, these appear to be the only bonds to display any great deviation from kinase-to-kinase. The virtual ligands were prepared using Discovery Studio (Dockings were performed with an exhaustiveness of 256 within a search space measuring 22×24×28, centered at (11, 90.5, 57.5) x,y,z.
We modeled each of the 20 common amino acids, along with addition of phosphotyrosine, as carboxyesters of dasatinib to establish the feasibility of our approach. This small collection of potential derivatives could be conveniently synthesized and represent a variety of sizes and physical properties while keeping computational cost low (by virtue of few rotatable bonds). Combining accuracy and ease of use, we chose Autodock Vina to perform our in silico modeling. Autodock Vina’s performance in dockings has been relatively well established, and its algorithms require no assumptions to be made concerning the ionization of side-chains.22–23
MD Simulations
MD simulations of the protein kinase crystal structures and docked complexes (each of 200 ns duration) were carried out using NAMD 2.6 package39 with the CHARMM27 force field40–41 and the explicit TIP3P water model. The employed MD protocol is consistent with the overall setup that was described in details in our earlier studies42. An NPT production simulation was run on the equilibrated structures for 500 ns keeping the temperature at 300 K and constant pressure (1 atm) using Langevin piston coupling algorithm.
Binding Free Energy Calculations
The binding free energy if inhibitor-kinase binding was calculated using MM-GBSA approach43–45 In this approach the binding free energy ΔGbind is written as the sum of the gas phase contribution ΔGMM, the solvation free energy ΔGsolv, and an entropic contribution −TΔS
| (1) |
The brackets <> denote an average of these contributions calculated over the MD trajectories. The gas-phase contribution <ΔGMM> to the binding free energy is the difference in the molecular mechanics energy of the complex and the isolated protein and ligand. This contribution is the sum of the differences in the internal energies ΔEintra, the van der Waals interaction energy ΔEvdw, and the electrostatic interaction energy ΔEelec:
| (2) |
| (3) |
where Ebond is the energy of the bonded terms (bonds, angles, dihedral angles, and improper angles) of a given molecule; Evdw is the van der Waals energy of the molecule; and Eelec is the electrostatic energy of the molecule. These contributions are calculated according to the CHARMM22 molecular mechanics force field.
The solvation free energy ΔGsolv is the difference between the solvation energy of the complex and solvation free energies of the isolated protein and ligand:
| (4) |
| (5) |
The solvation free energy of a molecule is given as the sum of nonpolar and polar contributions. The nonpolar contribution is computed using the solvent accessible surface are (SASA) model and give as where the parameter σ=0.0072 kcal/(mol*Ǻ2). The electrostatic contribution to the solvation free energy was calculated using the analytical generalized Born (GB) model implemented in CHARMM. This model is known to reproduce accurately the solvation free energies calculated by solving the Poisson equations. All energy terms were calculated for 10,000 frames regularly separated by 20 ps along the 200ns trajectory performed for the complex.
The entropy contribution consists of translational ΔStrans, rotational ΔSrot and vibrational ΔSvib components:
| (6) |
The vibrational entropy terms were computed using normal mode analysis that yields better convergence than the quasiharmomic analysis from MD trajectories. The VIBRAN module of the CHARMM program was used to calculate and diagonalize the force constant matrix for the normal mode vectors and frequencies determination. The normal modes were calculated on minimized average structures obtained from MD simulations. The minimization was performed using the Newton–Raphson minimization algorithm, using the same cutoff scheme and constraints as for the normal mode calculations. Since the normal mode analysis is computationally demanding, −TΔS was averaged over only 500 frames extracted from the 200 ns MD trajectories. All energy terms are calculated using single 200 ns trajectories of the kinase-inhibitor complexes. This is followed by separation of the complexes into isolated protein and ligand structures and subsequent minimization of the isolated molecules without conducting additional simulations of the individual protein. With this simplified approach, the difference in the internal energy of the isolated molecules upon complexation is neglected and generally yields better convergence of binding free energies.
Protein Expression and Purification
Enzymes and protein substrate were prepared as previously described.29, 35 Briefly, vectors for expressing Abl, CrkL, or Csk as GST-fusions and the 6xHis-tagged Src catalytic domain (SrcCata) were made and cloned into appropriate E. coli systems. For each, 1-liter LB cultures were grown at 37°C until an OD600 of about 1 was reached and then induced at RT with 0.4 mM isopropyl β-d-thiogalactopyranoside while shaking at 250 rpm for 4–6 h. Cells were harvested at 4°C via centrifugation at 7,000 × g. Pellets were either stored at −20°C or immediately re-suspended in appropriate lysis buffer: PBS + β-mercaptoethanol, pH 7.3 for GST-fusions and 50 mM HEPES + 200mM NaCl + 10 mM imidazole + 0.01% triton x-100, pH 8.0 for 6xHis-proteins. Following 4 × 15 sec sonication in 12 mL glass tubes on ice, lysates were cleared by centrifuge for 10 min at 22,000 × g and incubated with 1 mL agarose resin at 4 °C for 1hr. This suspension was then loaded into a column, washed with lysis buffer and eluted with either 50 mM Tris + 10 mM GSH, for GST-fusions, or 50 mM HEPES + 200 mM imidazole to release 6xHis-tagged protein. All samples were quantified using standard Bradford assays and analyzed by SDS-PAGE/coomassie staining to check purity. When purified from the phosphatase system and assayed at these concentrations, autophosphorylation of the enzymes which can often throw-off these sorts of assays (before and during the assay) is minimal.
Kinase Assays and IC50 Measurements
All kinase activity and inhibition assays were performed in duplicate at least three times. Three replicates of each assay were performed in duplicate. Enzyme activity and inhibition were characterized using the following methods, adapted from previous work.46–47 Csk and SrcCata activity levels were measured from incubations with polyE4Y (1 mg/mL) and CrkL (0.5 mg/mL) was used as a substrate for Abl. All assays were performed between 20 and 500 fM concentrations of enzyme. In 50 μL kinase assay, the final concentration will be between 0.4 and 10 nM. Dasatinib and derivatives were dissolved in 10% DMSO/90% kinase buffer solution (KB10) and diluted in series as needed. In screening assay reactions, multiple inhibitors were used at a fixed concentration for relative inhibition comparisons. IC50 measurement assays, however, had a single inhibitor, but each reaction varied in the compound’s concentration. In assays requiring pre-treatment, undiluted stocks of the inhibitors were incubated at RT with microsomal preparations to a final concentration of 12.5 μg/mL.
In each assay, a series of 50 μL duplicate reactions were setup at 30 °C to contain the appropriate inhibitor concentration, substrate, and enzyme in 75 mM EPPS, pH 8.0 + 200 μM [γ-32P]-ATP (~1000 dpm/pmol) + 12 mM MgCl2 + 5% glycerol + 0.005% Triton X-100. The reactions were stopped after 20 min incubation by blotting onto filter paper strips prior to precipitation and washing in hot 5% trichloroacetic acid. The strips were then sorted and separated into vials of scintillation fluid for signal quantification in a Beckman Coulter LS-6500. From these counts, enzyme turnover was calculated for each reaction. Three replicates of each assay were performed in duplicate. The standard errors range were in the range of 0.034–0.095 nM for Src, 0.041–0.242 nM for Csk, and 0.041–0.109 nM for Abl.
Binding Assays
To determine the binding of compound 26 (-G-Flu) toward the kinase-deficient Src catalytic domain (kdSrcCata), fluorescence polarization (FP) binding assays were carried out as described previously.48–49 The enzyme is a kinase-deficient form of Src catalytic domain. Inactivation of the kinase activity was due to a Lys295Met mutation that destroys the catalytic activity but does not affect its substrate binding. We assume that the purified kinase-deficient Src catalytic domain is homogeneous in its activation state. The concentration was calculated based on total protein that was apparently pure judged by SDS-PAGE and coomassie blue staining.
Specifically, a series of kdSrcCata dilutions were made, ranging from 62.5 – 1,000 nM (and an additional blank). Each dilution contained 400 nM compound 26 (-G-Flu) in 50 mM Tris, pH 8.0. FP was then detected using a PerkinElmer LS55 Luminescence Spectrophotometer at 25 °C. Wavelengths utilized were 485 nM for excitation and 530 nM for emission. The net change in FP was plotted as a function of kdSrcCata concentration and fit to the following equation: . Where FPmax is the maximum polarization value at saturation and KD is the dissociation constant of compound 26 (-G-Flu) binding to kdSrcCata. Regression analysis was carried out in software (www.labfit.net).
To determine the binding of unlabeled dasatinib, its ability to compete against the fluorogenic compound for binding to kdSrcCata was determined by an FP competition binding assay.49 This assay setup differs from the above in that each tube had a 50 nM concentration of compound 26 (-G-Flu), 125 nM kdSrcCata (held constant), and concentrations of the unlabeled dasatinib from 200 – 1,600 nM. The competing ligand’s KD was then determined by plotting the FP as a function of the increasing concentration of the unlabeled compound and fitting the curve to the equation: , where KD1 is the dissociation constant of compound 26 (-G-Flu), as determined above, and KD2 is the dissociation constant of the unlabeled dasatinib. A is a conversion factor between the concentration of the probe-kinase complex and the FP value. [kdSrcCata]t and [Probe]t were total kinase and compound 26 (-G-Flu) concentrations. [Competitor]t is the total concentration of unlabeled, competing ligand.
Cytotoxicity Assays
Human B cell leukemia cell line BV-173 (CSC-C0203) was purchased from Creative Bioarray and human bone marrow leukemia cell line K562 (CCL-243), and pancreatic epithelioid carcinoma cell line Panc-1 (CRL-1469) were obtained from American Type Culture Collection. BV-173 cells were cultured using RPMI 1640 with 20% FBS. K562 cells were cultured using iscove’s modified dulbecco’s medium (IMDM) with 10% FBS. Panc-1 cells were cultured using Dulbecco’s modified eagle medium (DMEM) with 10% FBS. 100 μg/ml penicillin and 100 μg/ml streptomycin were added to all of the culture media. All the Cultures were maintained at 37 °C in a humidified atmosphere of 5% CO2 and 95% air. Test compounds 1, 7, 13, 15, 16, 21, and 24 were diluted with DMSO in 10-dose and 5 fold serial dilutions starting at 20 mM. The reference compound staurosporine was diluted with DMSO in 10-dose and 3 fold serial dilutions starting at 10 mM. 125 nL of 1, 7, 13, 15, 16, 21, and 24 or 25 nL of staurosporine was delivered from the source plate to the wells of 384-well cell culture plates by Echo 550 Liquid Handler. 25 μL of culture medium containing 2000 of BV-173, K562, or Panc-1 cells was added to the wells of the 384-well culture plate. The plates were incubated at 37 °C in a humidified atmosphere of 5% CO2 and 95% air for 72 hours. 4 μl of MTT Dye Solution was added to each well. The plates were incubated at 37 °C in a CO2 incubator for 4 h. After incubation, the formazan was produced in the cells. 25 μl of Solubilization/Stop Solution was added to each well and the plates were incubated for another 1 hour at 37 °C in a CO2 incubator to dissolve the formazan crystals. The absorbance of each sample was measured at 590 nm using Envision 2104 Multilabel Reader (PerkinElmer, Santa Clara, CA). The cell viability was determined based on the quantification of the color intensity in each culture well. The IC50 curves were plotted and the IC50 values were calculated using the GraphPad Prism 4 program based on a sigmoidal dose-response equation. All the data points are average of duplicate experiments.
Supplementary Material
Acknowledgments
We acknowledge the financial support from National Center for Research Resources, NIH, Grant number 1 P20 RR16457 for sponsoring the core facility.
Abbreviations
- ALL
Acute lymphoblastic leukemia
- BCR
Breakpoint Cluster Region
- CML
chronic myeloid leukemia
- DMSO-d6
dimethyl-d6 sulfoxide
- DMF
N,N-dimethylformamide
- EDC
N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride
- HPLC
high pressure liquid chromatography
- HOBt
hydroxybenzotriazole
- HRMS
high resolution mass spectrometry
- NMM
N-methylmorpholine
- MD
molecular dynamics
- Ph+
Philadelphia chromosome-positive
- PTKs
Protein tyrosine kinases
- SFK
Src Family Kinase
- TKI
Tyrosine kinase inhibitor
Footnotes
Supporting Information for this article is given via a link at the end of the document.
References
- 1.Zhang J, Yang PL, et al. Nat Rev Cancer. 2009;9(1):28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jilaveanu LB, Zito CR, et al. Pigment Cell Melanoma Res. 2011;24(2):386–389. doi: 10.1111/j.1755-148X.2011.00835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shah NP, Tran C, et al. Science. 2004;305(5682):399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 4.FDA Consum. 2006;40(6):5. [PubMed] [Google Scholar]
- 5.SPRYCEL® (dasatinib) [package insert] 2006. (REV 2012) [Google Scholar]
- 6.Liu-Dumlao T, Kantarjian H, et al. Curr Oncol Rep. 2012;14(5):387–394. doi: 10.1007/s11912-012-0247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berman HM, Westbrook J, et al. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tokarski JS, Newitt JA, et al. Cancer Res. 2006;66(11):5790–5797. doi: 10.1158/0008-5472.CAN-05-4187. [DOI] [PubMed] [Google Scholar]
- 9.Getlik M, Grutter C, et al. J Med Chem. 2009;52(13):3915–3926. doi: 10.1021/jm9002928. [DOI] [PubMed] [Google Scholar]
- 10.Williams NK, Lucet IS, et al. J Biol Chem. 2009;284(1):284–291. doi: 10.1074/jbc.M807850200. [DOI] [PubMed] [Google Scholar]
- 11.Marcotte DJ, Liu YT, et al. Protein Sci. 2010;19(3):429–439. doi: 10.1002/pro.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farenc C, Celie PH, et al. FEBS Lett. 2011;585(22):3593–3599. doi: 10.1016/j.febslet.2011.10.028. [DOI] [PubMed] [Google Scholar]
- 13.Muckelbauer J, Sack JS, et al. Chem Biol Drug Des. 2011;78(5):739–748. doi: 10.1111/j.1747-0285.2011.01230.x. [DOI] [PubMed] [Google Scholar]
- 14.Rothweiler U, Åberg E, et al. J Mol Biol. 2011;411(2):474–485. doi: 10.1016/j.jmb.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 15.Lee S, Lin X, et al. Proc Natl Acad Sci U S A. 2003;100(25):14707–14712. doi: 10.1073/pnas.2534493100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee S, Ayrapetov MK, et al. J Biol Chem. 2006;281(12):8183–8189. doi: 10.1074/jbc.M508120200. [DOI] [PubMed] [Google Scholar]
- 17.Rix U, Hantschel O, et al. Blood. 2007;110(12):4055–4063. doi: 10.1182/blood-2007-07-102061. [DOI] [PubMed] [Google Scholar]
- 18.Zhou G, Sylvester JE, et al. Anal Biochem. 2011;408(1):5–11. doi: 10.1016/j.ab.2010.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi H, Zhang CJ, et al. Journal of the American Chemical Society. 2012;134(6):3001–3014. doi: 10.1021/ja208518u. [DOI] [PubMed] [Google Scholar]
- 20.Fabian MA, Biggs WH, 3rd, et al. Nat Biotechnol. 2005;23(3):329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 21.Karaman MW, Herrgard S, et al. Nat Biotechnol. 2008;26(1):127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 22.Trott O, Olson AJ. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houston DR, Walkinshaw MD. J Chem Inf Model. 2013;53(2):384–390. doi: 10.1021/ci300399w. [DOI] [PubMed] [Google Scholar]
- 24.Shirazi AN, Tiwari RK, et al. Mol Pharmaceut. 2014;11(10):3631–3641. doi: 10.1021/mp500364a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng Y, Prusoff WH. Biochemical pharmacology. 1973;22(23):3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 26.Cer RZ, Mudunuri U, et al. Nucleic Acids Res. 2009;37(Web Server issue):W441–445. doi: 10.1093/nar/gkp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor SS, Kornev AP. Trends Biochem Sci. 2011;36(2):65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meharena HS, Chang P, et al. PLoS Biol. 2013;11(10):e1001680. doi: 10.1371/journal.pbio.1001680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sobolev V, Sorokine A, et al. Bioinformatics. 1999;15(4):327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- 30.Gambacorti-Passerini C, le Coutre P, et al. Blood Cells Mol Dis. 1997;23(3):380–394. doi: 10.1006/bcmd.1997.0155. [DOI] [PubMed] [Google Scholar]
- 31.Chang Q, Jorgensen C, et al. Br J Cancer. 2008;99(7):1074–1082. doi: 10.1038/sj.bjc.6604676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hantschel O, Rix U, et al. Proc Natl Acad Sci U S A. 2007;104(33):13283–13288. doi: 10.1073/pnas.0702654104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rix U, Gridling M, et al. Methods Mol Biol. 2012;803:25–38. doi: 10.1007/978-1-61779-364-6_3. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Rix U, et al. Nat Chem Biol. 2010;6(4):291–299. doi: 10.1038/nchembio.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paunescu E, Clavel CM, et al. ACS Med Chem Lett. 2015;6(3):313–317. doi: 10.1021/ml500496u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marti-Renom MA, Stuart AC, et al. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 37.Fiser A, Do RK, et al. Protein Sci. 2000;9(9):1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernandez-Fuentes N, Zhai J, et al. Nucleic Acids Res. 2006;34(Web Server issue):W173–176. doi: 10.1093/nar/gkl113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phillips JC, Braun R, et al. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacKerell AD, Bashford D, et al. J Phys Chem B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 41.MacKerell AD, Jr, Banavali N, et al. Biopolymers. 2000;56(4):257–265. doi: 10.1002/1097-0282(2000)56:4<257::AID-BIP10029>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 42.James KA, Verkhivker GM. PLoS One. 2014;9(11):e113488. doi: 10.1371/journal.pone.0113488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J, Wolf RM, et al. J Comput Chem. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 44.Kollman PA, Massova I, et al. Acc Chem Res. 2000;33(12):889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 45.Srinivasan TECJ, Cieplak P, Kollman PA, Case DA. J Am Chem Soc. 1998;120:9401–9409. [Google Scholar]
- 46.Huang K, Wang YH, et al. J Mol Biol. 2009;386(4):1066–1077. doi: 10.1016/j.jmb.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kemble DJ, Wang YH, et al. Biochemistry. 2006;45(49):14749–14754. doi: 10.1021/bi061664+. [DOI] [PubMed] [Google Scholar]
- 48.Tiwari R, Brown A, et al. Biochimie. 2010;92(9):1153–1163. doi: 10.1016/j.biochi.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 49.Ayrapetov MK, Nam NH, et al. J Biol Chem. 2005;280(27):25780–25787. doi: 10.1074/jbc.M504022200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.