Abstract
TMEM173 encodes MPYS/STING, is an innate immune sensor for cyclic dinucleotides (CDNs) playing a critical role in infection, inflammation, and cancer. The R71H-G230A-R293Q (HAQ) of TMEM173 is the second most common human TMEM173 allele. Here, using data from 1000Genome Project, we found that homozygous HAQ individuals account for ~16.1% of East Asians and ~2.8% of Europeans while Africans have no homozygous HAQ individuals. Using B cells from homozygous HAQ carriers, we found, surprisingly, that HAQ/HAQ carriers express extremely low MPYS protein and have decreased TMEM173 transcript. Consequently, the HAQ/HAQ B cells do not respond to CDNs. We subsequently generated an HAQ knock-in mouse expressing mouse-equivalent of the HAQ allele (mHAQ). The mHAQ mouse has decreased MPYS protein in B cells, T cells, Ly6Chi monocytes, bone-marrow-derived DC and lung tissue. The mHAQ mouse also does not respond to CDNs in vitro and in vivo. Lastly, Pneumovax®23 whose efficacy depends on TMEM173, is less effective in the mHAQ mice than the WT mice. We conclude that HAQ is a null TMEM173 allele. Our findings have a significant impact on research related to MPYS-mediated human diseases and medicine.
Introduction
Early detection of invasive pathogens is achieved by germline encoded innate immune sensors. TMEM173 encodes an endoplasmic reticulum (ER) associated molecule MPYS (also known as MITA and STING)(1–3). MPYS is a cytosolic sensor for cyclic dinucleotides (CDNs) including bacterial CDNs, cyclic di-AMP (CDA), cyclic di-GMP (CDG), and mammalian CDN 2′5′-3′5′-cyclic GMP-AMP (2′3′-cGAMP) generated during cytosolic DNA sensing(4–6). Consequently, MPYS is critical for host defense against DNA viruses(7), RNA viruses(7, 8), intracellular bacteria(9, 10) and extracellular bacteria(11, 12) in mice. MPYS also plays a key role in the development of auto-inflammatory diseases in mice(13–15) and STING-associated vasculopathy with onset in infancy (SAVI) in humans(16, 17). Last, there are on-going efforts to develop MPYS/STING-targeting immunotherapy for cancer and infectious diseases(11, 18–22).
We first showed that human TMEM173 gene has significant heterogeneity (23). We identified R232 of TMEM173, not H232, as the most prevalent allele (wt) in the human population(23). However, we found that only ~50% of Americans are R232/R232(23). We further identified HAQ, which contains three non-synonymous SNPs, R71H-G230A-R293Q, as the second most common human TMEM173 allele and estimated that ~3% of Americans are homozygous for HAQ(23). Transiently overexpressing HAQ in 293T cells leads to >90% decrease of type I IFN production, the hallmark function of MPYS/STING(23). 293T cells stably transfected with HAQ also have decreased response to CDN stimulation (6, 24). Here, we studied the endogenous function of the HAQ allele using human cells from homozygous HAQ carriers and the mHAQ knock-in mouse. We discovered, unexpectedly, that the HAQ TMEM173 has decreased protein expression (~90%) and did not respond to CDN stimulation in vivo and in vitro.
Materials and Methods
Generation of an HAQ-MPYS knock-in mice
The linearized targeting vector (Figure S3A), which covers ~10 kb of the genomic region in MPYS locus on mouse chromosome 18, was transfected into JM8A3.N1 embryonic stem (ES) cells originated from the C57BL/6 strain, followed by the selection for neomycin positive and diphtheria toxin (DTA) negative clones. Targeted clones were screened by PCR. Positive ES clone was subjected to the generation of chimera mice by injection using C57BL/6J blastocysts as the host. The male chimeras (chimerism >95% determined by coat color) were mated with C57BL/6J female mice for germline transmission. Successful germline transmission was confirmed by PCR-sequencing (Figure S3B). The heterozygous mice were bred to Actin-flpase mice (The Jackson Laboratory, B6.Cg-Tg(ACTFLPe)9205Dym/J) (Figure S3A) to remove the neo gene and make the HAQ-MPYS knock-in mouse. Animals were generated at the National Jewish Health Mouse Genetics Core Facility. Animal care and handling was performed according to institutional animal care and use committee guidelines.
Mice
Six- to twelve-week-old mice, both males and females, were used for all experiments. MPYS−/− mice (Tmem173<tm1Camb>) were described previously(25). All mice were on a C57BL/6 background. Mice were housed and bred in the Animal Research Facility at Albany Medical College and the University of Florida. All experiments with mice were performed by the regulations and approval of the Institutional Animal Care and Use Committee from Albany Medical College or the University of Florida.
Reagent
The following reagent was obtained through BEI Resources, NIAID, NIH: Streptococcus pneumonia Family 1, Clade 2 Pneumococcal Surface Protein A (PspA UAB055) with C-Terminal Histidine Tag, Recombinant from Escherichia coli, NR-33178.
Data Mining
Human TMEM173 genotype information was obtained from 1000Genome Project (Phase III, http://browser.1000genomes.org/index.html). Human B cells with the corresponding TMEM173 genotypes were obtained from Coriell Cell Repositories (https://catalog.coriell.org/) and cultured in RPMI 1640 with 15% FCS, 2mM L-glutamine, 37°C under 5% CO2. Information related to TMEM173 gene expression was obtained from The Genotype-Tissue Expression (GTEx) project (http://www.gtexportal.org/home/).
Human B cells Activation by CDNs
For CDA (Invivogen, cat#vac-cda), CDG(Invivogen, cat#vac-cdg) and 2′3′-cGAMP (Invivogen, cat#vac-cga23) activation, human B cells were harvested and suspended (5×106 cells/ml) in transporter buffer (26) (110mM potassium acetate, 5mM sodium acetate, 2mM magnesium acetate, 1mM EGTA, 2mM DTT, 20mM HEPE pH 7.3 and protease-inhibitor cocktail (Biotool, cat# B14011)) with 10μg/ml digitonin (Calbiochem®, cat# 300410) in the presence or absence of CDNs (10μg/ml). Cells were cultured at 37°C for 10min in 24 well plate. Afterward, cells were harvested and resuspended in the human B cell culture medium at 5×106 cells/ml, and cultured with or without CDNs (10μg/ml) for 5hrs. Human IFNβ was measured in cell supernatant by ELISA (PBL Bioscience, cat#41415).
To measure IRF3 nuclear translocation, cells were harvested at the end of 5hr incubation. Nuclear fraction was isolated as previous reported (27) and run on a 10% Mini-PROTEAN® TGX gel (BioRAD, CAT#456-1035). Abs used for western blot are IRF3 Ab (CellSignaling, cat# 43025), α-rabbit IgG-HRP (CellSignaling, cat#7074s), α-mouse IgG-HRP (CellSignaling, cat#7076s), cyclophilinB Ab (CellSignaling, cat#76952s) and Tubulin Ab (Rockland, cat#200301-880) and rabbit anti-MPYS polyclonal Ab (3).
To measure IRF3 phosphorylation, cells were harvested at the end of 5hr incubation and lysed in the RIPA buffer as previous reported (27) and run on a 10% Mini-PROTEAN® TGX gel (BioRAD, CAT#456-1035) and probed for anti-p-IRF3 (s396) (CellSignaling, cat# 4D4G).
For RpRp-ssCDA (Biolog, cat#c118-001) activation, cells were suspended in human B cell medium at 5×106 cells/ml. 5μg/ml RpRp-ssCDA was added directly into a medium for 5hrs. Afterward, IRF3 activation was examined as above.
Q-PCR to Determine TMEM173 mRNA in Human B cells
Human B cells (1.2×106) were harvested and lysed in 350μl of RLT sample buffer with 40μM dithiothreitol. Total RNA was extracted using the RNeasy Plus Mini kit (Qiagen, cat#74134) and reverse-transcribed using the high capacity reverse transcription kit (Applied Biosystems). Quantitative PCR (Q-PCR) was carried out on a StepOnePlusTM instrument (Applied Biosystems) using the following primers and probes: human αActin (Fwd:5′-TCACCCACACTGTGCCCATCTACG-3′,Rev:5′-CAGCGGAACCGCTCATT GCCAATG-3′) and SYBER-Green human TMEM173 (BioRad, cat# 10025636, Assay ID: qHsaCID0010565). Gene expression was normalized to Actin expression and relative expression of the respective gene in untreated cells.
Semi-Quantitative PCR to Amplify Full-length Human TMEM173 gene
Total RNA was extracted from human B cells using the RNeasy Plus Mini kit (Qiagen, cat#74134). Total cDNA was made using the Superscript™ IV First-Strand Synthesis System (Invitrogen, #18091050). Full-length human TMEM173 gene (1379bp) was amplified with following primers: TMEM173-For: 5′-TTGGCTGAGTGTGTGGAGTC-3′; TMEM173-Rev: 5′-CAGTCCAGAGGCTTGGAGAC-3′. Human GAPDH primers (R&D Systems, # RDP39) were used to amplify the GAPDH cDNA as a control.
BMDM and BMDC Activation
Bone marrow cells were cultured in RPMI (Invitrogen, cat#11965118) with 10% FBS, 2mM L-glutamine, 1mM Sodium pyruvate, 10mM HEPES buffer, 1% Non-essential amino acids, 50μM 2-Mercaptoethanol, 1% Pen/Strep, 20ng/ml GM-CSF (Kingfisher, cat# RP0407M) or 20ng/ml M-CSF (Kingfisher, cat# RP0462M). The medium was changed at day 3 and 6. At day 6, cells (1×106) were transferred to a 24-well plate with fresh medium. Cells were activated at day 7 with 10μg/ml CDA, CDG, 2′3′-cGAMP or 5μg/ml Rp-Rp-ssCDA in culture directly. Mouse IFNβ was measured in culture supernatant after 5hrs by ELISA (PBL Bioscience, cat#42410). Separately, BMDM and BMDC were activated with 5μg/ml HSV DNA (Invivogen, cat# tlrl-hsv60n) and Vaccinia virus DNA (Invivogen, cat# tlrl-vav70n) transfected with lipofectamine®2000(27) and mouse IFNβ was measured in culture supernatant after 5hrs by ELISA. Alternatively, BMDC were activated with Heat kill streptococcus pneumonia (HKSP) (108c.f.u/ml) (Invivogen, cat# tlrl-hksp), LPS from Salmonella (25ng/ml) (Sigma, cat# L7261), Imiquimod (4ng/ml) (Invivogen, cat# tlrl-imqs) or CpG-ODN2395 (8ng/ml) (Invivogen, cat# tlrl-2395). Mouse TNFα and IFNβ were measured in culture supernatant after 5hrs by ELISA.
In vivo CDN Activation
Mice were intranasally administered 5μg 2′3′-cGAMP (Invivogen, cat#vac-cga23), then sacrificed after 5hrs by CO2 asphyxiation(11). Lungs were perfused with cold PBS. The harvested lungs were washed with PBS once, then stored in 0.7ml Tissue protein extraction reagent (T-PER) (Thermo Scientific, cat#78510) containing protease inhibitors (Roche, cat#11836153001) at −80°C. Later, the lung was thawed on ice and homogenized with Minilys® (Precellys, 5,000 RPM for 30sec) using Precellys lysing kit (Precellys, cat# KT03961). Lung homogenates were transferred to a 1.5ml tube and spun at 14,000g for 30min at 4°C. The supernatant was collected and analyzed for cytokine production.
Cytokine concentrations were measured by ELISA kits from eBioscience. The ELISA kits used were IL-5 (cat#88-7054), IL-12/p70 (cat#88-7921), IL-13 (cat#88-7137), IL-17A (cat#88-7371), TNF-α (cat#88-7324), IFN-λ (cat#88-7284), and IFN-γ (cat#88-7314).
Intranasal CDN Immunization
Groups of mice (4 per group) were intranasally vaccinated with 5μg 2′3′-cGAMP adjuvanted PspA (2μg, BEI Resources) or PspA alone(11). Mice were immunized twice at 14 days interval. For intranasal vaccination, animals were anesthetized using isoflurane in an E–Z Anesthesia system (Euthanex Corp, Palmer, PA). PspA, with or without 2′3-cGAMP was administered in 20μl saline. Sera, BALF, and nasal washes were collected 14 days after the last immunization. The PspA-specific Abs were determined by ELISA. Secondary Abs used were anti-mouse IgG1-HRP (Southern Biotech, cat#1070-05), anti-mouse IgG2C-HRP (Southern Biotech, cat#1079-05), and anti-mouse IgA-HRP (Southern Biotech, cat#1040-05). To determine Ag-specific Th response, splenocytes from PspA or 2′3-cGAMP + PspA immunized mice were stimulated with 5μg/ml PspA for four days in culture. Th1, Th2, and Th17 cytokines were measured in the supernatant by ELISA.
Pneumovax®23 Immunization
A group of mice (four mice per group for the Ab experiment and ten mice per group for the survival experiment) was intramuscularly administered with 0.125μg of Pneumovax® 23 (Merck, cat#7002681601) in 50μl Ultrapure PBS (Amresco, cat#K812) or PBS alone. Blood was collected, before and after immunization at indicated time. Anti-PPS2 and PPS3 IgM were determined by ELISA. Following reagents were used Pneumococcal polysaccharide Type 3 (PPS3) (ATCC® 31-X, ID # 61810463), Pneumococcal polysaccharide Type 2 (PPS2) (ATCC® 500-X,ID# 63406999), 1× ELISA assay diluent (eBioscience, REF # 00-4202-43) and goat anti-Mouse IgM HRP (cat # 1020-05, SouthernBiotech). One month after the immunization, mice were challenged (i.n) with S. pneumonia (A66.1 strain, serotype 3, ~106 c.f.u in 50μl PBS). Animal health was monitored for eight days.
Human PBMC Experiments
The study was approved by the Ethics Committee of the Charité University Medicine Berlin, and participants gave informed written consent. Genomic DNA from individuals was isolated and genotyped for analysis of the HAQ haplotype. Three individuals carrying the HAQ haplotype in homozygosity and 3 carrying R232 (WT) TMEM173 were identified.
DNA from buccal swabs was extracted using a DNA mini kit (Qiagen). Genotyping was performed by PCR using fluorescence-labeled hybridization FRET probes and melting curve analysis employing the LightCyler 480TM (Roche Diagnostics). Genotyping of the tmem173 SNP R71H was carried out using the following primer and probe: F-primer (rs11554776 S) ggagtgacacacgttgg, R-primer (Rs11554776 A) gcctagctgaggagctg, probe (rs11554776 C): ctggagtgga-XI-tgtggcgcag-PH. Primer and probes for the tmem173 SNP R293Q were as follows: F-primer (rs7380824 F): accctggtaggcaatga, R-primer (rs7380824 R): gcttagtctggtcttcctcttac, sensor probe (rs7380824 C): cctcaagtgtccggcagaagagtt-FL, anchor probe (Anc rs7380824): 640-ggcctgctcaagcctatcctcccgg-PH.
Peripheral blood samples were drawn in 50 ml EDTA-coated syringes, and PBMCs were isolated by density gradient centrifugation using sterile-filtered Histopaque®-1077 (Sigma-Aldrich Chemie Gmbh). Cells were plated at a density of 1.2×106 cell/well in a 24-well format and stimulated with 0.4 or 2 μg/well of Rp, Rp-ssCDA.
Total RNA was isolated from PBMCs lysates using the PerfectPure RNA Cultured Cell Kit (5prime GmbH) and reverse-transcribed using the high capacity reverse transcription kit (Applied Biosystems). Quantitative PCR (Q-PCR) was carried out on an ABI 7300 instrument (Applied Biosystems) using the following primers and probes: ifnb: F-primer: ccaacaagtgtctcctccaaatt, R-primer: gtaggaatccaagcaagtgtagct, probe: FAM-tgttgtgcttctccactacagctctttcca-TAMRA. Analysis of tmen173 expression was performed with a TaqMan gene expression assay Hs00736958_m1 (Applied Biosystems). Gene expression was normalized to GAPDH expression and relative expression of the respective gene in untreated cells.
Statistical Analysis
All data are expressed as means ± SEM. Statistical significance was evaluated using Prism 5.0 software to perform a Student’s t-test (unpaired, two-tailed) for comparison between mean values.
Results
Homozygous HAQ individuals are common in non-Africans
We previously estimated that ~3% of Americans are HAQ/HAQ (23). To expand this knowledge to other ethnic groups, we extracted TMEM173 genotypes data from the 1000Genome Project (Phase III). Among the five ethnic groups defined in the 1000Genome Project, we found that HAQ/HAQ is most common in East Asian (~16.07%), followed by South American (~7.78%), South Asian (~6.75%) and European (~2.78%) (Table 1). Surprisingly, no homozygous HAQ individual is found in Africans (Table 1). Instead, ~4.39% of Africans are AQ/AQ (G230A-R293Q), which is not found in non-Africans (Table 1). We concluded that human TMEM173 gene has not only great heterogeneity but also show significant population stratification.
Table 1.
Homozygous HAQ individuals are common in non-Africans1.
| European | ||
|---|---|---|
| Genotype | Carriers | Population Frequency |
| R232/R232 | 251 | 0.499 |
| R232/H232 | 99 | 0.1968 |
| HAQ/R232 | 97 | 0.1928 |
| HAQ/H232 | 22 | 0.0437 |
| H232/H232 | 14 | 0.0278 |
| HAQ/HAQ | 14 | 0.0278 |
| AQ/R232 | 3 | 0.006 |
| A230/R232 | 1 | 0.002 |
| AQ/AQ | 1 | 0.002 |
| AQ/HAQ | 1 | 0.002 |
| Total | 503 | 1 |
| East Asian | ||
|---|---|---|
| Genotype | Carriers | Population Frequency |
| HAQ/R232 | 173 | 0.3433 |
| R232/R232 | 111 | 0.2202 |
| HAQ/HAQ | 81 | 0.1607 |
| HAQ/H232 | 66 | 0.131 |
| R232/H232 | 58 | 0.1151 |
| H232/H232 | 9 | 0.0179 |
| AQ/R232 | 3 | 0.006 |
| AQ/HAQ | 1 | 0.002 |
| HA/HAQ | 1 | 0.002 |
| AQ/H232 | 1 | 0.002 |
| Total | 504 | 1 |
| African | ||
|---|---|---|
| Genotype | Carriers | Population Frequency |
| R232/R232 | 255 | 0.3858 |
| AQ/R232 | 184 | 0.2784 |
| R232/H232 | 88 | 0.1331 |
| AQ/H232 | 33 | 0.0499 |
| AQ/AQ | 29 | 0.0439 |
| Q293/R232 | 23 | 0.0348 |
| AQ/Q293 | 14 | 0.0212 |
| HAQ/R232 | 10 | 0.0152 |
| H232/H232 | 7 | 0.0106 |
| Q293/H232 | 7 | 0.0106 |
| AQ/HAQ | 4 | 0.0061 |
| HAQ/H232 | 2 | 0.003 |
| Q293/Q293 | 2 | 0.003 |
| A230-H232/H232 | 1 | 0.0015 |
| A230/H232 | 1 | 0.0015 |
| AQ/A230 | 1 | 0.0015 |
| Total | 661 | 1 |
| South American | ||
|---|---|---|
| Genotype | Carriers | Population Frequency |
| R232/R232 | 106 | 0.3055 |
| HAQ/R232 | 103 | 0.2968 |
| R232/H232 | 56 | 0.1614 |
| HAQ/HAQ | 27 | 0.0778 |
| HAQ/H232 | 24 | 0.0692 |
| H232/H232 | 11 | 0.0317 |
| AQ/R232 | 5 | 0.0144 |
| HA/HAQ | 3 | 0.0086 |
| AQ/HAQ | 3 | 0.0086 |
| A230/R232 | 2 | 0.0058 |
| HAQ/A230 | 2 | 0.0058 |
| Q293/R232 | 2 | 0.0058 |
| HA/R232 | 1 | 0.0029 |
| AQ/H232 | 1 | 0.0029 |
| A230/H232 | 1 | 0.0029 |
| Total | 347 | 1 |
| South Asian | ||
|---|---|---|
| Genotype | Carriers | Population Frequency |
| R232/R232 | 201 | 0.411 |
| HAQ/R232 | 159 | 0.3251 |
| R232/H232 | 60 | 0.1227 |
| HAQ/HAQ | 33 | 0.0675 |
| HAQ/H232 | 28 | 0.0573 |
| AQ/R232 | 3 | 0.0061 |
| H232/H232 | 3 | 0.0061 |
| H71/R232 | 1 | 0.002 |
| HA/HAQ | 1 | 0.002 |
| Total | 489 | 1 |
All TMEM173 genotypes found in each of the 5 ethnic groups in the 1000Genome Project (Phase III) were summarized. Homozygous HAQ individuals are colored in Green. The two other non-functional genotypes: HAQ/H232 (light yellow) and H232/H232 (dark yellow) were also colored. Homozygous HAQ individuals are absent in African population. Instead, they have the AQ/AQ (blue). Notably, heterozygous HAQ (HAQ/R232, yellow) is the most common TMEM173 genotype in East Asian population and the second most common genotype in South American and South Asian population.
Homozygous HAQ B cells have very low MPY protein expression compared to the R232 B cells
To study the function of HAQ, we obtained EBV-transformed human B cells from homozygous HAQ individuals identified in the 1000Genome Project. These cells are distributed by National Human Genome Research Institute (NHGRI) Repository at Coriell Institute. They express B-cell surface markers IgM and HLA-DR (Figure S1A). Notably, these cells also express B cell activation markers CD80, CD86 and CD69 (Figure S1A). The expression level of these markers is similar between R232 (wt) and HAQ B cells (Figure S1A).
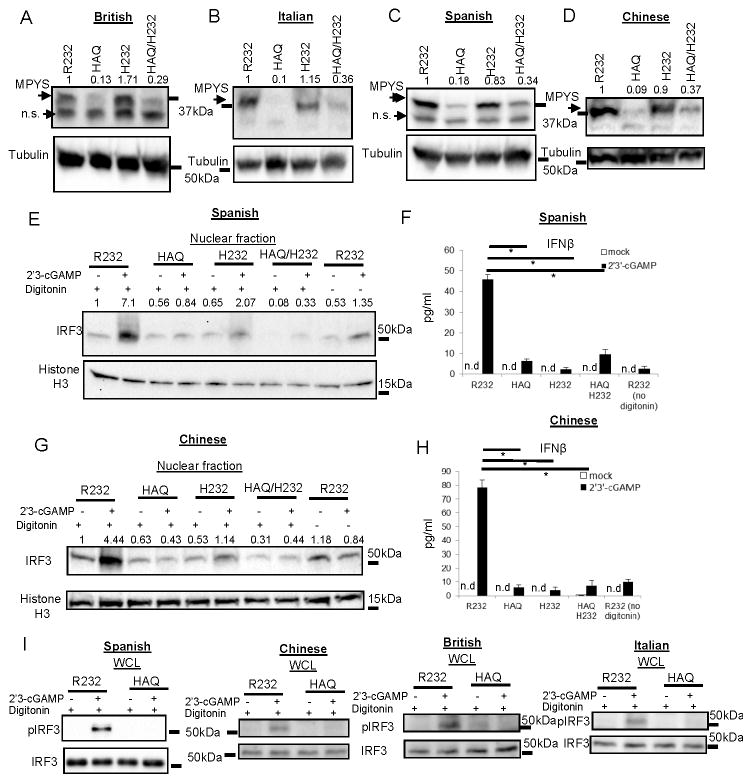
We next examined MPYS expression in these cells. Surprisingly, we found that the homozygous HAQ B cells from different ethnic groups have very low MPYS protein (Figure 1A, 1B, 1C & 1D). No MPYS protein was detected in the cell debris (Figure S1B) excluding the possibility that HAQ protein somehow may be insoluble or aggregate.
Figure 1. Homozygous HAQ human B cells have very low MPYS protein expression compared to the R232 B cells and do not respond to natural CDNs.
A–D. R232/R232, HAQ/HAQ, H232/H232 and HAQ/H232 human B cells from indicated ethnic groups were lysed in RIPA buffer and probed for MPYS expression using the rabbit anti-mouse MPYS Ab (n>3). E&G. R232/R232, HAQ/HAQ, H232/H232 and HAQ/H232 human B cells from indicated ethnic groups were activated with 2′3′-cGAMP (10μg/ml) for 5 h as described in Materials and Methods. Nuclear fractions were isolated. Samples were run on a SDS-PAGE gel and probed with the indicated Abs (n=3). F&H. Human IFNβ was measured in cell supernatant from E&G by ELISA (n=3). I. R232/R232, HAQ/HAQ human B cells from indicated ethnic groups were activated with 2′3′-cGAMP (10μg/ml) for 5 h as in E&G. Cells were lysed in the RIPA buffer. Whole cell lysate (WCL) were run on a SDS-PAGE gel and probed with the indicated Abs (n=2). Graph present means ± SEM from three independent experiments. The significance is represented by an asterisk, where p<0.05. n.s, nonspecific. n.d, not detected.
We have been using this rabbit anti-MPYS Ab since we initially identify MPYS in 2008 and its specificity has been well documented by the literature (3, 9, 23, 25). Nevertheless, to exclude the possibility that our anti-MPYS Ab may not recognize the HAQ MPYS, we cloned the HAQ TMEM173 transcript from the homozygous HAQ human B cells and expressed it in 293T cells, which lack the endogenous MPYS expression(23). Our anti-MPYS Ab staining showed a similar expression of the HAQ and R232 of MPYS in the 293T cells (Figure S1C), which indicated that our anti-MPYS Ab recognizes the HAQ of MPYS as good as the R232 of MPYS. We thus concluded that the low MPYS staining in the HAQ B cells (Figure 1A~1D) is indeed an indication of low MPYS protein expression.
We also compared the MPYS level in these 293T transfectants with our human B cells. We found that the endogenous MPYS level in human B cells is ~50-fold lower than that of 293T transfectants (Figure S1C), which suggested that overexpressing TMEM173 in 293T cells likely masked the expression difference between the endogenous R232 and HAQ of TMEM173.
Homozygous HAQ human B cells are defective in response to natural CDNs
MPYS senses natural CDNs, including the bacterial CDN CDA, CDG and mammalian CDN 2′3′-cGAMP(4, 5, 28, 29). We hypothesized that HAQ/HAQ cells would not respond to these CDNs due to the low MPYS expression.
Directly adding CDN in the human B cells culture did not activate these cells (Figure 1E &1G, Figure S2A, S2C, S2E & 2F, no digitonin). To deliver CDN into the cytosol, human B cells were reversibly permeabilized with digitonin in the presence of 2′3′-cGAMP. Activation of MPYS by CDNs leads to phosphorylation and nuclear translocation of IRF3 and subsequently IFNβ production. In Spanish and Chinese samples, 2′3′-cGAMP activates IRF3 translocation and IFNβ production in R232/R232 individuals but not the HAQ/HAQ B cells (Figure 1E–1H). Furthermore, 2′3′-cGAMP did not induce IRF3 phosphorylation in the HAQ/HAQ cells from Chinese, Spanish, British and Italian (Figure 1I). Similar observations were made in HAQ/HAQ samples in response to CDA and CDG (Figure S2A–2F).
CDN stimulation also activates MPYS-dependent NF-κB signaling(1, 27, 30). However, we found that these B cells have constitutively activated NF-κB as indicated by the presence of nuclear RelA and RelB (Figure S1D). CDN activation, which increases nuclear IRF3, did not further increase nuclear RelA or RelB (Figure S1D). This is consistent with our observation that these B cells have an activated phenotype (Figure S1A).
Homozygous HAQ human B cells are defective in response to synthetic CDN
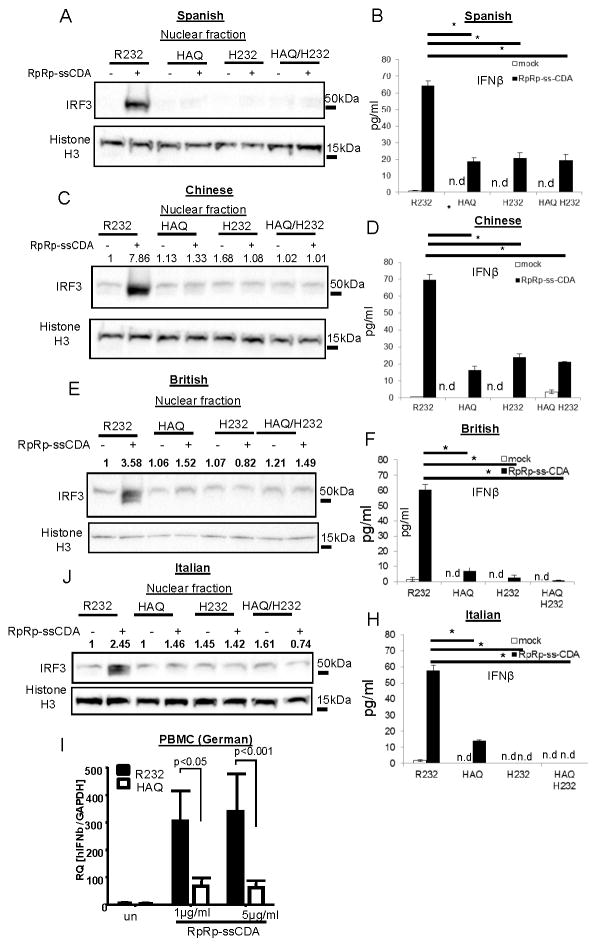
Recently, a synthetic CDN, RpRp-ssCDA, was showed to activate all major human TMEM173 variants overexpressed in 293T cells (18, 31). We next examined this synthetic CDN in human HAQ/HAQ B cells. First, we found, surprisingly, that RpRp-ssCDA can activate human B cells in medium without the need of permeabilization (Figure 2A, 2C, 2E & 2J). Second, HAQ/HAQ cells from British, Italian, Spanish and Chinese, are all defective in IRF3 activation and IFNβ production in response to RpRp-ssCDA (Figure 2A–2H). Lastly, PBMC from three German HAQ/HAQ individuals also had a defective IFNβ response to RpRp-ssCDA compared to the R232/R232 individuals (Figure 2I). RpRp-ssCDA can induce Type I IFN production in the Goldenticket mouse, which lacks detectable MPYS/STING protein (18), which may explain the residual IFNβ by RpRp-ssCDA in some samples. We concluded that homozygous HAQ B cells are defective in response to the synthetic CDN RpRp-ssCDA.
Figure 2. Homozygous HAQ human B cells are defective in response to synthetic CDN RpRp-ssCDA.
A,C,E,J. R232/R232, HAQ/HAQ, H232/H232 and HAQ/H232 human B cells from indicated ethnic groups were activated with RpRp-ssCDA (5μg/ml) for 5 h in culture. Nuclear fractions were isolated, run on a SDS-PAGE gel and probed with the indicated Abs (n=3). B,D,F,H. Human IFNβ was measured in cell supernatant from A,C E,J by ELISA (n=3). I. PBMCs from three homozygous HAQ and R232 German were stimulated with RpRp-ssCDA. Relative expression of ifnb was determined by q-PCR (n=3). Graph present means ± SEM from three independent experiments. The significance is represented by an asterisk, where p<0.05. n.d, not detected.
Homozygous H232 and the HAQ/H232 human B cells are defective in response to natural and synthetic CDNs
The H232 of MPYS has low binding affinity for CDNs (6). We found that MPYS expression in H232/H232 human B cells is similar to the R232/R232 while the HAQ/H232 B cells have decreased MPYS expression likely due to the presence of the HAQ allele (Figure 1A, 1B, 1C & 1D). We next examined their CDN responses. We found that both the homozygous H232 and the HAQ/H232 B cells did not have IRF3 nuclear translocation and IFNβ production in response to 2′3′-cGAMP, CDA, CDG or RpRp-ssCDA (Figure 1E–1H) (Figure S2) (Figure 2). We concluded that similar to the homozygous HAQ B cells, homozygous H232, and the HAQ/H232 B cells are also defective in response to CDN.
Establish an HAQ mouse model
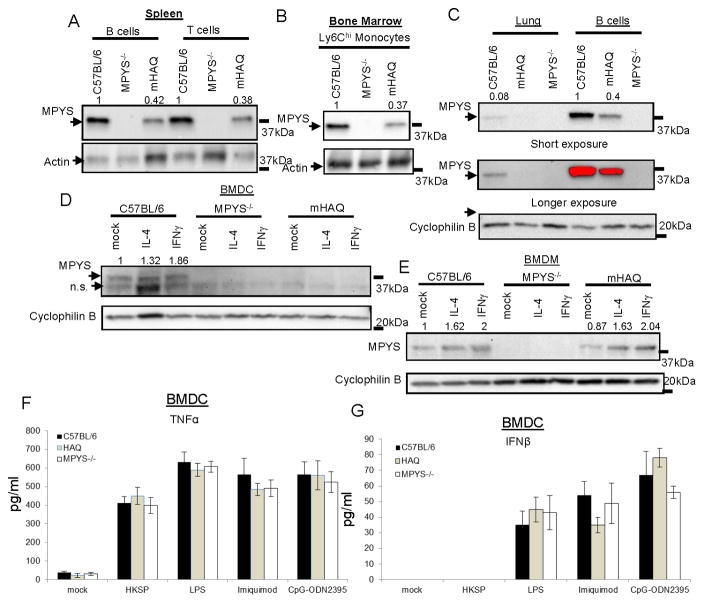
Mouse and human MPYS proteins are 82% homologous (2). To understand the in vivo significance of the HAQ of TMEM173, we generated an HAQ knock-in mouse (mHAQ). This knock-in mouse contains mouse equivalent of the HAQ mutations: C71H, I229A, and R292Q (Figure S3A). The presence of these three mutations was confirmed by sequencing (Figure S3B). Similar to the human HAQ B cell, we found that MPYS expression is also decreased in mHAQ spleen B cells (Figure 3A), which suggested that the mHAQ mouse recapitulates the main feature of the human HAQ.
Figure 3. The HAQ knock-in mouse (mHAQ) has decreased MPYS expression in multiple tissues.
A–C. Various types of cells from the C57BL/6, HAQ, and MPYS−/− mice were lysed in RIPA buffer and probed for indicated Abs as in Figure 1 (n=3). D–E. BMDC or BMDM from the C57BL/6, mHAQ, and MPYS−/− mice were treated with IL-4 (40ng/ml) or IFNγ (40ng/ml) or mock (PBS) overnight. Cells were lysed in RIPA buffer and probed for indicated Abs as in Figure 1 (n=3). F–G. BMDC from C57BL/6, MPYS−/− or mHAQ were stimulated with HKSP (107c.f.u/ml), LPS (20ng/ml), imiquimod (4ng/ml) or CpG-ODN2395 (8ng/ml) for 5hrs. TNFα (F) and IFNβ (G) were measured in the cell supernatant (n=3). Graph present means ± SEM from three independent experiments.
The establishment of the mHAQ mouse allowed us to examine HAQ expression and function beyond the B cells. Indeed, we found that MPYS expression is decreased in mHAQ spleen T cells (Figure 3A) and mHAQ bone marrow Ly6Chi monocytes (Figure 3B). The defect is more pronounced in the mHAQ lung (Figure 3C) and mHAQ BMDC (Figure 3D) where MPYS expression is not detectable. Notably, in naïve BMDM, IFNγ differentiated M1 macrophage or IL-4 differentiated M2 macrophage, MPYS expression is similar between the WT and mHAQ mice (Figure 3E), which may indicate a macrophage-specific regulation of MPYS protein expression.
The mHAQ mouse is defective in response to CDNs in vitro and in vivo
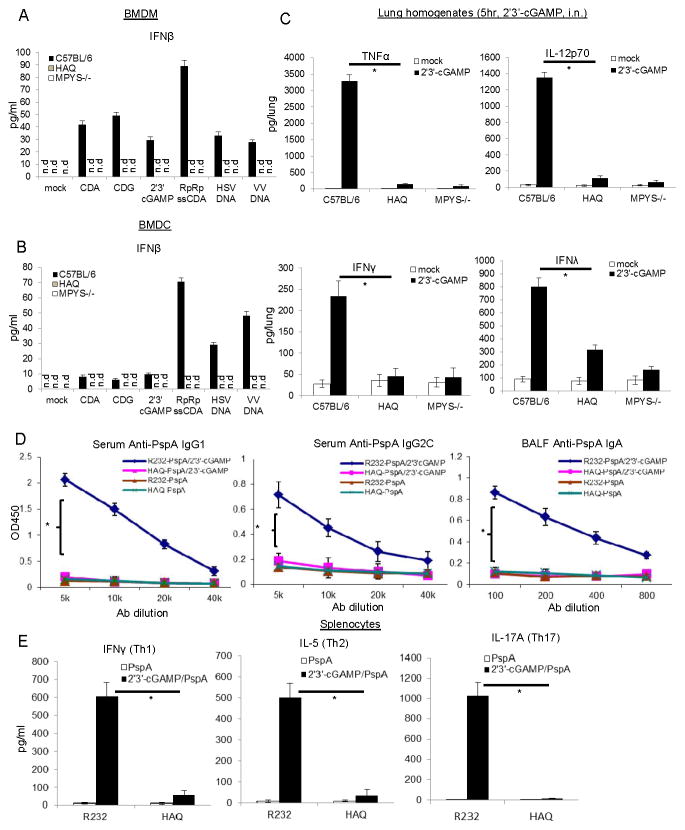
Human HAQ B cells are non-responsive to CDNs (Figure 1 and S2). We next examined CDNs response in BMDC and BMDM from the mHAQ mouse. As expected, both the mHAQ BMDM and BMDC did not produce IFNβ in response to CDA, CDG, 2′3′-cGAMP or RpRp-ssCDA (Figure 4A & 4B). The mHAQ BMDM and BMDC also did not make IFNβ in response to transfected Herpes Simplex Virus DNA or Vaccinia virus DNA (Figure 3A & 3B). As a control, BMDC from mHAQ mice has similar TNFα (Figure 3F) and IFNβ (Figure 3G) production as the WT mice when stimulated with TLR2 ligand Heat-killed streptococcus pneumonia (HKSP), TLR4 ligand LPS, TLR7 ligand imiquimod, and TLR9 ligand CpG-ODN2395.
Figure 4. mHAQ mouse does not respond to CDNs in vitro and in vivo.
A&B. BMDM or BMDC from C57BL/6, mHAQ and MPYS−/− mice were activated by 10μg/ml of CDA, CDG, 2′3′-cGAMP or 5μg/ml RpRp-ssCDA, HSV-DNA, VV-DNA for 5hrs. Mouse IFNβ was measured in cell supernatant (n=3). C. C57BL/6, mHAQ and MPYS−/− mice were treated (i.n.) with saline or 2′3′-cGAMP (5μg) for 5hrs. Cytokines were determined in lung homogenates by ELISA (n=3). D. WT littermate (R232) and mHAQ mice were immunized (i.n.) with PspA (2μg) alone or together with 5μg 2′3′-cGAMP as described in Material and Methods. Anti-PspA IgG1, IgG2C, and IgA were measured by ELISA (n=3). E. Splenocytes from PspA or 2′3′-cGAMP + PspA immunized mice were stimulated with 5μg/ml PspA for 4 days in culture. Cytokines were measured in the supernatant by ELISA (n=3). Graph present means ± SEM from three independent experiments. The significance is represented by an asterisk, where p<0.05. n.d, not detected.
We next examined the in vivo CDN responses in the mHAQ mouse. Intranasal administration of CDN elicits rapid cytokine productions in the lung that is important for the mucosal adjuvant activity of CDNs (11). We found that intranasal administration of 2′3′-cGAMP did not elicit lung production of TNFα, IL-12p70, IFNγ or IFNλ in the mHAQ mouse (Figure 4C). We further examined the mucosal adjuvant activity of 2′3′-cGAMP in the mHAQ mouse(32). As expected, 2′3′-cGAMP did not induce Ag-specific Ab or Th response in the mHAQ mouse (Figure 4D&4E). We concluded that the mHAQ mouse does not respond to CDN in vivo and in vitro.
Pneumovax®23 is less effective in the mHAQ mouse than the WT mice
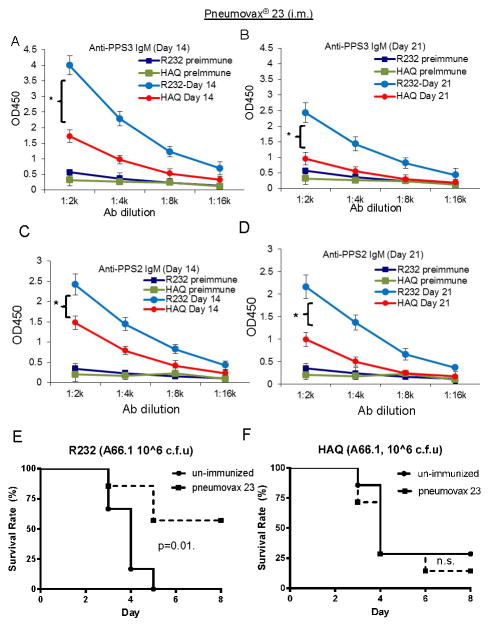
The CDNs-MPYS/STING activation in B cells is required for polysaccharide-based vaccine activity such as Pneumovax®23 (33). Since mHAQ mice do not have functional CDNs-MPYS pathway (Figure 4), we hypothesized that Pneumovax®23 would not be effective in the mHAQ mouse. Indeed, upon intramuscular Pneumovax®23 immunization, the mHAQ mice have lower anti-PPS3 IgM (Figure 5A & 5B) and anti-Pneumococcal polysaccharide Type 2 (PPS2) IgM (Figure 5C & 5D) production than the WT mice at day 14 and 21.
Figure 5. Pneumovax®23 is less effective in mHAQ mice.
A–D. WT littermates (R232) and mHAQ mice were immunized (i.m.) with Pneumovax®23 (0.125μg in 50μl saline). Anti-PPS3 (A–B) and anti-PPS2 (C–D) IgM Ab was determined at day 14 and 21 post immunization as well as pre-immunization (n=3). E–F. WT littermates (R232) or mHAQ mice were given (i.m.) saline or Pneumovax®23 as in A–D. One month post-immunization, mice were infected (i.n.) with S.pneumoniae (A66.1 strain, ~ 1.0×106 c.f.u.). Mice health was monitored for 8 days (n=2). Graph present means ± SEM from three independent experiments. The significance is represented by an asterisk, where p<0.05.
To examine the protective immunity of Pneumovax®23 in the mHAQ mouse, we challenged vaccinated mice with the A66.1 strain, an invasive strain of S. pneumococcus. Consistent with the Ab results, Pneumovax®23 protected WT mice from the A66.1 S. pneumococcus infection (Figure 5E) but not the mHAQ mouse (Figure 5F). We concluded that Pneumovax®23 is not effective in the mHAQ mouse.
Homozygous HAQ B cells have decreased TMEM173 transcript
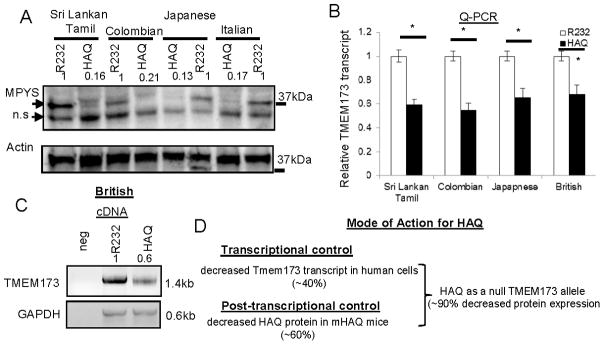
We next ask why the human HAQ allele has low MPYS protein expression. We examined the TMEM173 mRNA level in the homozygous HAQ B cells. Surprisingly, we found that HAQ B cells from Sri Lankan Tamil, Colombian, Japanese and Italian that have low MPYS expression (Figure 6A), all have ~40% lower TMEM173 mRNA than their R232/R232 ethnic controls (Figure 6B). Semi-quantitative PCR also show that the full-length human TMEM173 transcript (~1.4kb) is decreased in the homozygous HAQ B cells compared to their R232 counterparts (Figure 6C). We concluded that human HAQ B cells had decreased TMEM173 transcript.
Figure 6. Homozygous HAQ human B cells have decreased TMEM173 transcript.
A. MPYS expression was determined by western blot in homozygous HAQ or R232 human B cells from indicated ethnic groups as Figure 1 (n>3). B. TMEM173 mRNA was measured by Q-PCR in the homozygous HAQ or R232 human B cells (n=3). C. Full-length human TMEM173 cDNA was amplified from homozygous HAQ and R232 human B cells as described in Materials and Methods. D. Model for the transcriptional and post-transcriptional control of the HAQ expression. Graph present means ± SEM from three independent experiments. The significance is represented by an asterisk, where p<0.05.
We next asked if the homozygous HAQ individuals have decreased TMEM173 transcript in tissues other than B cells. To answer that, we mined data from The Genotype-Tissue Expression (GTEx) database, which compiles data regarding human gene expression related to genetic variations. We focused on the SNPs that affect the TMEM173 transcript. We found that all three HAQ SNPs, rs11554776(R71H) -rs78233829(G230A) -rs7380824(R293Q), are associated with decreased TMEM173 transcript with highly significant p values as low as 10−19, 10−22 and 10−23 (Table 2). Furthermore, this decreased TMEM173 transcript in HAQ individuals can be found in non-B cell dominant tissues such as Artery, fibroblasts, lung, Thyroid and Esophagus (Table 2). Thus, homozygous HAQ individuals have decreased TMEM173 in tissues other than B cells.
Table 2.
HAQ/HAQ individuals have decreased TMEM173 transcript in various tissues1.
| R71H | |||
| rs11554776 | 2.50E-19 | −0.7 | Artery - Aorta |
| rs11554776 | 1.50E-16 | −0.44 | Artery - Tibial |
| rs11554776 | 3.70E-13 | −0.45 | Cells - Transformed fibroblasts |
| rs11554776 | 9.10E-10 | −0.33 | Thyroid |
| rs11554776 | 9.70E-10 | −0.44 | Lung |
| rs11554776 | 1.90E-09 | −0.36 | Esophagus - Muscularis |
| rs11554776 | 1.40E-08 | −0.57 | Heart - Atrial Appendage |
| rs11554776 | 6.50E-08 | −0.23 | Adipose - Subcutaneous |
| rs11554776 | 0.0000012 | −0.25 | Nerve - Tibial |
| rs11554776 | 0.0000047 | −0.16 | Skin - Sun Exposed (Lower leg) |
| rs11554776 | 0.0000092 | −0.31 | Breast - Mammary Tissue |
| G230A | |||
| rs78233829 | 1.50E-22 | −0.46 | Artery - Tibial |
| rs78233829 | 1.10E-17 | −0.63 | Artery - Aorta |
| rs78233829 | 3.70E-15 | −0.43 | Cells - Transformed fibroblasts |
| rs78233829 | 4.50E-11 | −0.35 | Esophagus - Muscularis |
| rs78233829 | 5.60E-11 | −0.42 | Lung |
| rs78233829 | 1.30E-10 | −0.31 | Thyroid |
| rs78233829 | 1.20E-09 | −0.23 | Adipose - Subcutaneous |
| rs78233829 | 6.00E-09 | −0.26 | Nerve - Tibial |
| rs78233829 | 8.60E-09 | −0.5 | Heart - Atrial Appendage |
| rs78233829 | 6.80E-08 | −0.17 | Skin - Sun Exposed (Lower leg) |
| rs78233829 | 0.0000039 | −0.28 | Pancreas |
| rs78233829 | 0.0000054 | −0.3 | Breast - Mammary Tissue |
| rs78233829 | 0.000014 | −0.17 | Muscle - Skeletal |
| R293Q | |||
| rs7380824 | 8.90E-23 | −0.45 | Artery - Tibial |
| rs7380824 | 2.50E-19 | −0.66 | Artery - Aorta |
| rs7380824 | 4.30E-15 | −0.42 | Cells - Transformed fibroblasts |
| rs7380824 | 2.90E-11 | −0.35 | Esophagus - Muscularis |
| rs7380824 | 3.90E-11 | −0.42 | Lung |
| rs7380824 | 1.70E-10 | −0.25 | Adipose - Subcutaneous |
| rs7380824 | 2.80E-10 | −0.3 | Thyroid |
| rs7380824 | 3.50E-09 | −0.5 | Heart - Atrial Appendage |
| rs7380824 | 4.20E-09 | −0.26 | Nerve - Tibial |
| rs7380824 | 6.20E-08 | −0.17 | Skin - Sun Exposed (Lower leg) |
| rs7380824 | 0.0000029 | −0.31 | Breast - Mammary Tissue |
| rs7380824 | 0.000003 | −0.29 | Pancreas |
| rs7380824 | 0.0000062 | −0.49 | Artery - Coronary |
| rs7380824 | 0.0000077 | −0.21 | Esophagus - Mucosa |
| rs7380824 | 0.000009 | −0.18 | Muscle - Skeletal |
Data were compiled from The Genotype-Tissue Expression Database (GTEx).
Discussion
The common human HAQ TMEM173 allele was first identified and characterized by us in 2011(23). We characterized the HAQ as a loss-of-function TMEM173 allele because it loses >90% of the ability to stimulate IFNβ production when transiently overexpressed in the 293T cells, a hallmark function of MPYS/STING(23). In 2013, Diner, E.J., et.al.(5) found that the THP-1 cell, a human monocytic cell line originated from a Japanese (34) have the HAQ of TMEM173. However, it is not clear if the THP-1 cells are homozygous or heterozygous for HAQ. Also in 2013, Yi, G., et. al found that 293T cells stably expressing the HAQ can respond to CDN, albeit weaker than the R232 of TMEM173 (24). Using 293T cells stable transfectants to study HAQ function, nevertheless, could be misleading. The reasons are (i) the level of MPYS is 50-fold higher in the 293T cells transfectants than the endogenous MPYS (Figure S1C). (ii) Human HAQ cells have decreased TMEM173 transcript (Figure 6). This feature of the HAQ is lost when expressing the HAQ cDNA in the 293T cells. In the current report, we used homozygous HAQ human B cells from multiple ethnic groups showed that homozygous HAQ B cells have very low MPYS expression compared to the R232 B cells and do not respond to CDN in vitro. Furthermore, PBMCs from 3 homozygous HAQ German have decreased CDN response compared to the R232 German. Lastly, an HAQ knock-in mouse has decreased MPYS expression and did not respond to CDN in vitro and in vivo. Thus, HAQ is indeed a loss-of-function human TMEM173 allele likely due to its extremely low protein expression.
Two other TMEM173 genotypes, HAQ/H232, and H232/H232 also did not respond to CDN (Figure 1 & 2). The underlying molecular mechanisms are likely different. The H232/H232 B cells, unlike the HAQ/HAQ, have similar MPYS expression as the R232/R232 B cells. Previous studies found that the H232 of MPYS binds CDN (Kd ~5.3μM) much poorly than the R232 of MPYS (Kd ~0.11μM) (6). Consequently, the H232 is severely defective in response to CDN stimulation when expressed in 293T cells (6, 29). Here, we verified this observation in our homozygous H232 human B cells (Figure 1). The HAQ/H232 B cells have the HAQ allele, which contributes to its low MPYS expression (Figure 1), and the non-functional H232 allele. Together, they lead to the unresponsiveness to CDN in the HAQ/H232 B cells.
It is worth noting that in all, the HAQ/HAQ, HAQ/H232 and H232/H232 genotypes, consist of ~10% of Europeans and ~31% of East Asians (Table 1). This is significant because there are tremendous interests to develop MPYS/STING-targeting immunotherapies for cancers and infectious diseases (18, 19, 21, 22, 35). It will be especially challenging to develop MPYS/STING-targeting immunotherapies for the homozygous HAQ individuals because of their extremely low MPYS protein expression. Indeed, we showed that the licensed pneumococcal vaccines Pneumovax®23 is less effective in the mHAQ mouse than the WT mice (Figure 5). Fu, J et.al., did show that the synthetic CDN RpRp-ssCDA activates PBMCs similarly in HAQ/HAQ and R232/R232 donors (18). However, the PBMC was from 1 single HAQ/HAQ donor (18). Here, we used three homozygous German HAQ individuals and found they were defective in response to RpRp-ssCDA (Figure 2I). Future development of MPYS-targeting immunotherapies must adopt the concept of personalized medicine.
Surprisingly, we found that the synthetic CDN RpRp-ssCDA is membrane permeable. Natural CDNs have two phosphate groups preventing it from directly passing through the cell membrane. To activate MPYS, which is inside cells, investigators have to use transfection or membrane permeabilizing reagents to deliver CDN to the cytosol. We previously showed that, in vivo, only pinocytosis-efficient cells such as macrophage and dendritic cells, can directly take up CDG and be activated (11). The observation that RpRp-ssCDA is cell-permeable makes it a very attractive CDN to direct activate cells that are not pinocytosis-efficient such as B cells.
Last, the null phenotype of the HAQ allele is likely a result of the decreased TMEM173 transcript and the amino acids changes (R71H-G230A-R293Q) in the HAQ protein (Figure 6D). The MPYS protein level is down ~60% in the mHAQ knock-in mouse. Previously, an I199N change in the mouse TMEM173 gene leads to a complete loss of the STING/MPYS protein expression(36). Thus, amino acid changes in MPYS protein can impact its expression. MPYS expression can also be regulated at a transcriptional level. Mouse and human TMEM173 genes have conserved STAT1 binding sites(37). Type I and II treatments increase mouse and human TMEM173 expression via an STAT-1 dependent mechanism (37). Nevertheless, treating homozygous human HAQ B cells with IFNγ did not restore MPYS protein expression (data not shown). Further studies are needed to reveal the mechanisms by which human TMEM173 expression is controlled on the transcriptional and post-transcriptional level.
In summary, we found that human HAQ, the second most common TMEM173 allele, is a null allele. The mouse model of HAQ, the mHAQ knock-in mouse, are not protected by Pneumovax®23. Future studies need to be done to determine the impact and mechanisms by which HAQ, as a loss-of-function common TMEM173 allele, influence human diseases, and medicines. Our HAQ knock-in mouse will be especially valuable in this endeavor.
Supplementary Material
Acknowledgments
This work was supported by National Institute of Allergy and Infectious Diseases Grant 1R56AI110606, 1R01AI110606, R21AI099346-Subcontract and 1R21AI125999 (to L.J.), the Gary and Janis Grover Young Scientist Award (to L.J.), the Deutsche Forschungsgemeinschaft (OP 86/10-1) and SFB-TR84 (to B.O.), and GRK1673 (to J.S.R.M. and B.O).
Abbreviations
- HAQ
R71H-G230A-R293Q
- CDN
cyclic dinucleotide
- CDG
Cyclic di-GMP
- CDA
Cyclic di-AMP
- 2′3′-cGAMP
GMP-AMP, 2′5-3′5′-cyclic
- PspA
Pneumococcal surface protein A
References
- 1.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, Shu HB. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Jin L, Waterman PM, Jonscher KR, Short CM, Reisdorph NA, Cambier JC. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol Cell Biol. 2008;28:5014–5026. doi: 10.1128/MCB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao P, Ascano M, Zillinger T, Wang W, Dai P, Serganov AA, Gaffney BL, Shuman S, Jones RA, Deng L, Hartmann G, Barchet W, Tuschl T, Patel DJ. Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell. 2013;154:748–762. doi: 10.1016/j.cell.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holm CK, Rahbek SH, Gad HH, Bak RO, Jakobsen MR, Jiang Z, Hansen AL, Jensen SK, Sun C, Thomsen MK, Laustsen A, Nielsen CG, Severinsen K, Xiong Y, Burdette DL, Hornung V, Lebbink RJ, Duch M, Fitzgerald KA, Bahrami S, Mikkelsen JG, Hartmann R, Paludan SR. Influenza A virus targets a cGAS-independent STING pathway that controls enveloped RNA viruses. Nat Commun. 2016;7:10680. doi: 10.1038/ncomms10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin L, Getahun A, Knowles HM, Mogan J, Akerlund LJ, Packard TA, Perraud AL, Cambier JC. STING/MPYS mediates host defense against Listeria monocytogenes infection by regulating Ly6C(hi) monocyte migration. J Immunol. 2013;190:2835–2843. doi: 10.4049/jimmunol.1201788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Archer KA, Durack J, Portnoy DA. STING-dependent type I IFN production inhibits cell-mediated immunity to Listeria monocytogenes. PLoS Pathog. 2014;10:e1003861. doi: 10.1371/journal.ppat.1003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blaauboer SM, Mansouri S, Tucker HR, Wang HL, Gabrielle VD, Jin L. The mucosal adjuvant cyclic di-GMP enhances antigen uptake and selectively activates pinocytosis-efficient cells in vivo. Elife. 2015;4 doi: 10.7554/eLife.06670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koppe U, Hogner K, Doehn JM, Muller HC, Witzenrath M, Gutbier B, Bauer S, Pribyl T, Hammerschmidt S, Lohmeyer J, Suttorp N, Herold S, Opitz B. Streptococcus pneumoniae stimulates a STING- and IFN regulatory factor 3-dependent type I IFN production in macrophages, which regulates RANTES production in macrophages, cocultured alveolar epithelial cells, and mouse lungs. J Immunol. 2012;188:811–817. doi: 10.4049/jimmunol.1004143. [DOI] [PubMed] [Google Scholar]
- 13.Ahn J, Gutman D, Saijo S, Barber GN. STING manifests self DNA-dependent inflammatory disease. Proc Natl Acad Sci U S A. 2012;109:19386–19391. doi: 10.1073/pnas.1215006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gall A, Treuting P, Elkon KB, Loo YM, Gale M, Jr, Barber GN, Stetson DB. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity. 2012;36:120–131. doi: 10.1016/j.immuni.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma S, Campbell AM, Chan J, Schattgen SA, Orlowski GM, Nayar R, Huyler AH, Nundel K, Mohan C, Berg LJ, Shlomchik MJ, Marshak-Rothstein A, Fitzgerald KA. Suppression of systemic autoimmunity by the innate immune adaptor STING. Proc Natl Acad Sci U S A. 2015;112:E710–717. doi: 10.1073/pnas.1420217112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Jesus AA, Marrero B, Yang D, Ramsey SE, Montealegre Sanchez GA, Tenbrock K, Wittkowski H, Jones OY, Kuehn HS, Lee CC, DiMattia MA, Cowen EW, Gonzalez B, Palmer I, DiGiovanna JJ, Biancotto A, Kim H, Tsai WL, Trier AM, Huang Y, Stone DL, Hill S, Kim HJ, St Hilaire C, Gurprasad S, Plass N, Chapelle D, Horkayne-Szakaly I, Foell D, Barysenka A, Candotti F, Holland SM, Hughes JD, Mehmet H, Issekutz AC, Raffeld M, McElwee J, Fontana JR, Minniti CP, Moir S, Kastner DL, Gadina M, Steven AC, Wingfield PT, Brooks SR, Rosenzweig SD, Fleisher TA, Deng Z, Boehm M, Paller AS, Goldbach-Mansky R. Activated STING in a vascular and pulmonary syndrome. N Engl J Med. 2014;371:507–518. doi: 10.1056/NEJMoa1312625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeremiah N, Neven B, Gentili M, Callebaut I, Maschalidi S, Stolzenberg MC, Goudin N, Fremond ML, Nitschke P, Molina TJ, Blanche S, Picard C, Rice GI, Crow YJ, Manel N, Fischer A, Bader-Meunier B, Rieux-Laucat F. Inherited STING-activating mutation underlies a familial inflammatory syndrome with lupus-like manifestations. J Clin Invest. 2014;124:5516–5520. doi: 10.1172/JCI79100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu J, Kanne DB, Leong M, Glickman LH, McWhirter SM, Lemmens E, Mechette K, Leong JJ, Lauer P, Liu W, Sivick KE, Zeng Q, Soares KC, Zheng L, Portnoy DA, Woodward JJ, Pardoll DM, Dubensky TW, Jr, Kim Y. STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci Transl Med. 2015;7:283ra252. doi: 10.1126/scitranslmed.aaa4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Celis E. STING activator c-di-GMP enhances the anti-tumor effects of peptide vaccines in melanoma-bearing mice. Cancer Immunol Immunother. 2015;64:1057–1066. doi: 10.1007/s00262-015-1713-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carroll EC, Jin L, Mori A, Munoz-Wolf N, Oleszycka E, Moran HB, Mansouri S, McEntee CP, Lambe E, Agger EM, Andersen P, Cunningham C, Hertzog P, Fitzgerald KA, Bowie AG, Lavelle EC. The Vaccine Adjuvant Chitosan Promotes Cellular Immunity via DNA Sensor cGAS-STING-Dependent Induction of Type I Interferons. Immunity. 2016;44:597–608. doi: 10.1016/j.immuni.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng L, Liang H, Xu M, Yang X, Burnette B, Arina A, Li XD, Mauceri H, Beckett M, Darga T, Huang X, Gajewski TF, Chen ZJ, Fu YX, Weichselbaum RR. STING-Dependent Cytosolic DNA Sensing Promotes Radiation-Induced Type I Interferon-Dependent Antitumor Immunity in Immunogenic Tumors. Immunity. 2014;41:843–852. doi: 10.1016/j.immuni.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woo SR, Fuertes MB, Corrales L, Spranger S, Furdyna MJ, Leung MY, Duggan R, Wang Y, Barber GN, Fitzgerald KA, Alegre ML, Gajewski TF. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41:830–842. doi: 10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin L, Xu LG, Yang IV, Davidson EJ, Schwartz DA, Wurfel MM, Cambier JC. Identification and characterization of a loss-of-function human MPYS variant. Genes Immun. 2011;12:263–269. doi: 10.1038/gene.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yi G, Brendel VP, Shu C, Li P, Palanathan S, Cheng Kao C. Single nucleotide polymorphisms of human STING can affect innate immune response to cyclic dinucleotides. PLoS One. 2013;8:e77846. doi: 10.1371/journal.pone.0077846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin L, Hill KK, Filak H, Mogan J, Knowles H, Zhang B, Perraud AL, Cambier JC, Lenz LL. MPYS is required for IFN response factor 3 activation and type I IFN production in the response of cultured phagocytes to bacterial second messengers cyclic-di-AMP and cyclic-di-GMP. J Immunol. 2011;187:2595–2601. doi: 10.4049/jimmunol.1100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyamoto K, Yamashita T, Tsukiyama T, Kitamura N, Minami N, Yamada M, Imai H. Reversible membrane permeabilization of mammalian cells treated with digitonin and its use for inducing nuclear reprogramming by Xenopus egg extracts. Cloning Stem Cells. 2008;10:535–542. doi: 10.1089/clo.2008.0020. [DOI] [PubMed] [Google Scholar]
- 27.Blaauboer SM, Gabrielle VD, Jin L. MPYS/STING-mediated TNF-alpha, not type I IFN, is essential for the mucosal adjuvant activity of (3′-5′)-cyclic-di-guanosine-monophosphate in vivo. J Immunol. 2014;192:492–502. doi: 10.4049/jimmunol.1301812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, Tuschl T, Patel DJ. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abe T, Barber GN. Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-kappaB activation through TBK1. J Virol. 2014;88:5328–5341. doi: 10.1128/JVI.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corrales L, Glickman LH, McWhirter SM, Kanne DB, Sivick KE, Katibah GE, Woo SR, Lemmens E, Banda T, Leong JJ, Metchette K, Dubensky TW, Jr, Gajewski TF. Direct Activation of STING in the Tumor Microenvironment Leads to Potent and Systemic Tumor Regression and Immunity. Cell Rep. 2015;11:1018–1030. doi: 10.1016/j.celrep.2015.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skrnjug I, Guzman CA, Rueckert C. Cyclic GMP-AMP displays mucosal adjuvant activity in mice. PLoS One. 2014;9:e110150. doi: 10.1371/journal.pone.0110150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng M, Hu Z, Shi X, Li X, Zhan X, Li XD, Wang J, Choi JH, Wang KW, Purrington T, Tang M, Fina M, DeBerardinis RJ, Moresco EM, Pedersen G, McInerney GM, Karlsson Hedestam GB, Chen ZJ, Beutler B. MAVS, cGAS, and endogenous retroviruses in T-independent B cell responses. Science. 2014;346:1486–1492. doi: 10.1126/science.346.6216.1486. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Tsuchiya S, Yamabe M, Yamaguchi Y, Kobayashi Y, Konno T, Tada K. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1) Int J Cancer. 1980;26:171–176. doi: 10.1002/ijc.2910260208. [DOI] [PubMed] [Google Scholar]
- 35.Temizoz B, Kuroda E, Ohata K, Jounai N, Ozasa K, Kobiyama K, Aoshi T, Ishii KJ. TLR9 and STING agonists synergistically induce innate and adaptive type-II IFN. Eur J Immunol. 2015;45:1159–1169. doi: 10.1002/eji.201445132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma F, Li B, Yu Y, Iyer SS, Sun M, Cheng G. Positive feedback regulation of type I interferon by the interferon-stimulated gene STING. EMBO Rep. 2015;16:202–212. doi: 10.15252/embr.201439366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.