Abstract
Aims
To assess the prevalence of ROS1 rearrangements in a retrospective and prospective diagnostic Australian cohort and evaluate the effectiveness of immunohistochemical screening.
Methods
A retrospective cohort of 278 early stage lung adenocarcinomas and an additional 104 prospective NSCLC cases referred for routine molecular testing were evaluated. ROS1 immunohistochemistry (IHC) was performed (D4D6 clone, Cell Signaling Technology) on all cases as well as fluorescence in situ hybridisation (FISH) using the ZytoVision and Abbott Molecular ROS1 FISH probes, with ≥15% of cells with split signals considered positive for rearrangement.
Results
Eighty eight cases (32%) from the retrospective cohort showed staining by ROS1 IHC, and one case (0.4%) showed ROS1 rearrangement by FISH. Nineteen of the prospective diagnostic cases showed ROS1 IHC staining of which 12 (12%) cases were confirmed as ROS1 rearranged by FISH. There were no ROS1 rearranged cases that showed no expression of ROS1 with IHC. The ROS1 rearranged cases in the prospective cohort were all EGFR wildtype and ALK rearrangement negative. The sensitivity of ROS1 IHC in the retrospective cohort was 100% and specificity was 76%.
Conclusions
ROS1 rearrangements are rare events in lung adenocarcinomas. Selection of cases for ROS1 FISH testing, by excluding EGFR/ALK positive cases and use of IHC to screen for potentially positive cases can be used to enrich for the likelihood of a identifying a ROS1 rearranged lung cancer and prevent the need to undertake expensive and time consuming FISH testing in all cases.
Keywords: c-ros oncogene 1 (ROS1), non-small cell lung cancer (NSCLC), immunohistochemistry (IHC), fluorescence in situ hybridisation (FISH)
Introduction
A new paradigm of targeted therapies has emerged for non-small cell lung cancer following the discovery of a number of targetable, generally mutually exclusive driver mutations such as those involving epidermal growth factor receptor (EGFR), anaplastic lymphoma kinase (ALK), and c-ros oncogene 1 (ROS1). Like ALK, rearrangements involving ROS1 are strongly predictive of response to the inhibitor crizotinib1.
ROS1 is the oncogene product of the avian sarcoma RNA tumour virus2–4 that encodes a transmembrane tyrosine kinase receptor from the insulin receptor subfamily, and has high homology with the intracellular kinase domain and ATP binding site of ALK. Activation of ROS1 leads to signalling through downstream oncogenic pathways including PI3Kinase/Akt, MTOR and RAS-MAPK/ERK pathways 5–7. Rearranged or activated ROS1 has been shown to have transforming potential in nude mice with NSCLC8. In addition, ROS1 gene rearrangements have also been identified in other malignancies such as glioblastomas9, cholangiocarcinoma10–12, ovarian cancer13, gastric cancer14 and colorectal cancer15.
Driver mutations involving rearrangement of the ROS1 gene have recently been described in NSCLC and act as a target for tyrosine kinase inhibitors. ROS1 rearrangements have been identified in 1–3% of NSCLC8,16,17, but at higher rates in young, never-smoker, lung adenocarcinoma patients17. Due to the high homology between the kinase domains of ROS1 and ALK, ALK inhibitors were tested on ROS1 positive cell lines and tumours and were found to be inhibitory18. ROS1 rearranged tumours determined by break apart fluorescence in situ hybridisation (FISH) testing were added to eligibility criteria for the PROFILE 1001 study, a phase I study evaluating the ALK, MET/ROS1 tyrosine kinase inhibitor crizotinib, which reported an overall objective response rate of 72% in 50 ROS1 positive patients19. Preliminary studies suggest ROS1 rearranged patients treated with crizotinib may have longer median response duration compared with ALK rearranged patients, perhaps due to crizotinib having a higher binding efficiency and potency to inhibit ROS119.
Although some ROS1 fusion partners are intrachromosomal involving the long (q) arm of chromosome 6, most partners occur on other chromosomes20. A total of 12 ROS1 fusion variants have been identified, with fusion partners that include: SLC34-A2, CD74, TMP3, SDC4, EZR, LRIG3, GOPC (FIG), KDELR2 and CCDC68,21,22. Importantly, all fusions include the receptor tyrosine kinase domain of ROS18.
Although ROS1 rearranged lung cancers show promise as targetable tumours, there is a significant challenge in determining the best way to identify this rare alteration, often in small biopsy samples with limited tissue available for analysis. A number of studies show that ROS1 immunohistochemistry can be utilised in conjunction with FISH to reveal ROS1 rearrangements in NSCLC23–31. The finding that some cases with ROS1 rearrangement show weak ROS1 immunoreactivity has led a number of authors to conclude that although diffuse-positive ROS1 IHC staining with moderate-strong intensity is more commonly associated with ROS1 rearrangement, this is not always the case23,24,26. Furthermore, strongly positive ROS1 IHC cases can sometimes be ROS1 FISH negative23–25,27,28.
IHC screening for ALK rearrangements in NSCLC has been established as an effective technique to identify ALK positive tumours32–37. An ALK IHC assay has also received FDA approval as a diagnostic companion test for screening for ALK rearrangements in the USA. We aimed to assess if a similar process of IHC screening could be useful to identify ROS1 rearrangements. In this study, we assessed the prevalence of ROS1 rearrangements in a retrospective cohort of Australian NSCLC comparing 2 techniques (IHC and FISH) and then applied the testing process in a prospective cohort of lung cancers referred for mutation assessment.
Materials & Methods
Patient Cohorts
A retrospective cohort of 278 resected stage I–III lung adenocarcinomas from Royal Prince Alfred Hospital (RPAH) and Concord Repatriation General Hospital between January 1990 and May 2002 were included in the study as previously described38–40. Formalin fixed paraffin embedded tissue was used to construct tissue microarrays to test for ROS1 expression and ROS1 gene rearrangement. EGFR, KRAS and ALK status had previously been assessed in this cohort as previously described [16,19]. 10% (29/278) harboured an activating EGFR mutation, 28% (77/278) harboured a KRAS mutation, 1% (3/278) had an ALK rearrangement and 26% were not mutation tested (72/278) (Table 1).
Table 1.
Clinicopathological characteristics of all cohorts
| Retrospective cohort (n=278) | Prospective cohort (n=104) | |||||
|---|---|---|---|---|---|---|
| ROS1 FISH | All cases (278) | ROS1 − (277) | ROS1 + (1) | All cases (104) | ROS1 − (92) | ROS1 + (12) |
| Age (years) | ||||||
| Median (range) | 69 (40–87) | 69 (40–87) | 66 | 62 (34–85) | 64 (34–85) | 59 (34–81) |
| Sex | ||||||
| Male | 167 (60%) | 167 (60%) | 0 (0%) | 42 (40%) | 39 (42%) | 3 (25%) |
| Female | 111 (40%) | 110 (40%) | 1 (100%) | 62 (60%) | 53 (58%) | 9 (75%) |
| Histology | ||||||
| Adenocarcinoma | 277 (99.6%) | 276 (99.7%) | 1 (100%) | 98 (94%) | 87 (95%) | 11 (92%) |
| Squamous cell carcinoma | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Large cell carcinoma | 0 (0%) | 0 (0%) | 0 (0%) | 3 (3%) | 3 (3%) | 0 (0%) |
| Adenosquamous | 1 (0.4%) | 1 (0.3%) | 0 (0%) | 3 (3%) | 2 (2%) | 1 (8%) |
| Smoking history | ||||||
| Never | 11 (4%) | 10 (3.6%) | 1 (100%) | 29 (28%) | 27 (29.4%) | 2 (17%) |
| Ever | 74 (26.6%) | 74 (26.7%) | 0 (0%) | 14 (13%) | 14 (15.2%) | 0 (0%) |
| Unknown | 193 (69.4%) | 193 (69.7%) | 0 (0%) | 61 (59%) | 51 (55.4%) | 10 (83%) |
| Mutation status | ||||||
| EGFR wildtype | 99 (36%) | 98 (35.4%) | 0 (0%) | 69 (66%) | 57 (62%) | 12 (100%) |
| EGFR mutant | 29 (10%) | 29 (10.5%) | 0 (0%) | 15 (14%) | 15 (16%) | 0 (0%) |
| Other mutation^ | 78 (28%) | 78 (28.1%) | 0 (0%) | 9 (9%) | 9 (10%) | 0 (0%) |
| Unknown | 72 (26%) | 72 (26%) | 1 (100%) | 11 (11%) | 11 (12%) | 0 (0%) |
| ROS1 IHC | ||||||
| Positive | 89 (32%) | 88 (32%) | 1 (100%) | 19 (18%) | 7 (8%) | 12 (100%) |
| Negative | 189 (68%) | 189 (68%) | 0 (0%) | 83 (80%) | 83 (90%) | 0 (0%) |
| Unable to be tested | 0 (0%) | 0 (0%) | 0 (0%) | 2 (2%) | 2 (2%) | 0 (0%) |
KRAS, PIK3CA, KIT, MET, FGFR1, HRAS, BRAF mutation or ALK rearrangement.
An additional cohort of 104 NSCLC cases referred for diagnostic molecular testing (EGFR, ALK or ROS1) at RPAH between November 2012 and May 2016 were also included in the study. Ethics approval was granted by the Sydney Local Health District Ethics Review Committee (X12-0313 and HREC/12/RPAH/479). All cases referred for EGFR mutation testing underwent ROS1 IHC if sufficient tissue was available. The 104 cases consisted of tumours found to be ROS1 IHC positive on screening, or were separately referred for ROS1 FISH testing at the request of the clinical team which included cases with enriched likelihood for ROS1 rearrangement (EGFR/KRAS−, ALK−, female never smoker). The cohort consisted of predominantly EGFR wildtype cases (66%), however 15 cases (14%) possessed an EGFR mutation, 5 cases (5%) had a KRAS mutation, 1 case (1%) had a KIT mutation and 1 case (1%) had a BRAF mutation (Table 1). Eleven cases (11%) had unknown mutation status. These cases were referred for ROS1 rearrangement testing as requested by the clinical team or selected due to clinical features or ROS1 IHC positivity. Sixty two cases were mutation tested at RPAH in parallel with diagnostic ROS1 testing (ROS1 IHC & FISH). Mutation testing was performed using the Oncocarta v1.0 and OncoFOCUS v3 on the Sequenom MassARRAY platform (Sequenom/Agena Bioscience, San Diego, CA). The other 42 cases had previously undergone EGFR testing elsewhere. ALK rearrangement status was negative for all cases except for two positive cases which were ALK FISH tested in parallel with ROS1 at the time due to clinician request. All cases in the diagnostic cohort were adenocarcinomas, except for three adenosquamous and three large cell carcinomas.
ROS1 Immunohistochemistry
Immunohistochemistry was performed on sections, cut at 4um, using the Cell Signaling Technology rabbit monoclonal ROS1 (D4D6) antibody (Danvers, MA, USA) at 1:50 dilution for 2 hours. Staining was performed using the UltraView DAB universal detection kit (Roche, Basel, Switzerland) including an Amplification Kit (Roche), and was performed on a Benchmark ULTRA autostainer (Roche). Positive controls included lung tumor confirmed by FISH to be positive for ROS1 rearrangement. Any cytoplasmic staining for ROS1 IHC in tumour cells was considered positive. Non-specific staining of macrophages and type II pneumocytes were disregarded. Percentage of cells expressing ROS1 and intensity of expression was also evaluated, in addition to H-scores (% positive cells x intensity [1 mild, 2 moderate, 3 strong staining]).
ROS1 Fluorescence In Situ Hybridisation
Interphase fluorescence in situ hybridisation (FISH) was performed in a NATA (National Association of Testing Authorities) accredited diagnostic laboratory for ROS1 rearrangement using the ZytoLight SPEC ROS1 Dual Colour Break Apart Probe (ZytoVision) and the LSI ROS1 (Tel) SpectrumOrange Probe and LSI ROS1 (Cen) SpectrumGreen Probe (Abbott Molecular). The ZytoVision FISH probe was used before the Abbott Molecular FISH probe was commercially available in March 2013. The retrospective cohort was analysed using the ZytoVision ROS1 FISH probe, and the prospective cohort utilised both FISH probes. In our experience both probes show equivalent performance, and both have been utilised and published in international cohorts23,26,27,43–45. FISH was performed following the manufacturers guidelines except that Invitrogen Pretreatment solution was used at 98–100°C for 20 minutes. Interphase signals were counted in at least 50 tumour nuclei per cases using an epifluorescence microscope (Zeiss). Cases were classified as ROS1 FISH positive if they showed ≥15% cells with split signals at least 2 signal distances apart or an isolated centromeric 3′ (green signal) pattern (as indicated by manufacturer and Mazieres et al45). The specific tissue region undergoing evaluation was verified by a pathologist and all FISH results were evaluated by a scientist and at least one expert pathologist with considerable experience reviewing lung cancer FISH (WC or SOT).
Results
All cases within the retrospective cohort were evaluated using ROS1 IHC and FISH. Two of the 104 cases in the prospective diagnostic cohort were unable to have ROS1 IHC testing due to limited material available. Both cases were ROS1 FISH negative.
Within the retrospective cohort a single case (0.4%) showed strong diffuse staining by ROS1 IHC, and this case was confirmed rearrangement positive by ROS1 FISH. This case was a poorly differentiated adenocarcinoma in a 66 year old female, which showed a growth pattern containing intracytoplasmic mucin. All other cases were ROS1 rearrangement negative by FISH. ROS1 IHC staining was present in 32% of cases (88/278), however most of these cases showed weak immunoreactivity (1+).
Nineteen cases (Table 1) in the prospective cohort showed ROS1 IHC staining. Twelve cases (12%) were confirmed as ROS1 rearranged (Table 1). These cases all showed 3+ immunoreactivity with H scores 270–300. The remaining seven IHC positive cases were ROS1 FISH negative. All of these IHC positive-FISH negative cases showed 3+ immunoreactivity except for one which was 2+. H scores ranged from 40–300.
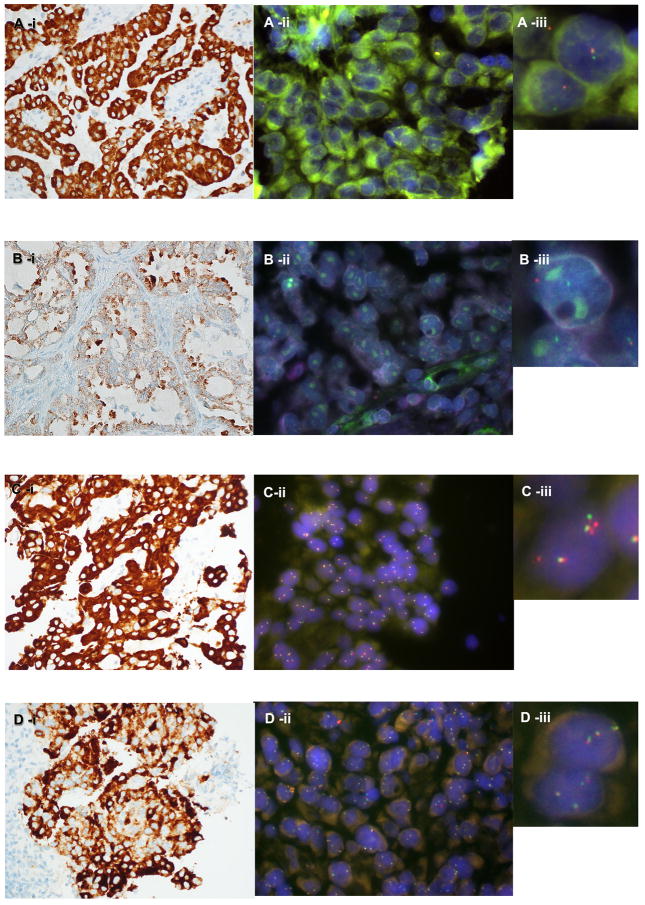
Four ROS1 rearranged cases showed a predominantly isolated 3′ green locus (5′ locus deletion) signal pattern (Table 2; One example is presented in Figure 1-A). A ROS1 IHC positive case (adenosquamous carcinoma in a 56yr old female, unknown smoking status, EGFR wildtype) that was confirmed rearranged with ROS1 FISH, showed a signal pattern consisting predominantly of a single set of split signals per cell, with loss of the accompanying set of fusion signals (Table 2; Figure 1-B). The other ROS1 rearranged case showed a predominantly classical split signal pattern (Table 2).
Table 2.
ROS1 IHC positive cases
| Case # | KRAS/EGFR/ALK status | ROS1 IHC - % cells positive | ROS1 IHC - Intensity | ROS1 IHC - H score (% x intensity) | ROS1 FISH |
|---|---|---|---|---|---|
| Retrospective Cohort | |||||
| 1 | KRAS & EGFR unknown/ALK− | 90% | 3+ | 244 | + (single set of split signals) |
| 2–81 | 23 KRAS+, 13 EGFR+, 4 PIK3CA+, 2 MET+, 1 ALK+, 1 BRAF+, 1 FGFR1+, 1 HRAS+, 6 unknown | 5–80% (mean =24) | 1+ | 5–80 (mean=24) | - |
| 82–90 | 3 KRAS+, 5 EGFR+, 0 unknown | 20–78 (mean 40) | 2+ | 40–120 (mean 79) | - |
| Prospective cohort | |||||
| 1 | All Negative | 100% | 3+ | 300 | + (classic split signal pattern) |
| 2 | All Negative | 100% | 3+ | 300 | + (classic split signal pattern) |
| 3 | All Negative | 100% | 3+ | 300 | + (single set of split signals) |
| 4 |
KRAS unknown EGFR−/ALK− |
100% | 3+ | 300 | + (classic split signal pattern) |
| 5 |
KRAS unknown EGFR−/ALK− |
100% | 3+ | 300 | + (isolated 3′ green signal pattern) |
| 6 | All Negative | 100% | 3+ | 300 | + (classic split signal pattern) |
| 7 | All Negative | 90% | 3+ | 270 | + (isolated 3′ green signal pattern) |
| 8 | All Negative | 100% | 3+ | 300 | + (classic split signal pattern) |
| 9 | All Negative | 100% | 3+ | 300 | + (classic split signal pattern) |
| 10 | All Negative | 90% | 3+ | 270 | +(isolated 3′ green signal pattern) |
| 11 | All Negative | 100% | 3+ | 300 | + (classic split signal pattern) |
| 12 | All Negative | 100% | 3+ | 300 | + (isolated 3′ green signal pattern) |
| 13 |
KRAS unknown EGFR−/ALK− |
100% | 3+ | 300 | − (isolated 5′ red signal pattern) |
| 14 | All Negative | 25% | 3+ | 75 | - |
| 15 | All Negative | 80% | 3+ | 280 | - |
| 16 | EGFR+/KRAS−/ALK− | 33% | 3+ | 99 | - |
| 17 | EGFR+/KRAS−/ALK− | 20% | 2+ | 40 | - |
| 18 |
KRAS unknown EGFR−/ALK− |
30% | 3+ | 90 | - |
| 19 |
KRAS unknown EGFR−/ALK− |
20% | 3+ | 60 | - |
Figure 1.
Examples of ROS1 expression and ROS1 rearrangement patterns observed: A-i) Strong ROS1 IHC staining, with ROS1 FISH A-ii) and A-iii) showing a predominantly positive isolated green signal pattern. B-i) Moderate-strong ROS1 IHC staining with ROS1 FISH B-ii) & B-iii) showing predominantly a single set of split signals. C-i) Strong ROS1 IHC staining, C-ii) & ROS1 FISH C-iii) showing a negative signal pattern of subtle isolated red signals. D-i) Strong ROS1 IHC staining, however ROS1 FISH D-ii) & D-iii) showed less than 15% of tumour nuclei with ROS1 rearrangement.
One of the ROS1 IHC positive - FISH negative cases (adenocarcinoma from a 62yr old female, non-smoker, EGFR wildtype) showed an atypical ROS1 FISH signal pattern of 26% isolated 5′ (red) signals (Table 2; Figure 1-C). Additionally, another strongly ROS1 IHC positive case (adenocarcinoma from a 62yr old male, non-smoker, EGFR wildtype) was ROS1 FISH negative (Table 2; Figure 1-D).
All ROS1 positive cases were adenocarcinomas except for one adenosquamous carcinoma.
The sensitivity of ROS1 IHC in the retrospective cohort was 100% and specificity was 76%; positive predictive value (PPV) was 1% and negative predictive value (NPV) was 100%. If only 3+ IHC staining was classified as positive, the sensitivity and specificity of ROS1 IHC in the retrospective cohort was 100% with positive predictive value (PPV) and negative predictive value (NPV) also at 100%.
The combined thirteen ROS1 rearranged cases were all EGFR wildtype (except for one case with unknown EGFR mutation status in the retrospective cohort) and were all ALK rearrangement negative. Two cases were non-smokers and the remaining ten cases had unknown smoking history.
Discussion
Compared with rearrangements involving ALK which are reported to occur in around 3% of patients with NSCLC46, ROS1 rearrangements have been reported to occur at slightly lower percentages (1%)46,47. This was somewhat validated in our retrospective cohort, which showed 0.4% of 278 cases with ROS1 rearrangement. As the prospective cohort of 104 cases was enriched with patients that were previously confirmed to be EGFR and ALK wildtype, the rate of ROS1 rearrangements was higher (12%). Despite the lower rate of ROS1 rearrangement, compared with that of ALK, the rate of response to targeted therapy appears slightly higher with a longer median duration of response and progression-free survival19, adding a valuable incentive to screen for ROS1 rearrangements.
Presumably due to the high sensitivity of ROS1 IHC, our retrospective and prospective cohorts showed a notable level of ROS1 immunoreactivity in cases that were negative for a ROS1 rearrangement by FISH. Although theoretically break apart FISH can detect all rearrangements involving the common breakpoint region, it has been suggested that it may be difficult to identify ROS1 fusions involving intrachromosomal rearrangements on the same chromosome, such as ROS1-EZR by breakapart FISH [16] due to close location of the split signals. ROS1 rearrangements involving GOPC (formally known as FIG) would not be detected by some break apart FISH assays as the 5′ probe overlaps or includes GOPC, which is only 134kb upstream [13]17. The ZytoVision ROS1 FISH probe is able to detect GOPC-ROS1 fusions, however as the Abbott Molecular ROS1 FISH probe is designed with the 5′ telomeric probe covering both ROS1 and GOPC genes, it is thought that the FISH probe cannot detect GOPC-ROS1 fusions. Fusions with GOPC however, are thought to make up only 3% of ROS1 fusion partners in NSCLC47, although further data is required to characterise the frequency of this fusion partner with certainty.
High ROS1 IHC expression without ROS1 FISH positivity may be caused by a number of factors. A fusion gene not revealed by FISH, such as a GOPC-ROS1 fusion, could be responsible for a subset of these cases – however the ZytoVision FISH probe which can detect this fusion, was used to confirm FISH results in all but 3 of these cases, where further tissue was not available for assessment. Alternative cryptic ROS1 rearrangements could also explain why some cases show high protein ROS1 expression with no detected ROS1 rearrangement by FISH. Alternative methods such as RT-PCR and sequencing20,26,31,43 have revealed ROS1 rearrangements in similar cases, and would be of great benefit when feasible diagnostically. In addition, activation of the ROS1 oncogene could take place independent of structural DNA aberrations at the ROS1 locus, and be epigenetically driven, such as by alternative transcript initiation - which has been documented for ALK48. Other mechanisms of RNA and protein conformational activation of ROS1 could also theoretically contribute, but have not yet manifested in studies so far.
During the early stage of this study, limited commercial options for ROS1 FISH probes were available, therefore we utilised the ZytoVision ROS1 break apart FISH probe before the Abbott Molecular ROS1 FISH probe was also available. Many studies have used either the Abbott Molecular ROS1 FISH probe set, or the ZytoVision probe set 23,26,27,43,44, including the EUROS1 cohort conducted in six European countries45.
Interestingly, Warth et al23 described in 1478 cases, ROS1 IHC positivity but not ROS1 rearrangement, was associated with prolonged overall survival. This suggests that ROS1 immunoreactivity may have more clinical significance than previously understood, however, its association with response to ROS1 inhibitors has not been investigated.
No false negative ROS1 IHC cases were observed in the retrospective cohort. The only ROS1 rearranged case in the retrospective cohort expressed ROS1 with 3+ intensity IHC, and similarly in the prospective cohort, all ROS1 rearranged cases expressed ROS1 with 3+ IHC intensity.
In our study ROS1 IHC identified all ROS1 rearranged tumours, as all ROS1 FISH-positive cases in our cohorts were immunoreactive. Our data suggests that IHC for ROS1 is a highly sensitive method to screen for ROS1 rearrangements with a very high negative predictive value but not as specific. As such, we recommend IHC screening, followed by FISH confirmation. While our results suggest FISH could be undertaken only on those cases with diffuse strong intensity IHC staining (3+ staining in at least 90% of tumour cells, H score ≥ 270), assessment of intensity is subjective and other studies have shown occasional FISH+ cases may be missed with such an approach24,26 so in our centre we have adopted an approach of FISH testing cases with any ROS1 IHC positivity. A similar approach has been shown to be effective in identifying ALK rearranged lung cancer32.
Compared with performing FISH as a routine screening method for ROS1 rearrangements in all EGFR/ALK-negative NSCLC cases, ROS1 IHC is preferable, due to the reduced cost and reduced labour involved in interpretation. Confirmatory FISH could be reserved for cases showing diffuse and moderately strong ROS1 IHC staining in centres where FISH testing is not readily available.
In conclusion, potentially targetable ROS1 gene rearrangements occur in a very small percentage of lung adenocarcinomas and are largely mutually exclusive with EGFR mutations and ALK rearrangements. Screening with IHC is highly sensitive, but not as specific, and may be a suitable method of reducing the number of cases requiring FISH to identify the ROS1 genetic abnormality. Selection of cases for ROS1 FISH testing such as exclusion of EGFR/ALK positive cases and use of ROS1 IHC to screen for potentially positive cases can be used to enrich for the likelihood of a identifying a ROS1 rearranged lung cancer and prevent the need to undertake expensive and time consuming FISH testing in all cases.
Acknowledgments
Clinical Professor Sandra O’Toole received funding from the CINSW, the Sydney Breast Cancer Foundation and the NHMRC. This research was also supported by generous donations from the Tag family foundation, Mr David Paradice, ICAP and the O’Sullivan family. Bob Li received funding from the NIH/NCI Cancer Center Support Grant P30 CA008748.
CIS performed the research and wrote the paper. BTL, NP, ML, AG, AL, SC and LH provided data and assisted with the manuscript. TNT assisted with cohort construction and assisted with the manuscript. TL performed the IHC and assisted with the manuscript. PYY assisted with cohort construction, provided data and assisted with the manuscript. BY assisted with mutation testing analysis and assisted with the manuscript. MRJKC assisted with cohort construction and assisted with the manuscript. SOT & WC analysed and interpreted the results, designed the study and assisted with the manuscript.
Footnotes
Conflicts of Interest: W. Cooper has participated in Lung Cancer Advisory Boards and has received honoraria from Pfizer Oncology. S. O’Toole has received honoraria from Roche, Pfizer, and Lilly Oncology. A. Gill has received honoraria from Pfizer Oncology and Astra Zeneca. N. Pavlakis has participated in Lung Cancer Advisory Boards for Pfizer, and received honoraria. The other authors state that they have no conflicts of interest.
References
- 1.Ou S-HI, Tan J, Yen Y, Soo RA. ROS1 as a “druggable” receptor tyrosine kinase: lessons learned from inhibiting the ALK pathway. Expert Rev Anticancer Ther. 2012 Apr;12(4):447–56. doi: 10.1586/era.12.17. [DOI] [PubMed] [Google Scholar]
- 2.Shibuya M, Hanafusa H, Balduzzi PC. Cellular sequences related to three new onc genes of avian sarcoma virus (fps, yes, and ros) and their expression in normal and transformed cells. J Virol. 1982 Apr;42(1):143–52. doi: 10.1128/jvi.42.1.143-152.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang LH, Hanafusa H, Notter MF, Balduzzi PC. Genetic structure and transforming sequence of avian sarcoma virus UR2. J Virol. 1982 Mar;41(3):833–41. doi: 10.1128/jvi.41.3.833-841.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balduzzi PC, Notter MF, Morgan HR, Shibuya M. Some biological properties of two new avian sarcoma viruses. J Virol. 1981 Oct;40(1):268–75. doi: 10.1128/jvi.40.1.268-275.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uttamsingh S, Zong CS, Wang L-H. Matrix-independent activation of phosphatidylinositol 3-kinase, Stat3, and cyclin A-associated Cdk2 Is essential for anchorage-independent growth of v-Ros-transformed chicken embryo fibroblasts. J Biol Chem. 2003 May 23;278(21):18798–810. doi: 10.1074/jbc.M211522200. [DOI] [PubMed] [Google Scholar]
- 6.Charest A, Wilker EW, McLaughlin ME, et al. ROS fusion tyrosine kinase activates a SH2 domain-containing phosphatase-2/phosphatidylinositol 3-kinase/mammalian target of rapamycin signaling axis to form glioblastoma in mice. Cancer Res. 2006 Aug 1;66(15):7473–81. doi: 10.1158/0008-5472.CAN-06-1193. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen KT, Zong CS, Uttamsingh S, et al. The role of phosphatidylinositol 3-kinase, rho family GTPases, and STAT3 in Ros-induced cell transformation. J Biol Chem. 2002 Mar 29;277(13):11107–15. doi: 10.1074/jbc.M108166200. [DOI] [PubMed] [Google Scholar]
- 8.Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012 Mar;18(3):378–81. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 9.Birchmeier C, Sharma S, Wigler M. Expression and rearrangement of the ROS1 gene in human glioblastoma cells. Proc Natl Acad Sci U S A. 1987 Dec;84(24):9270–4. doi: 10.1073/pnas.84.24.9270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu P, Wu Y, Sun L, Zuo Q, Shi M. ROS kinase fusions are not common in Chinese patients with cholangiocarcinoma. Nan Fang Yi Ke Da Xue Xue Bao. 2013 Apr;33(4):474–8. [PubMed] [Google Scholar]
- 11.Gu T-L, Deng X, Huang F, et al. Survey of tyrosine kinase signaling reveals ROS kinase fusions in human cholangiocarcinoma. PloS One. 2011;6(1):e15640. doi: 10.1371/journal.pone.0015640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peraldo Neia C, Cavalloni G, Balsamo A, et al. Screening for the FIG-ROS1 fusion in biliary tract carcinomas by nested PCR. Genes Chromosomes Cancer. 2014 Dec;53(12):1033–40. doi: 10.1002/gcc.22212. [DOI] [PubMed] [Google Scholar]
- 13.Birch AH, Arcand SL, Oros KK, et al. Chromosome 3 anomalies investigated by genome wide SNP analysis of benign, low malignant potential and low grade ovarian serous tumours. PloS One. 2011;6(12):e28250. doi: 10.1371/journal.pone.0028250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J, Lee SE, Kang SY, et al. Identification of ROS1 rearrangement in gastric adenocarcinoma. Cancer. 2013 May 1;119(9):1627–35. doi: 10.1002/cncr.27967. [DOI] [PubMed] [Google Scholar]
- 15.Aisner DL, Nguyen TT, Paskulin DD, et al. ROS1 and ALK fusions in colorectal cancer, with evidence of intratumoral heterogeneity for molecular drivers. Mol Cancer Res MCR. 2014 Jan;12(1):111–8. doi: 10.1158/1541-7786.MCR-13-0479-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies KD, Le AT, Theodoro MF, et al. Identifying and targeting ROS1 gene fusions in non-small cell lung cancer. Clin Cancer Res Off J Am Assoc Cancer Res. 2012 Sep 1;18(17):4570–9. doi: 10.1158/1078-0432.CCR-12-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergethon K, Shaw AT, Ou S-HI, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol Off J Am Soc Clin Oncol. 2012 Mar 10;30(8):863–70. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanizaki J, Okamoto I, Okamoto K, et al. MET tyrosine kinase inhibitor crizotinib (PF-02341066) shows differential antitumor effects in non-small cell lung cancer according to MET alterations. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer. 2011 Oct;6(10):1624–31. doi: 10.1097/JTO.0b013e31822591e9. [DOI] [PubMed] [Google Scholar]
- 19.Shaw AT, Ou S-HI, Bang Y-J, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 2014 Nov 20;371(21):1963–71. doi: 10.1056/NEJMoa1406766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suehara Y, Arcila M, Wang L, et al. Identification of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions. Clin Cancer Res Off J Am Assoc Cancer Res. 2012 Dec 15;18(24):6599–608. doi: 10.1158/1078-0432.CCR-12-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rimkunas VM, Crosby KE, Li D, et al. Analysis of receptor tyrosine kinase ROS1-positive tumors in non-small cell lung cancer: identification of a FIG-ROS1 fusion. Clin Cancer Res Off J Am Assoc Cancer Res. 2012 Aug 15;18(16):4449–57. doi: 10.1158/1078-0432.CCR-11-3351. [DOI] [PubMed] [Google Scholar]
- 22.Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007 Dec 14;131(6):1190–203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 23.Warth A, Muley T, Dienemann H, et al. ROS1 expression and translocations in non-small-cell lung cancer: clinicopathological analysis of 1478 cases. Histopathology. 2014 Aug;65(2):187–94. doi: 10.1111/his.12379. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida A, Tsuta K, Wakai S, et al. Immunohistochemical detection of ROS1 is useful for identifying ROS1 rearrangements in lung cancers. Mod Pathol Off J U S Can Acad Pathol Inc. 2014 May;27(5):711–20. doi: 10.1038/modpathol.2013.192. [DOI] [PubMed] [Google Scholar]
- 25.Sholl LM, Sun H, Butaney M, et al. ROS1 immunohistochemistry for detection of ROS1-rearranged lung adenocarcinomas. Am J Surg Pathol. 2013 Sep;37(9):1441–9. doi: 10.1097/PAS.0b013e3182960fa7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shan L, Lian F, Guo L, et al. Detection of ROS1 gene rearrangement in lung adenocarcinoma: comparison of IHC, FISH and real-time RT-PCR. PloS One. 2015;10(3):e0120422. doi: 10.1371/journal.pone.0120422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mescam-Mancini L, Lantuéjoul S, Moro-Sibilot D, et al. On the relevance of a testing algorithm for the detection of ROS1-rearranged lung adenocarcinomas. Lung Cancer Amst Neth. 2014 Feb;83(2):168–73. doi: 10.1016/j.lungcan.2013.11.019. [DOI] [PubMed] [Google Scholar]
- 28.Cha YJ, Lee JS, Kim HR, et al. Screening of ROS1 rearrangements in lung adenocarcinoma by immunohistochemistry and comparison with ALK rearrangements. PloS One. 2014;9(7):e103333. doi: 10.1371/journal.pone.0103333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao B, Wei P, Liu Z, et al. Detection of lung adenocarcinoma with ROS1 rearrangement by IHC, FISH, and RT-PCR and analysis of its clinicopathologic features. OncoTargets Ther. 2016;9:131–8. doi: 10.2147/OTT.S94997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyle TA, Masago K, Ellison KE, Yatabe Y, Hirsch FR. ROS1 Immunohistochemistry Among Major Genotypes of Non-Small-Cell Lung Cancer. Clin Lung Cancer. 2014 Oct 24; doi: 10.1016/j.cllc.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee SE, Lee B, Hong M, et al. Comprehensive analysis of RET and ROS1 rearrangement in lung adenocarcinoma. Mod Pathol Off J U S Can Acad Pathol Inc. 2015 Apr;28(4):468–79. doi: 10.1038/modpathol.2014.107. [DOI] [PubMed] [Google Scholar]
- 32.Selinger CI, Rogers T-M, Russell PA, et al. Testing for ALK rearrangement in lung adenocarcinoma: a multicenter comparison of immunohistochemistry and fluorescent in situ hybridization. Mod Pathol Off J U S Can Acad Pathol Inc. 2013 Dec;26(12):1545–53. doi: 10.1038/modpathol.2013.87. [DOI] [PubMed] [Google Scholar]
- 33.Boland JM, Erdogan S, Vasmatzis G, et al. Anaplastic lymphoma kinase immunoreactivity correlates with ALK gene rearrangement and transcriptional up-regulation in non-small cell lung carcinomas. Hum Pathol. 2009 Aug;40(8):1152–8. doi: 10.1016/j.humpath.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 34.Camidge DR, Hirsch FR, Varella-Garcia M, Franklin WA. Finding ALK-positive lung cancer: what are we really looking for? J Thorac Oncol Off Publ Int Assoc Study Lung Cancer. 2011 Mar;6(3):411–3. doi: 10.1097/JTO.0b013e31820cf068. [DOI] [PubMed] [Google Scholar]
- 35.Houang M, Toon CW, Clarkson A, et al. Reflex ALK immunohistochemistry is feasible and highly specific for ALK gene rearrangements in lung cancer. Pathology (Phila) 2014 Aug;46(5):383–8. doi: 10.1097/PAT.0000000000000114. [DOI] [PubMed] [Google Scholar]
- 36.Mino-Kenudson M, Chirieac LR, Law K, et al. A novel, highly sensitive antibody allows for the routine detection of ALK-rearranged lung adenocarcinomas by standard immunohistochemistry. Clin Cancer Res Off J Am Assoc Cancer Res. 2010 Mar 1;16(5):1561–71. doi: 10.1158/1078-0432.CCR-09-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takeuchi K, Choi YL, Togashi Y, et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res Off J Am Assoc Cancer Res. 2009 May 1;15(9):3143–9. doi: 10.1158/1078-0432.CCR-08-3248. [DOI] [PubMed] [Google Scholar]
- 38.Yip PY, Yu B, Cooper WA, et al. Patterns of DNA mutations and ALK rearrangement in resected node negative lung adenocarcinoma. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer. 2013 Apr;8(4):408–14. doi: 10.1097/JTO.0b013e318283558e. [DOI] [PubMed] [Google Scholar]
- 39.Selinger CI, Cooper WA, Al-Sohaily S, et al. Loss of special AT-rich binding protein 1 expression is a marker of poor survival in lung cancer. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer. 2011 Jul;6(7):1179–89. doi: 10.1097/JTO.0b013e31821b4ce0. [DOI] [PubMed] [Google Scholar]
- 40.Cooper WA, Kohonen-Corish MRJ, Chan C, et al. Prognostic significance of DNA repair proteins MLH1, MSH2 and MGMT expression in non-small-cell lung cancer and precursor lesions. Histopathology. 2008 Apr;52(5):613–22. doi: 10.1111/j.1365-2559.2008.02999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tran TN, Selinger CI, Yu B, et al. Alterations of insulin-like growth factor-1 receptor gene copy number and protein expression are common in non-small cell lung cancer. J Clin Pathol. 2014 Nov;67(11):985–91. doi: 10.1136/jclinpath-2014-202347. [DOI] [PubMed] [Google Scholar]
- 42.Tran TN, Selinger CI, Kohonen-Corish MRJ, et al. Alterations of MET Gene Copy Number and Protein Expression in Primary Non-Small-Cell Lung Cancer and Corresponding Nodal Metastases. Clin Lung Cancer. 2015 Aug 18; doi: 10.1016/j.cllc.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Lira ME, Choi Y-L, Lim SM, et al. A single-tube multiplexed assay for detecting ALK, ROS1, and RET fusions in lung cancer. J Mol Diagn JMD. 2014 Mar;16(2):229–43. doi: 10.1016/j.jmoldx.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 44.Scheffler M, Schultheis A, Teixido C, et al. ROS1 rearrangements in lung adenocarcinoma: prognostic impact, therapeutic options and genetic variability. Oncotarget. 2015 Apr 30;6(12):10577–85. doi: 10.18632/oncotarget.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mazières J, Zalcman G, Crinò L, et al. Crizotinib therapy for advanced lung adenocarcinoma and a ROS1 rearrangement: results from the EUROS1 cohort. J Clin Oncol Off J Am Soc Clin Oncol. 2015 Mar 20;33(9):992–9. doi: 10.1200/JCO.2014.58.3302. [DOI] [PubMed] [Google Scholar]
- 46.Rosell R, Karachaliou N, Wolf J, Ou S-HI. ALK and ROS1 non-small-cell lung cancer: two molecular subgroups sensitive to targeted therapy. Lancet Respir Med. 2014 Dec;2(12):966–8. doi: 10.1016/S2213-2600(14)70259-0. [DOI] [PubMed] [Google Scholar]
- 47.Gainor JF, Shaw AT. Novel targets in non-small cell lung cancer: ROS1 and RET fusions. The Oncologist. 2013;18(7):865–75. doi: 10.1634/theoncologist.2013-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiesner T, Lee W, Obenauf AC, et al. Alternative transcription initiation leads to expression of a novel ALK isoform in cancer. Nature. 2015 Oct 15;526(7573):453–7. doi: 10.1038/nature15258. [DOI] [PMC free article] [PubMed] [Google Scholar]