Abstract
Optimization of a protein’s pharmaceutical properties is usually carried out by rational design and/or directed evolution. Here we test an alternative approach based on ancestral sequence reconstruction. Using available genomic sequence data on coagulation factor VIII and predictive models of molecular evolution, we engineer protein variants with improved activity, stability. biosynthesis potential, and reduced inhibition by clinical anti-drug antibodies. In principle, this approach can be applied to any protein drug based on a conserved gene sequence.
Many protein drugs possess suboptimal pharmacological properties yet are not amenable to standard biotechnology optimization approaches. However, the endogenous proteins these drugs are based on generally are thought to have selectively evolved over the past half-billion years to optimize functionality within the context of current or ancestral species-specific physiology. This inferred protein design landscape represents an unexplored avenue for protein drug optimization that is accessible through ancestral sequence reconstruction (ASR).
Factor VIII (FVIII) is an essential component of blood coagulation pathways, and its deficiency results in hemophilia A, the most common severe bleeding disorder. The human (h) F8 gene was first described in 1984, and by the early 1990’s several recombinant hFVIII biologics were in clinical use. FVIII infusion therapy converts this otherwise lethal disease into a clinically manageable condition, but current hFVIII biologics have several important limitations, including poor biosynthetic efficiency, short half-life, and potent immunogenicity. Despite intensive academic and commercial efforts, traditional rational design and directed evolution approaches have not yielded substantially improved FVIII biologics.
We1–4 and others5 have shown that extant FVIII orthologs have molecular, cellular and immune recognition properties that vary between species. This diversity may represent adaptive traits acquired throughout natural selection to promote hemostatic balance. For example, during bipedal hominid evolution, thrombosis-related mortality could have exerted selective pressure on plasma FVIII toward reduced coagulation activity. Irrespective of the validity of such evolutionary hypotheses, some ortholog-specific properties are likely to be beneficial from a pharmaceutical perspective. Previously, we exploited extant ortholog diversity to identify and engineer pharmacologically beneficial sequence determinants6, 7. This ortholog-scanning approach led to the creation of a highly expressed human/porcine hybrid FVIII named ET3 with 149 (11%) porcine amino acids. However, the combinatorial complexity associated with identifying the multiple, non-linear amino acid determinants blocked further humanization. The inability to sufficiently ‘humanize’ ortholog-hybrid molecules with xenogeneic sequences, similarly to what is done for monoclonal antibody biologics, is a major limitation of ortholog-scanning for pharmaceutical development.
ASR entails the prediction of ancient DNA and protein sequences based on information from extant sequences8. In contrast to ortholog scanning, it provides a higher-resolution mapping through comparisons of sequential phylogeny branches, and infers novel sequences with high potential for intended biomolecular functions as they are predicted to have once existed. This aspect of uniform functionality distinguishes ASR from all other protein drug design approaches. Advances in custom DNA synthesis now facilitate laboratory ‘resurrection’ and characterization of inferred ancestral proteins9. Recent examples of ASR applications include examination of ancestral enzyme promiscuity and functional diversification10, 11, study of ancient receptor-ligand interactions12, resurrection of a human pseudogene13, and definition of an oncology drug mechanism14. As an approach to pharmaceutical bioengineering, ASR requires only genome information, modest computing resources, and analysis of a limited set of recombinant ancestral protein variants.
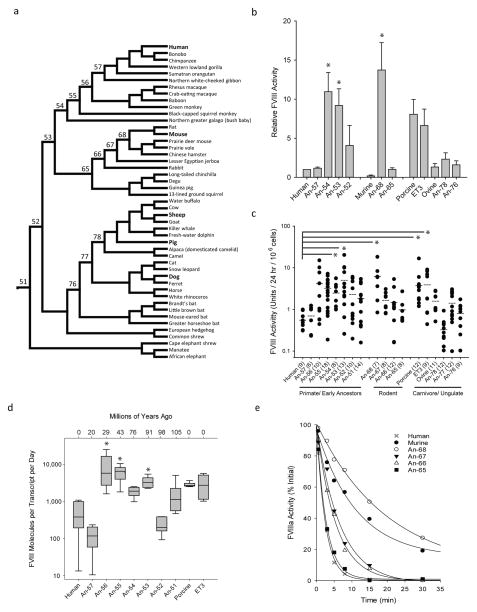
We performed FVIII ASR to infer a mammalian evolutionary tree as described previously15 (Fig. 1a and Supplemental Fig. 1). Available extant sequence data was sufficient to accurately extend the phylogenetic tree and infer ancestral (An)-FVIII sequences beyond the class Mammalia. We synthesized inferred An-FVIII cDNAs de novo and expressed the An-FVIII proteins in cell lines commonly used for recombinant FVIII manufacturing. Consistent with the hypothesis of plasma FVIII activity levels decreasing during hominid evolution, several inferred early mammalian An-FVIII (one early rodent, one early primate and one common ancestor to both) demonstrated protein expression rates ranging from 9 – 14-fold higher to those of hFVIII, and at least equivalent to the most efficiently produced recombinant FVIII molecules described previously (i.e. pFVIII and ET3; Fig. 1b and c). We examined An-FVIII constructs along the hominid lineage for differentials in pre-, post- and/or co-translational biosynthesis, and show that An-53, An-55 and An-56 display significantly higher FVIII production per steady-state transcript than hFVIII (Fig 1d and Supplemental Note). The mechanisms supporting increased expression of ancestral FVIII proteins are unclear, but previous reports that describe non-hFVIII residues in pFVIII and ET3 that confer increased secretion efficiency by reducing the engagement of unfolded protein response pathways offer a possible explanation2, 6, 16. Other processes that can improve An-FVIII biosynthesis include changes in protein synthesis or secretion efficiency, all predictably beneficial to recombinant FVIII protein production and gene therapy applications.
Fig. 1. An-FVIII phylogeny, recombinant productivity and cofactor stability analysis.
a) A FVIII evolutionary tree and ancestral FVIII sequences were inferred from extant genomic data. The numbers shown represent the node designations for resurrected An-FVIII molecules. b) Recombinant An-FVIII production rates were determined in transient (b) and stable (c) mammalian cell expression systems. Error bars indicate sample standard deviation (s.d.), closed circles represent individual clones (with sample sizOKe in parentheses and mean as a dashed line) and asterisks highlight comparisons where P < 0.05. d) Biosynthetic efficiency was determined by comparison of the FVIII secretion rate to steady state mRNA transcript levels. Again, asterisks denote comparisons to hFVIII where P < 0.05. e) Recombinant FVIII preparations (1 nM) were activated with thrombin and residual activity was measured over time.
FVIII circulates as a pro-cofactor in a heterodimeric, A1-A2-B/ap-A3-C1-C2 domain structure. Upon proteolytic activation, the essential A2 domain quickly dissociates and FVIII becomes inactive—a natural negative feedback mechanism that prevents long-lasting hFVIII activity (Supplemental Fig. 2). Mutations that further decrease A2 stability in activated hFVIII result in reduction of coagulation potential and a mild bleeding phenotype in humans17. We hypothesized previously that activated hFVIII evolved to its current state of rapid A2 dissociation to reduce thrombotic risk7. The pharmacological correlate of this hypothesis is that stabilization of activated FVIII should result in a more potent FVIII product, an hypothesis that was supported by data from a preclinical study of murine hemophilia A18 which showed increased cofactor activity in an in vitro thrombin generation assay and greater protection from hemostatic challenge in vivo following tail transection. To assess the stability of thrombin-activated An-FVIII molecules, we monitored proteolytic activation of purified recombinant preparations for residual cofactor activity (Supplemental Fig. 3 – 4 and Table 1). Contrary to the initial hypothesis, we observed rapid decay in activated hFVIII in all primate/hominid activated An-FVIII molecules, including all ancestral proteins back to the last common mammalian ancestor. In the rodent lineage, early ancestors An-66 and -67 displayed modestly prolonged decay rates, with a t1/2 of 3.8 and 4.4 min respectively, progressing towards the fully-extended murine activated FVIII-like with t1/2 of 15.6 min observed for An-68 (Fig. 1e). These data suggest that the improved stability of murine activated FVIII is the result of adaptive evolutionary steps in a separate direction from the common mammalian ancestor.
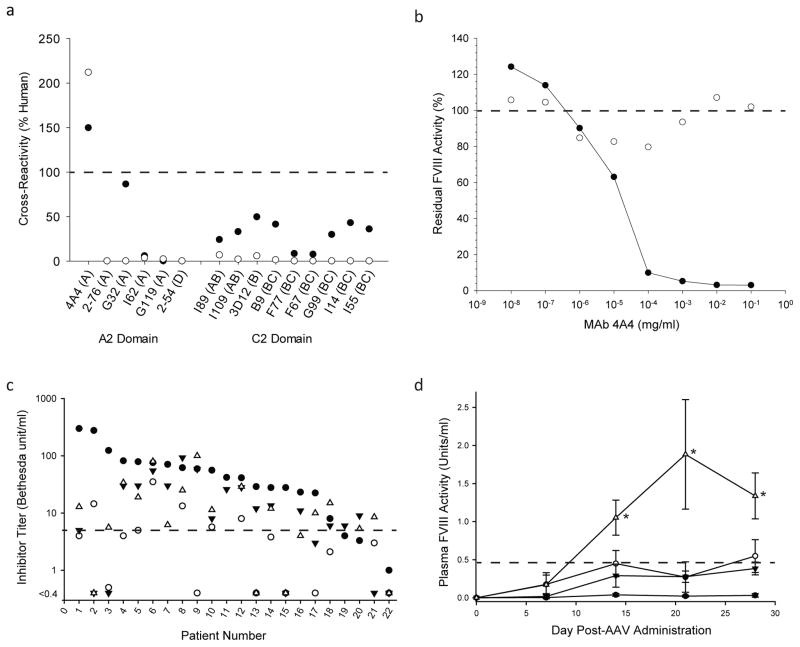
In addition to production cost, the greatest obstacle to treatment of hemophilia A is the development and clinical management of anti-FVIII inhibitory antibodies. Using well-characterized anti-hFVIII murine monoclonal antibodies (MAbs) that are potent inhibitors of activated FVIII activity and display specificity for the dominant epitopes inhibited by plasma from patients with hemophilia A, we first examined An-FVIII antigenicity profiles (Fig. 2a). Despite sharing 95% identity to hFVIII, An-53 displays markedly reduced cross-reactivity to MAbs with known epitopes near or overlapping clinically-relevant epitopes in the A2 and C2 domains. All group A anti-hFVIII MAbs bind a common epitope spanning residues 484-508, and only MAbs 4A4 and G32 demonstrated a noticeable cross-reactivity to An-5319. Using ASR, it was possible to map and eliminate binding to MAb 4A4 by replacing a single amino acid, E434V (Fig. 2b and Supplemental Fig. 5). We then assessed the inhibition of An-53 and An-68 by hemophilia A patient plasma (which contains polyclonal anti-FVIII IgG antibodies) and observed a reduction in inhibition of >75% when compared with hFVIII. Furthermore, in 10 of 25 plasmas, the residual inhibition of the two ancestral species was either below the level of detection or within a range where therapeutic efficacy of FVIII is predicted (Fig. 2c and Supplementary Table 2). Comparison of predicted B-cell epitopes within hFVIII, ET3 and An-53 by in silico methods showed that ET3 stood apart from An-53 and hFVIII, whose profiles were virtually indistinguishable. These findings support the ability of ASR to guide recombinant protein bioengineering and humanization (Supplemental Fig. 6).
Fig. 2. An-FVIII immune safety and in vivo pharmacology studies.
a) Cross-reactivity of inhibitory anti-hFVIII MAbs to An-53 (closed circles) and An-68 (open circles) was determined via direct ELISA. b) Concentration dependent inhibition of An-53 (closed circles) and An-53 E434V (open circles) by MAb 4A4 was studied using a modified Bethesda assay. c) Inhibition of An-53 (inverted closed triangles), An-68 (open triangles), porcine FVIII (open circles) and hFVIII (closed circles) by hemophilia A inhibitor patient plasma was measured using a modified Bethesda assay. The dashed line indicates a threshold beyond which the clinical benefit of FVIII infusion therapy is insufficient (≤ 5 BU/mL). d) Hemophilia A mice were injected with 2×1011 vg/kg AAV8 particles encoding hFVIII (closed circles), the bioengineered codop-hFVIII-V3 variant (inverted closed triangles), or An-53 (open triangles) all under transcriptional control of an identical liver-specific promoter (n = 4 per cohort). For comparison, a ten-fold higher dose (2×1012 vg/kg) of hFVIII encoding AAV8 vector (open circles) was administered to a separate cohort. Plasma FVIII activity measurements revealed significantly increased FVIII levels of An-53 compared to the equivalent dose of codop-hFVIII-V3 or either dose of hFVIII.AAV8 at weeks 2–4 (as denoted by asterisk, P < 0.001). The dashed line marks the transition from hemophilia A classification to normal FVIII levels (below and above 0.45 units/ml, respectively).
Based on favorable in vitro productivity, biochemical stability and immunogenicity data, we undertook in vivo pharmacology studies to assess the therapeutic potential of An-53 and An-68 as recombinant protein therapeutics or transgene components of a gene therapy product. ED50 estimates of 89 and 47 units/kg, were obtained for intravenously-delivered recombinant An-53 and An-68, respectively (Supplemental Table 3). A possible explanation for the apparent higher potency of An-68 is the greater stability of cofactor activity shown in Fig. 1e. The major limitation to clinical translation of gene therapy for hemophilia A is the low-level biosynthesis of hFVIII at safe vector doses as vector-directed immune responses and insertional mutagenesis represent dose-limiting toxicities for AAV and lentiviral vectors, respectively. Furthermore, current clinical gene therapy vector manufacture is inefficient compared to other biologics and the vector products costs at or near these dose limits may not be commercially feasible. Based on the enhanced in vitro biosynthesis data obtained for An-53, and on its higher degree of sequence similarity to hFVIII (95% identity), we compared the performance of An-53 to ET3 and hFVIII in two gene transfer settings, hydrodynamic plasmid DNA infusion and intravenous AAV vector, for delivery of liver-directed FVIII expression cassettes in hemophilia A mice. Both approaches successfully led to higher plasma FVIII activity levels with An-53 compared with ET3 or hFVIII transgenes (Fig. 2d and Supplemental Figs. 7 – 12).
In conclusion, we used ASR to identify FVIII pharmaceutical candidates with superior properties as compared with current hFVIII biologics. These properties included biosynthetic efficiency, specific activity, stability and immune reactivity. ASR is a widely accessible strategy that utilizes both known and unknown natural protein diversity to rapidly probe a protein design space that has already been refined by natural selection for beneficial properties.
ONLINE MATERIALS AND METHODS
Materials
An-FVIII cDNAs were codon optimized with human genome or liver-specific codon bias for in vitro and in vivo studies, respectively, and synthesized de novo by GenScript Biotech Corporation (Piscataway, NJ). SP-Sepharose, Source-Q chromatography resins, and Tricorn columns were purchased from GE Healthcare Life Sciences (Marlborough, MA). Lipofectamine 2000, Power SYBR® PCR Master Mix, RNAlater®, reverse transcriptase and RNase inhibitor were purchased from ThermoFisher (Grand Island, NY). HEK 293T/17 cells were purchased from ATCC (Manassas, VA), BHK-M cells have been previously reported and are described further in a subsequent section 1, 2. Serum-free AIM-V medium, Opti-mem®, and DMEM were purchased from Gibco®, ThermoFisher. Geneticin (G418) was purchased from Invitrogen, ThermoFisher. RNA and DNA isolation kits were purchased from Qiagen (Valencia, CA). Double stranded DNA fragments (gBlocks) were purchase from Integrated DNA Technologies (Coralville, IA). Polyethylenimine, Tween-80, and factor Xa chromogenic substrate were purchased from Sigma-Aldrich (St. Louis, MO). SDS-PAGE gels, protein standard, alkaline phosphatase substrate kit (AP pNPP), and Poly-Prep® columns were purchased from Bio-Rad (Hercules, CA). Pooled citrated normal plasma (FACT) and FVIII-deficient plasma were purchased from George King Biomedical (Overland Park, KS). Automated APTT reagent was purchased from Trinity Biotech (Wicklow, Ireland). Chromagenix FVIII Coatest was purchased from diaPharma (West Chester, OH). Purified factor X and human alpha-thrombin were purchased from Enzyme Research Laboratories (South Bend, IN). Factor IXa and phospholipid vesicles (PCPS) were generated and purified as previously described 7, 20, 21. Cell culture flasks, Costar ELISA and EIA/RIA plates were purchased from Corning®, Sigma-Aldrich. Streptavidin-alkaline phosphate conjugate was purchased from Jackson Immuno Research (West Grove, PA). TransIT®-EE delivery solution was purchased from Mirus (Madison, WI). FVIII domain specific monoclonal antibodies were generated and purified as previously described 19, 22, 23. B-domain deleted recombinant oFVIII, pFVIII, ET3 and hFVIII were generated and purified as described previously 2, 3, 6, 24. De-identified congenital and acquired hemophilia A inhibitor patient plasma samples were provided from the Emory Inhibitor Bank. The plasmas were collected in accordance with Emory IRB protocol no. IRB00006290. AAV8-FVIII vectors were prepared by ViGene Biosciences Inc. (Rockville, MD).
Animal studies
All animal studies were performed under the guidelines set by the Emory University Institutional Animal Care and Use Committee. Exon 16-disrupted hemophilia A mice (E16) have been described previously 25. Upon receipt of these animals from American Red Cross, the animals were backcrossed onto a C57BL/6J background. The mice are maintained as a breeding colony in the homozygous (F8−/−) and hemizygous (F8−/Y) state. For the hydrodynamic injection studies described, both male and female mice between 8 – 12 weeks of age were utilized without subjective bias. For the AAV gene transfer studies, only male mice were used as they are transduced more efficiently than female mice 26. Randomization of the mice in each experiment was performed using the www.random.org “Random Sequence Generator” function. DNA and protein samples were blinded to the experimenter prior to administration to the randomized animals. Based on statistical power estimates (alpha = 0.05, power = 0.8, sigma = 0.3), and assuming expression differentials similar to those observed in vitro (i.e. 1 unit/mL) in the current study or in vivo in previously published studies, a minimum of 3 mice per group would be needed to determine differences. Total animal numbers (n) for each experiment are indicated.
Cell lines
The modified baby hamster kidney (BHK-M) cell line used in the current study has been described by our group previously. This cell line was used for the commercial development of recombinant porcine FVIII and was submitted by our colleague, Dr. Pete Lollar, to American Type Culture Collection (ATCC) and is available as catalog reference – BHK-M; hamster (PTA-4506). The human embryonic kidney 293T/17 cell line (ATCC CRL-11268) was obtained directly from ATCC.
An-FVIII sequence inference
An-FVIII sequence reconstruction was performed as described previously 15, 27. Briefly, 47 available mammalian FVIII sequences were aligned using MUSCLE and an evolutionary tree was inferred using MrBayes (Fig. 1a) 28. Ancestral sequences were inferred using both DNA and amino acid-based models in PAML VERSION 4.1 29. The multiple sequence alignment is electronically available upon request.
Biosynthetic efficiency of An-FVIII variants
HEK293T/17 and BHK-M cells were transfected with PEI or Lipofectamine 2000, respectively, in antibiotic-free media. Transiently transfected cells were washed with PBS and switched to serum-free media 48 hours after transfection and FVIII activity was measured by one-stage coagulation assay following an additional 24 h. Activity was normalized to human FVIII activity. Stable BHK-M clones were generated as previously described 1–3 and 24 – 36 clones of each FVIII construct were isolated for screening. Clones resistant to G418 but lacking detectable FVIII activity were discarded. FVIII activity was measured in serum-free media by one-stage coagulation assay and normalized to cell counts taken at the time of FVIII activity determination. RNA for subsequent steady-state transcript levels was collected following a phosphate-buffered saline (PBS) wash and storage at −80 °C in RNAlater®. Transcript levels were determined using one-step RT-PCR through interpolation to a plasmid DNA standard curve. Primers for transcript analysis are shown in Supplemental Table S4. To calculate the number of FVIII molecules generated per FVIII mRNA transcript, a generic FVIII specific activity estimate of 1 unit per 150 ng purified protein was utilized for all constructs.
Statistical analysis
All calculations were performed using SigmaPlot 13 software (Systat Software Inc., San Jose, CA). Cell-line FVIII expression comparisons made between An-FVIII constructs and hFVIII were performed using the Kruskal-Wallis one-way analysis of variance on ranks non-parametric test. Post hoc comparisons of the individual construct pairs were conducted using Dunnett’s test of multiple comparisons versus a control group. Comparisons of FVIII concentrations following AAV8 administration were performed each week using Holm-Sidak one-way analysis of variance with all pairwise comparisons.
Purification of An-FVIII variants
The BHK-M clones displaying the greatest rate of FVIII production were expanded into triple-flasks and FVIII was collected in serum-free media and stored at −20 °C with 0.05% sodium azide until purification. Cell debris was removed prior to storage by centrifugation at 1,000 rcf for 15 min. FVIII was purified by one- or two-stage ion exchange chromatography as previously described 2, 3. Elution fractions were analyzed for purity via SDS-PAGE and concentration was determined by one-stage coagulation. Specific activity of homogenous FVIII samples was determined by one-stage coagulation determination compared to λ280 nm following λ320 nm and buffer corrections. Purified FVIII was aliquoted and stored at −80 °C until use for biochemical characterization.
Non-proteolytic decay via A2 domain dissociation
Activated factor VIII (FVIIIa) was measured by chromogenic assay using purified human factor IXa, human factor X, and synthetic phospholipid vesicles as described previously 2, 6, 7, 30. Briefly, 1 nM FVIII was activated with 100 nM human thrombin for 15 s at room temperature. Desulfatohirudin (150 nM final) was added to stop the reaction and FVIII activity was measured at several time points.
Antigenicity and inhibition of An-FVIII variants
The generation and characterization of the anti-hFVIII MAbs used in the current study has been described previously 19, 22, 23, 31. Hemophilia A inhibitor patient plasma samples were obtained from the Emory Inhibitor Bank after approval by Institutional Review Board (IRB) and written informed consent from patients, plasma samples are drawn and banked in accordance with Emory IRB protocol no. IRB00006290. Sixty-four inhibitor plasmas (52 congenital and 12 acquired) were assayed via ELISA for anti-human Ig and cross-reactivity to porcine or ancestral FVIII. Samples with an anti-human titer > 20 were included and further analyzed for inhibitor titer by modified Bethesda assay if residual volumes permitted, yielding 21 congenital and 4 acquired samples. The cross-reactivity of monoclonal anti-human FVIII antibodies (MAbs) against ancestral FVIII was measured via direct ELISA as previously described 4. Briefly, 1.5 μg FVIII was adsorbed to an ELISA plate in 20 mM Bicine, 2 mM CaCl2 pH 9.0 for 2 h and blocked with 2% BSA in 20 mM HEPES, 150 mM NaCl, 2 mM CaCl2 at 4°C for at least 12 h. MAbs were selected due to A2 and C2 domain epitope recognition as well as anti-human inhibitor titers exceeding 1,000 Bethesda units (BU)/mg as previously determined 19, 23. Biotinylated MAb at 4 μg/mL was added to the well for one h at room temperature, followed by 1:10,000 dilution of strepavidin-conjugated alkaline phosphatase in blocking buffer. Absorbance at 405 nm was measured following addition of chromogenic substrate. Mean values of triplicates were recorded and normalized to anti-hFVIII signal as percent cross-reactivity. Inhibitory titers against each FVIII construct were determined by modified Bethesda Assay as described previously 4.
Disinhibition of E434V mutant by MAb 4A4
A de novo synthesized DNA fragment (gBlock) containing a single point mutation E434V was inserted into An-53 by enzymes NheI and BlpI and confirmed via Sanger sequencing. A polyclonal population of BHK-M cells producing the E434V mutant was generated and expanded for protein collection and subsequent inhibitor testing. Inhibitor titer of Mab 4A4 was conducted using a modified Bethesda assay 32. Briefly, E434V FVIII or purified An-53 was diluted to 0.8 – 1.2 unit/ml in conditioned serum-free supernatant from naïve BHK-M cells and buffered with 0.1 M imidazole. FVIII and MAb 4A4 were combined and incubated at 37°C for 2 h, and residual FVIII activity was determined via one-stage coagulation assay.
In silico prediction of FVIII B-cell epitopes
B-domain deleted FVIII amino acid sequences were analyzed for potential B cell epitopes using the using the BiprePred software program 33. Designated sensitivity and specificity thresholds were set to 0.35 and 1.3, respectively.
ED50 up-down efficacy determination
Hemostatic challenge was performed via tail transection as previously described 3, 34 and the ED50 was calculated by Dixon up-and-down method as previously described 35, 36. Briefly, hemophilia A E16 mice were injected with saline, recombinant ancestral FVIII at varying doses, or bolus hFVIII diluted in 0.9% saline via tail vein. Mouse tails were incubated at 37 °C for 15 min prior to challenge. FVIII doses were determined a priori and prepared immediately preceding injection. A bleeding event was defined as blood loss (g/kg) exceeding the standard deviation of wildtype mice in an identical challenge without infused FVIII (16 mg/g body weight). Following transection, blood was collected directly in conical tubes containing pre-warmed PBS and measured by change in mass after 40 min.
In vivo FVIII gene transfer
Codon-optimized human, ET3, and An-53 FVIII were subcloned into an AAV expression cassette incorporating the liver-directed hybrid liver promoter (HLP) promoter and a synthetic polyadenylation sequence described previously 37. Plasmid DNA was linearized outside the inverted terminal repeat (ITR) sequences and the DNA quality and quantity was assessed via gel electrophoresis prior to injection. Hydrodynamic injections were conducted as previously described 26. Briefly, mice were weighed prior to injection and varying doses of linear plasmid was diluted into Transit-EE hydrodynamic delivery solution totaling 10% body weight. Naked DNA was delivered to hemophilia A mice age 8 – 12 weeks by tail vein injection over the course of 5–8 seconds. Blood plasma was collected at several time points following injection and FVIII activity was measured by COATEST SP assay according to the manufacturer’s instructions using a standard curve generated from pooled citrated human plasma (FACT). In vivo specific activity of An-53 was determined by plasma activity compared to antigen levels determined by ELISA using cross-reactive anti-hFVIII murine monoclonal antibodies (MAbs) and purified recombinant An-53 as a standard. AAV8 vector was delivered intravenously via tail vein to 8 – 12 week old male hemophilia A mice at a dose of either 2 × 1011 or 2 × 1012 vector genomes (vg)/kg in a volume of 100 μl sterile PBS. Plasma samples were drawn weekly post AAV delivery and assayed for FVIII activity as described above.
Supplementary Material
Acknowledgments
This work was supported by funding from the National Institutes of Health/National Heart, Lung and Blood Institute (H.T.S., S.L.M., and C.B.D.), the Bayer Hemophilia Awards Program, Bayer HealthCare (C.B.D.) as well as a research partnership between Children’s Healthcare of Atlanta and the Georgia Institute of Technology (C.B.D. and E.A.G.). We also thank Ernest T. Parker for technical assistance with the in vivo mouse studies.
Footnotes
AUTHOR CONTRIBUTIONS
P.M.Z. designed and performed experiments, analyzed the data, and drafted the manuscript. H.C.B. performed gene transfer experiments and edited the manuscript. K.K. performed experiments. S.L.M. contributed reagents, designed experiments, analyzed data and edited the manuscript. H.T.S. conceived the project, designed experiments, analyzed data and edited the manuscript. E.A.G. performed ASR and edited the manuscript. C.B.D. conceived the project, designed experiments, analyzed data and drafted and edited the manuscript.
COMPETING FINANCIAL INTERESTS STATEMENT
Drs. Doering, Gaucher, Spencer and Zakas are inventors on a patent application describing ancestral FVIII technology filed by Emory University/Children’s Healthcare of Atlanta and Georgia Institute of Technology. Drs. Doering and Spencer are co-founders Expression Therapeutics and own equity in the company. Expression Therapeutics owns the intellectual property associated with ET3. The terms of this arrangement have been reviewed and approved by Emory University in accordance with its conflict of interest policies.
References
- 1.Doering C, et al. Expression and characterization of recombinant murine factor VIII. Thromb Haemost. 2002;88:450–458. [PubMed] [Google Scholar]
- 2.Doering CB, Healey JF, Parker ET, Barrow RT, Lollar P. High level expression of recombinant porcine coagulation factor VIII. J Biol Chem. 2002;277:38345–38349. doi: 10.1074/jbc.M206959200. [DOI] [PubMed] [Google Scholar]
- 3.Zakas PM, et al. Development and characterization of recombinant ovine coagulation factor VIII. PLoS One. 2012;7:e49481. doi: 10.1371/journal.pone.0049481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zakas PM, Vanijcharoenkarn K, Markovitz RC, Meeks SL, Doering CB. Expanding the ortholog approach for hemophilia treatment complicated by factor VIII inhibitors. J Thromb Haemost. 2015;13:72–81. doi: 10.1111/jth.12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sabatino DE, et al. Recombinant canine B-domain-deleted FVIII exhibits high specific activity and is safe in the canine hemophilia A model. Blood. 2009;114:4562–4565. doi: 10.1182/blood-2009-05-220327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doering CB, Healey JF, Parker ET, Barrow RT, Lollar P. Identification of porcine coagulation factor VIII domains responsible for high level expression via enhanced secretion. J Biol Chem. 2004;279:6546–6552. doi: 10.1074/jbc.M312451200. [DOI] [PubMed] [Google Scholar]
- 7.Parker ET, Doering CB, Lollar P. A1 subunit-mediated regulation of thrombin-activated factor VIII A2 subunit dissociation. J Biol Chem. 2006;281:13922–13930. doi: 10.1074/jbc.M513124200. [DOI] [PubMed] [Google Scholar]
- 8.Zuckerkandl E, Pauling L. Molecules as documents of evolutionary history. J Theor Biol. 1965;8:357–366. doi: 10.1016/0022-5193(65)90083-4. [DOI] [PubMed] [Google Scholar]
- 9.Merkl R, Sterner R. Ancestral protein reconstruction: techniques and applications. Biol Chem. 2016;397:1–21. doi: 10.1515/hsz-2015-0158. [DOI] [PubMed] [Google Scholar]
- 10.Risso VA, Gavira JA, Mejia-Carmona DF, Gaucher EA, Sanchez-Ruiz JM. Hyperstability and substrate promiscuity in laboratory resurrections of Precambrian beta-lactamases. J Am Chem Soc. 2013;135:2899–2902. doi: 10.1021/ja311630a. [DOI] [PubMed] [Google Scholar]
- 11.Ivarsson Y, Mackey AJ, Edalat M, Pearson WR, Mannervik B. Identification of residues in glutathione transferase capable of driving functional diversification in evolution. A novel approach to protein redesign. J Biol Chem. 2003;278:8733–8738. doi: 10.1074/jbc.M211776200. [DOI] [PubMed] [Google Scholar]
- 12.Harms MJ, et al. Biophysical mechanisms for large-effect mutations in the evolution of steroid hormone receptors. Proc Natl Acad Sci U S A. 2013;110:11475–11480. doi: 10.1073/pnas.1303930110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kratzer JT, et al. Evolutionary history and metabolic insights of ancient mammalian uricases. Proc Natl Acad Sci U S A. 2014;111:3763–3768. doi: 10.1073/pnas.1320393111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson C, et al. Kinase dynamics. Using ancient protein kinases to unravel a modern cancer drug’s mechanism. Science. 2015;347:882–886. doi: 10.1126/science.aaa1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaucher EA, Govindarajan S, Ganesh OK. Palaeotemperature trend for Precambrian life inferred from resurrected proteins. Nature. 2008;451:704–707. doi: 10.1038/nature06510. [DOI] [PubMed] [Google Scholar]
- 16.Brown HC, Gangadharan B, Doering CB. Enhanced biosynthesis of coagulation factor VIII through diminished engagement of the unfolded protein response. J Biol Chem. 2011;286:24451–24457. doi: 10.1074/jbc.M111.238758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pipe SW, Eickhorst AN, McKinley SH, Saenko EL, Kaufman RJ. Mild hemophilia A caused by increased rate of factor VIII A2 subunit dissociation: evidence for nonproteolytic inactivation of factor VIIIa in vivo. Blood. 1999;93:176–183. [PubMed] [Google Scholar]
- 18.Leong L, et al. Noncovalent stabilization of the factor VIII A2 domain enhances efficacy in hemophilia A mouse vascular injury models. Blood. 2015;125:392–398. doi: 10.1182/blood-2014-02-555656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Markovitz RC, Healey JF, Parker ET, Meeks SL, Lollar P. The diversity of the immune response to the A2 domain of human factor VIII. Blood. 2013;121:2785–2795. doi: 10.1182/blood-2012-09-456582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esmon CT, Lollar P. Involvement of thrombin anion-binding exosites 1 and 2 in the activation of factor V and factor VIII. J Biol Chem. 1996;271:13882–13887. doi: 10.1074/jbc.271.23.13882. [DOI] [PubMed] [Google Scholar]
- 21.Barrow RT, Parker ET, Krishnaswamy S, Lollar P. Inhibition by heparin of the human blood coagulation intrinsic pathway factor X activator. J Biol Chem. 1994;269:26796–26800. [PubMed] [Google Scholar]
- 22.Healey JF, et al. The humoral response to human factor VIII in hemophilia A mice. J Thromb Haemost. 2007;5:512–519. doi: 10.1111/j.1538-7836.2007.02373.x. [DOI] [PubMed] [Google Scholar]
- 23.Meeks SL, Healey JF, Parker ET, Barrow RT, Lollar P. Antihuman factor VIII C2 domain antibodies in hemophilia A mice recognize a functionally complex continuous spectrum of epitopes dominated by inhibitors of factor VIII activation. Blood. 2007;110:4234–4242. doi: 10.1182/blood-2007-06-096842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doering CB, et al. Expression and Characterization of Recombinant Murine Factor VIII. Thromb Haemost. 2002;88:450–458. [PubMed] [Google Scholar]
- 25.Bi L, et al. Targeted disruption of the mouse factor VIII gene produces a model of haemophilia A. Nat Genet. 1995;10:119–121. doi: 10.1038/ng0595-119. [DOI] [PubMed] [Google Scholar]
- 26.Brown HC, et al. Bioengineered coagulation factor VIII enables long-term correction of murine hemophilia A following liver-directed adeno-associated viral vector delivery. Mol Ther Methods Clin Dev. 2014;1:14036. doi: 10.1038/mtm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaucher EA, Thomson JM, Burgan MF, Benner SA. Inferring the palaeoenvironment of ancient bacteria on the basis of resurrected proteins. Nature. 2003;425:285–288. doi: 10.1038/nature01977. [DOI] [PubMed] [Google Scholar]
- 28.Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–2314. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- 29.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 30.Lollar P, Parker ET, Fay PJ. Coagulant properties of hybrid human/porcine factor VIII molecules. J Biol Chem. 1992;267:23652–23657. [PubMed] [Google Scholar]
- 31.Healey JF, et al. The comparative immunogenicity of human and porcine factor VIII in haemophilia A mice. Thromb Haemost. 2009;102:35–41. doi: 10.1160/TH08-12-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrow RT, Lollar P. Neutralization of antifactor VIII inhibitors by recombinant porcine factor VIII. J Thromb Haemost. 2006;4:2223–2229. doi: 10.1111/j.1538-7836.2006.02135.x. [DOI] [PubMed] [Google Scholar]
- 33.Larsen JE, Lund O, Nielsen M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006;2:2. doi: 10.1186/1745-7580-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spencer HT, et al. Lentiviral vector platform for production of bioengineered recombinant coagulation factor VIII. Mol Ther. 2011;19:302–309. doi: 10.1038/mt.2010.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parker ET, Lollar P. A quantitative measure of the efficacy of factor VIII in hemophilia A mice. Thromb Haemost. 2003;89:480–485. [PubMed] [Google Scholar]
- 36.Dixon WJ. Staircase bioassay: the up-and-down method. Neurosci Biobehav Rev. 1991;15:47–50. doi: 10.1016/s0149-7634(05)80090-9. [DOI] [PubMed] [Google Scholar]
- 37.McIntosh J, et al. Therapeutic levels of FVIII following a single peripheral vein administration of rAAV vector encoding a novel human factor VIII variant. Blood. 2013;121:3335–3344. doi: 10.1182/blood-2012-10-462200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.