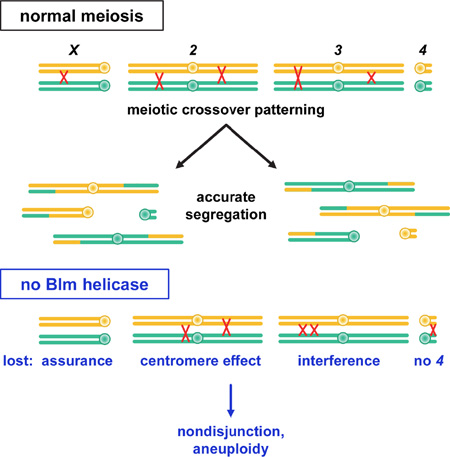
Summary
In most sexually reproducing organisms, crossover formation between homologous chromosomes is necessary for proper chromosome disjunction during meiosis I. During meiotic recombination, a subset of programmed DNA double-strand breaks (DSBs) are repaired as crossovers, with the remainder becoming noncrossovers [1]. Whether a repair intermediate is designated to become a crossover is a highly-regulated decision that integrates several crossover patterning processes, both along chromosome arms (interference and the centromere effect) and between chromosomes (crossover assurance) [2]. Because the mechanisms that generate crossover patterning have remained elusive for over a century, it has been difficult to assess the relationship between crossover patterning and meiotic chromosome behavior. We show here that meiotic crossover patterning is lost in Drosophila melanogaster mutants that lack the Bloom syndrome helicase. In the absence of interference and the centromere effect, crossovers are distributed more uniformly along chromosomes. Crossovers even occur on the small chromosome 4, which normally never has meiotic crossovers [3]. Regulated distribution of crossovers between chromosome pairs is also lost, resulting in an elevated frequency of homologs that do not receive a crossover, which in turn leads to elevated nondisjunction.
Keywords: Bloom syndrome helicase, crossover patterning, meiosis, meiotic recombination, crossover interference, crossover assurance, centromere effect, nondisjunction, Drosophila
Graphical Abstract
eTOC Blurb
Hatkevich et al. demonstrate that the Drosophila Bloom syndrome helicase chaperones double-strand break intermediates into the meiotic recombination pathway. In Blm mutants, the crossover/noncrossover decision is unregulated, leading to loss of crossover patterning and increased nondisjunction.
Results and Discussion
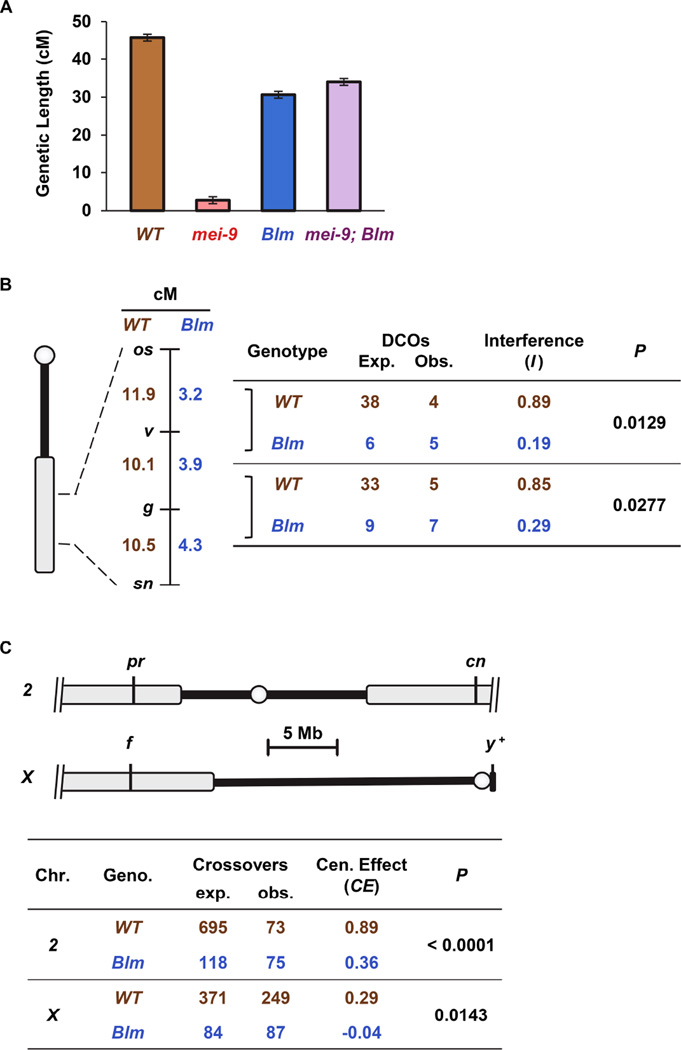
Crossover interference, discovered by Sturtevant more than 100 years ago [4], is a meiotic crossover patterning phenomenon in which the presence of a crossover in one interval reduces the probability of a crossover in an adjacent interval [5]. Studies in budding yeast, Arabidopsis, and mice revealed a subset of meiotic crossovers that do not show interference [5]. These “Class II crossovers” are generated through a different pathway than most (Class I) meiotic crossovers [6]. In budding yeast, single-locus hotspot assays show that meiotic crossovers generated in the absence of the Bloom syndrome helicase (Blm) ortholog Sgs1 are formed primarily or exclusively by the Class II pathway [7, 8]. This conclusion was partially derived from the observation that crossovers formed in sgs1 meiotic-null mutants are not dependent upon Mlh1, a component of the meiosis-specific, Class I Holliday junction resolvase. We asked whether Drosophila Blm is also required to populate the Class I pathway by determining whether crossovers generated in the absence of Blm are dependent upon MEI-9, the catalytic subunit of the presumptive Drosophila meiotic resolvase [9]. We measured crossovers in five adjacent intervals spanning most of 2L and part of 2R (for simplicity, referred to as 2L), a region comprising ~20% of the euchromatic genome. As in previous studies [10], loss of MEI-9 results in a >90% reduction in crossovers compared to wild-type flies (Figure 1A). While Blm single mutants exhibit ~30% decrease in crossovers on 2L [11], there is no additional reduction of crossovers in mei-9; Blm double mutants. Therefore, the crossovers that occur in Blm mutants do not require MEI-9, suggesting that they are generated through the Class II pathway.
Figure 1. Crossover patterning is lost in Blm mutants.
(A) Genetic length of the net – cn region in different genotypes. Bars are ±95% confidence intervals. Genotypes were wild-type (WT; n=4222), mei-9 (n=2433), Blm (n=1171), and mei-9; Blm (n=1074). For the entire dataset, refer to Figure S1 and Table S1.
(B) X chromosome interference. Crossovers were measured in three adjacent intervals in the middle of the X, as in the schematic (circle: centromere; black line: unassembled pericentromeric satellite sequences; gray: genome assembly). To the right of the schematic, genetic lengths of these intervals in wild-type flies (n=3088) and Blm mutants (n=4953) are shown. Stevens’ [13] measurement of interference (I) was calculated as (observed DCOs / expected DCOs); I = 1 indicates complete interference and I = 0 is no interference. For complete dataset, refer to Table S2.
(C) The centromere effect. Schematics show the centromere-spanning intervals assayed. Lines are as in Figure 1B. The table below shows CE values for wild-type flies and Blm mutants in centromere-spanning intervals of 2 and X. CE values were calculated as 1 − (observed crossovers/expected crossovers) in proximal f−y+ and pr-cn intervals. Expected was calculated as: total crossovers * (length of proximal interval / total length). Two-tailed Fisher’s exact test was used to determine significance between observed and expected CO values between specified genotypes. For the entire dataset, refer to Tables S1 and S3.
The original distinction between crossovers generated by the Class I and Class II pathways is that only the former exhibit crossover interference [12]. We measured crossovers in three adjacent intervals on the X chromosome and calculated Stevens’ [13] measure of interference (I = 1−[observed double crossovers / expected double crossovers]) between pairs of intervals. Interference was strong in wild-type flies (I= 0.89 and 0.85 for the two pairs of adjacent intervals), but was significantly reduced in Blm mutants (I = 0.19 and 0.29, Figure 1B). Thus, without Blm helicase, crossovers are not dependent on the Class I resolvase MEI-9 and interference among these crossovers is severely reduced or absent. This demonstrates that, as in S. cerevisiae, Drosophila Blm is required for generation of crossovers through the Class I pathway.
Given the loss of interference, we asked whether another important process that patterns crossovers along chromosomes arms – the centromere effect – is also lost in Blm mutants. This phenomenon, first reported by Beadle in 1932 [14], is the suppression of crossover formation in centromere-proximal euchromatin. To quantify the centromere effect, we devised a measure, CE, that is analogous to I as a measure of interference in that CE = 1−(observed/expected), where observed is the number of crossovers counted in the interval and expected is the number expected in a random distribution (see Supplemental Information for more details). In wild-type females, the interval between pr and cn, which spans the chromosome 2 centromere, has a CE of 0.89, consistent with a strong centromere effect (Figure 1C). In Blm mutants this is reduced to 0.36 (P < 0.0001). The centromere effect is much weaker on the X chromosome due to the larger block of heterochromatin that moves the euchromatin further from the centromere [15, 16] (Figure 1C). CE in a centromere-spanning interval on the X is 0.29 in wild-type flies but is reduced to −0.04 in Blm mutants (P =0.0143).
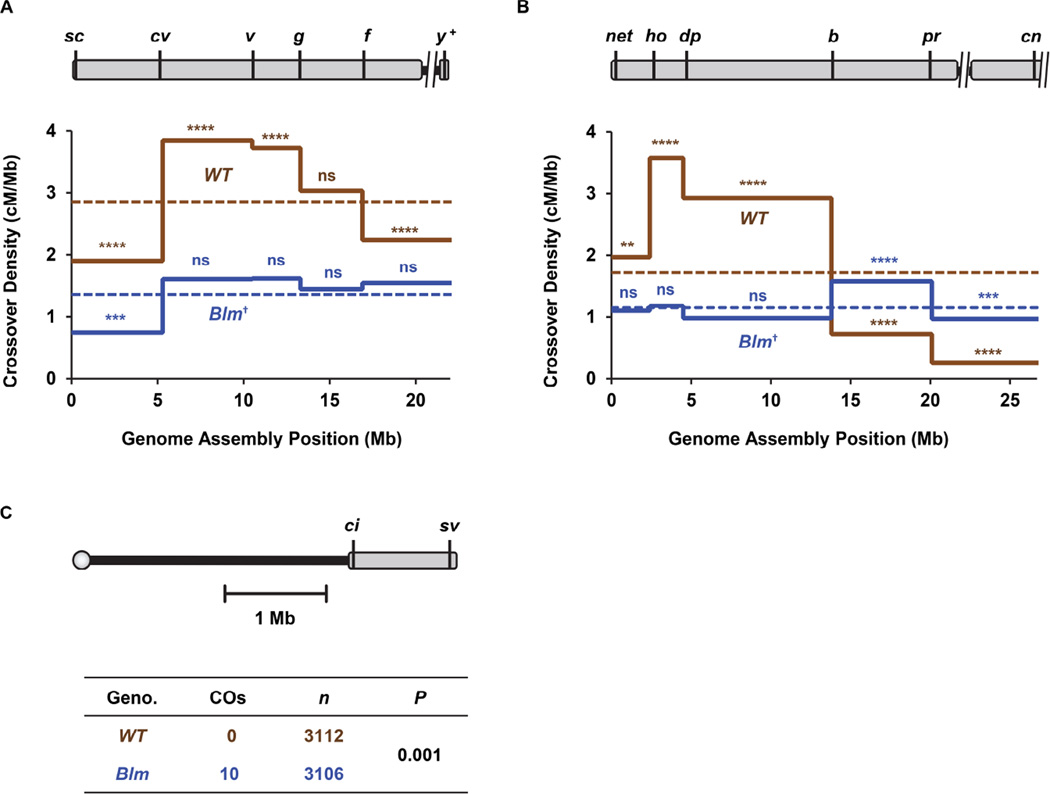
Loss of interference and the centromere effect in Blm mutants allows us to assess the consequences of loss of crossover patterning along chromosome arms. Because these crossover patterning processes are responsible for the overall crossover distribution along each chromosome arm [17], we first assessed the effect of these losses on crossover distribution along entire arms. In wild-type flies, genetic length is not proportional to physical length, with crossover density being higher in the middle of each arm [17, 18]. On both the X and 2L, crossover distribution in Blm mutants is significantly different from the wild-type distribution (P =0.0009 and P <0.0001, respectively) (Figure 2A–B). Instead, crossovers in Blm mutants appear to be distributed in a manner more proportional to physical length. In wild-type flies, nine of the ten intervals we examined have significantly different numbers of crossovers than expected if genetic distance is proportional to physical distance, but in Blm mutants only three intervals are significantly different than this expectation (Figure 2A–B). The deviations in three intervals in Blm mutants may reflect residual crossover patterning; however, the 2L crossover distributions in mei-9; Blm double mutants (Figure S1A) and in mutants carrying the helicase-dead allele BlmE866K (Figure S1C) are even more closely proportional to physical length, suggesting that the departures from proportionality in Blm null mutants may be an effect of strain background.
Figure 2. Intrachromosomal effects of loss of crossover patterning.
(A) Crossover distribution on the X chromosome. Schematic at the top is as in Figure 1 except that unassembled satellite sequences and the centromere are not included in density calculations. Locations of markers used to score crossovers are indicated. The graph below shows crossover density in each interval in wild-type flies (n=2179) and in Blm mutants (n=1099). Dotted line is mean density across the entire region assayed. Indicators of statistical significance are for chi-squared tests on observed number of crossovers versus the expected number if crossover number is proportional to physical size: ns, P >0.05; ***P <0.001; ****P <0.0001 after correction for multiple comparisons. †P <0.001 indicates significance of Blm distribution across all intervals as compared to wild-type, as determined by G-test of goodness of fit. For complete dataset, refer to Table S3.
(B) Crossover distribution on 2L. Schematic and graph are as in panel A. n=4222 for wild type and 1171 for Blm. **P <0.003; †P < 0.0001 for overall distribution in Blm mutants compared to wild-type, (G-test). For complete dataset, refer to Table S1. Transposable elements were excluded from physical lengths in these analyses. See Figure S4 for details.
(C) Crossovers on chromosome 4. Schematic is as in panels A and B, but the scale is different. The table below shows the number of flies scored and the number of crossovers detected. For details on parental and recombinant classes, please refer to Table S4. Crossover density on the X, 2L, and 4 are shown in Figure S2A.
We next examined a particularly extreme case of crossover patterning: the absence of crossovers on the small chromosome 4 of Drosophila melanogaster. There are never crossovers on this chromosome in wild-type females [3], but there have been reports of conditions that do result in crossovers. Grell [19] induced crossovers on 4 through heat-shock, but it is not known whether these are meiotic or mitotic. Sandler and Szauter [20] observed crossovers in mei-218 mutants, but others were unable to repeat this [17]. Osborne [21] found crossovers in 4-derived sequences when they were translocated to chromosome 3. This result suggests that the absence of crossovers on 4 may be a consequence of crossover patterning processes. Support for this idea came from whole-genome sequencing that revealed the presence of noncrossover gene conversion on 4 [22], indicating that DSBs are made on 4, and therefore it is the repair process that is regulated to prevent crossovers.
We scored recombination between two markers near opposite ends of the genome sequence assembly of 4 (Figure 2C). As expected, we did not recover any crossovers between these markers in wild-type females (n = 3112 progeny); however, in Blm mutants we recovered 10 crossovers among 3106 progeny (P = 0.001, two-tailed Fisher’s exact test). Blm mutants have spontaneous mitotic crossovers in the male germline [23]. To ensure that the crossovers we observed are meiotic, we eliminated meiotic DSBs; we did not observe any crossovers in this case (Tables S1 and S2). We conclude that the absence of crossovers on chromosome 4 in wild-type females is a result of active meiotic crossover patterning processes that are intertwined with the Class I crossover pathway. This is most likely due to the centromere effect, consistent with the observation that crossovers occur in 4 sequences that are translocated to a genomic region further from the centromere [21]. Interference should not be applicable to 4 because there are no initial crossover designations to discourage nearby additional designations.
Although crossovers in Blm mutants are distributed approximately evenly along X and 2L and also occur on chromosome 4, average crossover density is not the same between these chromosomes (Figure S2A). In both wild-type females and Blm mutants, crossover density is higher on the X than on 2L, and is lower still on chromosome 4 in Blm mutants. Possible explanations for this include different DSB densities, different strengths of crossover patterning (e.g., the weaker centromere effect on the X compared to chromosome 2), and residual crossover patterning in Blm mutants.
The results above show that crossover patterning along chromosomes is lost or severely reduced in Blm mutants. Crossover patterning also occurs between chromosomes. Darlington and Dark [24] reported that in a grasshopper species with a large range of chromosome sizes, every pair of homologous chromosomes always had at least one chiasma (the cytological manifestation of a crossover), called the obligate chiasma [25]. The occurrence of an obligate chiasma suggests that there is an active process, referred to as crossover assurance, that monitors the designation of crossovers on each chromosome. To determine whether loss of Blm affects crossover assurance, we compared the observed and expected frequencies of E0 tetrads (homologous chromosome pairs with no crossovers). In wild-type flies, the observed E0 frequency for the X chromosome (0.112) is less than half the frequency expected based on Poisson distribution (0.285, P <0.0001) (Figure 3A), indicating that crossover assurance is present, but it is not absolute. Crossover assurance is significantly reduced or absent in Blm mutants (P <0.0001 compared to wild type), resulting in the observed E0 frequency (0.514) being similar to the expected frequency (0.550).
Figure 3. Loss of interchromosomal crossover patterning in Blm mutants.
(A) The expected (based on Poisson distribution) and observed frequency of E0 X chromosomes in wild type and in Blm mutants. Bars are 95% confidence intervals. ***P <0.0001 based on two-tailed Fisher’s exact test of numbers observed and expected. For a similar analysis of chromosome 2L, please see Figure S3.
(B) Classification of nondisjoined chromosomes. The top schematic represents heterozygous X chromosomes in Blm mutant females. Below are structures of chromosomes recovered in daughters that inherited two maternal X chromosomes. All were meiosis I nondisjunction based on the centromere-linked marker (y+). 33 were non-recombinant; one had a crossover between f and y+ (arbitrarily drawn within the euchromatin). The frequency of non-exchange (E0) X chromosomes among those that failed to disjoin (0.97) is significantly different than among those X chromosomes that disjoined correctly (0.51) (P =0.0005).
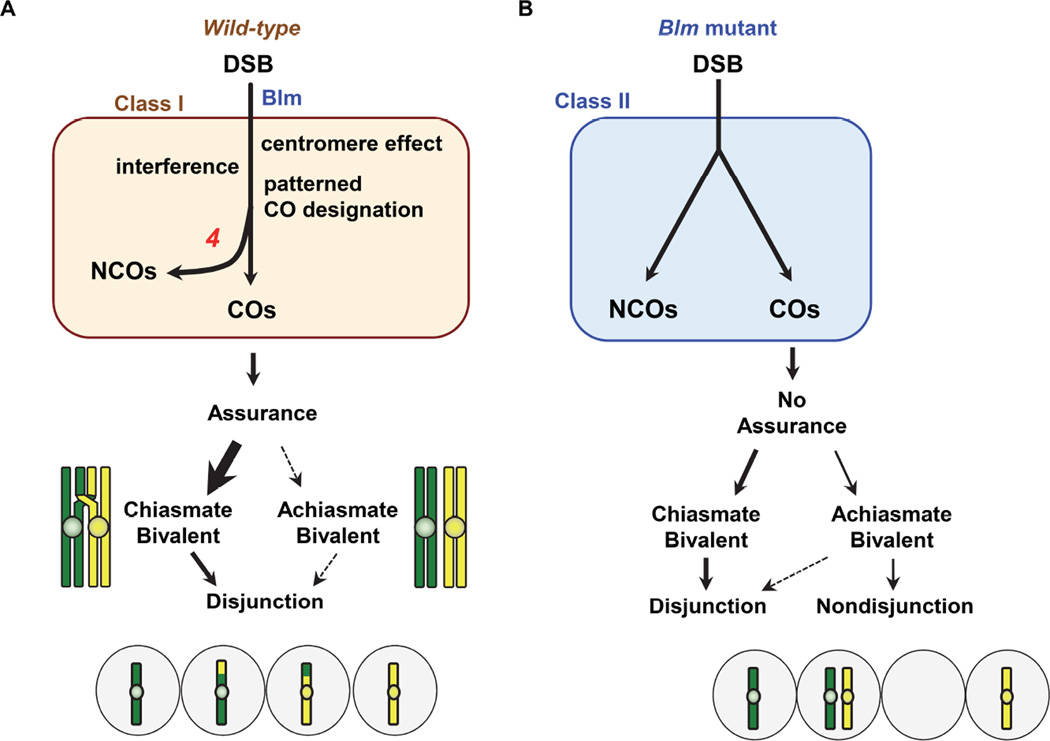
The results described above reveal that three major aspects of crossover patterning that occur along and among chromosomes – interference, the centromere effect, and assurance – are significantly decreased or eliminated when Blm helicase is absent. This suggests an inability to make or execute the crossover/noncrossover decision. Mapping of noncrossover gene conversion events in wild-type flies through whole-genome sequencing [22, 26] reveals a flat distribution along each of the major chromosome arms, similar to the distribution of crossovers in Blm mutants (Figures 2 and S2). Miller et al. [26] showed that noncrossovers do not participate in interference and are not subject to the centromere effect. These findings suggest that DSBs are evenly distributed along each arm, at least at the Mb scales at which we mapped crossovers. In wild-type flies, crossover patterning processes act on this distribution such that events in the central regions of the major chromosome arms have a higher probability of being designated to become crossovers, and those on chromosome 4 are never so designated. Regulated crossover designation is lost in Blm mutants, and as a result every DSB repair event has the same probability of becoming a crossover, regardless of where along the chromosome it is located.
Our results argue that meiotic DSB repair in Blm mutants occurs outside of the predominant meiotic recombination pathway, and that this results in loss of regulated crossover designation and loss of patterning. What are the consequences of these losses on meiosis? In Blm mutants, nondisjunction of the X chromosome is elevated 30-fold [23]. In wild-type females, most X chromosomes that nondisjoin did not have any crossovers, had only a single crossover that was distal, or had a centromere-proximal crossover [27]. We analyzed X chromosomes that nondisjoined in Blm mutants. In 33 of 34 cases, the nondisjoined chromosomes had no crossovers; the remaining case had a single crossover in the most centromere-proximal interval (Figure 3B).
Most X nondisjunction in Blm mutants occurs between chromosomes that did not experience a crossover. The incidence of E0 X chromosomes is elevated in Blm mutants due to a combination of decreased crossover frequency and loss of assurance (Figure 3A). To separate these effects, we analyzed Blm rec double mutants. REC, the Drosophila ortholog of MCM8, is required in the Class I crossover pathway [11, 28]. Crossovers are greatly reduced in rec single mutants, but are elevated above wild-type levels in Blm rec double mutants [11] (Figure S3A). The reasons for this elevation are unknown, but may be related to the poorly understood role of REC in the noncrossover pathway [28]. Despite the elevated crossover frequency, nondisjunction rates are similar in Blm mutants and Blm rec double mutants [11]. Like Blm single mutants, Blm rec double mutants exhibit a loss of interference, the centromere effect, and crossover assurance, and crossovers occur on chromosome 4 (Figures S3B–D). These results argue that the elevated nondisjunction seen in Blm mutants is due primarily to loss of crossover patterning.
Interference, the centromere effect, and the obligate chiasma were all described more than 80 years ago [4, 14, 24], but the mechanisms behind these phenomena remain unknown. These phenomena are entwined in the Class I crossover pathway, but it is unclear whether they are generated independently within this pathway or are merely different manifestations of a single regulatory process. Mathematical modeling has suggested that an obligatory crossover is ensured by a combination of interference and other features of the Class I pathway, so these processes may be inter-dependent [2, 29]. The centromere effect may be an augmentation that reinforces interference by pushing crossovers toward the middle of the arm [30]. However, since the telomere effect in Drosophila is far weaker than the centromere effect (Figure 1B), it seems likely that the centromere effect is an independent phenomenon that functions to prevent proximal crossovers, presumably because these can induce nondisjunction [27]. We identified only a single case of a proximal crossover in the set of nondisjoined chromosomes we analyzed, but this is a significant increase from the frequency in wild-type females (one case in ~2900 progeny in Blm mutants, compared to six cases from ~600,000 progeny of wild-type females [27]; P =0.0109).
The meiotic function of Drosophila Blm appears to be similar to the role of S. cerevisiae Sgs1 in allowing recombination intermediates to populate the Class I crossover pathway [7, 8], but this is not conserved in some other species. In Arabidopsis, redundant Blm paralogs prevent Class II crossovers, perhaps by promoting noncrossover repair, but are not required for Class I crossovers[31]. The C. elegans ortholog, HIM-6, does have a role in making Class I crossovers [32]; however, this occurs after normal crossover designation. This is not unlike Drosophila mei-9 mutants, where crossover designation is intact but crossover formation is impaired, resulting in the few crossovers that are made having a wild-type distribution [10] (Figure S1B).
In summary, we have assessed the importance of crossover patterning in meiosis by exploiting the loss of patterning in Drosophila mutants lacking Blm helicase. In wild-type females, the primary meiotic recombination pathway incorporates the centromere effect and interference to promote patterned designation of which events will become crossovers (Figure 4A). Strong crossover assurance means that most homologous chromosomes have a crossover that ensures their disjunction, but the few achiasmate pairs are still segregated accurately by the achiasmate segregation system. Blm is essential for entrance into this meiosis-specific Class I repair pathway; in Blm mutants, repair instead occurs through the Class II pathway (Figure 4B). These crossovers lead to chiasmata that are competent to promote accurate disjunction. However, because crossover patterning is lost there is an elevated frequency of chromosomes at risk for nondisjunction (primarily achiasmate chromosomes, but possibly also chromosomes with very proximal crossovers).
Figure 4. Crossover patterning promotes proper disjunction.
(A) In wild-type flies, crossovers (COs) are produced almost exclusively by the Class I pathway. Entry into this pathway requires Blm helicase activity. Interference and the centromere effect impact which intermediates will be designated to become crossovers. Repair intermediates not selected to become crossovers are repaired into noncrossovers (NCOs). On chromosome 4, crossover patterning processes prevent all recombination intermediates from earning the crossover designation, so all are repaired as noncrossovers (indicated by red 4). Highly regulated crossover patterning ensures that each bivalent receives at least one crossover designation, leading to crossover assurance. Assurance is not absolute in Drosophila, so some achiasmate bivalents remain; however, the achiasmate segregation pathway ensures accurate disjunction of these.
(B) In Blm mutant flies, the Class I pathway is not populated, so all DSBs are repaired by the backup Class II pathway. Because the Class II pathway lacks crossover patterning, every DSB has a fixed probability of become a crossover, regardless of genomic location. Crossover assurance is absent, leading to an elevation in achiasmate bivalents. The achiasmate segregation system cannot compensate for the high number of E0 bivalents, so nondisjunction is elevated.
Supplementary Material
Highlights.
Blm is essential for the primary meiotic recombination pathway in Drosophila
In Blm mutants, interference and other types of crossover patterning are lost
In Blm mutants, meiotic crossovers are made on chromosome 4
Loss of regulated crossover designation leads to elevated nondisjunction
Acknowledgments
We thank K.N. Crown, L.P. Morris, D. Rognstad, and G. Copenhaver for their helpful comments and insightful suggestions regarding this manuscript. We thank J. Comeron for sharing his noncrossover data. This work was supported by grants from the NIGMS to JS (1R01GM061252 and 1R35GM118127). TH was supported in part by NIH grants 5T32GM007092 and 1F31AG055157. KPK and AMW were supported in part by NIH grant P20GM103499. All data are in the Supplementary Information.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplemental information includes Supplemental Experimental Procedures, four Supplemental Figures, and four Supplemental Tables.
Author Contributions
TH, KPK, SM, MAH, and AMW collected data. TH, KPK, MAH, and JS analyzed data. TH, KPK, and JS wrote the manuscript. TH and KPK contributed equally. All authors read and approved the final manuscript.
The authors declare that they have no competing interests.
References
- 1.Lake CM, Hawley RS. Becoming a crossover-competent DSB. Semin Cell Dev Biol. 2016;54:117–125. doi: 10.1016/j.semcdb.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 2.Wang S, Zickler D, Kleckner N, Zhang L. Meiotic crossover patterns: Obligatory crossover, interference and homeostasis in a single process. Cell Cycle. 2015;14:305–314. doi: 10.4161/15384101.2014.991185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bridges CB. The mutants and linkage data of chromosome four of Drosophila melanogaster. Виологический Журнал. 1935;4:401–420. [Google Scholar]
- 4.Sturtevant AH. The linear arrangement of six sex-linked factors in Drosophila as shown by their mode of association. J Exp Biol. 1913;14:43–59. [Google Scholar]
- 5.Berchowitz LE, Copenhaver GP. Genetic interference: don't stand so close to me. Curr Genomics. 2010;11:91–102. doi: 10.2174/138920210790886835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohl KP, Sekelsky J. Meiotic and mitotic recombination in meiosis. Genetics. 2013;194:327–334. doi: 10.1534/genetics.113.150581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Muyt A, Jessop L, Kolar E, Sourirajan A, Chen J, Dayani Y, Lichten M. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol Cell. 2012;46:43–53. doi: 10.1016/j.molcel.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zakharyevich K, Tang S, Ma Y, Hunter N. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell. 2012;149:334–347. doi: 10.1016/j.cell.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sekelsky JJ, McKim KS, Chin GM, Hawley RS. The Drosophila meiotic recombination gene mei-9 encodes a homologue of the yeast excision repair protein Rad1. Genetics. 1995;141:619–627. doi: 10.1093/genetics/141.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker BS, Carpenter ATC. Genetic analysis of sex chromosomal meiotic mutants in Drosophila melanogaster. Genetics. 1972;71:255–286. doi: 10.1093/genetics/71.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohl KP, Jones CD, Sekelsky J. Evolution of an MCM complex in flies that promotes meiotic crossovers by blocking BLM helicase. Science. 2012;338:1363–1365. doi: 10.1126/science.1228190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zalevsky J, MacQueen AJ, Duffy JB, Kemphues KJ, Villeneuve AM. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics. 1999;153:1271–1283. doi: 10.1093/genetics/153.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stevens WL. The analysis of interference. J Genet. 1936;32:51–64. [Google Scholar]
- 14.Beadle GW. A possible influence of the spindle fibre on crossing-over in Drosophila. Proc. Natl. Acad. Sci. USA. 1932;18:160–165. doi: 10.1073/pnas.18.2.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mather K. Crossing over and heterochromatin in the X chromosome of Drosophila melanogaster. Genetics. 1939;24:413–435. doi: 10.1093/genetics/24.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto M, Miklos GL. Genetic studies on heterochromatin in Drosophila melanogaster and their implications for the functions of satellite DNA. Chromosoma. 1978;66:71–98. doi: 10.1007/BF00285817. [DOI] [PubMed] [Google Scholar]
- 17.Ashburner M, Golic KG, Hawley RS. Drosophila: A Laboratory Handbook. 2nd. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 2005. [Google Scholar]
- 18.Painter TS, Muller HJ. Parallel cytology and genetics of induced translocations and deletions in Drosophila. J. Hered. 1929;20:287–298. [Google Scholar]
- 19.Grell RF. Heat-induced exchange in the fourth chromosome of diploid females of Drosophila melanogaster. Genetics. 1971;69:523–527. doi: 10.1093/genetics/69.4.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandler L, Szauter P. The effect of recombination-defective meiotic mutants on fourth-chromosome crossing over in Drosophila melanogaster. Genetics. 1978;90:699–712. doi: 10.1093/genetics/90.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osborne JD. Department of Biology Sciences, Volume M.S. Edmonton, Alberta, Canada: University of Alberta; 1998. Crossing over in a T(1;4) translocation in Drosophila melanogaster; p. 94. [Google Scholar]
- 22.Comeron JM, Ratnappan R, Bailin S. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 2012;8:e1002905. doi: 10.1371/journal.pgen.1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McVey M, Andersen SL, Broze Y, Sekelsky J. Multiple functions of Drosophila BLM helicase in maintenance of genome stability. Genetics. 2007;176:1979–1992. doi: 10.1534/genetics.106.070052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darlington CD, Dark SOS. The origin and behaviour of chiasmata, II. Stenobothrus parallelus. Cytologia. 1932;3:169–185. [Google Scholar]
- 25.Owen ARG. A possible interpretation of the apparent interference across the centromere found by Callan and Montalenti in Culex pipiens. Heredity (Edinb) 1949;3:357–367. doi: 10.1038/hdy.1949.26. [DOI] [PubMed] [Google Scholar]
- 26.Miller DE, Smith CB, Yeganeh Kazemi N, Cockrell AJ, Arvanitakis AV, Blumenstiel JP, Jaspersen SL, Hawley RS. Whole-genome analysis of individual meiotic events in Drosophila melanogaster reveals that noncrossover gene conversions are insensitive to interference and the centromere effect. Genetics. 2016;203:159–171. doi: 10.1534/genetics.115.186486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koehler KE, Boulton CL, Collins HE, French RL, Herman KC, Lacefield SM, Madden LD, Schuetz CD, Hawley RS. Spontaneous X chromosome MI and MII nondisjunction events in Drosophila melanogaster oocytes have different recombinational histories. Nat. Genet. 1996;14:406–414. doi: 10.1038/ng1296-406. [DOI] [PubMed] [Google Scholar]
- 28.Blanton HL, Radford SJ, McMahan S, Kearney HM, Ibrahim JG, Sekelsky J. REC, Drosophila MCM8, drives formation of meiotic crossovers. PLoS Genet. 2005;1:e40. doi: 10.1371/journal.pgen.0010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Liang Z, Hutchinson J, Kleckner N. Crossover patterning by the beam-film model: analysis and implications. PLoS Genet. 2014;10:e1004042. doi: 10.1371/journal.pgen.1004042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleckner N, Zickler D, Jones GH, Dekker J, Padmore R, Henle J, Hutchinson J. A mechanical basis for chromosome function. Proc. Natl. Acad. Sci. USA. 2004;101:12592–12597. doi: 10.1073/pnas.0402724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seguela-Arnaud M, Crismani W, Larcheveque C, Mazel J, Froger N, Choinard S, Lemhemdi A, Macaisne N, Van Leene J, Gevaert K, et al. Multiple mechanisms limit meiotic crossovers: TOP3alpha and two BLM homologs antagonize crossovers in parallel to FANCM. Proc. Natl. Acad. Sci. USA. 2015;112:4713–4718. doi: 10.1073/pnas.1423107112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schvarzstein M, Pattabiraman D, Libuda DE, Ramadugu A, Tam A, Martinez-Perez E, Roelens B, Zawadzki KA, Yokoo R, Rosu S, et al. DNA helicase HIM-6/BLM both promotes MutSgamma-dependent crossovers and antagonizes MutSgamma-independent interhomolog associations during Caenorhabditis elegans meiosis. Genetics. 2014;198:193–207. doi: 10.1534/genetics.114.161513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.