Abstract
Background
Adenomatous polyps are the most common precursor to colorectal cancer (CRC), the second leading cause of cancer death in the United States. We sought to learn more about early events of carcinogenesis by investigating shifts in the gut microbiota of patients with adenomas.
Methods
We analyzed 16S rRNA gene sequences from the fecal microbiota of patients with adenomas (n=233) and without (n=547).
Results
Multiple taxa were significantly more abundant in patients with adenomas, including Bilophila, Desulfovibrio, pro-inflammatory bacteria in the genus Mogibacterium, and multiple Bacteroidetes species. Patients without adenomas had greater abundances of Veillonella, Firmicutes (Order Clostridia), and Actinobacteria (family Bifidobacteriales). Our findings were consistent with previously reported shifts in the gut microbiota of CRC patients. Importantly, the altered adenoma profile is predicted to increase primary and secondary bile acid production, as well as starch, sucrose, lipid, and phenylpropanoid metabolism.
Conclusions
These data hint that increased sugar, protein, and lipid metabolism along with increased bile acid production could promote a colonic environment that supports the growth of bile-tolerant microbes such as Bilophilia and Desulfovibrio. In turn, these microbes may produce genotoxic or inflammatory metabolites such as H2S and secondary bile acids, which could play a role in catalyzing adenoma development and eventually CRC.
Impact
This study suggests a plausible biological mechanism to explain the links between shifts in the microbiota and CRC. This represents a first step toward resolving the complex interactions that shape the adenoma-carcinoma sequence of CRC and may facilitate personalized therapeutics focused on the microbiota.
Keywords: colorectal cancer, gut microbiota, adenoma, bile acids, H2S
1. Introduction
Adenomatous polyps, or adenomas, have long been recognized as a critical precursor to colorectal cancer (CRC) (1, 2), the second leading cause of cancer deaths in the United States (3). Although screening (4–6) and lifestyle (7–10) play important roles in CRC prevention, identifying a causal mechanism of mutagenesis is essential to understand the adenoma-carcinoma sequence and to develop new and personalized prevention strategies. The gut microbiota has recently been implicated in adenoma and CRC pathogenesis (11, 12) and offers a promising avenue for personalized prevention (13). Importantly, many of the risk factors for CRC—including diet (high red meat / high fat / low fiber) (8, 14), obesity (15), physical activity (10), smoking (7), and alcohol use (9)—also have significant effects on the gut microbial community (16). Because the gut microbiota alters the metabolic environment of the host, it may directly or indirectly influence mutagenesis rates (11, 17), and thus carcinogenesis.
Previous studies on the microbiome of individuals with adenomas have identified many microbes associated with these particular polyps (Table 1). However, most of these studies lack functional analyses necessary to suggest a mechanistic link between microbiota, adenoma development, and carcinogenesis. Microbial functionality, which can be predicted based on microbial genomes, provides greater insight into the microbial ecology of the colon by not only indicating what taxa are differentially abundant, but also the putative function of these taxa (18). Without functional analyses, it is difficult to elucidate the role of microbes in the adenoma-carcinoma sequence because microbial taxa associated with adenomas and CRC vary widely by study (11, 12). Additionally, many subject cohorts are relatively underpowered, ranging in size from 6 to 67 individuals with adenomas (see Table 1), making it even more difficult to identify subtle microbial or functional changes that may be underlying adenoma/CRC pathogenesis. Moreover, meta-analysis on these data is particularly challenging due to multiple biases attributed to extraction methods (19), PCR regions (20), and collection protocols (21). As such, a well-powered study with a uniform collection/extraction protocols and functional analyses is needed to more definitively probe the link between the microbial community and adenoma development.
Table 1.
Microbial taxa associated with adenomas in previous studies.
| Study | Number Individuals with Adenomas |
Number Individuals without Adenomas |
Microbial Taxa Enriched in Individuals with Adenomas |
Microbial Taxa Enriched in Individuals without Adenomas |
Reference |
|---|---|---|---|---|---|
| 1 | 20 | 20 | increased microbial diversity within the Clostridium leptum and C. coccoides subgroups |
Scanlan et al. 2008 | |
| 2 | 21 | 23 | Proteobacteria Dorea spp. Faecalibacterium spp. |
Bacteroidetes Bacteroides spp. Coprococcus spp |
Shen et al. 2010 |
| 3 | 33 | 38 | TM7 Cyanobacteria Verrucomicrobia Acidovorax Aquabacterium Cloacibacterium Helicobacter Lactococcus Lactobacillus Pseudomonas |
Streptococcus | Sanapareddy et al. 2012 |
| 4 | 6 | 6 | Firmicutes Proteobacteria |
Bacteroides | Brim et al. 2013 |
| 5 | 47 | 47 |
Enterococcus Streptococcus Bacteroidetes |
Clostridium Roseburia Eubacterium |
Chen et al. 2013 |
| 6 | 67 | 48 | Fusobacterium | McCoy et al. 2013 | |
| 7 | 11 | 10 |
Bifidobacterium Fusobacterium Enterobacteriaceae Akkermansia Blautia |
Methanobacteriales Methanobrevibacterium Faecalibacterium |
Mira-Pascual et al. 2014 |
| 8 | 15 | 15 |
Bifidobacterium Eubacteria |
Nugent et al. 2014 | |
| 9 | 30 | 30 | Ruminococcaceae Clostridium Pseudomonas Porphyromonadaceae |
Bacteroides Lachnospiraceae Clostridales Clostridium |
Zackular et al. 2014 |
| 10 | 20 | 15 | Proteobacteria Gammaproteobacteria |
Goedert 2015 |
In this study, we compared the fecal microbiota of patients with (n=233) and without adenomas (n=547). Our aim was twofold: to determine whether gut microbial communities can be used to predict the presence of adenomas and to elucidate the microbial ecology underlying the adenoma-carcinoma sequence. Here we report significant shifts in the gut microbiota composition of patients with adenomas and use these changes and their predicted functional consequences to propose a model linking diet, gut microbes, and the development of adenomas, the precursors to CRC.
2. Materials and Methods
2.1 Subject enrollment
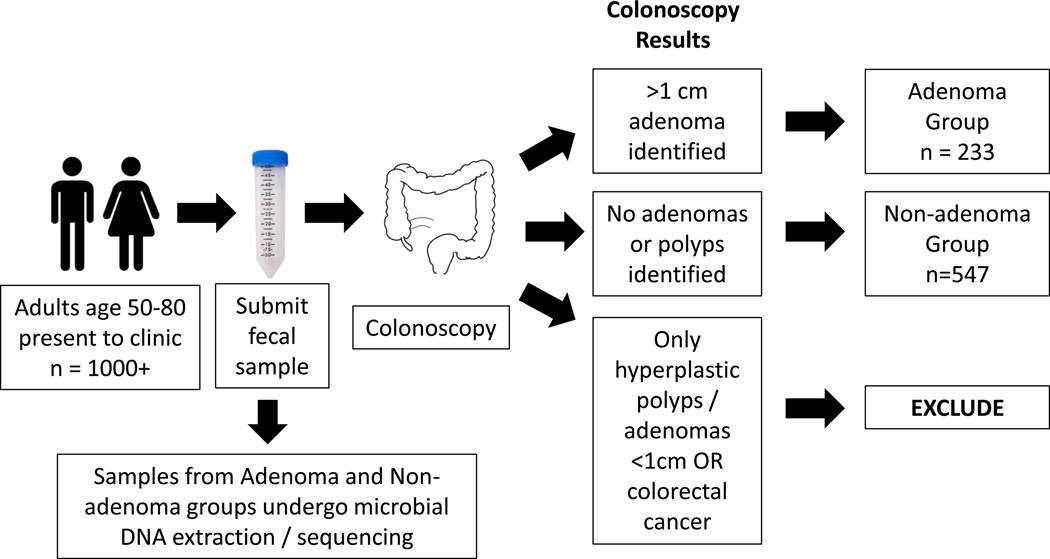
Fecal samples (n=780) were selected from a freezer archive of stools collected without preservative buffer. All stool samples came from patients presenting for standard screening colonoscopy between 2001–2005 at multiple medical centers, including the Mayo Clinic, Rochester, MN; Kaiser Permanente in Sacramento and Oakland, CA; Oregon Health & Science University, Portland, OR; University of Colorado Health Sciences Center, Denver, CO; Roswell Park Cancer Institute, Buffalo, NY; Indiana University Medical Center, Indianapolis, IN; and other North Central Cancer Treatment Group institutions (22). All patients were 50–80 years old and were voluntarily enrolled prior to presenting for colonoscopy (Fig. 1). Exclusion criteria for the original study comprised premenopausal women, hematochezia or melena within the month prior to enrollment, prior colorectal resection, coagulopathy or anticoagulant use, chemotherapy within 3 months of enrollment, contraindications to colonoscopy, inability to desist from therapeutic doses of nonsteroidal anti-inflammatory drugs (NSAIDS), aerodigestive cancer within 5 years of enrollment, a fecal occult blood test within the year prior to enrollment, and colorectal evaluation (e.g., sigmoidoscopy or colonoscopy) within 10 years of enrollment. Patients at high risk for CRC—including patients with familial adenomatous polyposis, cancer syndromes, inflammatory bowel disease, prior CRC or adenomas, or ≥2 first-degree relatives with CRC—were also excluded.
Figure 1.
Subject enrollment flowchart.
Standard diagnostic colonoscopies were performed on all patients and included intravenous sedation (unless otherwise requested); inspection of the colonic mucosal surfaces up to the point of the cecum; and lesion assessment, including recording the location, size, number, and architecture all polypoid lesions. All polyps/lesions removed from the colon were submitted for histological classification and reviewed by the same pathologist. Fecal samples from patients in which at least one adenoma > 1 cm was identified were included in the “adenoma” group. Fecal samples from patients with no polyps were included in the “non-adenoma” group. Fecal samples from patients who were diagnosed with CRC were excluded from analysis.
Approval for this study was granted by the Mayo Clinic’s Institutional Review Board. Fecal samples were collected under protocol #15-004021, from patients who had previously enrolled under protocol #532-00, undergone standard screening colonoscopies, and given consent for the use of their samples in future research studies.
2.2 Sample collection and processing
Fecal samples were self-collected by patients after enrollment and up to 3 months prior to bowel preparation and colonoscopy. Samples were collected in a bucket container mounted to a toilet seat. Promptly after defecation, whole stools were express shipped on ice in insulated containers to a central lab where they were immediately archived at −80°C until further processing. Samples received >48 hours after defecation were disqualified. In preparation for DNA extraction, a 4-mm biopsy punch (Miltex, York, PA, USA) was used to collect a core sample from the still-frozen feces. The frozen fecal core was immediately transferred into Chemagic lysis buffer (PerkinElmer, Baesweiler, Germany). DNA extraction was performed on a Chemagic MSM I (PerkinElmer), using the Chemagic DNA blood special kit (PerkinElmer). DNA quantification and amplification was performed as previously described (23). The 16S rRNA sequencing library was constructed at the University of Minnesota Genomics Center, and sequencing was performed at the Mayo Clinic Medical Genomics Facility, on a MiSeq using a MiSeq Reagent Kit v3 (2 × 300, 600 cycles, Illumina Inc., San Diego, CA, USA).
2.3 Sequence processing
After sequencing, adapter-primer sequences were removed from reads as previously described (23). Sequences were then processed via the IM-TORNADO bioinformatics pipeline, using a 97% identity threshold to assign operational taxonomic units (OTUs) (24). Paired R1 and R2 reads were analyzed. In total, 17,579,026 reads passed quality control. Singleton OTUs as well as samples with less than 2,000 reads were removed. Sequencing data are available at SRA Study accession SRP070783.
2.4 Statistical analyses
2.4.1 α-diversity and β-diversity
To compare the microbial communities of the adenoma and non-adenoma groups, we summarized microbiota data using both α-diversity and β-diversity measures. Two α-diversity metrics were used, the observed OTU number and the Shannon index. The observed OTU number reflects species richness, whereas the Shannon index places more weight on species evenness. β-diversity, by contrast, indicates the shared diversity between bacterial populations in terms of ecological distance; different distance metrics provide distinctive views of community structure. Two β-diversity measures, unweighted and weighted UniFrac distances, were calculated using the OTU table and a phylogenetic tree (with the “GUniFrac” function in the R package GUniFrac) (16). The unweighted UniFrac reflects differences in community membership (i.e., the presence or absence of an OTU), whereas the weighted UniFrac mainly captures differences in abundance. Rarefaction was performed on the OTU table before calculating the distances.
To assess the association between adenoma status and α-diversity, we fitted a linear regression model to the α-diversity metrics after rarefaction, adjusting for technical covariates such as sequencing batch. A Wald test was used to determine significance. To assess the association between adenoma status and β-diversity measures, we used the recently proposed MiRKAT, which is a kernel-based association test based on ecological distance matrices (25). We also used MiRKAT to assess the relationship between polyp characteristics (size, number, location, architecture, histology) and β-diversity measures. In individuals with multiple polyps, a single polyp location was chosen at random, and the most severe architecture and histology per patient were selected for analysis. MiRKAT produces analytic p-values for individual distance metrics, as well as a permutation-based omnibus p-value that combines multiple distance metrics, for a more robust and powerful assessment of significance. For the omnibus test, significance was assessed using 1,000 permutations, and the covariate - sequencing batch - was adjusted if necessary. Ordination plots were generated using principal coordinate analysis as implemented in R (“cmdscale” function in the R ‘vegan’ package).
2.4.2 Differential abundance analysis
We conducted differential abundance analysis at the phylum, class, order, family, and genus levels, and we filtered out taxa with prevalence less than 10%. We normalized the count data into relative abundances (proportions) by dividing by the total read count; taxa with a maximum proportion less than 0.2% were excluded from testing to reduce the number of the tests. To identify differentially abundant taxa while accommodating covariates (e.g., sequencing batch) and the non-normality of the count data, we used a permutation test in which a regular linear model was fitted, with taxa proportion data as the outcome variable. To reduce the effects of outliers, taxa proportion data was square-root transformed. Statistical significance was assessed using 1,000 permutations with the F-stat as the test statistic. False discovery rate (FDR) control (B-H procedure, ‘p.adjust’ in standard R packages) was used to correct for multiple testing, and FDR-adjusted p-values or q-values less than 0.2 were considered significant. This q-value cutoff was chosen to avoid missing important taxa with small effect sizes and is a significance threshold frequently utilized in human microbiome studies (26, 27). To quantify the effect size of the differential taxa, we used the fold change of the mean relative abundance between the normal and adenoma groups.
2.4.3. Predictive modeling based on random forests
The machine learning algorithm random forests (RF) was used to predict adenoma status based on the microbiota profile (genus-level proportion data) using default parameters of the R implementation of the algorithm (28). The RF algorithm, due to its non-parametric assumptions, can detect both linear and nonlinear effects and potential taxon-taxon interactions, thereby identifying the taxa that best discriminate between groups. Boruta variable selection was applied to select the most discriminatory taxa based on importance values produced by RF (29). The Boruta method spikes abundance data with “shadow” taxa, which are shuffled versions of real taxa. This enables us to assess whether the importance of a given taxon is significant, that is, whether the importance is discernible from the effects that arise from random fluctuations (shadow taxa). We then assessed the ability of the Boruta-selected taxa to predict adenoma status using the receiver operating characteristic (ROC) curve, which was estimated using the 0.632+ bootstrap method to more accurately assess error rates (30).
2.4.4. Functional data analysis
PICRUSt was used to infer the abundance of functional categories (KEGG metabolic pathways and COG functional groups) based on the 16S rRNA data, and differential abundance analysis was performed using the same permutation test that was used for the taxon analysis (18). No prevalence-based filtering was applied before differential abundance testing, since most of the functional categories are shared across subjects. All statistical analyses were performed in R 3.0.2 (R Development Core Team, Vienna, Austria).
3. Results
Cases (“adenoma” group) comprised 233 patients with at least one large adenoma (≥ 1 cm); controls included 547 patients with no polyps on colonoscopy (“non-adenoma” group). The groups did not differ with regard to the potential confounders of age, sex, race, history of smoking, history of cancer, or diagnosis of CRC or polyps in first-degree relatives (Table 2).
Table 2.
Demographics of the adenoma and non-adenoma groups.
| Adenoma (n=233) |
Non-adenoma (n=547) |
p-value | |
|---|---|---|---|
| Age (mean, SD) | 66.5, 6.9 | 66.5, 6.9 | 0.60 |
| Sex (n, %) | 0.98 | ||
| Female | 100 (42.9) | 237 (43.3) | |
| Male | 133 (57.1) | 310 (56.7) | |
| Race (n, %) | 0.30 | ||
| White | 223 (95.7) | 503 (92) | |
| Black | 3 (1.3) | 14 (2.6) | |
| Hispanic | 4 (1.7) | 14 (2.6) | |
| Asian | 0 (0) | 8 (1.5) | |
| Native American | 0 (0) | 2 (0.5) | |
| Other / Unknown | 3 (1.3) | 5 (0.9) | |
| Ever smoker (n, %) | 138 (59.2) | 310 (56.7) | 0.56 |
|
History of cancer, any type (n, %) |
44 (18.9) | 116 (21.2) | 0.52 |
|
First degree relative with colorectal cancer (n, %) |
35 (15) | 89 (16.3) | 0.74 |
|
First degree relative with polyps (n, %) |
25 (10.7) | 47 (8.6) | 0.82 |
The overall composition of the groups’ gut microbial communities appeared similar at the levels of phylum, family, and genus (Supplementary Fig. S1A). The groups did not differ significantly in terms of microbial species richness (P=0.21) or diversity (Shannon Index; P=0.23) (Supplementary Fig. S1B and S1C). Neither did they cluster in PCoA plots using unweighted or weighted UniFrac distance metrics (Supplementary Fig. S1D and S1E). However, our large sample size allowed us to detect small yet statistically significant differences in microbial composition between the adenoma and non-adenoma groups (MiRKAT omnibus P=0.032). No differences in microbial composition were detected based on polyp size, architecture, or location, but polyp number was significant (MiRKAT omnibus P=0.035) and histology (hyperplastic, low grade dysplasia, or high grade dysplasia) was marginally significant (MiRKAT omnibus P=0.091; Supplementary Table S1).
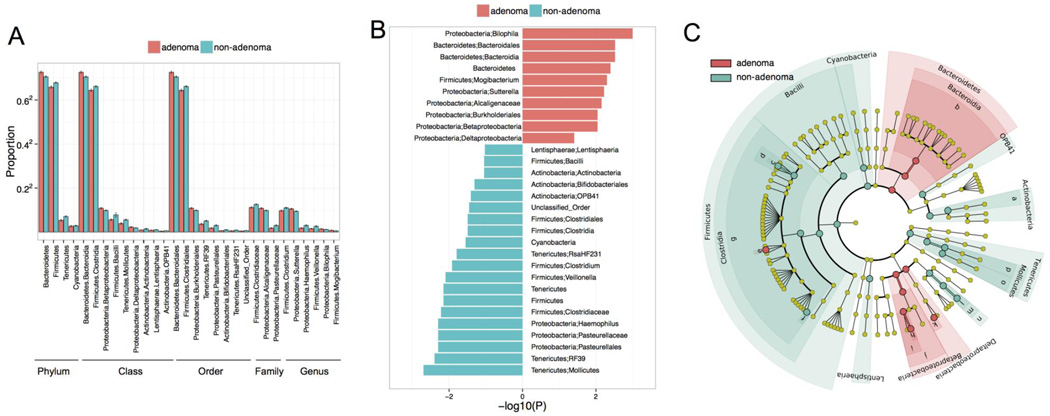
Next, we identified 31 specific taxa that differed in abundance between patients with and without adenomas (Fig. 2 q <0.2) Taxa that were more abundant in the adenoma group included multiple OTUs in the Bacteroidetes phyla and Deltaproteobacteria class—including OTUs in the Bilophila, Desulfovibrio, Sutterella, and Mogibacterium genera. Taxa more common in the non-adenoma group included Firmicutes, such as OTUs in the Clostridia class and Veillonella genus, as well as OTUs in the Bifidobacteriales order and Haemophilus genus. Despite moderate effect sizes (fold change range: 1.06–2.77), these significant results indicate that the microbiota in the adenoma group systematically differs from the non-adenoma group.
Figure 2.
Thirty-one taxa differ in abundance between patients with and without adenomas. A) Relative abundance of OTUs in each group, across taxonomic levels. B) −log(P value) of these taxa’s differential abundance. C) Cladogram of the taxa that differed between groups.
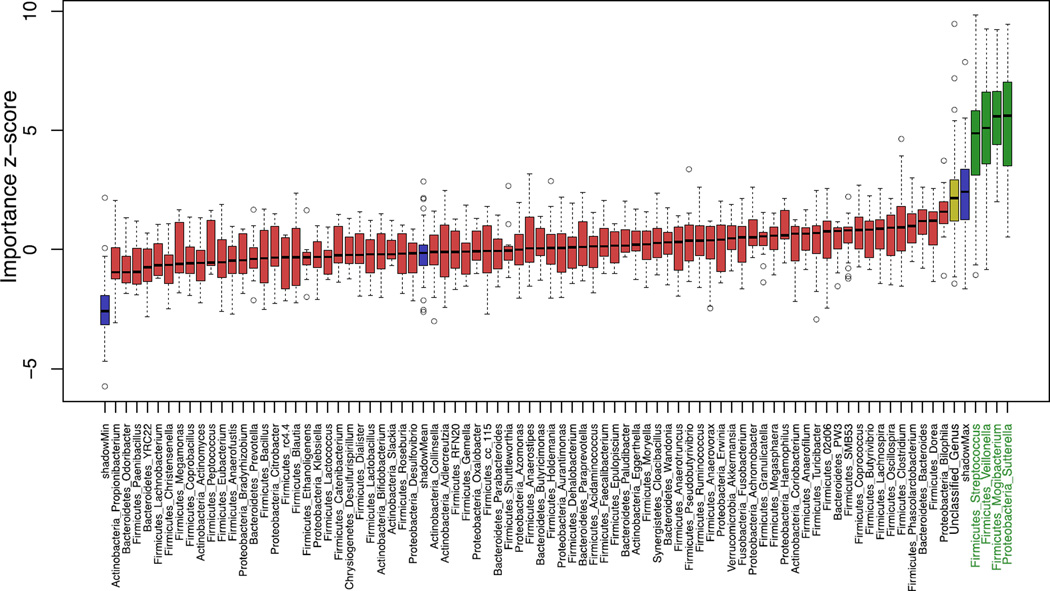
We next assessed the utility of the gut microbiota as a clinical biomarker for adenomas using random forests-based prediction. Boruta feature selection was used to select the most predictive taxa to improve prediction. Of the 31 taxa identified by differential abundance testing, the Boruta algorithm identified four genera that significantly predicted adenoma status: Streptococcus and Veillonella, which were enriched in the non-adenoma group, and Mogibacterium and Sutterella, which were enriched in the adenoma group (Fig. 3; for heat map see Supplementary Fig. S2). The Bilophila genus was also more predictive than most other genera included in this analysis; however, this genera did not exceed the threshold for significance. An ROC curve generated with the four significantly predictive taxa resulted in an area under the curve (AUC) of 0.6599 (Fig. S3; DeLong test, p = 0.001). Although significant, this level of sensitivity/specificity is too low for consideration as a clinical biomarker for adenomas. Thus, this analysis indicates that although the abundance of Streptococcus, Veillonella, Mogibacterium, and Sutterella is not sufficient to reliably identify samples from patients with adenomas, the levels of these genera are consistently altered in their respective groups.
Figure 3.
Based on the results of a random forests (RF) algorithm, four taxa significantly predict adenomatous polyp status: Streptococcus, Veillonella, Mogibacterium, and Sutterella. The four taxa that are significant predictors are shown in green. Blue boxplots correspond to minimal, average and maximum Z score of a shadow taxa. Red, yellow and green boxplots represent Z scores of rejected, tentative and confirmed taxa respectively.
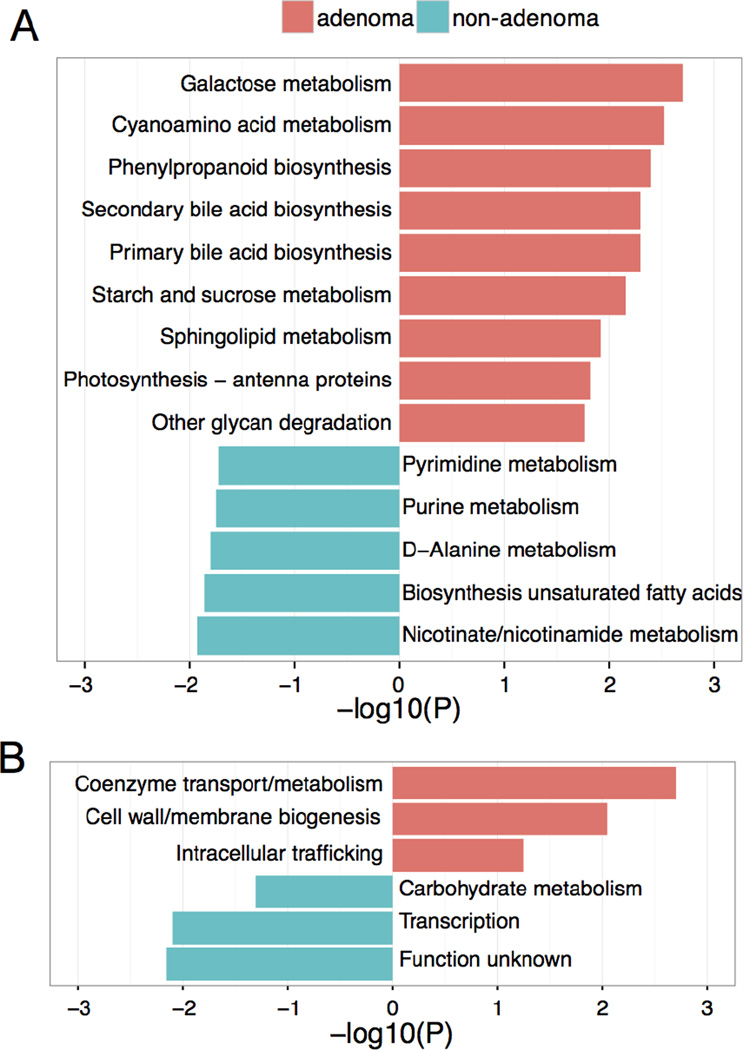
To determine whether the taxonomic differences between the groups’ microbiota corresponded to functional changes, we performed a predictive functional analysis of the 16S rRNA sequences present (Fig. 4, q<0.2). PICRUSt analyses predicted that the adenoma group’s microbiota exhibits increased primary and secondary bile acid synthesis; increased galactose, starch and sucrose, and sphingolipid metabolism; and increased phenylpropanoid biosynthesis. By contrast, the non-adenoma group’s microbiota is predicted to exhibit increased biosynthesis of unsaturated fatty acids and increased purine, pyrimidine, D-Alanine, nicotinate, and nicotinamide metabolism.
Figure 4.
Functional differences, predicted using 16S sequencing data, between the gut microbial communities of patients with and without adenomas, A) Pink bars represent the −log(P value) of KEGG metabolic pathways predicted to be more common among the microbiota of individuals with adenomatous polyps. Turquoise bars represent the effect sizes of functions predicted to be more common among the microbiota of individuals without polyps. B) Summary of the log(P value) of COG groups predicted to differ between the groups; colors as in (A).
4. Discussion
In this study, we report significant differences in the microbial composition of individuals with adenomas. We also observe differences based on polyp number and histology but not size, architecture, or polyp location, suggesting that microbial communities associated with polyps change (or are detectable) with some but not all aspects of polyp severity. We identified 31 taxa that were differentially abundant among patients with and without adenomas, and four of these taxa were significantly predictive of adenoma status, although they could not be used to reliably classify samples. Based on the 16S sequences present in each group, we also identified putative metabolic shifts between the microbiota of the adenoma and non-adenoma groups.
Links with CRC have already been reported in many of the taxa we identified as differentially abundant in individuals with adenomas. This suggests that changes in the microbial community associated with adenomas may represent early events in the pathway leading to CRC. For example, we identified increased levels of Bilophila, Desulfovibrio, Bacteroidetes, and Mogibacterium in individuals with adenomas. Both Bilophila and Desulfovibrio produce genotoxic hydrogen sulfide (H2S) as an end product of anaerobic respiration (31–33) and have been associated with CRC in other studies (34, 35). In addition, multiple studies have reported elevated proportions of Bacteroidetes in patients with adenomas (36, 37) or CRC (38, 39) (but not all; see (40)). Bacteroides fragilis, in particular, causes colitis-associated carcinogenesis (41). Finally, Mogibacterium is an oral bacterium associated with periodontal disease and root canal infections, and it, too, has been linked to CRC (42–44).
Other taxa differentially abundant in individuals with adenomas are also plausible contributors to carcinogenesis. For example, Sutterella, a genus highly predictive of adenoma status (Fig. 3), may play a role in inflammation, as it has been linked to active colitis in a mouse model of inflammatory bowel disease (45). Gastrointestinal inflammation has been strongly linked to CRC pathogenesis (11). By contrast, Veillonella, also highly predictive of adenoma status but enriched in patients without adenomas, may exert a protective role in the colon (46) along with other taxa enriched in this group, including Firmicutes and Actinobacteria (family Bifidobacteriales) (47). Notably, we did not identify an enrichment of Fusobacterium or Porphyromonadaceae in individuals with adenomas, as reported in other studies (39, 48, 49). This may have been due to differences in study populations, fecal collection and preservation techniques (21, 50), library preparation (51), or primers and sequencing platforms (52, 53).
We evaluated microbial alterations in tandem with predictive functional differences identified by PICRUST. In an analysis of Human Microbiome Project data, PICRUST produced an average correlation of 0.8 between predicted functions and actual functions identified through deep metagenomic sequencing (18). Additionally, PICRUST produced more accurate and reliable functional predictions than shallow metagenomic sequencing (18). Despite these strengths of PICRUST, predictive functions should be examined with care, as genomes and functions of the microbes present in a given sample may differ from the genomes and functions upon which PICRUST builds its predictions. The results from our predictive functional analysis suggest a link the between microbial shifts observed in individuals with adenomas to metabolic pathways that have previously been associated with dietary risk factors common in a Western diet. The adenoma microbiota was characterized by putative functional groups associated with galactose, sphingolipid, and starch/sucrose metabolism, as well as phenylpropanoid biosynthesis. Importantly, diets high in dairy result in increased galactose metabolism; diets high in fat result in increased lipid/sphingolipid metabolism (54); diets high in refined starches and sugars lead to increased starch/sucrose metabolism (55); and diets high in protein result in increased phenylpropanoid biosynthesis (56). Diets high in animal fat and protein also lead to increased BA production (57). Interestingly, the adenoma microbiota is predicted to display increased levels of primary and secondary bile acid (BA) synthesis. These functional predictions suggest that individuals with adenomas are consuming diets higher in fat, sugar, starch, protein, and dairy than non-adenoma individuals. These findings are consistent with multiple epidemiological studies, which have drawn links between a Western diet (high in fat, dairy, meat, and sugars) and the incidence of adenomas (58–60). This suggests a potential link between diet and the molecular mechanisms involved in adenoma pathogenesis.
We propose the following mechanism linking diet, the microbiota, and the adenoma-carcinoma sequence: Diets high in fat and protein increase production of primary BAs, which help digest and absorb lipids in the small intestine (61, 62). This promotes the growth of bile-tolerant bacteria such as Bilophila and some species of Desulfovibrio. Blooms of these species may increase the production of genotoxic metabolites such as H2S (61, 62). In addition, the colon microbiota can deconjugate primary bile acids to form secondary BAs (62, 63), and some of these secondary BAs, such as lithocholic and deoxycholic acid, have cytotoxic and genotoxic effects (62–66). Elevated levels of secondary BAs and pro-inflammatory bacteria such as Mogibacterium and Sutterella may result in the perfect storm of DNA damage and inflammation, leading to adenoma development and eventually malignant transformation.
Several limitations of our study warrant mention. Three include a lack of information on participants’ diet, body mass index (BMI), and recent antibiotic use. Without dietary information, we cannot confirm that the adenoma group consumed a diet higher in sugar, animal fat, and protein; although, previous studies have indicated a link between Western diet and adenomas (58, 59). Additionally, we are also unable to determine whether BMI acts as a confounder; it is certainly possible, as the gut microbiome of obese individuals differs significantly from the microbiome of lean individuals (67), and higher BMI has been associated with adenoma development (68). However, obese / high BMI phenotypes are commonly associated with increased relative abundances of microbes in the phylum Firmicutes while lean phenotypes are associated with increased abundances of Bacteroidetes phylum microbes (67, 69). In our study, individuals with adenomas had increased abundances of Bacteroidetes microbes while individuals without adenomas had increased abundances of Firmicutes phylum microbes. This is opposite to what we would have expected if BMI was the main driver of adenoma development; thus, we suggest BMI was not a strong confounder in our data set. Lack of antibiotic data prevents us from excluding or analyzing data based on antibiotic use, which can dramatically alter the gut microbiota (70); although, we have no a priori reason to believe that either group would exhibit increased antibiotic use in relation to the other. Finally, the cross-sectional nature of our data does not allow us to parse correlation versus causation between microbial alterations and adenoma status. While our results show that observed microbial changes lack the specificity and sensitivity to serve as a clinical biomarker for adenomas, these findings provide important insights into mechanisms that may be driving adenoma development.
This study represents the largest study on microbial communities associated with adenomas to date. This robust data set allowed us to detect subtle microbial changes that may be key to understanding how a healthy colon develops adenomas, which can then transform into carcinomas. We also adjusted our analyses for multiple comparisons, which not all studies on adenoma microbiota opt to do (37, 71, 72). Sample collection is another strength of this study. All fecal samples from individuals in the adenoma and non-adenoma groups were shipped on ice and received and frozen at -80°C within 48 hours of defecation. Previous studies have demonstrated that fecal microbial communities stored at ambient temperatures for up to 24 hours, are relatively unaffected (21), and no significant changes in microbial diversity or composition are detected in fecal samples stored at 4°C for up to 72 hours (73). Additionally, long term storage of fecal samples at -80°C seems to have little effect on overall microbial composition (50, 74); although, no study, to our knowledge, has examined fecal preservation in samples over 10 years old, as is the case with samples in this study. Notably, we only examined fecal microbiota and not the mucosal-associated microbiota, which has been reported to differ in composition and diversity (75). Every individual sampled in this study underwent a complete colonoscopy with full visualization of the colon from rectum to cecum, and colonoscopy is regarded as the most robust reference standard for presence or absence of polyps. Polyps removed during colonoscopies were all reviewed and classified by the same pathologist. Finally, our study included predictive functional analyses based on the microbial communities of the adenoma and non-adenoma groups. Functional analyses have not been performed on previous adenoma datasets, and this effort suggested key insights as to how the host and microbial community may be interacting within the context of adenoma development.
In conclusion, we have shown that the composition of the gut microbiota in individuals with adenomas differs significantly from that of healthy individuals and resembles the microbiota of individuals with CRC. Moreover, we suggest that these shifts may be consistent with the effects of the Western diet and are predicted to result in metabolic changes that could increase rates of cellular damage and mutagenesis in the gut. Collectively, our findings support a proposed model in which diet alters the microbial composition of our gastrointestinal tract, leading to an environment conducive to the development of adenomas, and potentially CRC. Future studies are needed to assess the effects of diet on the metabolic environment of the gut and the microbial community. Genotoxic metabolites such as H2S and secondary bile acids should also be examined in relation to adenoma and carcinoma development. Identifying key interactions between diet, microbial community, and metabolites that catalyze the adenoma-carcinoma sequence will give us a basis for personalized therapeutics aimed at preventing CRC.
Supplementary Material
Acknowledgments
The authors thank all members of the Chia Laboratory for their input and efforts on this project. We acknowledge Kristin Harper for her thoughtful suggestions on this manuscript. We also thank the reviewers for their comments and advice on this manuscript.
Source and number of grants / funding for each author
Mayo Clinic Center for Individualized Medicine: Vanessa L. Hale, Jun Chen, Nicholas Chia
Gerstner Family Career Development Award: Jun Chen
Fred C Andersen Foundation: Heidi Nelson, Nicholas Chia
National Institutes of Health (NIH), R01 CA 179243: Vanessa L. Hale, Nicholas Chia
National Cancer Institute (NIH), U01 CA 89389 and R01 CA 71680: David Ahlquist
Exact Sciences: Tracy C. Yab, David Ahlquist
Mayo Clinic: David Ahlquist
National Cancer Institute (NIH), R01 CA 170357: Lisa Boardman
Conflicts of interest
Mayo Clinic has licensed technology to Exact Sciences. Dr. David Ahlquist and Tracy C. Yab are co-inventors of licensed technology and share in related royalties to Mayo Clinic according to institutional policy. Dr. Ahlquist serves as scientific advisor to and research collaborator with Exact Sciences.
Dr. Dennis J. Ahnen received an honoraria from Ambry Genetics and serves as a scientific advisor to Cancer Prevention Pharmaceuticals.
Abbreviations
- AUC
area under curve
- BMI
body mass index
- CRC
colorectal cancer
- BA
bile acid
- FDR
false discovery rate
- OTU
operational taxonomic unit
- RF
random forests
- ROC
receiver operating characteristic
References
- 1.Fenoglio CM, Lane N. The anatomical precursor of colorectal carcinoma. Cancer. 1974;34:819–823. doi: 10.1002/1097-0142(197409)34:3+<819::aid-cncr2820340706>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 2.Leslie A, Carey FA, Pratt NR, Steele RJC. The colorectal adenoma–carcinoma sequence. Br J Surg. 2002;89:845–860. doi: 10.1046/j.1365-2168.2002.02120.x. [DOI] [PubMed] [Google Scholar]
- 3.American Cancer Society. Colorectal cancer facts & figures 2014–2016. 2014 Available from: http://www.cancer.org/acs/groups/content/documents/document/acspc-042280.pdf. [Google Scholar]
- 4.Zauber AG, Winawer SJ, O'Brien MJ, Lansdorp-Vogelaar I, van Ballegooijen M, Hankey BF, et al. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N Engl J of Med. 2012;366:687–696. doi: 10.1056/NEJMoa1100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atkin WS, Morson BC, Cuzick J. Long-term risk of colorectal cancer after excision of rectosigmoid adenomas. N Engl J Med. 1992;326:658–662. doi: 10.1056/NEJM199203053261002. [DOI] [PubMed] [Google Scholar]
- 6.Winawer SJ, Zauber AG, Ho MN, O'Brien MJ, Gottlieb LS, Sternberg SS, et al. Prevention of colorectal cancer by colonoscopic polypectomy. N Engl J Med. 1993;329:1977–1981. doi: 10.1056/NEJM199312303292701. [DOI] [PubMed] [Google Scholar]
- 7.Chao A, Thun MJ, Jacobs EJ, Henley SJ, Rodriguez C, Calle EE. Cigarette smoking and colorectal cancer mortality in the cancer prevention study II. J Natl Cancer Inst. 2000;92:1888–1896. doi: 10.1093/jnci/92.23.1888. [DOI] [PubMed] [Google Scholar]
- 8.Bouvard V, Loomis D, Guyton KZ, Grosse Y, Ghissassi FE, Benbrahim-Tallaa L, et al. Carcinogenicity of consumption of red and processed meat. Lancet Oncol. 2015;16:1599–1600. doi: 10.1016/S1470-2045(15)00444-1. [DOI] [PubMed] [Google Scholar]
- 9.Fedirko V, Tramacere I, Bagnardi V, Rota M, Scotti L, Islami F, et al. Alcohol drinking and colorectal cancer risk: an overall and dose–response meta-analysis of published studies. Annals Oncol. 2011 doi: 10.1093/annonc/mdq653. [DOI] [PubMed] [Google Scholar]
- 10.Slattery ML. Physical activity and colorectal cancer. Sports Med. 2012;34:239–252. doi: 10.2165/00007256-200434040-00004. [DOI] [PubMed] [Google Scholar]
- 11.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 12.Dulal S, Keku TO. Gut microbiome and colorectal adenomas. Cancer jJ. 2014;20:225–231. doi: 10.1097/PPO.0000000000000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sonnenburg JL, Fischbach MA. Community health care: Therapeutic opportunities in the human microbiome. Sci Transl Med. 2011;3:78ps12–78ps12. doi: 10.1126/scitranslmed.3001626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Keefe SJD, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun. 2015;6 doi: 10.1038/ncomms7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bardou M, Barkun AN, Martel M. Obesity and colorectal cancer. Gut. 2013;62:933–947. doi: 10.1136/gutjnl-2013-304701. [DOI] [PubMed] [Google Scholar]
- 16.Chen J, Bittinger K, Charlson ES, Hoffmann C, Lewis J, Wu GD, et al. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinform. 2012;28:2106–2113. doi: 10.1093/bioinformatics/bts342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0070803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotech. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner Mackenzie B, Waite DW, Taylor MW. Evaluating variation in human gut microbiota profiles due to DNA extraction method and inter-subject differences. Frontiers in Microbiology. 2015;6:130. doi: 10.3389/fmicb.2015.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeraldo P, Chia N, Goldenfeld N. On the suitability of short reads of 16S rRNA for phylogeny-based analyses in environmental surveys. Environmental Microbiology. 2011;13:3000–3009. doi: 10.1111/j.1462-2920.2011.02577.x. [DOI] [PubMed] [Google Scholar]
- 21.Sinha R, Chen J, Amir A, Vogtmann E, shi J, Inman KS, et al. Collecting fecal samples for microbiome analyses in epidemiology studies. Cancer Epidemiol Biomark Prev. 2015 doi: 10.1158/1055-9965.EPI-15-0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahlquist DA, Sargent DJ, Loprinzi CL, Levin TR, Rex DK, Ahnen DJ, et al. Stool DNA and Occult Blood Testing for Screen Detection of Colorectal Neoplasia. Annals of Internal Medicine. 2008;149:441–450. doi: 10.7326/0003-4819-149-7-200810070-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baldwin EA, Walther-Antonio M, MacLean AM, Gohl DM, Beckman KB, Chen J, et al. Persistent microbial dysbiosis in preterm premature rupture of membranes from onset until delivery. PeerJ. 2015;3:e1398. doi: 10.7717/peerj.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeraldo P, Kalari K, Chen X, Bhavsar J, Mangalam A, White B, et al. IM-TORNADO: A tool for comparison of 16S reads from paired-end libraries. PLoS ONE. 2014;9:e114804. doi: 10.1371/journal.pone.0114804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao N, Chen J, Carroll Ian M, Ringel-Kulka T, Epstein Michael P, Zhou H, et al. Testing in microbiome-profiling studies with MiRKAT, the microbiome regression-based kernel association test. Am J Hum Genetics. 2015;96:797–807. doi: 10.1016/j.ajhg.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aagaard K, Petrosino J, Keitel W, Watson M, Katancik J, Garcia N, et al. The Human Microbiome Project strategy for comprehensive sampling of the human microbiome and why it matters. FASEB J. 2013;27:1012–1022. doi: 10.1096/fj.12-220806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 29.Kursa MB, Rudnicki WR. Feature selection with the boruta package. J Stat Softw. 2010;36:1–13. [Google Scholar]
- 30.Efron B, Tibshirani R. Improvements on cross-validation: The .632+ bootstrap method. J Am Stat Assoc. 1997;92:548–560. [Google Scholar]
- 31.Warren YA, Citron DM, Merriam CV, Goldstein EJC. Biochemical differentiation and comparison of Desulfovibrio species and Other phenotypically similar genera. J Clin Microbiol. 2005;43:4041–4045. doi: 10.1128/JCM.43.8.4041-4045.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baron EJ. Bergey's Manual of Systematics of Archaea and Bacteria. John Wiley & Sons, Ltd; 2015. Bilophila. [Google Scholar]
- 33.Attene-Ramos MS, Nava GM, Muellner MG, Wagner ED, Plewa MJ, Gaskins HR. DNA damage and toxicogenomic analyses of hydrogen sulfide in human intestinal epithelial FHs 74 Int cells. Environ Mol Mutagen. 2010;51:304–314. doi: 10.1002/em.20546. [DOI] [PubMed] [Google Scholar]
- 34.Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS ONE. 2012;7:e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yazici C, Wolf PG, Carroll TP, Mutlu E, Xicola RM, Llor X, et al. 511 Bilophila wadsworthia is more abundant in the colonic microbiome of colorectal cancer cases compared to healthy controls. Gastroenterol. 2015;148:S-100. [Google Scholar]
- 36.Brim H, Yooseph S, Zoetendal EG, Lee E, Torralbo M, Laiyemo AO, et al. Microbiome analysis of stool samples from African Americans with colon polyps. PLoS ONE. 2013;8:e81352. doi: 10.1371/journal.pone.0081352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen H-M, Yu Y-N, Wang J-L, Lin Y-W, Kong X, Yang C-Q, et al. Decreased dietary fiber intake and structural alteration of gut microbiota in patients with advanced colorectal adenoma. Am J Clin Nutr. 2013;97:1044–1052. doi: 10.3945/ajcn.112.046607. [DOI] [PubMed] [Google Scholar]
- 38.Wu N, Yang X, Zhang R, Li J, Xiao X, Hu Y, et al. Dysbiosis signature of fecal microbiota in colorectal cancer Ppatients. Microb Ecol. 2013;66:462–470. doi: 10.1007/s00248-013-0245-9. [DOI] [PubMed] [Google Scholar]
- 39.Zackular JP, Baxter NT, Iverson KD, Sadler WD, Petrosino JF, Chen GY, et al. The gut microbiome modulates colon tumorigenesis. mBio. 2013;4:13. doi: 10.1128/mBio.00692-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen XJ, Rawls JF, Randall T, Burcal L, Mpande CN, Jenkins N, et al. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes. 2010;1:138–147. doi: 10.4161/gmic.1.3.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kostic AD, Chun E, Meyerson M, Garrett WS. Microbes and inflammation in colorectal cancer. Cancer Immunol Res. 2013;1:150157. doi: 10.1158/2326-6066.CIR-13-0101. [DOI] [PubMed] [Google Scholar]
- 42.Candela M, Turroni S, Biagi E, Carbonero F, Rampelli S, Fiorentini C, et al. Inflammation and colorectal cancer, when microbiota-host mutualism breaks. World J Gastroenterol. 2014;20:908–922. doi: 10.3748/wjg.v20.i4.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakazawa F, Sato M, Poco SE, Hashimura T, Ikeda T, Kalfas S, et al. Description of Mogibacterium pumilum gen. nov., sp. nov. and Mogibacterium vescum gen. nov., sp. nov., and reclassification of Eubacterium timidum (Holdeman et al. 1980) as Mogibacterium timidum gen. nov., comb. nov. Intl J Syst Evol Microbiol. 2000;50:679–688. doi: 10.1099/00207713-50-2-679. [DOI] [PubMed] [Google Scholar]
- 44.Casarin RCV, Saito D, Santos VR, Pimentel SP, Duarte PM, Casati MZ, et al. Detection of Mogibacterium timidum in subgingival biofilm of aggressive and non-diabetic and diabetic chronic periodontitis patients. Braz J Microbiol. 2012;43:931–937. doi: 10.1590/S1517-838220120003000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rooks MG, Veiga P, Wardwell-Scott LH, Tickle T, Segata N, Michaud M, et al. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J. 2014;8:1403–1417. doi: 10.1038/ismej.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steinhart CE, Doyle ME, Cochrane BA. Food Safety 1995. New York: Marcel Dekker, Inc.; 1995. [Google Scholar]
- 47.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nat. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 48.McCoy AN, Araujo-Perez F, Azcarate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium is associated with colorectal adenomas. PLoS ONE. 2013;8:e53653. doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mira-Pascual L, Cabrera-Rubio R, Ocon S, Costales P, Parra A, Suarez A, et al. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J Gastroenterol. 2014;50:167–179. doi: 10.1007/s00535-014-0963-x. [DOI] [PubMed] [Google Scholar]
- 50.Hale VL, Tan CL, Knight R, Amato KR. Effect of preservation method on spider monkey (Ateles geoffroyi) fecal microbiota over 8 weeks. J Microbiol Methods. 2015;113:1626. doi: 10.1016/j.mimet.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 51.Jones MB, Highlander SK, Anderson EL, Li W, Dayrit M, Klitgord N, et al. Library preparation methodology can influence genomic and functional predictions in human microbiome research. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:14024–14029. doi: 10.1073/pnas.1519288112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamady M, Knight R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Research. 2009;19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clooney AG, Fouhy F, Sleator RD, O' Driscoll A, Stanton C, Cotter PD, et al. Comparing Apples and Oranges?: Next Generation Sequencing and Its Impact on Microbiome Analysis. PLoS ONE. 2016;11:e0148028. doi: 10.1371/journal.pone.0148028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi S, Snider AJ. Sphingolipids in high fat diet and obesity-related diseases. Mediat Inflam. 2015;2015:12. doi: 10.1155/2015/520618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rampelli S, Schnorr Stephanie L, Consolandi C, Turroni S, Severgnini M, Peano C, et al. Metagenome sequencing of the Hadza hunter-gatherer gut microbiota. Curr Biol. 2015;25:1682–1693. doi: 10.1016/j.cub.2015.04.055. [DOI] [PubMed] [Google Scholar]
- 56.Russell WR, Duncan SH, Scobbie L, Duncan G, Cantlay L, Calder AG, et al. Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol Nutr Food Res. 2013;57:523–535. doi: 10.1002/mnfr.201200594. [DOI] [PubMed] [Google Scholar]
- 57.Reddy BS. Diet and excretion of bile acids. Cancer Res. 1981;41:3766–3768. [PubMed] [Google Scholar]
- 58.Makambi KH, Agurs-Collins T, Bright-Gbebry M, Rosenberg L, Palmer JR, Adams-Campbell LL. Dietary patterns and the risk of colorectal adenomas: the Black women's health study. Cancer Epidemiol Biomark Prev. 2011;20:818–825. doi: 10.1158/1055-9965.EPI-10-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu K, Hu FB, Fuchs C, Rimm EB, Willett WC, Giovannucci E. Dietary patterns and risk of colon cancer and adenoma in a cohort of men (United States) Cancer Causes Control. 2004;15:853–8562. doi: 10.1007/s10552-004-1809-2. [DOI] [PubMed] [Google Scholar]
- 60.Giovannucci E, Stampfer MJ, Colditz G, Rimm EB, Willett WC. Relationship of diet to risk of colorectal adenoma in men. J Natl Cancer Inst. 1992;84:91–98. doi: 10.1093/jnci/84.2.91. [DOI] [PubMed] [Google Scholar]
- 61.Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nat. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ridlon JM, Wolf PG, Gaskins HR. Taurocholic acid metabolism by gut microbes and colon cancer. Gut Microbes. 2016;7:201–215. doi: 10.1080/19490976.2016.1150414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nagengast FM, Grubben MJ, van Munster IP. Role of bile acids in colorectal carcinogenesis. Eur J Cancer. 1995;31A doi: 10.1016/0959-8049(95)00216-6. [DOI] [PubMed] [Google Scholar]
- 64.Bernstein C, Holubec H, Bhattacharyya AK, Nguyen H, Payne CM, Zaitlin B, et al. Carcinogenicity of deoxycholate, a secondary bile acid. Arch Toxicol. 2011;85:863–871. doi: 10.1007/s00204-011-0648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Payne CM, Bernstein C, Dvorak K, Bernstein H. Hydrophobic bile acids, genomic instability, Darwinian selection, and colon carcinogenesis. Clin Exp Gastroenterol. 2008;1 doi: 10.2147/ceg.s4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bernstein H, Bernstein C, Payne CM, Dvorak K. Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J Gastroenterol. 2009;15 doi: 10.3748/wjg.15.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nat. 2006;444:1027–1131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 68.Okabayashi K, Ashrafian H, Hasegawa H, Yoo J-H, Patel VM, Harling L, et al. Body mass index category as a risk factor for colorectal adenomas: A systematic review and meta-analysis. Am J Gastroenterol. 2012;107:1175–1185. doi: 10.1038/ajg.2012.180. [DOI] [PubMed] [Google Scholar]
- 69.Ferrer M, Ruiz A, Lanza F, Haange S-B, Oberbach A, Till H, et al. Microbiota from the distal guts of lean and obese adolescents exhibit partial functional redundancy besides clear differences in community structure. Environ Microbiol. 2013;15:211–226. doi: 10.1111/j.1462-2920.2012.02845.x. [DOI] [PubMed] [Google Scholar]
- 70.Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiol. 2010;156:3216–3223. doi: 10.1099/mic.0.040618-0. [DOI] [PubMed] [Google Scholar]
- 71.Zackular JP, Rogers MAM, Ruffin MT, Schloss PD. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res. 2014;7:1112–1121. doi: 10.1158/1940-6207.CAPR-14-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goedert JJ, Gong Y, Hua X, Zhong H, He Y, Peng P, et al. Fecal microbiota characteristics of patients with colorectal adenoma detected by screening: A population-based study. EBioMed. 2015;2:597–603. doi: 10.1016/j.ebiom.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Choo JM, Leong LEX, Rogers GB. Sample storage conditions significantly influence faecal microbiome profiles. Scientific Reports. 2015;5:16350. doi: 10.1038/srep16350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carroll IM, Ringel-Kulka T, Siddle JP, Klaenhammer TR, Ringel Y. Characterization of the fecal microbiota using high-throughput sequencing reveals a stable microbial community during storage. PloS ONE. 2012;7:e46953. doi: 10.1371/journal.pone.0046953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ringel Y, Maharshak N, Ringel-Kulka T, Wolber EA, Sartor RB, Carroll IM. High throughput sequencing reveals distinct microbial populations within the mucosal and luminal niches in healthy individuals. Gut Microbes. 2015;6:173–181. doi: 10.1080/19490976.2015.1044711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.