Abstract
This study aimed to investigate the genes and pathways that respond to heat stress in Holstein bull calves exposed to severe ranges of temperature and humidity. A total of ten animals from 4 to 6 months of age were subjected to heat stress at 37 °C and 90 % humidity for 12 h. Skin and rectal temperatures were measured before and after heat stress; while no correlation was found between them before heat stress, a moderate correlation was detected after heat stress, confirming rectal temperature to be a better barometer for monitoring heat stress. RNAseq analysis identified 8567 genes to be differentially regulated, out of which 465 genes were significantly upregulated (≥2-fold, P < 0.05) and 49 genes were significantly downregulated (≤2-fold, P < 0.05) in response to heat stress. Significant terms and pathways enriched in response to heat stress included chaperones, cochaperones, cellular response to heat stress, phosphorylation, kinase activation, immune response, apoptosis, Toll-like receptor signaling pathway, Pi3K/AKT activation, protein processing in endoplasmic reticulum, interferon signaling, pathways in cancer, estrogen signaling pathway, and MAPK signaling pathway. The differentially expressed genes were validated by quantitative real-time PCR analysis, which confirmed the tendency of the expression. The genes and pathways identified in this analysis extend our understanding of transcriptional response to heat stress and their likely functioning in adapting the animal to hyperthermic stress. The identified genes could be used as candidate genes for association studies to select and breed animals with improved heat tolerance.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-016-0739-8) contains supplementary material, which is available to authorized users.
Keywords: Heat stress, RNAseq, KEGG, Transcriptome, Differentially expressed genes
Introduction
Heat stress (HS) is one of the major factors that cause a significant reduction in production and reproduction rates in Holstein cattle (West 2003). Even though modification of the environment in farms can relieve the cattle from HS, the added costs involved in it together with increasing temperature due to global warming could lead to economic losses and will become a major issue for farmers around the world. The economic impact of HS on global livestock production is thought to be greater than $1.2 billion (Baumgard and Rhoads 2013). Therefore, selection of animals genetically less affected by thermal stress may help to increase production throughout the hot season and the first step involved in this is to identify genes that specifically respond to HS. Traditional breeding programs have limitations in terms of collecting precise records for thermal states of individuals, and usually try to identify animals that are resistant to HS by measuring reduction in certain production traits as critical indicators of heat tolerance under high heat conditions (Kadzere et al. 2002; West 2003). However, these indicators mask the actual effects of HS, and therefore, identification of genetic changes against environmental factors is necessary. Thus, a precise and efficient method of measuring heat tolerance on a genetic basis becomes important. Recent technologies with detection systems for causative genes and pathways under various experimental circumstances can help to identify target genomic regions, and it is possible to detect major genes and the pathways they are involved in HS response in animals (Collier et al. 2006; Sakatani et al. 2012). Study of genetic factors for HS in dairy cattle should be valuable to evaluate the availability of genetically superior animals that can respond better to changing temperatures without any significant drop in production rate and also for identifying target genes and genomic region for breeding genetically superior animal with increased tolerance to HS.
Animals respond to HS through an evolutionarily conserved process by modulating protein activation and differential gene expression (Lindquist and Craig 1988; Feder and Hofmann 1999; Kregel 2002); however, very little information is available regarding the complete set of genes that are influenced by HS because of the complex physiological actions of genes in vivo. Early studies had revealed that a genetic network of heat shock transcription factors is responsible for various responses to HS (Pirkkala et al. 2001; Page et al. 2006). In addition, the central role of heat shock proteins (HSPs) has been described as protecting hyperthermia, circulatory shock, and cerebral ischemia during heat stroke (Lee et al. 2006). Gene expression profiling of blood provides an opportunity to clarify the response of the animal to heat stress on a global level, and moreover, it will result in identification of target genes that accurately reflects not only the physiological state of the animal but also the response of the animal through the activation of various cellular processes involving various cell types. This study aimed to identify and catalogue genes and genetic pathways that respond to heat stress in Holstein bull calves using RNA sequencing (RNA-seq) of whole blood and to categorize transcription factors (TFs) and genes that can be used as candidate genes for breeding animals with superior thermal tolerance.
Materials and methods
Animals
Experimental procedures were approved by the ethics and welfare committee of the National Institute of Animal Science (NIAS) in Korea. The animals registered in the national database were a product of the standard breeding program described in the guidelines provided by NIAS. Animals were fed a diet formulated to meet nutritional requirements according to National Research Council with 68 % total digestible nutrients (TDN), 15 % crude protein, and hay (Dactylis glomerata L.). During the heat stress treatment, water and fresh hay were provided ad libitum. A total of ten bull calves, which weighed an average of 126 ± 5 kg and were between 4 to 6 months of age were selected from the dairy cattle division at NIAS. The process used to select animals for this study focused on reducing variation in genetic backgrounds among individuals, and therefore, the animals used in the study were the progeny of three sires (Supplementary File 1). To normalize the environmental effects, animals were placed in an open barn for 3 days without any restrictions for hay and water consumption and behavior of individual animals was monitored. After finishing medical checks, the animals were placed in an environmentally controlled chamber at NIAS.
HS experimental setup
Before the experiment began, the environmentally controlled chamber was tested with increasing temperature and humidity around the targeted severe region. When animals were first placed in the environmentally controlled chamber in individual pens, the ambient temperature was 22 °C with 60 % humidity at 18:00 hours on the day before experiment. The first setting of temperature and humidity was the same as the ambient temperature outside the building. The experiment was started at 35 °C and 90 % humidity at 09:00 hours on the day of experiment. Temperature was increased linearly to minimize experimental heat shocks for 1 h to reach the target stressful regions, and the actual severe heat stress was begun at 10:00 hours. The intensity of stress, which was continued from 10:00 to 19:00 hours, was based on temperature-humidity index (THI) level which was calculated as THI = (dry-bulb temperature, °C) + (0.36 dew point temperature, °C) + (41.2). The targeted severe stress range was set for the THI level 90 to 93, which corresponded to 35∼37.78 °C and 75∼95 % humidity. The THI values leading to severe stress are shown in Supplementary File 2.
The skin temperature was measured every 1.5 h with a temp-gun (Hitachi, USA) on the left side of the shoulder and neck for a total of three measurements, maintaining a gap of approximately 60 cm between human and the animal to prevent any human influence on the skin temperature. In addition, rectal temperature was measured before heat stress (BHS) and after heat stress (AHS). To minimize experimental errors, two experts measured skin and rectal temperature, and they shared their data and points of measurement.
A total of eight video cameras were set to record animal behavior throughout the experimental periods. Since the animals were subjected to severe stress levels, we feared the animals might collapse during the experiment or might not drink sufficient water so staff members stood by to provide any assistance if and when any irregular animal behavior or unexpected situations arose and also to make sure the animals consumed water and feed. The blood samples were collected before the experiment began (09:00 hours) and at the end of the stress period (19:00 hours). Ten milliliters of blood samples was taken from the jugular vein and placed in 50-ml falcon tubes without anticoagulants and were labeled with the animal ID and immediately stored in liquid nitrogen. The reason for collecting blood at only two time points, i.e., before and after heat stress, was to avoid any stress that might be caused due to repeated drawing of blood.
RNA preparation and sequencing
The whole blood samples were transferred to the laboratory of the Animal Genomics and Bioinformatics Division of NIAS. Frozen blood (approximately 2 g) was homogenized and isolated using TRIzol Reagent (Invitrogen, USA), and the RNA was cleaned using the RNeasy Midi Kit (Qiagen, USA) with DNase digestions according to the manufacturer’s guidelines. Integrity of the RNA was assessed using a 2100 Bioanalyzer and RNA 6000 Nano LabChip kit (Agilent Technologies, USA). Only RNA with a RIN value greater than 9.0 was used for library construction. RNA concentration was determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, USA).
A library was constructed using reagents provided in the ILLUMINA TruSeq™ RNA sample preparation kit. The first step in the workflow involved purifying the poly-A containing mRNA molecules using oligo-dT-attached magnetic beads. The cleaved RNA fragments were reverse-transcribed into first-strand complementary DNA (cDNA) using reverse transcriptase and random primers. This was followed by second-strand cDNA synthesis using DNA polymerase I and RNaseH (Invitrogene, USA). These cDNA fragments then went through an end repair process, with addition of a single A base and ligation of the adapters. The products were then purified and enriched with PCR to create the final cDNA library. The sequencing was carried out on an ILLUMINA HiSeq 2500 sequencer following the vendor’s protocol. The library construction and sequencing were performed by Macrogen Inc. (Korea).
Data handling procedures
The analysis involved quality control, transcript assembly, abundance estimation, test for differential expression, and regulation of RNA-seq samples. FastQC (version v0.10.0) was used to check the quality of the raw sequence data from high-throughput sequencing pipelines. TopHat program (v2.0.11) and Bowtie2 (v2.1.0) were used to align the RNA sequences and to map the sequence. Aligned RNA-seq reads were assembled into a parsimonious set of transcripts. Cufflinks (v2.1.1) was used to estimate the relative abundance of transcripts based on how many reads supported each one, taking into account biases in library preparation protocols. Expression data was statistically analyzed in R. Functional annotation and pathway analysis was carried out using a combination of gProfiler (Reimand et al. 2016) and Gene Ontology (Ashburner et al. 2000), and KEGG pathways (Kanehisa and Goto 2000; Kanehisa et al. 2015) implemented in ClueGO in Cytoscape v3.2 (Bindea et al. 2009). A hypergeometric test with Benjamin and Hochberg false discovery rate (FDR) was performed using the default parameters implemented in ClueGO. Genes that were 2-fold upregulated or downregulated and P <0.05 were considered to be significantly differentially expressed. A text mining approach using eGIFT (Tudor et al. 2010) was also performed to understand the functional role of the DEGs from published abstracts (Sun et al. 2015) and to find out the cell types to which the DEGs belong. Animal Transcription Factor Database (Zhang et al. 2012) was used to indentify TFs, transcriptional cofactors, and chromatin modulators.
Real-time PCR validation
Quantitative reverse transcription PCR (qRT-PCR) was performed using gene-specific primers (Table 1) and the Fast SYBR green master mix (Applied Biosystems) on an ABI 7500 Real Time PCR system following the manufacturer’s direction. A total of ten genes that were more than 2-fold differentially expressed were analyzed. ATP5B (ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide) and hydroxymethylbilane synthase (HMBS) were tested for use as endogenous controls. Primers for ATP5B were CATCG TGGCG GTCAT TGG and AATGG TCCTT ACTGT GCTCT C and for HMBS were TGCTT CCTCC TGGCT TCAC and GTTCC TACCA CACTC TTCTC TG. The expression of ATP5B and HMBS was checked for stability using GeNORM (http://medgen.ugent.be/%7Ejvdesomp/genorm) against same concentration of RNA from different samples, and HMBS was found to be more stable between samples and was used for normalization of the expression data.
Table 1.
Primers used for qRT-PCR validation of RNAseq data
| Gene | Accession no. | Forward 5′→3′ | Reverse 5′→3′ |
|---|---|---|---|
| SPDEF | NM_001081578 | TGAAACATGCCCTTGCCAGACCT | ACAGCCCTACCTCCCTCCATCCTC |
| HBM | NM_001083768 | CGCGCCCACATAACACAGGT | TCGGCAAGCGGGCTCAGGACT |
| MZT2B | NM_001099204 | GCTGCGGCGGAAGAAGGTGCTGAG | CGCGGCCGTGGGATCCTGGGAGTC |
| ADIF | NM_001114513 | ACAGCGGTTCAGCAAGTGGT | AGGAATATAGGTTTTAATGAGGTG |
| SELM | NM_001163171 | GGCCACCGCTTTGAGGAAC | CAGGCGGGAGGTGGGGGAGTGG |
| HBP1 | NM_001046196.2 | GTGAGTCGGAATCTGGCATT | TGCCATTCCTTATTGCTTCC |
| MYADM | NM_001075252 | CAGAAGCATGGCGGTGAGC | CTGGGGAGAAGCGGGGATTAGG |
| PTPRC | NM_001206523 | ATGATCAACAGCTTCTTATTAGTC | ATTCCGTCCTGGGTTTTATCCTGA |
| HSPH1 | NM_001075302 | AGTAGAAACCACGCTGCTCCTT | CTCCACCATAGACGCTGTAGA |
| HSPA1A | NM_203322 | GCTTCCGCAGACCCGCTATC | ACCTTGCCGTGCTGGAACA |
Results and discussion
In general, temperatures above the thermal neutral zone, which is significantly influenced by ambient temperature, wind and humidity, affect production, and reproduction rates of animals. Therefore, management techniques to protect animals from heat stress in hot seasons are necessary to minimize economic losses in farms due to heat stress. The physiological changes caused by heat are associated with phenotypic factors such as density and thickness of hair coat, sweat gland density, skin color, and length and color of hair (Olson et al. 2003; da Silva et al. 2003; Klungland and Våge 2003; Mariasegaram et al. 2007). Several environmental factors influence HS, one of the most important of which is air flow over the skin surface (Gebremedhin and Wu 2001; Olson et al. 2006). Thus, eliminating environmental variation around animals can help to characterize genes, which are specifically triggered in response to HS. As our study tried to fix environmental factors, the identified genes may be used as genetic indicators for selecting animals against heat stress.
Changes in skin and rectal temperature in response to heat stress
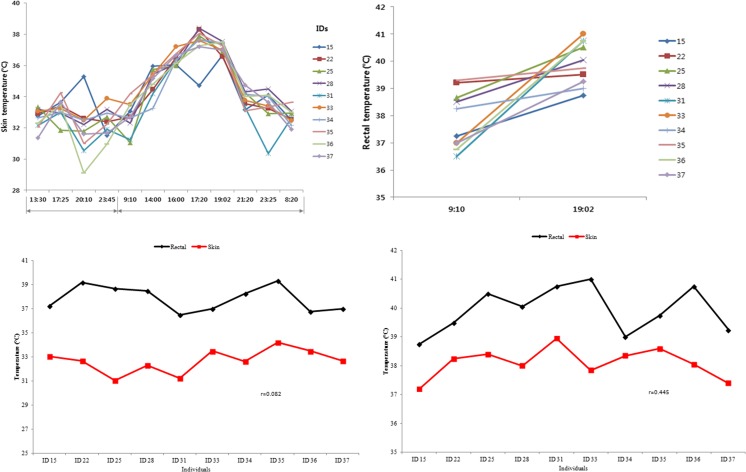
It has been reported that mild heat stress begins at a THI of 77 for dairy heifers (St-Pierre et al. 2003). Therefore, in this study, in order to subject the animal to severe ranges of heat stress, a THI of 90–95 was set (Supplemental File. 1). The skin temperature was measured at four different time points, i.e., at 13:30, 17:25, 20:10, and 23:45 hours the day before subjecting the animals to HS in order to determine the changing patterns of body temperature under normal circumstances (Fig. 1a, b). No tendency for increasing or decreasing skin temperatures were detected between 13:30 and 23:05 hours on the day prior to the HS experiment day. On the other hand, measurements of skin temperature on the day of the HS experiment showed increasing patterns for all individuals from 09:10 to 19:02 hours. The highest skin temperature was recorded in animal number 22 (approximately 38.5 °C), whereas animal number 15 presented the lowest temperature (approximately 35 °C) at 17:20 hours. Berman (2005) had reported that when the body temperature of animals reaches 35 °C, it triggers and activates the responding system for heat stress in dairy cows. Rectal temperature was also elevated for all individuals from 09:10 to 19:02 hours (Fig. 1c, d). Animal numbers 33, 36, and 31 showed rapidly increasing rectal temperatures, whereas animals 22, 35, and 34 showed little increment in temperatures between BHS and AHS. The animals 33 and 15 recorded the highest (41.3 °C) and lowest (38.7 °C) rectal temperatures, respectively, at 19:02 hours. A low correlation (r = 0.082) between rectal and skin temperatures was estimated at BHS (Fig. 1c), whereas a moderate correlation (r = 0.445) was detected at AHS. In general, rectal temperature is a barometer for actual body temperature, as skin temperature is affected by the environment, but in order to avoid any stress due to physical insertion of a temperature probe inside the animal and since the experiment was carried out inside an environmentally controlled chamber, rectal temperatures were measured only at two time points. Skin temperature is considered as not a good indicator, due to various nonmeasurable factors that may cause low correlations between skin and rectal temperature. Umphrey et al. (2001) reported that skin temperature was lowly correlated with rectal temperature (r = −0.022) and respiration rate (r = − 0.086). However, in contrast to previous reports, we found a moderate correlation (r = 0.445) between skin and rectal temperature following heat stress (Fig. 1d), suggesting that conducting the experiment in a controlled environment seems to have removed variables other than temperature and humidity leading to a moderate correlation in the skin and rectal temperature.
Fig. 1.
Measurements of skin and rectal temperatures for Holstein calves (a, b) during the course of the experiment; c, d correlation between measurements of skin and rectal temperature before heat stress (09:00 hours) and after heat stress (19:02 hours)
These results show that the increase in skin and rectal temperature were consistent with changes in environmental temperature. Though variance in skin and rectal temperatures for individual animals were detected, this was expected, as variation in body temperature may be due to physiological differences among individuals that are related to activities of glands (Olson et al. 2006), or due to different genetic backgrounds, that adapt easily to new environments. These results showed that the body temperature had reached severe HS levels in all the animals.
RNA sequencing summary
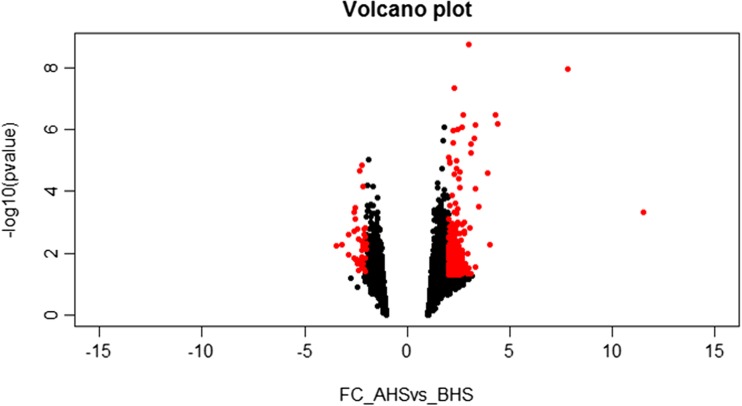
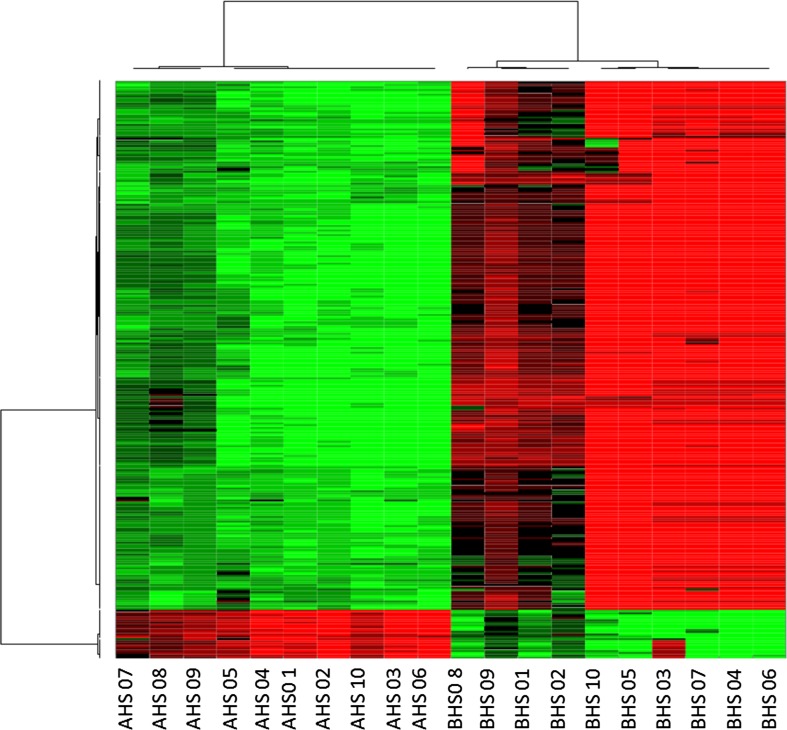
The RNA-seq analysis showed that the total read bases and the total read pairs of the samples increased significantly except in animals 31 and 34 from BHS to AHS (Table 2). Over 154 million reads were generated across 20 samples. The analysis identified 8567 genes (Supplementary File 3) to respond to heat stress, out of which 465 upregulated and 49 downregulated genes were identified as significantly differentially expressed (DEGs) (≥ or ≤2-fold and P < 0.05) (Fig. 2). Hierarchical clustering (Fig. 3) of the DEGs showed that the samples clustered based on condition (heat vs control).
Table 2.
Summary sequencing statistics
| ID | Before heat stress (BHS) | After heat stress (AHS) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total read bases | Total read pairs | GC (%) | Q20 (%) | Q30 (%) | Total read bases | Total read pairs | GC (%) | Q20 (%) | Q30 (%) | |
| 15 | 8,302,856,298 | 82,206,498 | 50.77 | 93.87 | 87.41 | 9,077,010,390 | 89,871,390 | 49.68 | 94.25 | 87.12 |
| 22 | 7,664,272,688 | 75,883,888 | 50.24 | 94.88 | 88.72 | 7,978,351,378 | 78,993,578 | 49.36 | 94.48 | 87.57 |
| 25 | 7,146,502,450 | 70,757,450 | 49.65 | 94.54 | 88.31 | 8,012,177,288 | 79,328,488 | 49.01 | 94.43 | 87.54 |
| 28 | 7,614,621,492 | 75,392,292 | 49.92 | 94.96 | 88.85 | 8,627,660,380 | 85,422,380 | 49.18 | 94.43 | 87.57 |
| 31 | 7,462,123,612 | 73,882,412 | 50.07 | 93.83 | 87.74 | 6,831,782,006 | 67,641,406 | 49.71 | 95.02 | 88.57 |
| 33 | 6,738,018,656 | 66,713,056 | 49.92 | 92.03 | 85.64 | 8,067,930,500 | 79,880,500 | 49.59 | 94.51 | 87.69 |
| 34 | 8,231,919,352 | 81,504,152 | 49.97 | 94.04 | 86.73 | 7,862,032,304 | 77,841,904 | 51.08 | 94.05 | 87.09 |
| 35 | 7,711,182,138 | 76,348,338 | 50.74 | 94.78 | 88.12 | 9,761,067,634 | 96,644,234 | 50.79 | 94.87 | 88.17 |
| 36 | 7,131,010,262 | 70,604,062 | 53.01 | 90.75 | 82.88 | 7,788,998,396 | 77,118,796 | 50.76 | 94.28 | 87.34 |
| 37 | 6,472,468,244 | 64,083,844 | 50.34 | 94.64 | 88.45 | 7,286,197,974 | 72,140,574 | 48.61 | 94.51 | 88.08 |
Fig. 2.
Volcano plot showing differentially expressed genes between AHS and BHS; the dots in red are genes that were considered significant FC ≤ or ≥2-fold and P value <0.05 (color figure online)
Fig. 3.
Hierarchical clustering of all the expressed genes before (BHS) and after (AHS) heat stress. Red corresponds to downregulated gene product, and green corresponds to upregulated gene product (color figure online)
Real-time PCR validation
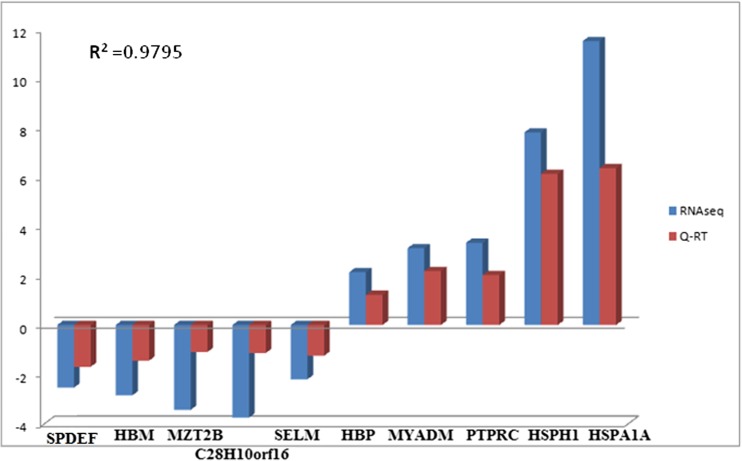
The RNAseq transcriptome data was validated using quantitative RT-PCR analysis. The expression levels of ten genes were analyzed between control and heat stressed samples (Table 1). The analysis showed that the tendency of gene expression was concordant with the RNAseq result, though the absolute fold changes differed between qRT-PCR and RNAseq. A correlation (r 2) of 0.968 was yielded between the q-RT PCR ΔCT and LOG2FC value of the RNAseq analysis (Fig. 4); this was similar to what was observed in several other studies (Nagalakshmi et al. 2008; Core et al. 2008; Camarena et al. 2010; Sun et al. 2015).
Fig. 4.
Expression levels of selected genes from RNAseq analysis (blue bar) and their validation by qRT-PCR (red bar). R 2 is the correlation of expression levels between the two methods (color figure online)
Functional annotation and classification
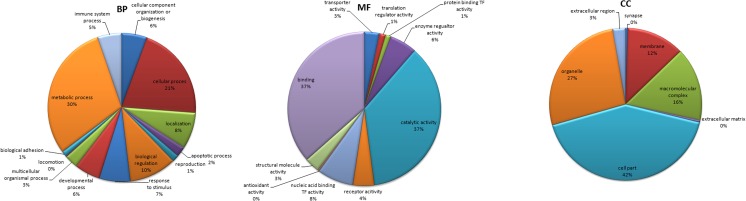
The DEGs were then functionally annotated and grouped based on Gene Ontology (GO) terms. The top ten upregulated genes after heat stress, were predominated by genes belonging to heat shock transcription factors (molecular chaperones) (Supplementary File 3). The top five upregulated genes were heat shock 70 kDa protein 1A (HSPA1A), heat shock 105 kDa/110 kDa protein 1 (HSPH1), heat shock 70 kDa protein 8 (HSPA8), DnaJ (Hsp40) homolog, subfamily A, member 1 (DNAJA1), and CDC-like kinase 1 (CDK1), while the top five downregulated genes were interferon-induced mitotic spindle organizing protein 2B (MZT2B), chromosome 28 open reading frame, human C10orf116 transmembrane (C10orf116), hemoglobin, mu (HBM), nucleoside diphosphate linked moiety X (nudix)-type motif 14 (NUDT14), and ybeY metallopeptidase (YBEY). The GO terms were enriched for biological process, molecular function, and cellular component. The DEGs were primarily located in the protoplast (42 %), organelle (27 %) and macromoleular complex (16 %). Most of the genes participated in metabolic process, catalytic activity, transcription factor activity, enzyme regulatory activity, protein binding, apoptotic process, response to stimulus, cellular process, and immune system process (Fig. 5). A complete list of GO terms and the genes involved in them are given in the Supplementary File 4.
Fig. 5.
Summary of GO terms for biological process (BP), molecular function (MF), and cellular component (CC) ontologies for upregulated and downregulated gene products in response to heat stress. Chart labels are GO terms, and the percentage indicates the total percentage of the DEGs involved in that process
Heat stress resulted in the activation of heat shock factors and factors involved in protein folding
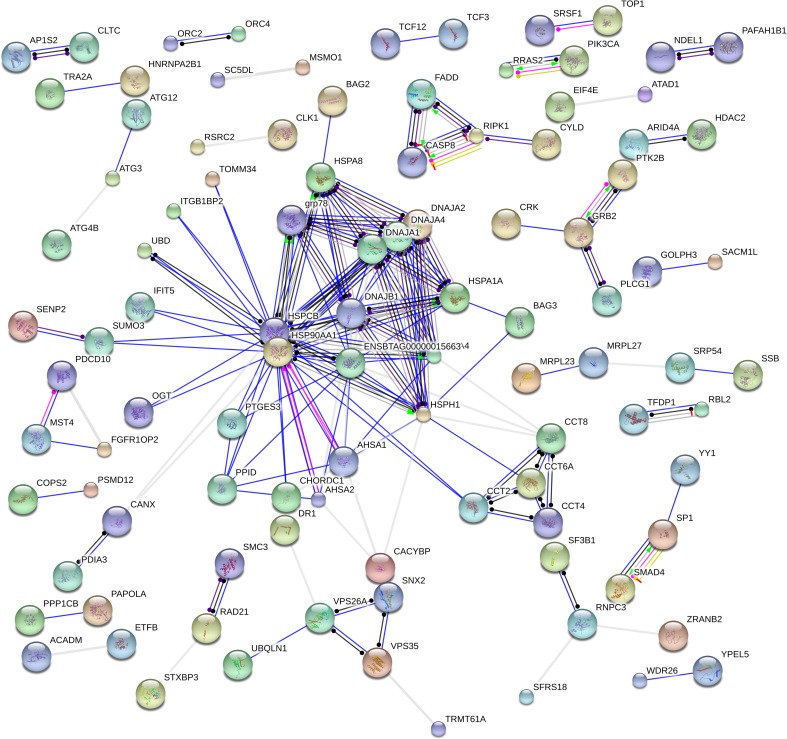
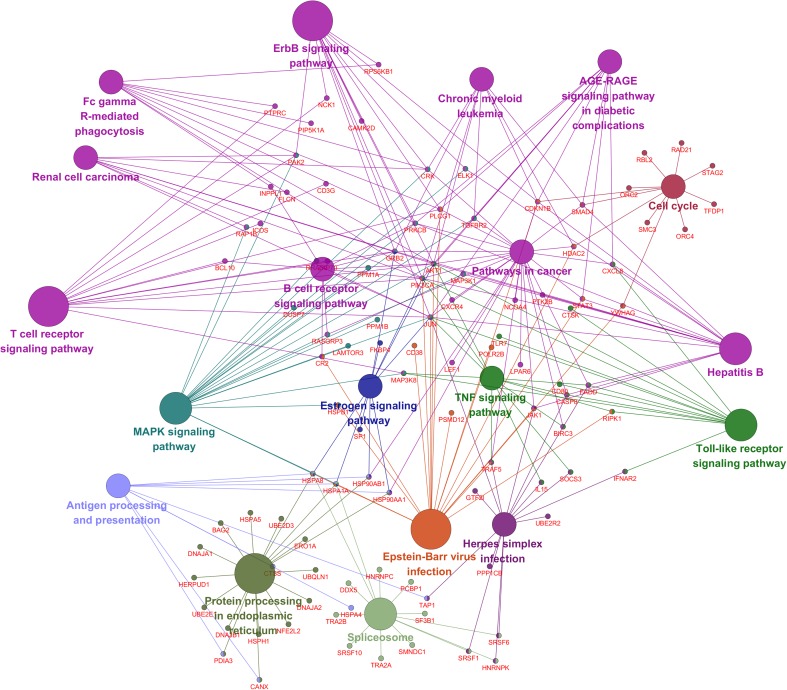
Heat is a proteotoxic stress and causes denatured proteins which can become cytotoxic by forming aggregates (Fink 1999; Liu et al. 2013). The function of many heat shock factors (HSFs) are to act as chaperones assisting in protein folding thereby avoiding protein aggregation (Lindquist and Craig 1988; Hightower 1991; Moseley 1997) which results in protein homeostasis during cellular response to HS. In all, 20 genes encoding molecular chaperones were upregulated in response to HS (Table 3). These included members of HSP70 family (HSPA1A, HSPA4, HSPA5, and HSPA8) and subfamily HSP110 (HSPH1), HSP40 (DNAJA1, DNAJB1, DNAJA2) family. HSP40 regulates the ATPase activity of HSP70 by interacting with the J domain of the HSP70 proteins. These genes also confer thermo-tolerance to cells on exposure to HS (De Maio 1999). Two members of the HSP90 family was also upregulated (HSP90AB1, HSP90AA1) and so was cofactor AHSA1 which interacts with HSP90AA1 as an activator, while the HSP90s assists in protein folding and protein stabilization (Panaretou et al. 2002). STIP1, a member of HSP70-HSP90 organizing protein (HOP) family which functions as a cochaperone that reversibly links together HSP70 and HSP90 protein chaperones (Odunuga et al. 2004) and prostaglandin E synthase 3 (PTGES3) which functions as a cochaperone, along with HSP90 were also upregulated. HS results in an increase in cytotoxic protein in the endoplasmic reticulum (ER), HERPUD1 which was found to be upregulated in this study functions in processing and degradation of these cytotoxic protein by participating in ER-associated degradation (ERAD) (Nogalska et al. 2006). Foldases are required for protein folding (Nagradova 2007); they catalyze protein folding by isomerizing peptide bonds with peptidyl-prolyl 4 hydroxylase which results in rearrangement of disulfide bonds (Wilkinson and Gilbert 2004); in this study, protein disulfide isomerase family A member 3 (PDIA3) was found to be elevated in response to HS. PDIA3, also known as GRP58, localizes to ER, and it interacts with calnexin (CANX) to modulate the folding of newly formed glycoproteins; CANX was also found to be elevated in this study. Looking at the function of these proteins, it looks like CANX, PDIA3, and HERPUD1 could closely interact in response to HS. Other than these, PTGES3, which is required for proper functioning of glucocorticoids and other steroid receptors (Freeman and Yamamoto 2002), and TP53INP1, which positively regulates autophagy (Okamura et al. 2001), were also upregulated. SERPINH1 a cochaperone which is involved in collagen biosynthesis was the least modulated chaperone (2-fold change) in response to HS, relative to the other chaperones reported above. A protein-protein interaction network analysis showed that the HSPs and cochaperones were strongly related and mostly coexpressed (Fig. 6). Several genes involved in the pathway for protein processing in the ER were also identified in this study to be differentially expressed (Fig. 7). Though the activation of HSPs in response to heat stress is very well characterized, polymorphism in these genes could be associated with milk yield in lactating cattle (Li et al. 2011; Liu et al. 2011); moreover, the expression of these genes could also be verified for their effect on milk yield in lactating cows in the hot season.
Table 3.
List of chaperones differentially expressed in this study
| Gene symbol | Entrez gene ID | Fold change |
|---|---|---|
| HSPA1A | 282254 | 11.52979 |
| HSPH1 | 507165 | 7.819208 |
| DNAJA1 | 528862 | 4.436372 |
| HSPA8 | 281831 | 4.294902 |
| STIP1 | 617109 | 3.915504 |
| DNAJB1 | 538426 | 3.321045 |
| HSP90AB1 | 767874 | 3.295318 |
| HSP90AA1 | 281832 | 2.673493 |
| ST13 | 510494 | 2.56736 |
| AHSA1 | 539220 | 2.497629 |
| PDIA3 | 281803 | 2.486621 |
| PTGES3 | 493638 | 2.484664 |
| HERPUD1 | 613577 | 2.412679 |
| HSPB1 | 516099 | 2.354337 |
| CANX | 407129 | 2.264448 |
| TP53INP1 | 782667 | 2.228985 |
| HSPA4 | 536558 | 2.197612 |
| DNAJA2 | 360006 | 2.043753 |
| HSPA5 | 415113 | 2.01403 |
| SERPINH1 | 510850 | 2.008764 |
Fig. 6.
Gene interaction network of DEGs analyzed using STRING protein database. Nodes are the genes, and the edges are the interaction between the nodes. The color of the edge indicates the type of interaction
Fig. 7.
A network map of pathways significantly enriched after heat stress. The nodes are the pathways, and edges connect the genes involved in the pathway
Heat stress regulates genes involved in immune response and immunity-related signaling pathways
Heat stress has a severe effect on the health of animals and compromises immune function (Strong et al. 2015) and has also been reported to lead to leaky guts in ruminants eliciting an inflammatory response (Kahl et al. 2015). In all, 84 genes involving in immune response or immune-related response were differentially expressed in response to HS (Supplementary File 3). HS or hyperthermia also results in hyperinsulinemia which results in the metabolic profile of HS subjected cattle to be similar to an immune stimulated system; this hyperinsulinemia is thought to be associated with an endotoxin, lipopolysaccharide (LPS), an abundant glycolipid of the outermembrane of gram-negative bacteria, taxol or Hsp60 (Asea 2008). Increased levels of LPS has also been reported in several other heat stressed species like human, poultry, pigs, and rodents (Mani et al. 2012). Heat stress has also been reported to lead to an increase in the expression of Toll-like receptors (TLRs) (Ju et al. 2014) especially with the increased expression of Hsp70 which specially binds to TLR2 and TLR4 and induces immunoregulatory effects such as cytokine and chemokine release by a process known as chaperokine activity. Several TLR genes were found to be 1.5- to 3-fold upregulated in this study; these included TLR1, TLR2, TLR3, TLR4, TLR6, TLR7, TLR8, TLR9, TLR10, out of which TLR7 was 3-fold upregulated. TLRs are generally grouped into two main groups based on the recognition of pathogen-associated molecular patterns (PAMPS) from invading agents: the first group comprises TLR1, TLR2, TLR4, and TLR6 and recognizes PAMPS from lipids; the second group consists of TLR3, TLR7, TLR8, and TLR9 that recognizes PAMPS from nucleic acids (Brikos and O’Neill 2008). TLR signaling leads to the activation of innate and adaptive immunity. Moreover, HS was found to activate TLR signaling pathway (Fig. 7) with the expression of genes like CD80, CASP8, RIPK1, IFNAR2, and BIRC3 found to be elevated in response to HS. The TLR signaling pathway is known to be activated in response to HS (Eicher et al. 2004; Zhou et al. 2005). Several interleukin genes such as NFIL3, IL8, and IL15 expressions were also increased in response to HS. TLR activation and dysregulation of cytokine expression in response to heat stress has been reported to lead to immunosupression and increased susceptibility to antigenic challenge in pigs (Ju et al. 2014), suggesting that heat stress could make the animal susceptible to pathogens. Other than TLR signaling pathways, several other immune-related pathways (Supplemental 4) were also activated in response to HS; these included T cell receptor signaling pathway, B cell receptor signaling pathway, Fc gamma R mediated phagocytosis, ErbB signaling pathway, antigen processing and presentation, and some viral infection-related pathways like Herpes simplex infection pathway, Epstein-Barr virus infection, and several pathways involved in cancer and MAPK signaling pathways were also activated in response to HS (Fig. 7). HSPs, while playing an important role in the stimulation of innate immunity through TLRs, also activates T cell and B cell lymphocytes (Breloer et al. 2001; Moré et al. 2001); moreover, HSPs can directly activate the adaptive immune T lymphocytes in a TLR2-dependent and TLR4-independent manner, while Hsp60 activates B cells via TLR4 recruitment (Zanin-Zhorov et al. 2003; Cohen-Sfady et al. 2005; Osterloh et al. 2008).
Effect of heat stress on fertility through MAPK signaling pathway
Heat stress has been recognized as a leading cause for subfertility in farm animals due to sperm damage (Hansen 2009). MAPK signal transduction, which is activated in response to several environmental stressors, promotes the inhibition of cell growth and apoptosis (Wada and Penninger 2004); specifically, MAPK signaling pathway has been shown to be involved in heat-induced sperm damage (Rahman et al. 2014). Eighteen genes belonging to the MAPK signaling pathways were differentially expressed (Fig. 7) in response to HS; these included DUSP7, PPM1A, GRB2, PRKCB, JUN, CRK, and HSP90AA.
Several transcription factors, transcriptional cofactors, and chromatin remodeling factors involved in cell cycle inhibition and tumor suppression were activated in response to heat stress
Thermal stress induces anomalies in cell function (Sonna et al. 2002), which includes inhibition of protein synthesis, and defects in protein structure and elevated heat load will elicit changes at the level of transcription factors (Collier et al. 2008). The effect of HS on transcriptional regulation of protein synthesis has been reported in several species (Buckley et al. 2006; Liu et al. 2013). Protein synthesis is regulated by transcriptional regulation, either by transcription factor binding or by changing the structure of chromatin by chromatin remodeling factors; the maintenance of chromatin structure is of vital importance for organismal homeostasis (Kokavec et al. 2007), while transcriptional cofactors are required for activator-dependent or activator-regulated transcription and for transmitting regulatory signals between gene-specific activators and the transcriptional machinery (Thomas and Chiang 2006). In this study, 31 TFs, 13 transcriptional cofactors, and 6 chromatin remodeling factors were found to be differentially expressed (Table 4). Several TFs involved in the regulation of cell cycle such as ARID4A, ELF4, TFCP2, TFDP1, and HBP1 were upregulated while SPDEF was downregulated. ARID4A is involved in cell cycle arrest at G1/G0 stage by repressing the transcription of E2F-regulated genes by forming a complex with retinoblastoma protein, thereby regulating cell proliferation (Chen et al. 2003). ELF4 encodes a protein that functions in the activation of IL3, IL8, PRF1, and CSF2 and in cell cycle arrest in naive CD8+ cells. The expression of TFCP2, which is involved in the regulation of cellular and viral promoters, cell cycle regulation, and also in cell survival (Santhekadur et al. 2012) was elevated, while SPDEF, which encodes a ETS family TF and functions in the activation of an androgen-independent transactivator of prostate-specific antigen (PSA) promoter and is strongly correlated with the development of several types of tumor, was found to be downregulated in response to HS. The expressions of several TFs, such as SMAD4, STAT3, IRF2, HBP1, ELF4, and ARID4A, involved in tumor suppression were also elevated. Moreover, cofactors such as RBL2, SETD8, and RNF2 which plays important roles in cell cycle inhibition were also significantly upregulated. Chromatin modifiers involved in epigenetic regulation of cell cycle such as HDAC2 and KDM3A (Lee et al. 2014) were also upregulated.
Table 4.
List of TFs, transcriptional cofactors, and chromatin remodeling factors differentially expressed in this study
| Gene symbol | Entrez gene ID | Fold change |
|---|---|---|
| Transcription factors | ||
| JUN | 280831 | 2.663937 |
| IRF2 | 337916 | 2.533374 |
| NFE2L2 | 497024 | 2.073162 |
| SPDEF | 497620 | −2.54385 |
| ELF4 | 504514 | 2.125593 |
| ELF1 | 505251 | 2.677581 |
| NFIL3 | 506097 | 2.014033 |
| ATF1 | 506967 | 1.957237 |
| STAT3 | 508541 | 1.954277 |
| ZBTB7B | 509019 | 2.318676 |
| TCF12 | 509039 | 1.992964 |
| TFCP2 | 509448 | 2.330451 |
| BAZ2A | 509799 | 2.063852 |
| TGIF1 | 510050 | 2.389241 |
| ARID3A | 511283 | 2.118166 |
| HBP1 | 515320 | 2.143308 |
| TCF3 | 530616 | 2.378868 |
| ARID4A | 531515 | 2.109133 |
| ADNP | 533757 | 2.051073 |
| TFDP1 | 534579 | 2.156619 |
| GTF2I | 534669 | 2.139872 |
| ZBTB40 | 534714 | 2.021622 |
| LEF1 | 535399 | 2.332525 |
| MIER1 | 538742 | 2.11698 |
| ZBTB18 | 538793 | 2.16256 |
| VEZF1 | 539120 | 2.139921 |
| HHEX | 539542 | 2.073288 |
| ZNF644 | 539923 | 2.192416 |
| SMAD4 | 540248 | 2.085932 |
| SP1 | 540741 | 2.325516 |
| ELK1 | 786886 | 2.096515 |
| Chromatin remodeling factors | ||
| CBX3 | 1E+08 | 2.025144 |
| HDAC2 | 407223 | 2.079579 |
| MSL3 | 515220 | 1.978494 |
| KDM3A | 536073 | 1.973173 |
| MORF4L2 | 538442 | 2.652781 |
| BAZ1A | 540621 | 2.028949 |
| Transcriptional cofactors | ||
| DNMT3B | 1.01E+08 | 1.960636 |
| PSIP1 | 282011 | 2.129971 |
| PRKCB | 282325 | 2.29706 |
| ATN1 | 513125 | 2.306827 |
| BANP | 513446 | 2.458954 |
| NCOA4 | 525329 | 2.101791 |
| KMT5A | 532622 | 2.399096 |
| RBL2 | 533294 | 2.491395 |
| DDX5 | 533700 | 1.962994 |
| RNF2 | 540090 | 2.086213 |
| BCL10 | 540824 | 2.03278 |
| CCT4 | 613336 | 2.310648 |
| TMF1 | 616786 | 2.023018 |
Apoptosis
Sixty-four genes involved in apoptotic processes were found to be differentially expressed in this study. The activation of HSF in response to thermal stress protects the cell from apoptosis, but when the stress is too strong or conversely, the failure to activate and maintain the protective response results in the activation of signaling cascades that ultimately results in cell death pathways (Perkins and Gilmore 2006; Weston and Davis 2007). Apoptosis along with cell cycle arrest is a major mechanism by which tumor formation is inhibited (Li et al. 2012).
Taken together, the activation of several cancer pathways and the expression of the tumor suppressors, the cell cycle arrestors, along with the activation of such a large number of genes involved in apoptosis and the expression of large number of genes belonging to macrophage lineage (Table 5) suggests that severe cell anomaly and DNA damage has taken place due to thermal stress. Gu et al. (2014) recently showed that HS can induce apoptosis by activating p53-mediated mitochondrial pathways; three genes involved in p53 signaling pathway (CASP8, MDM-X, and CYCLIN G) were differentially expressed in this study. Several genes, such as FADD, CASP8, BRIC4, JUN, PI3K, and AKT3, which play a critical role in inducing apoptosis through TNF and PI3K-Akt signaling pathways, were also differentially expressed.
Table 5.
Putative cell types to which the identified DEGs belong as revealed by eGIFT cell type analysis.
| iTERM (cell type) | Gene symbol |
|---|---|
| B cell | ZAP70, RHOH, TCF3, CD38, LCP1, NFIL3, CR2, GPR183, TCF12, BIRC3, PTPRC, TRAF5, MS4A1 |
| Lymphocyte | RHOH, MAT2B, CXCR4, TCF3, NFIL3, CD38, LEF1, DOCK10, CD80, WIPF1, IGJ, TAP1, CD69, SLAMF6, CD3G, CR2, ZBTB7B, ELF1, IL15, TRAF5, IDO1, MS4A1, PTPRC, BCL10, CX3CR1, GIMAP4 |
| T cell | PLCG1, TCF3, LAMP3, CD38, LEF1, ZAP70, CTSS, RHOH, IDO1, MS4A1, PTPRC, TCF12, ZBTB7B, ELF1, CD80, IL15 |
| Monocytes | CD80, IL15, TLR7, CXCR4, CD69, DNAJA1, CX3CR1 |
| NK cell | IL15, CX3CR1, PDIA3, NFIL3, SLAMF6, ELF4, CD69 |
| Adipocyte | RETN, RAB10, PSPH, ST13, RAB18, FFAR3 |
| Hepatocyte | TM4SF5, CDO1, MARK2, MAT2B, HDGF, P2RY13, UBD, MYLIP |
| Fibroblast | TGFBR2, SERPINH1, ETFB, FGFR1OP2, ELK1, LAMP3, GRB2, PLCG1, RBL2, WIPF1, HDGF, G3BP1, SLC38A2, PTPN12, CRK, NCK1, TIAL1 |
| Myocyte | HSPB1, CDKN1B, PRKCB, RPS6KB1, RAB10, CTSS, ELK1, STIM1, AKT1, PTK2B, SLC38A2 |
Putative cell types which contributes to the gene expression changes
Blood is a complex mix of cells, and various cell types must be contributing to the gene expression changes observed in this study. An attempt was made to classify the cell types based on the expressed genes using a text mining approach (Sun et al. 2015; Tudor et al. 2010) (Table 5). The result shows that most of the DEGs belonged to the lymphocytes, particularly B cell, T cell, and NK cell, followed by fibroblasts, monocytes, adipocytes, and hepatocytes. The increased expression of genes belonging to these cell types also suggest that their relative proportion in blood might have increased in response to heat stress; however, further studies are needed to validate this statement.
Conclusion
In this study, the effect of heat stress on Holstein bull calves has been examined by analyzing their transcriptome response to HS using RNAseq technology, and major biological process and pathways impacted by HS have been identified. We have also identified TFs that are impacted by HS. Three major response seems to be elicited in response to HS; initially, there is an elevated expression of chaperones and heat shock genes that acts to prevent protein aggregation and misfolding, thereby helping in cell survival; extracellular presence of HSPs triggers various immune system activation; and finally, the continued exposure to severe stress leads to expression of cell cycle arrestors, tumor suppressors, and genes involved in apoptotic processes to prevent tumorigenesis. The genes and TFs identified in this study can be used as candidates for improving thermal tolerance through breeding by marker-assisted selection program, thereby avoiding the use of expensive cooling technologies and also minimize production loss due to raising temperature which will not only result in agricultural profitability but also improve animal well-being.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PNG 24 kb)
(DOCX 42 kb)
(XLSX 3577 kb)
(XLSX 31 kb)
Acknowledgments
This work contributes to the internal project “Cooperative Research Program for Agriculture Science & Technology Development (ID PJ01005002)” and was supported by the National Institute of Animal Science in Rural Development Administration of Korea. Krishnamoorthy Srikanth was supported by 2015–2016 Postdoctoral Fellowship Program of National Institute of Animal Science, Rural Development Administration, Republic of Korea.
Compliance with ethical standards
Experimental procedures were approved by the ethics and welfare committee of the National Institute of Animal Science (NIAS) in Korea.
References
- Asea A (2008) Heat shock proteins and toll-like receptors. In: Toll-like receptors (TLRs) and innate immunity. Springer, pp 111–127 [DOI] [PubMed]
- Ashburner M, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgard LH, Rhoads RP., Jr Effects of heat stress on postabsorptive metabolism and energetics. Annu Rev Anim Biosci. 2013;1:311–337. doi: 10.1146/annurev-animal-031412-103644. [DOI] [PubMed] [Google Scholar]
- Berman A (2005) Estimates of heat stress relief needs for Holstein dairy cows. J Anim Sci 83(6):1377 [DOI] [PubMed]
- Bindea G, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breloer M, Dorner B, Moré SH, Roderian T, Fleischer B, Av B. Heat shock proteins as “danger signals”: eukaryotic Hsp60 enhances and accelerates antigen-specific IFN-γ production in T cells. Eur J Immunol. 2001;31:2051–2059. doi: 10.1002/1521-4141(200107)31:7<2051::AID-IMMU2051>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Brikos C, O’Neill LA (2008) Signalling of toll-like receptors. In: Toll-like receptors (TLRs) and innate immunity. Springer, pp 21–50
- Buckley BA, Gracey AY, Somero GN. The cellular response to heat stress in the goby Gillichthys mirabilis: a cDNA microarray and protein-level analysis. J Exp Biol. 2006;209:2660–2677. doi: 10.1242/jeb.02292. [DOI] [PubMed] [Google Scholar]
- Camarena L, Bruno V, Euskirchen G, Poggio S, Snyder M. Molecular mechanisms of ethanol-induced pathogenesis revealed by RNA-sequencing. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-F, et al. Retinoblastoma protein (pRB) was significantly phosphorylated through a Ras-to-MAPK pathway in mutant K-ras stably transfected human adrenocortical cells. DNA Cell Biol. 2003;22:657–664. doi: 10.1089/104454903770238139. [DOI] [PubMed] [Google Scholar]
- Cohen-Sfady M, et al. Heat shock protein 60 activates B cells via the TLR4-MyD88 pathway. J Immunol. 2005;175:3594–3602. doi: 10.4049/jimmunol.175.6.3594. [DOI] [PubMed] [Google Scholar]
- Collier RJ, Collier J, Rhoads R, Baumgard L. Invited review: genes involved in the bovine heat stress response. J Dairy Sci. 2008;91:445–454. doi: 10.3168/jds.2007-0540. [DOI] [PubMed] [Google Scholar]
- Collier RJ, Stiening CM, Pollard BC, VanBaale MJ, Baumgard LH, Gentry PC, Coussens PM. Use of gene expression microarrays for evaluating environmental stress tolerance at the cellular level in cattle. J Anim Sci. 2006;84:E1–E13. doi: 10.2527/2006.8413_supplE1x. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva RG, La Scala JN, Tonhati H. Radiative properties of the skin and haircoat of cattle and other animals. Trans ASAE. 2003;46:913. [Google Scholar]
- De Maio A. Heat shock proteins: facts, thoughts, and dreams. Shock. 1999;11:1–12. doi: 10.1097/00024382-199901000-00001. [DOI] [PubMed] [Google Scholar]
- Eicher S, McMunn K, Hammon H, Donkin S. Toll-like receptors 2 and 4, and acute phase cytokine gene expression in dexamethasone and growth hormone treated dairy calves. Vet Immunol Immunopathol. 2004;98:115–125. doi: 10.1016/j.vetimm.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- Fink AL. Chaperone-mediated protein folding. Physiol Rev. 1999;79:425–449. doi: 10.1152/physrev.1999.79.2.425. [DOI] [PubMed] [Google Scholar]
- Freeman BC, Yamamoto KR. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science. 2002;296:2232–2235. doi: 10.1126/science.1073051. [DOI] [PubMed] [Google Scholar]
- Gebremedhin KG, Wu B. A model of evaporative cooling of wet skin surface and fur layer. J Therm Biol. 2001;26:537–545. doi: 10.1016/S0306-4565(00)00048-6. [DOI] [Google Scholar]
- Gu ZT, Wang H, Li L, Liu YS, Deng XB, Huo SF, Yuan FF, Liu ZF, Tong HS, Su L (2014) Heat stress induces apoptosis through transcription-independent p53-mediated mitochondrial pathways in human umbilical vein endothelial cell. Sc Rep 4:4469. doi:10.1038/srep04469 [DOI] [PMC free article] [PubMed]
- Hansen PJ. Effects of heat stress on mammalian reproduction. Philos T Roy Soc B. 2009;364:3341–3350. doi: 10.1098/rstb.2009.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hightower LE. Heat shock, stress proteins, chaperones, and proteotoxicity. Cell. 1991;66:191–197. doi: 10.1016/0092-8674(91)90611-2. [DOI] [PubMed] [Google Scholar]
- Ju X, Xu H, Yong Y, An L, Jiao P, Liao M. Heat stress upregulation of Toll-like receptors 2/4 and acute inflammatory cytokines in peripheral blood mononuclear cell (PBMC) of Bama miniature pigs: an in vivo and in vitro study. Animal. 2014;8:1462. doi: 10.1017/S1751731114001268. [DOI] [PubMed] [Google Scholar]
- Kadzere CT, Murphy MR, Silanikove N, Maltz E. Heat stress in lactating dairy cows: a review. Livest Prod Sci. 2002;77:59–91. doi: 10.1016/S0301-6226(01)00330-X. [DOI] [Google Scholar]
- Kahl S, Elsasser T, Rhoads R, Collier RJ, Baumgard L. Environmental heat stress modulates thyroid status and its response to repeated endotoxin challenge in steers. Domest Anim Endocrinol. 2015;52:43–50. doi: 10.1016/j.domaniend.2015.02.001. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M (2015) KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res :gkv1070 [DOI] [PMC free article] [PubMed]
- Klungland H, Våge DI. Pigmentary switches in domestic animal species. Ann N Y Acad Sci. 2003;994:331–338. doi: 10.1111/j.1749-6632.2003.tb03197.x. [DOI] [PubMed] [Google Scholar]
- Kokavec J, Podskocova J, Zavadil J, Stopka T. Chromatin remodeling and SWI/SNF2 factors in human disease. Front Biosci. 2007;13:6126–6134. doi: 10.2741/3142. [DOI] [PubMed] [Google Scholar]
- Kregel KC. Invited review: heat shock proteins: modifying factors in physiological stress responses and acquired thermotolerance. J Appl Physiol. 2002;92:2177–2186. doi: 10.1152/japplphysiol.01267.2001. [DOI] [PubMed] [Google Scholar]
- Lee WC, Wen HC, Chang CP, Chen MY, Lin M-T. Heat shock protein 72 overexpression protects against hyperthermia, circulatory shock, and cerebral ischemia during heatstroke. J Appl Physiol. 2006;100:2073–2082. doi: 10.1152/japplphysiol.01433.2005. [DOI] [PubMed] [Google Scholar]
- Lee Y-H, et al. Antitumor effects in hepatocarcinoma of isoform-selective inhibition of HDAC2. Cancer Res. 2014;74:4752–4761. doi: 10.1158/0008-5472.CAN-13-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149:1269–1283. doi: 10.1016/j.cell.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Han J, Du F, Ju Z, Huang J, Wang J, Li R, Wang C, Zhong J. Novel SNPs in HSP70A1A gene and the association of polymorphisms with thermo tolerance traits and tissue specific expression in Chinese Holstein cattle. Mol Biol Rep. 2011;38:2657–2663. doi: 10.1007/s11033-010-0407-5. [DOI] [PubMed] [Google Scholar]
- Lindquist S, Craig E. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- Liu Y, Li D, Li H, Zhou X, Wang G. A novel SNP of the ATP1A1 gene is associated with heat tolerance traits in dairy cows. Mol Biol Rep. 2011;38:83–88. doi: 10.1007/s11033-010-0080-8. [DOI] [PubMed] [Google Scholar]
- Liu S, et al. RNA-Seq reveals expression signatures of genes involved in oxygen transport, protein synthesis, folding, and degradation in response to heat stress in catfish. Physiol Genomics. 2013;45:462–476. doi: 10.1152/physiolgenomics.00026.2013. [DOI] [PubMed] [Google Scholar]
- Mani V, Weber TE, Baumgard LH, Gabler NK. Growth and development symposium: endotoxin, inflammation, and intestinal function in livestock. J Anim Sci. 2012;90:1452–1465. doi: 10.2527/jas.2011-4627. [DOI] [PubMed] [Google Scholar]
- Mariasegaram M, Chase C, Chaparro J, Olson T, Brenneman R, Niedz R. The slick hair coat locus maps to chromosome 20 in Senepol-derived cattle. Anim Genet. 2007;38:54–59. doi: 10.1111/j.1365-2052.2007.01560.x. [DOI] [PubMed] [Google Scholar]
- Moré SH, Breloer M, von Bonin A. Eukaryotic heat shock proteins as molecular links in innate and adaptive immune responses: Hsp60-mediated activation of cytotoxic T cells. Int Immunol. 2001;13:1121–1127. doi: 10.1093/intimm/13.9.1121. [DOI] [PubMed] [Google Scholar]
- Moseley PL. Heat shock proteins and heat adaptation of the whole organism. J Appl Physiol. 1997;83:1413–1417. doi: 10.1152/jappl.1997.83.5.1413. [DOI] [PubMed] [Google Scholar]
- Nagradova N. Enzymes catalyzing protein folding and their cellular functions. Curr Protein Petpt Sci. 2007;8:273–282. doi: 10.2174/138920307780831866. [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogalska A, Engel WK, McFerrin J, Kokame K, Komano H, Askanas V. Homocysteine-induced endoplasmic reticulum protein (Herp) is up-regulated in sporadic inclusion-body myositis and in endoplasmic reticulum stress-induced cultured human muscle fibers. J Neurochem. 2006;96:1491–1499. doi: 10.1111/j.1471-4159.2006.03668.x. [DOI] [PubMed] [Google Scholar]
- Odunuga O, Longshaw VM, Blatch GL. Hop: more than an Hsp70/Hsp90 adaptor protein. Bioessays. 2004;26:1058–1068. doi: 10.1002/bies.20107. [DOI] [PubMed] [Google Scholar]
- Okamura Y, et al. The extra domain A of fibronectin activates Toll-like receptor 4. J Biol Chem. 2001;276:10229–10233. doi: 10.1074/jbc.M100099200. [DOI] [PubMed] [Google Scholar]
- Olson TA, Chase Jr CC, Lucena C, Godoy E, Zuniga A, Collier RJ (2006) Effect of hair characteristics on the adaptation of cattle to warm climates. In, 2006. Instituto Prociência, pp 16–07
- Olson TA, Lucena C, Chase CC, Hammond AC. Evidence of a major gene influencing hair length and heat tolerance in cattle. J Anim Sci. 2003;81:80–90. doi: 10.2527/2003.81180x. [DOI] [PubMed] [Google Scholar]
- Osterloh A, Veit A, Gessner A, Fleischer B, Breloer M. Hsp60-mediated T cell stimulation is independent of TLR4 and IL-12. Int Immunol. 2008;20:433–443. doi: 10.1093/intimm/dxn003. [DOI] [PubMed] [Google Scholar]
- Page TJ, Sikder D, Yang L, Pluta L, Wolfinger RD, Kodadek T, Thomas RS. Genome-wide analysis of human HSF1 signaling reveals a transcriptional program linked to cellular adaptation and survival. Mol Biosyst. 2006;2:627–639. doi: 10.1039/b606129j. [DOI] [PubMed] [Google Scholar]
- Panaretou B, et al. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol Cell. 2002;10:1307–1318. doi: 10.1016/S1097-2765(02)00785-2. [DOI] [PubMed] [Google Scholar]
- Perkins N, Gilmore T. Good cop, bad cop: the different faces of NF-κB. Cell Death Differ. 2006;13:759–772. doi: 10.1038/sj.cdd.4401838. [DOI] [PubMed] [Google Scholar]
- Pirkkala L, Nykänen P, Sistonen LEA. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001;15:1118–1131. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- Rahman MB, Kamal MM, Rijsselaere T, Vandaele L, Shamsuddin M, Van Soom A. Altered chromatin condensation of heat-stressed spermatozoa perturbs the dynamics of DNA methylation reprogramming in the paternal genome after in vitro fertilisation in cattle. Reprod Fertil Dev. 2014;26:1107–1116. doi: 10.1071/RD13218. [DOI] [PubMed] [Google Scholar]
- Reimand J, Arak T, Adler P, Kolberg L, Reisberg S, Peterson H, Vilo J (2016) g: Profiler—a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res :gkw199 [DOI] [PMC free article] [PubMed]
- Sakatani M, Balboula AZ, Yamanaka K, Takahashi M. Effect of summer heat environment on body temperature, estrous cycles and blood antioxidant levels in Japanese Black cow. Anim Sci. 2012;83:394–402. doi: 10.1111/j.1740-0929.2011.00967.x. [DOI] [PubMed] [Google Scholar]
- Santhekadur PK, et al. The transcription factor LSF: a novel oncogene for hepatocellular carcinoma. Am J Cancer Res. 2012;2:269. [PMC free article] [PubMed] [Google Scholar]
- Sonna LA, Fujita J, Gaffin SL, Lilly CM. Invited review: effects of heat and cold stress on mammalian gene expression. J Appl Physiol. 2002;92:1725–1742. doi: 10.1152/japplphysiol.01143.2001. [DOI] [PubMed] [Google Scholar]
- St-Pierre NR, Cobanov B, Schnitkey G. Economic losses from heat stress by US livestock industries. J Dairy Sci. 2003;86:E52–E77. doi: 10.3168/jds.S0022-0302(03)74040-5. [DOI] [Google Scholar]
- Strong R, Silva E, Cheng H, Eicher S. Acute brief heat stress in late gestation alters neonatal calf innate immune functions. J Dairy Sci. 2015;98:7771–7783. doi: 10.3168/jds.2015-9591. [DOI] [PubMed] [Google Scholar]
- Sun L, et al. Transcriptome response to heat stress in a chicken hepatocellular carcinoma cell line. Cell Stress Chaperones. 2015;20:939–950. doi: 10.1007/s12192-015-0621-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MC, Chiang C-M. The general transcription machinery and general cofactors. Crit Rev Biochem Mol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- Tudor CO, Schmidt CJ, Vijay-Shanker K. eGIFT: mining gene information from the literature. BMC Bioinformatics. 2010;11:418. doi: 10.1186/1471-2105-11-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umphrey JE, Moss BR, Wilcox CJ, Van Horn HH (2001) Interrelationships in Lactating Holsteins of Rectal and Skin Temperatures, Milk Yield and Composition, Dry Matter Intake, Body Weight, and Feed Efficiency in Summer in Alabama. J Dairy Sci 84(12):2680–2685 [DOI] [PubMed]
- Wada T, Penninger JM. Mitogen-activated protein kinases in apoptosis regulation. Oncogene. 2004;23:2838–2849. doi: 10.1038/sj.onc.1207556. [DOI] [PubMed] [Google Scholar]
- West JW. Effects of heat-stress on production in dairy cattle. J Dairy Sci. 2003;86:2131–2144. doi: 10.3168/jds.S0022-0302(03)73803-X. [DOI] [PubMed] [Google Scholar]
- Weston CR, Davis RJ. The JNK signal transduction pathway. Curr Opin Cell Biol. 2007;19:142–149. doi: 10.1016/j.ceb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Wilkinson B, Gilbert HF (2004) Protein disulfide isomerase. Biochim Biophys Acta Proteins Proteomics 1699(1–2):35–44 [DOI] [PubMed]
- Zanin-Zhorov A, Nussbaum G, Franitza S, Cohen IR, Lider O. T cells respond to heat shock protein 60 via TLR2: activation of adhesion and inhibition of chemokine receptors. FASEB J. 2003;17:1567–1569. doi: 10.1096/fj.02-1139fje. [DOI] [PubMed] [Google Scholar]
- Zhang H-M, Chen H, Liu W, Liu H, Gong J, Wang H, Guo A-Y. AnimalTFDB: a comprehensive animal transcription factor database. Nucleic Acids Res. 2012;40:D144–D149. doi: 10.1093/nar/gkr965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, An H, Xu H, Liu S, Cao X. Heat shock upregulates expression of Toll-like receptor-2 and Toll-like receptor-4 in human monocytes via p38 kinase signal pathway. Immunology. 2005;114:522–530. doi: 10.1111/j.1365-2567.2004.02112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PNG 24 kb)
(DOCX 42 kb)
(XLSX 3577 kb)
(XLSX 31 kb)