Abstract
Extracellular heat shock protein 70 (HSP70) is recognized by receptors on the plasma membrane, such as Toll-like receptor 4 (TLR4), TLR2, CD14, and CD40. This leads to activation of nuclear factor-kappa B (NF-κB), release of pro-inflammatory cytokines, enhancement of the phagocytic activity of innate immune cells, and stimulation of antigen-specific responses. However, the specific characteristics of HSP70 binding are still unknown, and all HSP70 receptors have not yet been described. Putative models for HSP70 complexation to the receptor for advanced glycation endproducts (RAGEs), considering both ADP- and ATP-bound states of HSP70, were obtained through molecular docking and interaction energy calculations. This interaction was detected and visualized by a proximity fluorescence-based assay in A549 cells and further analyzed by normal mode analyses of the docking complexes. The interacting energy of the complexes showed that the most favored docking situation occurs between HSP70 ATP-bound and RAGE in its monomeric state. The fluorescence proximity assay presented a higher number of detected spots in the HSP70 ATP treatment, corroborating with the computational result. Normal-mode analyses showed no conformational deformability in the interacting interface of the complexes. Results were compared with previous findings in which oxidized HSP70 was shown to be responsible for the differential modulation of macrophage activation, which could result from a signaling pathway triggered by RAGE binding. Our data provide important insights into the characteristics of HSP70 binding and receptor interactions, as well as putative models with conserved residues on the interface area, which could be useful for future site-directed mutagenesis studies.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-016-0746-9) contains supplementary material, which is available to authorized users.
Keywords: Heat shock protein 70, Receptor of advanced glycation end products, Signal transduction pathways, Molecular docking, Complexation
Introduction
Within the past decade, extracellular heat shock protein 70 (HSP70) was shown to bind with high affinity to the plasma membrane of antigen-presenting cells (APCs), eliciting a rapid intracellular Ca+2 flux and activating the transcription factor nuclear factor-kappa B (NF-κB). These events are followed by substantial release of various pro-inflammatory cytokines, including tumor necrosis factor (TNF)-α, interleukin (IL)-1β, IL-6, and IL-12, from APCs through Toll-like receptor (TLR) 2-, TLR4-, and CD-14-dependent processes (Asea et al. 2000a). HSP70 can activate various cells of the immune system, including natural-killer (NK) cells (Gross et al. 2003), APCs (Asea et al. 2002), macrophages, peripheral monocytes (Asea et al. 2000a, b), and B lymphocytes (Arnold-Schild et al. 1999). In addition, this interaction and the resulting signal transduction cascade are also mediated by the scavenger receptor CD36 (Delneste et al. 2002) and the costimulatory molecule CD40 (Becker 2002). The CD40/HSP70 interaction appears to be critical to B-lymphocytes, where this interaction promotes the phosphorylation of p38 and subsequent release of TNF-α and interferon (IFN)-γ (Asea 2003). It is also known that these chaperones are targeted by anti-inflammatory regulatory T-cells (Tregs). After an inflammatory event, the enhanced levels of HSP70 enable the immune system to target Tregs to specifically act on the inflammation sites. This phenomenon has been observed in experimental models with both microbial and endogenous heat shock proteins (van Eden 2015). However, the specific characteristics of surface-bound HSP70 receptors are not known, and all potential receptors have not yet been identified. Despite this lack of clear data regarding HSP70 receptors, we do know that this interaction is important because it results in the chaperokine-induced transduction of signals important for the survival of the host against pathogen infection.
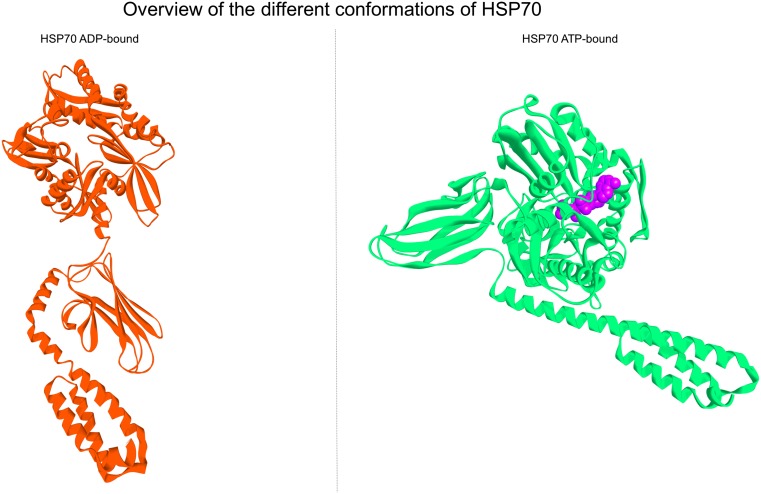
HSP70 is composed by two distinct domains: the N-terminal nucleotide binding domain (NBD), which has ATPase activity, and the C-terminal protein substrate binding domain (SBD), which has two subdomains: a peptide-binding pocket and a helical lid. When the chaperone is in its ATP-bound state, the helical lid is open, allowing the substrate to enter the substrate-binding pocket. The ATP is then hydrolyzed to ADP, and the helical lid closes tightly upon the substrate. To release the substrate, the ADP molecule must be substituted with ATP, which results in subsequent opening of the helical lid. When a new peptide substrate is presented, the ATP is again hydrolyzed, continuing the chaperone binding-release cycle (Sousa 2012). This cycle is partially illustrated by the side by side depiction of the PDB files of 2KHO and 4B9Q (Fig. 1).
Fig. 1.
Overview of the different conformations of HSP70. Left panel shows the depiction of PDB file 2KHO, which represents the HSP70 APO state or ADP-bound; right panel shows the depiction of the pdb file 4B9Q, which represents the HSP70 ATP state. When HSP70 is bound to an ATP molecule, the SBD lid opens, the NBD becomes more contracted and the overall conformation more compact. Upon ATP hydrolysis to ADP, the SBD lid closes, capturing the substrate. This leads to the relaxation of NBD and extension of the overall conformation
In multiple studies, researchers have attempted to assign the biological functions of HSP70 to either the NBD or the SBD. Becker and coworkers showed that HSP70 from different sources binds to different sites on CD40 through different regions of the protein; that is, human recombinant HSP70 binds through the NBD while DnaK (the homolog HSP70 from Escherichia coli) binds through the SBD. Moreover, they showed that this interaction depends on the ADP/ATP cycle (Becker 2002). In another work, hepatocytes treated with the truncated 3′ peptide (SBD) were found to have higher activation parameters when compared with cells treated with the 5′ peptide (NDB) (Galloway et al. 2008). These differences can be attributed to the different receptors involved; furthermore, the biological effects observed in these studies have always been found to be stronger when the cells are treated with the full-length protein rather than only the NBD or the SBD. However, there are no structural data at the atomic level describing the complexation of HSP70 with its target receptors.
A recent study showed that HSP70 could act as a ligand for the receptor of advanced glycation endproducts (RAGEs) (Ruan et al. 2010). RAGE is a multiligand, immunoglobulin-type transmembrane protein that functions as a pattern-recognition receptor (PRR) for the innate immune system. RAGE was initially described based on its affinity for advanced glycation endproducts (AGEs) and was later found to bind a vast array of non-glycated endogenous peptides, both soluble and membrane bound, involved in the host response against tissue damage, infections, and inflammatory processes; these other ligands include HGMB1, S100/calgranulin family members, lipopolysaccharide (LPS), and others (Bucciarelli et al. 2002). Increased expression and activation of RAGE affect a variety of pathological conditions, including diabetes and inflammation. Binding of RAGE induces the activation of multiple signaling pathways that may vary depending on the ligand, cell, and tissue microenvironment and thus mediates diverse cellular responses; activation of NF-κB and mitogen-activated protein kinase (MAPK) is among the key pathways activated by RAGE. These intracellular pathways can lead to production of inflammatory cytokines, activation of proteases, and oxidative stress (Schmidt et al. 2000).
In this study, we developed a putative model of the interaction between HSP70 and RAGE through a series of molecular docking simulations, their interaction energy and conformational changes of the complexes by normal mode analyses. Since few studies have considered the ADP/ATP cycle in this context, we performed docking simulations between RAGE and HSP70 in the ADP/APO or ATP state. Moreover, we confirmed our results using a fluorescence proximity assay on a cell line culture.
Experimental procedures
Chemicals
Purified recombinant human HSP70 (SRP5190) was purchased from Sigma-Aldrich. Antibodies against RAGE were bought from AbCam (Cambridge, UK). Antibodies against HSP70 were purchased from Sigma (St. Louis, MO, USA).
Cell culture
The human non-small cell lung cancer (NSCLC) cell line A549 was grown in RPMI-1640 containing 10% fetal bovine serum (FBS) and maintained at 37 °C in an atmosphere containing 5% CO2. The media was supplemented with 1% penicillin/streptomycin. Cells were trypsinized after reaching 70–90% confluence, counted, and plated.
Protein proximity ligand assay
The detection and visualization of the interaction between HSP70 and RAGE were carried out using a Duolink In Situ kit from Sigma-Aldrich. This assay facilitated the detection, visualization, and quantification of protein interactions in cell samples prepared for microscopy based on the proximity of fluorescent probes to the target primary antibodies. Cells were seeded in 12-well plates with a cover slip on the bottom of each well and then treated with 10 μg HSP70 or 10 μg HSP70 pretreated with 10 μg ATP. After 30 min, the cells were fixed with 4% paraformaldehyde for 30 min. Blocking solution was then added, and cells were incubated for 30 min at 37 °C. Samples were incubated overnight in a humidified chamber with the primary antibodies (anti-RAGE at a 1:2000 dilution; and anti-HSP70 at a 1:2500 dilution). PLA probes were added, and the samples were incubated in a humidified chamber for 60 min at 37 °C. Next, a ligation-ligase solution was added, and the samples were incubated for 30 min at 37 °C. Finally, an amplification-polymerase solution was added, and the samples were incubated for 100 min at 37 °C.
Preparation for imaging
Cover slips were mounted by inversion on a regular microscope slide using a minimal volume of Duolink In Situ Mounting Media with DAPI. Slides were analyzed on a confocal microscope using a ×40 objective. DAPI stained the cell nuclei in blue, whereas the fluorophore from the Detection Reagent kit stained the interaction spots in red, with an excitation of 495 nm and an emission of 624 nm. Images were obtained with ZEN Lite digital imaging software from Carl Zeiss Microscopy GmbH.
Morphometric and intensity of fluorescence analysis
Measurements of intensity of fluorescence and morphometric analysis of cytoplasm and nucleus were obtained using the ImageJ software (NIH, Bethesda, MD), and the intensity of fluorescence/cytoplasm/nucleus ratio of the cells was used as quantification of RAGE-HSP70 interaction.
Molecular docking, structural analyses, and interaction energy calculation
The protein structures under PDB IDs 2KHO (Bertelsen et al. 2009) and 4B9Q (Kityk et al. 2012) were employed as ligands in the molecular docking calculations, corresponding to HSP70 in the ADP/APO and ATP states, respectively. The protein structures under PDB IDs 3CJJ (Koch et al. 2010) and 4LP5 (Yatime and Andersen 2013) were employed as the receptors, corresponding to the V and C1 domains of RAGE (the ligand-binding domain) and to the V, C1, and C2 domains of RAGE, respectively. Additionally, the oligomeric form of RAGE was also retrieved from PDB ID 4LP5. The PQR files used to calculate the electrostatic potential surface of HSP70 were generated by the webserver PDB2PQR (Dolinsky et al. 2004) using the PDB 4B9Q, with the surface potentials being calculated by APBS (Baker et al. 2001), colored and visualized using UCSF Chimera (Pettersen et al. 2004). In order to increase the diversity of docking solutions, calculations were performed on four different macromolecular docking programs: PatchDock (Schneidman-Duhovny et al. 2005), Hex (Macindoe et al. 2010), GRAMM-X (Tovchigrechko and Vakser 2006), and PIPER via ClusPro 2.0 (Comeau et al. 2004; Kozakov et al. 2006). All docking calculations were performed without restrictions; that is, HSP70 protein was free to search the entire RAGE surface for its preferential binding site. Docking results were clustered by their own docking scores and then submitted to an interaction energy calculation by FoldX (Schymkowitz et al. 2005). In this step, we applied the commands Optimize to carry out a quick optimization of the structure, moving all side chains slightly to eliminate small Van der Waals’ clashes, and RepairPDB, which identifies residues with poor torsion angles, Van der Waals’ clashes, or inappropriate total energy and repairs them. The results of each method were used to refine all the docking solutions, allowing us to choose one docking program and one docking structure for each situation (i.e., HSP70 ADP bound to RAGE and HSP70 ATP bound to RAGE).
Normal mode analyses
The selected docking solutions were subjected to normal mode analyses using different methods for assessing major potential conformational changes in the complex: elNémo (Suhre and Sanejouand 2004), iMODS (Lopez-Blanco et al. 2014), and ProDy via the Normal Mode Wizard plug-in for VMD (Bakan et al. 2011). Default parameters recommended for each program were employed. To observe possible conformational transitions between different bound and apo states, only the protein coordinates were considered in the calculations. For clarity, we analyzed the first normal mode from each situation and choose the results from iMODS to be represented. The images from each mode are depicted in BIOVIA Discovery Studio 3.5 (Biovia 2015), and the proteins were colored according to the average isotropic displacement which represents the contribution of each residue to the deformability of the system, where blue corresponds to less flexible regions and red corresponds to more flexible regions. In addition, the ribbon size was also represented to match less flexible or more flexible regions, representing the deformability of the structure. For each structure, we created a corresponding scatter plot of the deformability of each residue.
Predictions on the thermostability effect of mutations in the HSP70-RAGE docking complex
The mutation energy calculation was used to evaluate the effect of single-point mutations on the binding affinity in the HSP70-RAGE docking complex. Mutagenesis was performed using the Pssm command of FoldX, which mutates each selected residue (selected according to the docking results and analyses of the residues present at the interacting interface) to alanine. The stabilities were calculated using the Stability command of FoldX, and ΔΔG values are computed by subtracting the energy of the WT from that of the mutant. Values were interpreted according to Frappier et al. (2015) (Frappier and Najmanovich 2014) as stabilizing mutations (ΔΔG < −0.5 kcal/mol), neutral mutations (ΔΔG [−0.5,0.5] kcal/mol), and destabilizing mutations (ΔΔG > 0.5 kcal/mol).
Results
In order to obtain structural evidence to support the hypothesis that HSP70 binds to RAGE, we first compared the amino acid sequences and secondary structures of known RAGE ligands, such as S100B, HMGB1, glycated albumin, and MAC1 protein, with that of HSP70 in both the ADP- and ATP-bound states (PDBs 2KHO and 4B9Q, respectively). No substantial similarities were found, consistent with a previous study (Fritz 2011). However, as shown in this previous work (40), RAGE ligands are predominantly negatively charged molecules. We then calculated the electrostatic surface potential of HSP70 in its different conformations and found it to be negative (Fig. S1), similar to the abovementioned RAGE ligands. Therefore, in order to determine the relationship between the electrostatic properties of HSP70 and its three-dimensional geometry, we performed a series of molecular docking experiments to investigate the possible interaction between the two molecules.
Docking experiments were performed considering the PDB IDs 2KHO for HSP70 in the ADP-bound state and 4B9Q for HSP70 in the ATP-bound state; 3CJJ was used for monomeric RAGE, and 4LP5 was used for oligomeric RAGE with four different macromolecular docking programs: GRAMM-X, PatchDock, Hex, and PIPER via ClusPro 2.0. All docking calculations were performed without restrictions. Docking results were clustered by their docking scores, and the best solution was then submitted to an interaction energy calculation by FoldX. While each program has its own specific strategies, which may make comparisons difficult, their different results may also represent different points on the potential energy surface associated with the complex, which is depth normalized through FoldX.
The results of each method are shown in Supplemental Table 1 and were used to refine all the docking solutions, allowing us to choose one docking program and one docking structure for each situation based on the lowest energy model or the most stable theoretical interaction. A similar approach to choose docking solutions was successfully demonstrated in a previous study (Varecha et al. 2012). We choose to analyze the calculations with the repaired PDBs in order to correct possible errors remaining from the docking force field and from the X-ray structure. These errors could result in non-standard angles or distances, i.e., biased conformations arising from the non-physiological conditions under which the structure was determined. Moreover, in some cases, these errors could represent average rotamers representing two or more true rotamers in equilibrium (Schymkowitz et al. 2005).
Based on the resulting interaction energy, we choose the docking solutions of the simulations performed on the ClusPro 2.0 server for further analysis, allowing us to establish a putative model for HSP70 and RAGE complexation. Tables 1, 2, 3, and 4 show the values for the first ten docking solutions and an in-depth analyses of the interacting interface between HSP70 and RAGE, which contemplates the number of interacting residues, their interface area, and the number of potential hydrogen bonds, non-covalent and non-bonded interactions. This information was later used to identify conserved residues in both HSP70 ADP and ATP-bound, as well as in RAGE and RAGE in its oligomeric form.
Table 1.
Interacting surface information of the best docking solutions generated by ClusPro 2.0
| Interacting surface information of HSP70 ATP-bound and RAGE | ||||||||
|---|---|---|---|---|---|---|---|---|
| Interaction energy (kcal/mol) | Interface area A (Å2) | Interface area B (Å2) | No. of interacating residues A | No. of interacting residues B | Salt bridges | Hydrogen bounds | Non-bounded contacts | |
| Mode1.00 | −9.44961 | 914 | 874 | 19 | 23 | 4 | 21 | 135 |
| Mode1.01 | −26.4293 | 1397 | 1331 | 26 | 34 | 6 | 19 | 198 |
| Mode1.02 | −11.6721 | 1136 | 1084 | 19 | 22 | 8 | 16 | 106 |
| Mode1.03 | −23.9613 | 1953 | 1902 | 33 | 43 | 7 | 34 | 243 |
| Mode1.04 | −15.463 | 1187 | 1222 | 26 | 25 | 7 | 25 | 192 |
| Mode1.05 | −15.9767 | 1572 | 1462 | 29 | 29 | 8 | 23 | 194 |
| Mode1.06 | −18.2888 | 1331 | 1269 | 24 | 27 | 6 | 14 | 138 |
| Mode1.07 | −15.8098 | 1338 | 1194 | 25 | 33 | 5 | 20 | 143 |
| Mode1.08 | −7.3837 | 1201 | 1057 | 20 | 26 | 4 | 19 | 156 |
| Mode1.09 | −2.42895 | 1654 | 1513 | 25 | 32 | 7 | 21 | 160 |
| Mean | −14.686 | 1368.3 | 1290.8 | 24.6 | 29.4 | 6.2 | 21.2 | 166.5 |
Chain A represents HSP70 ATP-bound (PDB/4B9Q) and chain B represents RAGE in the monomeric form (PDB/3CJJ)
Table 2.
Interacting surface information of the best docking solutions generated by ClusPro 2.0
| Interacting surface information of HSP70 ATP-bound and RAGE oligomeric | ||||||||
|---|---|---|---|---|---|---|---|---|
| Interaction energy (kcal/mol) | Interface area A (Å2) | Interface area B (Å2) | No. of interacating residues A | No. of interacting residues B | Salt bridges | Hydrogen bounds | Non-bounded contacts | |
| Mode1.00 | −22.3043 | 1695 | 1500 | 22 | 34 | 11 | 32 | 207 |
| Mode1.01 | −7.36259 | 1483 | 1518 | 30 | 26 | 7 | 22 | 199 |
| Mode1.02 | −13.4254 | 1304 | 1271 | 17 | 25 | 5 | 23 | 152 |
| Mode1.03 | −4.89881 | 1056 | 997 | 17 | 24 | 11 | 22 | 161 |
| Mode1.04 | −11.0521 | 1328 | 1115 | 18 | 30 | 6 | 21 | 124 |
| Mode1.05 | −17.934 | 1342 | 1191 | 22 | 29 | 12 | 26 | 179 |
| Mode1.06 | −7.39764 | 1203 | 1145 | 25 | 22 | 8 | 18 | 142 |
| Mode1.07 | −20.0421 | 1474 | 1422 | 27 | 30 | 9 | 28 | 199 |
| Mode1.08 | −3.08451 | 1142 | 1064 | 20 | 23 | 6 | 15 | 154 |
| Mode1.09 | −14.6118 | 1326 | 1230 | 29 | 31 | 11 | 23 | 203 |
| Mean | −12.211325 | 1335.3 | 1245.3 | 22.7 | 27.4 | 8.6 | 23 | 172 |
Chain A represents RAGE in the oligomeric form (PDB.4LP5) and chain B represents HSP70 ATP-bound (PDB.4B9Q)
Table 3.
Interacting surface information of the best docking solutions generated by ClusPro 2.0
| Interacting surface information of HSP70 ADP-bound and RAGE | ||||||||
|---|---|---|---|---|---|---|---|---|
| Interaction energy (kcal/mol) | Interface area A (Å2) | Interface area B (Å2) | No. of interacting residues A | No of interacting residues B | Salt bridges | Hydrogen bounds | Non-bounded contracts | |
| Mode1.00 | −13.9867 | 1527 | 1467 | 27 | 31 | 9 | 23 | 154 |
| Mode1.01 | −14.9640 | 1317 | 1249 | 24 | 28 | 7 | 27 | 173 |
| Mode1.02 | −13.5191 | 1175 | 1124 | 19 | 22 | 2 | 16 | 154 |
| Mode1.03 | −10.0052 | 1244 | 1182 | 18 | 25 | 4 | 14 | 125 |
| Mode1.04 | −17.3068 | 1194 | 1082 | 19 | 27 | 7 | 23 | 180 |
| Mode1.05 | −8.7484 | 1029 | 1115 | 22 | 19 | 6 | 12 | 119 |
| Mode1.06 | −13.9595 | 1699 | 1417 | 24 | 34 | 4 | 25 | 179 |
| Mode1.07 | −11.0688 | 989 | 1009 | 21 | 21 | 4 | 17 | 139 |
| Mode1.08 | −8.2834 | 1431 | 1551 | 28 | 28 | 7 | 21 | 235 |
| Mode1.09 | −13.1513 | 1438 | 1332 | 24 | 30 | 6 | 16 | 181 |
| Mean | −12.549 | 1304.3 | 1252.8 | 22.6 | 26.5 | 5.6 | 19.4 | 163.9 |
Chain A represents RAGE in the monomeric form (PDB/3CJJ) and chain B represents HSP70 ADP-bound (PDB/2KHO)
Table 4.
Interacting surface information of the best docking solutions generated by ClusPro 2.0
| Interacting surface information of HSP70 ADP-bound and RAGE oligomeric | ||||||||
|---|---|---|---|---|---|---|---|---|
| Interaction energy (kcal/mol) | Interface area A (Å2) | Interface area B (Å2) | No. of interacting residues A | No. of interacting residues B | Salt bridges | Hydrogen bounds | Non-bounded Contracts | |
| Mode1.00 | −18.4109 | 1610 | 1684 | 33 | 32 | 10 | 29 | 220 |
| Mode1.01 | −24.6075 | 1369 | 1372 | 30 | 26 | 11 | 33 | 186 |
| Mode1.02 | −15.4286 | 1256 | 1558 | 25 | 21 | 4 | 19 | 127 |
| Mode1.03 | −16.3374 | 1340 | 1203 | 25 | 33 | 4 | 23 | 145 |
| Mode1.04 | −16.2707 | 2212 | 2198 | 34 | 48 | 8 | 30 | 306 |
| Mode1.05 | −8.83572 | 1960 | 1721 | 28 | 44 | 12 | 32 | 254 |
| Mode1.06 | −9.42297 | 1834 | 1826 | 34 | 33 | 10 | 19 | 107 |
| Mode1.07 | −7.12172 | 1673 | 1610 | 35 | 31 | 4 | 19 | 157 |
| Mode1.08 | −13.2064 | 1622 | 1631 | 27 | 32 | 7 | 20 | 178 |
| Mode1.09 | −11.1987 | 1686 | 1670 | 36 | 37 | 4 | 23 | 202 |
| Mean | −14.084061 | 1656.2 | 1647.3 | 30.7 | 33.5 | 7.4 | 24.7 | 188.2 |
Chain A represents RAGE in the oligomeric form (PDB/4LP5) and chain B represents HSP70 ADP-bound (PDB/2KHO)
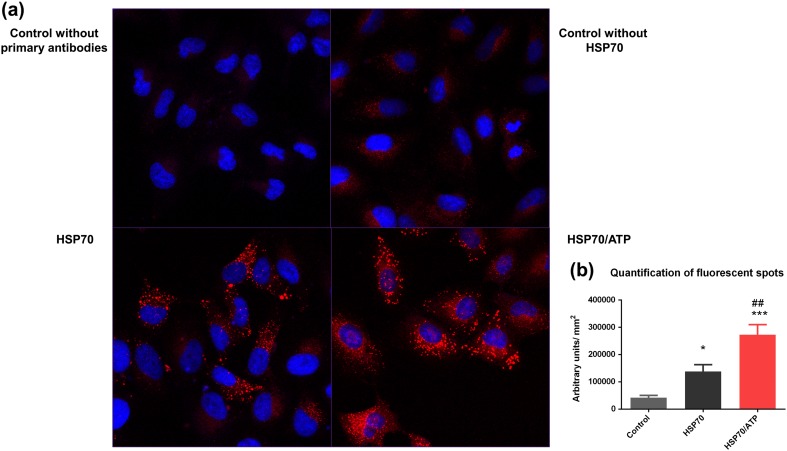
In order to confirm that the interaction occurred in vitro, we performed protein interaction assays (Duolink In Situ assays) based on the proximity of the two target proteins using A459 cells, which express high levels of RAGE (Nakano et al. 2006). In this assay, a pair of oligonucleotide-labeled secondary antibodies generates a signal only when the two PLA probes are bound in close proximity to both primary antibodies bound to the sample. Thus, the signal from each detected pair of PLA probes is visualized as a single fluorescent spot. Using confocal microscopy, we found that samples treated with HSP70 exhibited a large number of red fluorescent spots in comparison with the samples that were not incubated with HSP70. When comparing the two experimental groups, the one treated with HSP70 preincubated with ATP showed a higher number of red fluorescent spots in relation to the group treated with only HSP70, as can be seen by both confocal photomicrographs (Fig. 2a) and quantification of the red fluorescent spots (Fig. 2b). These results suggested that the specific conformation of the protein in the extracellular space may be important for the interaction with RAGE. The interaction with RAGE may be more probable and favored in this conformation, whereas in the ADP/APO conformation, the protein may interact with its other receptors with higher affinity, thus creating a possible mechanism that allows a shift in the later cellular signaling cascade.
Fig. 2.
Fluorescent protein proximity assay. HLA probe chain reacted, producing red fluorescent spots when both target proteins were within interacting range. a The top images show confocal microscopy analyses of technical control without the addition of primary antibodies and experimental control without the addition of eHSP70. The bottom images show confocal microscopy analyses of A549 cells treated with eHSP70 with or without ATP pretreatment. b Fluorescence intensity and morphometric analysis from control and treated cells. The cells were analyzed using the ratio between the intensity of red fluorescence and size of cytoplasm with the ImageJ software. Data are reported as means ± SEM. Statistically significant differences as determined by one-way ANOVA followed by Tukey’s test are indicated: *p < 0.05; ***p < 0.001 compared with control group; ##p < 0.01 compared with HSP70 group
After detecting and visualizing the interaction by confocal microscopy, we analyzed the best docking solutions (that is, the docking complexes with the lowest interaction energy values) via normal mode analyses in order to observe if the complex underwent any major conformational shifts. Normal mode analysis is a computational technique that allows us to analyze the dynamics of biological macromolecules. This method can provide insights into the large-scale conformational changes occurring in proteins, e.g., upon ligand binding or interaction and complexation, such as the proposed interaction between HSP70 and RAGE. The lowest modes are generally thought to dominantly contribute to the global and collective motions of the entire system (Bahar et al. 2010; Mahajan and Sanejouand 2015).
All files involved in the docking simulations were subjected to normal mode analyses prior to complexation, allowing their conformational profiles to be compared under different situations after the interaction. 2KHO, representing the HSP70 ADP-bound, showed a highly flexible region located in the linker between the NBD and the SBD. In addition, the first residues of the helical lid showed high flexibility, consistent with the observation that this region acts as a hinge and is responsible for the opening and closing of the lid over the β-sheet pocket (Fig. S2). The slowest mode also showed that the two domains could rotate in opposite directions, making the protein a not very likely ligand, at least in this conformation. In the model of HSP70 bound to an ATP molecule, the helical lid within the protein was open; however, the overall structure exhibited a more closed, tightened state. Flexible regions were located within the helical lid (Fig. S2).
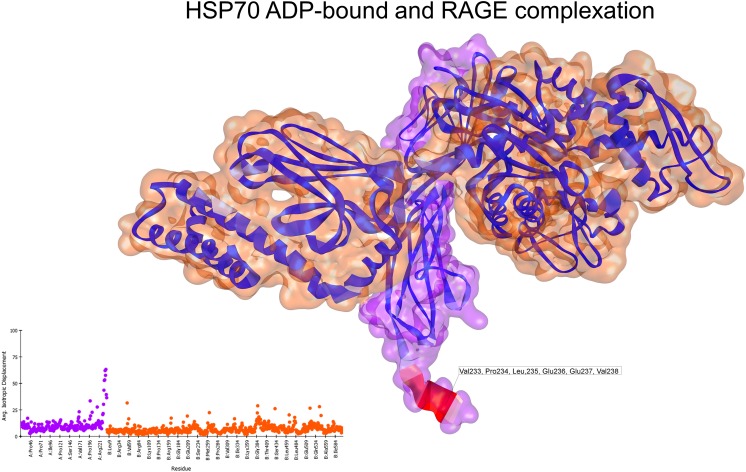
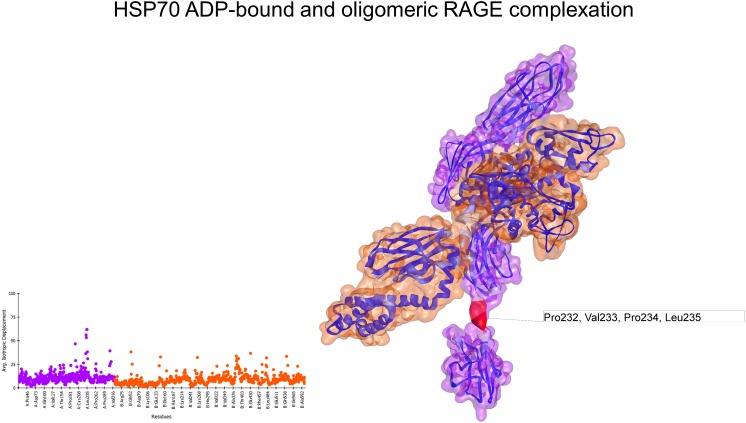
In its monomeric form (3CJJ), RAGE exhibited only a highly flexible region at the end of the C1 domain. On the other hand, the oligomeric form (4LP5) showed several spikes on the deformability profile of the residues; the most flexible residues were located in the region between the C1 and C2 domains and in the end of the C2 domain (Fig. S3). Normal mode analyses of the docked complex of ADP-bound HSP70 and monomeric RAGE showed that the regions with high flexibility in HSP70 in the ADP-bound state had reduced flexibility after interacting with RAGE. Additionally, the interaction interface showed no major conformational changes or movement (Fig. 3), with an associated interaction energy of −17.80 kcal/mol. In contrast, the complex between HSP70 bound to ADP and oligomeric RAGE exhibited several residues with moderate to high flexibility, as is shown in the depiction and deformability plot (Fig. 4). Nevertheless, the interaction energy (−26.42 kcal/mol) was lower than that of the ATP-bound HSP70 and monomeric RAGE.
Fig. 3.
Normal mode analyses for the complex 3CJJ-2KHO. Structures are shown according to their respective residue deformability. The color scale ranges from blue to red with increasing flexibility of residues. Every structure includes a corresponding scatter plot for residue deformability. Analysis of the interacting complex between the HSP70 APO state and monomeric RAGE showed that the most flexible regions of the ligand structure were relaxed after the interaction. RAGE surface is colored purple and HSP70 surface is colored orange. Residues that most contribute to the deformability of the system are indicated
Fig. 4.
Normal mode analyses for the complex 4LP5-2KHO. Structures are shown according to their respective residue deformability. The color scale ranges from blue to red with increasing flexibility of residues. Every structure includes a corresponding scatter plot for residue deformability. Both structures exhibited several spikes of residue deformability. RAGE surface is colored purple and HSP70 surface is colored orange. Residues that most contribute to the deformability of the system are indicated
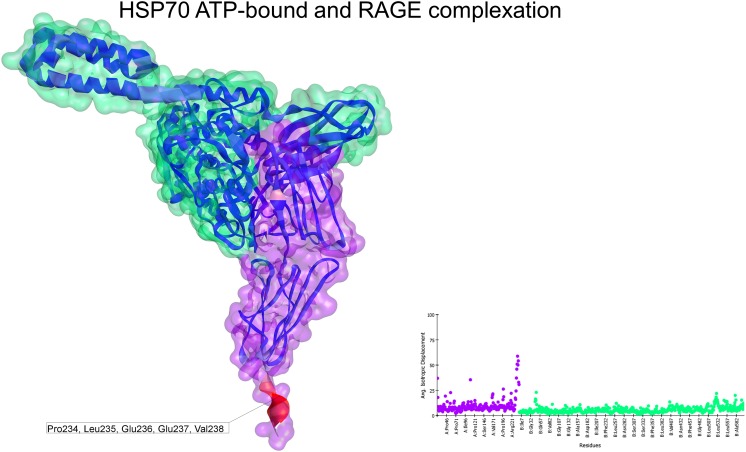
The slowest normal mode obtained from the docking between HSP70 in the ATP-bound state (4B9Q) and monomeric RAGE (3CJJ) exhibited an improved deformability plot. Although the helical lid of HSP70 and the C1 domain of RAGE were responsible for some degree of movement, the vast majority of the residues, including those located on the interacting interface, had less flexibility than that observed for the other combinations (Fig. 5). This docking solution had an interacting energy of −22.30 kcal/mol.
Fig. 5.
Normal mode analyses for the complex 3CJJ-4B9Q. Structures are shown according to their respective residue deformability. The color scale ranges from blue to red with increasing flexibility of residues. Every structure includes a corresponding scatter plot for residue deformability. There was a highly flexible region at the end of the C1 domain of the receptor and a moderately flexible region on the helical lid of HSP70. Overall, this complex exhibited a flattened profile of residue deformability and flexibility compared with the other complexes. Additionally, the interacting region was colored blue, indicating the relative stability of this complex. RAGE surface is colored purple and HSP70 surface is colored green. Residues that most contribute to the deformability of the system are indicated
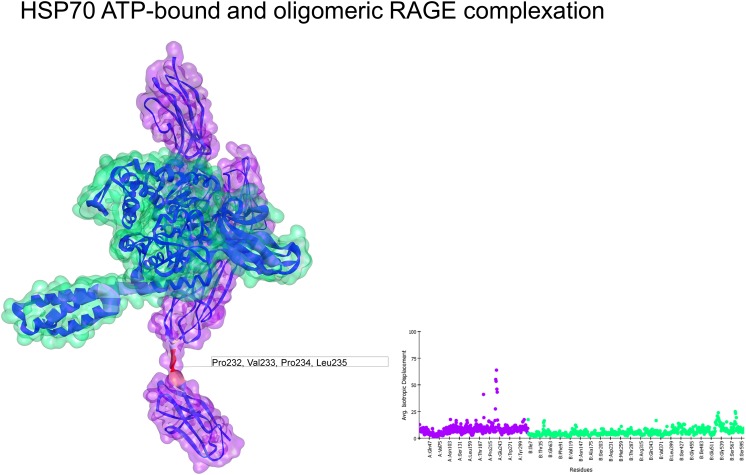
The structure 4LP5 had a deformability profile similar to that observed for the interaction with ADP-bound HSP70. Initially, the oligomeric conformation did not appear to be favorable for any complexation; the structure of HSP70 under these conditions had a higher deformability profile with some spikes in the flexibility of its residues. Moreover, the interaction energy (−26.42 kcal/mol) was lower than that of the monomeric form (Fig. 6).
Fig. 6.
Normal mode analyses for the complex 4LP5-4B9q. Structures are shown according to their respective residue deformability. The color scale ranges from blue to red with increasing flexibility of residues. Every structure includes a corresponding scatter plot for residue deformability. RAGE surface is colored purple and HSP70 surface is colored green. Residues that most contribute to the deformability of the system are indicated
RAGE residues GLN24, LYS37, ARG216, and ARG218 are present in all docking complexes analyzed, while HSP70 residues GLU206, GLN217, and ASN187 could be important for the interaction of the two proteins and could be targeted for site-directed mutagenesis. A complete list of the interacting residues that form potential hydrogen bonds for every docking situation is presented together with a representation of the structures on supplemental Figs. S4–S7.
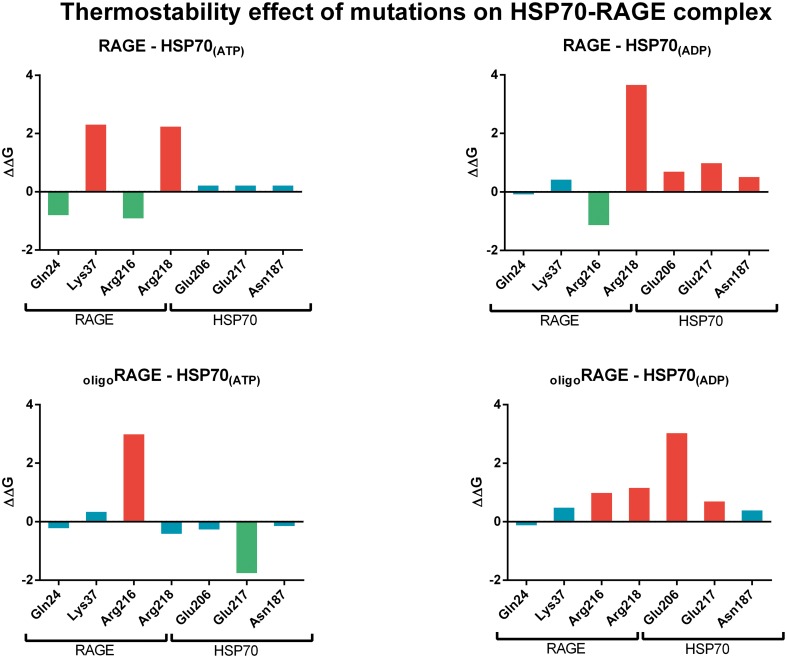
The effect of mutations on the thermostability of the HSP70-RAGE docking complex showed that some residues identified to be present on the interacting interface seemed to have an important role on the stability. Mutation of Lys37 and Arg218 of monomeric RAGE resulted on a destabilizing effect when interacting with HSP70 ATP-bound and Arg218 when interacting to HSP70 ADP-bound. Mutation of Arg216 of oligomeric RAGE resulted on a destabilizing effect when the receptor interacts with HSP70 ATP-bound, and mutation of Arg216 and Arg218 resulted on a destabilizing effect when the interaction occurs with HSP70 ADP-bound. HSP70 on its ATP-bound conformation has not been affected by destabilizing mutations on any of the residues, only a mutation on Glu217 resulted on a stabilizing effect when the chaperone interacted with oligomeric RAGE. In contrast, all amino acid mutations resulted on a destabilizing effect on HSP70 in the ADP-bound conformation, with the exception of Asn187 when the chaperone interacted with oligomeric RAGE (Fig. 7).
Fig. 7.
FoldX energy results and the effect on the thermostability of the HSP70-RAGE docking complex. Stabilizing mutations (ΔΔG < −0.5 kcal/mol) are represented by green-colored bars, neutral mutations (ΔΔG [−0.5,0.5] kcal/mol) by blue bars, and destabilizing mutations (ΔΔG > 0.5 kcal/mol) are represented by red-colored bars
Discussion
HSP70 was first identified many years ago (Ritossa 1962), and many studies have shown that this protein has a variety of functions in addition to its well-known activity as a chaperone protein. Recently, HSP70 has been shown to be present in serum after induction of different types of stress, such as heavy exercise, inflammation, septic shock, and trauma (Kindas-Mugge et al. 1993; Trautinger et al. 1996a, b). Initially, this extracellular HSP70 was thought to arise from necrotic disruption of cell membranes and extravasation of cytosolic proteins. However, later studies showed that HSP70 could be actively secreted from the intracellular environment to the extracellular space (Vega et al. 2008), suggesting that this protein may have a more prominent role in signal transduction during pathogenic processes.
In a recent study by our laboratory (Gelain et al. 2011), we showed that there is a correlation between clinical outcomes in patients with sepsis and the presence of HSP70 in their serum. Moreover, increased levels of oxidative stress were also associated with increased HSP70 levels in serum in patients who died. Our previous studies (Grunwald et al. 2014) showed that macrophages treated with preoxidized HSP70 exhibited a different pattern of activation compared with that of macrophages treated with HSP70, with lower cell proliferation and viability, lower levels of TNF-α release, and lower phagocytic activity. Similar results, including increased secretion of TNF-α and increased phagocytosis, have been observed in macrophages upon RAGE ligand binding (Fujiya et al. 2014; Ma et al. 2012; Tang et al. 2010), suggesting that RAGE may have unidentified interactions associated with specific signaling pathways.
In this study, we used in silico experiments to confirm the possible interaction between HSP70 and RAGE and to define the mechanism through which such a complexation occurs. The computational simulations provided herein served as guidelines for the additional steps in our experiment design, and the putative models obtained by molecular docking could improve our understanding of this interaction, acting as a baseline for site-directed mutagenesis studies until the exact complex may be obtained through X-ray crystallography. Despite its widespread use, docking has been considered an unreliable technique for the prediction of drug binding (Chen 2015), and macromolecular docking, such as the protein-protein binding predicted here, has improved greatly in the last decade (Musiani and Ciurli 2015; Park et al. 2015).
Theoretically, it is possible for HSP70 in both the ADP/APO and ATP states to complex with RAGE; normal mode analyses showed that the interaction interface did not undergo any major conformational shifts and that the residues in this area did not exhibit elevated flexibility. When these simulations were compared with experiments in actual cells, increased numbers of fluorescent-interacting spots were observed when HSP70 was added in the presence of ATP compared with that when HSP70 was added alone. This is consistent with the observed lowest calculated interacting energy for ATP-bound HSP70 and oligomeric RAGE (PDB IDs 4B9Q and 4PL5, respectively). From these observations, we can form hypotheses regarding the conditions under which such an interaction could occur. RAGE has four disulfide bonds when in the monomeric form and ten disulfide bonds in the oligomeric form. In the case of 4LP5, oligomerization occurs between the V domains of the receptor, meaning that the soluble form could interact and complex under the right conditions. This oligomerization can be extended to more than two molecules, increasing the interacting site and creating a larger area capable of interacting with larger proteins, such as HSP70. The abovementioned disulfide bonds could play an important role under pathological conditions involving excessive oxidative stress because these structures could help to stabilize some interactions and particular conformations. Moreover, in all situations, we observed the involvement of different domains of HSP70 when the protein interacted with the receptor.
Our results not only supported the observations made by Ruan and collaborators (12), who showed that there may be an interaction between RAGE and HSP70, but also showed that this interaction occurred within a cell culture model. Thus, our findings demonstrated that the ADP/ATP cycle may be important for mediating the affinity and stability of the interaction and that the different domains of HSP70 may contribute to the strength of the signaling response initiated upon RAGE binding.
This new interaction could have important implications in future studies of the pathological conditions associated with high levels of RAGE expression and the presence of HSP70 in the extracellular medium and serum in response to some form of injury or stress.
Electronic supplementary materials
(DOCX 4606 kb)
Acknowledgements
This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS), CAPES, and Propesq-UFRGS. The authors thank the Electron Microscopy Center of the Federal University of Rio Grande do Sul for their assistance with the microscopy analyses. The authors also thank Mr. Henrique Biehl for his technical assistance.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Author contribution
JCFM and HV conceived and coordinated the study. DPG helped conceive the idea. MSG designed, performed, and analyzed all the experiments, as well as wrote the manuscript and prepared all the figures. RLB provided technical assistance for normal mode analyses and contributed with the writing of the manuscript. CSS and LH provided technical assistance and contributed to the preparation of Fig. 2. All authors reviewed the results and approved the final version of the manuscript.
References
- Arnold-Schild D, Hanau D, Spehner D, Schmid C, Rammensee HG, de la Salle H, Schild H. Cutting edge: receptor-mediated endocytosis of heat shock proteins by professional antigen-presenting cells. J Immunol. 1999;162(7):3757–3760. [PubMed] [Google Scholar]
- Asea A. Chaperokine-induced signal transduction pathways. Exerc Immunol Rev. 2003;9:25–33. [PMC free article] [PubMed] [Google Scholar]
- Asea A, Kabingu E, Stevenson MA, Calderwood SK. HSP70 peptidem bearing and peptide-negative preparations act as chaperokines. Cell Stress Chaperones. 2000;5(5):425–431. doi: 10.1379/1466-1268(2000)005<0425:HPBAPN>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asea A, Kraeft SK, Kurt-Jones EA, Stevenson MA, Chen LB, Finberg RW, Koo GC, Calderwood SK. HSP70 stimulates cytokine production through a CD14-dependant pathway, demonstrating its dual role as a chaperone and cytokine. Nat Med. 2000;6:435–442. doi: 10.1038/74697. [DOI] [PubMed] [Google Scholar]
- Asea A, Rehli M, Kabingu E, Boch JA, Bare O, Auron PE, Stevenson MA, Calderwood SK. Novel signal transduction pathway utilized by extracellular HSP70: role of toll-like receptor (TLR) 2 and TLR4. J Biol Chem. 2002;277(17):15028–15034. doi: 10.1074/jbc.M200497200. [DOI] [PubMed] [Google Scholar]
- Bahar I, Lezon TR, Bakan A, Shrivastava IH. Normal mode analysis of biomolecular structures: functional mechanisms of membrane proteins. Chem Rev. 2010;110(3):1463–1497. doi: 10.1021/cr900095e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakan A, Meireles LM, Bahar I. ProDy: protein dynamics inferred from theory and experiments. Bioinformatics. 2011;27(11):1575–1577. doi: 10.1093/bioinformatics/btr168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T. CD40, an extracellular receptor for binding and uptake of Hsp70-peptide complexes. J Cell Biol. 2002;158(7):1277–1285. doi: 10.1083/jcb.200208083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ER. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc Natl Acad Sci U S A. 2009;106(21):8471–8476. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biovia, D. S. (2015). Discovery studio modeling environment (version 4.5). San Diego
- Bucciarelli LG, Wendt T, Rong L, Lalla E, Hofmann MA, Goova MT, Taguchi A, Yan SF, Yan SD, Stern DM, Schmidt AM. RAGE is a multiligand receptor of the immunoglobulin superfamily: implications for homeostasis and chronic disease. Cell Mol Life Sci. 2002;59(7):1117–1128. doi: 10.1007/s00018-002-8491-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC. Beware of docking! Trends Pharmacol Sci. 2015;36(2):78–95. doi: 10.1016/j.tips.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: a fully automated algorithm for protein-protein docking. Nucleic Acids Res. 2004;32(Web Server issue):W96–W99. doi: 10.1093/nar/gkh354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delneste Y, Magistrelli G, Gauchat J, Haeuw J, Aubry J, Nakamura K, Kawakami-Honda N, Goetsch L, Sawamura T, Bonnefoy J, Jeannin P. Involvement of LOX-1 in dendritic cell-mediated antigen cross-presentation. Immunity. 2002;17(3):353–362. doi: 10.1016/S1074-7613(02)00388-6. [DOI] [PubMed] [Google Scholar]
- Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32(Web Server issue):W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frappier V, Najmanovich RJ. A coarse-grained elastic network atom contact model and its use in the simulation of protein dynamics and the prediction of the effect of mutations. PLoS Comput Biol. 2014;10(4):e1003569. doi: 10.1371/journal.pcbi.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frappier V, Chartier M, Najmanovich RJ. ENCoM server: exploring protein conformational space and the effect of mutations on protein function and stability. Nucleic Acids Res. 2015;43(W1):W395–W400. doi: 10.1093/nar/gkv343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz G. RAGE: a single receptor fits multiple ligands. Trends Biochem Sci. 2011;36(12):625–632. doi: 10.1016/j.tibs.2011.08.008. [DOI] [PubMed] [Google Scholar]
- Fujiya A, Nagasaki H, Seino Y, Okawa T, Kato J, Fukami A, Himeno T, Uenishi E, Tsunekawa S, Kamiya H, Nakamura J, Oiso Y, Hamada Y. The role of S100B in the interaction between adipocytes and macrophages. Obesity (Silver Spring) 2014;22(2):371–379. doi: 10.1002/oby.20532. [DOI] [PubMed] [Google Scholar]
- Galloway E, Shin T, Huber N, Eismann T, Kuboki S, Schuster R, Blanchard J, Wong HR, Lentsch AB. Activation of hepatocytes by extracellular heat shock protein 72. Am J Physiol Cell Physiol. 2008;295(2):C514–C520. doi: 10.1152/ajpcell.00032.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelain DP, de Bittencourt Pasquali MA, Comin MC, Grunwald MS, Ritter C, Tomasi CD, Alves SC, Quevedo J, Dal-Pizzol F, Moreira JC. Serum heat shock protein 70 levels, oxidant status, and mortality in sepsis. Shock. 2011;35(5):466–470. doi: 10.1097/SHK.0b013e31820fe704. [DOI] [PubMed] [Google Scholar]
- Gross C, Schmidt-Wolf IG, Nagaraj S, Gastpar R, Ellwart J, Kunz-Schughart LA, Multhoff G. Heat shock protein 70-reactivity is associated with increased cell surface density of CD94/CD56 on primary natural killer cells. Cell Stress Chaperones. 2003;8(4):348–360. doi: 10.1379/1466-1268(2003)008<0348:HSPRIA>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald MS, Pires AS, Zanotto-Filho A, Gasparotto J, Gelain DP, Demartini DR, Schöler CM, de Bittencourt PI, Jr, Moreira JC. The oxidation of HSP70 is associated with functional impairment and lack of stimulatory capacity. Cell Stress Chaperones. 2014;19(6):913–925. doi: 10.1007/s12192-014-0516-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindas-Mugge I, Hammerle AH, Frohlich I, Oismuller C, Micksche M, Trautinger F. Granulocytes of critically ill patients spontaneously express the 72 kD heat shock protein. Circ Shock. 1993;39(4):247–252. [PubMed] [Google Scholar]
- Kityk R, Kopp J, Sinning I, Mayer MP. Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol Cell. 2012;48(6):863–874. doi: 10.1016/j.molcel.2012.09.023. [DOI] [PubMed] [Google Scholar]
- Koch M, Chitayat S, Dattilo BM, Schiefner A, Diez J, Chazin WJ, Fritz G. Structural basis for ligand recognition and activation of RAGE. Structure. 2010;18(10):1342–1352. doi: 10.1016/j.str.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov D, Brenke R, Comeau SR, Vajda S. PIPER: an FFT-based protein docking program with pairwise potentials. Proteins. 2006;65(2):392–406. doi: 10.1002/prot.21117. [DOI] [PubMed] [Google Scholar]
- Lopez-Blanco JR, Aliaga JI, Quintana-Orti ES, Chacon P. iMODS: internal coordinates normal mode analysis server. Nucleic Acids Res. 2014;42(Web Server issue):W271–W276. doi: 10.1093/nar/gku339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Rai V, Hudson BI, Song F, Schmidt AM, Barile GR. RAGE binds C1q and enhances C1q-mediated phagocytosis. Cell Immunol. 2012;274(1–2):72–82. doi: 10.1016/j.cellimm.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Macindoe G, Mavridis L, Venkatraman V, Devignes MD, Ritchie DW. HexServer: an FFT-based protein docking server powered by graphics processors. Nucleic Acids Res. 2010;38(Web Server issue):W445–W449. doi: 10.1093/nar/gkq311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan S, Sanejouand YH. On the relationship between low-frequency normal modes and the large-scale conformational changes of proteins. Arch Biochem Biophys. 2015;567:59–65. doi: 10.1016/j.abb.2014.12.020. [DOI] [PubMed] [Google Scholar]
- Musiani F, Ciurli S. Evolution of macromolecular docking techniques: the case study of nickel and iron metabolism in pathogenic bacteria. Molecules. 2015;20(8):14265–14292. doi: 10.3390/molecules200814265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano N, Fukuhara-Takaki K, Jono T, Nakajou K, Eto N, Horiuchi S, Takeya M, Nagai R. Association of advanced glycation end products with A549 cells, a human pulmonary epithelial cell line, is mediated by a receptor distinct from the scavenger receptor family and RAGE. J Biochem. 2006;139(5):821–829. doi: 10.1093/jb/mvj092. [DOI] [PubMed] [Google Scholar]
- Park H, Lee H, Seok C. High-resolution protein-protein docking by global optimization: recent advances and future challenges. Curr Opin Struct Biol. 2015;35:24–31. doi: 10.1016/j.sbi.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Ritossa F. A new puffing pattern induced by temperature shock and DNP in drosophila. Experientia. 1962;18(12):571–573. doi: 10.1007/BF02172188. [DOI] [Google Scholar]
- Ruan BH, Li X, Winkler AR, Cunningham KM, Kuai J, Greco RM, Nocka KH, Fitz LJ, Wright JF, Pittman DD, Tan XY, Paulsen JE, Lin LL, Winkler DG. Complement C3a, CpG oligos, and DNA/C3a complex stimulate IFN-alpha pzcation end product-dependent manner. J Immunol. 2010;185(7):4213–4222. doi: 10.4049/jimmunol.1000863. [DOI] [PubMed] [Google Scholar]
- Schmidt AM, Hofmann M, Taguchi A, Yan SD, Stern DM. RAGE: a multiligand receptor contributing to the cellular response in diabetic vasculopathy and inflammation. Semin Thromb Hemost. 2000;26(5):485–493. doi: 10.1055/s-2000-13204. [DOI] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33(Web Server issue):W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33(Web Server issue):W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa R. A dancer caught midstep: the structure of ATP-bound Hsp70. Mol Cell. 2012;48(6):821–823. doi: 10.1016/j.molcel.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K, Sanejouand YH. ElNemo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 2004;32(Web Server issue):W610–W614. doi: 10.1093/nar/gkh368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Loze MT, Zeh HJ, Kang R. The redox protein HMGB1 regulates cell death and survival in cancer treatment. Autophagy. 2010;6(8):1181–1183. doi: 10.4161/auto.6.8.13367. [DOI] [PubMed] [Google Scholar]
- Tovchigrechko A, Vakser IA. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 2006;34(Web Server issue):W310–W314. doi: 10.1093/nar/gkl206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautinger F, Kindas-Mugge I, Knobler RM, Honigsmann H. Stress proteins in the cellular response to ultraviolet radiation. J Photochem Photobiol B. 1996;35(3):141–148. doi: 10.1016/S1011-1344(96)07344-7. [DOI] [PubMed] [Google Scholar]
- Trautinger F, Knobler RM, Honigsmann H, Mayr W, Kindas-Mugge I. Increased expression of the 72-kDa heat shock protein and reduced sunburn cell formation in human skin after local hyperthermia. J Invest Dermatol. 1996;107(3):442–443. doi: 10.1111/1523-1747.ep12365498. [DOI] [PubMed] [Google Scholar]
- van Eden W. Diet and the anti-inflammatory effect of heat shock proteins. Endocr Metab Immune Disord Drug Targets. 2015;15(1):31–36. doi: 10.2174/1871530314666140922145333. [DOI] [PubMed] [Google Scholar]
- Varecha M, Potesilova M, Matula P, Kozubek M. Endonuclease G interacts with histone H2B and DNA topoisomerase II alpha during apoptosis. Mol Cell Biochem. 2012;363(1–2):301–307. doi: 10.1007/s11010-011-1182-x. [DOI] [PubMed] [Google Scholar]
- Vega VL, Rodriguez-Silva M, Frey T, Gehrmann M, Diaz JC, Steinem C, Multhoff G, Arispe N, De Maio A. Hsp70 translocates into the plasma membrane after stress and is released into the extracellular environment in a membrane-associated form that activates macrophages. J Immunol. 2008;180(6):4299–4307. doi: 10.4049/jimmunol.180.6.4299. [DOI] [PubMed] [Google Scholar]
- Yatime L, Andersen GR. Structural insights into the oligomerization mode of the human receptor for advanced glycation end-products. FEBS J. 2013;280(24):6556–6568. doi: 10.1111/febs.12556. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 4606 kb)