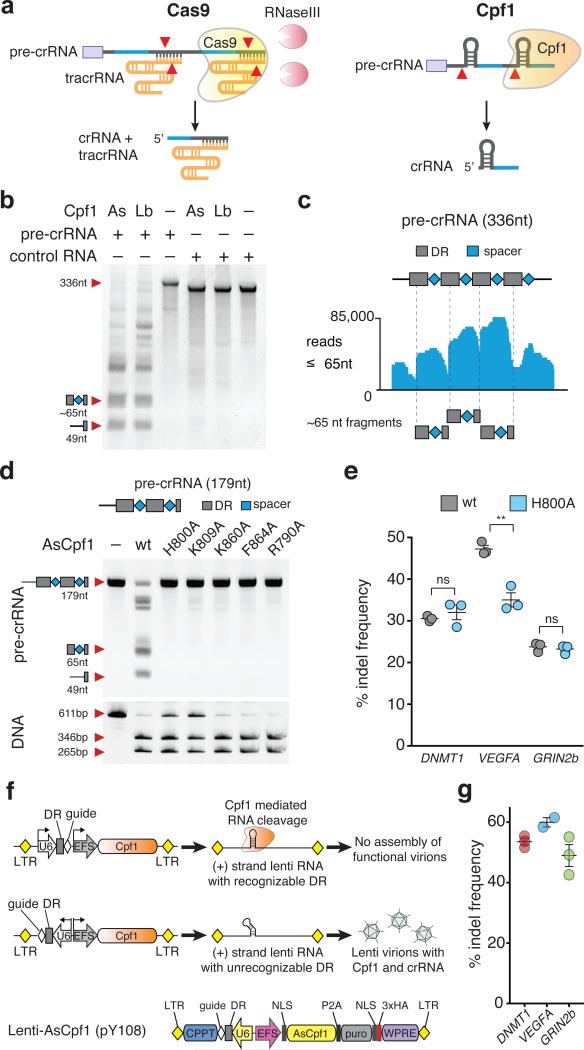
Although multiplex gene editing is possible with Cas9, it requires relatively large constructs or simultaneous delivery of multiple plasmids 7-11, both of which are problematic for multiplex screens or in vivo applications. In contrast, Cpf1 only requires one Pol III promoter to drive several small crRNAs (39nt per crRNA). We confirmed that Cpf1 alone is sufficient for array processing1, 2 using an artificial CRISPR pre-crRNA array consisting of four spacers separated by direct repeats (DRs) from the CRISPR locus of Francisella novicida (FnCpf1) and two Cpf1 orthologs with activity in mammalian cells, Acidaminococcus Cpf1 (AsCpf1) and Lachnospiraceae Cpf1 (LbCpf1) (Figure 1b and Supplementary figure 1). Small RNAseq showed that AsCpf1 cleavage products correlate to fragments resulting from cuts at the 5’ end of DR hairpins, identical to the cleavage pattern we observed in E.coli heterologously expressing FnCpf1 CRISPR systems1 (Figure 1c).
Figure 1. Cpf1 mediated processing of pre-crRNA is independent of DNA cleavage.
(a) Schematic of pre-crRNA processing for Cas9 and Cpf1. Cleavage sites indicated with red triangles. (b) In vitro processing of FnCpf1 pre-crRNA transcript (80 nM) with purified AsCpf1 or LbCpf1 protein (~320 nM), cropped gel image. (c) RNAseq analysis of FnCpf1 pre-crRNA cleavage products, as shown in (b). A high fraction of sequence reads smaller than 65nt are cleavage products of spacers flanked by DR sequences, cropped gel images. (d) Pre-crRNA (top) and DNA cleavage (bottom) mediated by AsCpf1 point mutants. H800A, K809A, K860A, F864A, and R790A fail to process precrRNA but retain DNA cleavage activity in vitro. 330 nM pre-crRNA was cleaved with 500 nM Cpf1 in 15 min and 25 nM DNA was cleaved with 165 nM Cpf1 in 30 min. (e) Indel frequencies mediated by AsCpf1H800A are comparable to wt AsCpf1, bars are mean of 3 technical replicates from one experiment, error bars are SEM. (Student t-test; ns = not significant; ** = p-value 0.003). (f) Schematic of lenti-Cpf1 construct with the U6::DR cassette in different orientations (top and middle), (+)-strand lenti RNA with recognizable DRs are susceptible to Cpf1 mediated degradation, preventing functional virion formation. Schematic of lenti-AsCpf1 (pY108) construct (bottom). (g) Indel frequencies analyzed by SURVEYOR nuclease assay after puromycin selection 10 days after transduction with lenti-AsCpf1 in HEK cells, bars are mean of 2 or 3 individual infections, error bars are SEM. U6, Pol III promoter; CMV, cytomegalovirus promoter; NLS, nuclear localization signal; HA, hemagglutinin tag; DR, direct repeat sequence; P2A, porcine teschovirus-1 2A self-cleaving peptide; LTR, long terminal repeat; WPRE, Woodchuck Hepatitis virus posttranscriptional regulatory element.
We further validated these results by generating AsCpf1 mutants that are unable to process arrays. Guided by the crystal structure of AsCpf13, we mutated five conserved amino acid residues likely to disrupt array processing (H800A, K809A, K860A, F864A, and R790A) 3. All mutations interfered with pre-crRNA processing but not DNA cleavage activity in vitro (Figure 1d and Supplementary figure 2a, b), an effect that was also observed for FnCpf12. AsCpf1 recognizes specific nucleotides at the 5’ flank of the DR stem loop. Substitution of these nucleotides weakens or abolishes RNA cleavage (Supplementary figure 3a). Dosage tests with the five AsCpf1 mutants revealed that mutants K809A, K860A, F864A, and R790A show pre-crRNA processing when used at high concentration (Supplementary figure 3b) or for extended incubation times (Supplementary figure 3c), but H800A was inactive regardless of dose and time.
We next tested if this mutant retains DNase activity in human embryonic kidney (HEK) 293T cells using three guides. Insertion/deletion (indel) frequency at the DNMT1 and GRIN2b loci were identical between wild-type and H800A AsCpf1, whereas indel frequencies at the VEGFA locus were higher in cells transfected with wild-type AsCpf1, demonstrating that the RNA and DNA cleavage activity can be separated in mammalian cells (Figure 1e).
Cpf1 mediated RNA cleavage needs to be considered when designing lenti-virus vectors for simultaneous expression of nuclease and guide (Figure 2f). Lenti virions carry a (+) strand RNA copy of the sequence flanked by long terminal repeats (LTR), allowing Cpf1 to bind and cleave at DR sequences. Hence, reversing the orientation of the DR is expected to result in (+) strand lenti RNAs not susceptible to Cpf1 mediated cleavage. We designed a lenti vector encoding AsCpf1 and a crRNA expression cassette. We transduced HEK293T cells with a MOI (multiplicity of infection) of <0.3 and analyzed indel frequencies in puromycin selected cells 10 days post infection. Using guides encoded on a reversed expression cassette targeting DNMT1, VEGFA, or GRIN2b resulted in robust indel formation for each targeted gene (Figure 2g).
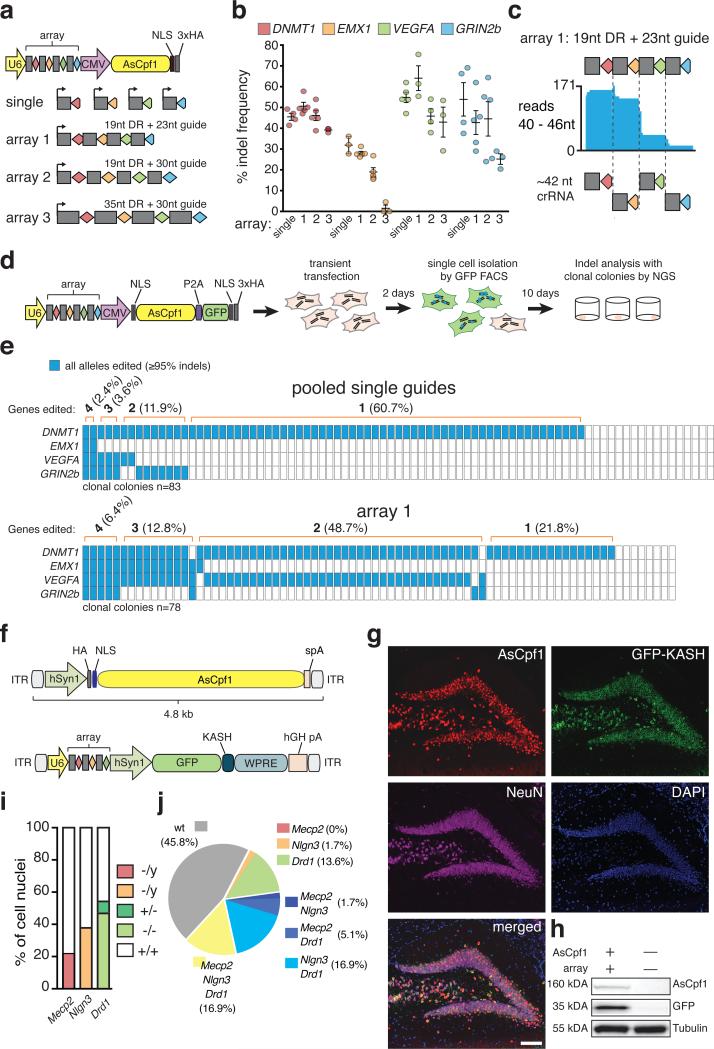
Figure 2. Cpf1-mediated multiplex gene editing in mammalian cells and mouse brain.
(a) Schematic of multiplex gene editing with AsCpf1, using a single plasmid approach. (b) Genome editing at four different genomic loci mediated by AsCpf1 with different versions of artificial CRISPR arrays (array-1, crRNAs in their mature form (19nt DR with 23nt guide); array-2, crRNAs are in an intermediate form (19nt DR with 30nt guide); array-3 crRNAs are in their unprocessed form (35nt DR with 30nt guides)). Indels were analyzed by SURVEYOR nuclease assay 3 days post transfection; bars are mean of two individual experiments with 3 to 5 technical replicates, error bars are SEM. (c) Small RNAseq reads from HEK cells transfected with AsCpf1 and array-1 show fragments corresponding to mature crRNA for each of the four guides. (d) Schematic for analysis of indel events in clonal colonies 48 hours after transient transfection. (e) Quantification of indel events measured by NGS in clonal colonies from HEK cells transiently transfected with pooled single guide plasmids or plasmid carrying array-1. Colonies were expanded for 10 days after sorting. Each column represents one clonal colony; blue rectangles indicate target genes with all alleles edited. (f) Schematic of AAV vector design for multiplex gene editing. Bottom: grey rectangles, direct repeat; diamonds, spacer (red: Mecp2, orange: Nlgn3, green: Drd1). (g) Immunostaining of dorsal DG 4 weeks after stereotactic AAV injection (Representative image of n = 4 mice). Brain sections were co-stained with anti-HA (red), anti-GFP (green) and anti-NeuN (magenta) antibodies. Nuclei were labeled with DAPI (blue). Scale bar: 100 um. (h) Western blot analysis of DG expressing HA-AsCpf1 and GFP-KASH (Representative blot from n = 4 mice). (i) Fraction of mono- and biallelic modifications of autosomal gene Drd1 is shown (Mecp2 and Nlgn3: x-chromosomal). (j) Analysis of multiplexing efficiency in individual cells. ITR, inverted terminal repeat; spA, synthetic polyadenylation signal; hSyn1, human synapsin 1 promoter; ANC1, Syne Homology nuclear transmembrane domain; hGH pA, human growth hormone polyadenylation signal;
We leveraged the simplicity of Cpf1 crRNA maturation to achieve multiplex genome editing in HEK293T cells using customized CRISPR arrays. We chose four guides targeting different genes (DNMT1, EMX1, VEGFA, and GRIN2b) and constructed three arrays with variant DR and guide lengths for expression of pre-crRNAs (Figure 2a). Indel events were detected at each targeted locus in cells transfected with array-1 or -2. However, the crRNA targeting EMX1 resulted in indel frequencies of <2% when expressed from array-3. Overall, array-1 performed best, with all guides showing indel levels comparable to those mediated by single crRNAs (Figure 2b). Furthermore, small RNAseq confirmed that autonomous, Cpf1-mediated pre-crRNA processing occurs in mammalian cells (Figure 2c). Using arrays with guides in different orders resulted in similar indel frequencies, suggesting that positioning within an array is not crucial for activity (Supplementary figure 4a, b).
To confirm that multiplex editing occurs within single cells, we generated AsCpf1-P2A-GFP constructs to enable fluorescence-activated cell sorting (FACS) of transduced single cells (Figure 2d) and clonal expansion. We used next generation deep sequencing (NGS) to compare edited loci within clonal colonies derived from cells transfected with either pooled single guides or array-1. Focusing on targeted genes edited at every locus (indels ≥95%) shows that multiplex editing occurs more frequently in colonies transfected with array-1 (6.4% all targets, 12.8% three targets, 48.7% two targets) than in pooled transfection (2.4% all targets, 3.6% three targets, 11.9% two targets).
We next tested multiplex genome editing in neurons using AsCpf1. We designed a gene-delivery system based on adeno-associated viral vectors (AAVs) for expression of AsCpf1. We generated a dual vector system in which AsCpf1 and the CRISPR-Cpf1 array were cloned separately (Figure 2f). We constructed a U6 promoter-driven Cpf1 array targeting the neuronal genes Mecp2, Nlgn3, and Drd1. This plasmid also included an green fluorescent protein (GFP) fused to the KASH nuclear transmembrane domain 4 to enable FACS of targeted cell nuclei 5.
We first transduced mouse primary cortical neurons in vitro and observed robust expression of AsCpf1 and GFP-KASH one week after viral delivery. SURVEYOR nuclease assay on purified neuronal DNA confirmed indel formations in all three targeted genes (Supplementary figure 5). Next, we tested whether AsCpf1 can be expressed in the brains of living mice for multiplex genome editing in vivo. We stereotactically injected our dual vector system in a 1:1 ratio into the hippocampal dentate gyrus (DG) of adult male mice. Four weeks after viral delivery we observed robust expression of AsCpf1 and GFP-KASH in the DG (Figure 2g, h). Consistent with previous studies 5, 6, we observed ~75% co-transduction efficiency of the dual viral vectors (Supplementary figure 2c). We isolated targeted DG cell nuclei by FACS (Supplementary figure 4) and quantified indel formation using NGS. We found indels in all three targeted loci with ~23%, ~38%, and ~51% indel formation in Mecp2, Nlgn3, and Drd1, respectively (Supplementary figure 4d, e). We quantified the effectiveness of biallelic disruption of the autosomal gene Drd1 and found ~47% of all sorted nuclei (i.e. ~87% of all Drd1-edited cells) harbored biallelic modifications (Figure 2i). Next, we quantified the multiplex targeting efficiency in single neuronal nuclei. Our results show that ~17% of all transduced neurons were modified in all three targeted loci (Figure 2j). Taken together, our results demonstrate the effectiveness of AAV-mediated delivery of AsCpf1 into the mammalian brain and simultaneous multi-gene targeting in vivo using a single array transcript.
Taken together, these data highlight the utility of Cpf1 array processing in designing simplified systems for in vivo multiplex gene editing. Although multiplex gene editing is possible with Cas9, it requires relatively large constructs or simultaneous delivery of multiple plasmids 7-11, both of which are problematic for multiplex screens or in vivo applications. In contrast, Cpf1 only requires one Pol III promoter to drive several small crRNAs (39nt per crRNA). Hence, this system has the potential to simplify guide RNA delivery for many genome editing applications where targeting of multiple genes is desirable.
Supplementary Material
Acknowledgements
We would like to thank F. A. Ran for helpful discussions and overall support, and Bas Cartigny and Jara van den Bogaerde for technical assistance, and the entire Zhang laboratory for support and advice. M.H was supported by the Human Frontiers Scientific Program. O.A.A. is supported by a Paul and Daisy Soros Fellowship and a Friends of the McGovern Institute Fellowship. J.S.G. is supported by a D.O.E. Computational Science Graduate Fellowship. E.D.G is supported by the National Institute of Biomedical Imaging and Bioengineering (NIBIB), of the National Institutes of Health (5T32EB1680). K.S. is supported by an NIH grant GM10407, Russian Science Foundation grant 14-14-00988, and Skoltech. J.v.d.O. is supported by Netherlands Organization for Scientific Research (NWO) through a TOP grant (714.015.001). F.Z. is supported by the NIH through NIMH (5DP1-MH100706 and 1R01-MH110049), the New York Stem Cell, Poitras, Simons, Paul G. Allen Family, and Vallee Foundations; and David R. Cheng, Tom Harriman, and B. Metcalfe. F.Z. is a New York Stem Cell Foundation Robertson Investigator. The authors plan to make the reagents widely available to the academic community through Addgene and to provide software tools via the Zhang lab website (www.genome-engineering.org).
Footnotes
Author Contributions
B.Z., M.H., J.v.d.O., and F.Z. conceived this study and designed the experiments. B.Z., M.H., P.M., Y.F., J.K., E.M.D., N.W., S.C., O.O.A., and J.S.G. conducted the experiments. K.S., J.v.d.O., F.Z. supervised this project. B.Z., M.H., J.v.d.O., and F.Z. wrote the manuscript with input from all authors.
Reference
- 1.Zetsche B, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fonfara I, Richter H, Bratovič M, Rhun A, Charpentier E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016 doi: 10.1038/nature17945. [DOI] [PubMed] [Google Scholar]
- 3.Yamano T, et al. Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA. Cell. 2016 doi: 10.1016/j.cell.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ostlund C, et al. Dynamics and molecular interactions of linker of nucleoskeleton and cytoskeleton (LINC) complex proteins. Journal of cell science. 2009;122:4099–4108. doi: 10.1242/jcs.057075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swiech L, et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol. 2015;33:102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Konermann S, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kabadi AM, Ousterout DG, Hilton IB, Gersbach CA. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Research. 2014;42 doi: 10.1093/nar/gku749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nissim L, Perli SD, Fridkin A, Perez-Pinera P, Lu TK. Multiplexed and Programmable Regulation of Gene Networks with an Integrated RNA and CRISPR/Cas Toolkit in Human Cells. Molecular Cell. 2014;54:698–710. doi: 10.1016/j.molcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakuma T, Nishikawa A, Kume S, Chayama K, Yamamoto T. Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Scientific Reports. 2014;4:5400. doi: 10.1038/srep05400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai SQ, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nature biotechnology. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie K, Minkenberg B, Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proceedings of the National Academy of Sciences. 2015;112:3570–3575. doi: 10.1073/pnas.1420294112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.