Abstract
MicroRNAs are increasingly recognized as epigenetic regulators for outcome of diverse infectious diseases and vaccination efficacy, but little information referring to this exists for malaria. This study investigates possible effects of both protective vaccination and P. chabaudi malaria on the miRNome of the liver as an effector against blood-stage malaria using miRNA microarrays and quantitative PCR. Plasmodium chabaudi blood-stage malaria takes a lethal outcome in female Balb/c mice, but a self-healing course after immunization with a non-infectious blood-stage vaccine. The liver robustly expresses 71 miRNA species at varying levels, among which 65 miRNA species respond to malaria evidenced as steadily increasing or decreasing expressions reaching highest or lowest levels toward the end of the crisis phase on day 11 p.i. in lethal malaria. Protective vaccination does not affect constitutive miRNA expression, but leads to significant (p < 0.05) changes in the expression of 41 miRNA species, however evidenced only during crisis. In vaccination-induced self-healing infections, 18 miRNA-species are up- and 14 miRNA-species are down-regulated by more than 50% during crisis in relation to non-vaccinated mice. Vaccination-induced self-healing and survival of otherwise lethal infections of P. chabaudi activate epigenetic miRNA-regulated remodeling processes in the liver manifesting themselves during crisis. Especially, liver regeneration is accelerated as suggested by upregulation of let-7a-5p, let-7b-5p, let-7c-5p, let-7d-5p, let-7f-5p, let-7g-5p, let-7i-5p, miR-26a, miR-122-5p, miR30a, miR27a, and mir-29a, whereas the up-regulated expression of miR-142-3p by more than 100% is compatible with the view of enhanced hepatic erythropoiesis, possibly at expense of megakaryopoiesis, during crisis of P. chabaudi blood-stage malaria.
Keywords: miRNA, liver, blood-stage malaria, Plasmodium chabaudi, protective vaccination
Introduction
The World Health Organization has reported about 214 million new cases of malaria with about 438,000 deaths worldwide in 2015 (WHO, 2015). A vaccine for an effective and reliable anti-malaria prophylaxis is not yet available, despite enormous efforts during the last 35 years (Halbroth and Draper, 2015; Hoffman et al., 2015; Miura, 2016). Until recently, the RTS,S/AS01 has been regarded as the most advanced vaccine candidate. Indeed, the European Medicines Agency has approved RTS,S/AS01 for immunizations of very young children, however the WHO has not recommended its inclusion in the Expanded Programme of Immunizations (Birkitt, 2016; Gosling and von Seidlein, 2016). This points to the urgency of more basic research to uncover those mechanisms of the host defense, which a vaccine has to activate to provide protection against malaria.
The experimental malaria P. chabaudi in mice shares several characteristics with P. falciparum, the most dangerous malaria species for humans (Longley et al., 2011; Stephens et al., 2012). It is a convenient model to study blood-stage malaria. The outcome of P. chabaudi blood-stage malaria is controlled by genes of the mouse-MHC, the H-2 complex, genes of the non-H-2 background, and environmental factors such as testosterone (Wunderlich et al., 1988d). Moreover, there is a vaccination technique available by which survival of mice, being naturally susceptible to malaria, can be raised from 0% to over 80% (Wunderlich et al., 1988a; Krücken et al., 2009). This technique, which uses a non-infectious vaccine consisting of surface membranes isolated from P. chabaudi-infected erythrocytes containing parasite-synthesized proteins (Wunderlich et al., 1988b,c; Fontaine et al., 2012), converts lethal blood-stage infections to take a self-healing course (Krücken et al., 2009). Upon challenging with 106 P. chabaudi-parasitized erythrocytes, all non-vaccinated mice succumb to malaria during the crisis phase of infection, whereas the majority of vaccinated mice survive the infections and generate long-lasting immune mechanisms against homologous re-challenge (Krücken et al., 2009).
The spleen is currently envisioned as the key effector organ against blood-stage malaria (Engwerda et al., 2005; Del Portillo et al., 2012). Indeed, the spleen is equipped with a unique system to eliminate senescent and aberrant erythrocytes including P.-infected erythrocytes. However, there is accumulating evidence that, also the liver, in particular the erythrophagocytotic Kupffer cells are able to remove senescent and other aberrant erythrocytes including P.-infected erythrocytes (Terpstra and van Berkel, 2000; Otogata et al., 2007; Delic et al., 2010; Lee et al., 2011; Wunderlich et al., 2014). In self-healing infections of P. chabaudi, it has been described that the liver dramatically increases its uptake of injected particles at peak parasitemia and during subsequent crisis, whereas, concomitantly, the spleen dramatically declines its uptake of injected particulate material including fluorescently labeled P. chabaudi-infected erythrocytes (Krücken et al., 2005, 2009). It is therefore rather likely that the liver, rather than the spleen, massively removes P. chabaudi-infected erythrocytes from circulation during crisis in vaccination-induced self-healing infections of P. chabaudi (Wunderlich et al., 2014). Very recently, the liver has been even described as the primary organ for a unique mechanism of rapid on-demand erythrocyte disposal (Theurl et al., 2016).
Current evidence demonstrates that miRNAs are critical for maintaining phenotype and functionality of the liver (Chen and Verfaillie, 2014), and that diverse diseases of the liver are associated with dysregulations in the miRNA signature (Szabo and Bala, 2013; Bandiera et al., 2015). MicroRNAs are non-coding single-stranded RNAs of 22 nts in length, which posttranscriptionally regulate gene expression by affecting both stability and translation of mRNAs. At least one third of human genes are estimated to be targeted by miRNAs (He and Hannon, 2004). MicroRNAs are involved in regulating epigenetic networks within cells and via exosomes between cells (Vyas and Dhawan, 2016). They are increasingly recognized as to be critical for the outcome of diverse infectious diseases (Bettencourt et al., 2016; Das et al., 2016; Verma et al., 2016). Also, miRNAs appear to be critical for efficacy of vaccination (Corral-Fernández et al., 2016; Shim et al., 2016; Wang et al., 2016). However, there is still little information available about host miRNAs in malaria, especially with respect to course and outcome of malaria infections and vaccination efficacy (Cohen et al., 2015; Rubio et al., 2016). At least, it is known that miRNAs of the liver are responsive to blood-stage infections of P. chabaudi malaria (Delic et al., 2011). The aim of this paper is therefore to analyze as to whether protective vaccination against P. chabaudi malaria leads to changes in the overall expression of miRNAs in the liver during self-healing infections of P. chabaudi in comparison to lethal infections in non-vaccinated female Balb/c mice.
Materials and methods
Mice
Balb/c mice were bred under specified pathogen-free conditions. For the experiments, only female Balb/c mice, approximately 10–14 weeks old, were used. They were housed in plastic cages, received a standard diet (Woehrlin, Bad Salzuflen, Germany) and water ad libitum. This study was carried out in strict accordance with the German law on animal protection. The keeping of mice and the experimental protocol of the study were officially approved by the State-controlled Committee on the Ethics of Animal Experiments and were regularly controlled, without being previously announced, by the local authorities. All efforts were made to minimize suffering of mice.
P. chabaudi malaria
A non-clonal line of P. chabaudi is used in our laboratory since 1982 (Wunderlich et al., 1982), which resembles to P. chabaudi chabaudi AS as described previously (Wunderlich et al., 2005; Krücken et al., 2009). Blood-stage infections were routinely maintained in outbred mice under sterile conditions by weekly passages of infected blood. Balb/c mice were i.p. challenged with 106 P. chabaudi-infected erythrocytes. Erythrocytes were counted in a Neubauer chamber and parasitemia was evaluated in Giemsa-stained smears from tail blood.
Vaccination
Mice were vaccinated as performed previously (Krücken et al., 2009). As a vaccine we have used host cell plasma membranes, isolated in the form of ghosts from P. chabaudi-parasitized red blood cells as described elsewhere (Wunderlich et al., 1985, 1987). Approximately 106 ghosts suspended in 100 μl Freund's complete adjuvant were injected on week 3 and week 1 before challenge with 106 P. chabaudi-parasitized erythrocytes (Krücken et al., 2009). Controls received only the adjuvant.
RNA isolation
Livers were removed from sacrificed mice, rapidly frozen in liquid nitrogen and stored at −80°C until use. Frozen livers were grounded in a mortar under liquid nirogen and aliquots were used to isolate total RNA using Trizol (Sigma-Aldrich). An additional cleaning step was performed by using the miRNeasy Kit (Qiagen). RNA quality and integrity were determined using the Agilent RNA 6000 Nano Kit on the Agilent 2100 Bioanalyzer platform (Agilent Technologies). RNA was quantified by measuring A260 nm on the ND-1000 Spectrophotometer (NanoDrop Technologies).
Labeling of miRNA
Labeling of the samples was performed as detailed in the “miRNA Microarray System with miRNA Complete Labeling and Hyb Kit” protocol (version 2.4, part number G4170-90011). In brief, 100 ng of each total RNA sample was used for the labeling step using the miRNA Complete Labeling and Hyb Kit (Agilent Technologies).
Hybridization of agilent microRNA microarrays
The used Agilent Mouse microRNA Microarrays 8 × 60 K v19 (Design ID 046065) contain 8 arrays per slide and one array is equipped with probes for detecting 1247 mouse miRNAs. Hybridization was performed according to the “miRNA Complete Labeling and Hyb Kit” protocol (version 2.4, part number G4170-90011) using the miRNA Complete Labeling and Hyb Kit (Agilent Technologies). In brief, Cy3-labeled RNA in hybridization buffer was hybridized overnight (20 h, 55°C) to the microarrays using Agilent's recommended hybridization chamber and oven. Thereafter, the microarrays were washed once with the Agilent Gene Expression Wash Buffer 1 for 5 min at room temperature and once by a second wash with preheated Agilent Gene Expression Wash Buffer 2 (37°C) for 5 min.
Scanning and data analysis
Agilent's Microarray Scanner System G2505C (Agilent Technologies) was used to detect fluorescence signals of the hybridized Agilent Microarrays. The Agilent Feature Extraction Software (FES) 10.7.3.1 was used to read out and process the microarray image files. For determination of differential miRNA expression, FES derived output data files were further analyzed using the GeneSpringGX (Version 12.6) analysis system (Agilent Technologies). Expression levels were log2 transformed and given in light units. If a miRNA is absent in the investigated sample the light units are automatically set by the FES to 0.1 light units, a value of −3.3 (log2 of 0.1) reflects therefore a non-expressed miRNA. Only those values were used with >100 lights units representing robustly expressed miRNAs. A heatmap was generated to visualize the expression levels of each miRNA (Spotfire, TIBCO Software Inc., Palo Alto, USA). The row and column dendrograms were clustered with the unweighted pair group method with arithmetic mean and Euclidean distance measure. Significant malaria-responsive and/or protective vaccination effects on miRNA expression were assessed using 2-way ANOVA. MicroRNAs with at least 2-fold deregulation and a P < 0.05 in any group-wise comparison were considered as to be differentially expressed. P values were corrected using Benjamini-Hochberg method for multiple testing (Benjamini and Hochberg, 1995). Raw data are publicly available at the EMBL-EBI Array Express repository (Array Express accession number: E-MTAB-5301).
Quantitative real-time PCR
TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems) and Megaplex™ RT Primers, Rodent Pool A and B (Applied Biosystems) were used to reverse transcribe miRNAs. Reactions were performed in triplicates using the following TaqMan® MicroRNA Assays (Applied Biosystems): let-7a-5p (assay ID: 00377), let-7c-5p (assay ID: 00379), miR-122-5p (assay ID: 002245), miR-142-3p (assay ID: 000464), miR-29b-3p (assay ID: 000413), miR-30c-5p (assay ID: 000419), miR-669n (assay ID: 197143_mat), miR-709 (assay ID: 001644), miR-92-3p (assay ID: 000430), miR-126-3p (assay ID: 002228). PCR reactions were performed with the TaqMan® gene expression master mix (Life Technologies) according manufacturer's protocol on a 7900HT real-time PCR System. The miRNA expression analysis was run on a SDS7900HT real time PCR system; raw Ct values were calculated using the SDS software v2.4 with automatic baseline and threshold settings. U6 snRNA (assay ID: 001973) was used for normalization. Fold change of expression was calculated using the comparative Ct method (2−ΔΔct) (Livak and Schmittgen, 2001). Data sets were analyzed for statistical significance using two-tailed unpaired heteroskedastic Student's t-test.
Results
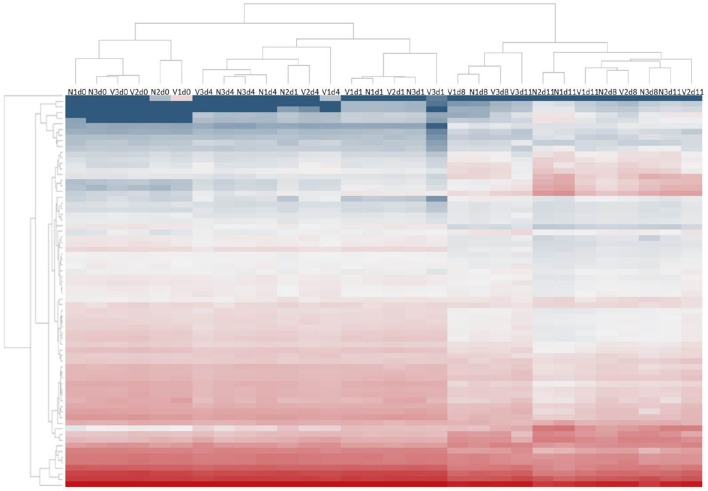
To explore possible changes of hepatic miRNome by protective vaccination and/or subsequent challenge infections with P. chabaudi blood-stage malaria, microarrays were used to analyze miRNA expression in livers from vaccination-protected (V) and non-protected non-vaccinated mice (N) before infection on day 0 p.i. (Vd0, Nd0), and during different phases of the infections, i.e., at the beginning of infection on day 1 p.i. (Vd1, Nd1), during the mid-precrisis phase on day 4 p.i. (Vd4, Nd4), at peak parasitemia on day 8 p.i. (Vd8, Nd8), and toward the end of the crisis phase on day 11 p.i. (Vd11, Nd11). The parasitemia is about the same in self-healing infections of vaccination-protected mice and in lethal infections of non-protected mice (Figure 1). Only at peak parasitemia, there is a significant lower parasitemia in self-healing infections (Figure 1). Figure 2 shows the heat map of miRNA expression profiles during self-healing and lethal infections in vaccination-protected and non-protected mice, respectively. The profiles on day 0, 1, and 4 p.i. cluster separately from those on day 8 and 11 p.i., respectively, which are closer together.
Figure 1.
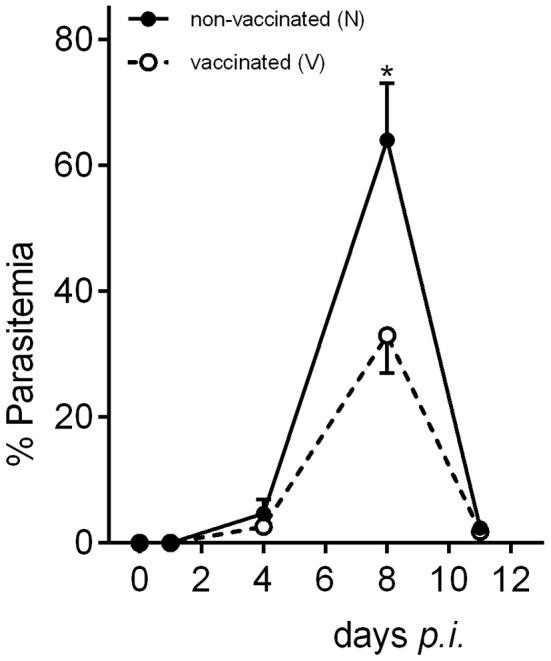
Course of parasitemia during self-healing infections of P. chabaudi in vaccination-protected Balb/c mice and lethal infections in non-vaccinated mice. Values represent means determined from three mice whose livers were taken to analyze miRNA expression. Bars represent half SD and star indicates significant difference (p < 0.01).
Figure 2.
Heatmap of expression levels of miRNAs in the liver of non-vaccinated (N) and vaccinated (V) mice infected with P. chabaudi on days 0, 1, 4, 8, and 11 p.i. Expression levels for each sample were hierarchically clustered. Log2 transformed expression levels range from −4 to 14 indicated by blue and red color, respectively.
Among the overall detectable 1247 miRNA species on a given array, 71 miRNA species are found to be robustly expressed (>100 light units) in the liver of both vaccinated and non-vaccinated mice. The most abundant expressions show the miRNAs miR-122-5p, miR-6366, miR-3963 and miR-5100, while relatively low expression is observed for the miRNAs miR-1196-5p, miR-468-3p and miR-669n. None of these miRNA species is affected by protective vaccination per se, evidenced as the expressions of the 71 miRNAs in vaccinated non-infected mice in comparison with the corresponding constitutive expressions in non-vaccinated mice. Upon infection with P. chabaudi, however, 65 miRNA species change their expression pattern, and 41 miRNA species out of the 65 malaria-responsive miRNAs significantly (p < 0.05) differ with respect to their expression levels in the liver between vaccinated and non-vaccinated mice. However, this difference is only evidenced toward the end of the crisis phase on day 11 p.i..
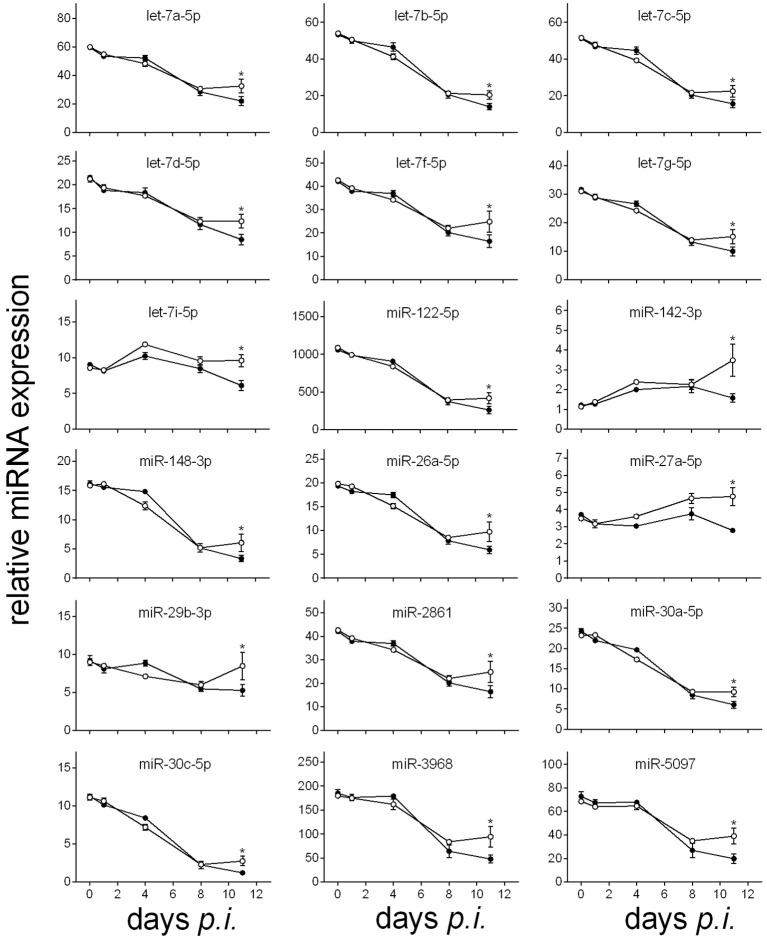
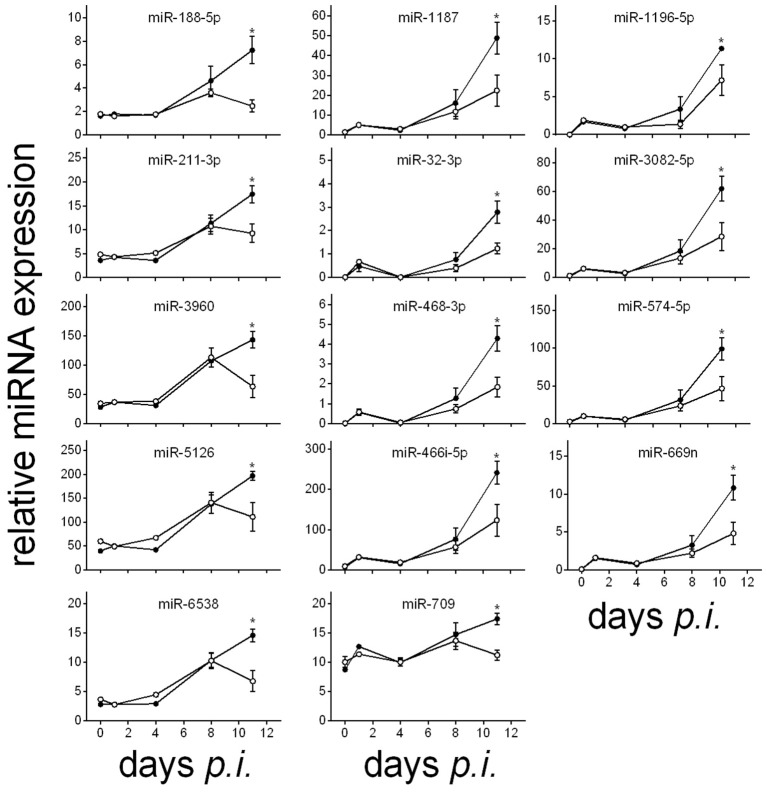
The lethal outcome of P. chabaudi malaria is associated with either increasing or decreasing expressions of miRNAs in the liver during the infections, reaching their minimal or maximal expression levels toward the end of the crisis phase on day 11 p.i. (cf. also Figures 3, 4). Decreasing expressions reveal the 18 miRNA species miR-122-5p, miR-142-3p, miR-148-3p, miR-26a-5p, miR-27a-5p, miR-29b-3p, miR-2861, miR30a/c-5p, miR-3968, miR-5097, and seven members of the let miRNA family (Figure 3), whereas increasing expressions occur for the 14 miRNAs miR-188-5p, miR1187, miR-1196-5p, miR-211-3p, miR-32-3p, miR-3082-5p, miR-3960, miR-466i-5p, miR-468-3p, miR-574-5p, miR-669n, miR-709, mir-5126, and miR-6538 (Figure 4). In vaccination-induced self-healing infections, however, the decreasing expression of the 18 miRNAs is stopped or even slightly turned to be up-regulated only by the end of crisis (Figure 3), whereas the increasing expression of the 14 miRNA species is impaired or even turned down (Figure 4). The up- or down-regulated expressions amount to more than 50% at the end of the crisis phase between vaccination-induced self-healing infections and lethal infections in non-vaccinated mice.
Figure 3.
Time course of relative miRNA expression with increased levels in the liver of vaccinated mice toward the end of the crisis phase of P. chabaudi infections in comparison to non-vaccinated mice. Relative miRNA expression was normalized to the mean constitutive expression of the corresponding miRNAs. Open circles are values of vaccinated mice and filled circles are those of non-vaccinated mice. Values are given in mean ± SD. Significant differences are indicated by *(P < 0.05).
Figure 4.
Time course of relative miRNA expression of significantly decreased hepatic miRNA levels in vaccinated mice toward the end of the crisis phase of P. chabaudi infections compared to non-vaccinated mice. Relative miRNA expression was normalized to the mean constitutive expression levels of the corresponding miRNAs. Open circles, vaccinated mice. Filled circles, non-vaccinated mice. Values represent means ± SD. Significant differences are indicated by *(P < 0.05).
Tables 1, 2 summarize the annotated functions of the 18 and 14 miRNA species, whose expression is either up- or down-regulated in the liver of vaccination-protected mice toward the end of the crisis phase of P. chabaudi infections in relation to non-vaccinated mice, respectively. Remarkably, all the seven up-regulated members of the let-7 miRNA family are known to be involved in liver regeneration.
Table 1.
Increased hepatic miRNA expression in vaccinated mice infected with P. chabaudi on day 11 p.i. (Vd11) compared to non-vaccinated mice infected with P. chabaudi on day 11 p.i. (Nd11).
| miRNA name | % increase Vd11 vs. Nd11 | Function | PMID |
|---|---|---|---|
| let-7a | +50 | Up-regulated during early phase of liver regeneration | 21574170, 18812516 |
| let-7b | +50 | Up-regulated during early phase of liver regeneration | 21574170, 18812516 |
| let-7c | +50 | Up-regulated during early phase of liver regeneration | 21574170, 18812516 |
| let-7d | +51 | Up-regulated during early phase of liver regeneration | 21574170, 18812516 |
| let-7f | +51 | Up-regulated during early phase of liver regeneration | 21574170, 18812516 |
| let-7g | +51 | Up-regulated during early phase of liver regeneration | 21574170, 18812516 |
| let-7i | +57 | Up-regulated during early phase of liver regeneration | 21574170, 18812516 |
| miR-122-5p | +60 | Abundantly expressed in liver; involved in cholesterol biosynthesis pathway; binds to the HCV genome and enhances viral translation and replication | 16258535,25574453, 16141076 |
| miR-142-3p | +121 | Orchestrates network of actin cytoskeleton during megakaryopiesis | 24859754 |
| miR-148a-3p | +81 | miR-148a-3p downregulates Met/Snail signaling pathway and thus inhibits the epithelial to mesenchymal transition (EMT) | 23532995 |
| miR-26a-5p | +63 | Involved in liver regeneration and hepatocyte proliferation; miR-26a expression reduced M-CSF expression and recruitment of macrophages in HCC | 26818545, 26021873 |
| miR-27a-3p | +71 | Involved in lung fibrosis | 26600197 |
| miR-29b-3p | +61 | Members of the miR-29 family are downregulated in HSCs in response to TNF and TGF-β signaling and suppress the transcription of ECM genes like collagen-1α1 | 20890893 |
| miR-2861 | +52 | Important physiological role in osteoblast differentiation and contributes to osteoporosis via its effect on osteoblasts | 19920351 |
| miR-30a-5p | +52 | Members of a network of miRNAs modifying the TGF-β-dependent regulation of extracellular matrix-related genes in HSCs in the manifestation and resolution of liver fibrosis | 26120970 |
| miR-30c-5p | +127 | Members of a network of miRNAs modifying the TGF-β-dependent regulation of extracellular matrix-related genes in HSCs in the manifestation and resolution of liver fibrosis | 26120970 |
| miR-3968 | +96 | Unknown | |
| miR-5097 | +98 | Unknown |
Table 2.
Decreased hepatic miRNA expression in vaccinated mice infected with P. chabaudi on day 11 p.i. (Vd11) compared to non-vaccinated mice infected with P. chabaudi on day 11 p.i. (Nd11).
| miRNA name | % Increase Vd11 vs. Nd11 | Function | PMID |
|---|---|---|---|
| miR-188-5p | −65 | Acts as a tumor suppressor in prostate caner | 25714029 |
| miR-1187 | −54 | Involved in hepatocyte apoptosis | 22266786 |
| miR-1196-5p | −37 | Unknown | |
| miR-211-3p | −55 | lncRNA-uc002kmd.1 regulates CD44 as a molecular decoy for miR211-3p | 26974151 |
| miR-32-3p | −54 | Acts a a tumor suppressor in NSCLC | 26229485 |
| miR-3082-5p | −55 | Unknown | |
| miR-3960 | −56 | miR-3960 regulated cellular growth and proliferation through a regulatory feedback loop with miR-2861, respnse to oxidative stress | 21324897, 26539117 |
| miR-466i-5p | −49 | Unknown | |
| miR-468-3p | −57 | Unknown | |
| miR-574-5p | −53 | Oncogene in various cancer types, incl. SCLC | 26587830 |
| miR-669n | −55 | Involved in control of LPS-induced macrophage activation | 26807181 |
| miR-709 | −35 | miR-709 may positively regulate invasion and metastasis of HCC through targeting GPC5 | 25818666 |
| miR-5126 | −43 | Unknown | |
| miR-6538 | −53 | Unknown |
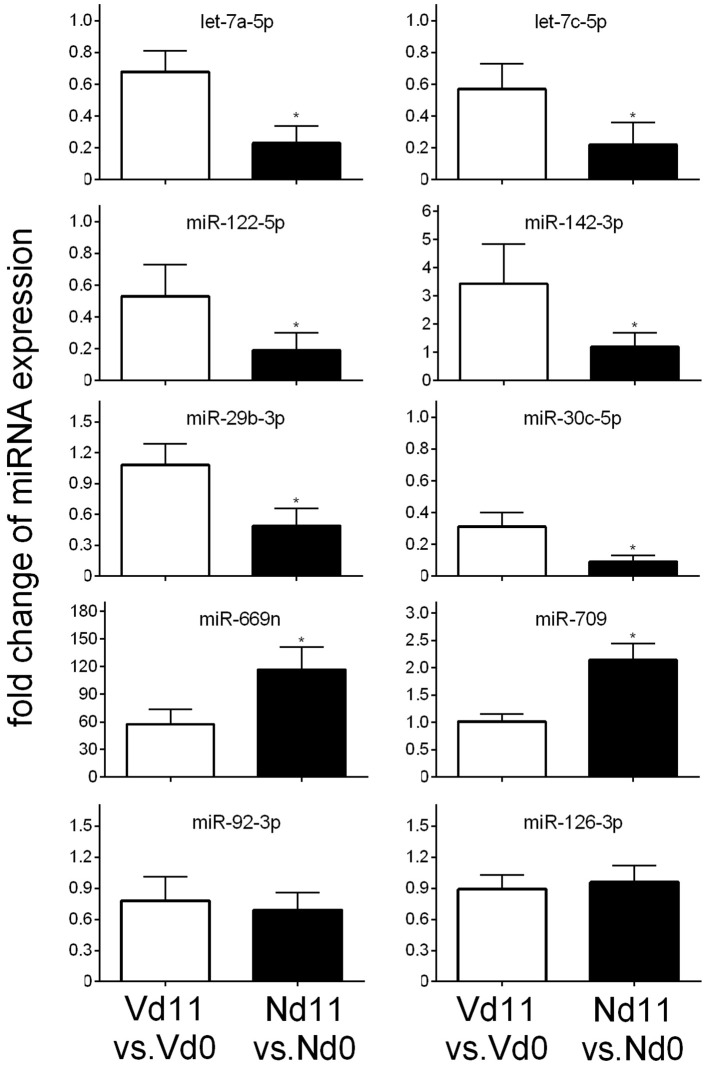
To further substantiate these differences in miRNA expression toward the end of the crisis phase, we have also performed quantitative PCR of arbitrarily selected miRNAs. Figure 5 shows that the miRNAs let-7a-5p, let-7c-5p, miR-122-5p, miR-142-3p, miR-29b-3p, and miR-30c-5p are up-regulated and the miRNA species miR-669n and miR-709 are down-regulated in livers of vaccination-induced self-healing infections on day 11 p.i. in comparison to lethal infections in non-vaccinated mice, thus confirming our microarray analyses. By contrast, the expressions of the malaria-responsive miR-92-3p and miR-126-3p are not significantly affected by infections, as it was also found by microarrays.
Figure 5.
Quantitative PCR of different miRNA species in the liver of vaccination-protected Balb/c mice infected with P. chabaudi on day 11 p.i. (Vd11) in relation to expression on day 0 p.i. (Vd0). Corresponding analyses for non-vaccinated mice (Nd11 vs. Nd0). Bars indicate half SD, (*) significant differences (p < 0.01).
Discussion
This is the first study providing evidence that protective vaccination against blood-stage malaria of P. chabaudi leads to changes in the expression of distinct miRNA species in the liver during subsequent homologous challenge infections. These changes, however, are evidenced only toward the end of the crisis phase on day 11 p.i. in relation to lethal infections in non-vaccinated mice. Lethal malaria induces increasing expressions of 18 miRNA species and decreasing expressions of 14 miRNA species during the precrisis phase reaching highest and lowest expression levels toward the end of the crisis phase. In vaccinated mice, however, the course of the decreasing and increasing expressions of the 18 and 14 miRNA species, respectively, is apparently stopped at peak parasitemia and even partly reversed during crisis, resulting in changes by more than 50 % in relation to non-vaccineted mice toward the end of the crisis phase. This suggests that vaccination-induced self-healing infections of P. chabaudi blood-stage malaria are associated with a remodeling of miRNA-controlled processes in the liver of Balb/c mice, which otherwise would lead to a lethal outcome of the infections during crisis.
Such a remodeling is presumably associated with an accelerated liver regeneration. Indeed, seven members of the let-7 miRNA family, found here to be up-regulated in the liver of vaccination-protected mice during crisis, have been all described to be involved in liver regeneration by targeting mRNAs of dicer and trbp (Forman et al., 2008; Shu et al., 2011). Also, the mir-26a up-regulated during crisis is known to be involved in liver regeneration and hepatocyte proliferation (Zhou et al., 2016). Moreover, the down-regulated expression levels of miR-1187 may reflect diminished hepatocyte apoptosis through targeting casp-8 (Yu et al., 2012). Furthermore, the down-regulated levels of miR-669n during crisis may conceivably contribute to accelerated liver regeneration by dampening malaria-induced Kupffer cell activation (Long et al., 2015). All these data support the view that liver regeneration may be accelerated in vaccination-protected mice during the crisis phase of self-healing infections of otherwise lethal P. chabaudi malaria. Accelerated liver regeneration in vaccinated mice is presumably not a direct effect of vaccination, but rather it may be associated with the process of self-healing, i.e., it may be an indirect consequence of the vaccination-induced conversion of lethal to self-healing infections of P. chabaudi.
Accelerated liver regeneration may contribute to overcome dysfunctions of the liver, presumably due to injuries induced by the P. chabaudi blood-stage infections. Indeed, the acute phase of P. chabaudi malaria has been described to be associated with pathological damages and heavy injuries in the liver with distant effects on other organs, as e.g., hepatoencephalopathy (Wunderlich et al., 2005; Delic et al., 2010). Even human patients suffering from malaria with P. falciparum and P. vivax have been shown to be associated with massive dys-functions of the liver (Ananad et al., 1992; Kochar et al., 2003; Nautyal et al., 2005; Rupani and Amarapurkar, 2009). Moreover, it is known that pathogenesis and diseases of the liver are in general associated with dys-regulated expressions of diverse miRNAs (Szabo and Bala, 2013; Chen and Verfaillie, 2014; Bandiera et al., 2015; Murakami and Kawada, 2016). For instance, injuries of the liver in HIV/HCV patients suffering from necroinflammation and portal hypertension have been recently reported to cause elevated levels of the miR-122, the most abundant miRNA species in the liver (Jansen et al., 2015). Our data, however, reveal continuously decreasing levels of miR-122-5p in the liver of both vaccinated and non-vaccinated mice during the precrisis phase of self-healing and lethal P. chabaudi infections, respectively. Only during the crisis phase, vaccination-protected mice exhibit elevated levels of miR-122-5p, whereas the corresponding levels further decrease in non-vaccinated mice with lethal infections. Similar courses of miRNA expression levels are here found for miR-30a known to be involved in manifestation and resolution of liver fibrosis (Roy et al., 2015), for miR-27a described to be involved in fibrosis (Cui et al., 2016), and for mir-29b known to suppress transcription of genes encoding for extracellular matrix proteins (cf. Table 1) (Roderburg et al., 2011; Lambrecht et al., 2015; Kitano and Bloomston, 2016). Thus, the dys-regulations of these different miRNA species may presumably due to the liver injuries induced by P. chabaudi blood-stage malaria occurring during precrisis in both vaccination-protected and non-vaccinated mice. However, the up-regulated levels of mir-122-5p, miR-30a, mir-27a, and miR-29, observed in vaccination-protected mice during crisis, may contribute to the accelerated liver regeneration suggested to occur in these mice.
Moreover, the present study shows that the expression level of mir-142-3p is significantly up-regulated during crisis by more than 100% in the liver of vaccinated mice as compared with non-vaccinated mice, i.e., this percental difference during crisis is by far higher than those found for all other malaria-responsive miRNA species in the liver. Recent evidence indicates that the miRNA-142 locus is important for the regulation of macrophage-related processes, as e.g., macrophage and dendritic cell differentiation (Fordham et al., 2015), regulation of cell migration (Kim et al., 2015), control of profibrogenic macrophage program (Su et al., 2015), role in colony-stimulating factor 1-induced monocyte differentiation into macrophages (Lagrange et al., 2013), prevention of macrophage differentiation during cancer-induced myelopoiesis (Sonda et al., 2013). The relevance of these data for the liver of vaccinated mice remains to be shown. Currently, however, the most unequivocal evidence demonstrates that mir-142-3p is critical for megakaryopoiesis (Chapnik et al., 2014). The miRNA-142-3p stringently controls specific cytoskeletal rearrangements required for maturation and function of megakaryocytes, and genetic deletion of miR-142-3p results in impaired megakaryocyte ablation, cytoskeletal dys-integrity, abnormal proplatelet formation and thrombocytopenia (Chapnik et al., 2014). In accordance, thrombocytopenia also occurs in experimental malaria including P. chabaudi malaria (Watier et al., 1992; Piguet et al., 2000, 2002; Gramaglia et al., 2005) and even represents a severe complication in human malaria caused by P. vivax and P. falciparum (Ansari et al., 2009; Lacerda et al., 2011; Gill et al., 2013; Gupta et al., 2013). Thus, our data showing up-regulation of mir-142-3p may suggest that protective vaccination affects hepatic megakaryopoiesis induced by P. chabaudi blood-stage malaria during crisis.
On the other hand, however, there is evidence that miR-142-3p is also involved in the regulation of erythropoiesis (Muhseen and Abbood, 2014). Erythropoiesis in turn is tightly coupled with megakaryopoiesis. Both erythropoiesis and megakaryopoiesis are controlled by the two transcription factors GATA1 and GFI1b (Fatica et al., 2006; Crispino and Weiss, 2014). Moreover, both megakaryocytic and erythroid cells are derived from common myeloid precursors (Fatica et al., 2006), and there exist even bipotent progenitor stem cells generating both erythroid and magakaryoctic cells (Debili et al., 1996; Papayannopoulou et al., 1996; Papayannopoulou and Kaushansky, 2016). However, there is also information available that erythropoiesis is enhanced at the expense of megakaryopoiesis (Bianchi et al., 2015). Under identical experimental conditions as described here, Al-Quraishy et al. (2016) have recently found that protective vaccination augments P. chabaudi-induced extramedullary hepatic erythropoiesis. It appears therefore plausible to assume that the up-regulation of mir-142-3p observed here during crisis is involved in enhanced erythropoiesis at the expense of megakaryopoiesis in the liver of vaccination-induced self-healing infections of P. chabaudi malaria (Al-Quraishy et al., 2016).
Collectively, our data suggest that vaccination-induced self-healing and survival of otherwise lethal blood-stage infections of P. chabaudi malaria activates epigenetic miRNA-regulated mechanisms in the liver, which accelerate liver regeneration and enhance hepatic erythropoiesis, possibly at the expense of megakaryopoiesis, during crisis of P. chabaudi blood-stage malaria.
Author contributions
MD, SA, and FW designed the study; MD, AA, and DD carried out the experiments and analyzed the data. All authors wrote and revised the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This project was funded by the National Plan for Science, Technology and Innovation (MAARIFAH), King Abdulaziz City for Science and Technology, Kingdom of Saudi Arabia, awarded number (13-B101206-02).
References
- Al-Quraishy S. A., Dkhil M. A., Abdel-Baki A. A., Delic D., Wunderlich F. (2016). Protective Vaccination against Blood-Stage Malaria of Plasmodium chabaudi: differential gene expression in the liver of Balb/c mice toward the end of crisis phase. Front. Microbiol. 7:1087. 10.3389/fmicb.2016.01087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananad A. C., Ramji C., Narula A. S., Singh W. (1992). Malarial hepatitis: a heterogenous syndrome. Natl. Med. J. India 5, 59–62. [PubMed] [Google Scholar]
- Ansari S., Khorano H. K., Abro A., Akhund I. A., Qureshi F. (2009). Thrombocytopenia in Plasmodium falciparum malaria. J. Ayub. Med. Coll. Abbottabad. 21, 145–147. [PubMed] [Google Scholar]
- Bandiera S., Pfeffer S., Baumert T. F., Zeisel M. B. (2015). miR-122 – A key factor and therapeutic target in liver disease. J. Hepatol. 62, 448–457. 10.1016/j.jhep.2014.10.004 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B. 57, 289–300. [Google Scholar]
- Bettencourt P., Pires D., Anes E. (2016). Immunomodulating microRNAs of mycobacterial infections. Tuberculosis (Edinb). 97, 1–7. 10.1016/j.tube.2015.12.004 [DOI] [PubMed] [Google Scholar]
- Bianchi E., Bulgarelli J., Ruberti S., Rontauroli S., Sacchi G., Norfo R., et al. (2015). MYB controls erythroid versus megakaryocyte lineage fate decision through the miR-486-3p-mediated downregulation of MAF. Cell Death Differ. 22, 1906–1921. 10.1038/cdd.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkitt A. J. (2016). Status of vaccine research and development of vaccines for malaria. Vaccine 34, 2915–2920. 10.1016/j.vaccine.2015.12.074 [DOI] [PubMed] [Google Scholar]
- Chapnik E., Rivkin N., Mildner A., Beck G., Pasvolsky P., Metzl-Raz E., et al. (2014). miR-142 orchestrates a network of action cytoskeleton regulators during megakaryopoiesis. eLIFE 3:e1964. 10.7554/eLife.01964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Verfaillie C. M. (2014). MicroRNAs: the fine modulators of liver development and function. Liver Int. 34, 976–990. 10.1111/liv.12496 [DOI] [PubMed] [Google Scholar]
- Cohen A., Combes V., Grau G. E. R. (2015). MicroRNAs and malaria – a dynamic interaction still incompletely understood. J. Neuroinfect. Dis. 5, 165. [PMC free article] [PubMed] [Google Scholar]
- Corral-Fernández N. E., Cortez-Espinosa N., Salgado-Bustamante M., Romano-Moreno S., Medellin-Garibay S. E., Solis-Rodríguez M., et al. (2016). Induction of transcription factors, miRNAs and cytokines involved in T lymphocyte differentiation in BCG-vaccinated subjects. Mol. Immunol. 77, 44–51. 10.1016/j.molimm.2016.07.006 [DOI] [PubMed] [Google Scholar]
- Crispino J. D., Weiss M. J. (2014). Erythro-megakaryocytic transcription factors associated with hereditary anemia. Blood 123, 3080–3088. 10.1182/blood-2014-01-453167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Banerjee S., Xie N., Ge J., Liu R. M., Matalon S., et al. (2016). MicroRNA-27a-3p is a negative regulator of lung fibrosis by targeting myofibroblast differentiation. Am. J. Respir. Cell Mol. Biol. 54, 843–852. 10.1165/rcmb.2015-0205OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K., Garnica O., Dhandayuthapani S. (2016). Modulation of host miRNAs by intracellular bacterial pathogens. Front. Cell Infect. Microbiol. 6:79. 10.3389/fcimb.2016.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debili N., Coulombel L., Croisille L., Katz A., Guichard J., Breton-Gorius J., et al. (1996). Characterization of a bipotent erythro-megakaryocytic progenitor in human bone marrow. Blood 88, 1284–1296. [PubMed] [Google Scholar]
- Delic D., Dkhil M., Al-Quraishy S., Wunderlich F. (2011). Hepatic miRNA expression reprogrammed by Plasmodium chabaudi malaria. Parsitol. Res. 108, 1111–1121. 10.1007/s00436-010-2152-z [DOI] [PubMed] [Google Scholar]
- Delic D., Warskulat U., Borsch E., Al-Qahtani S., Al-Guraishy S., Häussinger D., et al. (2010). Loss of ability to self-heal malaria upon taurine transporter deletion. Infect. Immun. 78, 1642–1649. 10.1128/IAI.01159-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Portillo H. A., Ferrer M., Brugat T., Martin-Jaular L., Langhorne J., Lacerda M. V. G. (2012). The role of the spleen in malaria. Cell Microbiol. 14, 343–355. 10.1111/j.1462-5822.2011.01741.x [DOI] [PubMed] [Google Scholar]
- Engwerda C. R., Beattie L., Amante F. H. (2005). The importance of the spleen in malaria. Trends Parasitol. 21, 75–80. 10.1016/j.pt.2004.11.008 [DOI] [PubMed] [Google Scholar]
- Fatica A., Rosa A., Fazi F., Ballarino M., Morlando M., De Angelis F. G., et al. (2006). MicroRNAs and hematopoietic differention. Cold Spring Harb. Symp. Quant Biol. 71, 205–210. 10.1101/sqb.2006.71.014 [DOI] [PubMed] [Google Scholar]
- Fontaine A., Bourdon S., Belghazi M., Pophillat M., Fourquet P., Granjeaud S., et al. (2012). Plasmodium falciparum infection-induced changes in erythrocyte membrane proteins. Parasitol. Res. 110, 545–556. 10.1007/s00436-011-2521-2 [DOI] [PubMed] [Google Scholar]
- Fordham J. B., Naqvi A. R., Nares S. (2015). Regulation of mi-24, miR-30b, and miR-142-3p during macrophage and dendritic cell differentiation potentiates innate immunity. J. Leukoc. Biol. 98, 195–207. 10.1189/jlb.1A1014-519RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman J. J., Legesse-Miller A., Coller H. A. (2008). A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc. Natl. Acad. Sci. U.S.A. 105, 14879–14884. 10.1073/pnas.0803230105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill M. K., Makkar M., Bhat S., Kaut T., Jain K., Dhir G. (2013). Thrombocytopenia in malaria and its correlation with different types of malaria. Trop. Med. Public Health 6, 197–200. 10.4103/1755-6783.116521 [DOI] [Google Scholar]
- Gosling R., von Seidlein L. (2016). The future of the RTS,S/AS01 malaria vaccine. An alternative development plan. PLoS Med. 13:e1001994. 10.1371/journal.pmed.1001994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramaglia I., Sahlin H., Nolan J. P., Frangos J. A., Intaglietta M., van der Heyde H. C. (2005). Cell- rather than antibody-mediated immunity leads to the development of profound thrombocytopenia during experimental Plasmodium berghei malaria. J. Immunol. 175, 7699–7707. 10.4049/jimmunol.175.11.7699 [DOI] [PubMed] [Google Scholar]
- Gupta N. K., Bansal S. B., Jain U. C., Sahare K. (2013). Study of thrombocytopenia in patients of malaria. Trop. Parasitol. 3, 58–61. 10.4103/2229-5070.113914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbroth B. R., Draper S. J. (2015). Recent developents in malaria vaccinology. Adv. Parasitol. 88, 1–49. 10.1016/bs.apar.2015.03.001 [DOI] [PubMed] [Google Scholar]
- He L., Hannon G. J. (2004). MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531. 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- Hoffman S. L., Vekemans J., Richie T. L., Duffy P. E. (2015). The march toward malaria vaccines. Am. J. Prev. Med. 49(6 Suppl. 4), S319–S333. 10.1016/j.amepre.2015.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen C., Reiberger T., Huang J., Eischeid H., Schierwagen R., Mandorfer M., et al. (2015). Circulating miRNA-122 levels are associated with hepatic necroinflammation and portal hypertension in HIV/HCV coinfection. PLoS ONE 10:e0116768. 10.1371/journal.pone.0116768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Yang D. K., Kim S., Kang H. (2015). miR-142-3p is a regulator of the TGFbeta-mediated vascular smooth muscle cell phenotype. J. Cell Biochem. 116, 2325–2333. 10.1002/jcb.25183 [DOI] [PubMed] [Google Scholar]
- Kitano M., Bloomston P. M. (2016). Hepatic stellate cells and microRNAs in pathogenesis of liver fibrosis. J. Clin. Med. 5:38. 10.3390/jcm5030038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochar D. K., Singh P., Agarwal P., Kochar S. K., Sareen P. K. (2003). Malarial hepatitis. J. Assoc. Physicians India 51, 1069–1072. [PubMed] [Google Scholar]
- Krücken J., Delic D., Pauen H., Wojtalla A., El-Khadragy M., Dkhil M. A., et al. (2009). Augmented particle trapping and attenuated inflammation in the liver by protective vaccination against Plasmodium chabaudi malaria. Malaria J. 8, 54–64. 10.1186/1475-2875-8-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krücken J., Mehnert L. I., Dkhil M. A., El-Khadragy M., Benten W. P. M., Mossmann H., et al. (2005). Massive destruction of malaria-parasitized red blood cells despite spleen closure. Infect. Immun. 73, 6390–6398. 10.1128/IAI.73.10.6390-6398.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacerda M. V., Mourão M. P., Coelho H. C., Santois J. B. (2011). Thrombocytopenia in malaria: who cares? Mem. Inst. Oswaldo. Cruz. 106(Suppl. 1), 52–63. 10.1590/S0074-02762011000900007 [DOI] [PubMed] [Google Scholar]
- Lagrange B., Martin R. Z., Droin N., Aucagne R., Paggetti J., Largeot A., et al. (2013). A role for miR-142-3p in colony-stimulating factor 1-induced monocyte differentiation into macrophages. Biochim. Biophys. Acta 1833, 1936–1946. 10.1016/j.bbamcr.2013.04.007 [DOI] [PubMed] [Google Scholar]
- Lambrecht J., Mannaerts I., van Grunsven L. A. (2015). The role of miRNAs in stress-responsive hepatic stellate cells during liver fibrosis. Front. Physiol. 6:209. 10.3389/fphys.2015.00209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. J., Park S. Y., Jung M. Y., Bae S. M., Kim I. S. (2011). Mechanism for phosphatidylserine-dependent erythrophagocytosis in mouse liver. Blood 117, 5215–5223. 10.1182/blood-2010-10-313239 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Long Y., Liu X., Wang N., Zhou H., Zheng J. (2015). Chloroquine attenuates LPS-mediated macrophage activation through miR-669n-regulated SENP6 protein translation. Am. J. Transl. Res. 7, 2335–2345. [PMC free article] [PubMed] [Google Scholar]
- Longley R., Smith C., Fortin A., Berghout J., McMorran B., Burgio G., et al. (2011). Host resistance to malaria: using mouse models to explore the host response. Mamm. Genome 22, 32–42. 10.1007/s00335-010-9302-6 [DOI] [PubMed] [Google Scholar]
- Miura K. (2016). Progress and prospects for blood-stage malaria vaccines. Expert. Rev. Vaccines 3, 1–17. 10.1586/14760584.2016.1141680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhseen Z. T., Abbood N. N. (2014). MiR-142-3p regulates cardiovascular system during zebra fish development. Cell Biol. 2, 49–59. 10.11648/j.cb.20140206.11 [DOI] [Google Scholar]
- Murakami Y., Kawada N. (2016). MicroRNAs in hepatic pathophysiology. Hepatol. Res. [Epub ahead of print]. 10.1111/hepr.12730. [DOI] [PubMed] [Google Scholar]
- Nautyal A., Singh S., Parameswaran G., DiSalle M. (2005). Hepatic dysfunction in a patient with Plasmodium vivax infection. Med. Gen. Med. 7, 8–9. [PMC free article] [PubMed] [Google Scholar]
- Otogata K., Kinoshita K., Fujii H., Sakabe M., Shiga R., Nakatani R., et al. (2007). Erythrophagocytosis by liver macrophages (Kupffer cells) promotes oxidative stress, inflammation, and fibrosis in a rabbit model od steatohepatitis: implications for the pathogenesis of human nonalcoholic steatohepatitis. Am. J. Pathol. 17, 967–980. 10.2353/ajpath.2007.060441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papayannopoulou T., Brice M., Farrer D., Kaushansky K. (1996). Insights into the cellular mechanisms of erythropoietin-thrombopoietin synergy. Exp. Hematol. 24, 660–669. [PubMed] [Google Scholar]
- Papayannopoulou T., Kaushansky K. (2016). Evolving insights into the synergy between erythropoietin and thrombopoietin and the bipotent erythroid/megakaryocytic progenitor cell. Exp. Hematol. 44, 664–668. 10.1016/j.exphem.2015.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piguet P. F., Da Laperrousaz C., Vesin C., Tacchini-Cottier F., Senaldi G., Grau G. E. (2000). Delayed mortality and attenuated thrombocytopenia associated with severe malaria in urokinase- and urokinase receptor-deficient mice. Infect. Immun. 68, 3822–3829. 10.1128/IAI.68.7.3822-3829.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piguet P. F., Kan C. D., Vesin C. (2002). Thrombocytopenia in an animal model of malaria is associated with an increased caspase-mediated death of thrombocytes. Apoptosis 7, 91–98. 10.1023/A:1014341611412 [DOI] [PubMed] [Google Scholar]
- Roderburg C., Urban G. W., Bettermann K., Vucur M., Zimmermann H., Schmidt S., et al. (2011). Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology 53, 209–218. 10.1002/hep.23922 [DOI] [PubMed] [Google Scholar]
- Roy S., Benz F., Vargas C. D., Vucur M., Gautheron J., Schneider A., et al. (2015). miR-30c and miR-193 are a part of the TGF-β-dependent regulatory network controlling extracellular matrix genes in liver fibrosis. J. Dig. Dis. 16, 513–524. 10.1111/1751-2980.12266 [DOI] [PubMed] [Google Scholar]
- Rubio M., Bassat Q., Estevill X., Mayor A. (2016). Tying malaria and microRNAs: from the biology to future diagnostic perspectives. Malar. J. 15, 167. 10.1186/s12936-016-1222-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupani A. B., Amarapurkar A. D. (2009). Hepatic changes in fatal malaria: an emerging problem. Ann. Trop. Med. Parasitol. 103, 119–127. 10.1179/136485909X385054 [DOI] [PubMed] [Google Scholar]
- Shim B. S., Wu W., Kyriakis C. S., Bakre A., Jorquera P. A., Perwitasan O., et al. (2016). MicroRNA-555 has potent antiviral properties against poliovirus. J. Gen. Virol. 97, 659–668. 10.1099/jgv.0.000372 [DOI] [PubMed] [Google Scholar]
- Shu J., Kren B. T., Xia Z., Wong P. Y., Li L., Hanse E. A., et al. (2011). Genomewide microRNA down-regulation as a negative feedback mechanism in the early phases of liver regeneration. Hepatology 54, 609–619. 10.1002/hep.24421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonda N., Simonato F., Peranzoni E., Cali B., Bortoluzzi S., Bisognin A., et al. (2013). miR142-3p prevents macrophage differentiation during cancer-induced myelopoiesis. Immunity 38, 1236–1249. 10.1016/j.immuni.2013.06.004 [DOI] [PubMed] [Google Scholar]
- Stephens R., Culleton R. L., Lamb T. J. (2012). The contribution of Plasmodium chabaudi to our understanding of malaria. Trends Parasitol. 28, 73–82. 10.1016/j.pt.2011.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Zhao Q., He C., Huang D., Liu J., Chen F., et al. (2015). miR-142-5p and miR130a-3p are regulated by IL-4 and Il-13 and control profibrogenic macrophage program. Nat. Commun. 6, 8523. 10.1038/ncomms9523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo G., Bala S. (2013). MicroRNAs in liver disease. Nat. Rev. Gastroenterol. Hepatol. 10, 542–552. 10.1038/nrgastro.2013.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terpstra V., van Berkel T. J. (2000). Scavenger receptors on liver Kupffer cells mediate the in vitro uptake of oxidatively damaged red blood cells in mice. Blood 15, 2157–2163. [PubMed] [Google Scholar]
- Theurl I., Hilgendorf I., Nairz M., Tymoszuk P., Haschka D., Asshoff M., et al. (2016). On-demand erythrocyte disposal and iron recycling requires transient macrophages in the liver. Nat. Med. 22, 945–951. 10.1038/nm.4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma P., Pandey R. K., Prajapati P., Prajypati V. K. (2016). Circulating microRNAs: potential and emerging biomarkers for diagnosis of human infectious diseases. Front. Microbiol. 7:1274. 10.3389/fmicb.2016.01274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas N., Dhawan J. (2016). Exosomes: mobile platforms for targeted and synergistic signaling across cell boundaries. Cell. Mol. Life Sci. [Epub ahead of print]. 10.1007/s00018-016-2413-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Li J., Zhang X., Wen Y., Wang X. Y., Yuan Z. (2016). A pilot study of microRNAs expression profile in serum and HBsAg particles: predictors of therapeutic vaccine efficacy in chronic hepatitis B patients. Medicine (Baltimore) 95:e2511. 10.1097/md.0000000000002511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watier H., Verwaerde C., Landau I., Werner E., Fontaine J., Capron A., et al. (1992). T-cell-dependent immunity and thrombocytopenia in rats infected with Plasmodium chabaudi. Infect. Immun. 60, 136–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2015). World Malaria Report 2015, Released in December 2015 by World Health Organization 2015, WHO Document Production Services, Geneva: Available online at: www.who.int/malaria/publications/world-malaria-report-2015/report/en [Google Scholar]
- Wunderlich F., Al-Quraishy S., Dkhil M. (2014). Liver-inherent immune system: its role in blood-stage malaria. Front. Microbiol. 5:559. 10.3389/fmicb.2014.00559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich F., Brenner H. H., Helwig M. (1988a). Plasmodium chabaudi malaria: protective immunization with surface membranes of erythrocytes infected with Plasmodium chabaudi. Infect. Immun. 56, 3326–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich F., Dkhil M. A., Mehnert L. I., Braun J. V., El-Khadragy M., Borsch E., et al. (2005). Testosterone responsiveness of spleen and liver in female lymphotoxin beta receptor-deficient mice resistant to blood-stage malaria. Microbes Infect. 7, 399–409. 10.1016/j.micinf.2004.11.016 [DOI] [PubMed] [Google Scholar]
- Wunderlich F., Helwig M., Schillinger G., Speth V. (1988b). Cryptic disposition of antigenic parasite proteins in plasma membranes of erythrocytes infected with Plasmodium chabaudi. Mol. Biochem. Parasitol. 30, 55–65. [DOI] [PubMed] [Google Scholar]
- Wunderlich F., Helwig M., Schillinger G., Speth V., Wiser M. F. (1988c). Expression of the parasite protein Pc90 in plasma membranes of erythrocytes infected with Plasmodium chabaudi. Eur. J. Cell Biol. 47, 157–164. [PubMed] [Google Scholar]
- Wunderlich F., Helwig M., Schillinger G., Vial H., Philippot J., Speth V. (1987). Isolation and characterization of parasites and host cell ghosts from erythrocytes infected with Plasmodium chabaudi. Mol. Biochem. Parasitol. 23, 103–115. 10.1016/0166-6851(87)90145-9 [DOI] [PubMed] [Google Scholar]
- Wunderlich F., Mossmann H., Helwig M., Schillinger G. (1988d). Resistance to Plasmodium chabaudi in B10 mice: influence of the H-2 complex and testosterone. Infect. Immun. 56, 2400–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich F., Schillinger G., Helwig M. (1985). Fractionation of Plasmodium chabaudi-infected erythrocytes into parasites and ghosts. Z. Parasitenkd. 71, 545–551. 10.1007/BF00928358 [DOI] [PubMed] [Google Scholar]
- Wunderlich F., Stüebig H., Köenigk E. (1982). Development of Plasmodium chabaudi in mouse red blood cells: structural properties of the host and parasite membranes. J. Protozool. 29, 60–66. 10.1111/j.1550-7408.1982.tb02880.x [DOI] [PubMed] [Google Scholar]
- Yu D. S., An F. M., Gong B. D., Xiang X. G., Lin L. Y., Wang H., et al. (2012). The regulatory role of microRNA-1187 in TNF-α-mediated hepatocyte apoptosis in acute liver failure. Int. J. Mol. Med. 29, 663–668. 10.3892/ijmm.2012.888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Ju W. Q., Yuan X. P., Zhu X. F., Wang D. P., He X. S. (2016). miR-26a regulates mouse hepatocyte proliferation via directly targeting the 3′ untranslated region of CCND2 and CCNE2. Hepatobiliary Pancreat. Dis. Int. 15, 65–72. 10.1016/S1499-3872(15)60383-6 [DOI] [PubMed] [Google Scholar]