Abstract
Aptamers are a promising class of affinity reagents because they are chemically synthesized, making them highly reproducible and distributable as sequence information rather than a physical entity. Although many high-quality aptamers have been previously reported, it is difficult to routinely generate aptamers that possess both high affinity and specificity. One of the reasons is because conventional aptamer selection can only be performed either for affinity (positive selection) or for specificity (negative selection), but not both simultaneously. In this work, we harness the ability of fluorescence activated cell sorting (FACS) to perform multi-color sorting to simultaneously screen for affinity and specificity at a throughput of 107 aptamers per hour. As proof of principle, we generated DNA aptamers for three diverse proteins that exhibit picomolar to low nanomolar affinity in human serum, and show that these aptamers are capable of outperforming high-quality monoclonal antibodies in a standard ELISA detection assay.
Keywords: Aptamers, Molecular Recognition, High-Throughput Screening
Aptamers are a promising class of affinity reagents as they are chemically synthesized and their discovery is performed completely in vitro rather than relying on in vivo biological processes, making them reproducible and potentially well suited for high-throughput discovery.[1,2] They are also thermostable, fold reversibly and can be distributed as sequence information rather than as a physical entity, thereby ensuring reproducible synthesis and facile dissemination throughout the research community. Previously, aptamers with extremely high affinities have been reported for proteins, small molecules, and even cell surfaces. Importantly, a number of investigators have also demonstrated that aptamers can exhibit exceptional specificity. For example, Ferapontova et al. and Jiang et al. reported aptamers that bind strongly to theophylin, but not to caffeine, a molecule differing by only a single methyl group[3,4]. However, obtaining aptamers that simultaneously possess both high affinity and specificity, on a routine basis, has been difficult using conventional aptamer discovery methodologies such as SELEX[5,6]. Among many reasons, one of the main challenge is due to the fact that conventional selection can only be performed either for affinity (positive selection) or for specificity (negative selection), but not both simultaneously.
Here, we describe an aptamer-discovery technique in which we can simultaneously screen for aptamer affinity and specificity in a high-throughput manner. Our multi-parameter particle display (MPPD) strategy builds on our previous work[7] that transforms solution-phase aptamers into “aptamer particles”, wherein each displays many copies of a single aptamer. When incubated with a fluorescent protein, each aptamer can be interrogated individually for its ability to bind the target using fluorescence-activated cell sorting (FACS). In this work, we harness the ability of FACS to perform multi-color sorting to simultaneously screen for affinity and specificity. By labeling the target and background with different colored fluorophores we can simultaneously measure the binding of each aptamer to the target as well as the background. This major advancement allows us to isolate the aptamer that achieves the best affinity and specificity at a throughput of 107 aptamers per hour. As a demonstration, we have selected DNA aptamers with excellent target specificity for three diverse protein targets that show picomolar to low nanomolar affinity in human serum. Furthermore, we show that these aptamers are capable of outperforming high-quality monoclonal antibodies in a standard ELISA detection assay in serum.
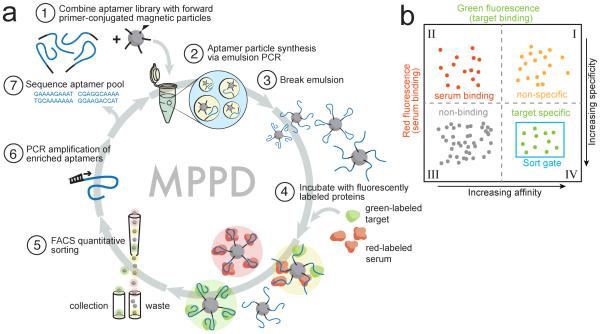
MPPD begins with the transformation of a solution-phase aptamer library into monoclonal aptamer particles through emulsion PCR, as described previously (Fig 1a)[7]. We started with a library of aptamers comprising 44 randomized nucleotides flanked by a pair of 18-nucleotide primer-binding sites (see Methods for sequences). We prepared water-in-oil emulsions with PCR reagents such that each water droplet contained a magnetic bead coated with the forward PCR primer, under conditions that maximize the statistical likelihood of having no more than one DNA template per droplet (Step 1)[7]. We then performed PCR amplification within the droplet, yielding a library of aptamer particles that each display ~2×105 copies of a single sequence on their surface (Step 2)[7]. The emulsion was then broken, and the aptamer particles were collected via magnetic separation after denaturation and release of the reverse strand (Step 3).
Figure 1. Overview of MPPD.
(a) We transform solution-phase aptamers into aptamer particles by covalently conjugating forward PCR primers to magnetic particles (1) and performing emulsion PCR (2) under conditions that produce monoclonal aptamer particles displaying many copies of a single sequence (3). These particles are incubated with target and non-target proteins labeled with distinct fluorophores (green and red, respectively) (4), then sorted using FACS (5) to isolate aptamers that exhibit high affinity and specificity. These aptamers are then PCR amplified for additional screening (6) or sequenced for further characterization (7). (b) Aptamer particles are sorted with two-dimensional FACS to selectively isolate aptamers with high green (high affinity) and low red (high specificity) fluorescence.
We tested MPPD by generating DNA aptamers for tumor necrosis factor α (TNF-α; MW=52kDa), an important marker of inflammation[8], in the complex medium of diluted human serum. We labeled TNF-α with a mAb conjugated to AlexaFluor-488 (green), and non-specifically labeled the serum proteins with AlexaFluor-647 (red) using a commercial kit (Step 4). Next, we used FACS to sort individual aptamer particles that simultaneously exhibit high affinity and specificity (Step 5). This entails discarding aptamers that exhibit either low green fluorescence (Fig. 1b, quadrants II and III), indicating low affinity for TNF-α, or high red fluorescence and high green fluorescence (quadrant I), indicating high target affinity but non-specific binding to serum proteins. We collected the aptamers with high green and low red fluorescence (quadrant IV), representing the desired population that bind TNF-α, with high affinity and specificity, and performed PCR on the sorted particles to generate an enriched pool for further screening (Step 6) or sequencing (Step 7).
Aptamer affinity and specificity are highly dependent on the screening stringency imposed by the concentration of the target [T] and competitors [C] in the sample. These two parameters can dramatically shift the binding of the aptamer population towards either the target or the competitor (see Supplemental Information and Fig S1 for details). MPPD enables us to monitor the on- and off-target binding behavior of each aptamer using FACS and identify the optimal screening stringency by varying [T] and [C]. Importantly, to minimize the time required for FACS screening while still ensuring that we do not compromise the diversity of the initial library, we performed one round of conventional bead-based selection starting with an initial library (~6×1014 molecules) to generate a ‘pre-enriched’ pool, which we used in the first round of MPPD. This allows for FACS screening to be performed at ~107 aptamer particles/hour to cover the entire sequence space of the enriched library in a reasonable time.
We performed two separate screens and compared the specificity of the resulting aptamers. For the ‘buffer-screen’, we performed four screening rounds in buffer with no competitors, starting with 100 nM TNF-α in Round 1 (Fig S2a). We isolated 0.19% of the population that exhibited the highest fluorescence, and then increased the screening stringency in subsequent rounds by decreasing [T]. Aptamer particles from Round 4 showed a high fluorescence signal even at 1 nM TNF-α (Fig S2), and we isolated 0.06% of the sorted aptamer particles to obtain the final buffer-screen pool. For the ‘serum-screen’, we performed four rounds of MPPD in diluted human serum. For Round 1, we used 100 nM TNF-α and 1% serum and sorted 0.08% of the aptamer particle population that exhibited high green and low red fluorescence (Fig S2 b). For subsequent rounds, we systematically decreased [T] while increasing the serum concentration [C]. By Round 4, the aptamer particles exhibited strong green fluorescence signal even with 1 nM TNF-α in 10% serum. This is remarkable since the total competitor protein concentration in 10% serum (~0.2 mM) exceeds that of TNF-α by roughly five orders of magnitude. We isolated 0.14% of the aptamer particles from Round 4 to obtain the final serum-screen pool.
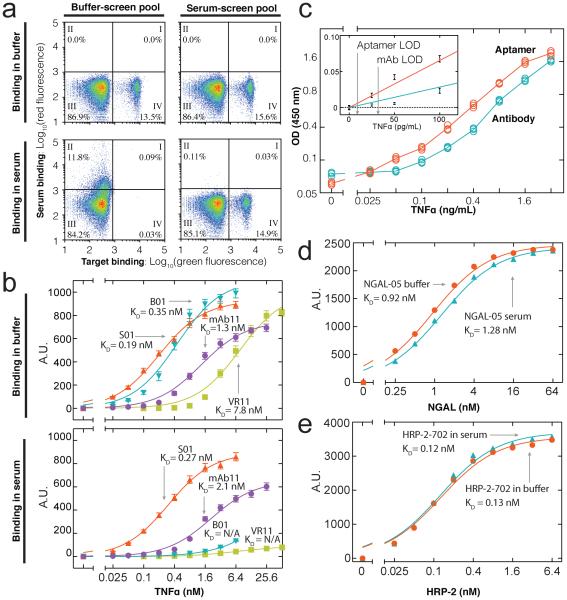
Although both pools showed comparable affinity for TNF-α in buffer (Fig. 2a, top), the buffer-screen aptamers showed poor specificity in serum (Fig 2a, bottom left). For example, when the buffer-screen aptamers were challenged with 1 nM TNF-α in 10% human serum, only 0.03% of the aptamer particles exhibited modest target binding and low serum binding (quadrant IV). In contrast, the serum-screen pool showed high specificity under identical conditions (Fig 2a, bottom right), with only a small reduction in the sorted aptamer population in serum (14.9%) versus buffer (15.6%). In this way, we show that MPPD can generate aptamers that specifically bind to TNF-α even among a vast excess of diverse competitor proteins.
Figure 2. MPPD isolates high affinity and specificity aptamers.
(a) Specificity of aptamers derived from buffer-screen (left column) and serum-screen (right column), incubated with 1 nM labeled TNF-α either in buffer (top) or 10% human serum (bottom). Both pools exhibit high affinity without competitors (Quadrant IV, top row), but only the serum-screen pool retains high affinity in serum (bottom row). (b) Binding curves and calculated effective affinities to TNF-α for the top-performing serum-screen (S01) and bufferscreen (B01) aptamers as well as a previously published TNF-α aptamer (VR11), and a commercial TNF-α antibody (mAb11). In buffer, S01 shows slightly higher affinity than B01. In serum, S01 exhibits specific, high affinity exceeding that of mAb11, whereas B01 and VR11 show negligible binding. (c) S01 achieves a superior limit of detection to mAb11 in ELISA assays performed in serum. We also used MPPD to isolate (d) an NGAL aptamer with a KD of 0.92 nM in buffer and 1.28 nM in serum and (e) an HRP-2 aptamer with a KD of 0.13 nM in buffer and 0.12 nM in serum.
We cloned individual aptamer sequences from both pools, and used a fluorescence-based binding assay to compare the equilibrium binding constant (KD) of 20 clones from each pool. Sequences and relative affinities of all aptamers are shown in Supplemental Information. In buffer, the highest-affinity aptamer from the serum-screen (S01) showed slightly higher affinity (KD= 0.19 nM) than its counterpart from the buffer-screen (B01; KD = 0.35 nM) (Fig 2b). Remarkably, the affinity of S01 was essentially unchanged in serum (KD = 0.27 nM) compared to that in buffer, clearly demonstrating its exquisite specificity. In contrast, B01 binding virtually disappeared in 10% serum, and we could not obtain a meaningful KD measurement.
We next compared S01 with VR11[9], a published TNF-α aptamer reported to possess good specificity, in that it does not bind TNF-β, a protein with ~30% sequence homology to TNF-α. VR11 exhibited an affinity for TNF-α of 7.8 nM in buffer, consistent with the reported value (7 nM), but showed minimal target affinity in serum (Fig 2b), suggesting that even an aptamer that can differentiate homologous targets may have insufficient specificity to recognize its target in a heterogeneous sample. S01 also exhibited superior performance to a monoclonal antibody currently used in high-sensitivity commercial TNF-α detection assays. mAb11 exhibited the highest affinity and specificity of the various different commercial kits we examined, with a KD of 1.3 nM in buffer and 2.1 nM in 10% serum (Fig 2d)—nearly an order of magnitude worse than that of S01 in the same conditions.
We also tested S01 as a potential tool for clinical molecular diagnostics in an enzyme-linked immunosorbent assay (ELISA). We selected the commercial ELISA kit that yielded the best limit of detection (LOD) in 10% serum, with LOD defined as the point on a linear fit of signal to concentration that reaches three times the standard deviation of the signal from a negative control. For comparison, we used mAb11 as the capture reagent and S01 as the detection reagent; these two reagents bind to different epitopes on TNF-α, as mAb11 was used to label TNF-α during MPPD screening. The commercial assay yielded a LOD of ~32 pg/ml, consistent with previously reported values[10], whereas our S01-based ELISA exhibited a LOD of ~9.2 pg/ml, an improvement of more than three-fold (Fig. 2c). This is particularly striking given that each detection antibody gains a boost in signal by having multiple (typically >5) biotin labels, allowing it to bind multiple streptavidin HRP reporters, whereas S01 is labeled with just a single biotin.
We next repeated the screening process for two additional protein targets that are chemically and structurally distinct from TNF-α as well each other. The first was neutrophil gelatinase-associated lipocalin (NGAL; MW = 25 kDa), a glycoprotein serum biomarker of renal injury, anemia, and cancer[11–13]. After five rounds of MPPD with decreasing target concentration and increasing competitor concentration, 28.9% of the aptamer particles showed strong binding in the presence of 500 pM NGAL in 10% serum (Fig S3a, right). The most abundant aptamer (NGAL-05) from this pool exhibited a KD of 920 pM in buffer and 1.28 nM in 10% serum (Fig. 2d). We performed a similar process for Plasmodium falciparum histidine-rich protein 2 (HRP-2; MW = 37 kDa), a biomarker for severity of infection and drug susceptibility for this pathogen[14–16]. HRP-2 is a particularly challenging target because it contains tandem repeats of positively-charged histidines that may non-specifically interact with the negatively-charged phosphate backbone of DNA[17]. After seven rounds of decreasing target concentration and increasing serum concentration (Fig. S3b, left), 25.47% of the aptamer population showed binding to 100 pM HRP-2 in 10% serum. The most abundant aptamer (HRP-2-702) from this pool showed high affinity and specificity for HRP-2, with a KD of 128 pM in buffer that was essentially unchanged (124 pM) in 10% serum (Fig. 2e).
In summary, we describe the MPPD aptamer discovery platform, which can simultaneously screen for affinity and specificity in complex mixtures (such as diluted serum), and yields reagents that can match or exceed the performance of high-quality commercial antibodies in an ELISA format. We show that our method is general - we performed the screen for three different target proteins (TNF-α, NGAL and HRP-2) in diluted serum and obtained DNA aptamers with affinities in picomolar to nanomolar range. Given that FACS instruments have become increasingly available in many research settings, we believe that our aptamer discovery method could be used by the broader research community for generating custom reagents for a wide range of biomedical applications.
Supplementary Material
ACKNOWLEDGMENTS
Research reported in this publication was supported by National Institutes of Health (R44GM109721), DARPA (N66001-14-2-4055), ARO (W911NF-10-2-0114) and the Garland Initiative.
Footnotes
Supporting Information
The Supporting Information is available free of charge at XXX.
Notes
J.W., J.Y., Q.Y., J.M., A.S., M.V., R.L., Q.G., B.S.F., H.T.S. are employees and/or shareholders of Aptitude Medical Systems Inc.
Contributor Information
Dr. Jinpeng Wang, Aptitude Medical Systems Inc. Santa Barbara, CA, 93105.
Jingwen Yu, Aptitude Medical Systems Inc. Santa Barbara, CA, 93105.
Dr. Qin Yang, Aptitude Medical Systems Inc. Santa Barbara, CA, 93105
John McDermott, Aptitude Medical Systems Inc. Santa Barbara, CA, 93105.
Alexander Scott, Aptitude Medical Systems Inc. Santa Barbara, CA, 93105.
Matthew Vukovich, Aptitude Medical Systems Inc. Santa Barbara, CA, 93105.
Remy Lagrois, Aptitude Medical Systems Inc. Santa Barbara, CA, 93105.
Dr. Qiang Gong, Aptitude Medical Systems Inc. Santa Barbara, CA, 93105
William Greenleaf, Department of Genetics, Stanford University, Stanford, CA 94305
Dr. Michael Eisenstein, Department of Electrical Engineering and Radiology, Stanford University, Stanford, CA 94305
Dr. B. Scott Ferguson, Aptitude Medical Systems Inc. Santa Barbara, CA, 93105.
H. Tom Soh, Department of Electrical Engineering and Radiology, Stanford University, Stanford, CA 94305.
REFERENCES
- [1].Cho EJ, Lee J-W, Ellington AD. Annu. Rev. Anal. Chem. (Palo Alto. Calif) 2009;2:241–64. doi: 10.1146/annurev.anchem.1.031207.112851. [DOI] [PubMed] [Google Scholar]
- [2].Keefe AD, Pai S, Ellington A. Nat. Rev. Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jiang H, Ling K, Tao X, Zhang Q. Biosens. Bioelectron. 2015;70:299–303. doi: 10.1016/j.bios.2015.03.054. [DOI] [PubMed] [Google Scholar]
- [4].Ferapontova EE, Olsen EM, Gothelf KV. J. Am. Chem. Soc. 2008;130:4256–4258. doi: 10.1021/ja711326b. [DOI] [PubMed] [Google Scholar]
- [5].Ellington a D., Szostak JW. Nature. 1990;346:818–22. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- [6].Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- [7].Wang J, Gong Q, Maheshwari N, Eisenstein M, Arcila ML, Kosik KS, Soh HT. Angew. Chemie - Int. Ed. 2014;53:4796–4801. doi: 10.1002/anie.201309334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Strieter RM, Kunkel SL, Bone RC. Crit. Care Med. 1993;21:S447–63. doi: 10.1097/00003246-199310001-00006. [DOI] [PubMed] [Google Scholar]
- [9].Orava EW, Jarvik N, Shek YL, Sidhu SS, Gariepy J. ACS Chem Biol. 2013;8:170–178. doi: 10.1021/cb3003557. [DOI] [PubMed] [Google Scholar]
- [10].Ettehadi P, Greaves MW, Wallach D, Aderka D, Camp RD. Clin. Exp. Immunol. 1994;96:146–51. doi: 10.1111/j.1365-2249.1994.tb06244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chakraborty S, Kaur S, Guha S, Batra SK. Biochim. Biophys. Acta - Rev. Cancer. 2012;1826:129–169. doi: 10.1016/j.bbcan.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ozkan S, Durukan P, Kavalci C, Duman A, Burak Sayhan M, Salt O, Ipekci A. Iran. Red Crescent Med. J. 2014;16 doi: 10.5812/ircmj.14133. DOI 10.5812/ircmj.14133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Mishra J, Dent C, Tarabishi R, Mitsnefes MM, Ma Q, Kelly C, Ruff SM, Zahedi K, Shao M, Bean J, et al. Lancet. 2005;365:1231–1238. doi: 10.1016/S0140-6736(05)74811-X. [DOI] [PubMed] [Google Scholar]
- [14].Noedl H, Wernsdorfer WH, Miller RS, Wongsrichanalai C. Antimicrob. Agents Chemother. 2002;46:1658–1664. doi: 10.1128/AAC.46.6.1658-1664.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Howard RJ, Uni S, Aikawa M, Aley SB, Leech JH, Lew AM, Wellems TE, Rener J, Taylor DW. J. Cell Biol. 1986;103:1269–1277. doi: 10.1083/jcb.103.4.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Grigg MJ, William T, Barber BE, Parameswaran U, Bird E, Piera K, Aziz A, Dhanaraj P, Yeo TW, Anstey NM. J. Clin. Microbiol. 2014;52:2053–2060. doi: 10.1128/JCM.00181-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ravetch JV, Feder R, Pavlovec A, Blobel G. Nature. 1984;312:616–620. doi: 10.1038/312616a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.