Abstract
Most balanced translocations are thought to result mechanistically from non-homologous endjoining (NHEJ) or, in rare cases of recurrent events, by nonallelic homologous recombination (NAHR). Here, we use low coverage mate pair whole genome sequencing to fine map rearrangement breakpoint junctions in both phenotypically normal and affected translocation carriers. In total, 46 junctions from 22 carriers of balanced translocations were characterized. Genes were disrupted in 48% of the breakpoints; recessive genes in four normal carriers and known dominant intellectual disability genes in three affected carriers. Finally, seven candidate disease genes were disrupted in five carriers with neurocognitive disabilities (SVOPL, SUSD1, TOX, NCALD, SLC4A10) and one XX-male carrier with Tourette syndrome (LYPD6, GPC5). Breakpoint junction analyses revealed microhomology and small templated insertions in a substantive fraction of the analyzed translocations (17.4%; n=4); an observation that was substantiated by reanalysis of 37 previously published translocation junctions. Microhomology associated with templated-insertions is a characteristic seen in the breakpoint junctions of rearrangements mediated by the error prone replication-based repair mechanisms (RBMs). Our data implicate that a mechanism involving template switching might contribute to the formation of at least 15% of the interchromosomal translocation events.
Keywords: balanced chromosomal aberration, reciprocal translocation, whole genome sequencing, microhomology, non-homologous end joining, replication-based repair mechanisms
INTRODUCTION
Using banded chromosomes, the overall incidence of balanced structural rearrangements without apparent cytogenetically observed gain or loss has been estimated to be 0.212 % in an unselected newborn population (Jacobs, et al., 1992). Such balanced chromosome aberrations (BCAs) may be subdivided into reciprocal translocations, Robertsonian translocations, pericentric inversions and paracentric inversions. Specifically, reciprocal translocations are observed in 0.09% of newborns; the majority of which are inherited from either parent. Approximately 20% of translocations occur de novo with an estimated mutational rate of 2.7×10−4 per gamete per generation (Jacobs, et al., 1992).
Only some 6% of de novo reciprocal translocations are thought to have an associated disease phenotype, recognized at birth or at a 1 year follow-up physical examination (Warburton, 1991). However, due to errors in meiotic recombination and malsegregation of the rearranged chromosomes, translocation carriers have a risk of recurrent abortions and having children with inherited unbalanced rearrangements. Importantly, translocations are of high interest in identifying the causes of new genetic disease (Higgins, et al., 2008; Hofmeister, et al., 2015); this requires sequencing of translocation breakpoint junctions in patient DNA samples to identify genes potentially mediating the observed clinical phenotype. The advent of massively parallel sequencing and affordable whole genome sequencing (WGS) allows rapid and cost-efficient detailed investigation of translocation events, but this technology remains rarely used in clinical cytogenetics practice. An important potential clinical application is the identification of de novo translocations in prenatal samples. The disruption of known disease genes or potential dysregulation through a position effect may provide a molecular diagnosis, allowing for better and more accurate clinical management of the translocation carrier and genetic counseling information for the family (Talkowski, et al., 2012a). Similarly, in symptomatic carriers the identification of the specific disease-causing gene may enable personalized therapeutic strategies.
Breakpoint junctional analyses of WGS data obtained from translocation carriers can provide insights into the potential molecular mechanisms of chromosome break and repair that cause the aberrations; observed ‘mutational signatures’ may allow inference as to potential mechanisms for formation. By studying the break-join events of the DNA molecule at the chromosomal junctions, the potential mechanism(s) underlying the rearrangement can often be inferred. For example, the presence of large segments of DNA homology flanking translocation junctions (usually > 200 bp) suggest nonallelic homologous recombination (NAHR) mechanism (Stankiewicz and Lupski, 2002) which mediates some interchromosomal recurrent balanced translocations (Giglio, et al., 2002; Hastings, et al., 2009b; Lupski, 2015; Ou, et al., 2011). In contrast, most previous publications of balanced translocations have shown a lack of both large homology and microhomology in the breakpoint junctions. This was proposed to indicate that canonical non-homologous end joining (c-NHEJ) is the major mechanism underlying the formation of balanced translocations (Chiang, et al., 2012). Canonical NHEJ is a repair mechanism that joins double-stranded DNA breaks with a high degree of precision but occasionally deleting or inserting a few random nucleotides during DNA processing before ligation (Pannunzio, et al., 2014). Replication-based mechanisms (RBMs), such as fork stalling and template switching/microhomology-mediated break-induced replication (FoSTeS/MMBIR) (Hastings, et al., 2009a; Lee, et al., 2007) can underlie the formation of many disease-causing nonrecurrent structural variants in humans (reviewed in (Abyzov, et al., 2015; Carvalho and Lupski, 2016; Conrad, et al., 2010; Stankiewicz and Lupski, 2010). Occasional intra and/or inter chromosomal template-switches during RBM repair can occur, due to involvement of a lower processivity polymerase, and result in complex rearrangements. The mutational signature that may be observed in the breakpoint junctions of RBM mediated rearrangements involves the presence of microhomology, small templated insertions at breakpoint junctions as well as inversion of large genomic segments accompanied by copy number gains (e.g. duplications and triplications) (Carvalho, et al., 2013; Carvalho, et al., 2011). RBMs have been suggested to contribute to the formation of BCAs during mitosis based on the observation of complex rearrangements associated with seemingly balanced translocations, for example, copy number neutral inversions and copy number loss (Chiang, et al., 2012; Hsiao, et al., 2014). Other examples of constitutional translocations formed by RBM are recurrent translocations involving palindromic AT-rich repeats (Kato, et al., 2012) and one chromosomal translocation t(14;17)(q32;q11.2) disrupting NF1 (MIM# 613113), that seem to have been generated by a mechanism involving fork stalling and a rereplication process (Hsiao, et al., 2014).
Here we used low coverage mate pair WGS followed by capillary-sequencing confirmation to pinpoint the breakpoint location of 46 breakpoints from 22 balanced translocation carriers in order to i) ascertain gene variations of potential clinical utility for genetic counseling and facilitate gene discovery, and ii) access the mutational signatures of translocation junctions and infer potential underling mechanism for formation. The cohort includes both clinically unaffected (n=8) and affected individuals (n=14), thus contrasting the makeup of benign and pathological events. Finally, we validated our ‘mutational signature’ findings by reanalyzing the junctional sequences from 37 previously published translocations.
MATERIALS AND METHODS
Subjects
The samples included in this study, were originally referred for chromosome analysis either at the Clinical Genetics department clinical laboratory at the Karolinska University Hospital, Stockholm or in one case (8480THO) at the Helsinki University Hospital. In the 22 individuals studied the chromosome analysis had identified a balanced chromosomal aberration that could also be detected by WGS. Individual 872-05Ö harbored two separate events (Hofmeister, et al., 2015) making the total number of translocations studied here 23. Ten individuals were referred for chromosome analysis due to amniocentesis, multiple miscarriages or the birth of a child with an unbalanced karyotype, eight of these individuals were unaffected and two had mild neurocognitive deficits. Twelve individuals were referred for chromosome analysis due to neurocognitive deficits and/or malformations (Table 1). A custom array comparative genomic hybridization analysis had been done in six individuals (872-05Ö, 109-06Ö, 232-07F, 2-03E, 337-01D, 29-03E) (Lindstrand, et al., 2010) and fluorescent in situ hybridization breakpoint mapping had been performed in twelve individuals (31-05E, 862-06Ö, 106-06Ö, 58-06Ö, 157-06Ö, 191-06-Ö, 263-06Ö, 175-06Ö, 872-05Ö, 887-05Ö, 109-06Ö and 232-07F) (data not shown). The local ethical boards in Stockholm, Sweden and in Helsinki, Finland approved the study. Karyotypes and phenotypic status are provided in Table 1.
Table 1.
Karyotypes and Mode of Ascertainment of included cases
| Case | Karyotype | Ascertainment | Inheritance | Phenotype Summary |
|---|---|---|---|---|
| 31-05E | 46,XY,t(14;22)(q24;q12) | Recurrent miscarriages | n.i. | Healthy |
| 862-06Ö | 46,XX,t(16;22)(p11;q13.1) | Parent of a 46,XX,der(22)t(16;22)(p11;q13.1) miscarriage | Paternal | Healthy |
| 106-06Ö | 46,XX,t(4;7)(q25;q22) | Recurrent miscarriages | n.i. | Healthy |
| 58-06Ö | 46,XY,t(3;12)(q23;q21) | Recurrent miscarriages | n.i. | Healthy |
| 157-06Ö | 46,XX,t(4;7)(q21;p15) | Recurrent miscarriages | Maternal | Healthy |
| 191-06Ö | 46,XY,t(2;3)(p13;p25) | Recurrent miscarriages | Mother not carrier; Father not tested |
Healthy |
| 263-06Ö | 46,XX,t(10;11)(q22;p15) | Recurrent miscarriages | Father not carrier; Mother not tested |
Healthy |
| 175-06Ö | 46,XX,t(10;15)(q23;q15) | Parent of a 46,XY,der(10)t(10;15)(q23;q15) miscarriage | Paternal | Healthy |
| 872-05Ö | 46,XX,t(1;8)(p22;q24), t(5;18)(p15;q11) |
Amniocentesis because of previously stillborn child | Maternal | Proband: Reading difficulties Mother: Reading difficulties |
| 887-05Ö | 46,XX,t(5;7)(q14;q34) | Amniocentesis because of advanced maternal age | Maternal | Proband: ADHD and Reading difficulties Mother: Reading difficulties |
| 109-06Ö | 46,XY,t(2;6)(q34;q21)dn | Affected phenotype | de novo | Autism, Epilepsy |
| 232-07F | 46,XY,t(3;7)(p33;p12) | Affected phenotype | Paternal | Proband: Mild ID, Autistic traits Father: Speech delay |
| 851-06Ö | 46,X,t(X;17)(p22.1;p13) | Affected phenotype | Maternal | Proband: Epilepsy, psychatric illness Mother: Epilepsy, psychatric illness |
| 8480THO | 46,XY,t(2;9)(q37.3;q32)dn | Affected phenotype | de novo | Autistic features, Epilepsy |
| 841-95D | 46,XY,t(2;21)(p13;p11.2)dn | Affected phenotype | de novo | Autism, ADHD |
| 155-90D | 46,XX,t(2;13)(q24;q33) | Affected phenotype | Mother not carrier; Father not tested |
XX male, tics, Tourettes |
| 2644-07D | 46,XY,t(4;8)(q21;q13)dn | Affected phenotype | de novo | DD, Autism, ADHD |
| 2587-07D | 46,XY,t(1;2)(q42;q31)dn | Affected phenotype | de novo | Vertebral anomaly |
| 2-03E | 46,XX,t(6;8)(q23;q24)dn | Affected phenotype | de novo | DD, Epilepsy |
| 337-01D | 46,XX,t(3;12)(q26.1;p11.2)dn | Affected phenotype | de novo | ID, Autism, Epilepsy |
| 782-95D | 46,XY,t(10;12)(q24;q13)dn | Affected phenotype | de novo | DD |
| 29-03E | 46,XY,t(9;16)(p21;q21)dn | Affected phenotype | de novo | Obesity, ID |
ADHD, Attention Deficit Hyperactivity Disorder; ID, Intellectual disability; DD, Developmental Delay
Finally, we included 37 previously published seemingly balanced translocations in which breakpoints junctions had already been characterized and junction fragment sequences were available for reanalysis (Supp. Table S1). For phenotypic data of these individuals we refer to the original publications (Chen, et al., 2008; Chiang, et al., 2012; Higgins, et al., 2008; Hsiao, et al., 2014; Schluth-Bolard, et al., 2013; Talkowski, et al., 2012a; Talkowski, et al., 2012b).
Mate pair whole genome sequencing
To pinpoint the exact genomic positions of the chromosome breaks we used low coverage mate pair (Collins and Weissman, 1984) whole genome sequencing. Libraries were prepared using Illumina's Nextera Mate Pair Sample Preparation Kit according to the manufacturer's instruction (Illumina Document # 15035209 Rev. D, May 2013). The workflow uses 1 ug of high quality DNA estimated from gel imaging and concentration measured using a Qubit HS fluorometer (ThermoFisher Scientific, Pittsburg PA). Gel purification was not used to allow for a higher spread of insert sizes and less laborious lab protocol. In brief, the method uses simultaneous fragmentation to approximately 2 kb and ligation of the circularization adapter using 4 ul of the nextera enzyme provided in the kit. After strand displacement to fill-in a remaining gap and reaction clean-up, the insert sizes are controlled using a Bioanalyzer with the DNA 12000 kit (Agilent Technologies, Santa Clara CA) and quantified using Qubit. Then 600 ng of product is circularized by ligation and non-circularized material is degraded enzymatically. Next, the circles are fragmented to 300 to 1000 bp using a Covaris S2 with T6 glass tubes (Covaris, Woburn, MA). The adapter containing fragments are magnetic bead purified using a biotin moiety on the adapter. The remaining DNA is then subjected to the Illumina library preparation procedure including end repair, A-tailing, barcoded adapter ligation, PCR and finally magnetic bead clean up using carboxylic acid coated beads (Dynabeads MyOne CA, ThermoFisher Scientific) automated on a Bravo Workstation setup B (Agilent Technologies) (Borgstrom, et al., 2011). The final libraries were quality controlled using Qubit and Bioanalyser, diluted to 10 nM and sequenced as two samples per lane on an Illumina 2500 sequencer (2×100 bp). A technical summary of the sequencing raw data is provided in Supp. Table S2 (size distribution mode 2 kb, coverage 4x).
The raw sequence reads were base called using CASAVA RTA 1.18 (http://support.illumina.com/sequencing/sequencing_software/casava.htm). Following Illumina guidelines for Mate Pair post processing, adapter sequences were removed using Trimmomatic v0.32 (Bolger, et al., 2014). Remaining pairs were aligned to the hg19 human reference genome sequence using the Burrows-Wheeler Alignment tool (BWA; MEM-algorithm, version 0.7.4-r385) (Li and Durbin, 2009) resulting in a 4X mapping coverage. Discordant read mapping was processed using FindTranslocations (https://github.com/vezzi/TIDDIT/releases/tag/v0.9), a publicly available open source code software developed in-house, implementing a sliding window analogue of a previously published procedure (Talkowski, et al., 2012a). Briefly, chromosomes are divided into overlapping windows, which are scored for discordant read pairs, and the discordant reads are investigated for connection to a common receiving cluster of reads elsewhere. The algorithm proceeds linearly by considering only links to later, uninvestigated positions. Sufficient read mapping quality and deviating mapped insert size - or different chromosomes - are inclusion criteria. If the number of reciprocal read pairs are above a threshold, and the read coverage in the two cluster windows is not excessive, an event is called, and quality information such as the fraction of reads in each mapping orientation is stored. The program has been used previously for the detection of structural variants from WGS data; both balanced (Hofmeister, et al., 2015) and unbalanced (Lieden, et al., 2014) as well as leukemic aberrations (Nord, et al., 2014). A window size of 10 kb, stepping of 1 kb and a minimum of 8 supporting read-pairs was used. Calls were then annotated with frequency of occurrence in a local database containing calls from 62 samples analyzed with the same WGS protocol. Split read analysis was not implemented in FindTranslocations version 0.9, and was carried out using BLAT (Kent, 2002) of reads showing soft clipping or supplementary alignments in the area pinpointed by read pair analysis. CNVnator (Abyzov, et al., 2011) was used to call CNVs. Custom scripts were used to visualize variations with Circos (Krzywinski, et al., 2009).
Breakpoint junction PCR
Primers flanking the junctions were designed approximately 1-2 kb away from the estimated breakpoint area. For Sanger sequencing, new primers were designed approximately 300-500 bp away from the estimated break. In some cases, the genomic environment required primers to be designed further away or closer to the break. Primer sequences are shown in Supp. Table S3. Breakpoint PCR was performed by standard methods using Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific, Pittsburg PA) and subjected to electrophoreses on a 1.5% agarose gel. To ensure specificity, a control sample of pooled genomic DNA from Promega (Madison, WI, USA) was run together with the patient samples. Specific products of expected size, not present in control samples, were Sanger sequenced. Sequences were aligned using BLAT (UCSC Genome Browser) (Kent, 2002) and visualized in CodonCode Aligner (CodonCode Corp., Dedham, MA).
Nomenclature
The description of chromosome aberrations has previously been governed by ISCN, and sequencing results according to the guidelines of HGVS nomenclature. The current introduction of large scale sequencing into the determination of chromosomal aberrations requires an updated nomenclature. Previous suggestions include a BLAST-result centric description (Ordulu, et al., 2014). Currently the nomenclature suggestion under discussion is SVD-WG004 (http://www.hgvs.org/mutnomen/comments004.html), a hybrid between ISCN and HGVS nomenclatures. For molecular karyotypes we use a previous version of this scheme as suggested by Peter Taschner (http://www.hgvs.org/mutnomen/SVtrans_HGVS2013_PT.pdf).
RESULTS
Low coverage whole genome sequencing detects balanced translocations at nucleotide-level resolution
To ascertain clinically relevant gene disruptions, detect small genomic imbalances below the level of resolution of the clinical cytogenetics techniques used, and glean inferences from breakpoint junctional events to elucidate the potential underlying mechanisms of rearrangement formation, we investigated known translocation carriers with low coverage mate pair WGS. In total, 46 junctions from 22 balanced translocation carriers were analyzed, including both individuals who were deemed phenotypically normal (n=8) and those with a clinical pathological phenotype (n=14) (Table 1).
All the cytogenetically defined and balanced translocations presented here were detected by WGS (Table 2). Using an in-house software, FindTranslocations, we detected an average of 130 structural variants per sample. These may include common polymorphic variations, patient personal genome structural differences from the reference genome assembly and potential experimental or computational analysis methods artifacts. Filtering these variants to remove recurring events in a database resulted in an average of 3.5 unique rare variant events per individual. In one third of the study subjects only the previously karyotyped novel junctions were identified. The remaining samples showed one or more putative additional unique structural variant event calls. Generally, the additional unique event calls were of lower quality, but provided signal that passed the computational filters. The vast majority are additional rare variant inter-chromosomal events, leaving on average 0.5 intra-chromosomal e.g. large inversions, deletions, or duplications per genome analyzed. Since the individuals included in this study had undergone rigorous karyotype analysis previously, the additional unique events observed in WGS data were considered as potential artifacts of the experimental methods and therefore not considered further.
Table 2.
Molecular karyotypes and gene disruptions in our cohort
| Case | Molecular karyotype (hg19) | Disrupted gene Breakpoint A |
Disrupted gene Breakpoint B |
|---|---|---|---|
| 31-05E | t(14;22)(q24;q12)(chr14:g.77200096_?∷chr22:g.?_33670453;chr22:g.33670154_?∷chr14:g.?_77202063) | LARGE1 (AR) | |
| 862-06Ö | t(16;22)(p11;q13.1)(ochr22:g.?_44818191∷chr16:g.?_23411495;chr22:g.44818097_?∷ochr16:g.23411178_?) | COG7 (AR) | |
| 106-06Ö | t(4;7)(q25;q31.1)(chr4:g.109487500∷chr7:g.108207030; chr7:g.108207034∷GTTG∷chr4:109487495) | THAP5 | |
| 58-06Ö | t(3;12)(q21.2;q22.4)(chr3:g.140432520_?∷chr12:g.?_78361967; chr12:g.78361576_?∷chr3:g.?_140433048) | NAV3 | |
| 157-06Ö | t(4;7)(q13.2;p14.3)(chr4:g.69061163∷ochr7:g.30238755; chr4:g.69061167∷chr7:g.30238757) | TMPRSS11BNL FTLP10 | |
| 191-06Ö | t(2;3)(p13;p25.1)(chr2:g.73661733∷chr3:g.15920865; chr3:g.15920804_?∷chr2:?_73661842) | ALMS1 (AR) | |
| 263-06Ö | t(10;11)(chr10:g.81408289∷T∷ochr11:g.7293897;ochr10:g.81408292∷chr11:g.7293897) | SYT9 | |
| 175-06Ö | t(10;15)(q23.1;q13.1)(chr10:g.85563120∷T∷chr15:g.28330698; chr15:g.28330697∷chr10∷g.85563132) | OCA2 (AR) | |
| 872-05Ö | t(1;8)(p31.3;q24.23)(ochr8:g.137813394∷chr1:g.78931227;chr8:g.137813393∷ochr1:g.78931219), t(5;18)(p15.2;q12.2)(ochr18:g.32849314∷chr5:g.11291110; chr18:g.32849312∷ochr5:g.11291110) |
CTNND2 (AD) | ZSCAN30 |
| 887-05Ö | t(5;7)(q14.1;q34)(chr5:g.78475862∷chr7:g.138295848; chr7:g.138295847∷chr5:g.78475863) | SVOPL | |
| 109-06Ö | t(2;6)(q34;q21)(chr2:g.212140397∷chr6:g.?_107800308; chr6:g.107800226∷chr2:g.212140479) | ||
| 232-07F | t(3;7)(p22.1;p11.2)(chr3:g.35670981∷AAAAG∷chr7:g.54888748; chr7:g.54888752∷chr3:g.35670986) | ||
| 851-06Ö | t(X;17)(p22.1;p13)(chr17:g.11479837∷chrX:g.30607945; chrX:g.30607932∷TATACCTTTATA∷chr17:g.11479842) | ||
| 8480THO | t(2;9)(chr2:g.241890512∷chr9:g.114876902; chr9:g.114876465_?∷chr2:g.?_241890544) | SUSD1 | |
| 841-95D | t(2;21)(p13.4; q21.1)(chr21:g.?_17525142∷chr2:g.?_72983886; chr21:g.17524360_?∷chr2:72983776_?) | EXOC6B (AD) | LINC00478 |
| 155-90D | t(2;13)(chr2:g.150242475∷chr13:g.93150076; chr13:g.93150075∷chr2:g.150242493) | LYPD6 | GPC5 |
| 2644-07D | t(4;8)(q21.23;q12.1)(chr4:g.85939427∷chr8:g.59822562; chr8:g.59822520∷chr4:g.85939429) | TOX | |
| 2587-07D | t(1;2)(q42;q31)(chr1:g.210823159∷chr2:g.162498672; chr2:g.162498662∷chr1:g.210823170) | HHAT | SLC4A10 |
| 2-03E | t(6;8)(q23;q24)(chr6:g.93938325∷chr8:g.103098296; chr8:g.103098297∷chr6:g.93938333), del(1p31)(chr1:g.69814692_81274032del) |
NCALD | |
| 337-01D | t(3;12)(q26.1;p11.2)(chr3:g.175570204∷chr12:g.13863014; chr3:g.?_175576921∷chr12:g.?_13863426) | GRIN2B (AD) | |
| 782-95D | t(10;12)(q24;q13)(chr10:g.113094447_?∷chr12:g.?_49092809; chr12:g.49091842_?∷chr10:g.?_113094574) | CCNT1 | |
| 29-03E | t(9;16)(p21;q21)(chr16:g.63446103∷TTGGC∷chr9:g.26190816; chr16:g.63445881∷CATC∷chr9:g.26190776) |
AR, autosomal recessive; AD, autosomal dominant; bold text indicates split read resolution; underlined text indicates known disease causing gene.
Of the 46 analyzed junctions, 32 (70%) breakpoints were delineated by WGS split read analysis. For the remaining 14 junctions, discordant read pairs mapped the breakpoints to within 2 kb. The detailed findings from chromosomal aberration to base pair resolution are shown in Figure 1 for one individual (862-06Ö). The exact findings from the mate pair sequencing of all the individuals are presented as molecular karyotypes in Table 2.
Figure 1. Molecular cytogenetic and genomic findings in subjecft 862-06Ö.
A) G-banded chromosomes showing the balanced translocation between chromosome 16 and chromosome 22 present in individual 862-06Ö (the aberrant chromosome shown to the left)
(B) FISH analysis with BAC clone RP11-350D02 (red) localized at chr16p11 (Ensemble, GRCh37). The signal is split between the two derivative chromosomes (der16 and der22).
(C) Circos plot illustrating the WGS results in individual 862-06Ö. Fusion events between chromosome 16 and 22, as predicted by FindTranslocations, from read pair mapping data are illustrated as grey lines. On chromosome 16, COG7 is disrupted by the breakpoint. The chromosomes are karyogram painted, chr22 in gray and chr16 in blue, with the centromeres shown shaded dark red. Copy number changes according to CNVnator are shown in the central ring, with a light red bar corresponding to low coverage and light green to high. As can be seen, short read sequence mapping does not cover the centromeric region or the heterochromatic 22p-arm. All copy number changes were evaluated as benign normal variation for this patient.
(D) Sanger sequencing traces showing the chromosomal junctions at the nucleotide level with der16 on top and der22 on the bottom. A two-nucleotide microhomology (TT) that may have originated from either parental chromosome is present in the der16 breakpoint junction (black box) and a clean break is present on der22 (black vertical line).
Gene disruptions are present in both phenotypically normal and clinically affected individuals
Genes were disrupted at the breakpoints to the same extent in both affected and unaffected individuals; 44% (7/16) and 47% (14/30) for affected and unaffected respectively (p=1.0, Fishers exact test). However, differences in the inheritance pattern of the disrupted genes in the two cohorts were observed: in the unaffected cohort 50% of disrupted gene loci were known disease genes, all in which disease traits are associated with a recessive inheritance pattern (i.e. LARGE1 (MIM# 603590), COG7 (MIM# 606978), ALMS1 (MIM# 606844), OCA2 (MIM# 611409)). In contrast, only three of the genes disrupted in the affected cohort were known disease causing, all linked to dominant neurodevelopmental disorders concordant with the phenotype observed (i.e. CTNND2 (MIM# 604275), EXOC6B (MIM# 607880), GRIN2B (MIM# 138252)) (Table 2). A systematic evaluation of all genes disrupted or in the vicinity (<250 kb) of the chromosomal breakpoints is provided in Supp. Table S4.
A clinically significant copy number variant (CNV) is present in one affected individual
To identify possible gene dose abnormalities that were not detected by the cytogenetic analysis we used the CNVnator software (Abyzov, et al., 2011) to analyze the mate pair whole genome data for deletion and duplications >= 2 kb. One clinically significant CNV was detected in individual 2-03E; an 11.4 Mb heterozygous deletion in chromosome 1p31 affecting 37 protein coding genes, previously described (de novo; patient 5 in (Lindstrand, et al., 2010)). The deletion was also apparent from discordant read pair analysis, and had split read information delineating it to single nucleotide resolution.
Mutational signatures underlying mechanisms of rearrangement formation
To confirm the WGS results, all junctions were defined at the nucleotide level by breakpoint PCR and Sanger sequencing (Supp. Figure S1, Supp. Figure S2). This enabled delineation of mutational signatures at the translocation junctions; this information could then be used to infer the potential underlying mechanisms for rearrangement formation.
None of the reported junctions were mediated by low-copy repeats or repetitive elements such as LINEs or SINEs located in distinct chromosome translocation substrates nor was there any evidence for palindromic AT rich repeats at the breakpoints. Two individuals had one out of two breakpoints mapped to repetitive elements, L1M4 in 191-06Ö/chr2 and L1Mb4 in 263-06Ö/chr10. In one case, 887-05Ö, the translocation involved two Alu elements (Figure 2), but as the derivative breakpoint junctions are characterized by very short homologies (3 nt of microhomology each) no fusion Alu was generated. Interestingly, in the same individual (887-05Ö), an intrachromosomal 1,579 bp deletion occurred 121 nucleotides upstream to the translocation break site on the derivative chromosome 5. The deletion involves two Alu elements that generate a fusion Alu (AluSx3-AluY) directly upstream of the translocation junction (Boone, et al., 2011; Boone, et al., 2014; Gu, et al., 2015; Mayle, et al., 2015). Remarkably, the same AluY involved in the fusion Alu created by the deletion is also involved in the translocation junction, and connects to an AluSz6 on chromosome 7, but no Alu fusion is generated by the translocation event as discussed above (Figure 2). This observation, an Alu–Alu rearrangement 121 nt upstream of the translocation, is consistent with the downstream translocation as an intrachromosomal template switching event (Table 3, Figure 2). Interchromosomal template-switches between non-homologous chromosomes have been demonstrated previously in both yeast (Smith, et al., 2007) and in humans (Carvalho, et al., 2015) despite in those cases they were not mediated by Alu repeats.
Figure 2. Evidence for template switching during translocation formation in individual 887-05Ö.
(A) A schematic overview of chr5q14.1 and 7q34 region and the structural events in individual 887-05Ö. The derivative 5 (der5) in blue and derivative 7 (der7) in green are aligned to chromosome 5 (top) and chromosome 7 (bottom). Alu elements are shown as grey boxes. The 1579 nt upstream deletion on der 5 is shown as a dashed blue line. Both deletion breakpoints as well as the translocation breakpoints are located in Alus.
(B) Sequence alignment of der5 to the corresponding regions on chromosome 5 and chromosome 7. The derivative chromosome sequences as well as the corresponding parental chromosome sequence are labeled in blue. The deletion is shown in lower case bold letters. Microhomology is highlighted in purple with the most plausible parental chromosome indicated by bold text. A 22 nt microhomology is present in the first slippage event (TSL 1) between the proximal and distal end of the der 5 upstream deletion. In the second event, chromosome 5 to chromosome 7 (TSL 2), a 3 bp microhomology is present.
(C) Der7 illustrated in green otherwise as in (B). A three-nucleotide microhomology is present in the junction.
Table 3.
Breakpoint junction characteristics
| Case | Microhomology | Genomic Deletion |
Genomic Duplication |
Insertion and SNVs | Additional Features |
|---|---|---|---|---|---|
| 31-05E | der14: TT der22: 0 |
chr14: 4 nt chr22: 1 nt |
chr14: 0 nt chr22: 0 nt |
der14: 0 der22: RIns GACG |
|
| 862-06Ö | der16: TTA der22: TT |
chr16: 4 nt chr22: 5 nt |
chr16: 0 nt chr22: 0 nt |
der16: TIns TTATAC der22: 0 |
|
| 106-06Ö | der4: T der7: 0 |
chr4: 0 nt chr7: 0 nt |
chr4: 6 nt chr7: 5 nt |
der4: Ins or SNV T der7: RIns GTT |
|
| 58-06Ö | der3: TG der12: AGT |
chr3: 5 nt chr12: 0 nt |
chr3: 0 nt chr12: 0 nt |
der3: 0 der12: 0 |
|
| 157-06Ö | der 4: TC der 7: AGT |
chr4: 4 nt chr7: 0 nt |
chr4: 0 nt chr7: 0 nt |
der 4: 0 der 7: 0 |
|
| 191-06Ö | der2: GT der3: 0 |
chr2: 0 nt chr3: 0 nt |
chr2: 3 nt chr3: 0 nt |
der2: 0 der3: SNV C |
|
| 263-06Ö | der10: 0 der11: 0 |
chr10: 2 nt chr11: 0 nt |
chr10: 0 nt chr11: 0 nt |
der10: Ins or SNV T der11: 0 |
|
| 175-06Ö | der10: GCTGT der15: GGCTGT |
chr10: 11 nt chr15: 0 nt |
chr10: 0 nt chr15: 0 nt |
der10: Ins or SNV T der15: 0 |
|
| 872-05Ö | der1: 0 der8: G |
chr1: 7 nt chr8: 0 nt |
chr1: 0 nt chr8: 0 nt |
der1: 0 der8: 0 |
|
| 872-05Ö | der5: G der18: G |
chr5: 0 nt chr18: 0 nt |
chr5: 0 nt chr18: 0 nt |
der5: 0 der18: SNV T |
|
| 887-05Ö | der5: GGC der7: GGC |
chr5: 0 nt chr7: 0 nt |
chr5: 0 nt chr7: 0 nt |
der5: 0 der7: 0 |
chr 5 upstream deletion: (AluSx3-AluY) |
| 109-06Ö | der2: CTA der6: 0 |
ch2: 10 nt chr6: 3 nt |
ch2: 0 nt chr6: 0 nt |
der2: 0 der6: 0 |
chr2: palCTAG |
| 232-07F | der3: 0 der7: 0 |
chr3: 0 nt chr7: 3 nt |
chr3: 10 nt chr7: 0 nt |
der3: 0 der7: del C + ins G |
|
| 851-06Ö | derX: TGGGG der17: 0 |
chrX: 9 nt chr17: 7 nt |
chrX: 0 nt chr17: 0 nt |
derX: 0 der17: TInsTATACC TTTATA |
|
| 8480THO | der2: CA der9: CA |
chr2: 0 nt chr9: 1 nt |
chr2: 0 nt chr9: 0 nt |
der2: 0 der9: 0 |
chr2: (TCCA)n |
| 841-95D | der2: TA der21: 0 |
chr2: 3 nt chr21: 0 nt |
chr2: 0 nt chr21: 5 nt |
der2: 0 der21: RIns AAAAA |
|
| 155-90D | der2: GTATG der13: T |
chr2: 22 nt chr13: 0 nt |
chr2: 0 nt chr13: 0 nt |
der2: 0 der13: 0 |
|
| 2644-07D | der4: 0 der8: 0 |
chr4: 1 nt chr8: 0 nt |
chr4: 0 nt chr8: 1 nt (G) |
der4: 0 der8: 0 |
|
| 2587-07D | der1: TATA der2: 0 |
chr1: 13 nt chr2: 5 nt |
chr1: 0 nt chr2: 0 nt |
der1: 0 der2: 0 |
|
| 2-03E | der6: TAAA der8:ATG |
chr6: 3 nt chr8: 0 nt |
chr6: 0 nt chr8: 0 nt |
der6: A>C (mosaic) der8: 0 |
chr6: palTTTTA/TAAAA chr8: palTTTAAA/TTTAAA |
| 337-01D | der 3: A der12: CTTTT |
chr3: 6594 nt chr12: 13 nt |
chr3: 5 nt chr12: 0 nt |
der3: 0 der12: RIns A, TIns TTTTAAAATGT |
|
| 782-95D | der10: CTGA der12: A |
chr10: 0 nt chr12: 4 nt |
chr10: 0 nt chr12: 0 nt |
der10: 0 der12: 0 |
|
| 29-03E | der9: CTT, ATA der16: ATA |
chr9: 29 nt chr16: 222 nt |
chr9: 0 nt chr16: 0 nt |
der9: 0 der16: TIns TTGGC |
chr, chromosome; SNV, single nucleotide variant; der, derivative chromosome; nt, nucleotides; jct, junction, RIns, random insertion; TIns, templated insertion, pal, palindrome; NHEJ, non-homologous end joining, RBM, Replication-based mechanism
We next aligned the junction fragments to both parental chromosomes and quantified differences with respect to the haploid reference genome including the presence of microhomology at the junction and a few base pair deletions/duplications, insertions and single nucleotide variants not present in dbSNP. The detailed translocation breakpoint features observed in our cohort are shown as Table 3.
Microhomology, i.e. nucleotide sequence found in both substrate sequences that is reduced to one copy at the breakpoint junction, ranging from 2 to 6 nt was observed in 25 out of 46 sequence junctions (54%) involving 17 of the 23 translocation events studied (74%) (Table 4). Deletions of a few base pairs (1-6594 bp, median 5 nt) were observed in 26 junctions from 19 patients (Table 3, Table 4). Nucleotide insertions varying from 1 to 12 nt were identified at 11 translocation junctions from 10 individuals (24% of junctions). Among those, at least 4 insertions likely originate from nearby genomic segments (< 200 nt) resulting in an overall fraction of templated insertions in the studied translocation carriers of 17.4% (Figure 3, Table 3, Table 4). Duplications of 1-10 nt were present in six individuals (Table 3, Table 4); all in conjunction with additional either insertions and/or single nucleotide variants (SNVs).
Table 4.
Breakpoint characteristics of reported and reanalyzed reciprocal chromosome translocations
| All translocations | Our cohort | Reanalysis cohort | |||||
|---|---|---|---|---|---|---|---|
| By translocation | Total number | 60 | 100% | 23 | 100% | 37 | 100% |
| Balanced (cytogenetically) | 60 | 100% | 23 | 100% | 37 | 100% | |
| By breakpoint | Total number | 120 | 100% | 46 | 100% | 74 | 100% |
| Balanced | 44 | 37% | 14 | 30% | 30 | 41% | |
| Microhomology, total | 70 | 58% | 32 | 70% | 38 | 51% | |
| <2 nt | 24 | 20% | 7 | 15% | 17 | 23% | |
| 2-20 nt | 46 | 38% | 25 | 54% | 21 | 28% | |
| Insertions, total | 34 | 28% | 11 | 24% | 23 | 31% | |
| <2 nt | 5 | 4% | 4 | 9% | 1 | 1% | |
| 2-20 nt | 26 | 22% | 7 | 15% | 19 | 26% | |
| >20 nt | 3 | 3% | 0 | 0% | 3 | 4% | |
| Templated Insertions | 19 | 16% | 4 | 9% | 15 | 20% | |
| Base deletions, total | 67 | 56% | 26 | 57% | 41 | 55% | |
| <2 nt | 8 | 7% | 3 | 7% | 5 | 7% | |
| 2-20 nt | 42 | 35% | 19 | 41% | 23 | 31% | |
| >20 nt | 8 | 7% | 3 | 7% | 5 | 7% | |
| >1000 nt | 9 | 8% | 1 | 2% | 8 | 11% | |
| Base duplications, total | 9 | 8% | 7 | 15% | 2 | 3% | |
| <2 nt | 1 | 1% | 1 | 2% | 0 | 0% | |
| 2-20 nt | 8 | 7% | 6 | 13% | 2 | 3% | |
nt, nucleotide
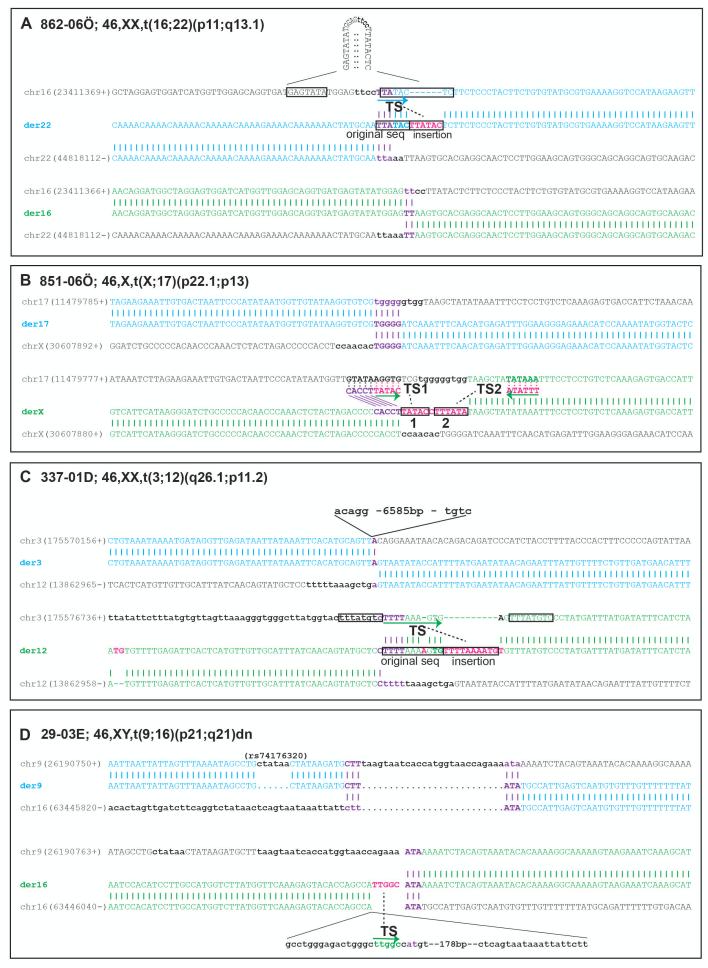
Figure 3. Four additional translocation carriers with evidence for templated insertions originating from nearby genomic segments.
Sequence alignments of the derivative chromosome sequences to the corresponding regions on the parental chromosomes. Deletions are shown in lower case bold letters. Microhomology is highlighted in purple with the most plausible parental chromosome indicated by bold text. Insertions and SNVs are shown in pink (A) Sequence alignments from case 862-06Ö. Derivative chromosome 22 (der22) is shown on top and derivative chromosome 16 (der16) on the bottom. The derivative chromosome sequences as well as the corresponding parental chromosome (chr16 and chr22) sequences are labeled in blue for der22 and in green for der16. Short deletions are present on both parental chromosomes, 4 nt on chr16 and 5 nt on chr22. On chr16 palindromic sequences are present on each side of the breakpoint. A six nucleotide (nt) insertion is present in the junction of der22 (TTATAC), likely due to template slippage (TSL) copying from the palindrome sequence using a 3 nt (TTA) microhomology. A 2 nt microhomology is present in der16.
(B) Sequence alignment from case 851-06Ö. Derivative chromosome 17 (der17) is shown on top and derivative chromosome X (derX) on the bottom. The derivative chromosome sequences as well as the corresponding parental chromosome (chr17 and chrX) sequences are labeled in blue for der17 and in green for derX. Short deletions are present on both parental chromosomes, 9 nt on chr17 and 7 nt on chrX. On der17 a 5 nt microhomology is present. A 12 nt insertion is present on der X that may originate from template switching to two different places on chr17 (- strand). In the TS1 a potential CACCT microhomology is present but for TS2 no microhomology could be observed.
(C) Sequence alignment from case 337-01D. Derivative chromosome 3 (der3) is shown on top and derivative chromosome 12 (der12) on the bottom. The derivative chromosome sequences as well as the corresponding parental chromosome (chr3 and chr12) sequences are labeled in blue for der3 and in green for der12. Deletions are present on both parental chromosomes, 6594 nt on chr3 and 13 nt on chr12. On der3 a 1 nt microhomology is present
A 10 nt insertion is present on der 12 that may have arisen through error prone backward slippage using a 4 nt (TTTT) microhomology.
(D) Sequence alignment from case 29-03E. Derivative chromosome 9 (der9) is shown on top and derivative chromosome 16 (der16) on the bottom. The derivative chromosome sequences as well as the corresponding parental chromosome (chr9 and chr16) sequences are labeled in blue for der9 and in green for der16. On der 9, 3 nt microhomologies are present on both sides of a 27 nt resection/deletion. On der16 the junction presents with a 222 nt deletion and the 5 bp insertion (TTGGC) originates from inside the deletion.
Novel SNVs not present in dbSNP, ExAC or 1000 Genomes were identified in six individuals (Table 3). In all cases, parental DNA was not available for origin studies or to determine if they occurred de novo concomitantly with the translocation event. In one individual (2-03E) a mosaic A>C SNV was detected adjacent to the der6 junction (Supp. Figure S1; Supp. Figure S2). The variant was confirmed with multiple rounds of PCR using different primers. The PCR product was then cloned and subsequent Sanger sequencing showed that 16/29 (55%) of clones carried the C allele.
Finally, to further investigate for potential mutational signatures in common with those observed in our study subjects, we reanalyzed 37 previously published seemingly balanced translocations with available junction fragment sequences (Chen, et al., 2008; Chiang, et al., 2012; Higgins, et al., 2008; Hsiao, et al., 2014; Schluth-Bolard, et al., 2013; Talkowski, et al., 2012a; Talkowski, et al., 2012b). Inversions, complex translocations and chromothripsis events were excluded from reanalysis. Two additional papers presented breakpoint positions for balanced translocations (Dong, et al., 2014; Suzuki, et al., 2014), however they did not report enough sequence information for reanalysis. In the reanalysis cohort, microhomology (2-6 nt) is present in 21 of the 74 chromosomal junctions (28%) and in 16 of the 37 translocation events (43%). Furthermore, templated insertions are observed in 15 junctions and 13 translocations (Figure 2, Figure 3, Supp. Figure S3, Table 3, Table 4, Supp. Table S1). By combining the data from our cases and the reanalysis cohort, junction sequences from 60 translocations were analyzed for breakpoint features. In aggregate, the combined data show that blunt ends are present in 36.7% of junctions (44/120) but only 17% of translocation events present with blunt ends on both chromosomal derivatives. Microhomology (>2 nt) was observed in 38.3% (46/120) of all the junctions. Finally, insertions are present in 34 junctions, and in 19 instances the inserted sequences have originated from local sequences making the overall incidence of templated insertions in 120 reciprocal translocation breakpoint junctions ~15.8%. The overall breakpoint characteristics are summarized in Table 4.
DISCUSSION
Gene disruptions are identified in both unaffected and affected carriers
We used low coverage mate pair whole genome sequencing to characterize the breakpoints of 23 translocations identified in a cohort of 22 carriers, both phenotypically normal (n=8) and abnormal individuals (n=14). In the phenotypically normal cohort four genes, in which biallelic pathological variants convey a disease trait with a recessive inheritance pattern, were disrupted making the BCA carriers also recessive carriers of the disease linked to the specific genes (31-05E, LARGE1; 862-06Ö, COG7; 191-06Ö, ALMS1; 175-06Ö; OCA2; Table 2, Supp. Table S4). This is in contrast to the phenotypically affected cohort where we identified the disruption of three known disease genes in which the disease traits are associated with an autosomal dominant inheritance pattern (described in detail in the next section). Disruption of recessive alleles has to be taken into consideration concerning reproduction and risk for disease in the offspring as structural variation can contribute to recessive disease carrier states (Boone, et al., 2013).
In the translocation carriers with a neurodevelopmental phenotype disrupted genes or genes in the vicinity of the breakpoint were manually curated and classified from A to G according to a predefined set of criteria on their likelihood of being causal for the phenotype (Supp. Table S4). In addition to the previously reported CTNND2 disruption segregating with reading difficulties (Hofmeister, et al., 2015) two disruptions of known disease genes were scored as likely causal (Class A; Supp. Table S4). First, in individual 337-01D a de novo translocation disrupts the glutamate receptor subunit GRIN2B, known to cause epileptic encephalopathy (MIM# 616139) (Lemke, et al., 2014) and autosomal dominant intellectual disability (ID) (MIM# 613970) (Endele, et al., 2010), concordant with the moderate ID, autism and epilepsy phenotype in our patient (Table 1, Supp. Table S4). Second, in case 841-95D, EXOC6B is disrupted on chromosome 2p13.2 in patient 841-95D with high functioning autism, ADHD and hypertelorism. Disruption of EXOC6B by a de novo balanced translocation was previously described in a patient with autistic traits, developmental delay, ID, epilepsy, aggressive behavior and various minor dysmorphic features (Fruhmesser, et al., 2013). In addition, a heterozygous de novo mosaic deletion of exons 2 -20 in EXOC6B was reported in a child with developmental delay, speech delay and minor dysmorphic features (Evers, et al., 2014). Overall, these studies provide evidence suggesting that the disruption of EXOC6B is causing the clinical phenotype in case 841-95D (Table 1, Supp. Table S4).
We further explored for potential regulatory effects caused by the translocations. Even though position effects have been reported as far as 1.3 Mb from the chromosomal breakpoints (Velagaleti, et al., 2005), in order to provide evidence for long range effects either the genotype-phenotype correlations need to be very specific or molecular evidence, such as a reduction in RNA and/or protein levels, should be shown. Since several hundred disease genes have been described in neurodevelopmental disorders (Gilissen, et al., 2014) and the relevant tissue was unavailable for functional studies we chose to focus on genes in the immediate breakpoint regions (250 kb upstream/downstream). In one affected individual (232-07F) no gene disruption was observed on either derivate, but the vicinity search showed that the chromosome 3 breakpoint was located 9.7 kb upstream of ARPP21 (MIM# 605488), a candidate ID-gene (Marangi, et al., 2013), possibly disrupting a gene regulating region or altering the genomic environment (Table 1, Supp. Table S4).
Finally, several potential candidate genes were disrupted in the affected cohort involved in various cellular functions and pathways such as calcium-ion binding (SUSD1; 8480THO) (Clark, et al., 2003), synaptic signaling (SVOPL (MIM# 611700); 887-05Ö) (Jacobsson, et al., 2007), Wnt/ß-catenin signaling (LYPD6 (MIM# 613359); 155-90D) (Ozhan, et al., 2013), hedgehog signaling (GPC5 (MIM# 602446); 155-90D) (Witt, et al., 2013) and mammalian corticogenesis (TOX (MIM# 606863); 2644-07D) (Artegiani, et al., 2015). None of these genes have been previously reported in human neurobehavioral syndromes and although identification of more individuals with mutations in these genes will be required to clearly determine causality, they present good candidates for further functional studies to investigate their role in neurodevelopmental disorders (Table 1, Supp. Table S4).
Evidence for template switching suggest that replicative mechanism may underlie a portion of balanced translocations
The detailed analysis of breakpoint junction sequences from 60 reciprocal translocations show mutational signatures consistent with an underlying mechanism involving template switching in approximately 16% of the junctions (Table 3, Table 4, Supp. Table S1).
Template switching, highlighted in breakpoint junctions by microhomology and templated insertions, is a hallmark feature of replicative repair. In humans, replicative errors (e.g. RBMs) give rise to complex genomic rearrangements (CGR) including gross copy-number gains, losses and inversions (Carvalho and Lupski, 2016; Lee, et al., 2007; Sakofsky, et al., 2015). Distinct from NHEJ that uses microhomology to facilitate ligation, in RBMs microhomology is used to prime and assist resumption and productive synthesis of a stalled/collapsed replication fork. Mostly, RBMs seem to result in formation of intrachromosomal structural variants but interchromosomal events resulting in complex copy number gain have also been reported (Carvalho, et al., 2015; Gu, et al., 2016).
Previous publications that have characterized non-recurrent balanced constitutional reciprocal translocations at the breakpoint level (Chen, et al., 2008; Chiang, et al., 2012; Dong, et al., 2014; Higgins, et al., 2008; Hsiao, et al., 2014; Schluth-Bolard, et al., 2013; Suzuki, et al., 2014; Talkowski, et al., 2012a; Talkowski, et al., 2012b) have suggested c-NHEJ as the predominant mechanism underlying translocations in humans (Chiang, et al., 2012). In the largest previous study, Chiang et al. state that most BCAs show little or no microhomology at the junctions. However, the detailed analysis of 81 breakpoints from simple translocations and chromosomal inversions showed both microhomology (1-20 nt, 31.3%) and insertions (20.5%) in a high fraction of junctions (Chiang, et al., 2012). In the same paper the authors also observed a surprisingly high fraction of BCAs with 3 or more breakpoints, many of which are accompanied by strand inversions. In the latter cases, the authors implicated that RBMs may have been involved in the formation of these complex interchromosomal rearrangements (Chiang, et al., 2012); in fact, such observations are indeed most consistent with iterative template switching during replicative repair (Carvalho and Lupski, 2016).
Recently, it was shown in embryonic mouse fibroblast that Pol theta is responsible for repair of DNA breaks in a context where c-NHEJ is defective (Ku70−/−) and that templated insertions from heterologous chromosomes are inserted in some of the junctions during this repair process. However, the authors also showed that Pol theta does not contribute to balanced translocations, at least in their model system, and that an alternative mechanism should be in place (Wyatt, et al., 2016). Additional support for the notion that mechanisms other than NHEJ and NAHR underlie the formation of some balanced translocations is provided by studies on human cells, ~6% of translocation junctions generated in vitro show the presence of templated insertions varying from 20 bp to several hundred bp (Ghezraoui, et al., 2014). Finally, chromosomal translocations have also been well studied in leukemia, where the same somatic events arise in multiple patients, and are indicative of prognosis and guide medical management. The recurrent translocation, t(9;22), seen in the formation of the Philadelphia chromosome, illustrates that replicative mechanisms may underlie balanced reciprocal translocations (Czuchlewski, et al., 2011).
In the translocations analyzed here, 37% of translocation junctions are precisely joined (blunt ends), consistent with c-NHEJ underlying a portion of the breakpoint junctions of balanced translocations. Nonetheless, 38% of the junctions we studied have 2 to 6 nt microhomology, some of them associated with templated insertions. Such templated insertions at the breakpoint junctions of structural variants may result from single or multiple iterative template switches during DNA repair processes that involve DNA replication (Carvalho and Lupski, 2016; Deem, et al., 2011; Hastings, et al., 2009a; Lee, et al., 2007; Sakofsky, et al., 2015). Another possibility that could explain some of the observed junction signatures is alternative non-homologous end-joining (alt-NHEJ). Alt-NHEJ is hypothesized to take over when components of c-NHEJ are absent, producing rearrangements with longer microhomology at the translocation junctions (Ghezraoui, et al., 2014).
In our original cohort, template switches seem to occur in at least five carriers (22.7%; 862-06Ö, 887-05Ö, 851-06Ö, 337-01D, 29-03E; Figure 2, Figure 3). All events, except the complex Alu mediated event in case 887-05Ö, can be interpreted as short-range backwards template switching resulting in the insertion of short segments within the same fork (< 200 nt). In our cohort, low-copy repeats and repetitive elements do not seem to mediate BCAs. Nonetheless, a deletion mediated by Alu elements generating a fusion Alu-Alu was observed nearby the translocation breakpoint of individual 887-05Ö. Events mediated by Alu generate Alu-Alu fusions (Boone, et al., 2014) and can also lead to formation of intrachromosomal complex structural variants (e.g. inverted triplications interspersed by duplications or DUPTRP/INV-DUP structures) (Gu, et al., 2015). The limited similarity provided by genome-wide extensive presence of Alu elements in the human genome is hypothesized to provide enough homology to allow template-switches to occur during RBMs (Gu, et al., 2015), a contention that is supported by recent yeast experiments that model human Alu-mediated deletions (Mayle, et al., 2015). Therefore, the presence of such a deletion nearby the translocation breakpoint supports a role for RBM in the formation of this translocation. Our hypothesis is that this intrachromosomal event was followed by an interchromosomal event but since we don't have access to parental DNA we cannot prove that the Alu-Alu deletion was formed concomitantly with the translocation.
End processing of the original chromosome breaks may give rise to short duplications and deletions
The junction analysis also confirmed previous observations that many seemingly balanced translocations are not balanced at the nucleotide level (Baptista, et al., 2008; Higgins, et al., 2008). We observed both the presence of not only deletions of a few base pairs but also short duplications (1-10 nt). To better understand this observation we reexamined the junction fragment sequences. The derivative translocation breakpoint junctions represent the rearrangement end-products and may enable us to infer some properties of the reactions by which the original double stranded breaks (DSBs) that gave rise to the translocation were resolved. For instance, upon double or single strand break, both 5′ and 3′ ends may be processed. In the MMBIR repair mechanism one ended, double stranded DNA breaks (oeDSB) can result from a collapsed fork after DNA replication proceeds through a nicked molecule. The 5′ ends undergoes resection to expose the 3′ end that will further anneal to a single stranded DNA that shares microhomology and resume replication (Hastings, et al., 2009a). Resection of the 5′ end is generally inhibited in NHEJ (Pannunzio, et al., 2014), but alt-NHEJ has been observed to present longer deletions than NHEJ (Ghezraoui, et al., 2014). If only one of the ends is processed there should be no loss of genetic information therefore the derivative products will present no copy number variation nearby the ligated junction (Figure 4A). Nonetheless, if both 5′ and 3′ ends are processed then deletions are expected (Figure 4A).
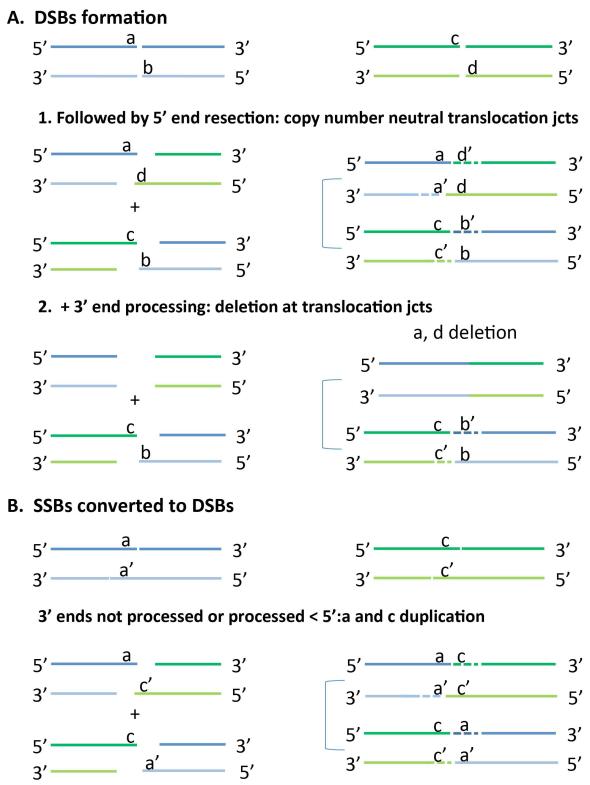
Figure 4. Schematic of double stranded break (DSB) and single stranded break (SSB) prior to formation of balanced translocations.
(A) The formation of two DSBs in distinct heterologous chromosomes is illustrated at the top in blue and green respectively. 1.) Left: DSB formation followed by 5′ resection. If 5' end resection occurs without 3′ end processing the translocation junctions (jcts) will be copy neutral (Right). 2.) If 5′ resection in addition to 3'end processing occurs at the breakpoints there will be formation of short deletions at the translocation jcts.
(B) SSB or nick formation on top as in A. If two nearby SSBs generated on opposite strands are converted into a DSB short duplications may be present at the translocation jcts. Small letters (a, b, c, d) indicate breakpoint segments. Gap filling is indicated by dashed lines with respective breakpoint segments indicated by primed letters (a’, b’, c’, d’).
In our original cohort, there was an overall lack of extensive processing of the ends as indicated by the short intrachromosomal distance between the endpoints in the broken chromosomes. The distance between the breakpoints located on the same chromosome range from 0 to 6,594 nt but the majority of chromosome ends (n=42) are less than 20 nt apart (median 2 nt, Table 3, Table 4). In all the analyzed translocations, copy number neutral junctions are observed in 44 out of 120 DSB ends (37%) indicating that those ends underwent either no or single end processing (Table 3, Table 4, Supp. Table S1). Interestingly, most of the breaks, i.e. 67 out of 120 (56%), are accompanied by a deletion that varies from 1 to 3,600,000 nt (median 1 nt). Although the occurrence of short deletions at the junctions is consistent with alt-NHEJ (Ghezraoui, et al., 2014), larger end processing is not consistent with this mechanism. Supporting this observation, the two carriers in our cohort (29-03E and 337-01D) with larger processed ends (222 nt and 6,594 nt) also have templated insertions at the junctions consistent with our hypothesis that RBM underlie formation of those translocations. However, since we lack inheritance data for both individuals, we cannot know apodictically for sure that the observed deletions have originated at the same time as the translocations.
Unexpectedly, 9 out of 120 breaks (7.5%) present with gain of genetic material, i.e. a short duplication varying from 1 to 14 nt. One possible explanation for such gains is by two nearby single-strand breaks (SSBs) or nicks that were generated in opposite strands, which can be converted into DSBs (Figure 4B). After processing and ligation of those overlapping, overhanging short segments to heterologous derivative chromosomes, duplication of the segments flanking the junctions can be observed (Figure 4B, Table 3; Table 4, Supp. Table S1).
In aggregate, breakpoint characterization of 60 balanced translocations reveal: i.) that both deletions and duplications of a few base pairs are frequently present in the chromosome junctions, and ii.) mutational signatures indicate an underling replicative mechanism involving templates switching in 16% of the junctions. These findings can help explain the observation that ~37% of apparently balanced translocations actually present with imbalances and cryptic rearrangements at or nearby the translocation junctions (Higgins, et al., 2008). Cryptic genomic imbalances in apparent balanced chromosomal aberrations have been associated with affected carriers and are less frequently observed in individuals without an associated clinical phenotype (Baptista, et al., 2008). The contribution of such variants for symptomatic carriers needs to be further assessed. Finally, the possibility that some translocations may have a mitotic origin influences genetic counseling. A de novo translocations showing mutational signatures indicative of a mitotic origin could have arisen either as a mitotic event in one parent, this individual will then be a low level mosaic with a higher recurrence risk, or in an early mitotic division after fertilization with no recurrence risk for the parents.
In conclusion, our studies highlight the importance of breakpoint resolution in the clinical molecular interpretation of chromosomal translocations. First, we show that the disruption of disease causing genes directly provides a molecular diagnosis to a subset of affected carriers. However, this is important also in the healthy carriers as we identify significant gene disruptions important to their own health and reproductive genetics. The disruption of a gene by a balanced chromosome break is a type of disease causing mutation that goes undetected by all the current methods used in genetic diagnostics except large scale sequencing. With 1/500 individuals being a carrier this most likely represents a highly underappreciated cause of disease especially taking into consideration that the resolution of a chromosome-banding assay is above 5-10 Mb (well illustrated by the cryptic 11 Mb deletion in individual 2-03E). We therefore propose that diagnostic WGS will be clinically important for characterizing balanced structural chromosomal variants in the investigation of diverse patient populations from monogenic disorders and infertility to neurocognitive diseases.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to the patients and their families for their cooperation and enthusiasm during this study.
Funding
This work was supported by the Swedish Research Council [2012-1526 to AL, and 2013-2603 to OM); The Swedish Society for Medical Research; the Marianne and Marcus Wallenberg foundation [2014.0084 to AL]; the Stockholm City Council; the Harald and Greta Jeanssons Foundation; the Ulf Lundahl memory fund through the Swedish Brain Foundation; the Nilsson Ehle donations and the Erik Rönnberg Foundation. Supported in part by US National Institutes of Health [HG006542] to the Baylor Hopkins Center for Mendelian Genomics. We also gratefully acknowledge the use of computer infrastructure resources at UPPMAX, projects b2011162 and b2014152.
Footnotes
CONFLICT OF INTEREST
J.R.L. has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, has stock options in Lasergen, Inc. is a member of the Scientific Advisory Board of Baylor Genetics, and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis (CMA) and clinical exome sequencing offered in the Baylor Genetics (BMGL: http://www.bmgl.com/BMGL/Default.aspx).
REFERENCES
- Abyzov A, Li S, Kim DR, Mohiyuddin M, Stutz AM, Parrish NF, Mu XJ, Clark W, Chen K, Hurles M, Korbel JO, Lam HY, et al. Analysis of deletion breakpoints from 1,092 humans reveals details of mutation mechanisms. Nat Commun. 2015;6:7256. doi: 10.1038/ncomms8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21(6):974–84. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artegiani B, de Jesus Domingues AM, Bragado Alonso S, Brandl E, Massalini S, Dahl A, Calegari F. Tox: a multifunctional transcription factor and novel regulator of mammalian corticogenesis. EMBO J. 2015;34(7):896–910. doi: 10.15252/embj.201490061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista J, Mercer C, Prigmore E, Gribble SM, Carter NP, Maloney V, Thomas NS, Jacobs PA, Crolla JA. Breakpoint mapping and array CGH in translocations: comparison of a phenotypically normal and an abnormal cohort. Am J Hum Genet. 2008;82(4):927–36. doi: 10.1016/j.ajhg.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone PM, Campbell IM, Baggett BC, Soens ZT, Rao MM, Hixson PM, Patel A, Bi W, Cheung SW, Lalani SR, Beaudet AL, Stankiewicz P, et al. Deletions of recessive disease genes: CNV contribution to carrier states and disease-causing alleles. Genome Res. 2013;23(9):1383–94. doi: 10.1101/gr.156075.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone PM, Liu P, Zhang F, Carvalho CM, Towne CF, Batish SD, Lupski JR. Alu-specific microhomology-mediated deletion of the final exon of SPAST in three unrelated subjects with hereditary spastic paraplegia. Genet Med. 2011;13(6):582–92. doi: 10.1097/GIM.0b013e3182106775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone PM, Yuan B, Campbell IM, Scull JC, Withers MA, Baggett BC, Beck CR, Shaw CJ, Stankiewicz P, Moretti P, Goodwin WE, Hein N, et al. The Alu-rich genomic architecture of SPAST predisposes to diverse and functionally distinct disease-associated CNV alleles. Am J Hum Genet. 2014;95(2):143–61. doi: 10.1016/j.ajhg.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgstrom E, Lundin S, Lundeberg J. Large scale library generation for high throughput sequencing. PLoS One. 2011;6(4):e19119. doi: 10.1371/journal.pone.0019119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Lupski JR. Mechanisms underlying structural variant formation in genomic disorders. Nat Rev Genet. 2016;17(4):224–38. doi: 10.1038/nrg.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Pehlivan D, Ramocki MB, Fang P, Alleva B, Franco LM, Belmont JW, Hastings PJ, Lupski JR. Replicative mechanisms for CNV formation are error prone. Nat Genet. 2013;45(11):1319–26. doi: 10.1038/ng.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Pfundt R, King DA, Lindsay SJ, Zuccherato LW, Macville MV, Liu P, Johnson D, Stankiewicz P, Brown CW, Study DDD, Shaw CA, et al. Absence of heterozygosity due to template switching during replicative rearrangements. Am J Hum Genet. 2015;96(4):555–64. doi: 10.1016/j.ajhg.2015.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Ramocki MB, Pehlivan D, Franco LM, Gonzaga-Jauregui C, Fang P, McCall A, Pivnick EK, Hines-Dowell S, Seaver LH, Friehling L, Lee S, et al. Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat Genet. 2011;43(11):1074–81. doi: 10.1038/ng.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Kalscheuer V, Tzschach A, Menzel C, Ullmann R, Schulz MH, Erdogan F, Li N, Kijas Z, Arkesteijn G, Pajares IL, Goetz-Sothmann M, et al. Mapping translocation breakpoints by next-generation sequencing. Genome Res. 2008;18(7):1143–9. doi: 10.1101/gr.076166.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Jacobsen JC, Ernst C, Hanscom C, Heilbut A, Blumenthal I, Mills RE, Kirby A, Lindgren AM, Rudiger SR, McLaughlan CJ, Bawden CS, et al. Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat Genet. 2012;44(4):390–7. S1. doi: 10.1038/ng.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark HF, Gurney AL, Abaya E, Baker K, Baldwin D, Brush J, Chen J, Chow B, Chui C, Crowley C, Currell B, Deuel B, et al. The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: a bioinformatics assessment. Genome Res. 2003;13(10):2265–70. doi: 10.1101/gr.1293003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Weissman SM. Directional cloning of DNA fragments at a large distance from an initial probe: a circularization method. Proc Natl Acad Sci U S A. 1984;81(21):6812–6. doi: 10.1073/pnas.81.21.6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Bird C, Blackburne B, Lindsay S, Mamanova L, Lee C, Turner DJ, Hurles ME. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat Genet. 2010;42(5):385–91. doi: 10.1038/ng.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czuchlewski DR, Farzanmehr H, Robinett S, Haines S, Reichard KK. t(9;22)(q34;q11.2) is a recurrent constitutional non-Robertsonian translocation and a rare cytogenetic mimic of chronic myeloid leukemia. Cancer Genet. 2011;204(10):572–6. doi: 10.1016/j.cancergen.2011.10.006. [DOI] [PubMed] [Google Scholar]
- Deem A, Keszthelyi A, Blackgrove T, Vayl A, Coffey B, Mathur R, Chabes A, Malkova A. Break-induced replication is highly inaccurate. PLoS Biol. 2011;9(2):e1000594. doi: 10.1371/journal.pbio.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, Jiang L, Yang C, Hu H, Wang X, Chen H, Choy KW, Hu H, Dong Y, Hu B, Xu J, Long Y, et al. A robust approach for blind detection of balanced chromosomal rearrangements with whole-genome low-coverage sequencing. Hum Mutat. 2014;35(5):625–36. doi: 10.1002/humu.22541. [DOI] [PubMed] [Google Scholar]
- Endele S, Rosenberger G, Geider K, Popp B, Tamer C, Stefanova I, Milh M, Kortum F, Fritsch A, Pientka FK, Hellenbroich Y, Kalscheuer VM, et al. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat Genet. 2010;42(11):1021–6. doi: 10.1038/ng.677. [DOI] [PubMed] [Google Scholar]
- Evers C, Maas B, Koch KA, Jauch A, Janssen JW, Sutter C, Parker MJ, Hinderhofer K, Moog U. Mosaic deletion of EXOC6B: further evidence for an important role of the exocyst complex in the pathogenesis of intellectual disability. Am J Med Genet A. 2014;164A(12):3088–94. doi: 10.1002/ajmg.a.36770. [DOI] [PubMed] [Google Scholar]
- Fruhmesser A, Blake J, Haberlandt E, Baying B, Raeder B, Runz H, Spreiz A, Fauth C, Benes V, Utermann G, Zschocke J, Kotzot D. Disruption of EXOC6B in a patient with developmental delay, epilepsy, and a de novo balanced t(2;8) translocation. Eur J Hum Genet. 2013;21(10):1177–80. doi: 10.1038/ejhg.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezraoui H, Piganeau M, Renouf B, Renaud JB, Sallmyr A, Ruis B, Oh S, Tomkinson AE, Hendrickson EA, Giovannangeli C, Jasin M, Brunet E. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol Cell. 2014;55(6):829–42. doi: 10.1016/j.molcel.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglio S, Calvari V, Gregato G, Gimelli G, Camanini S, Giorda R, Ragusa A, Guerneri S, Selicorni A, Stumm M, Tonnies H, Ventura M, et al. Heterozygous submicroscopic inversions involving olfactory receptor-gene clusters mediate the recurrent t(4;8)(p16;p23) translocation. Am J Hum Genet. 2002;71(2):276–85. doi: 10.1086/341610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilissen C, Hehir-Kwa JY, Thung DT, van de Vorst M, van Bon BW, Willemsen MH, Kwint M, Janssen IM, Hoischen A, Schenck A, Leach R, Klein R, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511(7509):344–7. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- Gu S, Yuan B, Campbell IM, Beck CR, Carvalho CM, Nagamani SC, Erez A, Patel A, Bacino CA, Shaw CA, Stankiewicz P, Cheung SW, et al. Alu-mediated diverse and complex pathogenic copy-number variants within human chromosome 17 at p13.3. Hum Mol Genet. 2015;24(14):4061–77. doi: 10.1093/hmg/ddv146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Szafranski P, Akdemir ZC, Yuan B, Cooper ML, Magriñá MA, Bacino CA, Lalani SR, Breman AM, Smith JL, Patel A, Son RH, et al. Mechanisms for Complex Chromosomal Insertions. PLoS Genet. 2016 doi: 10.1371/journal.pgen.1006446. (re-submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009a;5(1):e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009b;10(8):551–64. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins AW, Alkuraya FS, Bosco AF, Brown KK, Bruns GA, Donovan DJ, Eisenman R, Fan Y, Farra CG, Ferguson HL, Gusella JF, Harris DJ, et al. Characterization of apparently balanced chromosomal rearrangements from the developmental genome anatomy project. Am J Hum Genet. 2008;82(3):712–22. doi: 10.1016/j.ajhg.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmeister W, Nilsson D, Topa A, Anderlid BM, Darki F, Matsson H, Tapia Paez I, Klingberg T, Samuelsson L, Wirta V, Vezzi F, Kere J, et al. CTNND2-a candidate gene for reading problems and mild intellectual disability. J Med Genet. 2015;52(2):111–22. doi: 10.1136/jmedgenet-2014-102757. [DOI] [PubMed] [Google Scholar]
- Hsiao MC, Piotrowski A, Alexander J, Callens T, Fu C, Mikhail FM, Claes KB, Messiaen L. Palindrome-mediated and replication-dependent pathogenic structural rearrangements within the NF1 gene. Hum Mutat. 2014;35(7):891–8. doi: 10.1002/humu.22569. [DOI] [PubMed] [Google Scholar]
- Jacobs PA, Browne C, Gregson N, Joyce C, White H. Estimates of the frequency of chromosome abnormalities detectable in unselected newborns using moderate levels of banding. J Med Genet. 1992;29(2):103–8. doi: 10.1136/jmg.29.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsson JA, Haitina T, Lindblom J, Fredriksson R. Identification of six putative human transporters with structural similarity to the drug transporter SLC22 family. Genomics. 2007;90(5):595–609. doi: 10.1016/j.ygeno.2007.03.017. [DOI] [PubMed] [Google Scholar]
- Kato T, Kurahashi H, Emanuel BS. Chromosomal translocations and palindromic AT-rich repeats. Curr Opin Genet Dev. 2012;22(3):221–8. doi: 10.1016/j.gde.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12(4):656–64. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131(7):1235–47. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Lemke JR, Hendrickx R, Geider K, Laube B, Schwake M, Harvey RJ, James VM, Pepler A, Steiner I, Hortnagel K, Neidhardt J, Ruf S, et al. GRIN2B mutations in West syndrome and intellectual disability with focal epilepsy. Ann Neurol. 2014;75(1):147–54. doi: 10.1002/ana.24073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieden A, Kvarnung M, Nilssson D, Sahlin E, Lundberg ES. Intragenic duplication--a novel causative mechanism for SATB2-associated syndrome. Am J Med Genet A. 2014;164A(12):3083–7. doi: 10.1002/ajmg.a.36769. [DOI] [PubMed] [Google Scholar]
- Lindstrand A, Schoumans J, Gustavsson P, Hanemaaijer N, Malmgren H, Blennow E. Improved structural characterization of chromosomal breakpoints using high resolution custom array-CGH. Clin Genet. 2010;77(6):552–62. doi: 10.1111/j.1399-0004.2009.01341.x. [DOI] [PubMed] [Google Scholar]
- Lupski JR. Structural variation mutagenesis of the human genome: Impact on disease and evolution. Environ Mol Mutagen. 2015 doi: 10.1002/em.21943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marangi G, Orteschi D, Milano V, Mancano G, Zollino M. Interstitial deletion of 3p22.3p22.2 encompassing ARPP21 and CLASP2 is a potential pathogenic factor for a syndromic form of intellectual disability: a co-morbidity model with additional copy number variations in a large family. Am J Med Genet A. 2013;161A(11):2890–3. doi: 10.1002/ajmg.a.36257. [DOI] [PubMed] [Google Scholar]
- Mayle R, Campbell IM, Beck CR, Yu Y, Wilson M, Shaw CA, Bjergbaek L, Lupski JR, Ira G. DNA REPAIR. Mus81 and converging forks limit the mutagenicity of replication fork breakage. Science. 2015;349(6249):742–7. doi: 10.1126/science.aaa8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord KH, Lilljebjorn H, Vezzi F, Nilsson J, Magnusson L, Tayebwa J, de Jong D, Bovee JV, Hogendoorn PC, Szuhai K. GRM1 is upregulated through gene fusion and promoter swapping in chondromyxoid fibroma. Nat Genet. 2014;46(5):474–7. doi: 10.1038/ng.2927. [DOI] [PubMed] [Google Scholar]
- Ordulu Z, Wong KE, Currall BB, Ivanov AR, Pereira S, Althari S, Gusella JF, Talkowski ME, Morton CC. Describing sequencing results of structural chromosome rearrangements with a suggested next-generation cytogenetic nomenclature. Am J Hum Genet. 2014;94(5):695–709. doi: 10.1016/j.ajhg.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Z, Stankiewicz P, Xia Z, Breman AM, Dawson B, Wiszniewska J, Szafranski P, Cooper ML, Rao M, Shao L, South ST, Coleman K, et al. Observation and prediction of recurrent human translocations mediated by NAHR between nonhomologous chromosomes. Genome Res. 2011;21(1):33–46. doi: 10.1101/gr.111609.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozhan G, Sezgin E, Wehner D, Pfister AS, Kuhl SJ, Kagermeier-Schenk B, Kuhl M, Schwille P, Weidinger G. Lypd6 enhances Wnt/beta-catenin signaling by promoting Lrp6 phosphorylation in raft plasma membrane domains. Dev Cell. 2013;26(4):331–45. doi: 10.1016/j.devcel.2013.07.020. [DOI] [PubMed] [Google Scholar]
- Pannunzio NR, Li S, Watanabe G, Lieber MR. Non-homologous end joining often uses microhomology: implications for alternative end joining. DNA Repair (Amst) 2014;17:74–80. doi: 10.1016/j.dnarep.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakofsky CJ, Ayyar S, Deem AK, Chung WH, Ira G, Malkova A. Translesion Polymerases Drive Microhomology-Mediated Break-Induced Replication Leading to Complex Chromosomal Rearrangements. Mol Cell. 2015;60(6):860–72. doi: 10.1016/j.molcel.2015.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluth-Bolard C, Labalme A, Cordier MP, Till M, Nadeau G, Tevissen H, Lesca G, Boutry-Kryza N, Rossignol S, Rocas D, Dubruc E, Edery P, et al. Breakpoint mapping by next generation sequencing reveals causative gene disruption in patients carrying apparently balanced chromosome rearrangements with intellectual deficiency and/or congenital malformations. J Med Genet. 2013;50(3):144–50. doi: 10.1136/jmedgenet-2012-101351. [DOI] [PubMed] [Google Scholar]
- Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447(7140):102–5. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18(2):74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–55. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Tsurusaki Y, Nakashima M, Miyake N, Saitsu H, Takeda S, Matsumoto N. Precise detection of chromosomal translocation or inversion breakpoints by whole-genome sequencing. J Hum Genet. 2014;59(12):649–54. doi: 10.1038/jhg.2014.88. [DOI] [PubMed] [Google Scholar]
- Talkowski ME, Ordulu Z, Pillalamarri V, Benson CB, Blumenthal I, Connolly S, Hanscom C, Hussain N, Pereira S, Picker J, Rosenfeld JA, Shaffer LG, et al. Clinical diagnosis by whole-genome sequencing of a prenatal sample. N Engl J Med. 2012a;367(23):2226–32. doi: 10.1056/NEJMoa1208594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkowski ME, Rosenfeld JA, Blumenthal I, Pillalamarri V, Chiang C, Heilbut A, Ernst C, Hanscom C, Rossin E, Lindgren AM, Pereira S, Ruderfer D, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012b;149(3):525–37. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagaleti GV, Bien-Willner GA, Northup JK, Lockhart LH, Hawkins JC, Jalal SM, Withers M, Lupski JR, Stankiewicz P. Position effects due to chromosome breakpoints that map approximately 900 Kb upstream and approximately 1.3 Mb downstream of SOX9 in two patients with campomelic dysplasia. Am J Hum Genet. 2005;76(4):652–62. doi: 10.1086/429252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton D. De novo balanced chromosome rearrangements and extra marker chromosomes identified at prenatal diagnosis: clinical significance and distribution of breakpoints. Am J Hum Genet. 1991;49(5):995–1013. [PMC free article] [PubMed] [Google Scholar]
- Witt RM, Hecht ML, Pazyra-Murphy MF, Cohen SM, Noti C, van Kuppevelt TH, Fuller M, Chan JA, Hopwood JJ, Seeberger PH, Segal RA. Heparan sulfate proteoglycans containing a glypican 5 core and 2-O-sulfo-iduronic acid function as Sonic Hedgehog co-receptors to promote proliferation. J Biol Chem. 2013;288(36):26275–88. doi: 10.1074/jbc.M112.438937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt DW, Feng W, Conlin MP, Yousefzadeh MJ, Roberts SA, Mieczkowski P, Wood RD, Gupta GP, Ramsden DA. Essential Roles for Polymerase theta-Mediated End Joining in the Repair of Chromosome Breaks. Mol Cell. 2016;63(4):662–73. doi: 10.1016/j.molcel.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.