Abstract
YM155 (Sepantronium bromide) has been evaluated in clinical trials as a survivin suppressant but despite positive signals from early work later studies were negative. Clarifying the mechanism of action of YM155 is important to its further development. YM155 affects cells in a cell cycle specific manner. When cells are in G1, YM155 prevented their progression through S phase, leaving the cells at G1/S when exposed to YM155. Passage through mitosis from G2 is also defective following YM155 exposure. In this study, YM155 did not behave like a typical DNA intercalator in viscosity, circular dichroism (CD) and absorption spectroscopies studies. Additionally, molecular modeling experiments ruled out YM155 DNA interaction to produce DNA intercalation. We demonstrate that YM155 inhibited Top 2α decatenation and Top 1-mediated cleavage of DNA, suggesting that YM155 inhibits the enzyme function. Consistent with these findings, DNA double strand break repair was also inhibited by YM155.
Keywords: YM155, topoisomerase, replication, mitosis, repair
Topoisomerases are enzymes that disentangle DNA during DNA replication, transcription, repair and recombination processes [1]. While TOP1 releases the supercoiling and torsional tension of DNA introduced during the DNA replication and transcription processes by transiently cleaving and rejoining single strand of the DNA duplex, TOP2 catalyzes the breaking and rejoining of double strands of duplex DNA which allows the strands to pass through one another, thus altering the DNA topology. Irinotecan (CPT11) and topotecan are known TOP1-targeting agents in clinical use.[2] Drugs that target TOP2α can be divided into two broad classes. The first class is termed TOP2α poisons. Use of such agent leads to increased levels of TOP2α and DNA complexes. These drugs generate DNA lesions that include DNA strand breaks and protein covalently bound to DNA. TOP2α trapping and enzyme mediated DNA damage are thought to be the cause of cell death[3]. Active TOP2α poisons such as doxorubicin, etoposide and mitoxantrone belong to this class [4–6]. Resistance to these drugs is frequently associated with reduced expression of TOP2 isomers[7]. The second class is called catalytic inhibitor compound (CIC). They inhibit TOP2α enzyme activity but do not increase the levels of TOP2α and DNA covalent complexes usually. TOP2 CICs are thought to kill by depriving the essential enzyme activity of TOP2α[8].
YM155 is a small molecule inhibitor that emerged from a drug screen using a construct that consisted of a survivin promoter and a luciferase reporter and was described as a survivin suppressant [9]. It showed potent activities against a variety of cancer cells including non-small cell, breast, prostate and non-Hodgkin’s lymphoma in vitro and in animal models (for review, refer to [10]). Despite its potency and good tolerability in preclinical and early clinical studies, several phase II studies using YM155 either alone or in combination with other chemotherapy agents yielded negative results [11–14]. This failure may be a consequence of incorrect understanding of the molecular target and the resultant absence of patient population stratification. Indiscriminate trial of anticancer drug in mixed genetic background often results in trashing the drugs being studied [15].
Despite widespread acceptance as a survivin suppressant, YM155 efficacy does not correlate with survivin levels. In the Merkel cell model [16], YM155 was cytostatic instead of inducing apoptosis, which most of us would expect from a survivin suppressant. Interestingly, YM155 induced DNA damage and triggered DNA damage checkpoint signaling in our previous studies and others[17, 18]. The number of mutations in the YM155 resistant clones was ~10-fold of other drug resistant clones, suggesting that YM155 is highly mutagenic [19]. Analyses of mutations employing a technique combining the next generation sequencing with CRISPR/Cas9 based method failed to reveal a specific resistance mechanism possibly due to the large number of mutations YM155 induced. Interestingly, a genome-wide insertional mutagenesis screening using the KBM7 near haploid human cell line revealed that YM155 sensitivity depended on the expression of a solute carrier SLC35F2, which is highly expressed in a variety of cancer cell lines [20].
We show here that YM155 is a topoisomerase inhibitor. DNA topoisomerases play important roles in various cellular processes. Inhibition of DNA topoisomerase by YM155 could explain all phenomena observed during YM155 treatment. These include stalled DNA replication, decreased transcription, problems in mitosis, defects in DNA repair in addition to DNA damage, which triggers the DNA checkpoint signaling. This provides a unified mechanism of action for YM155 while forming contrast to reports defining YM155 as a DNA intercalator [19, 20]. Simultaneous elucidation of the molecular target of YM155 and marker for efficacy, when validated, will hopefully rejuvenate YM155 clinical research and lead to eventual use of this drug in cancer treatment. It is unknown at the moment whether TOP inhibitors that are not TOP poisons might be active anticancer agents [8]. YM155 has hopes to become the first non-topoisomerase poison in the clinical arena.
Results
YM155 suppress DNA replication, arrests cell cycle at the G1/S or G2/M
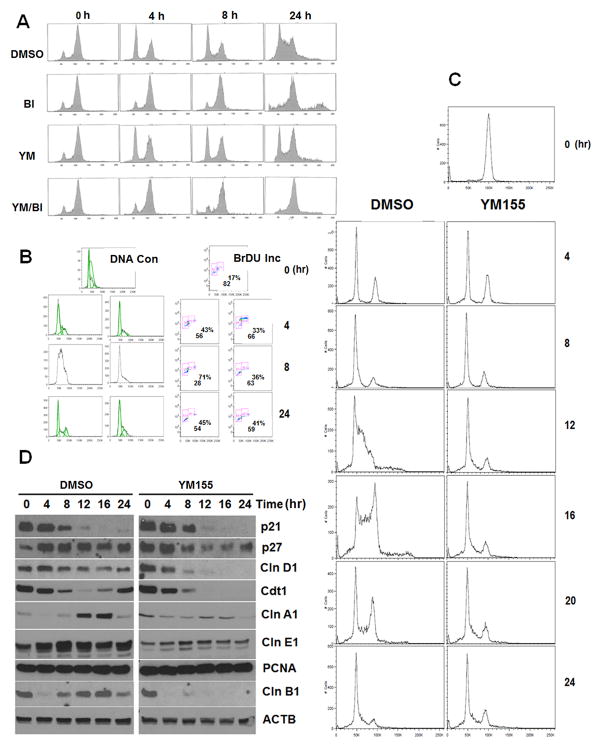
When unsynchronized H1299 cells growing in log phase were analyzed for cycle distribution, the majority of them were found in G1 with a 2N DNA content. When they were treated for 24 hours with BI6727 (volasertib, a small molecule PLK1 inhibitor), they arrested with a 4N content at M (polo arrest with 4N DNA content)[21]. However, the cell cycle distribution did not change when they were treated with either YM155 alone or in combination with BI6727 (data not shown). When cells arrested at M phase with BI6727 for 24 hours were released in medium containing DMSO, YM155, BI6727 or YM155 plus BI6727, cells in the DMSO group restarted the cell cycle, divided with a significantly bigger G1 peak after 4–8 hours, G1 and S peaks after 24 hours, indicating successful progression into the S phase. Cells in BI6727 (BI6727 added to RPMI1640 fresh at the same concentration used to arrest the cells at M) remained predominantly at M after 24 hours. Interestingly, we saw a sizable 8N peak in these cells, suggesting that some of these cells have come out of the M phase and went on to replicate their DNA before successful cytokinesis as previously reported [22]. When these cells were grown in medium containing YM155 alone, slightly less than half of all cells were able to progress into G1 phase from M similar to the DMSO group, indicating that these cells had no major problems transitioning from M to G1. The remaining cells however stayed in M suggesting these cells had problem moving forward, which is consistent with defects in chromosomal separation (see below). Noticeably also, cells grown in both BI6727 and YM155 had no 8N peak at all, suggesting that cells were not able to replicate their DNA in the presence of YM155 (Figure 1a). All these above suggested that YM155 arrested the cell cycle at G1/S boundary or G2/M since they could not finish DNA replication and may have problems to separate their chromosomes. This could occur when topoisomerase activity is impaired [23, 24]. To test this hypothesis more vigorously, we synchronized H1299 cells by treating with mimosine for 24 hours which arrests cells in the G1/S border.[25] These cells were shifted to medium containing either DMSO or YM155 after mimosine was washed out. Cell cycle profile and DNA replication were followed by BrdU incorporation at 4, 8 and 24 hours. While the percentage of S phase cells increased by 26.6% and 54.3% after 4 and 8 hours respectively in the DMSO group, the S phase cells in the YM155 group had only modest increase to 16.3% and 19.2% after 4 and 8 hours respectively. Even after 24 hours, there was only a 23% increase, consistent with a blockage in G1/S phase in the YM155 group (Figure 1b). Since some cells blocked with mimosine might have already entered into S phase, we further experimented with cells arrested at mitosis by nocodazole which arrests cells at G2/M by inhibiting microtubule polymerization.[26] When these cells were shifted from nocodazole containing medium to one containing DMSO or YM155, cells in the DMSO group progressed synchronously to G1 after 8 hours. These cells then progress further through S and into G2/M 12 and 16 hours later. In contrast, YM155 prevented the cells from further progressing into the S phase. Compared with BI6727 arrested cells (Figure 1a), only a small proportion of cells failed to exit M completely after 24 hours (Figure 1c), the majority of cells experienced no problem of M/G1 transition possibly due to the later arrest point of nocodazole compared with BI6727. Topoisomerases are presumably no longer needed. At the protein levels, Western blot analyses of nocodazole arrested cells released into YM155 were consistent with the fact that YM155 prevented progression of cells into the S phase (Fig 1d). Cyclin A1 expression which correlates with the initiation of S phase did not accumulate significantly at 24 hours after nocodazole-treated cells were released into medium containing YM155, suggesting that these cells were not progressing into the S phase. Additionally, the expression of p21, p27, PCNA and cyclin E1 were not affected, but YM155 prevented cyclin D1, Cdt1, cyclin B1 from accumulating again after the first drop.
Figure 1.
YM155 arrests cell cycle at G1/S or G2/M. H1299 non-small cell lung cancer cells were treated with volasertib (BI6727, 40 nM) to arrest cell cycle at G2/M before releasing into the media containing DMSO, BI6727 (40 nM), YM155 (40 nM) or YM/BI (40 nM each). Flow cytometry was performed at 4, 8, 24 hours later to determine the nuclear DNA contents (A). H1299 cells were synchronized with mimosine at G1/S. They were then released into media containing either DMSO (left panel) or YM155 (40 nM, right) on top. DNA content (DNA con) or BrdU incorporation (BrdU Inc) were monitored overtime at 4, 8, 24 hour. The percentages represented in the BrdU boxes are cells at S phase (upper) and G1 (lower) respectively (B). H1299 cells are arrested in M phase by nocodazole (50ng/ml) before released into medium containing DMSO (left column) or YM155 (40 nM, right) 4, 8, 12, 16, 20 and 24 hours. Passage through M and S were followed by flow cytometry (C). Cells were arrested in G1/S by mimosine (200 μM). Cells are released then into media with DMSO (left) or YM155 (40 nM, right). Cell cycle protein levels were followed with Western Blot at 0, 4, 8, 12, 16, 24 hours. Beta actin (ACTB) was used as loading control (D).
YM155 does not form a complex with free DNA
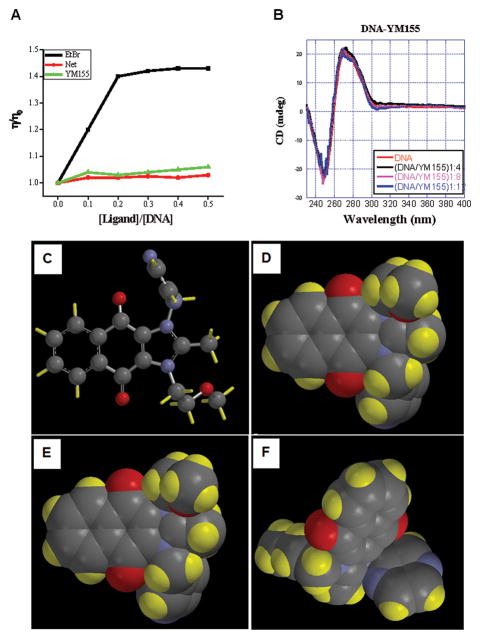
The positively charged planar scaffold of YM155 would also favor intercalation with DNA. Therefore it is possible that YM155 functions as a DNA intercalator and introduces positive supercoils in the DNA. The DNA is subsequently relaxed by topoisomerase I resulting in DNA damage and checkpoint signaling like a typical DNA intercalating agent ethidium bromide (EtBr). We therefore decided to test whether YM155 can function as a DNA intercalator in a viscosity study using EtBr as a control. As shown in Figure 2a, increasing concentrations of EtBr binding to DNA clearly increased the viscosity of the EtBr-DNA complex, which plateaued after the ratio reached 1:5. YM155 however failed to increase the viscosity of the DNA even when the YM155 concentration reached up to 100 μM. Molecular interactions can be detected by studying circular dichroism spectroscopy (CD). We also investigated whether YM155 behaves like a DNA chealator to induce circular dichoism. Normally, DNA minor groove binders always induce a large induced CD signal while intercalators give a more varied response. However, increasing concentration of YM155 up to 60 μM failed to induce a CD spectral change (Figure 2B), suggesting that YM155 is not a significant minor groove binder or an intercalator.
Figure 2.
YM155 does not bind DNA (A) Viscometric titrations of calf thymus DNA with ethidium bromide (EtBr), Netropsin (Net), and YM155. The data are the average of three separate readings. Where η and η0 represent the viscosity of the solution containing ligand-DNA complex and free DNA respectively. (B) Circular dichroism spectra of YM155 with hairpin duplex DNA at different DNA-drug ratios. The experiments were conducted in Tris-HCl buffer (50 mM Tris-HCl, 100 mM NaCl, 1mM EDTA, pH = 7.4) at 25 °C. (C–F) Energy minimized structures of YM155 at the 6–31G* (p,d) level of theory by Spartan 10 software. YM155 is represented as a ball and stick model (C), and space filling model with front (D), back (E), down (F) side view with carbon in grey, hydrogen in yellow, oxygen in red and nitrogen in blue.
To back up our conclusion (that YM155 is not a minor groove binder), we performed molecular modeling experiments. Not surprisingly, we found that the compound phenyl and alkyl substituents are out of the plane of the larger ring system and block both intercalation and minor groove binding of the compound to DNA (Figure 2c–f). Therefore, our data is not consistent with the hypothesis that YM155 functions as a DNA chelator in contrast to previous publications [19, 20].
YM155 inhibits TOP2α and TOP1 activity
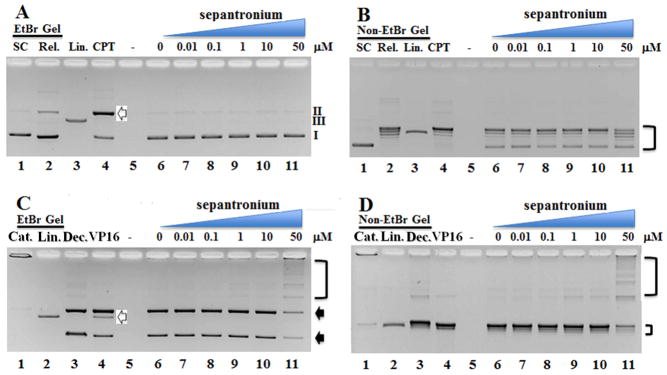
Topoisomerases change DNA topology, adjust DNA supercoiling by introducing single (TOP1) and double strand (TOP2) DNA breaks before resealing them during DNA replication, transcription, DNA repair, recombination and chromosomal separation. YM155 exposure led to inhibition of DNA replication (above) and transcription suppression and possibly chromosomal separation (above). We thus tested whether YM155 might interfere with topoisomerase(s) function. We first assayed topoisomerase I plasmid cleavage activity in the presence of YM155 first. In the EtBr gel (Figure 3a), TOP 1 was able to cut the supercoiled DNA (form I, lane 1) and lead to the appearance of open circular DNA (relaxed DNA, form II, lane 2). CPT, a well-known TOP1 poison formed intermediate with the plasmid DNA and led to increase in open circular DNA level and decrease in supercoiled DNA (lane 4). Increasing concentrations of YM155 failed to increase the amount of open circular DNA up to 50 μM, suggesting that YM155 is not a TOP1 poison like CPT. In the non-EtBr gel (Figure 3b), CPT treatment led to disappearance of supercoiled pHOT plasmid DNA, increased the amount of open circular DNA, forming relaxed pHOT topoisomer ladder (lane 4). YM155 neither decreased the amount of supercoiled DNA nor increase the amount of open circular pHOT with increasing concentrations. It indeed inhibited the formation of open circular DNA (lane 11, figure 3b). It should be pointed out that the distribution of topoisomers was shifted in lane 11 (parenthesis on right). To characterize the interaction between TOP2α and YM155, we carried out a TOP2α assay. In the EtBr gel assay (Figure 3c), TOP2α was able to convert catenated kDNA (did not enter into the gel due to size, lane 1) into decatenated DNA (lane 3). The presence of a known TOP2α poison etoposide (VP16) led to the appearance of linear DNA complex (lane 2). YM155, did not increase the amount of linear DNA complex as did etoposide. Instead, it significantly inhibited the decatenation of kDNA (lane 11, parenthesis). This strongly suggests that YM155 is suppressing the function of the enzyme. The same scenario was true when the assay was done in non-EtBr (Figure 3d). YM155 was hindering the decatenation process generating less decatenated DNA (lane 11, parenthesis).
Figure 3.
YM155 (sepantronium) does not stimulate TOP1 or TOP2α activity. Unlike CPT (irinotecan, a TOP1 poison, lane 4 white arrow, 50 μM), increasing concentrations of YM155 did not increase the amount of open circular DNA (form II) formed (A). The distribution of topoisomers was shifted in lane 11 (Figure 3B) indicating induced inhibition of TOP1 by YM155 in a dose dependent manner. SC: Supercoiled, Rel: Relaxed, Lin: Linear. Unlike VP16 (etoposide, a TOP2α poison at 50 μM, lane 4, white arrow), increasing concentrations of YM155 did not induce the formation of linear DNA. Instead, formation of nicked circular (up) and relaxed circular (down) from the kDNA was inhibited (black arrows and parenthesis on right) in EtBr gel (C) and Non-EtBr Gel (D). Cat: Catenated, Dec: Decatenate kDNA. Lane 4, 5–11 contained topoisomerase.
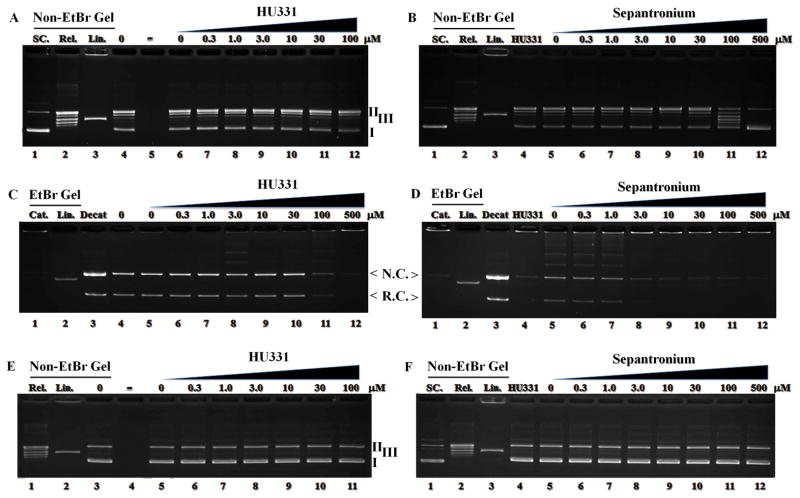
HU331 is a well characterized TOP2α ATPase inhibitor[27]. In further studies, it did not inhibit TOP1 activity when used up to 100 μM level (Figure 4A). Consistent with previous studies HU331 inhibited decatenation of TOP2α in a dose dependent manner with effects becoming obvious at 100 μM and reaching maximum at 500 μM (Figure 4C). The E. Coli. gyrase activity was also not affected (Figure 4E). Although E. Coli gyrase activity was not affected by YM155 (Figure 4F), YM155 inhibited TOP2α in a dose dependent fashion (Figure 4D). Compared to HU331, the activity was more potent which became obvious when 3.0 μM of YM155 was present. In addition, YM155 also inhibited TOP1 when the concentration reached 30 μM (Figure 4B).
Figure 4.
YM155 suppresses TOP1 and TOP2α but not E. Coli Gyrase activity YM155 activities against purified TOP1 (A, B), TOP2α (C, D) and E Coli Gyrase (E, F) were performed using H331, a known TOP2α CIC, as control. YM155 inhibited TOP1 (B) when the concentration increased whereas H331 did not (A). SC: supercoil, Rel: relax, Lin: Linear. Compared with HU331 (C, Left middle), YM155 inhibited TOP2α more potently leading to substantial decrease in the formation of nicked circular (N.C) and relaxed circular (R.C) kDNA (D, right middle). Cat: Catenated kDNA, Decat: Decatenated kDNA. Neither HU331 nor YM155 had activity against E. Coli Gyrase in E and F in the bottom panel. Topoisomerases were included in lane 4, 6–12 in A, B; lane 4–12 in C, D; lane 3, 5–11 in E and 4–12 in F.
YM155 delays DNA repair
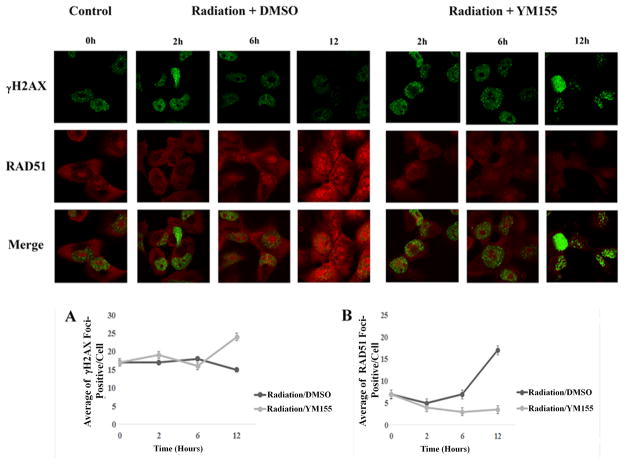
Topoisomerases are involved in DNA repair. Given that our results above were consistent with inhibited topoisomerase activity in the presence of YM155, we hypothesized that DNA repair was compromised in the presence of YM155. Ionized radiation results in DNA double strand breaks (DSBs). Active DSBs trigger phosphorylation of H2AX (γH2AX), forming foci in the nucleus. The persistence of foci signals continued presence of DSBs. On the other hand, RAD51 forms foci as homologous recombination repair mechanism starts to repair damaged DNA until the repair is done. Two, six, and twelve hours after H1299 cells were radiated with 2 Gy X-ray, cells were processed for γH2AX and RAD51 staining. In the absence of YM155, γH2AX foci (green) peaked 2 hours later and started to fade below the initial level after 12 hours. In the presence of YM155, the immunofluorescence continued unfaded, suggesting persistent high levels of DNA damage. RAD51 foci (red) peaked robustly after 12 hours in the presence of DMSO after DNA damage. However, significantly fewer RAD51 foci were present in these cells in the presence of YM155, suggesting inhibited DNA repair foci formation in these cells (Figure 5a). Counting for foci formation for γH2AX and RAD51 (Figure 5b) were consistent with the hypothesis that YM155 suppressed DNA repair while inflicting constant DNA damage.
Figure 5.
YM155 delays double strand break DNA repair H1299 cells were radiated with ironized radiation (2Gy) and are subsequently treated with DMSO or YM155 (40 nM) for 2, 6 and 12 hours. Cells were stained for γH2AX foci (green) to follow DNA damage and RAD51 foci (red) to follow the status of DNA repair. Quantitative analysis of γH2AX foci present as a function of time (A) or RAD51 foci (B).
Discussion
Identifying the mechanism of action of a new molecular entity poses a major challenge in drug discovery as well as in chemical biology research [28]. When the target of a drug is unknown, it is difficult to select the right type of cancer to treat, nearly impossible to improve its efficacy or reduce unanticipated toxicity. Rationale combination with other drugs is not possible. The cost of the abandoning drugs is contributing to the rapid increase of the drug price at 7.3% annually according to the 2003 publication [29].
We show here that YM155 targets DNA topoisomerases. Use of YM155 clearly blocked progression through S and M phase (G1/S, G2/M arrest). YM155 also induced DNA damage, checkpoint signaling and caused delay in DNA double strand break repair. In vitro assay of topoisomerase activity showed that YM155 potently suppressed TOP2α’s ability to decatenate kDNA and TOP1 cleavage of plasmid DNA. This explains why the luciferase activity under the control of a survivin promoter was severely diminished in the original discovery screening since YM155 is expected to reduce DNA transcription profoundly when both TOP1 and TOP2α activity are compromised. This also potentially explains YM155 GI50 is at nanomole level in vivo whereas micromole concentration was needed to completely block the topoisomerase activity in our study. As a comparison, YM155 shown here is at least as good as if not more potent than etoposide in prior publications (average concentration of etoposide used 50 μM).
In the presence of YM155, DNA repair was defective. There were fewer RAD51 repair foci observed with YM155 (above). In addition to suppressed topoisomerase activity itself, inability to enter into the S phase is thought to have contributed since homologous repair occurs predominantly during the S phase. This is consistent with previous reports that YM155 boosted effects of cisplatin [30] and radiation therapy [31] in earlier studies. Our work thus unifies studies related to the mechanism of action of YM155 and suggests topoisomerase but not survivin as the molecular target of YM155. This study also contrasts earlier studies claiming YM155 as DNA intercalator [20].
As stated earlier, TOP2 targeting agents fall into two categories, the poisons and the CICs. It is thought that TOP2α poisons effectively block DNA replication and transcription leading to increase in the levels of double strand breaks and protein covalently bound to DNA resulting in an increase in enzyme mediated DNA damage. On the other hand, CICs, on short term exposure, do not stimulate enzyme mediated DNA damage and they do not induce DNA damage response [32] except long term although most CICs studied so far are not specific except one [33] and are very weak compounds. It is not clear here exactly how YM155 targets TOP1 and TOP2α, and YM155 could be a competitive ATPase inhibitor. Alternatively, YM155 could compete out DNA binding sites on topoisomerases so these enzymes lose the ability to bind DNA. Since undecatenated DNA is believed to be monitored by the DNA damage/unreplicated DNA checkpoint [34], YM155 exposure is expected to cause DNA damage.
In clinical oncology, two things are crucial for bringing a drug to the clinical arena. One is having a clear molecular target and another is knowledge of the marker for sensitivity/resistance. Therefore it becomes much easier when a molecular marker that is both prognostic and predictive of response is known. Now that YM155 has a well-defined molecular target (as topoisomerase) and SLC35F2 looks like a promising marker for YM155 sensitivity/resistance. Rationale combination may start here. For example we may combine PLK1 inhibitor with YM155 in lung cancer since PLK1 also participates in the repair of TOP2α mediated DNA damage[17]. In a preselected patient population, combination use of two or more drugs that act synergistically in those cancers highly expressing TOP2α are expected to make a huge difference in the results. So far preclinical data predict that YM155 would be useful in patients with breast, prostate, lung cancer and mantle cell lymphoma[35], in which topoisomerase poison have made important contributions to treatment. Interestingly, anaplastic thyroid cancer that expresses TOP2α five-fold higher than differentiated thyroid cancer (Personal data) was exquisitely sensitive to YM155 [36]. Uncertainty of the molecular target and lack of rationale combination in an unselected population were likely responsible for the failure of YM155 in recent clinical trials.
Materials and Methods
Cell line, antibodies and chemicals
H1299 was purchased from American Type Culture Collection (ATCC) and cultured in RPMI 1640 (Invitrogen) with 5% FBS (Hyclone), at 37°C in 10% CO2. YM155 (S1130) and BI6727 (S2235) were purchased from Selleck Chemicals. L-mimosine, nocodazole and 5-BrdU were products of Sigma-Aldrich. All antibodies for Western Blot analysis of cell cycle proteins were from Cell Signaling Technology including p21, p27, Cyclin D1, Cdt1, Cyclin A1, Cyclin E1, PCNA and Cyclin B1. Actin B antibody was purchased from Sigma. Anti-BrdU-APC was from eBioscience. HU331 was purchased from Abcam (ab120922).
Cell Cycle Analysis and BrdU incorporation
Cell cycle was synchronized with either BI6727 or mimosine or nocodazole. Analyses were performed using standard flow cytometry procedures following either propidium iodide staining alone[37], or together with BrdU incorporation. These cells were then stained with anti-BrdU-APC (eBioscience, San Diego, CA, USA), as described previously [38]. Cells were analyzed using a LSRII flow cytometer. All flow cytometry data were analyzed using FlowJo software (Tree Star, Ashland, OR).
Western blot
Proteins were isolated using standard procedures. Whole-cell lysates containing 50 μg of proteins, were separated by SDS-PAGE and immunoblotted with the following specific antibodies. Antibodies were used at 1:1,000 as recommended by the manufacturer. Immunoblot was performed using standard protocols as described previously.
Viscometric Titrations
Viscometric titrations were performed using a Cannon-Ubbelohde semi-micro-viscometer, submerged in a water-bath to maintain a constant temperature at 25 °C as previously described[39]. The concentration of calf thymus DNA was measured spectrophotometrically. and a 10 mL, 200 μM (bp) solution of calf thymus DNA in Tris-HCl buffer (50 mM Tris-HCl, 100 mM NaCl, and 1 mM EDTA at pH = 7.4) was used in viscometric titrations. Incremental addition of compounds with thorough mixing from concentrated stock solutions was used to titrate the DNA in the viscometer. The final volume did not increase more than 2% of the initial volume during the titration. Each addition was followed by an equilibration time of 20 min before the flow times were measured. The intrinsic viscosities were proportional to the flow times of the solutions and calculated as given in literature [40].
| (1) |
where η is the viscosity of the solution containing ligand bound DNA, ηo is the viscosity of the solution containing free DNA, t is the flow time of solution containing ligand bound DNA, to is the viscosity of the solution containing the free DNA and tb is the flow time of buffer.
Circular Dichroism Spectroscopy (CD)
Circular dichroism experiments were performed on a Jasco J-810 CD spectrometer in a 1 cm quartz cell at 25 °C. The hairpin duplex DNA (5 μM) sequence in buffer (50 mM Tris-HCl, 100 mM NaCl, 1 mM EDTA, pH 7.4) was added to the cell prior to the experiment and then the compound was added to the hairpin DNA solution and incubated for 10 min to achieve equilibrium. For each titration point four spectra were averaged from 500 to 220 nm with scan speed of 50 nm/min, and a response time of 1 sec. Buffer subtracted graphs were created using the Kaleidagraph software.
Structural Model Calculation Methods
Molecular modeling studies were initiated with conformational analysis of the tested compound with a molecular mechanics MMFF approximation level with the Spartan 10 software package (Wavefunction, Inc.) [41]. The software package was employed to optimize the final geometry by using ab initio calculations with density functional theory (DFT), B3LYP at the 6–31G* approximation level.
Assay for TOP1, TOP2α and E Coli Gyrase
Topoisomerases and bacterial Gyrase assay were performed using TOPOGEN enzyme Kit (TG1018, TG1001 and TG2000G1). TOP1 assay was described previously[42]. Briefly, 200 ng of pHOT DNA (supercoiled, relaxed or linear) was assayed in the presence of 10x buffer, 1 unit/reaction of purified human TOP1, CPT (control) or various concentrations of YM155 (0–100 μM). The reaction mixtures were incubated at 37°C for 30 min. The reactions were stopped with stop buffer. The mixtures were treated with protein kinase at 37°C for 15 min, DNA extracted by phenol. The extracted DNAs were divided into half and loaded onto 1% agarose gel with or without ethidium bromide. Electrophoresis was carried out at 50 volts for 45–60 min before staining when necessary and photographed. HU331 was used as a TOP CIC control.
For TOP2α assay [43], reactions mixtures that contained 10x reaction buffer, 4 units/reaction of purified human TOP2α, 200 ng of kDNA, VP16 (control) or various concentrations YM155 (0–100 μM) were set up and incubated at 37°C for 30 min. The reaction was stopped and treated with protein kinase at 37°C for 15 min, phenol extracted before divided into halves and loaded onto 1% agarose gel with or without ethidium bromide. The gels were run at 50 volts for 45–60 min before photograph after staining when needed.
E. Coli Bacterial gyrase assays [44] were performed in a mixture containing 8 units of purified bacterial gyrase, 5x reaction buffer, 200 ng of relaxed pHOT, HU331 or YM155 at various concentrations at 37°C for 30 min. The reaction mixtures were stopped, treated with protein kinase and extracted by the phenol before loading. They were run under the same condition as above before pictures taken.
Immunofluorescence microscopy
The immunofluorescence staining was performed as described previously[45]. Briefly, cells were plated onto poly-L-lysine treated glass coverslips and cultured for 24 hours in RPMI1640 medium with 10% FBS and Penicillin-Streptomycin to allow cells to adhere to the substrate. Cells were radiated with 2 Gy in a radiator before the cultural medium were changed to RPMI1640 with either DMSO or YM155 (0 hour). At 0, 2, 4, 12 hours, coverslips were rinsed twice with PBS for 5 min. Cells were fixed in fixative solution, permeabilized with 0.1% triton in PBS for 10 min at room temperature, and blocked with 3% ovalbumin in PBS (blocking solution). Primary antibodies (γH2AX from Cell Signaling Technology) and RAD51 (Novus Biologicals) were diluted at 1:200 (or as recommended by the manufacturer) in blocking solution and incubated overnight at 4°C. Cell were rinse 3 times with PBS for 5 min, incubated for 30 minutes at 37°C with the respective secondary antibodies diluted 1:200 in blocking buffer. Confocal images were obtained using an Axiovert 200 M Zeiss microscope equipped with Zen 2009 software (Carl Zeiss Microscopy, Gottingen, Germany). Number of foci per cell were counted using Image J (NIH) software. Data analysis and statistical differences were determined with Student’s t-test using the Excel software.
Acknowledgments
This work is partially supported by the institution startup fund to Dr. Carsten Schroeder. Biophysical studies supported by NIH GM111749 (W.D.W). We thank Lynsey Ekema for expert assistance in illustration in Fig. 6.
Figure 6.
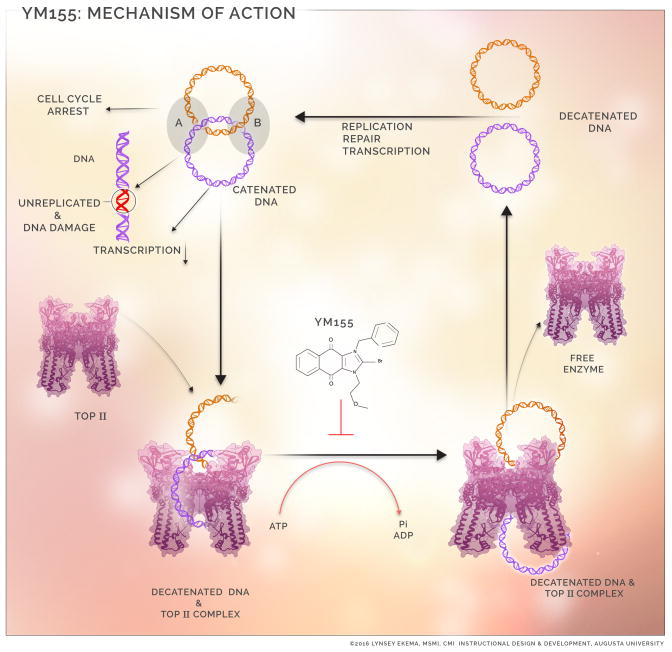
Schematic description of the mechanism of action of YM155. Catenated DNAs are frequently encountered during DNA replication, repair, transcription and chromosomal separation. Continued presence of such sends signals through checkpoint cascade resulting in cell cycle arrest, unreplicated/damaged DNA, decreased DNA transcription and unseparated chromosome. Progression of the above processes depend on prompt resolution of catenated DNA at junction A and B. YM155 impairs TOP2α mediated DNA decatenation.
Footnotes
Conflict of interests: The authors declare no conflict of interests.
References
- 1.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annual review of biochemistry. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 2.Streltsov SA. Action models for the antitumor drug camptothecin: formation of alkali-labile complex with DNA and inhibition of human DNA topoisomerase I. Journal of biomolecular structure & dynamics. 2002;20(3):447–454. doi: 10.1080/07391102.2002.10506863. [DOI] [PubMed] [Google Scholar]
- 3.Kaufmann SH. Cell death induced by topoisomerase-targeted drugs: more questions than answers. Biochimica et biophysica acta. 1998;1400(1–3):195–211. doi: 10.1016/s0167-4781(98)00136-5. [DOI] [PubMed] [Google Scholar]
- 4.Tewey KM, Rowe TC, Yang L, Halligan BD, Liu LF. Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science. 1984;226(4673):466–468. doi: 10.1126/science.6093249. [DOI] [PubMed] [Google Scholar]
- 5.Chen GL, Yang L, Rowe TC, Halligan BD, Tewey KM, Liu LF. Nonintercalative antitumor drugs interfere with the breakage-reunion reaction of mammalian DNA topoisomerase II. The Journal of biological chemistry. 1984;259(21):13560–13566. [PubMed] [Google Scholar]
- 6.Burden DA, Osheroff N. Mechanism of action of eukaryotic topoisomerase II and drugs targeted to the enzyme. Biochimica et biophysica acta. 1998;1400(1–3):139–154. doi: 10.1016/s0167-4781(98)00132-8. [DOI] [PubMed] [Google Scholar]
- 7.Walker JV, Nitiss JL. DNA topoisomerase II as a target for cancer chemotherapy. Cancer investigation. 2002;20(4):570–589. doi: 10.1081/cnv-120002156. [DOI] [PubMed] [Google Scholar]
- 8.Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nature reviews Cancer. 2009;9(5):338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakahara T, Kita A, Yamanaka K, Mori M, Amino N, Takeuchi M, Tominaga F, Hatakeyama S, Kinoyama I, Matsuhisa A, Kudoh M, Sasamata M. YM155, a novel small-molecule survivin suppressant, induces regression of established human hormone-refractory prostate tumor xenografts. Cancer research. 2007;67(17):8014–8021. doi: 10.1158/0008-5472.CAN-07-1343. [DOI] [PubMed] [Google Scholar]
- 10.Rauch A, Hennig D, Schafer C, Wirth M, Marx C, Heinzel T, Schneider G, Kramer OH. Survivin and YM155: how faithful is the liaison? Biochimica et biophysica acta. 2014;1845(2):202–220. doi: 10.1016/j.bbcan.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Kelly RJ, Thomas A, Rajan A, Chun G, Lopez–Chavez A, Szabo E, Spencer S, Carter CA, Guha U, Khozin S, Poondru S, Van Sant C, Keating A, Steinberg SM, Figg W, Giaccone G. A phase I/II study of sepantronium bromide (YM155, survivin suppressor) with paclitaxel and carboplatin in patients with advanced non-small-cell lung cancer. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2013;24(10):2601–2606. doi: 10.1093/annonc/mdt249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kudchadkar R, Ernst S, Chmielowski B, Redman BG, Steinberg J, Keating A, Jie F, Chen C, Gonzalez R, Weber J. A phase 2, multicenter, open-label study of sepantronium bromide (YM155) plus docetaxel in patients with stage III (unresectable) or stage IV melanoma. Cancer medicine. 2015;4(5):643–650. doi: 10.1002/cam4.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clemens MR, Gladkov OA, Gartner E, Vladimirov V, Crown J, Steinberg J, Jie F, Keating A. Phase II, multicenter, open-label, randomized study of YM155 plus docetaxel as first-line treatment in patients with HER2-negative metastatic breast cancer. Breast cancer research and treatment. 2015;149(1):171–179. doi: 10.1007/s10549-014-3238-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papadopoulos KP, Lopez-Jimenez J, Smith SE, Steinberg J, Keating A, Sasse C, Jie F, Thyss A. A multicenter phase II study of sepantronium bromide (YM155) plus rituximab in patients with relapsed aggressive B-cell Non-Hodgkin lymphoma. Leukemia & lymphoma. 2016:1–8. doi: 10.3109/10428194.2015.1113275. [DOI] [PubMed] [Google Scholar]
- 15.Chang A, Parikh P, Thongprasert S, Tan EH, Perng RP, Ganzon D, Yang CH, Tsao CJ, Watkins C, Botwood N, Thatcher N. Gefitinib (IRESSA) in patients of Asian origin with refractory advanced non-small cell lung cancer: subset analysis from the ISEL study. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2006;1(8):847–855. [PubMed] [Google Scholar]
- 16.Arora R, Shuda M, Guastafierro A, Feng H, Toptan T, Tolstov Y, Normolle D, Vollmer LL, Vogt A, Domling A, Brodsky JL, Chang Y, Moore PS. Survivin is a therapeutic target in Merkel cell carcinoma. Science translational medicine. 2012;4(133):133ra156. doi: 10.1126/scitranslmed.3003713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong M, Ren M, Silva J, Kennedy T, Choi J, Cowell JK, Hao Z. Sepantronium is a DNA damaging agent that synergizes with PLK1 inhibitor volasertib. American journal of cancer research. 2014;4(2):135–147. [PMC free article] [PubMed] [Google Scholar]
- 18.Glaros TG, Stockwin LH, Mullendore ME, Smith B, Morrison BL, Newton DL. The “survivin suppressants” NSC 80467 and YM155 induce a DNA damage response. Cancer chemotherapy and pharmacology. 2012;70(1):207–212. doi: 10.1007/s00280-012-1868-0. [DOI] [PubMed] [Google Scholar]
- 19.Kasap C, Elemento O, Kapoor TM. DrugTargetSeqR: a genomics- and CRISPR-Cas9-based method to analyze drug targets. Nature chemical biology. 2014;10(8):626–628. doi: 10.1038/nchembio.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winter GE, Radic B, Mayor-Ruiz C, Blomen VA, Trefzer C, Kandasamy RK, Huber KV, Gridling M, Chen D, Klampfl T, Kralovics R, Kubicek S, Fernandez-Capetillo O, Brummelkamp TR, Superti-Furga G. The solute carrier SLC35F2 enables YM155-mediated DNA damage toxicity. Nature chemical biology. 2014;10(9):768–773. doi: 10.1038/nchembio.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudolph D, Steegmaier M, Hoffmann M, Grauert M, Baum A, Quant J, Haslinger C, Garin-Chesa P, Adolf GR. BI 6727, a Polo-like kinase inhibitor with improved pharmacokinetic profile and broad antitumor activity. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15(9):3094–3102. doi: 10.1158/1078-0432.CCR-08-2445. [DOI] [PubMed] [Google Scholar]
- 22.Barr FA, Sillje HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nature reviews Molecular cell biology. 2004;5(6):429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 23.Chang CJ, Goulding S, Earnshaw WC, Carmena M. RNAi analysis reveals an unexpected role for topoisomerase II in chromosome arm congression to a metaphase plate. Journal of cell science. 2003;116(Pt 23):4715–4726. doi: 10.1242/jcs.00797. [DOI] [PubMed] [Google Scholar]
- 24.Coelho PA, Queiroz-Machado J, Carmo AM, Moutinho-Pereira S, Maiato H, Sunkel CE. Dual role of topoisomerase II in centromere resolution and aurora B activity. PLoS biology. 2008;6(8):e207. doi: 10.1371/journal.pbio.0060207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lalande M. A reversible arrest point in the late G1 phase of the mammalian cell cycle. Experimental cell research. 1990;186(2):332–339. doi: 10.1016/0014-4827(90)90313-y. [DOI] [PubMed] [Google Scholar]
- 26.Harper JV. Synchronization of cell populations in G1/S and G2/M phases of the cell cycle. Methods in molecular biology. 2005;296:157–166. doi: 10.1385/1-59259-857-9:157. [DOI] [PubMed] [Google Scholar]
- 27.Regal KM, Mercer SL, Deweese JE. HU-331 is a catalytic inhibitor of topoisomerase IIalpha. Chemical research in toxicology. 2014;27(12):2044–2051. doi: 10.1021/tx500245m. [DOI] [PubMed] [Google Scholar]
- 28.Schenone M, Dancik V, Wagner BK, Clemons PA. Target identification and mechanism of action in chemical biology and drug discovery. Nature chemical biology. 2013;9(4):232–240. doi: 10.1038/nchembio.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DiMasi JA, Hansen RW, Grabowski HG. The price of innovation: new estimates of drug development costs. Journal of health economics. 2003;22(2):151–185. doi: 10.1016/S0167-6296(02)00126-1. [DOI] [PubMed] [Google Scholar]
- 30.Iwasa T, Okamoto I, Takezawa K, Yamanaka K, Nakahara T, Kita A, Koutoku H, Sasamata M, Hatashita E, Yamada Y, Kuwata K, Fukuoka M, Nakagawa K. Marked anti-tumour activity of the combination of YM155, a novel survivin suppressant, and platinum-based drugs. British journal of cancer. 2010;103(1):36–42. doi: 10.1038/sj.bjc.6605713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akamatsu Y, Murayama Y, Yamada T, Nakazaki T, Tsutsui Y, Ohta K, Iwasaki H. Molecular characterization of the role of the Schizosaccharomyces pombe nip1+/ctp1+ gene in DNA double-strand break repair in association with the Mre11-Rad50-Nbs1 complex. Molecular and cellular biology. 2008;28(11):3639–3651. doi: 10.1128/MCB.01828-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skoufias DA, Lacroix FB, Andreassen PR, Wilson L, Margolis RL. Inhibition of DNA decatenation, but not DNA damage, arrests cells at metaphase. Molecular cell. 2004;15(6):977–990. doi: 10.1016/j.molcel.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 33.Wang L, Eastmond DA. Catalytic inhibitors of topoisomerase II are DNA-damaging agents: induction of chromosomal damage by merbarone and ICRF-187. Environmental and molecular mutagenesis. 2002;39(4):348–356. doi: 10.1002/em.10072. [DOI] [PubMed] [Google Scholar]
- 34.Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246(4930):629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 35.Ali Y, Abd Hamid S. Human topoisomerase II alpha as a prognostic biomarker in cancer chemotherapy. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015 doi: 10.1007/s13277-015-4270-9. [DOI] [PubMed] [Google Scholar]
- 36.Mehta A, Zhang L, Boufraqech M, Liu-Chittenden Y, Zhang Y, Patel D, Davis S, Rosenberg A, Ylaya K, Aufforth R, Li Z, Shen M, Kebebew E. Inhibition of Survivin with YM155 Induces Durable Tumor Response in Anaplastic Thyroid Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21(18):4123–4132. doi: 10.1158/1078-0432.CCR-14-3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren M, Qin H, Ren R, Cowell JK. Ponatinib suppresses the development of myeloid and lymphoid malignancies associated with FGFR1 abnormalities. Leukemia. 2013;27(1):32–40. doi: 10.1038/leu.2012.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Darzynkiewicz Z, Juan G. Analysis of DNA content and BrdU incorporation. In: Paul Robinson J managing editor, et al., editors. Current protocols in cytometry / editorial board. Unit 7. Chapter 7. 2001. p. 7. [DOI] [PubMed] [Google Scholar]
- 39.Wilson WD, Wang YH, Kusuma S, Chandrasekaran S, Boykin DW. The effect of intercalator structure on binding strength and base-pair specificity in DNA interactions. Biophysical chemistry. 1986;24(2):101–109. doi: 10.1016/0301-4622(86)80003-5. [DOI] [PubMed] [Google Scholar]
- 40.Secco F, Venturini M, Biver T, Sanchez F, Prado-Gotor R, Grueso E. Solvent effects on the kinetics of the interaction of 1-pyrenecarboxaldehyde with calf thymus DNA. The journal of physical chemistry B. 2010;114(13):4686–4691. doi: 10.1021/jp910411c. [DOI] [PubMed] [Google Scholar]
- 41.Varvaresou A, Iakovou K. Molecular modeling study of intercalation complexes of tricyclic carboxamides with d(CCGGCGCCGG)(2) and d(CGCGAATTCGCG)(2) Journal of molecular modeling. 2011;17(8):2041–2050. doi: 10.1007/s00894-010-0891-5. [DOI] [PubMed] [Google Scholar]
- 42.Dexheimer TS, Pommier Y. DNA cleavage assay for the identification of topoisomerase I inhibitors. Nature protocols. 2008;3(11):1736–1750. doi: 10.1038/nprot.2008.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bower JJ, Karaca GF, Zhou Y, Simpson DA, Cordeiro-Stone M, Kaufmann WK. Topoisomerase IIalpha maintains genomic stability through decatenation G(2) checkpoint signaling. Oncogene. 2010;29(34):4787–4799. doi: 10.1038/onc.2010.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phillips JW, Goetz MA, Smith SK, Zink DL, Polishook J, Onishi R, Salowe S, Wiltsie J, Allocco J, Sigmund J, Dorso K, Lee S, Skwish S, de la Cruz M, Martin J, Vicente F, et al. Discovery of kibdelomycin, a potent new class of bacterial type II topoisomerase inhibitor by chemical-genetic profiling in Staphylococcus aureus. Chemistry & biology. 2011;18(8):955–965. doi: 10.1016/j.chembiol.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 45.Silva J, Sharma S, Cowell JK. Homozygous Deletion of the LGI1 Gene in Mice Leads to Developmental Abnormalities Resulting in Cortical Dysplasia. Brain pathology. 2015;25(5):587–597. doi: 10.1111/bpa.12225. [DOI] [PMC free article] [PubMed] [Google Scholar]