Abstract
Background
Rotavirus (RV) is the leading cause of diarrhea-related death in children worldwide and 95% of RV-associated deaths occur in Africa and Asia where RV vaccines (RVVs) have lower efficacy. We hypothesize that differences in intestinal microbiome composition correlate with the decreased RVV efficacy observed in poor settings.
Methods
We conducted a nested, case-control study comparing prevaccination, fecal microbiome compositions between 6-week old, matched RVV responders and nonresponders in rural Ghana. These infants' microbiomes were then compared with 154 age-matched, healthy Dutch infants' microbiomes, assumed to be RVV responders. Fecal microbiome analysis was performed in all groups using the Human Intestinal Tract Chip.
Results
We analyzed findings in 78 Ghanaian infants, including 39 RVV responder and nonresponder pairs. The overall microbiome composition was significantly different between RVV responders and nonresponders (FDR, 0.12), and Ghanaian responders were more similar to Dutch infants than nonresponders (P = .002). RVV response correlated with an increased abundance of Streptococcus bovis and a decreased abundance of the Bacteroidetes phylum in comparisons between both Ghanaian RVV responders and nonresponders (FDR, 0.008 vs 0.003) and Dutch infants and Ghanaian nonresponders (FDR, 0.002 vs 0.009).
Conclusions
The intestinal microbiome composition correlates significantly with RVV immunogenicity and may contribute to the diminished RVV immunogenicity observed in developing countries.
Keywords: rotavirus vaccine, intestinal microbiome, mucosal immunity
(See the editorial commentary by Iturriza-Gómara and Cunliffe on pages 8–10.)
Rotavirus (RV) is the leading cause of diarrhea-related death in children worldwide, with 95% of RV deaths occurring in low-income countries in Africa and Asia [1]. Oral RV vaccines (RVVs) have the potential to dramatically reduce the morbidity and mortality caused by RV infection, but RVVs demonstrate significantly lower efficacy in low-income countries [2]. Large clinical efficacy studies showed a combined vaccine efficacy against severe RV gastroenteritis ranging from 48% to 64% for both Rotarix and RotaTeq vaccines in Africa and Asia [3–5]. Emerging effectiveness data in Africa provides similar estimates of RVV protection [6]. This compares to an observed efficacy of 85%–98% against severe RV in trials in wealthier countries in Latin America and Europe [7–10].
Understanding the pathophysiologic mechanism driving this diminished efficacy in developing countries is critical, because even small improvements in vaccine efficacy could increase the number of children's lives saved by the vaccine by hundreds of thousands over the next 15 years [11]. There are several hypotheses as to why oral RVVs are underperforming in Africa and Asia [12]. These include interference with the first dose of coadministered oral poliovirus vaccine, RVV immune response suppression through high prevaccination levels of serum immunoblogulin (Ig) G, including transplacentally derived IgG, high levels of breast milk–derived RV-specific IgA, and HLA blood group antigen type [13–16]. However, none of these explanations have adequately and sufficiently explained the underperformance of RVV in developing countries, where vaccine efficacy can dip even lower than 50% in some settings. One underexplored hypothesis is that the intestinal microbiome may be modulating an infant's immune response to the enteric RVV [17]. We hypothesized that the composition of the intestinal microbiome is correlated with RVV response, that RVV responders have different intestinal microbes as compared with nonresponders and that these dissimilarities may contribute to the decreased efficacy of RVV found in resource-poor settings. To test these hypotheses, we conducted a nested, case-control study in Navrongo, Ghana, comparing the differences in intestinal microbiome composition and diversity between RVV seroconverters and nonseroconverters after vaccination with the Rotarix vaccine. We then compared these infants' microbiomes with those from a large group of age-matched healthy infant from the Netherlands, where RVV response is postulated to be high, similar to responses observed in other Northern European countries [10, 18].
METHODS
Study Design and Participants
Ghanaian Infants
The original trial within which this trial was nested was conducted in Navrongo, a rural setting in Northern Ghana where >70% of the population belong to the lowest wealth quintile in Ghana, in 2012. The neonatal and mortality rates are 24 and 46 deaths per 1000 live births, respectively [19].
All participating infants were healthy infants with a birth weight > 2000 g and/or a gestational age >38 weeks. The infants were enrolled at 6 weeks of age in a previously reported phase IV randomized clinical trial conducted in 2012 in Navrongo to evaluate the immunogenicity of the Rotarix vaccine after different dosing schedules (at age 6 and 10 weeks, 10 and 14 weeks, or 6, 10, and 14 weeks) (NCT01575197, clinicaltrials.gov) [20]. In this trial, all infants received concomitant standard Expanded Program on Immunization vaccinations, including the trivalent oral poliovirus vaccine and pentavalent vaccine (diphtheria, tetanus, whole-cell pertussis, hepatitis B, and Haemophilus influenza type).
Only infants from the 6- and 10-week and the 6-, 10-, and 14-week dose arms of the clinical trial were included in this microbiome study. A serum samples was collected before the receipt of the first dose of vaccine (at 6 weeks of age) and serum was collected again approximately 4 weeks after the last dose of vaccine (at age 14 or 18 weeks, depending on the study arm) for anti-RV IgA antibody measurements. Fecal samples were collected immediately before vaccination, at 6 weeks of age.
Infants were included in the study if during the original study their guardians had consented to additional testing of specimens in RV-vaccine related studies. Inclusion criteria further mandated that a baseline fecal sample was available and that there was no evidence of natural RV infection before vaccination (prevaccination IgA level ≥20 IU/mL). An IgA level ≥20 IU/mL was considered an indication of seroconversion and a surrogate marker for RVV protection against severe RV gastroenteritis [21].
Participating infants were then grouped as either RVV responders (postvaccination anti-RV IgA antibody ≥20 IU/mL) or RVV nonresponders (postvaccination anti-RV IgA antibody <20 IU/mL) and matched by hand in a 1:1 ratio using the following ranked variables: number and timing of doses of vaccine received (at age 6 and 10 weeks or 6, 10, and 14 weeks), sex, age at vaccination, RV season (defined as the date of the serum IgA level obtained 28 days after the last vaccination, between 1 December 2012 and 1 March 2013), ethnicity, height and weight at enrollment (including underweight, stunting, and wasting Z scores), and whether the infant regurgitated the vaccine after administration. The exact mode of delivery data and breastfeeding practices data are not known for the original Ghanaian study population. However, all infants in Ghana were delivered in the study hospital where >95% of deliveries are vaginal, and almost all infants delivered vaginally are breastfed. The nested trials were approved by the institutional review board of the Noguchi Memorial Institute for Medical Research and the research was conducted in accordance with good clinical practice guidelines.
Dutch Infants
In parallel, we compared the microbiome of both the Ghanaian responder and nonresponder infants with those in a cohort of healthy, age-matched Dutch control infants. The Dutch infants had not received RVV, but were assumed to be RVV responders, in line with ample clinical trial data demonstrating a >90% RVV seroconversion rate in Northern European countries [7, 10]. Fecal samples for these control infants were collected at approximately 30 days of age as part of a previously published study (the Bibo study) in which mothers and their children were followed up beginning with the third trimester of pregnancy [22, 23]. Pregnant women were recruited through midwife practices in Nijmegen and surrounding areas in the Netherlands. All parents provided written informed consent for participation in the study. The study was approved by the ethical committee of the faculty of social sciences at the Radboud University in Nijmegen. The infants' microbiomes were analyzed in the study using an identical protocol based on the Human Intestinal Tract Chip (HITChip) phylogenetic microarray [22]. Consent had been given for further use of the samples, which were entirely anonymous in our analysis.
Laboratory Evaluations
Assays
Anti-RV IgA antibody was measured in serum using an enzyme-linked immunosorbent assay described elsewhere, with values expressed in international units per milliliter [24, 25].
Fecal Microbiome and Enteric Pathogen Analysis
In Ghana, fecal samples were collected by community health workers in infants' homes, transported in a cool box, and frozen to −20°C within 24–48 hours of collection. All samples were stored in 3% glycerol in frost-free freezers. Routine temperature monitoring did not indicate any freeze-thaw cycles. In the Netherlands, fecal samples were collected by parents at home and immediately stored at −20°C. The samples were transported in coolers with freezing cartridges or dry ice for further storage at −80°C. These procedures are both considered adequate for fecal microbiome analysis [26].
The fecal samples were analyzed by means of HITChip microarrays in duplicate, as described elsewhere [27]. In brief, total DNA was extracted from the fecal material by a repeated bead beating procedure using a modified protocol for the QiaAmp DNA MiniStool Kit (Qiagen), also as described elsewhere [28]. The 16S ribosomal RNA gene was amplified using primers that enabled incorporation of T7 promoter sequence at the 5-terminus of the amplicon. RNA was transcribed with amino-allyl modified nucleotides that were later coupled to cyanine (Cy) 3 or Cy5 dyes. Labeled RNA was fragmented, and 2 samples, each carrying a different dye, were hybridized in duplicate to the HITChip microarrays.
The HITChip microarray is a comprehensive and highly reproducible phylogenetic microarray that enables the parallel profiling and the semiquantitative analysis of >1100 phylotypes representing all major intestinal phyla grouped in 130 genuslike groups described for the human intestinal microbiome [27]. This high-throughput technique has been benchmarked with ultradeep pyrosequencing of 16S ribosomal RNA amplicons and next-generation parallel sequencing of intestinal metagenomes [29, 30]. After microarray hybridization, a sample was accepted only if 2 independent hybridizations (labeled with Cy5 and Cy3) correlated significantly (>95% Pearson correlation). This highly reproducible microbiome analysis allowed for direct comparison of all samples described in this study.
Statistical Analysis
A χ2 test was used to determine statistically significant differences in baseline characteristics between Ghanaian RVV responders and nonresponders. The Shannon diversity index was used to measure the diversity of the microbiome per sample, including richness and evenness, using the hybridization signal of all probes included in the HITChip microarray [31]. Paired 2-tailed Student t tests were used to evaluate statistical significance.
Comprehensive multivariate statistical analyses were performed using Canoco 5.0 software for Windows [32]. Genus-level principal coordinate analysis and redundancy analyses were performed to evaluate differences in the overall microbial composition between the Ghanaian study groups (Ghanaian RVV responders and nonresponders). The 130 genuslike bacterial groups targeted by the HITChip microarray were used as biological variables, and the matching variables named above were used as background variables and visualized by means of inverse distance-weighted interpolation of the variable values over the component space. Monte Carlo permutation testing was used to assess the significance of the effect of these variables in the data set.
Generalized linear models were used to identify individual bacterial phyla and genuslike groups (class for Firmicutes) associated with RV seroconversion, as differing significantly either between Ghanaian nonresponders and responders or between the Ghanaian nonresponders and Dutch infants. P values corrected for the false discovery rate (FDR) were used to correct for multiple testing [33]. Bacterial taxa, whose log-transformed relative abundance was differed significantly (FDR-corrected P < .10) between the Ghanaian nonresponders and responders or between the Ghanaian nonresponders and Dutch infants, were considered correlated with RVV immunogenicity. These statistical analyses were performed with the program R [34]. To evaluate the similarity between Ghanaian infants (nonresponders and responders) and Dutch infants, we generated Pearson correlation scores and then performed an analysis of variance between Ghanaian nonresponders and responders.
RESULTS
Ghanaian Infants
A total of 234 infants (of which 74 were RVV responders) with prevaccination fecal samples were available from the original Ghana clinical trials and eligible for this study. A total of 52 RV responders were successfully matched to 52 nonresponders in a 1:1 ratio based on the predefined matching criteria. Of those, a total of 78 samples (equaling 39 per-protocol matched pairs) had sufficient DNA of good enough quality to be successfully analyzed using the HITChip pipeline.
No significant differences between the vaccine responders and nonresponders in Ghana were identified for any of the measured variables and baseline characteristics as described in Table 1. The diversity index (Shannon index) of the intestinal microbiome did not differ between infants who were RVV responders and those who were nonresponders (mean [SD], 4.40 [0.24] vs 4.41 [0.25], respectively; P = .87).
Table 1.
Baseline Characteristics and Matching Characteristics of the 78 Infants Enrolled in the Nested Study in Ghana and Differences Between RVV Nonresponders and Responders as Determined With χ2 Tests
| Infants, No.
(%)a |
|||
|---|---|---|---|
| Characteristic | RVV Nonresponders | RVV Responders | P Value |
| Total | 39/78 (50) | 39/78 (50) | … |
| Sex | >.99 | ||
| Male | 14/39 (36) | 14/39 (36) | |
| Female | 25/39 (64) | 25/39 (64) | |
| Ethnicity | >.99 | ||
| Nankam | 15/39 (38) | 15/39 (38) | |
| Kassem | 24/39 (62) | 24/39 (62) | |
| Age at vaccination mean (SD), d | 42 (0.47) | 43 (1.44) | >.99 |
| Vaccination schedule | >.99 | ||
| Arm 2: age 6 and 10 wk | 12/39 (31) | 12/39 (31) | |
| Arm 3: age 6, 10, and 14 wk | 27/39 (69) | 27/39 (69) | |
| RV season | .36 | ||
| Yes | 31/39 (79) | 34/39 (87) | |
| No | 8/39 (21) | 5/39 (13) | |
| Malnutrition | .95 | ||
| Underweight | 1/39 (3) | 0/39 (0) | .31 |
| Stunting | 2/39 (5) | 3/39 (8) | .64 |
| Wasting | 1/39 (3) | 1/39 (3) | >.99 |
| Wasting | 1/39 (3) | 1/39 (3) | >.99 |
Abbreviations: RV, rotavirus; RVV, RV vaccine.
a Data represent No. (%) of infants unless otherwise specified.
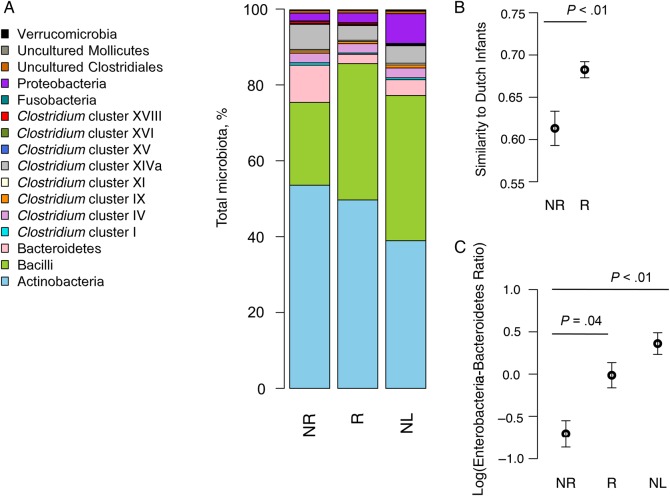
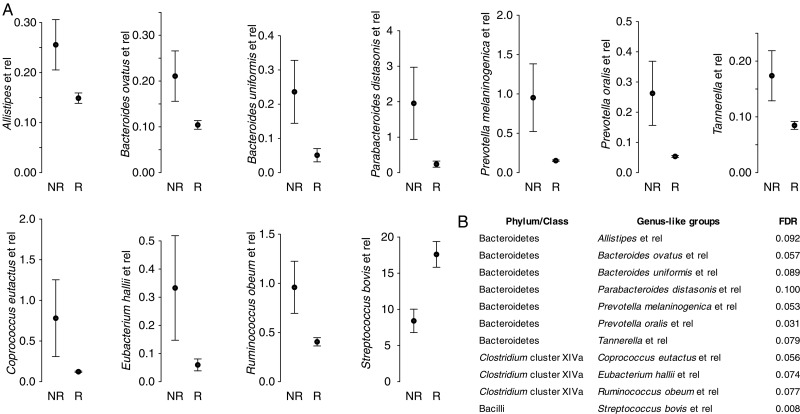
In Ghana, a high abundance of the Bacteroidetes phylum (FDR, 0.003), particularly several bacteria related to Bacteroides and Prevotella species, were significantly correlated with a lack of RVV response (Figures 1 and 2). Conversely, the Bacilli phylum (FDR, 0.027) correlated significantly with RVV response, specifically bacteria related to Streptococcus bovis (FDR = 0.008) (Figures 1 and 2).
Figure 1.
Overall microbiome composition. A, Bar plot of the percentage of each phylum in the total microbiome composition for the Ghanaian rotavirus vaccine (RVV) nonresponders (NR) and responders (R) and Dutch infants (NL). B, Ghanaian RVV responders are significantly more similar to Dutch infants than nonresponders. Y-axis shows mean similarity score as calculated by means of Pearson correlation. C, Ghanaian RVV responders and Dutch infants have a significantly higher enterobacteria-Bacteroidetes ratio than Ghanaian nonresponders. Y-axis shows the log-transformed ratio of all species of enterobacteria to all species of Bacteroidetes.
Figure 2.
A, Comparison of the relative abundance (median signal intensity) of all bacterial genuslike groups with a statistically significant (false discovery rate [FDR] <0.1) difference in abundance between Ghanaian rotavirus vaccine (RVV) responders (R) and nonresponders (NR). Plots show means with standard errors. B, The FDR for each of the significantly different genera.
We further evaluated the high abundance of bacteria in the Bacteroidetes phylum in nonresponder infants. Recent literature has suggested that many bacteria in the Bacteroidetes phylum have a less immunogenic lipopolysaccharide (LPS) than Enterobacteriacae, such as Escherichia coli [35]. We therefore calculated an enterobacteria-Bacteroidetes ratio to provide a rough estimate of the presence of toxigenic LPS in the microbiome and compared the ratios between all groups. The enterobacteria-Bacteroidetes ratio was significantly higher in the Ghanaian responder infants (P = .04) than in nonresponder infants (Figure 1C).
Spearman correlation analyses were performed to assess whether the actual titer of the IgA—as opposed to RV seroconversion as a binary variable (≥20 IU/mL)—was correlated with specific bacterial groups. The Spearman correlation results showed that 33 genuslike bacterial groups were significantly correlated with RVV response (defined as an FDR <0.200). As with the results obtained when using IgA as a binary variable, bacteria related to Streptococcus bovis (FDR, 0.070) were the only bacteria whose abundance were correlated significantly (FDR, <0.1) with increased RVV titers. All other bacterial groups were significantly associated with lower RVV titers, and findings in only 1 bacterial group (bacteria related to Xanthomonadaceae) differed from those of the statistical analysis using IgA as a binary variable. Therefore, the Spearman correlation analysis using absolute IgA titers both mirrored and confirmed the findings attained when evaluating IgA as a binary variable with 20 IU/mL as a cutoff point. (Supplementary Figure 1).
Finally, the genuslike principal coordinate analysis indicated that the main variable differentiating the microbiome composition between the Ghanaian responders and nonresponders was RVV seroconversion, as calculated with Monte Carlo permutation testing (P = .01; FDR 0.12). (Supplementary Figure 2). None of the other environmental variables that were tested correlated with significant microbiome differences.
Ghanaian Infants Compared With Dutch Infants
The Ghanaian infant microbiome compositions were then compared with the fecal microbiomes of 154 healthy Dutch infants. Of these Dutch infants, 62% were still receiving breast milk at 4 weeks of age, 42% were female, and 93% had been delivered vaginally; their mean (SD) birth weight was 3619 (457) g. When the Dutch infants were compared with the Ghanaian infants, their overall microbiome composition was significantly more similar to that in the Ghanaian RVV responders than to that in nonresponders (P = .002; Figure 1B).
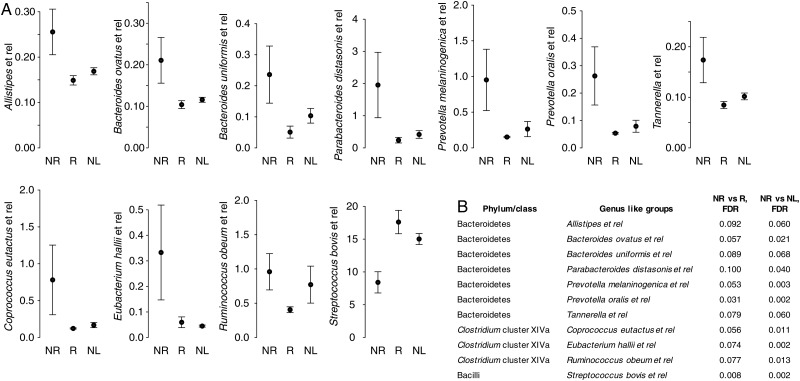
We then evaluated which phyla and bacterial genus groups differed significantly between Dutch infants and Ghanaian RVV nonresponders (see Supplementary Table 1) and subsequently assessed which of these bacterial groups also differed significantly in the comparison between Ghanaian RVV responders and nonresponders (Figure 3). At the phylum level, bacilli were significantly more abundant in both Ghanaian RVV responders and Dutch infants than in nonresponder infants (FDR for Dutch vs Ghanaian RVV nonresponders, 0.0002 ). The Bacteroidetes phylum was also significantly more abundant in Ghanaian nonresponders than both the Ghanaian responders and the Dutch infants (FDR, 0.006).
Figure 3.
A, Comparison of the relative abundance (median signal intensity) of all bacterial genuslike groups with a statistically significant (false discovery rate [FDR], < 0.1) difference in abundance between both (1) Ghanaian rotavirus vaccine (RVV) responders (R) and nonresponders (NR) and (2) Dutch infants (NL) and Ghanaian RVV nonresponders (NR). Plots show means with standard errors. B, FDRs for the significantly different genera.
At the genus level, all bacterial groups that differed significantly between Ghanaian nonresponders and responders (FDR, <0.1) also differed significantly between Ghanaian nonresponders and Dutch infants. Bacteria related to Streptococcus bovis were more abundant in both Dutch infants and Ghanaian responders than in the Ghanaian infants without an RVV immune response (FDR, 0.0018). In parallel, several bacteria from the Bacteroidetes phylum (FDR, 0.002), particularly the Bacteroides and Prevotella genera, were more abundant in the Ghanaian nonresponders than in the Dutch infants (all Figure 3). Finally, the enterobacteria-Bacteroidetes ratio was also significantly higher (P < .01) in Dutch infants than in Ghanaian infants without an RVV immune response (Figure 1C).
DISCUSSION
This study of Ghanaian infants demonstrates that the prevaccination intestinal microbiome differs significantly—on a genuslike, phylum, and overall composition level—between RVV responders and nonresponders in a rural, low-income setting in sub-Saharan Africa and that these microbiome differences are robustly recapitulated when Dutch infants are compared with RVV nonresponders.
RVV response was positively associated with the Bacilli phylum, specifically bacteria related to Streptococcus bovis. Several bacterial groups were significantly associated with a lack of RVV response—namely, bacteria belonging to the Bacteroidetes phlyum, especially several bacteria related to species from the Bacteroides and Prevotella genera.
The high abundance of Bacteroidetes in the microbiome of infants not responding to RVV is intriguing. Certain species from that phylum have been shown to express a form of LPS that is functionally and structurally different from the LPS expressed in some proteobacteria, such as E. coli [35, 36]. LPS is present in the outer membrane of most gram-negative bacteria and is a strong immunogenic stimulator of the innate immune system [37]. LPS structure varies from species to species, and variation—particularly in the lipid A core of LPS—can influence LPS' immunostimulatory capacity [38]. LPS derived from several Bacteroidetes species has recently been shown to have an impaired or even immune-inhibitory capacity to stimulate inflammatory cytokines in vitro when compared with LPS derived from E. coli [35].
Although our study could not measure infants' microbiome expression of a less toxigenic LPS, we chose to roughly evaluate this phenomenon by comparing the ratio of all enterobacteria to Bacteroidetes in each infant group. We did not compare specific enterobacteria or Bacteroidetes species because of some cross-hybridization at the species level in the HITChip microarray. This enterobacteria-Bacteroidetes ratio was significantly increased in both the Ghanaian RVV responders and Dutch infants compared with the nonresponders, raising the possibility that early microbiome colonization with bacteria expressing a less toxigenic or perhaps inhibitory form of LPS might be down-regulating innate immune responses to the live attenuated RV contained in the vaccine. Alternatively, more toxic LPS might be having an adjuvant effect on RVV responses in those Ghanaian infants capable of eliciting an immunogenic response to RVV.
Interestingly, in the Ghanaian responders, bacteria related to Streptococcus bovis was significantly correlated with RV response. The Streptococcus bovis group belongs to the S. bovis–Streptococcus equinus complex, which, like bacteria with toxigenic LPS, such as the E. coli, Serratia, and Klebsiella groups, can cross from being commensals to being opportunistic pathogens and whose cell surface proteins can trigger inflammatory responses [39]. Although speculative, these S. bovis–S. equinus complex bacteria could also be priming the immune system or acting as natural vaccine adjuvants.
An alternative hypothesis, given that the vaccines contain live attenuated virus, is that these bacteria might be expressing blood group antigens or glycans needed for RV replication, as has recently been demonstrated with norovirus and RV [40, 41]. Unfortunately, FUT2 secretor status, which is epidemiologically associated with RV gastroenteritis, is unknown among these infants.
This study has some limitations, restricting the strength of our findings. One of the most important is that, for the cohort of Dutch infants, we do not have RVV immunogenicity data and are predicting RVV response from randomized control studies showing >90% RVV efficacy in Europe [7, 10]. Because these were all healthy, at-term infants, we do predict that they would mount immune responses to RVV but are unable to demonstrate this. In addition, the study lacks specific data on breastfeeding and delivery practices for the infants in Ghana, and these may be confounders in our study results. Nevertheless, because >95% of deliveries are vaginal and all infants delivered vaginally are breastfed in this cohort, we do not expect mode of delivery and breastfeeding practices to be significant confounders in this study population. We also were not able to match our infants in Ghana for maternal antibodies to RV, such as breast milk anti-RV IgA and transplacentally acquired anti-G1 RV IgG antibody. High levels of maternally derived antibodies have been shown to diminish RVV immunogenicity, which could be contributing to the lack of response observed in our cohort and confounding the results [14, 15].
As another limitation, we used anti-RV IgA response as a correlate of vaccine protection, which can be an imperfect surrogate for vaccine efficacy [21]. Clinical vaccine efficacy would require larger sample sizes and follow-up. In addition, the intestinal microbiome is a complex ecosystem, and bacterial populations are in constant interplay with other intestinal inhabitants and pathogens, such as archae and eukaryotic microbes. This analysis of the microbiome did not evaluate the intestinal virome, fungiome, or parasites, and these could be immunologic mediators and influence bacterial populations. Finally, the associations between microbiome and RVV response are correlative and not causative and taken at a single time-point—directly before vaccination in a very young infants with high variability in their microbiome population. Nevertheless, our study used an identical, standardized, and reproducible technique to measure the intestinal microbiome in 2 geographic locations, and matching for numerous demographic variables in Ghana helps minimize possible study cofounders. As a consequence, our work is a springboard to understanding the mechanistic relationships between the intestinal microbiome and oral RVV responses in vulnerable infant populations and a call for more research to inform the design of evidence-based interventions to improve RVV efficacy worldwide.
Supplementary Data
Supplementary materials are available at http://jid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Supplementary Material
Notes
Acknowledgments. This study is dedicated to Professor Joseph (Joep) Lange, who died before its completion. It would not have been possible without his initiative, support, and inspiration. We acknowledge and thank all the families who participated in this study and the staff members of the RVV trial team in Ghana for their work in conducting this study; Monica McNeal and her staff at the Laboratory of Specialized Clinical Studies, Cincinnati Children's Hospital, who performed all the immunoglobulin testing for the original dosing studies; Duncan Steele for his contribution to the original dosing study in Ghana; and Lauren Gazley for her contribution to the original dosing study in Ghana and data management support for this research in Ghana.
Disclaimer. The findings and conclusions in this report are those of the authors and do not necessarily represent the official positions of policies of PATH, the Bill & Melinda Gates Foundation, the US Centers for Disease Control and Prevention, or the Netherlands Organization for Scientific Research.
Financial support. This publication is based on research funded in part by PATH through the Bill & Melinda Gates Foundation (grant OPP 1017334 to PATH) and the Netherlands Organization for Scientific Research (Spinoza grant 2008 to W. M. d. V.).
Potential conflict of interest. C. G. has received honoraria for participating in advisory boards and speaking at international conferences from GlaxoSmithKline, Merck, and Sanofi PasteurMSD. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Tate JE, Burton AH, Boschi-Pinto C et al. . 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect Dis 2012; 12:136–41. [DOI] [PubMed] [Google Scholar]
- 2. Fischer Walker CL, Black RE. Rotavirus vaccine and diarrhea mortality: quantifying regional variation in effect size. BMC Public Health 2011; 11(Suppl 3):S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Armah GE, Sow SO, Breiman RF et al. . Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in sub-Saharan Africa: a randomised, double-blind, placebo-controlled trial. Lancet 2010; 376:606–14. [DOI] [PubMed] [Google Scholar]
- 4. Madhi SA, Cunliffe NA, Steele D et al. . Effect of human rotavirus vaccine on severe diarrhea in African infants. N Engl J Med 2010; 362:289–98. [DOI] [PubMed] [Google Scholar]
- 5. Zaman K, Dang DA, Victor JC et al. . Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in Asia: a randomised, double-blind, placebo-controlled trial. Lancet 2010; 376:615–23. [DOI] [PubMed] [Google Scholar]
- 6. Baar-Zeev N, Kapanda L, Tate JE et al. . Effectiveness of a monovalent rotavirus vaccine in infants in Malawi after programmatic roll-out: an observational and case-control study. Lancet Infect Dis 2015; 15:422–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ruiz-Palacios GM, Pérez-Schael I, Velázquez FR et al. . Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N Engl J Med 2006; 354:11–22. [DOI] [PubMed] [Google Scholar]
- 8. Linhares AC, Velázquez FR, Pérez-Schael I et al. . Efficacy and safety of an oral live attenuated human rotavirus vaccine against rotavirus gastroenteritis during the first 2 years of life in Latin American infants: a randomised, double-blind, placebo-controlled phase III study. Lancet 2008; 371:1181–9. [DOI] [PubMed] [Google Scholar]
- 9. Vesikari T, Karvonen A, Prymula R et al. . Efficacy of human rotavirus vaccine against rotavirus gastroenteritis during the first 2 years of life in European infants: randomised, double-blind controlled study. Lancet 2007; 370:1757–63. [DOI] [PubMed] [Google Scholar]
- 10. Vesikari T, Matson DO, Dennehy P et al. . Safety and efficacy of a pentavalent human-bovine (WC3) reassortant rotavirus vaccine. N Engl J Med 2006; 354:23–33. [DOI] [PubMed] [Google Scholar]
- 11. Atherly D, Dreibelbis R, Parashar UD, Levin C, Wecker J, Rheingans RD. Rotavirus vaccination: cost-effectiveness and impact on child mortality in developing countries. J Infect Dis 2009; 200(suppl 1):S28–38. [DOI] [PubMed] [Google Scholar]
- 12. Clarke E, Desselberger U. Correlates of protection against human rotavirus disease and the factors influencing protection in low-income settings. Mucosal Immunology 2015; 8:1–17. [DOI] [PubMed] [Google Scholar]
- 13. Emperador DM, Velasquez DE, Estivariz CF et al. . Interference of monovalent, bivalent, and trivalent oral poliovirus vaccines on monovalent rotavirus vaccine immunogenicity in rural Bangladesh. Clin Infect Dis 2016; 62:150–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Moon S-S, Groome MJ, Velasquez DE et al. . Prevaccination rotavirus serum IgG and IgA are associated with lower immunogenicity of live, oral human rotavirus vaccine in South African infants. Clin Infect Dis 2016; 62:157–65. [DOI] [PubMed] [Google Scholar]
- 15. Chilengi R, Simuyandi M, Beach L et al. . Association of maternal immunity with rotavirus vaccine immunogenicity in Zambian infants. PLoS One 2016; 11:e0150100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nordgren J, Sharma S, Bucardo F et al. . Both Lewis and secretor status mediate susceptibility to rotavirus infections in a rotavirus genotype-dependent manner. Clin Infect Dis 2014; 59:1567–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Isolauri E, Joensuu J, Suomalainen H, Luomala M, Vesikari T. Improved immunogenicity of oral D x RRV reassortant rotavirus vaccine by Lactobacillus casei GG. Vaccine 1995; 13:310–2. [DOI] [PubMed] [Google Scholar]
- 18. Ruiz-Palacios GM, Guerrero ML, Bautista-Márquez A et al. . Dose response and efficacy of a live, attenuated human rotavirus vaccine in Mexican infants. Pediatrics 2007; 120:e253–61. [DOI] [PubMed] [Google Scholar]
- 19. Ghana Statistical Service, Ghana Health Service, and ICF International. Ghana demographic and health survey, 2014. Rockville, MD: GSS, GHS, and ICF International, 2015. Available at: https://dhsprogram.com/pubs/pdf/FR307/FR307.pdf. Accessed 20 June 2016.
- 20. Armah G, Lewis KDC, Cortese MM et al. . A randomized controlled trial of the impact of alternative dosing schedules on the immune response to human rotavirus vaccine in rural Ghanaian infants. J Infect Dis 2016; 213:1678–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Patel M, Glass RI, Jiang B, Santosham M, Lopman B, Parashar U. A systematic review of anti-rotavirus serum IgA antibody titer as a potential correlate of rotavirus vaccine efficacy. J Infect Dis 2013; 208:284–94. [DOI] [PubMed] [Google Scholar]
- 22. de Weerth C, Fuentes S, Puylaert P, De Vos WM. Intestinal microbiota of infants with colic: development and specific signatures. Pediatrics 2014; 131:e550–8. [DOI] [PubMed] [Google Scholar]
- 23. Zijlmans MAC, Korpela K, Riksen-Walraven JM, De Vos WM, de Weerth C. Maternal prenatal stress is associated with the infant intestinal microbiota. Psychoneuroendocrinology 2015; 53:233–45. [DOI] [PubMed] [Google Scholar]
- 24. Bernstein DI, Smith VE, Sherwood JR et al. . Safety and immunogenicity of live, attenuated human rotavirus vaccine 89-12. Vaccine 1997; 16:381–7. [DOI] [PubMed] [Google Scholar]
- 25. Ward RL, Bernstein DI, Shukla R et al. . Protection of adults rechallenged with a human rotavirus. J Infect Dis 1990; 161:440–5. [DOI] [PubMed] [Google Scholar]
- 26. Tedjo DI, Jonkers DMAE, Savelkoul PH et al. . The effect of sampling and storage on the fecal microbiota composition in healthy and diseased subjects. PLoS One 2015; 10:e0126685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rajilić-Stojanović M, Heilig HGHJ, Molenaar D et al. . Development and application of the Human Intestinal Tract Chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ Microbiol 2009; 11:1736–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Salonen A, Nikkilä J, Jalanka-Tuovinen J et al. . Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: Effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J Microbiol Methods 2010; 81:127–34. [DOI] [PubMed] [Google Scholar]
- 29. Claesson MJ, O'Sullivan O, Wang Q et al. . Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 2009; 4:e6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arumugam M, Raes J, Pelletier E et al. . Enterotypes of the human gut microbiome. Nature 2011; 473:174–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Haegeman B, Hamelin JEROM, Moriarty J, Neal P, Dushoff J, Weitz JS. Robust estimation of microbial diversity in theory and in practice. ISME J 2013; 7:1092–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Leps J, Smilauer P. Multivariate analysis of ecological data using CANOCO. Cambridge, United Kingdom: Cambridge University Press, 2003. [Google Scholar]
- 33. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing on JSTOR. J R Stat Soc Ser B 1995; 57:289–300. [Google Scholar]
- 34. The R Project for Statistical Computing. 2016. https://www.r-project.org . [Google Scholar]
- 35. Vatanen T, Kostic AD, d'Hennezel E et al. . Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 2016; 165:842–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hofstad T, Sveen K, Dahlén G. Chemical composition, serological reactivity and endotoxicity of lipopolysaccharides extracted in different ways from Bacteroides fragilis, Bacteroides melaninogenicus and Bacteroides oralis. Acta Pathol Microbiol Scand B 1977; 85:262–70. [DOI] [PubMed] [Google Scholar]
- 37. Alexander C, Rietschel ET. Bacterial lipopolysaccharides and innate immunity. J Endotoxin Res 2001; 7:167–202. [PubMed] [Google Scholar]
- 38. Needham BD, Carroll SM, Giles DK, Georgiou G, Whiteley M, Trent MS. Modulating the innate immune response by combinatorial engineering of endotoxin. PNAS 2013; 110:1464–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jans C, Meile L, Lacroix C, Stevens MJA. Genomics, evolution, and molecular epidemiology of the Streptococcus bovis/Streptococcus equinus complex (SBSEC). Infect Genet Evol 2015; 33:419–36. [DOI] [PubMed] [Google Scholar]
- 40. Jones MK, Watanabe M, Zhu S et al. . Enteric bacteria promote human and mouse norovirus infection of B cells. Science 2014; 346:755–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Payne DC, Currier RL, Staat MA et al. . Epidemiologic association between FUT2 secretor status and severe rotavirus gastroenteritis in children in the United States. JAMA Pediatr 2015; 169:1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.