Abstract
The strain Marseille-P2749T (= CSUR P2749 = DSM 103085) was isolated as part of culturomics study from a liquid duodenum sample from a French man. Bacterial cells were Gram-negative bacilli, fusiform shaped and non–spore forming, and they grew in microaerophilic and anaerobic atmosphere. Its genome is 1 809 169 bp long and contains 1646 protein-coding genes. The DNA G+C content was 27.33 mol%. This strain exhibited a 95.9% sequence similarity with Fusobacterium periodonticum, the phylogenetically closest species with standing in nomenclature. Strain Marseille-P2749T is suggested to be a novel species belonging to the genus Fusobacterium, for which the name Fusobacterium massiliense sp. nov. is proposed.
Keywords: Culturomics, Fusobacterium massiliense, gastrointestinal microbiome, genomics, taxonomy
Introduction
The human microbiome study is an ongoing revolution in health and disease understanding [1]. Metagenomics is currently regarded as the reference method to study the gut microbiota, but culturomics is a complementary approach allowing the cultivation of new species [2]. Indeed, advances in culture [3] and identification by amplification and sequencing of the 16S rRNA [4] combined with routine matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) analyses [5], have resulted in a dramatic increase of bacterial species identified by culture [6].
Most studies showing the relationship between human diseases and the digestive microbiota have been made from stool sample analysis. However, analysis of faeces does not appear to be the best method to examine the gastrointestinal microbiota: the bacterial composition and concentration are different between stools and the duodenum [7]. We thus compared the culturomes of the stomach, duodenum, ileum, and right and left colon from six patients undergoing upper endoscopy and colonoscopy for various indications. While doing so, we isolated the strain Marseille-P2749T from the duodenum of a patient with iron deficiency. This strain belongs to the genus Fusobacterium of the family Fusobacteriaceae.
The genus Fusobacterium included 20 species inhabiting the oral cavity and digestive tract of animals or humans [8]. Fusobacteria are Gram negative, anaerobic, nonmotile and nonsporulating [9]. Fusobacterium nucleatum is the type species of this genus and the most frequently isolated from humans [10], and Fusobacterium periodonticum was first isolated in 1983 from the human oral cavity [11]. However, our strain did not fulfil the usual criteria to be assigned to a previously known species of this genus.
The definition of a bacterial species is debatable [12]. The classic definition proposes to define a new species by DNA-DNA hybridization (DDH), a nonreproducible and expensive method that is possibly only in a few laboratories. Among others, we proposed since 2012 to systematically add the MALDI-TOF spectrum and the genome sequence to define a new species [13]. We called this approach taxonogenomics. Here the main characteristics of strain Marseille-P2749T (= CSUR P2749 = DSM 103085) are reported with a phylogenetic and phenotypic analysis, along with a description of the complete genome.
Materials and Methods
Sample information
Using the culturomics approach [14], as a part of the human microbiome study, strain Marseille-P2749T was first cultivated from the duodenum liquid sample of a 60-year-old man living in Marseille who underwent digestive endoscopy to diagnose iron-deficiency anaemia. A signed informed consent was obtained from the patient at the gastroenterology service of the Hôpital Nord (Marseille, France), and the ethics committee of the Institut Fédératif de Recherche IFR48 validated the study under number 2016-010.
Strain isolation and identification by MALDI-TOF MS and 16S rRNA sequencing
After collection in sterile conditions during an upper endoscopic examination, the sample was immediately preincubated in a blood culture bottle (BD BACTEC, Plus Anaerobic/F Media; Becton Dickinson, Le Pont de Claix, France) previously enriched with 5 mL of sheep’s blood, 0.2 μm filtered rumen and three antioxidants (uric acid 0.1 g/L, glutathione 0.1 g/L and ascorbate 1 g/L; Sigma-Aldrich, Saint-Quentin Fallavier, France). The initial growth of strain Marseille-P2749T was obtained in anaerobic conditions (AnaeroGen Compact; Oxoid, Thermo Scientific, Dardilly, France) after a 1-day preincubation and a 1-day incubation at 37°C of this liquid medium seeded on a 5% sheep’s blood–enriched Columbia agar (Columbia agar + 5% sheep’s blood; bioMérieux, Marcy l’Etoile, France).
Colonies obtained were then identified with our systematic MALDI-TOF MS screening on a Microflex spectrometer (Bruker Daltonics, Bremen, Germany) as previously described [15]. When a colony was unidentifiable, a 16S rRNA gene amplification was performed by standard PCR using fD1-rP2 universal primers (Eurogentec, Angers, France) as previously described [16]. The complete 16S rRNA gene was then sequenced using internal primers (536F, 536R, 800F, 800R, 1050F, 1050R) as previously described [17]. Sequences were finally corrected using the Codon Code Aligner software (http://www.codoncode.com) and then BLASTed (Basic Local Alignment Search Tool) against the GenBank nucleotide collection (nr/nt) or against GenBank 16S ribosomal RNA sequences (http://blast.ncbi.nlm.nih.gov/Blast.cgi). After the 16S rRNA gene sequence comparison, the strain is considered as a new species if the percentage of similarity is ≤98.7% [18].
Phylogenetic classification
A custom Python script was used to automatically retrieve all species from the same family of strain Marseille-P2749T and download 16S sequences from National Center for Biotechnology Information (NCBI) by parsing NCBI results and the NCBI taxonomy page. It only keeps sequences from type strains found on the List of Prokaryotic Names With Standing in Nomenclature (LPSN) website (http://www.bacterio.net/). The phylogenetic tree was constructed using MEGA6 (Molecular Evolutionary Genetics Analysis) software [19].
Phenotypic characteristics
Different growth conditions were tested on a 5% sheep’s blood–enriched Columbia agar (bioMérieux) for strain Marseille-P2749T. Five temperatures (room temperature, 28, 37, 45 and 55°C) and three atmospheres (anaerobic (AnaeroGen Compact; Oxoid, Thermo Scientific), microaerophilic (CampyGen Compact; Oxoid, Thermo Scientific) and aerobic in a plastic pouch to maintain a humid atmosphere) were realized. Tolerance of this strain to salt (10, 15 and 20%) and pH (pH 5, 5.5, 6, 6.5, 7, 7.5 and 8) was also managed under an anaerobic atmosphere. For the sporulation test, a heat shock at 80°C for 20 minutes was performed. Then Gram staining, motility and sporulation were observed using a light microscope (DM1000; Leica Microsystems, Nanterre, France).
Cell size was determined by transmission electron microscopy. Cells were fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer for 1 hour at 4°C. A drop of cell suspension was deposited for 5 minutes on glow-discharged formvar carbon film on 400 mesh nickel grids (FCF400-Ni; Electron Microscopy Sciences (EMS), Hatfield, PA, USA). The grids were dried on blotting paper, and the cells were negatively stained for 10 seconds with 1% ammonium molybdate solution in filtered water at room temperature. Electron micrographs were acquired with a Tecnai G20 Cryo transmission electron microscope (FEI Company, Limeil-Brevannes, France) operated at 200 keV.
Biochemical characterization
Catalase and oxidase tests (BD Catalase Reagent and BD Oxidase Reagent; Becton Dickinson) were performed. Moreover, biochemical traits and enzyme activities were analyzed using API Gallery systems (API 20A, API 50CH and API ZYM test strips; bioMérieux) according to the manufacturer’s instructions. A fresh culture of strain Marseille-P2749T was used to perform all tests.
Cellular fatty acid methyl ester (FAME) analysis was performed by gas chromatography/mass spectrometry (GC/MS) as previously described [20]. Two samples were prepared with 40 mg of bacterial biomass per tube collected from eight culture plates. GC/MS analyses were carried out as described before [21]. FAME were separated using an Elite 5-MS column and monitored by mass spectrometry (Clarus 500-SQ 8 S; PerkinElmer, Courtaboeuf, France). Spectral database search was performed using MS Search 2.0 operated with the Standard Reference Database 1A (National Institute of Standards and Technology, Gaithersburg, MD, USA) and the FAMEs mass spectral database (Wiley, Chichester, UK).
Antibiotic susceptibility was tested using a disk diffusion method [22] on Mueller-Hinton E agar (bioMérieux). Seventeen antibiotics were tested (i2a, Montpellier, France): vancomycin 30 μg, colistin 50 μg, gentamycin 15 μg, amoxicillin 25 μg, amoxicillin/clavulanic acid 30 μg, penicillin 10 U, clindamycin 15 μg, metronidazole 4 μg, trimethoprim/sulfamethoxazole 25 μg, oxacillin 5 μg, imipenem 10 μg, tobramycin 10 μg, fosfomycin 50 μg, ceftriaxone 30 μg, rifampicin 30 μg, doxycycline 30 μg and erythromycin 15 μg.
Genome sequencing
Genomic DNA (gDNA) of strain Marseille-P2749T was sequenced using MiSeq technology (Illumina, San Diego, CA, USA) with mate pair strategy. The gDNA was barcoded and mixed with 11 other projects with the Nextera Mate Pair sample prep kit (Illumina), then quantified by Qubit assay with the high-sensitivity kit (Life Technologies, Carlsbad, CA, USA) to 61.6 ng/μL. Using the Nextera Mate Pair Illumina guide, the Mate Pair library was prepared with 1.5 μg of fragmented and tagged genomic DNA sample, with a Mate Pair junction adapter. The pattern of fragmentation was validated on an Agilent 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA, USA) with a DNA 7500 labchip. The DNA fragments were ranged in size from 1.5 to 11 kb with an optimal size of 6.13 kb [23]. No size selection was performed, and 441.5 ng of tagmented fragments were circularized. The circularized DNA was mechanically sheared with Optima on a bimodal curve at 896 and 1663 bp on the Covaris device S2 in T6 tubes (Covaris, Woburn, MA, USA). The library profile was visualized on a High Sensitivity Bioanalyzer LabChip (Agilent Technologies), and the final concentration library was measured at 24.52 nmol/L. The libraries were normalized at 2 nM and pooled. After a denaturation step and dilution at 15 pM, the pool of libraries was loaded. Automated cluster generation and sequencing run were performed in a single 39-hour run with a 2 × 251 bp read length.
Total information of 7.2 Gb was obtained from a 765K/mm2 cluster density with a cluster passing quality control filters of 94.7% (14 162 000 passing filter paired reads). Within this run, the index representation for strain Marseille-P2749T was determined to be 7.18%. The 1 012 974 paired reads were trimmed then assembled into six scaffolds.
Genome annotation, assembly and comparison
Open reading frame (ORFs) were predicted using Prodigal (http://prodigal.ornl.gov/) with default parameters. However, the predicted ORFs were excluded if they spanned a sequencing gap region. The predicted bacterial protein sequences were searched against the GenBank [24] and Clusters of Orthologous Groups database (COGs) databases using BLASTP. The tRNAs and rRNAs were predicted using the tRNAScan-SE [25] and RNAmmer [26] tools respectively. Signal peptides and numbers of transmembrane helices were predicted using SignalP [27] and TMHMM [28] respectively. Mobile genetic elements were predicted using PHAST [29] and RAST [30]. ORFans were identified if their BLASTP E value was lower than 1e-03 for an alignment length greater than 80 amino acids. If alignment lengths were smaller than 80 amino acids, an E value of 1e-05 was used. Such parameter thresholds have already been used in previous works to define ORFans. Artemis [31] and DNA Plotter [32] were used for data management and visualization of genomic features respectively. The Mauve 2.3.1 alignment tool was used for multiple genomic sequence alignment [33].
The mean level of nucleotide sequence similarity at the genome level between strain Marseille-P2749T and other bacteria (Fusobacterium perfoetens ATCC 29250T JHXW00000000, Fusobacterium nucleatum ATCC 49256T CP003700, Fusobacterium varium ATCC 8501T ACIE00000000, Fusobacterium mortiferum ATCC 25557T ACDB00000000, Fusobacterium russii ATCC 25533T ARMK00000000 and Ilyobacter polytropus DSM 2926T CP002281) was estimated using average genomic identity of orthologous gene sequences (AGIOS) software [13]. Digital DDH, which exhibits high correlation with DDH [34], was also performed to evaluate genomic similarity with the closest strains (http://ggdc.dsmz.de/).
Results
Phylogenetic analysis
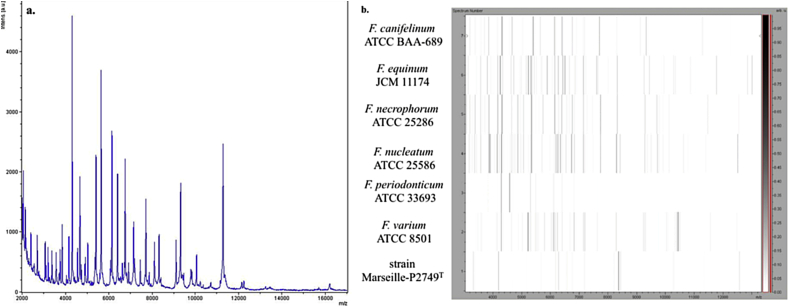
After a 1-day preincubation period at 37°C in a blood culture bottle with sheep’s blood, filtered rumen and antioxidants, and a 1-day anaerobic incubation period of cultivation of this enriched liquid medium seeded on Colombia agar with 5% sheep’s blood, the strain Marseille-P2749T was first isolated. MALDI-TOF MS was unable to provide reliable identification for colonies from this strain because its spectrum was not part of the database (Fig. 1(a)). The phylogenetic analysis, based on 16S rRNA, showed that strain Marseille-P2749T exhibited a 95.9% sequence similarity [35] with Fusobacterium periodonticum strain ATCC 33693T (GenBank accession no. X55405), the phylogenetically closest species with standing in nomenclature.
Fig. 1.
MALDI-TOF MS analysis of strain Marseille-P2749T. (a) Reference mass spectrum of strain Marseille-P2749T. (b) Gel view comparing strain Marseille-P2749T to other close species. Gel view displays raw spectra of loaded spectrum files arranged in pseudo-gel-like look. x-axis records m/z value. Left y-axis displays running spectrum number originating from subsequent spectra loading. Peak intensity is expressed by greyscale scheme code. Colour bar and right y-axis indicate relation between colour peak displayed and peak intensity in arbitrary units. Displayed species are indicated at left. MALDI-TOF MS, matrix-assisted desorption ionization–time of flight mass spectrometry.
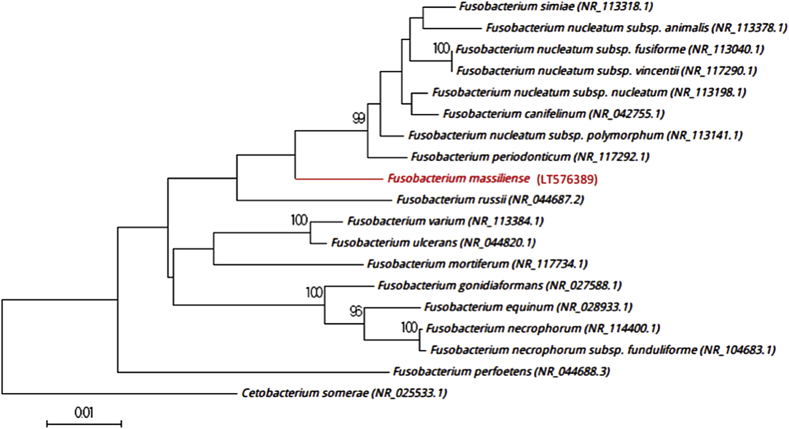
Because the 16S rRNA sequence divergence was >1.3% from its phylogenetically closest species with standing in nomenclature [36], we propose the creation of a new species which putatively classifies it as a member of the Fusobacterium genus within the Fusobacteriaceae family in the Fusobacteria phylum. The phylogenetic tree shows the position of strain Marseille-P2749T relative to other phylogenetically close neighbours, based on the 16S rRNA gene sequence (Fig. 2). The 16S rRNA sequence of strain Marseille-P2749T was deposited in European Molecular Biology Laboratory–European Bioinformatics Institute (EMBL-EBI) under accession number LT576389 (Table 1). To compare the spectrum of strain Marseille-P2749T and the spectra of other related bacteria, a gel view was performed (Fig. 1(b)).
Fig. 2.
Phylogenetic tree showing position of strain Marseille-P2749T relative to other phylogenetically close neighbours. Sequences were aligned using Muscle 3.8.31 with default parameters, and phylogenetic inferences were obtained using neighbour-joining method with 500 bootstrap replicates within MEGA6 software. Only bootstraps >95% are shown. Scale bar represents 1% nucleotide sequence divergence.
Table 1.
Percentage 16S rRNA similarities within Fusobacterium genus: 1) Strain Marseille-P2749T, 2) F. periodonticum ATCC 33693T, 3) F. nucleatum ATCC 25586T, 4) F. canifelinum ATCC BAA-689T, 5) F. simiae ATCC 33568T, 6) F. russii ATCC 25533T, 7) F. equinum JCM 11174T and 8) F. necrophorum ATCC 25286T
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
|---|---|---|---|---|---|---|---|---|
| 1 | 100 | 95.9 | 95.8 | 95.8 | 95.4 | 94.7 | 94.6 | 94.1 |
| 2 | 100 | 97.2 | 97.2 | 96.6 | 93.6 | 93.1 | 92.8 | |
| 3 | 100 | 99.4 | 98.6 | 93.9 | 92.9 | 92.6 | ||
| 4 | 100 | 98.1 | 93.7 | 92.7 | 92.5 | |||
| 5 | 100 | 93.7 | 92.4 | 92.1 | ||||
| 6 | 100 | 93.4 | 93.1 | |||||
| 7 | 100 | 98.2 | ||||||
| 8 | 100 |
Phenotypic analysis
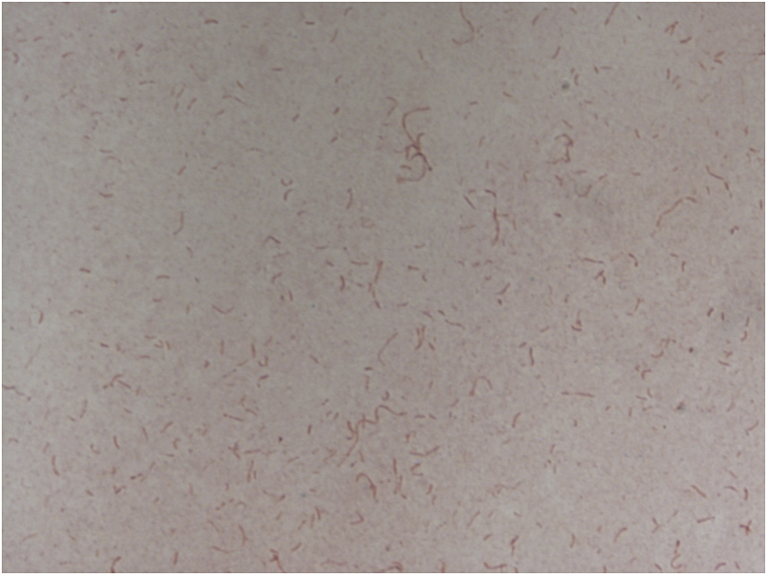
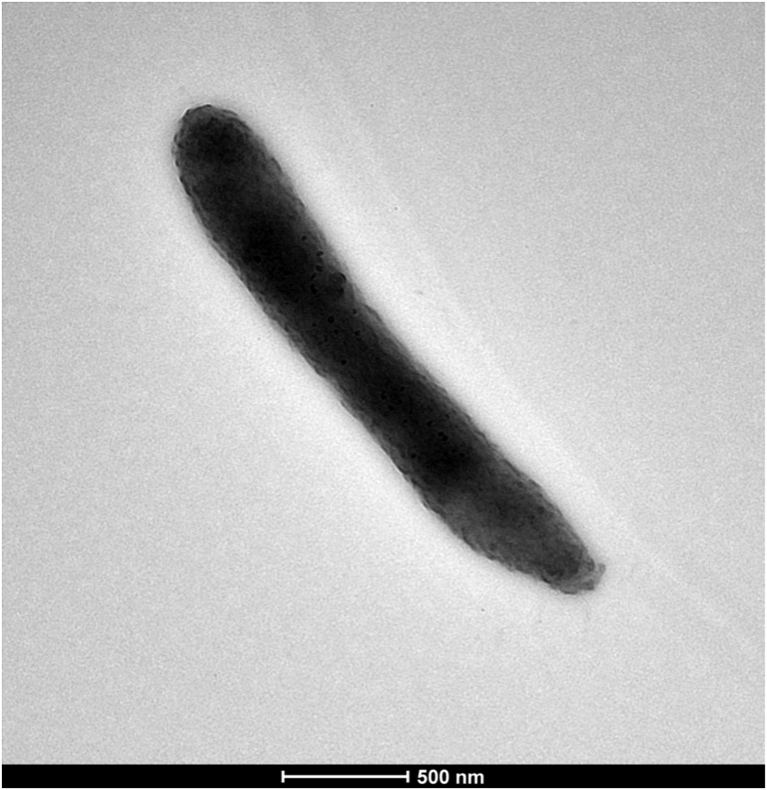
The optimum growth of strain Marseille-P2749T was observed after a 48 hours’ incubation at 37°C on a 5% sheep’s blood–enriched Colombia agar at pH 7 and under anaerobic atmosphere. This strain was also able to grow in microaerophilic conditions, from 28 to 37°C or from pH 5.5 to pH 8. Growth was not observed in salty environments. Colonies were circular, white and opalescent; they measured 0.6 to 0.8 mm in diameter. Cells were Gram-negative bacilli and fusiform shaped (Fig. 3), with a length ranging from 1.7 to 2.5 μm and a width ranging from 0.3 to 0.4 μm (Fig. 4). This strain was nonmotile and non–spore forming. Moreover, it was catalase and oxidase negative. Classification and general features of strain Marseille-P2749T are summarized in Table 2.
Fig. 3.
Gram staining of strain Marseille-P2749T.
Fig. 4.
Transmission electron microscopy of strain Marseille-P2749T with Tecnai G20 Cryo microscope (FEI Company) at operating voltage of 200 keV. Scale bar = 500 nm.
Table 2.
Classification and general features of Fusobacterium massiliense strain Marseille-P2749T
| Property | Term |
|---|---|
| Current classification | Domain: Bacteria |
| Phylum: Fusobacteria | |
| Class: Fusobacteria | |
| Order: Fusobacteriales | |
| Family: Fusobacteriaceae | |
| Genus: Fusobacterium | |
| Species: Fusobacterium massiliense | |
| Marseille-P2749T | |
| Gram stain | Gram-negative bacillus |
| Cell shape | Fusiform shaped |
| Motility | Nonmotile |
| Sporulation | Non–endospore forming |
| Oxygen requirement | Anaerobic and microaerophilic |
| Temperature range | 28 to 37°C |
| Optimum temperature | 37°C |
| pH | 5.5 to 8 |
| Optimum pH | 7 |
| Salinity | 0 g/L |
| Optimum salinity | 0 g/L |
| Habitat | Human gut: Duodenum |
| Pathogenicity | Unknown |
| Isolation | Duodenum |
Biochemical characterization
Using the API 50CH Gallery system, positive reactions were observed for esculin (ferric citrate), d-arabinose, d-raffinose, gentiobiose, d-tagatose and potassium 5-ketogluconate, whereas negative reactions were observed for fructose, glucose, lactose, maltose and mannose. The API ZYM system showed positive reactions for alkaline phosphatase, acid phosphatase, esterase (C4), lipase esterase (C8), α-chymotrypsin and naphtol-AS-BI-phosphohydrolase, but negative reactions for β-galactosidase, β-glucuronidase and N-acetyl-glucosaminidase. Moreover, in the API 20A system, the strain showed indole production but not for gelatin. These characteristics are compared to other strains of Fusobacterium in Table 3.
Table 3.
Characteristics of Fusobacterium species: 1) strain Marseille-P2749T, 2) F. periodonticum ATCC 33693T, 3) F. nucleatum ATCC 25586T, 4) F. canifelinum ATCC BAA-689T, 5) F. simiae ATCC 33568T, 6) F. russii ATCC 25533T, 7) F. equinum JCM 11174T and 8) F. necrophorum ATCC 25286T
| Property | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| Oxygen requirement | − | − | − | − | − | − | − | − |
| Gram stain | − | − | − | − | − | − | − | − |
| Motility | − | − | − | − | − | − | − | − |
| Spore formation | − | − | − | − | − | − | − | − |
| Production of: | ||||||||
| Catalase | − | − | − | − | − | − | − | − |
| Oxidase | − | − | − | − | − | − | − | − |
| Urease | − | − | − | − | − | − | − | + |
| Gelatin | − | − | − | − | − | − | − | − |
| Indole | + | + | + | + | + | − | + | + |
| β-Galactosidase | − | − | − | − | − | − | − | − |
| β-Glucuronidase | − | − | − | − | − | − | − | − |
| N-acetyl-glucosaminidase | − | − | − | − | − | − | − | − |
| Alkaline–phosphatase | + | − | − | − | − | + | − | + |
| Acid–phosphatase | + | − | − | − | − | + | + | + |
| Esterase (C4) | + | − | − | − | + | − | + | + |
| Lipase esterase (C8) | + | − | − | − | − | − | + | + |
| Utilization of: | ||||||||
| Esculin | + | − | − | − | − | − | − | − |
| d-Mannose | − | − | − | − | − | − | − | − |
| d-Glucose | − | + | − | − | + | − | − | − |
| d-Fructose | − | + | + | + | + | − | − | − |
| d-Maltose | − | − | − | − | − | − | − | − |
| d-Lactose | − | − | − | − | − | − | − | − |
| d-Raffinose | + | − | − | − | − | − | − | − |
| Original source | HD | HOC | HOC | COC, DOC | MOC | HNO | EOC | HOC, COC |
+, positive result; −, negative result; COC, cat oral cavity; DOC, dog oral cavity; EOC, horse oral cavity; HD, human duodenum; HNO, human nonoral; HOC, human oral cavity; MOC, monkey oral cavity.
The major fatty acid found for this strain was hexadecanoic acid (40%). Two specific 3-hydroxy fatty acids were described: 14:0 3-OH (9%) and 16:0 3-OH (5%). An unusual branched unsaturated fatty acid was also detected: 15:1n5 anteiso (1%). Cellular fatty acid composition of strain Marseille-P2749T is compared to other Fusobacterium [37] in Table 4.
Table 4.
Cellular fatty acid composition (mean peak area percentage) of Fusobacterium species: 1) strain Marseille-P2749T, 2) F. periodonticum ATCC 33693T, 3) F. nucleatum ATCC 25586T, 4) F. canifelinum ATCC BAA-689T, 5) F. russii ATCC 25533T and 6) F. necrophorum ATCC 25286T
| Fatty acid | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| 4:0 | 2.6 ± 1.7 | NA | NA | NA | NA | NA |
| 12:0 | TR | 1.03 | 1.3 ± 0.25 | 2.38 ± 0.66 | 5.0 ± 1.14 | 6.0 ± 2.08 |
| 14:0 | 18.1 ± 4.5 | 20.17 | 26.1 ± 3.69 | NA | 11.7 ± 2.25 | 16.7 ± 3.14 |
| 14:0 3-OH | 8.9 ± 3.3 | NA | NA | NA | NA | NA |
| 15:1n5 anteiso | 1.1 ± 0.4 | NA | NA | NA | NA | NA |
| 15:0 | TR | NA | NA | NA | NA | NA |
| 16:1n7 | 9.9 ± 1.8 | 0.75 | 0.7 ± 0.17 | NA | 8.1 ± 0.63 | 5.3 ± 0.53 |
| 16:1n9 | 1.5 ± 0.4 | 12.86 | 20.3 ± 4.29 | NA | 15.7 ± 1.20 | 19.7 ± 1.53 |
| 16:0 | 40.2 ± 6.3 | 18.61 | 21.8 ± 4.50 | NA | 35.7 ± 0.93 | 16.4 ± 1.88 |
| 16:0 3-OH | 5.4 ± 0.3 | 5.43 | 4.1 ± 0.56 | 3.48 ± 0.30 | NA | 1.8 ± 1.13 |
| 18:1n9 | 5.4 ± 2.3 | 4.69 | 0.5 ± 0.22 | NA | 7.1 ± 1.37 | 9.2 ± 1.85 |
| 18:2n6 | 2.2 ± 0.7 | NA | NA | NA | NA | NA |
| 18:0 | 2.3 ± 1.9 | 3.79 | NA | NA | NA | NA |
| 18:1n7 | 1.0 ± 0.6 | NA | NA | NA | NA | NA |
16:0, hexadecanoic acid; 14:0, tetradecanoic acid; 16:1n7, 9-hexadecenoic acid; 14:0 3-OH, 3-hydroxy-tetradecanoic acid; 16:0 3-OH, 3-hydroxy-hexadecanoic acid; 18:1n9, 9-octadecenoic acid; 4:0, butanoic acid; 18:0, octadecanoic acid; 18:2n6, 9,12-octadecadienoic acid; 16:1n9, 7-hexadecenoic acid; 15:1n5 anteiso, 12-methyltetradec-9-enoic acid; 18:1n7, 11-octadecenoic acid; 12:0, dodecanoic acid; 15:0, Pentadecanoic acid; NA, not available; TR, trace amounts <1%.
Strain Marseille-P2749T was susceptible to colistin, amoxicillin, amoxicillin/clavulanic acid, penicillin, clindamycin, metronidazole, oxacillin, imipenem, tobramycin, ceftriaxone, rifampicin and doxycycline, whereas it was resistant to vancomycin, gentamicin, trimethoprim/sulfamethoxazole, fosfomycin and erythromycin.
Genome analysis
The genome length is 1 809 169 bp with 27.33 mol% G+C content (Table 5 and Fig. 5), and it is composed of six scaffolds (seven contigs). Of the 1703 genes, 1646 were protein-coding genes (96.65%) and 57 were RNAs (48 were tRNA genes and nine were rRNA genes). A total of 1274 genes (77.39%) were assigned as putative function (by COGs or by nonredundant protein sequence BLAST), and 27 genes (1.64%) were associated with ORFans. No gene was associated with resistance, and four genes were associated with polyketide synthase or nonribosomal peptide synthase. Table 6 and Fig. 6 show the genes’ distribution into COGs functional categories for strain Marseille-P2749T. The genome sequence was deposited in EMBL-EBI under accession number FMJA01000000.
Table 5.
Nucleotide content and gene count levels of the genome of strain Marseille-P2749T
| Attribute | Value | % of totala |
|---|---|---|
| Size (bp) | 1 809 169 | 100 |
| G+C content (bp) | 494 380 | 27.33 |
| Coding region (bp) | 1 617 050 | 89.38 |
| Total of genes | 1703 | 100 |
| Protein-coding genes | 1646 | 96.65 |
| RNA genes | 57 | 3.35 |
| Genes associated with function prediction | 1274 | 77.39 |
| Genes assigned to COGs | 1123 | 68.22 |
| Genes associated with ORFans | 27 | 1.64 |
| Genes with peptide signals | 208 | 12.63 |
| Genes associated with PKS or NRPS | 4 | 0.24 |
| Genes associated with resistance genes | 0 | 0 |
| Genes associated with bacteriocin genes | 10 | 0.61 |
| Genes associated with toxin/antitoxin | 43 | 2.61 |
| Genes associated with virulence | 264 | 16.04 |
| Genes associated with mobilome | 605 | 36.76 |
| Genes with paralogues (E value 1e-10) | 189 | 11.48 |
| Genes with paralogues (E value 1e-25) | 105 | 6.37 |
| Genes larger than 5000 nucleotides | 6 | 0 |
COGs, Clusters of Orthologous Groups database; NRPS, nonribosomal peptide synthase; PKS, polyketide synthase.
Total is based on either size of genome in base pairs or total number of protein-coding genes in annotated genome.
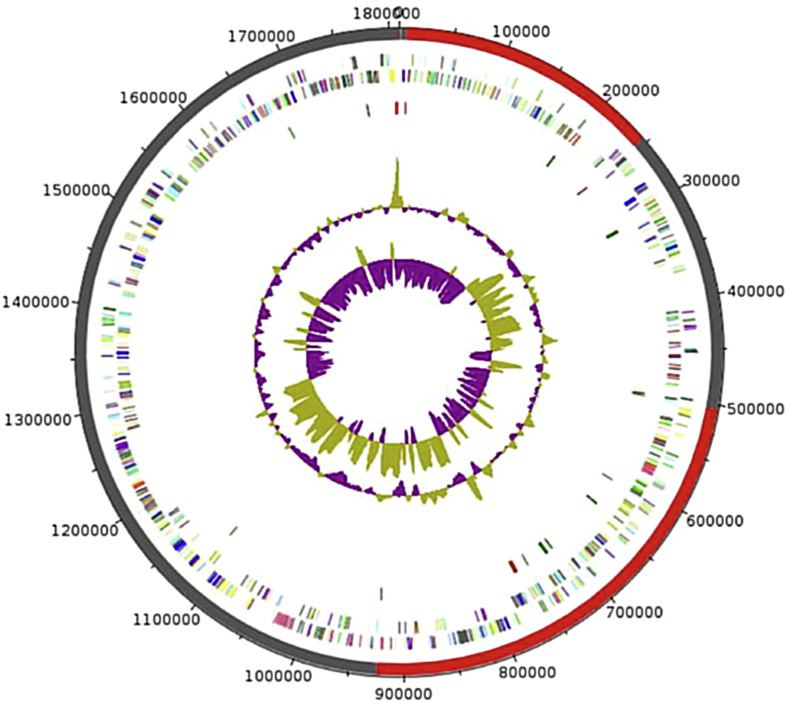
Fig. 5.
Graphical circular map of strain of chromosome of Marseille-P2749T. From outside to center: outer two circles show ORF oriented forward (coloured by COGs categories) and backwards (coloured by COGs categories) respectively. Third circle marks tRNA genes (green). Fourth circle shows G+C% content plot. Innermost circle shows GC skew; purple indicates negative values and olive positive values. COGs, Clusters of Orthologous Groups database; ORF, open reading frame.
Table 6.
Genes associated with the 25 COGs functional categories of strain Marseille-P2749T
| Code | Value | % Value | Description |
|---|---|---|---|
| J | 173 | 10.51 | Translation |
| A | 0 | 0 | RNA processing and modification |
| K | 50 | 3.03 | Transcription |
| L | 58 | 3.52 | Replication, recombination and repair |
| B | 0 | 0 | Chromatin structure and dynamics |
| D | 21 | 1.28 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0 | Nuclear structure |
| V | 51 | 3.10 | Defense mechanisms |
| T | 34 | 2.07 | Signal transduction mechanisms |
| M | 86 | 5.22 | Cell wall/membrane biogenesis |
| N | 8 | 0.49 | Cell motility |
| Z | 0 | 0 | Cytoskeleton |
| W | 3 | 0.18 | Extracellular structures |
| U | 27 | 1.64 | Intracellular trafficking and secretion |
| O | 43 | 2.61 | Posttranslational modification, protein turnover, chaperones |
| X | 16 | 0.97 | Mobilome: prophages, transposons |
| C | 65 | 3.95 | Energy production and conversion |
| G | 71 | 4.31 | Carbohydrate transport and metabolism |
| E | 144 | 8.75 | Amino acid transport and metabolism |
| F | 45 | 2.73 | Nucleotide transport and metabolism |
| H | 85 | 5.16 | Coenzyme transport and metabolism |
| I | 48 | 2.92 | Lipid transport and metabolism |
| P | 61 | 3.71 | Inorganic ion transport and metabolism |
| Q | 11 | 0.67 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 90 | 5.47 | General function prediction only |
| S | 45 | 2.73 | Unknown function |
| — | 523 | 31.77 | Not in COGs |
COGs, Clusters of Orthologous Groups database.
Fig. 6.
Distribution of functional classes of predicted genes according to COGs of proteins from strain Marseille-P2749T. COGs, Clusters of Orthologous Groups database.
Genome comparison
The draft genome sequence of strain Marseille-P2749T (1.81 Mb) was smaller than all the compared genomes: Fusobacterium russii ATCC 25533T (1.94 Mb), Fusobacterium perfoetens ATCC 29250T (2.10 Mb), Fusobacterium nucleatum ATCC 49256T (2.12 Mb), Fusobacterium mortiferum ATCC 25557T (2.67 Mb), Ilyobacter polytropus DSM 2926T (3.13 Mb) and Fusobacterium varium ATCC 8501T (3.30 Mb). The G+C content of strain Marseille-P2749T (27.33%) was higher than that of F. perfoetens (25.97%) but smaller than the other compared genomes: F. nucleatum (27.34%), F. russii (28.64%), F. mortiferum (29.07%), F. varium (29.18%) and I. polytropus (34.37%). The gene content of strain Marseille-P2749T (1646) was also smaller than all compared genomes: F. russii (1764), F. perfoetens (1949), F. nucleatum (2250), F. mortiferum (2538), I. polytropus (2880) and F. varium (3008).
Strain Marseille-P2749T shared 993 orthologous genes with F. russii, 912 with F. varium, 880 with F. mortiferum, 855 with F. nucleatum, 823 with F. perfoetens and 811 with I. polytropus. The distribution of genes into COGs categories was similar in all compared genomes (Fig. 6). The AGIOS value ranged from 57.46 to 76.55% when the closest species were compared. For strain Marseille-P2749T, the AGIOS value ranged from 57.70% with I. polytropus to 76.55% with F. russii (Table 7). The DDH value was of 26.10% (23.8–28.6%) with F. nucleatum, 26.00% (23.7–28.5%) with F. periodonticum, 21.30% (19.0–23.7%) with F. russii, 21.10% (18.8–23.5%) with I. polytropus, 20.90% (18.6–23.3%) with F. mortiferum, 20.60% (18.4–23.1%) with F. varium and 18.00% (15.9–20.4%) with F. perfoetens (Table 8).
Table 7.
Orthologous proteins shared between genomes (upper right), average percentage similarity of nucleotides corresponding to orthologous proteins shared between genomes (lower left) and number of proteins per genome (bold)
| Strain Marseille-P2749T | Fusobacterium russii | Fusobacterium varium | Fusobacterium mortiferum | Fusobacterium nucleatum | Fusobacterium perfoetens | Ilyobacter polytropus | |
|---|---|---|---|---|---|---|---|
| Strain Marseille-P2749T | 1646 | 993 | 912 | 880 | 855 | 823 | 811 |
| F. russii | 76.55 | 1764 | 931 | 886 | 854 | 834 | 793 |
| F. varium | 67.50 | 66.76 | 3008 | 1267 | 775 | 1095 | 1054 |
| F. mortiferum | 68.48 | 67.49 | 69.27 | 2538 | 750 | 1069 | 1040 |
| F. nucleatum | 64.13 | 62.35 | 60.12 | 59.37 | 2250 | 690 | 685 |
| F. perfoetens | 71.57 | 70.53 | 66.96 | 68.84 | 59.89 | 1949 | 823 |
| I. polytropus | 57.70 | 57.46 | 58.37 | 58.99 | 59.09 | 58.05 | 2880 |
Table 8.
Pairwise comparison of strain Marseille-P2749T with other species using GGDC, formula 2 (DDH estimates based on identities/HSP length)a
| Strain Marseille-P2749 | Fusobacterium nucleatum | Fusobacterium periodonticum | Fusobacterium russii | Ilyobacter polytropus | Fusobacterium mortiferum | Fusobacterium varium | Fusobacterium perfoetens | |
|---|---|---|---|---|---|---|---|---|
| Strain Marseille-P2749 | 100 (100–100) | 26.10 (23.8–28.6) | 26.00 (23.7–28.5) | 21.30 (19.0–23.7) | 21.10 (18.8–23.5) | 20.90 (18.6–23.3) | 20.60 (18.4–23.1) | 18.00 (15.9–20.4) |
| F. nucleatum | 100 (100–100) | 30.30 (27.9–32.8) | 21.00 (18.8–23.4) | 23.60 (21.3–26.0) | 21.30 (19.1–23.7) | 20.10 (17.9–22.5) | 19.10 (16.9–21.4) | |
| F. periodonticum | 100 (100–100) | 21.10 (18.9–23.5) | 22.90 (20.7–25.4) | 20.90 (18.7–23.4) | 21.30 (19.1–23.7) | 19.20 (17.0–21.6) | ||
| F. russii | 100 (100–100) | 18.40 (16.3–20.8) | 19.30 (17.1–21.7) | 18.70 (16.5–21.1) | 19.70 (17.5–22.1) | |||
| I. polytropus | 100 (100–100) | 22.40 (20.1–24.8) | 22.30 (20.0–24.7) | 19.50 (17.3–21.9) | ||||
| F. mortiferum | 100 (100–100) | 20.40 (18.2–22.9) | 22.40 (20.1–24.8) | |||||
| F. varium | 100 (100–100) | 18.20 (16.0–20.6) | ||||||
| F. perfoetens | 100 (100–100) |
DDH, DNA-DNA hybridization; GGDC, Genome-to-Genome Distance Calculator; HSP, high-scoring segment pairs.
Confidence intervals indicate inherent uncertainly in estimating DDH values from intergenomic distances based on models derived from empirical test data sets (which are always limited in size). These results are in accordance with 16S rRNA (Fig. 1(b)) and phylogenomic analyses as well as GGDC result.
The probability of error to say that this strain is a new species (DDH >70%) compared to Fusobacterium periodonticum ATCC 33693T, the type strain of the closest phylogenetic neighbour based on 16S rRNA similarity (95.9%), was of 0.02% (GGDC2.1 formula 2; DSMZ website (http://ggdc.dsmz.de/distcalc2.php)).
Conclusion
On the basis of the phylogenetic, phenotypic, biochemical and genomic evidence presented, the creation of a novel species of Fusobacterium is proposed, with the name Fusobacterium massiliense sp. nov., the type strain of which is Marseille-P2749. This bacterium was first isolated from the duodenum of a 60-year-old man as part of the culturomics study of the gut microbiota from stored samples taken from the gastrointestinal tract.
Description of Fusobacterium massiliense sp. nov.
Fusobacterium massiliense (mas.si.li.en′se, L. neut. n. massiliense, ‘of Massilia,’ the Latin name of Marseille, France, where the type strain was first isolated). Cells are Gram-negative bacilli and fusiform shaped, with a length ranging from 1.7 to 2.5 μm and a width ranging from 0.3 to 0.4 μm. This strain exhibited no catalase and no oxidase activities. Fusobacterium massiliense is nonmotile and non–spore forming. Colonies are circular, white and opalescent, with a diameter of 0.6 to 0.8 mm on Colombia agar enriched with 5% sheep’s blood after 48 hours’ growth. Optimum growth occurred at 37°C and pH 7 in an anaerobic atmosphere. Microaerophilic growth was observed. Using the API 50CH Gallery system, positive reactions were observed for esculin, d-arabinose, d-raffinose, gentiobiose, d-tagatose and potassium 5-ketogluconate. The API ZYM system showed positive reactions for alkaline phosphatase, acid phosphatase, esterase (C4), lipase esterase (C8), α-chymotrypsin and naphtol-AS-BI-phosphohydrolase. Moreover, in the API 20A system, the strain showed indole production. The major fatty acid is hexadecanoic acid. Cells are susceptible to colistin, amoxicillin, amoxicillin/clavulanic acid, penicillin, clindamycin, metronidazole, oxacillin, imipenem, tobramycin, ceftriaxone, rifampicin and doxycycline. The length of the genome is of 1 809 169 bp, and the G+C content is 27.33 mol%. The accession number of the 16S rRNA gene sequence deposited in EMBL-EBI is LT576389, and the accession number of the genome sequence is FMJA01000000. The type strain Marseille-P2749T (= CSUR P2749 = DSM 103085) was first isolated from the duodenum liquid sample of a 60-year-old man. The habitat of this microorganism is the human gut.
Acknowledgements
The authors thank the Xegen Company (www.xegen.fr/) for automating the genomic annotation process. The authors also thank M. Lardière for English-language review. This study was funded by the Fondation Méditerranée Infection.
Conflict of Interest
None declared.
References
- 1.Turnbaugh P.J., Ley R.E., Hamady M., Fraser-Liggett C.M., Knight R., Gordon J.I. The human microbiome project. Nature. 2007;449(7164):804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lagier J.C., Hugon P., Khelaifia S., Fournier P.E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagier J.C., Edouard S., Pagnier I., Mediannikov O., Drancourt M., Raoult D. Current and past strategies for bacterial culture in clinical microbiology. Clin Microbiol Rev. 2015;28:208–236. doi: 10.1128/CMR.00110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drancourt M., Bollet C., Carlioz A., Martelin R., Gayral J.P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000;38:3623–3630. doi: 10.1128/jcm.38.10.3623-3630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seng P., Drancourt M., Gouriet F., La Scola B., Fournier P.E., Rolain J.M. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 6.Hugon P., Dufour J.C., Colson P., Fournier P.E., Sallah K., Raoult D. A comprehensive repertoire of prokaryotic species identified in human beings. Lancet Infect Dis. 2015;15:1211–1219. doi: 10.1016/S1473-3099(15)00293-5. [DOI] [PubMed] [Google Scholar]
- 7.Raoult D., Henrissat B. Are stool samples suitable for studying the link between gut microbiota and obesity? Eur J Epidemiol. 2014;29:307–309. doi: 10.1007/s10654-014-9905-4. [DOI] [PubMed] [Google Scholar]
- 8.Citron D.M. Update on the taxonomy and clinical aspects of the genus Fusobacterium. Clin Infect Dis. 2002;35(Suppl 1):S22–S27. doi: 10.1086/341916. [DOI] [PubMed] [Google Scholar]
- 9.Conrads G., Citron D.M., Mutters R., Jang S., Goldstein E.J.C. Fusobacterium canifelinum sp. nov., from the oral cavity of cats and dogs. Syst Appl Microbiol. 2004;27:407–413. doi: 10.1078/0723202041438509. [DOI] [PubMed] [Google Scholar]
- 10.Bolstad A.I., Jensen H.B., Bakken V. Taxonomy, biology, and periodontal aspects of Fusobacterium nucleatum. Clin Microbiol Rev. 1996;9:55–71. doi: 10.1128/cmr.9.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slots J., Potts T.V., Mashimo P.A. Fusobacterium periodonticum, a new species from the human oral cavity. J Dent Res. 1983;62:960–963. doi: 10.1177/00220345830620090901. [DOI] [PubMed] [Google Scholar]
- 12.Stackebrandt E., Frederiksen W., Garrity G.M., Grimont P.A.D., Kämpfer P., Maiden M.C.J. Report of the Ad Hoc Committee for the Re-evaluation of the Species Definition in Bacteriology. Int J Syst Evol Microbiol. 2002;52(Pt 3):1043–1047. doi: 10.1099/00207713-52-3-1043. [DOI] [PubMed] [Google Scholar]
- 13.Ramasamy D., Mishra A.K., Lagier J.C., Padhmanabhan R., Rossi M., Sentausa E. A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int J Syst Evol Microbiol. 2014;64(Pt 2):384–391. doi: 10.1099/ijs.0.057091-0. [DOI] [PubMed] [Google Scholar]
- 14.Lagier J.C., Armougom F., Million M., Hugon P., Pagnier I., Robert C. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 15.Seng P., Abat C., Rolain J.M., Colson P., Lagier J.C., Gouriet F. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol. 2013;51:2182–2194. doi: 10.1128/JCM.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.La Scola B., Fournier P.E., Raoult D. Burden of emerging anaerobes in the MALDI-TOF and 16S rRNA gene sequencing era. Anaerobe. 2011;17:106–112. doi: 10.1016/j.anaerobe.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 17.Morel A.S., Dubourg G., Prudent E., Edouard S., Gouriet F., Casalta J.P. Complementarity between targeted real-time specific PCR and conventional broad-range 16S rDNA PCR in the syndrome-driven diagnosis of infectious diseases. Eur J Clin Microbiol Infect Dis. 2014;34:561–570. doi: 10.1007/s10096-014-2263-z. [DOI] [PubMed] [Google Scholar]
- 18.Lagier J.C., El Karkouri K., Nguyen T.T., Armougom F., Raoult D., Fournier P.E. Non-contiguous finished genome sequence and description of Anaerococcus senegalensis sp. nov. Stand Genomic Sci. 2012;6:116–125. doi: 10.4056/sigs.2415480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sasser M., Kunitsky C., Jackoway G., Ezzell J.W., Teska J.D., Harper B. Identification of Bacillus anthracis from culture using gas chromatographic analysis of fatty acid methyl esters. J AOAC Int. 2005;88:178–181. [PubMed] [Google Scholar]
- 21.Dione N., Sankar S.A., Lagier J.C., Khelaifia S., Michele C., Armstrong N. Genome sequence and description of Anaerosalibacter massiliensis sp. nov. New Microb New Infect. 2016;10:66–76. doi: 10.1016/j.nmni.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Page S., van Belkum A., Fulchiron C., Huguet R., Raoult D., Rolain J.M. Evaluation of the PREVI® Isola automated seeder system compared to reference manual inoculation for antibiotic susceptibility testing by the disk diffusion method. Eur J Clin Microbiol. 2015;34:1859–1869. doi: 10.1007/s10096-015-2424-8. [DOI] [PubMed] [Google Scholar]
- 23.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol J Comput Mol Cell Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J. GenBank. Nucleic Acids Res. 2013;41(Database issue):D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lagesen K., Hallin P., Rødland E.A., Staerfeldt H.H., Rognes T., Ussery D.W. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bendtsen J.D., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 28.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Y., Liang Y., Lynch K.H., Dennis J.J., Wishart D.S. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39(Web Server issue):W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M.A. Artemis: sequence visualization and annotation. Bioinforma. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 32.Carver T., Thomson N., Bleasby A., Berriman M., Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinforma. 2009;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darling A.C.E., Mau B., Blattner F.R., Perna N.T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Auch A.F., von Jan M., Klenk H.P., Göker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci. 2010;2:117–134. doi: 10.4056/sigs.531120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim M., Oh H.S., Park S.C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64(Pt 2):346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 36.Huson D.H., Auch A.F., Qi J., Schuster S.C. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tunér K., Baron E.J., Summanen P., Finegold S.M. Cellular fatty acids in Fusobacterium species as a tool for identification. J Clin Microbiol. 1992;30:3225–3229. doi: 10.1128/jcm.30.12.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]