Abstract
Most melanomas are driven by BRAF(V600E)-activating mutations, while nevi harboring the same mutations have growth arrest. Although decreased p16 expression has been associated with melanoma formation, in recent work, p15 represented a primary effector of oncogene-induced senescence in nevomelanocytes that was diminished in melanomas. This study determined whether decreased p15 levels represent a general biomarker for the transition from nevus to melanoma. We performed p15 and p16 IHC analyses on a random series of nevi and melanomas. Staining was evaluated and graded for percentage and intensity to determine the H score. For real-time quantitative RT-PCR analysis of p15, RNA was extracted from FFPE sections from 14 nevus and melanoma samples via macrodissection. A two-sided t-test was used to evaluate between-group differences in mean H scores and qΔCt values. p15 Expression was significantly increased in melanocytic nevi compared with melanomas (mean H scores, 254.8 versus 132.3; P < 0.001). On p15 staining, the H score differential was greater than that with p16 staining [122.5 (P < 0.001) and 64.8 (P = 0.055), respectively]. Real-time quantitative RT-PCR analysis revealed a lower mean qΔCt value in melanomas, consistent with lower p15 expression (P = 0.018). Together, these data support the hypothesis that decreased p15 expression is a robust biomarker for distinguishing nevus from melanoma.
BRAF is a well-established proto-oncogene that is involved in the pathogenesis of melanoma.1, 2, 3, 4 Serine/threonine protein kinase B-raf (BRAF) is a component of the ERK1/2-signaling pathway, which regulates cellular proliferation, differentiation, and survival in response to various mitogenic, stress, and inflammatory stimuli.4 Mutations in BRAF that lead to constitutive activation of this pathway are examples of driver mutations found in many cancers and are best established in melanomas.1 The most common BRAF(V600E) mutation makes up >90% of BRAF mutations in melanoma.4 However, the presence of BRAF mutations in 82% of nevi indicates that such a mutation alone is insufficient for conferring malignancy.5
Clinically, most nevi grow until they reach 3 to 5 mm, at which time nevomelanocytes typically exit the proliferative growth phase and enter a state of growth arrest.6 Although most nevi remain growth-arrested (or even regress in advanced age), a small percentage regain the ability to proliferate and form melanomas.7, 8, 9 In some studies, approximately 50% of melanomas are seen arising from benign nevi; therefore, it is logical to infer that BRAF(V600E) oncogene-induced senescence (OIS) in nevomelanocytes is reversible.10, 11
OIS is promoted by increased expression of cyclin-dependent kinase (CDK) inhibitors p16 and p15, encoded by the CDKN2A and CDKN2B loci, respectively, on chromosomal locus 9p21.1 Altered p16 expression has been implicated in both sporadic and germline mutant–mediated melanoma.12, 13, 14 Studies of OIS have found increased expression of p16 in senescent nevi melanocytes but also that p16 alone was insufficient for maintenance of senescence.6, 15, 16 CDK inhibitors p15 and p16 belong to the INK (inhibitors of kinases) family and act by binding to CDK4, thus preventing its ability to associate with cyclin D, which in turn inhibits CDK4/cyclin D–mediated phosphorylation of retinoblastoma-associated protein and prevents G1–S phase progression.15, 17, 18
CDKN2A is part of a gene cluster with CDKN2B, which encodes p15, and therefore the common CDKN2A deletion in human cancer is typically associated with deletion of CDKN2B.19 The Cancer Genome Atlas analysis of 278 melanomas demonstrated that 93% of melanomas with homozygous deletions of CDKN2A also have homozygous deletions of CDKN2B likely related to the close proximity of these loci on 9p21.3 Although rare, it has also been documented in The Cancer Genome Atlas series, and in additional cases, that CDKN2B is deleted while CDKN2A remains intact.20, 21 These data suggest a significant role of p15 in melanomagenesis.
A recent study has established p15 as an important mediator of OIS in melanocytes and nevomelanocytes.11 In that study, primary melanocytes were transduced with p15 or p16 alone for assessment of whether the CDK inhibitors were powerful enough to induce OIS in isolation. The p15-expressing melanocytes entered a state of OIS while the p16-expressing melanocytes continued to proliferate, although the rate was decreased compared with those in controls.11
OIS can be overcome by additional mutations in tumor suppressor genes or their targets.1, 11 For example, progression to melanoma can occur with p15 loss or the development of p15-resistant CDK mutations such as CDK4(R24C).11
In this current study, we test the hypothesis that p15 is a robust biomarker associated with the transition from nevus to melanoma. Nevus and melanoma tissue sections were subjected to p15 and p16 immunostaining, and the intensity and distribution of staining were used for calculating an H score. Additionally, real-time quantitative RT-PCR (RT-qPCR) for p15 was performed to determine the relative levels of p15 expression in melanomas and nevi. The data showed that p15 immunohistochemistry (IHC) staining was a more robust biomarker for distinguishing nevi from melanomas than was p16. The findings from RT-qPCR analysis of p15 in nevi and melanomas support this conclusion by demonstrating decreased p15 transcript in melanomas. Together, these data suggest that a decreased p15 level is a reliable biomarker of the nevus–melanoma transition and that p15 immunostaining or RT-qPCR analysis may have diagnostic utility in distinguishing benign melanocytic lesions from melanoma.
Materials and Methods
Histologic Examination and IHC Analysis
Sample usage was covered under an Institutional Review Board–approved protocol (808225) at the University of Pennsylvania (Philadelphia, PA). Formalin-fixed and paraffin-embedded (FFPE) skin tissues from nevi and melanomas were cut, mounted on glass slides, and subjected to IHC analysis of p15 and p16 protein expression. For p15 staining, tissue sections of nevi and melanomas were screened using a peroxidase/diaminobenzidine complex method and detection kit (Abcam, Cambridge, MA). Briefly, tissue sections were deparaffinized and rehydrated. A heat-induced antigen retrieval was performed at sub-boiling temperature for 10 minutes using 10 mmol/L citrate buffer (pH 6.0); to quench endogenous peroxidase, tissues were incubated in 3% H2O2 for 14 minutes at room temperature. Tissue sections were incubated with primary antibody to p15 (1:200; catalog number ab53034; Abcam) overnight at 4°C. The secondary antibody was a goat anti-rabbit horseradish peroxidase supplied with the kit (Abcam) and incubated for 20 minutes at room temperature according to instructions. After multiple washes in Tris-buffered saline, the antigen–antibody complex was visualized with diaminobenzidine solution. The tissues were counterstained with hematoxylin, dehydrated, and coverslipped. The cells immunoreactive to p15 were analyzed under an Axiophot microscope (Carl Zeiss, Oberkochen, Germany) coupled with a DFC450 digital camera (Leica Microsystems, Buffalo Grove, IL).
In a similar manner, FFPE sections of nevus and melanoma tissue samples were stained using a peroxidase/diaminobenzidine complex method as provided by the CINtec Histology Kit (Ventana Medical Systems, Tucson, AZ), optimal for the detection of the p16INK4a antigen through the use of a commercially available mouse anti-human antibody, E6H4 (Roche Diagnostics, Indianapolis, IN). An external control from a tissue sample negative for p16INK4a was stained to verify the validity of the IHC assay as well as to compare against IHC-stained nevus and melanoma samples.
Slides were reviewed in a blinded manner by two independent investigators (L.A.T. and J.T.S.) and scored based on level of intensity and extent of staining. The epidermis was used as an internal positive control for p15, and control slides were used for p16 to designate a reference score of 4 on a 5-point scale (0 indicates no visible staining; 4 indicates highest level of staining detected for that antigen). An H score was calculated as the product of the degree of intensity (0 to 4) and the percentage of the lesion staining at that level, and these products from each lesion were summed.
Six p16 cases (two nevi and four melanomas) were not included in the analysis due to inadequate controls.
Purification of Total RNA from FFPE Tissue Sections
Hematoxylin and eosin–stained template slides were used for direct dissection of tissue from the glass-mounted samples using stainless steel, disposable scalpels (catalog number 11; Exel International Medical Products, St. Petersburg, FL), and the dissectate was pooled in 1.5-mL DNA LoBind tubes (Eppendorf, Hamburg, Germany). Purified RNA was then isolated as described in the protocol of the RNeasy FFPE Kit (Qiagen, Valencia, CA) for total RNA from microdissected FFPE tissue sections. Purified RNA was collected from each sample and stored at −80°C.
RNA Quality Assessment and Quantification
A Qubit 3.0 Fluorometer (Thermo Fisher Scientific, Rochester, NY) was used for determining the quantity of purified RNA through a high-sensitivity RNA assay. The quality of the purified RNA was determined using the Agilent 2100 Bioanalyzer system (Agilent Technologies, Santa Clara, CA). The RNA samples demonstrated RNA integrity values (Agilent) ranging from 2.0 to 5.1 (on a scale of 0 to 10).
Reverse Transcription
Approximately 91.8 ng of purified RNA was used as a template for cDNA synthesis, which was performed using the random-primer QuantiTect Reverse Transcription Kit (Qiagen). cDNA samples were stored at −20°C before RT-qPCR.
RT-qPCR
The QuantiFast SYBR Green PCR Kit (Qiagen) was used for RT-qPCR analysis. Two QuantiTect primer pairs were used: CDKN2B (p15), HS_CDKN2B_1_SG (a 72-bp amplicon-length primer assay) and GAPDH, a standardization transcript cataloged as HS_GAPDH_1_SG (a 95-bp amplicon length primer assay). The RT-qPCR reaction was performed using a 96-well plate on a ViiA 7 real-time PCR system (Thermo Fisher Scientific, Rochester, NY).
Statistical Analysis
Stata software version 13 (Stata Inc., College Station, TX) was used for data analysis. The t-test, Wilcoxon rank-sum statistic, and logistical regressions were used for assessing differences among groups. P < 0.05 was considered significant.
For analysis of H scores, 58 samples were stained for p15 and p16. Two of these samples were misdiagnoses, which were excluded from statistical analysis given decreased cellularity on recuts, leaving 56 (29 nevi, 27 melanoma). For each sample, the data reflected diagnosis (melanoma, nevus), p15 H score, and p16 H score. Totals of 27 nevus and 26 melanoma samples (n = 53) were successfully stained for p15, while 25 nevus and 22 melanoma samples (n = 47) were successfully stained for p16.
For the p15 and p16 staining, the mean H scores for the melanoma and nevus groups were compared using an unpaired t-test, with the null hypotheses of H score equality between groups. Variances were not constrained to be equal between groups.
A two-sample Wilcoxon rank-sum statistic was used for confirming these results. Finally, multivariate logistic regression was used for assessing whether p15 or p16 H scores were independently associated with diagnosis.
The X2 statistic was used for selecting optimal threshold H scores for hypothetical diagnostic tests; SEMs for these diagnostic tests were determined using a bootstrap procedure with 1000 bootstrap replications. In addition, the sample split was performed independently for each bootstrap replication, and the point estimates reported represent bootstrap means.
RT-qPCR was performed on 28 cDNA samples (14 nevi and 14 melanomas). Relative levels of p15 transcript were calculated as the difference between p15 and GAPDH Ct values, raised to the second power (2ΔΔCt), yielding a value of qΔCt for each observation. Before equality in mean concentrations was tested, Tukey outlier analysis was performed, based on observed paradoxically large concentrations of p15 in some melanoma samples.22 This analysis involved calculating the 25th and 75th quartiles of the subsamples, as well as the difference between these two values (IQR). The value of 1.5 × IQR was added to the 75th percentile and subtracted from the 25th percentile, to yield the range outside of which outliers would be excluded. This procedure resulted in the exclusion of 3 melanoma samples, leaving 11 melanoma and 14 nevus samples.
Mean qΔCt values from the melanoma and nevus groups were compared using an unpaired t-test, with the null hypothesis of equality between groups. Variances were not constrained to be equal between groups.
Results
H Scores for p15 and p16 Immunostaining of Nevi and Melanoma
FFPE tissue sections of nevi and melanomas were stained to detect p15 and p16 (Figure 1). The stained sections were evaluated for the intensity and extent of staining. H scores were determined for p15 and p16 in each lesion that had appropriate controls. Two-sided t-tests were used for assessing the differences in p15 and p16 expression in nevi and melanomas. The difference in mean p15 H scores between melanomas (132.3) and nevi (254.8) of 122.5 was significant (P < 0.001), with increased staining present in the nevus set (Figure 2A). The difference in mean p16 H scores between melanomas (100.5) and nevi (165.3) was 64.8 (P = 0.055) (Figure 2A). Wilcoxon rank-sum tests were run to confirm these results, showing that both p15 (P = 0.001) and p16 (P = 0.0096) were significantly different between the two diagnostic groups. Logistic regression was used for testing the ability of both p15 and p16 H scores to detect melanoma in a multivariate context. The logistic regression showed an odds ratio of 0.979 for p15 H score (P = 0.002), while p16 did not contribute significantly (P = 0.426) (Figure 2A). These data are depicted in Figure 1.
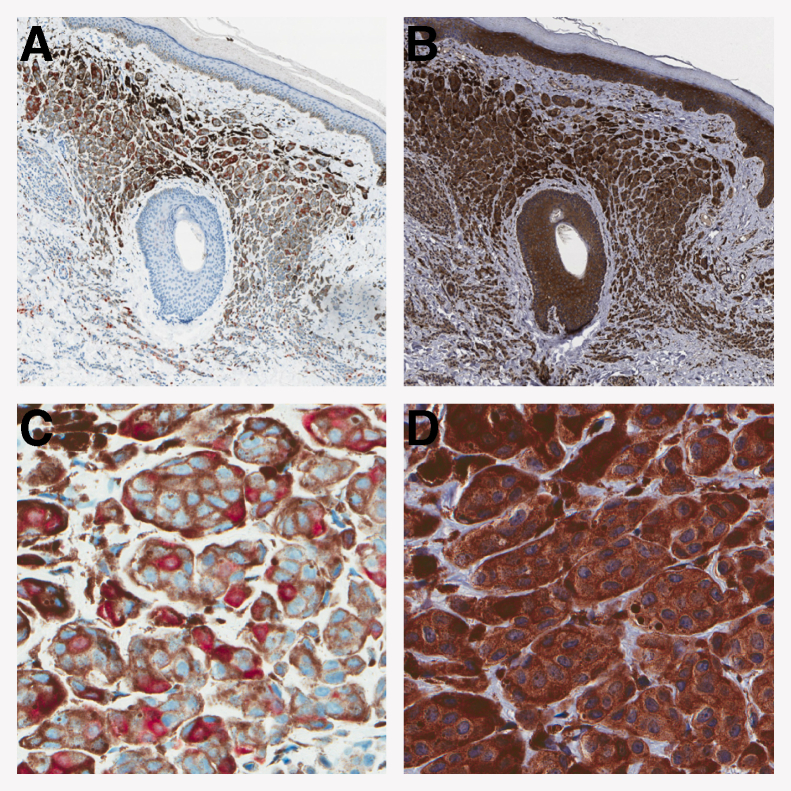
Figure 1.
Representative staining for p16 and p15 in a nevus. Nevus was stained for p16 (A and C) and p15 (B and D). Note the uniform, prominent staining seen with p15 and the internal controls of the epidermis and the hair follicle. C and D: Higher magnification contrasts the consistent p15 staining pattern with the classic p16 checkerboard pattern. Original magnification: ×25 (A and B); ×200 (C and D).
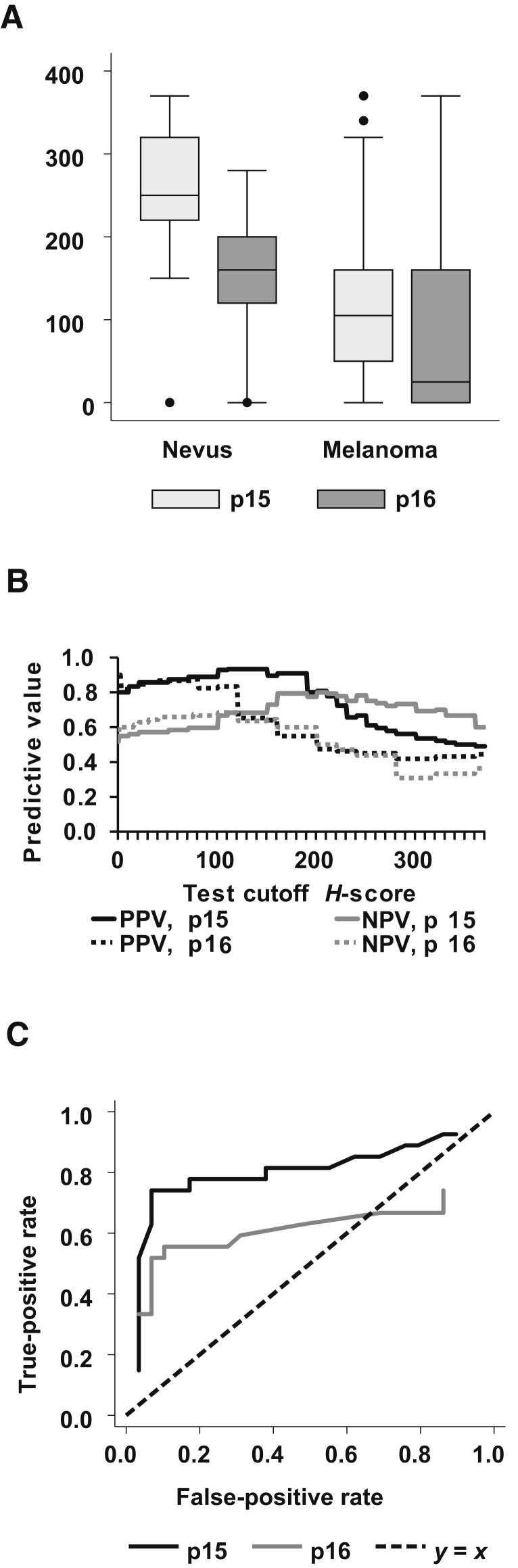
Figure 2.
A: Box-and-whisker plots of p15 and p16 H scores, by diagnosis. Whiskers mark the most extreme points within 1.5 × interquartile range (IQR); dots, points outside of the IQR. B: Positive predictive values (PPV) and negative predictive values (NPV), by cutoff points for H scores of the immunohistochemistry–stained slides. Each point on the x axis represents a different test threshold (H score) for detecting melanoma. The test result is positive if the H score value lies below the cutoff point. C: Receiver operating characteristic curve shows that the tradeoff between false-positive rate (x axis) and true-positive rate (y axis) as the test threshold H score is varied. The top left point represents a perfect test, with a 100% true-positive rate and a 0% false-positive rate. The dashed line represents a coin flip. Points above and to the left of the dashed line represent informative tests. Data are expressed as medians (lines) [interquartile ranges (boxes)] ± SEM and outliers (A).
Both p15 and p16 H scores were lower, on average, in melanomas compared with nevi. However, p16 H score variability was somewhat greater in melanomas than was p15 H score variability, yet the difference in mean p15 H score between nevi and melanomas was greater. This finding shows why statistical significance was achieved.
To determine the value of p15 and p16 H scores as diagnostic tools, we calculated the positive and negative predictive values associated with H score test cutoffs between 0 and 370 (Figure 2B). Cutoffs between 160 and 200 for p15 showed positive predictive values of about 91% and negative predictive values of nearly 80%. These values were stronger than those from the best available test using p16 H score.
The receiver operating characteristic curve depicts the tradeoff between test sensitivity and specificity as the test threshold varies (Figure 2C). Regardless of which false-positive rate was considered, there was a p15 H score–based test that provided a higher true-positive rate.
To validate the biomarker, the original cohort was randomly split into two: one cohort for selecting an optimal threshold, and another cohort for validation. The bootstrap method replicated this procedure 1000 times to resample from the original sample and to reselect cohorts. From this result, the mean threshold values, validation statistics, and SEMs were calculated. The findings from this analysis show that by randomly redrawing half of the sample, the mean optimal test threshold H scores were 191.3 for p15 and 108.6 for p16 (Supplemental Table S1). For p15 staining, the following validation statistics were obtained: positive predictive value, 0.84; negative predictive value, 0.78; specificity, 0.86; and sensitivity, 0.75 (Supplemental Table S1). In contrast, p16 staining yielded a positive predictive value of 0.77; negative predictive value, 0.65; specificity, 0.82; and sensitivity, 0.54 (P < 0.001 for all values). By all four statistical measures, the p15 H score produced a superior test.
RT-qPCR Analysis of p15 in Nevi and Melanomas
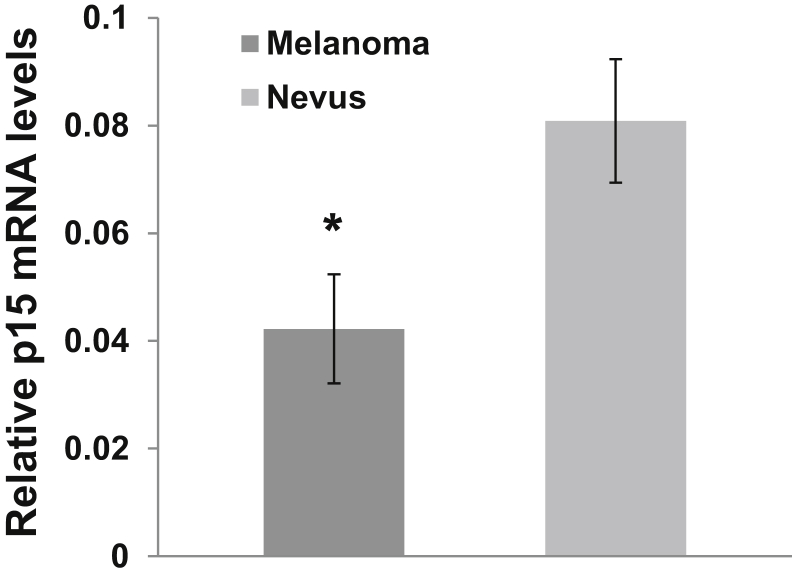
To determine whether p15 was differentially expressed between nevi and melanomas, RT-qPCR analysis of p15 transcript was performed on cDNA samples derived from lesional RNA. The relative levels of p15 transcript were determined using the 2ΔCt method using GADPH as a control. Melanoma cDNA samples from p15 transcripts demonstrated a means ± SEM qΔCt of 0.0422 ± 0.010, while nevi had a mean qΔCt of 0.0808 ± 0.011 (Figure 3). The difference in the qΔCt values between melanomas and nevi was statistically significant, with P < 0.05 (Figure 2).
Figure 3.
Relative levels of p15 transcript in melanomas and nevi. RNA was isolated from macrodissected melanomas and nevi, and cDNA was synthesized for real-time quantitative RT-PCR analysis of p15 transcript. Data are expressed as means ± SEM qΔCt of the p15 products in the melanoma and nevus samples, using glyceraldehyde-3-phosphate dehydrogenase as the control. n = 11 melanomas; n = 14 nevi. ∗P < 0.05 versus nevi.
Discussion
The current study supports the hypothesis that p15 expression is a promising biomarker distinguishing nevus from melanoma. This work shows that staining for p15 may be a diagnostic tool useful for evaluating nevi and melanomas. Results showed a strong and robust relationship between p15 H score and the diagnosis of melanoma, with melanomas showing substantially lower p15 expression than did nevi. Compared with the current standard, p16, this marker shows additional promise. The increased staining for p16 in nevi compared with melanoma was consistent with findings from the literature, although in the current study the difference was not significant (P = 0.055).23 After conditioning on p15 H score in a multivariate logistic model, the p16 H score provided no additional discernible information.
A binary test for melanoma based on H score uses a p15 H score cutoff of 191.3, with a mean positive predictive value of 84% and a negative predictive value of 78%, a mean specificity of 86%, and a mean sensitivity of 75%, dominating any such test based on p16 H score. IHC staining for Wilms tumor protein in melanomas and the lack of staining in nevi had demonstrated utility in distinguishing melanomas from nevi.24 In that study, which used cutoff points similar to those in the current study, Wilms tumor protein exhibited a positive predictive value of 92.6% and a negative predictive value of 51.0%, values less robust than those of p15. Despite this difference, it would be interesting to determine whether analyses of p15 and Wilms tumor protein together would provide added benefit over the use of either marker alone. Further studies of p15 will help to validate this IHC stain, but the current data are encouraging.
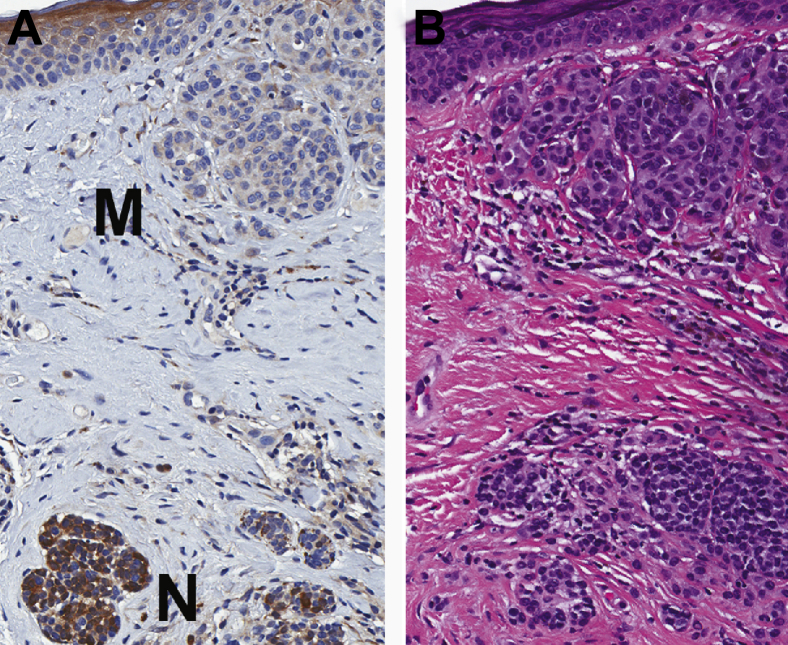
Qualitatively, the p15 stain seemed especially beneficial in cases of melanomas and nevomelanocytic precursor lesions. In the six cases reviewed, there was a clear difference in the strong nevic staining and the much lighter–stained melanoma cells (Figure 4).
Figure 4.
Representative IHC analysis staining of p15 in a melanoma arising in a nevus. A: p15 Staining of a melanoma (M) arising from a nevus (N). Note the intense staining present in the nevus versus minimal staining in the melanoma. B: The corresponding area of the hematoxylin and eosin–stained section. Original magnification, ×50.
RT-qPCR analysis demonstrated that, on average, melanomas showed lower expression of p15 than did nevi, which corresponds to the IHC data. These data suggest that decreased p15 staining in melanomas may be secondary to decreased mRNA levels. These findings are consistent with those from a prior study that correlated higher p16 mRNA levels with stronger p16 IHC staining in nevi compared with melanomas.23 The p15 IHC and RT-qPCR data indicate that mechanisms regulating p15 transcript or mRNA stability may play a role in melanoma formation.
While the activating BRAF(V600E) mutation appears sufficient for inducing transient cell proliferation resulting in nevi, this growth phase is followed by proliferative arrest that typically persists for a lifetime.5, 6, 11 Although these growth-arrested melanocytes can exhibit senescent features, the mitotic arrest is not permanent, since almost half of melanomas likely arise from nevi that have acquired additional changes that restore proliferative capacity.10, 25, 26 Genetic changes associated with melanoma formation include inactivation of tumor suppressors such as CDK inhibitors 2A/B, phosphatase and tensin homolog, and cellular tumor antigen p53. Activation of mitotic drivers such as GTPase NRas, protein kinase B, mast/stem cell growth factor receptor Kit, epidermal growth factor receptor, or serine/threonine protein kinases B-raf/c-Raf dimers has also been found.13, 27 Ongoing research will determine the role that p15 plays in melanomagenesis and how genetic and epigenetic mechanisms regulate levels of this important cell-cycle regulator.
Footnotes
Supported by the Department of Dermatology, Perelman School of Medicine, University of Pennsylvania; Dermatology Foundation Career Development Award (E.Y.C.); NIH grant RO1 CA-163566 (T.W.R.); and NIH grant RO1 CA-165836 (J.T.S.). This work was performed entirely outside of official time and did not employ the use of any government resources (K.H.).
Disclosures: None declared.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2016.08.009.
Supplemental Data
References
- 1.Bastian B.C. The molecular pathology of melanoma: an integrated taxonomy of melanocytic neoplasia. Annu Rev Pathol. 2014;9:239–271. doi: 10.1146/annurev-pathol-012513-104658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies H., Bignell G.R., Cox C., Stephens P., Edkins S., Clegg S. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 3.Garnett M.J., Marais R. Guilty as charged: B-RAF is a human oncogene. Cancer Cell. 2004;6:313–392. doi: 10.1016/j.ccr.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 4.Gill M., Celebi J.T. B-RAF and melanocytic neoplasia. J Am Acad Dermatol. 2005;53:108–114. doi: 10.1016/j.jaad.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 5.Pollock P.M., Harper U.L., Hansen K.S., Yudt L.M., Stark M., Robbins C.M., Moses T.Y., Hostetter G., Wagner U., Kakareka J., Salem G., Pohida T., Heenan P., Duray P., Kallioniemi O., Hayward N.K., Trent J.M., Meltzer P.S. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 6.Michaloglou C., Vredeveld L.C., Soengas M.S., Denoyelle C., Kuilman T., van der Horst C.M., Majoor D.M., Shay J.W., Mooi W.J., Peeper D.S. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 7.Marks R., Dorevitch A.P., Mason G. Do all melanomas come from “moles”? A study of the histological association between melanocytic naevi and melanoma. Australas J Dermatol. 1990;31:77–80. doi: 10.1111/j.1440-0960.1990.tb00656.x. [DOI] [PubMed] [Google Scholar]
- 8.Bevona C., Goggins W., Quinn T., Fullerton J., Tsao H. Cutaneous melanomas associated with nevi. Arch Dermatol. 2003;139:1620–1624. doi: 10.1001/archderm.139.12.1620. [discussion 1624] [DOI] [PubMed] [Google Scholar]
- 9.Rivers J.K. Is there more than one road to melanoma? Lancet. 2004;363:728–730. doi: 10.1016/S0140-6736(04)15649-3. [DOI] [PubMed] [Google Scholar]
- 10.Tsao H., Bevona C., Goggins W., Quinn T. The transformation rate of moles (melanocytic nevi) into cutaneous melanoma: a population-based estimate. Arch Dermatol. 2003;139:282–288. doi: 10.1001/archderm.139.3.282. [DOI] [PubMed] [Google Scholar]
- 11.McNeal A.S., Liu K., Nakhate V., Natale C.A., Duperret E.K., Capell B.C., Dentchev T., Berger S.L., Herlyn M., Seykora J.T., Ridky T.W. CDKN2B loss promotes progression from benign melanocytic nevus to melanoma. Cancer Discov. 2015;5:1072–1085. doi: 10.1158/2159-8290.CD-15-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chin L., Garraway L.A., Fisher D.E. Malignant melanoma: genetics and therapeutics in the genomic era. Genes Dev. 2006;20:2149–2182. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- 13.Hodis E., Watson I.R., Kryukov G.V., Arold S.T., Imielinski M., Theurillat J.P. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruis N.A., van der Velden P.A., Sandkuijl L.A., Prins D.E., Weaver-Feldhaus J., Kamb A., Bergman W., Frants R.R. Homozygotes for CDKN2 (p16) germline mutation in Dutch familial melanoma kindreds. Nat Genet. 1995;10:351–353. doi: 10.1038/ng0795-351. [DOI] [PubMed] [Google Scholar]
- 15.Roh M.R., Eliades P., Gupta S., Tsao H. Genetics of melanocytic nevi. Pigment Cell Melanoma Res. 2015;28:661–672. doi: 10.1111/pcmr.12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haferkamp S., Scurr L.L., Becker T.M., Frausto M., Kefford R.F., Rizos H. Oncogene-induced senescence does not require the p16(INK4a) or p14ARF melanoma tumor suppressors. J Invest Dermatol. 2009;129:1983–1991. doi: 10.1038/jid.2009.5. [DOI] [PubMed] [Google Scholar]
- 17.Serrano M., Hannon G.J., Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 18.Jeffrey P.D., Tong L., Pavletich N.P. Structural basis of inhibition of CDK-cyclin complexes by INK4 inhibitors. Genes Dev. 2000;14:3115–3125. doi: 10.1101/gad.851100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., Antipin Y., Reva B., Goldberg A.P., Sander C., Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glendening J.M., Flores J.F., Walker G.J., Stone S., Albino A.P., Fountain J.W. Homozygous loss of the p15INK4B gene (and not the p16INK4 gene) during tumor progression in a sporadic melanoma patient. Cancer Res. 1995;55:5531–5535. [PubMed] [Google Scholar]
- 21.Robinson W.A., Elefanty A.G., Hersey P. Expression of the tumour suppressor genes p15 and p16 in malignant melanoma. Melanoma Res. 1996;6:285–289. doi: 10.1097/00008390-199608000-00002. [DOI] [PubMed] [Google Scholar]
- 22.Tukey J.W. Addison-Wesley; Boston, MA: 1977. Exploratory Data Analysis. [Google Scholar]
- 23.Keller-Melchior R., Schmidt R., Piepkorn M. Expression of the tumor suppressor gene product p16INK4 in benign and malignant melanocytic lesions. J Invest Dermatol. 1998;110:932–938. doi: 10.1046/j.1523-1747.1998.00211.x. [DOI] [PubMed] [Google Scholar]
- 24.Perry B.N., Cohen C., Govindarajan B., Cotsonis G., Arbiser J.L. Wilms tumor 1 expression present in most melanomas but nearly absent in nevi. Arch Dermatol. 2006;142:1031–1034. doi: 10.1001/archderm.142.8.1031. [DOI] [PubMed] [Google Scholar]
- 25.Gruber S.B., Barnhill R.L., Stenn K.S., Roush G.C. Nevomelanocytic proliferations in association with cutaneous malignant melanoma: a multivariate analysis. J Am Acad Dermatol. 1989;21:773–780. doi: 10.1016/s0190-9622(89)70253-x. [DOI] [PubMed] [Google Scholar]
- 26.Skender-Kalnenas T.M., English D.R., Heenan P.J. Benign melanocytic lesions: risk markers or precursors of cutaneous melanoma? J Am Acad Dermatol. 1995;33:1000–1007. doi: 10.1016/0190-9622(95)90294-5. [DOI] [PubMed] [Google Scholar]
- 27.Shain A.H., Yeh I., Kovalyshyn I., Sriharan A., Talevich E., Gagnon A., Dummer R., North J., Pincus L., Ruben B., Rickaby W., D'Arrigo C., Robson A., Bastian B.C. The genetic evolution of melanoma from precursor lesions. N Engl J Med. 2015;373:1926–1936. doi: 10.1056/NEJMoa1502583. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.