Abstract
Increased polyamine synthesis is known to play an important role in prostate cancer. We aimed to explore its functional significance in prostate tumor initiation and its link to androgen receptor (AR) signaling. For this purpose, we generated a new cell line derived from normal epithelial prostate cells (RWPE-1) with overexpression of ornithine decarboxylase (ODC) and used it for in vitro and in vivo experiments. We then comprehensively analyzed the expression of the main metabolic enzymes of the polyamine pathway and spermine abundance in 120 well-characterized cases of human prostate cancer and high-grade prostate intraepithelial neoplasia (HGPIN). Herein, we show that the ODC-overexpressing prostate cells underwent malignant transformation, revealing that ODC is sufficient for de novo tumor initiation in 94% of injected mice. This oncogenic capacity was acquired through alteration of critical signaling networks, including AR, EIF2, and mTOR/MAPK. RNA silencing experiments revealed the link between AR signaling and polyamine metabolism. Human prostate cancers consistently demonstrated up-regulation of the main polyamine enzymes analyzed (ODC, polyamine oxidase, and spermine synthase) and reduction of spermine. This phenotype was also dominant in HGPIN, rendering it a new biomarker of malignant transformation. In summary, we report that ODC plays a key role in prostate tumorigenesis and that the polyamine pathway is altered as early as HGPIN.
The polyamine pathway has been reported to play an essential role in cell growth and mitochondrial functions.1, 2, 3, 4 The ornithine decarboxylase (ODC) enzyme, encoded by the ODC1 gene, has been identified as the critical rate-limiting enzyme in this metabolic pathway.5, 6 It has been previously reported that ODC is a transcriptional target of the androgen receptor (AR),7 with its potential oncogenic properties being ascribed to the pleiotropic effects of AR-responsive biology in prostate cancer, including aberrant metabolic activities.
In a previous report describing ODC overexpression in prostate cancer, the number of prostate cancer cases examined was limited compared to the numbers of normal prostate and benign prostatic hyperplasia tissue samples assessed.6 Normal prostatic glands have high levels of polyamines, mainly spermine, whereas in tumor tissue, spermine levels are reduced.8 Studies by our group and others using noninvasive proton magnetic resonance spectroscopy have shown that the level of polyamines is higher than the choline level significantly more often in benign prostate tissue than in prostate cancer.9, 10, 11, 12
The chain of molecular events involved in tumor initiation [ie, the transformation of normal epithelium to high-grade prostate intraepithelial neoplasia (HGPIN) and to invasive cancer] has been difficult to analyze in the entire prostate gland because of the multifocal nature and heterogeneity of prostate tumors.13 Several candidate genes and pathways have been implicated in prostate cancer development and progression, including PTEN, p27/KIP1, and Nkx3.1 tumor suppressor genes, as well as the oncogenes c-myc and Bcl-2.14, 15, 16 Gene fusions involving TMPRSS2 and several ETS family members have been found in a high percentage of prostate cancers, and it has been suggested that they may play an important role in prostate tumorigenesis.17 These gene fusions have been found in at least 16% of HGPIN lesions, and all of the HGPIN lesions with these gene fusions were associated with cancer that shared the same fusion pattern; however, 60% of TMPRSS2-ERG fusion prostate cancers had associated fusion-negative HGPIN.18 Recently, genomic alterations observed in prostate cancer genomes were found to be direct descendants of HGPIN, although more genetic changes are required for progression.19 Hence, there is a critical need to explore the implications of polyamines in prostate tumor initiation and to reveal the mechanisms by which the main enzymes elicit their tumorigenic effects.
Given the importance of ODC in prostate cancer, we generated a new normal prostate cell line overexpressing ODC to mechanistically elucidate, for the first time, ODC's potential oncogenic role. This study is also the first to comprehensively analyze the expression of the main metabolic enzymes and abundance of the critical amine of the polyamine pathway in a large and well-annotated cohort of patients with prostate cancer, including cases of HGPIN.
Materials and Methods
Cell Culture
The RWPE1 cell line was purchased from ATCC (Manassas, VA) and maintained in RPMI 1640 medium with 20% fetal bovine serum, according to manufacturer's protocols, at 37°C in a humidified air atmosphere at 5% CO2. RWPE1 cells were generated by immortalization with HPV16 in 1997 by Bello et al20 and characterized as nontumorigenic and noninvasive. The metastatic prostate cancer cells CWR22, LNCaP, and VCaP were purchased from ATCC and maintained in RPMI 1640 medium and Dulbecco's modified Eagle's medium with 10% fetal bovine serum at 37°C in a humidified air atmosphere at 5% CO2.
Gene Silencing and Transient and Stable Transfection and Infection
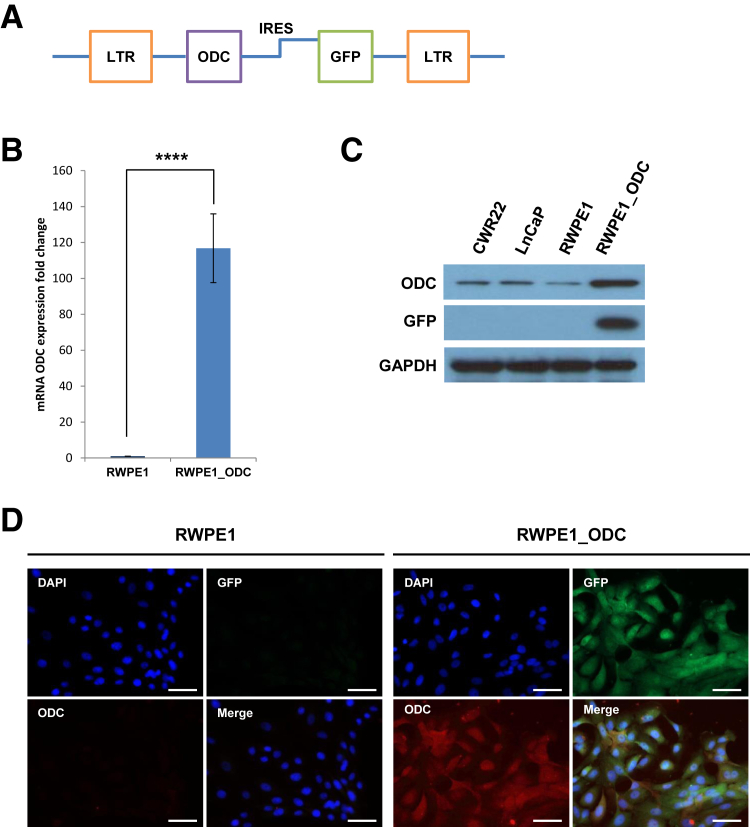
We generated a stable cell line (named RWPE1_ODC) overexpressing ODC. ODC cDNA [catalog number MHS1011-7509233 Human MGC-verified FL Cdna (IRAU) clone ID: 5088190; Thermo Fisher Scientific, Lafayette, CO] was cloned into a SFG-CBR IRES green fluorescent protein vector (courtesy of Dr. Vladimir Ponomarev) (Figure 1A), as previously described.21 The 293-GPG cells (courtesy of Dr. Vladimir Ponomarev, Memorial Sloan Kettering Cancer Center, New York, NY) were used as a retroviral packaging system and were transfected using x-tremeGENE 9 DNA Transfection Reagent (Roche, Indianapolis, IN). The newly generated cells were then sorted for green fluorescent protein, and overexpression of ODC in the stable RWPE1_ODC cell line at the mRNA level was assessed by quantitative RT-PCR (RT-qPCR) (Figure 1B), and at the protein level by immunoblot and immunofluorescence (Figure 1, C and D).
Figure 1.
Generation and validation of a stable cell line overexpressing ornithine decarboxylase (ODC). A: Schematic representation of the construct generated to overexpress ODC in RWPE-1 cells. B: Quantification of three independent experiments of the relative fold increase of ODC mRNA levels in the RWPE1_ODC compared to the parental cells (t-test). C: Representative immunoblot of ODC and green fluorescent protein (GFP) in RWPE1 and RWPE1_ODC compared to prostate cancer cell lines CWR22 and LnCaP. D: Representative immunofluorescence analysis of ODC and GFP. RWPE1, parental cell line; RWPE1_ODC, stable cell line overexpressing ODC. n = 3 for each cell line for experiment (B). ∗∗∗∗P < 0.0001. Scale bars = 100 μm (D). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IRES, internal ribosome entry site; LTR, long terminal repeats.
To address the functional link between ODC and AR, we transiently infected the prostate cancer cells LNCaP and VCaP with two independent siRNA duplexes targeted to human ODC-1 (SASI_Hs01_00223603 and SASI_Hs01_00223604; Sigma-Aldrich, St. Louis, MO), and one with siGenome nontarget siRNA (D-001210-02-20; GE Dharmacon, Lafayette, CO). A total of 1 × 105 cells were seeded in 6-well plates and cultured in media with 10% charcoal-stripped fetal bovine serum for 48 hours. Transfections were then performed using Lipofectamine RNAiMAX (Invitrogen, Life Technologies, Grand Island, NY) with 50 nmol/L of indicated siRNA in Opti-MEM reduced serum media. Cells were recovered into media with charcoal-stripped fetal bovine serum at 12 hours after transfection. At 48 hours after transfection, cells were treated with 10 nmol/L dihydrotestosterone (Sigma-Aldrich), followed by RT-qPCR after 6 hours of treatment and immunoblotting after 24 hours of treatment.
RNA Extraction
RNA was extracted from cell lines using the RNeasy Mini Kit and QIAshredder (Qiagen, Valencia, CA), following the manufacturer's protocol, to perform both RT-qPCR and gene expression analyses.
RT-qPCR
First, strand cDNA was synthesized using Oligo dT and the SuperScript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA), following the manufacturer's instructions. RT-qPCR was performed on a Mastercycler epgradient S realplex (Eppendorf, Enfield, CT) in accordance with the manufacturer's instructions. In a final volume of 20 μL, the RT-qPCR contained 10 μL of 2× QuantiTect SYBR Green PCR Master Mix (Qiagen, Valencia, CA), 1 μL of cDNA, and 2 μL of the primers. The primers used are summarized in Table 1. The thermal cycling conditions included the following: initial denaturation at 95°C for 15 minutes, followed by 40 cycles of denaturation at 94°C for 30 seconds, annealing at 60°C for 40 seconds, and elongation at 72°C for 30 seconds. The PCR products were analyzed by melting curve analysis to confirm that no by-products were formed. The relative concentrations of the PCR products derived from the target gene were calculated using realplex software version 2.0 (Eppendorf) and the ΔΔCT method. All samples were run in triplicate and were normalized against actin (n = 3 in each of the three experiments performed).
Table 1.
List of Primers Used for RT-qPCR
| Gene | Primer | Sequence |
|---|---|---|
| ODC | Forward | 5′-ACGTTCAATGGCTTCCAGAG-3′ |
| Reverse | 5′-ATTGCTGCATGAGTTGCCACGCA-3′ | |
| Probe | 5′-ATGTGATGTCAGGGCCTGC-3′ | |
| PSA | Forward | 5′-ACGCTGGACAGGGGGCAAAAG-3′ |
| Reverse | 5′-GGGCAGGGCACATGGTTCACT-3′ | |
| TMPRSS2 | Forward | 5′-CAGGAGTGTACGGGAATGTGATGGT-3′ |
| Reverse | 5′-GATTAGCCGTCTGCCCTCATTTGT-3′ | |
| FKBP5 | Forward | 5′-TCTCATGTCTCCCCAGTTCC-3′ |
| Reverse | 5′-TTCTGGCTTTCACGTCTGTG-3′ | |
| AR | Forward | 5′-CAGTGGATGGGCTGAAAAAT-3′ |
| Reverse | 5′-GGAGCTTGGTGAGCTGGTAG-3′ | |
| ERG | Forward | 5′-CACGAACGAGCGCAGAGTTAT-3′ |
| Reverse | 5′-CTGTACTCCATAGCGTAGGATCT-3′ | |
| RPLP0 | Forward | 5′-GCTTCCTGGAGGGTGTCC-3′ |
| Reverse | 5′-GGACTCGTTTGTACCCGTTG-3′ |
RT-qPCR, quantitative RT-PCR.
Gene Expression Analyses
Whole-genome gene expression profiling was performed by the Genomics CORE using the HumanHT-12 Expression BeadChip (Illumina Inc., San Diego, CA), following manufacturer's instructions, in RWPE1_P and RWPE1_ODC (n = 3 for each cell line). The bioinformatics analyses were performed using the Bioconductor package version 3.4 (Roswell Park Cancer Institute, Buffalo, NY), TIGR Multiple Experiment Viewer software version 4.0,22 and Significance Analysis of Microarrays software version 4.0 (Standford University, Stanford, CA). Genes differentially expressed were assessed by the DAVID Bioinformatics Resources for enrichment analysis of gene ontology categories. Gene ontology categories enriched on the highest hierarchical level (level 5 or higher) at statistical significance (P < 0.01) were taken into consideration. Pathway analyses were performed using the Ingenuity System. Data are publicly available through the Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo; accession number GSE82179).
Immunoblot Analysis
For immunoblotting, cells were lysed in radioimmunoprecipitation assay lysis buffer (Boston BioProducts, Ashland, MA), including protease inhibitor cocktail (Roche). Proteins were resolved by 4% to 20% SDS-PAGE and transferred to a nitrocellulose membrane. Blots were blocked with 5% nonfat dry milk and probed with primary antibodies as follows: green fluorescent protein (rabbit polyclonal; Abcam, Cambridge, MA), β-actin (mouse monoclonal, clone AC-15; Sigma-Aldrich), p44/42 mitogen-activated protein kinase (MAPK; rabbit monoclonal, clone 137F5; Cell Signaling Technology, Danvers, MA), phospho-p44/42 MAPK (Thr202/Tyr204, rabbit monoclonal, clone 20G11; Cell Signaling Technology), mammalian target of rapamycin (mTOR; rabbit monoclonal, clone 7C10; Cell Signaling Technology), phospho-mTOR (Ser2448, rabbit polyclonal; Cell Signaling Technology), p70S6 kinase (rabbit polyclonal; Cell Signaling Technology), phospho-4E-BP1 (Thr37/46, rabbit monoclonal, clone 236B4; Cell Signaling Technology), erythroblast transformation-specific–related gene (rabbit monoclonal, clone EPR3864; Abcam), AR (N-20, rabbit polyclonal; Santa Cruz Biotechnology Inc., Dallas, TX), prostate-specific antigen (PSA; K92110R; Meridian Life Science, Memphis, TN), as well as the same antibodies against ODC, polyamine oxidase (PAO), spermine synthase (SPSY), and spermine used for immunofluorescence (see below). Horseradish peroxidase–conjugated anti-rabbit and anti-mouse secondary antibodies (GE Healthcare Life Sciences, Piscataway, NJ) were used. Reactive bands were visualized using electrochemiluminescence plus Western blotting detection reagents (GE Healthcare Life Sciences).
DFMO Treatment
Cells were treated with d,l-α-difluoromethylornithine (DFMO; Santa Cruz Biotechnology Inc.) at 100 μmol/L (suspended in distilled sterile water) for up to 72 hours to inhibit ODC activity, as previously described.23 Proliferation assays were performed as described below with cells in regular medium and cells with DFMO.
Putrescine Treatment
RWPE-1 cells were treated with putrescine (Sigma-Aldrich) at 100 μmol/L (suspended in distilled water), as previously described.24 Proliferation and invasion assays were performed as explained below with RWPE-1 cells in regular medium and medium with putrescine.
Cell Proliferation Assay
A total of 2 × 103 cells were seeded in a 96-well plate. Cell viability was determined at 24, 48, 72, 96, 120, and 144 hours by using the CellTiter 96 Aqueous One Solution Cell Proliferation Assay kit (Promega, Madison, WI), according to the manufacturer's protocol. Absorbance was measured at 490 and 655 nm (used as a background reading) in a Synergy H1 Hybrid multimode microplate reader (Biotek, Winooski, VT). Data are presented as the mean value of three separate experiments for sixtuplicate wells (n = 6 for each time point for cell line in each of the three experiments).
Cell Cycle Analysis
For both cell lines, 1 × 106 cells were seeded in a 10-cm plate and allowed to grow for 24 hours. Cells were then harvested, pelleted, and fixed with 70% ethanol for 12 hours and washed with 1× phosphate-buffered saline. Cells were stained with propidium iodide solution at 5% in Triton–phosphate-buffered saline for 20 minutes at room temperature, and analyzed in a BD LSRII flow cytometer (BD Biosciences, San Jose, CA). Cell cycle phases were analyzed with the FlowJo software version 9.9.4 (TreeStar, Ashland, OR) in three independent experiments (n = 3 for each cell line in each of the three experiments).
In Vitro Invasion Assay
In vitro invasion assays were performed using BD BioCoat Matrigel Invasion Chambers (8-μm pore; BD Biosciences, San Jose, CA) in 24-well tissue culture plates, according to manufacturer's protocol. Cells were placed in serum-free media 24 hours before invasion assay set-up. Then, 5 × 105 cells were seeded per well and allowed to invade toward the underside of the chamber for 48 hours at 37°C. After incubation, Calcein AM solution (BD Biosciences) was added, and fluorescence of invaded cells was read at a wavelength of 494 to 517 nm (excitation/emission) on a SpectraMax M5e Multimode microplate reader (Molecular Devices, Sunnyvale, CA). Membranes were then stained with DAPI, and cell invasion was assessed using a Fluorescent Nikon Eclipse 50i microscope (Morrell Instruments Co., Inc., Melville, NY). Experiments were performed three times in duplicates (n = 6 for each time point for cell line in each of the three experiments).
Mouse Experiments
All assays were performed on 5- to 6-week-old male nonobese diabetic severe combined immunodeficiency γ mice purchased from Jackson Laboratories (Bar Harbor, ME). Animal use and care followed institutional guidelines established by Icahn School of Medicine at Mount Sinai and its Institutional Animal Care and Use Committee. For the tumor xenograft experiments, 1 × 106 RWPE1_P and RWPE1_ODC cells were injected in nonobese diabetic severe combined immunodeficiency γ mice. Three independent experiments were performed, including eight mice for each arm (parental and ODC-overexpressing cells) and mice were injected twice, in the right and left flanks (n = 16 for each cell line in each of the three experiments). Tumors were allowed to develop for up to 6 months or whenever they reached a size of 1 cm3. If a tumor developed in one flank to the size limit, we surgically removed it and let the mouse be alive up to 6 months. Tumors were measured twice every week with a caliper, and histologically analyzed after sacrificing the mice after 6 months, using cell injection, resection, and fixation with 10% formalin.
Study Design for Human Samples
A sample size calculation was performed for the differential expression of four biomarkers (three enzymes and spermine) across the two groups (normal and diseased). A minimum desired fold difference of 1.5 at a power level of 0.9 (rather than the traditional 0.8) was used, as a robust result for the biomarker was desired. Varying degrees of SD were modeled up to a maximum of 1.5, suggesting a sample size range of 35 to 78 samples in each group. A total of 128 prostate cancer patients who underwent radical prostatectomy between January 1991 and December 2004 and who had not received any adjuvant therapy were included in the study. The protocol was approved by the institutional review board and was compliant with the Health Insurance Portability and Accountability Act. Follow-up clinical information was available from 120 (93.75%) patients (Table 2). The primary outcome measure was the time (in months) to biochemical recurrence [defined as two consecutive detectable increasing PSA levels (>0.2 ng/mL) 4 weeks or more after surgery] or specific mortality after surgery.
Table 2.
Patient Clinicopathological Characteristics (n = 120)
| Characteristics | Value |
|---|---|
| Age at diagnosis, years∗ | 60.8 (40–74) |
| Preoperative PSA, ng/mL∗ | 11.3 (0.9–118.0) |
| Preoperative PSA, risk stratification†‡ | |
| <10 ng/mL | 75 (65.2) |
| 10–20 ng/mL | 26 (22.6) |
| >20 ng/mL | 14 (12.2) |
| Preoperative PSA, dichotomous†‡ | |
| <20 ng/mL | 101 (87.8) |
| ≥20 ng/mL | 14 (12.2) |
| Surgical Gleason score† | |
| 5 | 7 (5.9) |
| 6 | 28 (23.3) |
| 7 | 58 (48.3) |
| 8 | 21 (17.5) |
| 9 | 6 (5.0) |
| Surgical Gleason score, risk stratification† | |
| ≤6 | 35 (29.2) |
| 7 | 58 (48.3) |
| ≥8 | 27 (22.5) |
| Surgical Gleason score, dichotomous† | |
| ≤7 | 93 (77.5) |
| >7 | 27 (22.5) |
| Pathological stage† | |
| pT2 | 53 (44.2) |
| pT3 | 62 (51.6) |
| pT4 | 5 (4.2) |
| Pathological stage, risk stratification† | |
| ≤pT2a | 4 (3.3) |
| pT2b-pT2c | 49 (40.9) |
| ≥pT3 | 67 (55.8) |
| Pathological stage, dichotomous† | |
| Organ confined | 53 (44.2) |
| Non–organ confined | 67 (55.8) |
| BCR†§ | 48 (45.7) |
| BCR time, months∗§ | 62.0 (3.0–187.0) |
| OS time, months∗¶ | 103.6 (3.0–222.0) |
BCR, biochemical recurrence; OS, overall survival; PSA, prostate-specific antigen.
Data are given as mean (range).
Data are given as number (percentage).
Data from five patients were missing.
BCR data were available in 105 patients.
OS data were available in 117 patients.
Tissue Microarrays of Prostate Samples and Patient Cohort
Four tissue microarrays were built following institutional review board–approved protocol. Sections from formalin-fixed, paraffin-embedded prostatectomy specimens were stained with hematoxylin and eosin and reviewed to identify viable, morphologically representative areas of normal prostate glands and acinar adenocarcinoma from which needle core samples could be taken. From each specimen, triplicate tissue cores with diameters of 0.6 mm were punched and arrayed onto a recipient paraffin block using a precision instrument (Beecher Instruments Inc., Sun Prairie, WI). Sections (5 μm thick) of these tissue microarray blocks were stained with hematoxylin and eosin and used for immunofluorescence analysis. These tissue microarrays included acinar prostate adenocarcinoma (n = 120), and matched normal prostate glands (n = 103) as well as additional matched HGPIN (n = 18).
Immunofluorescence Analysis
Immunofluorescence analysis was performed blindly by an experienced uropathologist (M.C.-M.) on sections from these four tissue microarrays following standard staining procedure. Briefly, sections (5 μm thick) were deparaffinized and then submitted to antigen retrieval by steamer treatment for 15 minutes in 10 mmol/L citrate buffer at pH 6.0. Subsequently, sections were incubated in 10% normal donkey serum for 30 minutes, followed by primary antibody incubation overnight at 4°C. Then, slides were incubated with fluorescent-labeled anti-rabbit and anti-mouse immunoglobulins at a 1:200 dilution for 30 minutes (Invitrogen). DAPI was used as nuclear counterstain. Primary antibodies to detect critical enzymes and amines of the polyamine pathway included ODC (mouse monoclonal, clone ODC-29; Abcam), SPSY (rabbit polyclonal; Santa Cruz Biotechnology Inc.), PAO (mouse monoclonal, clone C-3; Santa Cruz Biotechnology Inc.), and spermine (rabbit polyclonal; Abcam). To confirm the sensitivity and specificity of antibodies, positive and negative controls were run in parallel on paraffin-embedded tissue sections. ODC expression was characterized by a fine granular cytoplasmic staining, whereas PAO, SPSY, and spermine showed a diffuse cytoplasmic distribution. The immunoreactivity for each antibody was scored considering the percentage of cells displaying a positive immunostaining profile [from undetectable (0%) to homogeneous expression (100%)]. Average values of the representative cores from each arrayed sample were obtained, and a positive cutoff for statistical analyses was determined at 10% protein expression, which is commonly used as a standard cutoff for immunohistochemical analyses. Slides were analyzed with a Fluorescent Nikon Eclipse 50i microscope (Nikon Instruments, Melville, NY) equipped with a SPOT Insight2MP Mosaic camera and SPOT Advanced software version 5.0 (Diagnostic Instruments, Sterling Heights, MI).
Statistical Analysis
Statistical analyses were performed with SPSS version 19.0 (SPSS Inc., Chicago, IL). For the in vitro analyses, data were displayed as a mean value of three different experiments, and t-test was used to assess significance. Genes significantly dysregulated in RWPE1_ODC cells in comparison to RWPE1 parental lines were identified by using the Limma test P < 0.05 and greater than twofold change. To analyze the associations between protein expression values and clinicopathological variables, we used a t-test when one variable was continuous and the other qualitative and a χ2 test or Fisher exact test when both variables were qualitative. The associations of molecular and clinicopathological variables with biochemical recurrence-free survival and overall survival were assessed using the log-rank test. Survival curves were plotted using standard Kaplan-Meier method. A two-sided value of P < 0.05 was considered statistically significant.
Results
Overexpression of ODC Is Sufficient for Malignant Transformation of Prostate Epithelial Cells
We first analyzed the polyamine pathway in two commercially available metastatic prostate cancer cell lines (CWR22 and LNCap) and the immortalized normal epithelial prostate cell line (RWPE1). Levels of the polyamine enzymes ODC, PAO, and SPSY were higher in the two cancer cell lines than in the normal counterpart, whereas spermine abundance showed the opposite trend (Figure 2A). Because of the critical relevance of ODC, we centered our study on this enzyme to further elucidate its potential role in prostatic tumor initiation and progression. Using RWPE1 cells, we generated a stable cell line overexpressing ODC, which we termed RWPE1_ODC. On stable transfection, RWPE1_ODC cells displayed high levels of ODC compared to their parental counterpart, both at the mRNA level (>100-fold increase) (Figure 1B) and the protein level (>20-fold increase) (Figure 1, C and D).
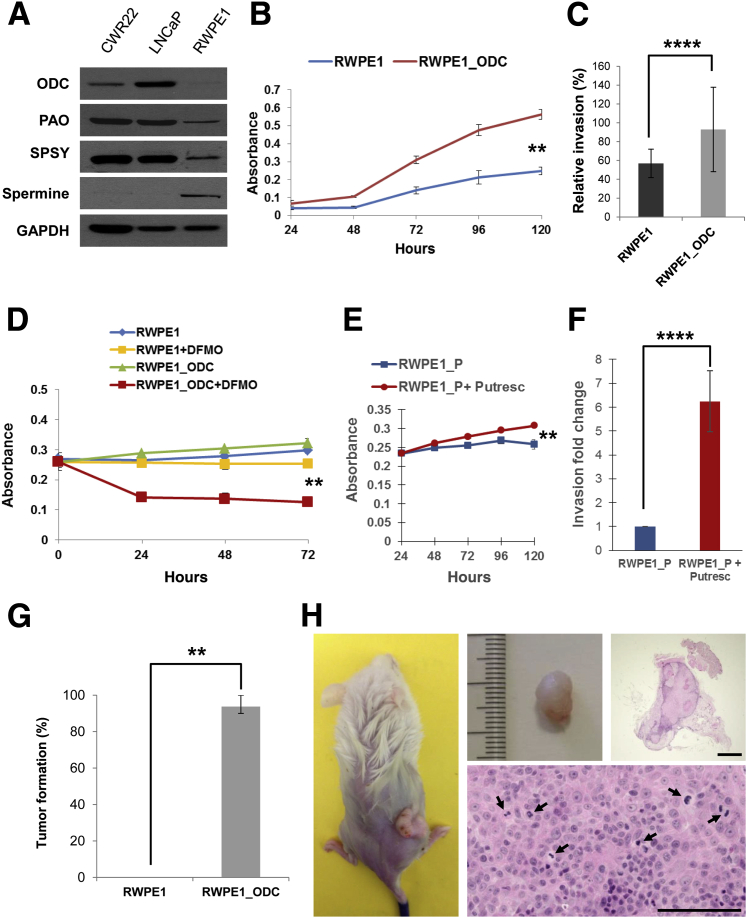
Figure 2.
ODC is critical for malignant transformation in prostate cells. A: Representative immunoblot illustrating the expression of the main enzymes of the polyamine pathway and spermine abundance in the normal RWPE1 cell line, compared to the prostate cancer cell lines CWR22 and LnCaP. B: Quantification of proliferation by MTS assay at 24, 48, 72, 96, and 120 hours after seeding. Three independent experiments for each time point are represented (t-test). C: Quantification of three independent experiments of Matrigel invasion assay (t-test). D: Quantification of proliferation by MTS assay at 24, 48, and 72 hours after seeding and treating with d,l-α-difluoromethylornithine (DFMO). Three independent experiments for each time point are represented (t-test). E: Quantification of proliferation by MTS assay at 24, 48, 96, and 120 hours after seeding and treating with 100 μmol/L of putrescine. Three independent experiments for each time point are represented (t-test). F: Quantification of Matrigel invasion assay comparing RWPE1_P with regular medium or with putrescine treatment. Three independent experiments for each time point are represented (t-test). G: Tumor initiation experiments showed that RWPE1_ODC cells had a high de novo tumor initiation capacity (tumors formed in 45 of 48 injection sites for the RWPE1_ODC cells). Three independent experiments were performed (t-test). H: Gross and microscopic appearance of a representative tumor that formed after s.c. injection of 1 × 106 RWPE1_ODC cells in immunodeficient mice. Arrows point to mitotic figures observed in the tumor. n = 6 for each time point and cell line for experiment (B–F); n = 16 for each cell line (G). ∗∗P < 0.01, ∗∗∗∗P < 0.0001. Scale bars = 200 μm (H). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MTS, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium; ODC, ornithine decarboxylase; PAO, polyamine oxidase; SPSY, spermine synthase.
Proliferative activity was assessed by MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium)] assay, and, as expected, RWPE1_ODC cells showed a significantly higher proliferative index than the parental line (P < 0.01) (Figure 2B). However, as would be expected of diploid populations, parental and RWPE1_ODC cells showed no significant differences on flow cytometry analysis (data not shown). After performing Matrigel invasion assays, we observed that the ODC-overexpressing cells displayed a significantly higher invasive ability (Figure 2C), thus confirming that RWPE1_ODC cells had malignant features.
To determine whether ODC was the critical factor involved in the malignant phenotype, we treated RWPE1_P and RWPE1_ODC cells with a well-known ODC inhibitor (DFMO). This experiment confirmed that the increase in proliferation was because of ODC overexpression, since treated RWPE1_ODC cells showed a significant decrease in proliferation (Figure 2D), whereas the effect in parental cells was much lower. Furthermore, we observed that these malignant features could be phenocopied by putrescine addition, since RWPE1_P cells treated with 100 μmol/L of putrescine showed a significant increase in cell proliferation (Figure 2E) as well as in Matrigel invasion (Figure 2F). This malignant phenotype was subsequently tested by s.c. injecting 1 × 106 cells in 24 nonobese diabetic severe combined immunodeficiency γ mice and monitoring them over a 6-month period. As previously described by the group that generated this normal prostate line, RWPE1 cells were nontumorigenic,20 whereas tumor was palpated in 94% (45/48) of mice after injection with RWPE1_ODC cells (Figure 2G). The mean time to palpation was 48.0 days (range, 29.0 to 81.0 days). Histological analyses revealed that tumors corresponded to high-grade carcinomas and were morphologically characterized by increased mitotic activity and prominent nuclear atypia, including the presence of macronucleoli (Figure 2H). Taken together, these observations validate the hypothesis that the polyamine metabolic pathway is significantly involved in prostate tumorigenesis, and that ODC overexpression is sufficient for tumor initiation.
ODC Malignant Transformation Is Linked to Specific Genetic Alterations
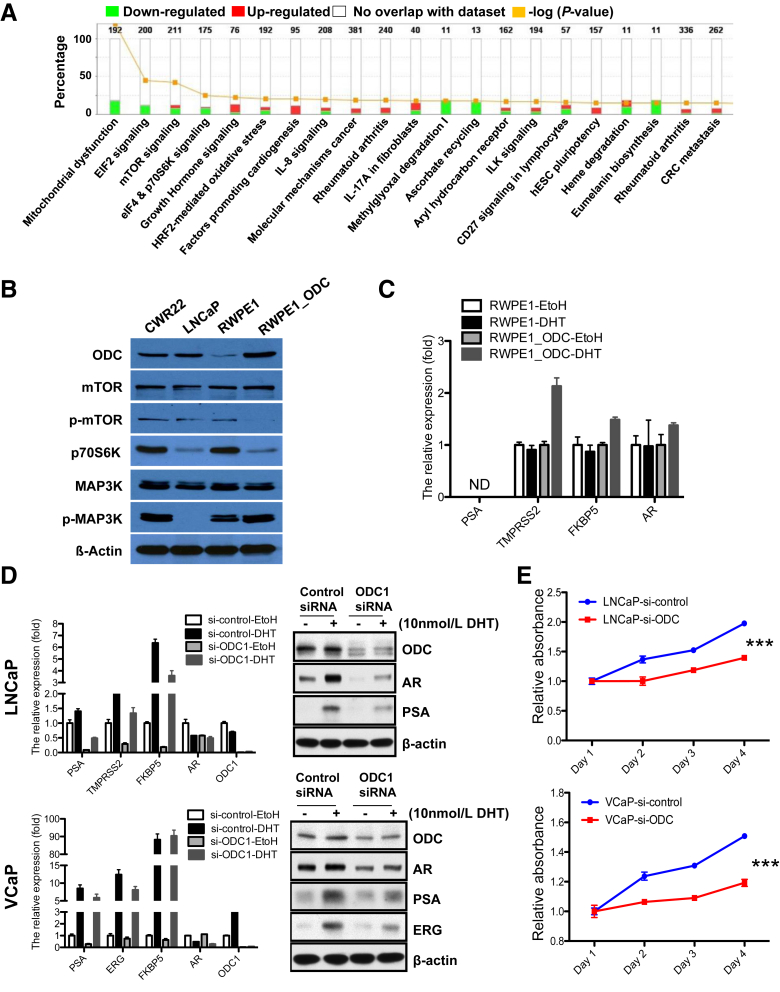
To further characterize the altered genetic pathways after ODC overexpression, we performed gene expression analyses of RWPE1_ODC and parental RWPE1 cells. These studies revealed 113 up-regulated and 234 down-regulated genes in RWPE1_ODC cells (limma test, P < 0.05 and greater than twofold change) (Supplemental Figure S1A). The top 60 up-regulated and down-regulated genes between RWPE1 and RWPE1_ODC are listed in Table 3. As expected, the gene ontology analysis for biological processes showed that many metabolic processes were up-regulated in the cells with an activated polyamine pathway (Figure 3A). Some of the genes that were up-regulated, such as MAPK and MMP13, are associated with increased metabolism and invasion, whereas down-regulated genes included those associated with mitochondrial dysfunction and signaling pathways involved in malignant transformation, such as EIF2 and mTOR (Supplemental Figure S1, B–D). These findings were confirmed at the protein level (Figure 3B). More specifically, p-mTOR and p-70S6kinase expression levels were lower in the RWPE1_ODC cells than in the parental cells. In contrast, p-MAP3K expression was slightly higher in the ODC-overexpressing cells, with protein levels similar to the CWR22 prostate cancer cells. Interestingly, both total mTOR and total MAP3K remained unchanged. Altogether, these results demonstrate, for the first time, that activation of the metabolic pathway is sufficient for prostate tumor initiation, and is linked to specific alterations involving signaling networks critical for maintenance of cellular homeostasis (Supplemental Figure S2A).25
Table 3.
List of the Top 60 Genes of the 113 Up-Regulated and 234 Down-Regulated Genes between RWPE1 and RWPE1_ODC
| Up-regulated genes |
Down-regulated genes |
||||
|---|---|---|---|---|---|
| Gene name | P value | Fold change | Gene name | P value | Fold change |
| HCG2P7 | 0.043362 | 8.699880252 | APTX | 3.52×10-9 | −7.05842823 |
| TXNL2 | 0.049122 | 7.829825014 | ITGB4 | 0.046939 | −6.66785551 |
| AP3M1 | 0.039578 | 6.289572722 | C20ORF7 | 0.049135 | −6.01590958 |
| TRAF3IP2 | 4.29×10-9 | 6.10987277 | TMEM218 | 0.037714 | −5.00290573 |
| MAP3K4 | 8.58×10-9 | 5.673337574 | MFAP2 | 0.040073 | −4.87076243 |
| ARFGAP3 | 2.24×10-8 | 5.611857728 | TSC22D3 | 0.000163 | −4.70222855 |
| OLFM4 | 0.041942 | 4.942136094 | ALAD | 0.029013 | −4.5713245 |
| C10ORF61 | 0.042706 | 4.795673117 | SLC25A25 | 0.043912 | −4.49495234 |
| KIF18A | 5.53×10-7 | 4.119945152 | FBXL5 | 1.07×10-5 | −4.49288692 |
| KLK3 | 0.04459 | 3.973581652 | VEGFA | 0.048416 | −4.07647083 |
| BZW1 | 1.11×10-7 | 3.878693704 | STEAP1 | 0.042833 | −3.8271634 |
| CHKB | 0.03817 | 3.807354922 | SLC6A10P | 9.55×10-8 | −3.6042244 |
| C6ORF108 | 5.82×10-6 | 3.804026904 | ARTN | 5.21×10-5 | −3.59655048 |
| KRT19 | 9.32×10-7 | 3.433820975 | RNF5 | 0.049682 | −3.56543994 |
| HYAL2 | 0.022243 | 3.303191532 | MAP2K5 | 0.035137 | −3.56150353 |
| FAIM | 5.69×10-5 | 3.230679033 | NCRNA00152 | 0.028316 | −3.48459554 |
| TRAF6 | 0.045793 | 3.057030945 | STXBP1 | 5.91×10-7 | −3.37223318 |
| ENTPD3 | 2.08×10-7 | 2.976541027 | DENND4A | 0.042738 | −3.169925 |
| HEY2 | 0.005789 | 2.878693704 | MFI2 | 0.021025 | −3.1490915 |
| VAPA | 0.029251 | 2.865982652 | KIAA1704 | 0.029999 | −3.11726605 |
| SULT1E1 | 0.030167 | 2.827163403 | RPRML | 0.003449 | −2.82059075 |
| RTKN2 | 0.0059 | 2.800691192 | MARCH4 | 0.014532 | −2.63076619 |
| NUPL1 | 2.27×10-5 | 2.766902878 | MIR330 | 0.002721 | −2.52961947 |
| ADAM23 | 8.15×10-5 | 2.696855381 | HS.123511 | 0.005457 | −2.52153712 |
| GPX7 | 0.019206 | 2.521537121 | IFNK | 0.021435 | −2.4970148 |
| PTGS2 | 0.00074 | 2.451893572 | NGF | 0.009074 | −2.27517984 |
| GREM1 | 6.12×10-5 | 2.420843121 | SFTA1P | 0.006957 | −2.24853484 |
| AQP3 | 0.000148 | 2.31259023 | FAIM3 | 0.018611 | −2.1324503 |
| MMP13 | 0.0009 | 2.277909628 | FAM25G | 0.031209 | −2.12637729 |
| ARMCX2 | 0.008776 | 1.974988112 | HS.553310 | 0.005057 | −2.12261929 |
Figure 3.
Polyamine pathway activation regulates critical genetic pathways as well as androgen receptor (AR) signaling. A: Gene ontology pathway analysis of the most frequent and significantly up-regulated and down-regulated genes in RWPE1_ODC cells. B: Representative immunoblot of the most critical proteins [mammalian target of rapamycin (mTOR), p70S6K, and mitogen-activated protein kinase (MAPK)] confirmed the alteration in several genetic pathways involved in cancer at the protein level. C: Quantification of the relative fold increase of well-known AR target mRNA gene levels in the RWPE1_ODC compared to the parental cells (t-test). D: Knocking down of ornithine decarboxylase (ODC) in LNCaP and VCaP cells significantly reduces mRNA and protein expression levels of AR and well-known AR target genes by RT-qPCR and immunoblot, respectively (t-test). E: Proliferation activity is reduced in prostate cancer cells after knocking down of ODC. Three independent experiments were performed (t-test). n = 3 for each cell line for experiment (C and D); n = 6 for each time point and cell line (E). ∗∗∗P < 0.001. DHT, dihydrotestosterone; EtOH, ethanol; ERG, erythroblast transformation-specific–related gene; ND, not detected; PSA, prostate-specific antigen; si, small interfering RNA technology against.
ODC Gene Regulates AR Transcription Targets in Normal and Prostate Cancer Cells
To elucidate whether ODC regulates AR signaling in prostate cells, we analyzed a subset of AR-target genes (eg, PSA, TMPRSS2, FKBP5, and ERG) in RWPE1_P and RWPE1_ODC cells by RT-qPCR. These analyses revealed that these genes were significantly overexpressed in the RWPE1_ODC cells, only after Dihydrotestosterone treatment (Figure 3C). To further confirm this link, we transiently knocked down ODC in two AR-dependent prostate cancer cell lines (LNCaP and VCaP). These studies revealed that AR, as well as other well-known AR-target genes, was significantly down-regulated at both the transcript and protein levels in the small interfering RNA technology against–ODC cells (Figure 3D). Furthermore, when we knocked down ODC in these prostate cancer cells, their proliferative activity diminished significantly (Figure 3E). These results confirm that ODC and AR are mutually regulated and may imply that overexpression of the ODC protein causes malignant transformation through activation of the AR axis, which, in turn, can affect different pathways involved in cell proliferation, cell survival, and tumor invasion.
The Polyamine Pathway Is Up-Regulated in Prostate Cancer, as Early as HGPIN
We followed the above-mentioned mechanistic studies with translational analyses aimed at deciphering the clinical significance of an increased metabolic pathway in prostate cancer (Supplemental Figure S2B). We performed immunophenotyping of the patients' tissues using specific antibodies to ODC, PAO, SPSY, and spermine. Patient characteristics are summarized in Table 2. Fifty-seven patients (54%) had no evidence of biochemical recurrence, with a median follow-up of 62.0 months (range, 3.0 to 187.0 months). Of the patients who showed biochemical relapse after surgery, 19 (40%) died of the disease. At the time of analysis, overall survival was 76%, with a median follow-up of 103.6 months (range, 3.0 to 222 months). The analysis of clinicopathological characteristics in relation to biochemical recurrence-free survival revealed, as expected, that surgical Gleason score (P < 0.0001) and preoperative PSA levels (P = 0.034) were significantly associated with a worse prognosis, thus validating the studied cohort. However, the pathologic stage of tumors did not significantly correlate with biochemical recurrence-free survival (P = 0.400) (Supplemental Figure S3).
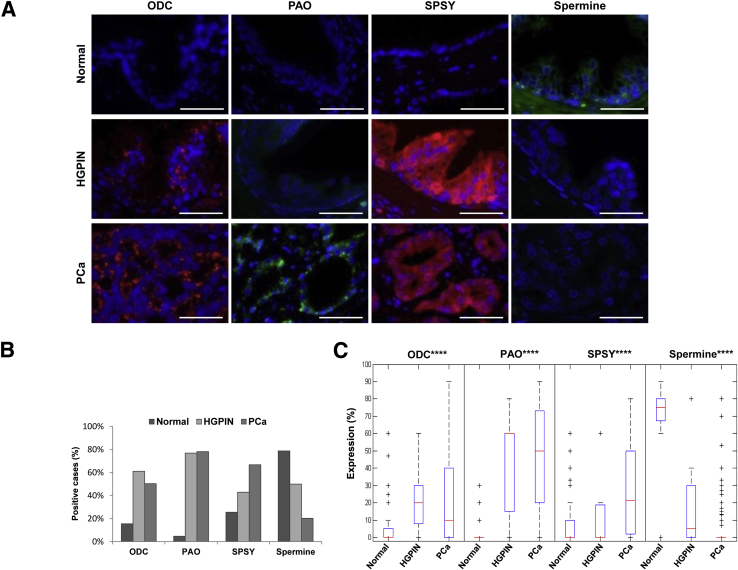
The immunophenotyping analyses revealed that expression of the polyamine enzymes was low in the epithelial cells lining normal prostate glands, whereas epithelial cells in malignant glands, including both HGPIN and prostate cancer samples, showed intense cytoplasmic expression of ODC, PAO, and SPSY (Figure 4A). Notably, spermine abundance showed the opposite trend, being high in normal tissue and almost absent in tumor areas (Figure 4, A and B, and Table 4). After quantification, mean expression values of the three enzymes in normal tissue corresponded to 6.7% ± 13.6% for ODC, 1.0% ± 5.1% for PAO, 7.4% ± 5.1% for SPSY, and 62.2% ± 33.3% for spermine abundance (Figure 4C). Notably, we observed that expression levels of the polyamine enzymes were significantly higher in HGPIN and prostate cancer samples than in normal prostate glands, which had significantly lower abundance of spermine (P < 0.0001) (Figure 4C).
Figure 4.
The polyamine pathway is activated in prostate cancer (PCa) and high-grade prostatic intraepithelial neoplasia (HGPIN) when compared to normal prostatic glands (Normal). A: Representative immunofluorescence analyses of PA pathway enzymes and spermine abundance in normal prostatic glands, HGPIN, and PCa. B: Bar diagram illustrates the percentage of samples with overexpression of PA pathway enzymes and down-regulation of spermine in normal prostate, HGPIN, and PCa (using a cutoff of 10%). C: Box-plot diagram illustrates expression levels of the main PA pathway enzymes and spermine abundance in normal prostate, HGPIN, and PCa. Data are expressed as means ± SD (analysis of variance test; C). ∗∗∗∗P < 0.0001 (analyses of variances test between the three different groups). Scale bars = 100 μm (A). ODC, ornithine decarboxylase; PA, polyamine; PAO, polyamine oxidase; SPSY, spermine synthase.
Table 4.
Samples with Overexpression of PA Pathway Enzymes and Down-Regulation of Spermine in Normal Prostate, HGPIN, and PCa (Using a Cutoff of 10%)
| Variable | Normal | HGPIN | PCa |
|---|---|---|---|
| ODC∗∗∗∗ | 15.5 | 61.1 | 50.4 |
| PAO∗∗∗∗ | 4.7 | 76.9 | 78.2 |
| SPSY∗∗∗∗ | 25.5 | 42.9 | 66.7 |
| Spermine∗∗∗∗ | 78.8 | 50.0 | 20.3 |
Data are given as percentage.
∗∗∗∗P < 0.0001.
HGPIN, high-grade prostate intraepithelial neoplasia; ODC, ornithine decarboxylase; PA, polyamine; PAO, polyamine oxidase; PCa, prostate cancer; SPSY, spermine synthase.
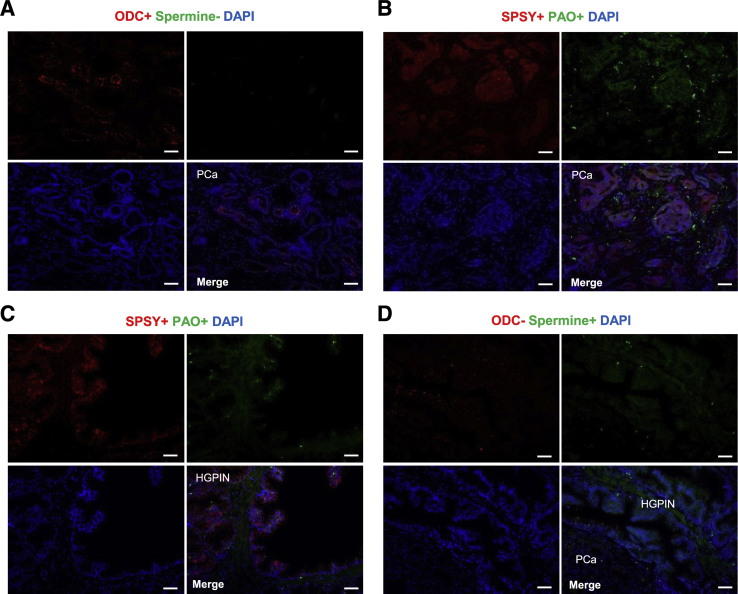
All enzymes showed a direct and significant correlation among themselves (Table 5). For example, of the tumors that were characterized by an ODC-positive phenotype, 72.2% expressed PAO (P < 0.0001) and 63.4% expressed SPSY (P < 0.001). Similarly, 52.3% of tumors with a PAO-positive phenotype overexpressed ODC (P < 0.0001), and 47.3% of tumors with a SPSY-positive phenotype overexpressed ODC (P < 0.001). Furthermore, spermine abundance correlated inversely with the expression of all of the analyzed polyamine pathway enzymes: loss of spermine was observed in 68.8% (P < 0.01), 75.7% (P < 0.0001), and 70.7% (P < 0.0001) of the ODC-positive, PAO-positive, and SPSY-positive tumors, respectively (Table 5). Summarizing these results, we can conclude that there was a consistent pattern of expression in the polyamine pathway (specifically, an overexpression of the three enzymes and loss of spermine) that was characteristic of malignant transformation (Figure 5, A and B), similar to previous reports studying mRNA expression.26, 27, 28 Notably, most of the analyzed HGPIN lesions showed the same immunophenotype as their adjacent prostate cancer lesions (Figure 5C), and only 12% of these HGPIN lesions were characterized by a different pattern, showing high abundance of spermine and lack of expression of ODC (Figure 5D).
Table 5.
PCa Cases with Specific Phenotypes Displayed
| Variable | ODC+ | PAO+ | SPSY+ | Spermine− |
|---|---|---|---|---|
| ODC+ | NA | 72.2∗∗∗∗ | 63.4∗∗∗ | 68.8∗∗ |
| PAO+ | 52.3∗∗∗∗ | NA | 66.0∗∗∗∗ | 75.7∗∗∗∗ |
| SPSY+ | 47.3∗∗∗ | 64.8∗∗∗∗ | NA | 70.7∗∗∗∗ |
| Spermine− | 44.5∗∗ | 67.8∗∗∗∗ | 59.8∗∗∗∗ | NA |
Data are given as percentage.
∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001.
NA, not applicable; ODC, ornithine decarboxylase; PAO, polyamine oxidase; PCa, prostate cancer; SPSY, spermine synthase.
Figure 5.
Prostate cancer is characterized by direct correlation between the expression of polyamine pathway enzymes and inverse correlation of these enzymes with spermine abundance. A–D: Representative immunofluorescence analysis of PA pathway enzymes' expression and spermine abundance in prostate cancer (PCa) and high-grade prostatic intraepithelial neoplasia [HGPIN; ornithine decarboxylase (ODC) and spermine in A and D; polyamine oxidase (PAO) and spermine synthase (SPSY) in B and C]. Scale bars = 100 μm (A–D). PA, polyamine.
Polyamine Pathway Immunophenotypes and Their Association with Clinicopathological Variables and Patient Outcome
When we examined the associations between the analyzed polyamine pathway enzymes/amine and the main clinicopathological variables (Supplemental Table S1), we observed that only the ODC-positive phenotype correlated significantly with Gleason score (P < 0.05) and pathological stage (P < 0.01), whereas the other enzymes and spermine abundance did not. Furthermore, we did not observe any association between polyamine pathway enzymes/amine and biochemical recurrence-free survival (Supplemental Figure S4A) or overall survival (Supplemental Figure S4B).
Discussion
The metabolic polyamine pathway has been previously reported to be critical for the maintenance of homeostatic processes, such as cell growth, apoptosis, mitochondrial function, autophagy, and gene regulation (Supplemental Figure S2A).25 More specifically, ODC, the gene coding for the rate-limiting enzyme of this pathway, has been reported to be a target gene in the pleiotropic androgen-responsive biology of prostate cancer.7, 28 This relationship is also observed in skeletal muscle, where ODC is up-regulated by the androgen receptor, promoting cell proliferation and delayed differentiation, both of which are features of tumor formation.29 Even though some authors have reported how polyamine pathway controls cell cycle,30 ODC functional contribution in prostate tumor initiation has not been fully explored to date.
Others have previously demonstrated the transformation capacity of ODC overexpression in fibroblasts1, 31, 32; nonetheless, this is the first study, to our knowledge, to functionally implicate ODC as a critical and sufficient enzyme in the malignant transformation of prostatic epithelium both in vitro and in vivo. Some earlier studies described that polyamine pathway gene expression signatures were linked to prostate cancer,26, 27 but only a few reports with small cohorts of patients have described the increase of ODC protein expression in prostate cancer as compared to normal prostate tissue.6, 16, 33 In our series of 120 well-characterized prostate cancer patients, we found ODC overexpression in 61% of the 18 HGPIN lesions and 50% of invasive prostate cancer samples analyzed. This suggests that the polyamine pathway enzymes could be involved in the early stages of carcinogenesis and could promote the progression from HGPIN to invasive carcinoma. Previously, preclinical imaging studies with positron emission tomography have explored the potential roles of different radiolabeled polyamines as prostate imaging agents.34, 35 Herein, we show, for the first time, that overexpression of ODC in a normal prostate cell line confers de novo tumor initiation capacity. Functionally, in comparison to its parental cell line, the new ODC-overexpressing cell line showed higher proliferation activity and higher invasion potential through the activation of signaling pathways commonly associated with cancer. Gene expression analysis comparing parental RWPE1 and RWPE1_ODC cells revealed that specific transcription factors and signaling pathways, including MAPK, CHKB, MMP13, mitochondrial dysfunction, EIF2, and mTOR, were altered in the RWPE1_ODC cells, and these findings were confirmed at the protein level.
These results demonstrate that prostate tumorigenesis may be promoted by the activation of the polyamine pathway through the above-mentioned cancer-related genes. Our findings in the ODC-overexpressing cells were consistent with literature showing that MAPK and CHKB were up-regulated in prostate cancer. The RAS/MAPK pathway is significantly elevated in both primary and metastatic prostate cancer, and its activation promotes aggressive tumor behavior, but there is no information about its implication in tumor initiation.36, 37 Although CHKB has been shown to be involved in carcinogenesis in different types of solid tumors, to our knowledge, it has not been studied in human prostate cancer.38 Different metalloproteinases, including MMP13, which has previously been linked to prostate cancer, were also up-regulated in the ODC-overexpressing cells in our study.39 Mitochondrial dysfunction and EIF2 and mTOR signaling pathways were found to be the most significantly altered genetic pathways in conjunction with the polyamine pathway's increased activity. Defects in mitochondrial function, which are important in cellular metabolism and apoptosis, have long been known to contribute to the development and progression of cancer.40 Similarly, EIF2 has been associated with hypoxia and metabolism in cancer cells, making it one of the most critical pathways involved in malignant transformation.41 The mTOR pathway has been extensively studied in prostate cancer and tumorigenesis.42, 43 Inhibition of mTORC1 has been shown to lead to MAPK pathway activation, which is consistent with the results we observed after polyamine pathway activation.44
Several gene alterations, such as decreased GSTP1 and Nkx3.1 expression, have been implicated in the malignant transformation of normal prostate epithelium to HGPIN.45 HGPIN is a precursor of invasive prostate carcinoma,46, 47 but little is known about the genetic events involved in the advancement to invasive disease,48, 49 a sequence of events that remains unclear.50 Few studies have shown specific genetic alterations in HGPIN, including in the SIRT1/S6K axis, mTOR-RAPTOR pathway, PEDF, and miR-21,51, 52, 53, 54 but much remains unexplained. In the present study, we show that the polyamine pathway plays a unique role in early prostate transformation, being already altered in HGPIN lesions; therefore, ODC, PAO, SPSY, and spermine could play an important role in improving early detection and risk stratification of prostate cancer.
A single study has validated tissue-based expression levels of spermine in prostate cancer by immunohistochemistry.8 In our study, spermine was abundant in normal prostatic glands (mean, 62.2%) and lost during malignant transformation, as early as HGPIN (mean, 18.8% in HGPIN and 5.8% in prostate cancer). Our results show that ODC, PAO, and SPSY levels are commonly up-regulated in HGPIN tissues as compared to normal prostate glands, with the expression levels for PAO expression being the highest of the three (mean, 5.1% in normal prostate tissue versus 41.2% in HGPIN). Notably, expression of these polyamine pathway enzymes in prostate cancer was similar to the expression of the same enzymes in HGPIN (eg, mean PAO level was 46.0% in prostate cancer), which further highlights the importance of these markers in prostate tumorigenesis. In a smaller patient cohort, Goodwin et al5 found that PAO was up-regulated in HGPIN and prostate cancer as compared to benign prostate tissue.
It is well known that the polyamine pathway can be successfully inhibited by specific enzyme inhibitors, such as DFMO for ODC. We showed that treatment of RWPE1-ODC cells with DFMO reduced the proliferation activity to levels similar to those seen in RWPE1 parental cells. Although a phase 2 clinical trial of DFMO with patients with a family history of prostate cancer but no previous personal history of prostate cancer has been reported,55 no trials have included patients with a diagnosis of HGPIN and no evidence of prostate cancer. Different chemopreventive clinical trials have been developed to control progression of HGPIN to prostate cancer using selenium, soy, vitamin E,56, 57 toremifene,58 or green tea extract,59, 60 with different success rates. Our findings suggest that DFMO treatment, which is usually associated with minimal toxicity,61 may prove valuable for reducing the development of prostate cancer in patients with a diagnosis of HGPIN and may play a crucial role in future chemopreventive clinical trials.
Acknowledgments
We thank Dr. Rong Wang and Dr. Georgia Dolios (Mass Spectrometry, Proteomics and Metabolomics Core Facility at the Icahn Institute) for performing the spermine mass spectrometry analyses in cell culture media and cell pellets (data not shown). These extra studies were performed to confirm the presence of spermine in cell extracts and media, as per Reviewers' comment. We also thank Sandhya George and Ada Muellner for editing the manuscript.
A.S.-D. designed the study and wrote the manuscript; M.C.-M. designed the study, developed the method, performed and supervised several of the in vitro and in vivo experiments, performed the analyses, and wrote the manuscript; M.C. performed all of the siRNA in vitro experiments; J.L. performed the overexpression and several of the in vitro experiments; N.G. performed several of the in vitro experiments and reviewed the manuscript; A.C.L. performed several of the in vitro and in vivo experiments and reviewed the manuscript; F.M.K. performed the sample size calculation and most of the statistical analyses; V.P. supervised the overexpression experiments; Z.Y. and W.Z. performed all of the gene expression analyses, provided the diagrams to illustrate the results, and uploaded the data in the Gene Expression Omnibus repository website; P.P.P. supervised the performance of several experiments and reviewed the manuscript; H.H. supervised the writing of the manuscript; and C.C.-C. designed the study, supervised all of the experiments, and wrote the manuscript.
C.C.-C. is the guarantor of this work and, as such, had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
Supported by the Margaret Dorrance Strawbridge Foundation, NIH/National Cancer Institute Cancer Center Support grant P30 CA008748 (Department of Radiology, Memorial Sloan Kettering Cancer Center), NIH grant R01-CA-076423 (A.S.-D. and H.H.), and NIH grant P01-CA-087497 (C.C.C. and M.C.-M.).
A.S.-D. and M.C.-M. contributed equally to this work.
Disclosures: None declared.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2016.08.021.
Supplemental Data
Heat and Ingenuity maps of the most significant altered genetic pathways. A: Heat map illustrating the differences in gene expression between RWPE1_parental and RWPE1_ODC cells. Ingenuity maps of mitochondrial dysfunction (B), EIF2 (C), and mTOR (D) signaling pathways, where up-regulated genes are displayed in pink and down-regulated genes in green. Graphs were generated with Ingenuity IPA tools. n = 3 for each of the cell line (A). EIF2, eukaryotic initiation factor 2; mTOR, mechanistic target of rapamycin; IPA, ingenuity pathway analysis.
Polyamine pathway and molecular mechanism of action in mammalian cells. A: Summary of the molecular mechanisms of action of polyamines. Highlighted in color (red up-regulated and green down-regulated) are the critical genes involved in various cellular functions (inside rectangles) affected by polyamine metabolism (adapted with permission from Aging by Minois et al24). B: Diagram illustrates the interconversion cycle of the main enzymes and amines of the polyamine pathway, which include ornithine decarboxylase (ODC), spermine synthase (SPSY), spermidine synthase (SPDSY), spermidine/spermine N1-acetyltransferase (SSAT), and polyamine oxidase (PAO). Highlighted in blue are the four markers analyzed in this study. 4E-BP, 4E-binding protein; eIF, eukaryotic translation initiation factor; MAPK, mitogen-activated protein kinase.
Validation of the prostate cancer patient cohort. Kaplan-Meier survival curves representing results for Gleason score (GS; A), preoperative prostate-specific antigen (Preop PSA; B), and pathological stage and biochemical recurrence-free survival (BRFS; C). n = 120.
No correlation is observed between the expression of ornithine decarboxylase (ODC), polyamine oxidase (PAO), and spermine synthase (SPSY), as well as spermine abundance with biochemical recurrence-free or overall survival. Kaplan-Meier survival curves representing results for ODC, PAO, SPSY, and spermine for biochemical recurrence-free survival (BRFS; A) and overall survival (OS; B).
References
- 1.Auvinen M., Paasinen A., Andersson L.C., Holtta E. Ornithine decarboxylase activity is critical for cell transformation. Nature. 1992;360:355–358. doi: 10.1038/360355a0. [DOI] [PubMed] [Google Scholar]
- 2.Cipolla B., Guille F., Moulinoux J.P., Quemener V., Staerman F., Corbel L., Lobel B. Polyamines and prostatic carcinoma: clinical and therapeutic implications. Eur Urol. 1993;24:124–131. doi: 10.1159/000474279. [DOI] [PubMed] [Google Scholar]
- 3.Gerner E.W., Meyskens F.L., Jr. Polyamines and cancer: old molecules, new understanding. Nat Rev Cancer. 2004;4:781–792. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- 4.Nowotarski S.L., Woster P.M., Casero R.A., Jr. Polyamines and cancer: implications for chemotherapy and chemoprevention. Expert Rev Mol Med. 2013;15:e3. doi: 10.1017/erm.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodwin A.C., Jadallah S., Toubaji A., Lecksell K., Hicks J.L., Kowalski J., Bova G.S., De Marzo A.M., Netto G.J., Casero R.A., Jr. Increased spermine oxidase expression in human prostate cancer and prostatic intraepithelial neoplasia tissues. Prostate. 2008;68:766–772. doi: 10.1002/pros.20735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young L., Salomon R., Au W., Allan C., Russell P., Dong Q. Ornithine decarboxylase (ODC) expression pattern in human prostate tissues and ODC transgenic mice. J Histochem Cytochem. 2006;54:223–229. doi: 10.1369/jhc.5A6672.2005. [DOI] [PubMed] [Google Scholar]
- 7.Bai G., Kasper S., Matusik R.J., Rennie P.S., Moshier J.A., Krongrad A. Androgen regulation of the human ornithine decarboxylase promoter in prostate cancer cells. J Androl. 1998;19:127–135. [PubMed] [Google Scholar]
- 8.Cohen R.J., Fujiwara K., Holland J.W., McNeal J.E. Polyamines in prostatic epithelial cells and adenocarcinoma: the effects of androgen blockade. Prostate. 2001;49:278–284. doi: 10.1002/pros.10023. [DOI] [PubMed] [Google Scholar]
- 9.Jung J.A., Coakley F.V., Vigneron D.B., Swanson M.G., Qayyum A., Weinberg V., Jones K.D., Carroll P.R., Kurhanewicz J. Prostate depiction at endorectal MR spectroscopic imaging: investigation of a standardized evaluation system. Radiology. 2004;233:701–708. doi: 10.1148/radiol.2333030672. [DOI] [PubMed] [Google Scholar]
- 10.Kurhanewicz J., Vigneron D.B., Hricak H., Narayan P., Carroll P., Nelson S.J. Three-dimensional H-1 MR spectroscopic imaging of the in situ human prostate with high (0.24-0.7-cm3) spatial resolution. Radiology. 1996;198:795–805. doi: 10.1148/radiology.198.3.8628874. [DOI] [PubMed] [Google Scholar]
- 11.Shukla-Dave A., Hricak H., Moskowitz C., Ishill N., Akin O., Kuroiwa K., Spector J., Kumar M., Reuter V.E., Koutcher J.A., Zakian K.L. Detection of prostate cancer with MR spectroscopic imaging: an expanded paradigm incorporating polyamines. Radiology. 2007;245:499–506. doi: 10.1148/radiol.2452062201. [DOI] [PubMed] [Google Scholar]
- 12.van der Graaf M., Schipper R.G., Oosterhof G.O., Schalken J.A., Verhofstad A.A., Heerschap A. Proton MR spectroscopy of prostatic tissue focused on the detection of spermine, a possible biomarker of malignant behavior in prostate cancer. MAGMA. 2000;10:153–159. doi: 10.1007/BF02590640. [DOI] [PubMed] [Google Scholar]
- 13.Konishi N., Shimada K., Ishida E., Nakamura M. Molecular pathology of prostate cancer. Pathol Int. 2005;55:531–539. doi: 10.1111/j.1440-1827.2005.01865.x. [DOI] [PubMed] [Google Scholar]
- 14.Abate-Shen C., Shen M.M. Molecular genetics of prostate cancer. Genes Dev. 2000;14:2410–2434. doi: 10.1101/gad.819500. [DOI] [PubMed] [Google Scholar]
- 15.Di Cristofano A., De Acetis M., Koff A., Cordon-Cardo C., Pandolfi P.P. Pten and p27KIP1 cooperate in prostate cancer tumor suppression in the mouse. Nat Genet. 2001;27:222–224. doi: 10.1038/84879. [DOI] [PubMed] [Google Scholar]
- 16.Tomlins S.A., Mehra R., Rhodes D.R., Cao X., Wang L., Dhanasekaran S.M., Kalyana-Sundaram S., Wei J.T., Rubin M.A., Pienta K.J., Shah R.B., Chinnaiyan A.M. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 17.Tomlins S.A., Rhodes D.R., Perner S., Dhanasekaran S.M., Mehra R., Sun X.W., Varambally S., Cao X., Tchinda J., Kuefer R., Lee C., Montie J.E., Shah R.B., Pienta K.J., Rubin M.A., Chinnaiyan A.M. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 18.Mosquera J.M., Perner S., Genega E.M., Sanda M., Hofer M.D., Mertz K.D., Paris P.L., Simko J., Bismar T.A., Ayala G., Shah R.B., Loda M., Rubin M.A. Characterization of TMPRSS2-ERG fusion high-grade prostatic intraepithelial neoplasia and potential clinical implications. Clin Cancer Res. 2008;14:3380–3385. doi: 10.1158/1078-0432.CCR-07-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung S.H., Shin S., Kim M.S., Baek I.P., Lee J.Y., Lee S.H., Kim T.M., Lee S.H., Chung Y.J. Genetic progression of high grade prostatic intraepithelial neoplasia to prostate cancer. Eur Urol. 2016;69:823–830. doi: 10.1016/j.eururo.2015.10.031. [DOI] [PubMed] [Google Scholar]
- 20.Bello D., Webber M.M., Kleinman H.K., Wartinger D.D., Rhim J.S. Androgen responsive adult human prostatic epithelial cell lines immortalized by human papillomavirus 18. Carcinogenesis. 1997;18:1215–1223. doi: 10.1093/carcin/18.6.1215. [DOI] [PubMed] [Google Scholar]
- 21.Likar Y., Zurita J., Dobrenkov K., Shenker L., Cai S., Neschadim A., Medin J.A., Sadelain M., Hricak H., Ponomarev V. A new pyrimidine-specific reporter gene: a mutated human deoxycytidine kinase suitable for PET during treatment with acycloguanosine-based cytotoxic drugs. J Nucl Med. 2010;51:1395–1403. doi: 10.2967/jnumed.109.074344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saeed A.I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., Sturn A., Snuffin M., Rezantsev A., Popov D., Ryltsov A., Kostukovich E., Borisovsky I., Liu Z., Vinsavich A., Trush V., Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 23.Tsai Y.H., Lin K.L., Huang Y.P., Hsu Y.C., Chen C.H., Chen Y., Sie M.H., Wang G.J., Lee M.J. Suppression of ornithine decarboxylase promotes osteogenic differentiation of human bone marrow-derived mesenchymal stem cells. FEBS Lett. 2015;589:2058–2065. doi: 10.1016/j.febslet.2015.06.023. [DOI] [PubMed] [Google Scholar]
- 24.Farriol M., Segovia-Silvestre T., Castellanos J.M., Venereo Y., Orta X. Role of putrescine in cell proliferation in a colon carcinoma cell line. Nutrition. 2001;17:934–938. doi: 10.1016/s0899-9007(01)00670-0. [DOI] [PubMed] [Google Scholar]
- 25.Minois N., Carmona-Gutierrez D., Madeo F. Polyamines in aging and disease. Aging (Albany NY) 2011;3:716–732. doi: 10.18632/aging.100361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bettuzzi S., Davalli P., Astancolle S., Carani C., Madeo B., Tampieri A., Corti A. Tumor progression is accompanied by significant changes in the levels of expression of polyamine metabolism regulatory genes and clusterin (sulfated glycoprotein 2) in human prostate cancer specimens. Cancer Res. 2000;60:28–34. [PubMed] [Google Scholar]
- 27.Bettuzzi S., Scaltriti M., Caporali A., Brausi M., D'Arca D., Astancolle S., Davalli P., Corti A. Successful prediction of prostate cancer recurrence by gene profiling in combination with clinical data: a 5-year follow-up study. Cancer Res. 2003;63:3469–3472. [PubMed] [Google Scholar]
- 28.Betts A.M., Waite I., Neal D.E., Robson C.N. Androgen regulation of ornithine decarboxylase in human prostatic cells identified using differential display. FEBS Lett. 1997;405:328–332. doi: 10.1016/s0014-5793(97)00209-3. [DOI] [PubMed] [Google Scholar]
- 29.Lee N.K., Skinner J.P., Zajac J.D., MacLean H.E. Ornithine decarboxylase is upregulated by the androgen receptor in skeletal muscle and regulates myoblast proliferation. Am J Physiol Endocrinol Metab. 2011;301:E172–E179. doi: 10.1152/ajpendo.00094.2011. [DOI] [PubMed] [Google Scholar]
- 30.Bettuzzi S., Davalli P., Astancolle S., Pinna C., Roncaglia R., Boraldi F., Tiozzo R., Sharrard M., Corti A. Coordinate changes of polyamine metabolism regulatory proteins during the cell cycle of normal human dermal fibroblasts. FEBS Lett. 1999;446:18–22. doi: 10.1016/s0014-5793(99)00182-9. [DOI] [PubMed] [Google Scholar]
- 31.Auvinen M., Laine A., Paasinen-Sohns A., Kangas A., Kangas L., Saksela O., Andersson L.C., Holtta E. Human ornithine decarboxylase-overproducing NIH3T3 cells induce rapidly growing, highly vascularized tumors in nude mice. Cancer Res. 1997;57:3016–3025. [PubMed] [Google Scholar]
- 32.Moshier J.A., Dosescu J., Skunca M., Luk G.D. Transformation of NIH/3T3 cells by ornithine decarboxylase overexpression. Cancer Res. 1993;53:2618–2622. [PubMed] [Google Scholar]
- 33.Mohan R.R., Challa A., Gupta S., Bostwick D.G., Ahmad N., Agarwal R., Marengo S.R., Amini S.B., Paras F., MacLennan G.T., Resnick M.I., Mukhtar H. Overexpression of ornithine decarboxylase in prostate cancer and prostatic fluid in humans. Clin Cancer Res. 1999;5:143–147. [PubMed] [Google Scholar]
- 34.Palakurthi S., McQuarrie S.A., Suresh M.R. Screening of 14C-polyamines in the AT3B-1 rat prostate tumor model: the search for a new PET prostate imaging agent. In Vivo. 2007;21:823–828. [PubMed] [Google Scholar]
- 35.Hwang D.R., Lang L.X., Mathias C.J., Kadmon D., Welch M.J. N-3-[18F]fluoropropylputrescine as potential PET imaging agent for prostate and prostate derived tumors. J Nucl Med. 1989;30:1205–1210. [PubMed] [Google Scholar]
- 36.Mukherjee R., McGuinness D.H., McCall P., Underwood M.A., Seywright M., Orange C., Edwards J. Upregulation of MAPK pathway is associated with survival in castrate-resistant prostate cancer. Br J Cancer. 2011;104:1920–1928. doi: 10.1038/bjc.2011.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mulholland D.J., Kobayashi N., Ruscetti M., Zhi A., Tran L.M., Huang J., Gleave M., Wu H. Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer Res. 2012;72:1878–1889. doi: 10.1158/0008-5472.CAN-11-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gallego-Ortega D., Gomez del Pulgar T., Valdes-Mora F., Cebrian A., Lacal J.C. Involvement of human choline kinase alpha and beta in carcinogenesis: a different role in lipid metabolism and biological functions. Adv Enzyme Regul. 2011;51:183–194. doi: 10.1016/j.advenzreg.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 39.Morgia G., Falsaperla M., Malaponte G., Madonia M., Indelicato M., Travali S., Mazzarino M.C. Matrix metalloproteinases as diagnostic (MMP-13) and prognostic (MMP-2, MMP-9) markers of prostate cancer. Urol Res. 2005;33:44–50. doi: 10.1007/s00240-004-0440-8. [DOI] [PubMed] [Google Scholar]
- 40.Carew J.S., Huang P. Mitochondrial defects in cancer. Mol Cancer. 2002;1:9. doi: 10.1186/1476-4598-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu K., Chan W., Heymach J., Wilkinson M., McConkey D.J. Control of HIF-1alpha expression by eIF2 alpha phosphorylation-mediated translational repression. Cancer Res. 2009;69:1836–1843. doi: 10.1158/0008-5472.CAN-08-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nardella C., Carracedo A., Alimonti A., Hobbs R.M., Clohessy J.G., Chen Z., Egia A., Fornari A., Fiorentino M., Loda M., Kozma S.C., Thomas G., Cordon-Cardo C., Pandolfi P.P. Differential requirement of mTOR in postmitotic tissues and tumorigenesis. Sci Signal. 2009;2:ra2. doi: 10.1126/scisignal.2000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nardella C., Chen Z., Salmena L., Carracedo A., Alimonti A., Egia A., Carver B., Gerald W., Cordon-Cardo C., Pandolfi P.P. Aberrant Rheb-mediated mTORC1 activation and Pten haploinsufficiency are cooperative oncogenic events. Genes Dev. 2008;22:2172–2177. doi: 10.1101/gad.1699608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carracedo A., Ma L., Teruya-Feldstein J., Rojo F., Salmena L., Alimonti A., Egia A., Sasaki A.T., Thomas G., Kozma S.C., Papa A., Nardella C., Cantley L.C., Baselga J., Pandolfi P.P. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118:3065–3074. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knudsen B.S., Vasioukhin V. Mechanisms of prostate cancer initiation and progression. Adv Cancer Res. 2010;109:1–50. doi: 10.1016/B978-0-12-380890-5.00001-6. [DOI] [PubMed] [Google Scholar]
- 46.Clouston D., Bolton D. In situ and intraductal epithelial proliferations of prostate: definitions and treatment implications: part 2: intraductal carcinoma and ductal adenocarcinoma of prostate. BJU Int. 2012;110(Suppl 4):22–24. doi: 10.1111/j.1464-410X.2012.11612.x. [DOI] [PubMed] [Google Scholar]
- 47.Montironi R., Mazzucchelli R., Lopez-Beltran A., Cheng L., Scarpelli M. Mechanisms of disease: high-grade prostatic intraepithelial neoplasia and other proposed preneoplastic lesions in the prostate. Nat Clin Pract Urol. 2007;4:321–332. doi: 10.1038/ncpuro0815. [DOI] [PubMed] [Google Scholar]
- 48.Bostwick D.G., Pacelli A., Lopez-Beltran A. Molecular biology of prostatic intraepithelial neoplasia. Prostate. 1996;29:117–134. doi: 10.1002/(SICI)1097-0045(199608)29:2<117::AID-PROS7>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 49.Haggman M.J., Macoska J.A., Wojno K.J., Oesterling J.E. The relationship between prostatic intraepithelial neoplasia and prostate cancer: critical issues. J Urol. 1997;158:12–22. doi: 10.1097/00005392-199707000-00004. [DOI] [PubMed] [Google Scholar]
- 50.DeMarzo A.M., Nelson W.G., Isaacs W.B., Epstein J.I. Pathological and molecular aspects of prostate cancer. Lancet. 2003;361:955–964. doi: 10.1016/S0140-6736(03)12779-1. [DOI] [PubMed] [Google Scholar]
- 51.Leite K.R., Tomiyama A., Reis S.T., Sousa-Canavez J.M., Sanudo A., Camara-Lopes L.H., Srougi M. MicroRNA expression profiles in the progression of prostate cancer-from high-grade prostate intraepithelial neoplasia to metastasis. Urol Oncol. 2013;31:796–801. doi: 10.1016/j.urolonc.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 52.Li G., Rivas P., Bedolla R., Thapa D., Reddick R.L., Ghosh R., Kumar A.P. Dietary resveratrol prevents development of high-grade prostatic intraepithelial neoplastic lesions: involvement of SIRT1/S6K axis. Cancer Prev Res (Phila) 2013;6:27–39. doi: 10.1158/1940-6207.CAPR-12-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Evren S., Dermen A., Lockwood G., Fleshner N., Sweet J. mTOR-RAPTOR and 14-3-3sigma immunohistochemical expression in high grade prostatic intraepithelial neoplasia and prostatic adenocarcinomas: a tissue microarray study. J Clin Pathol. 2011;64:683–688. doi: 10.1136/jclinpath-2011-200083. [DOI] [PubMed] [Google Scholar]
- 54.Qingyi Z., Lin Y., Junhong W., Jian S., Weizhou H., Long M., Zeyu S., Xiaojian G. Unfavorable prognostic value of human PEDF decreased in high-grade prostatic intraepithelial neoplasia: a differential proteomics approach. Cancer Invest. 2009;27:794–801. doi: 10.1080/07357900802175617. [DOI] [PubMed] [Google Scholar]
- 55.Simoneau A.R., Gerner E.W., Nagle R., Ziogas A., Fujikawa-Brooks S., Yerushalmi H., Ahlering T.E., Lieberman R., McLaren C.E., Anton-Culver H., Meyskens F.L., Jr. The effect of difluoromethylornithine on decreasing prostate size and polyamines in men: results of a year-long phase IIb randomized placebo-controlled chemoprevention trial. Cancer Epidemiol Biomarkers Prev. 2008;17:292–299. doi: 10.1158/1055-9965.EPI-07-0658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fleshner N.E., Kapusta L., Donnelly B., Tanguay S., Chin J., Hersey K., Farley A., Jansz K., Siemens D.R., Trpkov K., Lacombe L., Gleave M., Tu D., Parulekar W.R. Progression from high-grade prostatic intraepithelial neoplasia to cancer: a randomized trial of combination vitamin-E, soy, and selenium. J Clin Oncol. 2011;29:2386–2390. doi: 10.1200/JCO.2010.32.0994. [DOI] [PubMed] [Google Scholar]
- 57.Marshall J.R., Tangen C.M., Sakr W.A., Wood D.P., Jr., Berry D.L., Klein E.A., Lippman S.M., Parnes H.L., Alberts D.S., Jarrard D.F., Lee W.R., Gaziano J.M., Crawford E.D., Ely B., Ray M., Davis W., Minasian L.M., Thompson I.M., Jr. Phase III trial of selenium to prevent prostate cancer in men with high-grade prostatic intraepithelial neoplasia: SWOG S9917. Cancer Prev Res (Phila) 2011;4:1761–1769. doi: 10.1158/1940-6207.CAPR-10-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taneja S.S., Morton R., Barnette G., Sieber P., Hancock M.L., Steiner M. Prostate cancer diagnosis among men with isolated high-grade intraepithelial neoplasia enrolled onto a 3-year prospective phase III clinical trial of oral toremifene. J Clin Oncol. 2013;31:523–529. doi: 10.1200/JCO.2012.41.7634. [DOI] [PubMed] [Google Scholar]
- 59.Bettuzzi S., Brausi M., Rizzi F., Castagnetti G., Peracchia G., Corti A. Chemoprevention of human prostate cancer by oral administration of green tea catechins in volunteers with high-grade prostate intraepithelial neoplasia: a preliminary report from a one-year proof-of-principle study. Cancer Res. 2006;66:1234–1240. doi: 10.1158/0008-5472.CAN-05-1145. [DOI] [PubMed] [Google Scholar]
- 60.Brausi M., Rizzi F., Bettuzzi S. Chemoprevention of human prostate cancer by green tea catechins: two years later: a follow-up update. Eur Urol. 2008;54:472–473. doi: 10.1016/j.eururo.2008.03.100. [DOI] [PubMed] [Google Scholar]
- 61.Zell J.A., Pelot D., Chen W.P., McLaren C.E., Gerner E.W., Meyskens F.L. Risk of cardiovascular events in a randomized placebo-controlled, double-blind trial of difluoromethylornithine plus sulindac for the prevention of sporadic colorectal adenomas. Cancer Prev Res (Phila) 2009;2:209–212. doi: 10.1158/1940-6207.CAPR-08-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Heat and Ingenuity maps of the most significant altered genetic pathways. A: Heat map illustrating the differences in gene expression between RWPE1_parental and RWPE1_ODC cells. Ingenuity maps of mitochondrial dysfunction (B), EIF2 (C), and mTOR (D) signaling pathways, where up-regulated genes are displayed in pink and down-regulated genes in green. Graphs were generated with Ingenuity IPA tools. n = 3 for each of the cell line (A). EIF2, eukaryotic initiation factor 2; mTOR, mechanistic target of rapamycin; IPA, ingenuity pathway analysis.
Polyamine pathway and molecular mechanism of action in mammalian cells. A: Summary of the molecular mechanisms of action of polyamines. Highlighted in color (red up-regulated and green down-regulated) are the critical genes involved in various cellular functions (inside rectangles) affected by polyamine metabolism (adapted with permission from Aging by Minois et al24). B: Diagram illustrates the interconversion cycle of the main enzymes and amines of the polyamine pathway, which include ornithine decarboxylase (ODC), spermine synthase (SPSY), spermidine synthase (SPDSY), spermidine/spermine N1-acetyltransferase (SSAT), and polyamine oxidase (PAO). Highlighted in blue are the four markers analyzed in this study. 4E-BP, 4E-binding protein; eIF, eukaryotic translation initiation factor; MAPK, mitogen-activated protein kinase.
Validation of the prostate cancer patient cohort. Kaplan-Meier survival curves representing results for Gleason score (GS; A), preoperative prostate-specific antigen (Preop PSA; B), and pathological stage and biochemical recurrence-free survival (BRFS; C). n = 120.
No correlation is observed between the expression of ornithine decarboxylase (ODC), polyamine oxidase (PAO), and spermine synthase (SPSY), as well as spermine abundance with biochemical recurrence-free or overall survival. Kaplan-Meier survival curves representing results for ODC, PAO, SPSY, and spermine for biochemical recurrence-free survival (BRFS; A) and overall survival (OS; B).