Abstract
Next-generation sequencing (NGS) of immune receptors has become a standard tool to assess minimal residual disease (MRD) in patients treated for lymphoid malignancy, and it is being used to study the T-cell repertoire in many clinical settings. To better understanding the potential clinical utility and limitations of this application outside of MRD, we developed a BIOMED-2 primer–based NGS method and characterized its performance in controls and patients with graft-versus-host disease (GVHD) after allogeneic hematopoietic transplant. For controls and patients with GVHD, replicate sequencing of the same T-cell receptor β (TRB) libraries was highly reproducible. Higher variability was observed in sequencing of different TRB libraries made from the same DNA stock. Variability was increased in patients with GVHD compared with controls; patients with GVHD also had lower diversity than controls. In the T-cell repertoire of a healthy person, approximately 99.6% of the CDR3 clones were in low abundance, with frequency <10−3. A single library could identify >93% of the clones with frequency ≥10−3 in the repertoire. Sequencing in duplicate increased the average detection rate to >97%. This work demonstrates that NGS reliably and robustly characterizes TRB populations in healthy individuals and patients with GVHD with frequency ≥10−3 and provides a methodologic framework for applying NGS immune repertoire methods to clinical testing applications beyond MRD.
Study and analysis of T-cell receptor (TCR) repertoires are essential for better understanding the development and reconstitution of immune systems, for providing insights into the pathogenesis of immune disorders, and for diagnosing and assessing therapeutic interventions for diseases driven by T cells. Next-generation sequencing (NGS) technology has become a standard method for assessing minimal residual disease (MRD) in patients with lymphoid malignancies.1, 2, 3, 4, 5, 6, 7, 8 From a technical standpoint, MRD is a relatively simple application of NGS technology, essentially looking for known malignant or related somatically mutated clones; by combining replicate libraries, a sensitivity of 1 in 10−5 has been demonstrated for MRD. Beyond detecting MRD, NGS is also an innovative and promising approach to assess clonal populations of T cells in healthy people during immune system development, patients with suspected malignancy, response to targeted therapies, immune reconstitution in patients undergoing hematopoietic cell transplant, and autoimmune diseases.7, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25 However, assessment of the T-cell repertoire in individuals is a far more complex task than assessment of MRD. In consideration of extending NGS methods beyond MRD applications in clinical laboratories, we examined the opportunity and technical limitations of the widely used BIOMED-2 primer sets and the MiSeq system (Illumina Inc., San Diego, CA) as an NGS application to assess the TCR repertoire. We evaluated replicate testing of individual libraries (the same biological replicates) and replicate testing of different libraries prepared from the same DNA sample (different biological replicates). We tested a group of healthy individuals with intact immune systems and a group of patients with incomplete immune reconstitution and acute graft-versus-host disease (GVHD) after hematopoietic stem cell transplant (HSCT) by the following approach. Genomic DNA from peripheral blood mononuclear cell samples of patients with acute GVHD after HSCT and control individuals was extracted and was used to construct TCRβ (TRB) amplicon libraries. The libraries were sequenced at varying sequencing depths. We compared TRB repertoires detected from the same biological replicates in different sequencing runs and TRB repertoires detected from different biological replicates made from the same DNA stock in all groups. The goal was to obtain estimates of test performance, including reproducibility, sensitivity, underlying repertoire diversity, and the impact of higher-depth sequencing versus multiple lower-depth replicates on these parameters in the two groups.
Materials and Methods
Patient Characteristics
Baseline laboratory parameters of patients with GVHD and healthy individuals extracted from routine clinical laboratory tests are shown in Table 1.
Table 1.
Baseline Laboratory Parameters of Patients with GVHD and Controls Extracted from Routine Clinical Laboratory Tests
| Parameter | Controls (n = 10) | Patients with GVHD (n = 8) |
|---|---|---|
| WBC count, ×109/L | 6.20 ± 1.36 | 5.54 ± 4.54 |
| Hemoglobin, g/dL | 13.72 ± 1.16 | 9.69 ± 0.89∗ |
| Platelet count, ×109/L | 304.30 ± 45.87 | 112.00 ± 92.20∗ |
| Lymphocyte, ×109/L | 1.35 ± 0.45 | 0.40 ± 0.44∗ |
| Lymphocyte, % | 21.58 ± 4.67 | 8.20 ± 6.94∗ |
| Age, years | 54.60 (19–69) | 42.82 (23–61) |
| Days after transplant | NA | 48.43 ± 14.06∗ |
Data are expressed as means ± SD or means (range).
∗P < 0.0005 (unpaired t-test).
GVHD, graft-versus-host disease; NA, not applicable; WBC, white blood cell.
Genomic DNA Extraction
Genomic DNA was extracted from peripheral blood mononuclear cells of patients with GVHD (n = 8) and healthy individuals (n = 10) using an AllPrep DNA/RNA mini kit (Qiagen Inc., Valencia, CA) according to the manufacturer's instructions. Genomic DNA was stored at −80°C until use.
Library Preparation and Sequencing
Equal amounts (2.0 μg) of genomic DNA were used as templates to amplify completely rearranged TRB genes between V and J segments in TRB repertoires with a set of modified BIOMED-2 PCR primers optimized for the MiSeq system, including 23 V primers and 13 J primers in multiplex PCRs. Briefly, a set of PCR primers was designed based on each individual BIOMED-2 V and J primer for TRB genes by adding different numbers of random nucleotides at the 5′ end of each primer to increase the complexity of the amplified TRB repertoires prepared in the next steps for NGS (Table 2). The multiplex PCR conditions were the same as described in the standard BIOMED-2 PCR protocol.26 The DNA template was replaced with H2O as a negative control in the step of multiplex PCR. The amplified TRB amplicons were purified using the QIAquick PCR purification kit (Qiagen Inc., San Diego, CA). The same amounts of purified amplicons were subjected to library preparation with the TruSeq kit (Illumina Inc.) or the KAPA Hyper Prep kit (Kapa Biosystems, Boston, MA) following the manufacturers' instructions. Sequencing was performed using a MiSeq reagent 500-cycle V2 kit (Illumina Inc.) by paired-end 250 × 2 cycles. To assess the effect of various sequencing depths, we pooled 10 to 12 indexed libraries or 1 library in a sequencing run on the MiSeq instrument.
Table 2.
Modified BIOMED-2 PCR Primers
| Name of primer | Sequence of primer (forward) | Tube name |
|---|---|---|
| VB2-A | 5′-NNNNAACTATGTTTTGGTATCGTCA-3′ | A and B |
| VB2-B | 5′-NNNNNAACTATGTTTTGGTATCGTCA-3′ | A and B |
| VB4-A | 5′-NNNNCACGATGTTCTGGTACCGTCAGCA-3′ | A and B |
| VB4-B | 5′-NNNNNCACGATGTTCTGGTACCGTCAGCA-3′ | A and B |
| VB5/1-A | 5′-NNNNCAGTGTGTCCTGGTACCAACAG-3′ | A and B |
| VB5/1-B | 5′-NNNNNCAGTGTGTCCTGGTACCAACAG-3′ | A and B |
| VB6a/11-A | 5′-NNNNAACCCTTTATTGGTACCGACA-3′ | A and B |
| VB6a/11-B | 5′-NNNNNAACCCTTTATTGGTACCGACA-3′ | A and B |
| VB6b/25-A | 5′-NNNNATCCCTTTTTTGGTACCAACAG-3′ | A and B |
| VB6b/25-B | 5′-NNNNNATCCCTTTTTTGGTACCAACAG-3′ | A and B |
| VB6c-A | 5′-NNNNAACCCTTTATTGGTATCAACAG-3′ | A and B |
| VB6c-B | 5′-NNNNNAACCCTTTATTGGTATCAACAG-3′ | A and B |
| VB7-A | 5′-NNNNCGCTATGTATTGGTACAAGCA-3′ | A and B |
| VB7-B | 5′-NNNNNCGCTATGTATTGGTACAAGCA-3′ | A and B |
| VB8a-A | 5′-NNNNCTCCCGTTTTCTGGTACAGACAGAC-3′ | A and B |
| VB8a-B | 5′-NNNNNCTCCCGTTTTCTGGTACAGACAGAC-3′ | A and B |
| VB9-A | 5′-NNNNCGCTATGTATTGGTATAAACAG-3′ | A and B |
| VB9-B | 5′-NNNNNCGCTATGTATTGGTATAAACAG-3′ | A and B |
| VB10-A | 5′-NNNNTTATGTTTACTGGTATCGTAAGAAGC-3′ | A and B |
| VB10-B | 5′-NNNNNTTATGTTTACTGGTATCGTAAGAAGC-3′ | A and B |
| VB11-A | 5′-NNNNCAAAATGTACTGGTATCAACAA-3′ | A and B |
| VB11-B | 5′-NNNNNCAAAATGTACTGGTATCAACAA-3′ | A and B |
| VB2a/3/13a/15-A | 5′-NNNNATACATGTACTGGTATCGACAAGAC-3′ | A and B |
| VB2a/3/13a/15-B | 5′-NNNNNATACATGTACTGGTATCGACAAGAC-3′ | A and B |
| VB13b-A | 5′-NNNNGGCCATGTACTGGTATAGACAAG-3′ | A and B |
| VB13b-B | 5′-NNNNNGGCCATGTACTGGTATAGACAAG-3′ | A and B |
| VB13c/12B/14-A | 5′-NNNNGTATATGTCCTGGTATCGACAAGA-3′ | A and B |
| VB13c/12B/14-B | 5′-NNNNNGTATATGTCCTGGTATCGACAAGA-3′ | A and B |
| VB16-A | 5′-NNNNTAACCTTTATTGGTATCGACGTGT-3′ | A and B |
| VB16-B | 5′-NNNNNTAACCTTTATTGGTATCGACGTGT-3′ | A and B |
| VB17-A | 5′-NNNNGGCCATGTACTGGTACCGACA-3′ | A and B |
| VB17-B | 5′-NNNNNGGCCATGTACTGGTACCGACA-3′ | A and B |
| VB18-A | 5′-NNNNTCATGTTTACTGGTATCGGCAG-3′ | A and B |
| VB18-B | 5′-NNNNNTCATGTTTACTGGTATCGGCAG-3′ | A and B |
| VB19-A | 5′-NNNNTTATGTTTATTGGTATCAACAGAATCA-3′ | A and B |
| VB19-B | 5′-NNNNNTTATGTTTATTGGTATCAACAGAATCA-3′ | A and B |
| VB20-A | 5′-NNNNCAACCTATACTGGTACCGACA-3′ | A and B |
| VB20-B | 5′-NNNNNCAACCTATACTGGTACCGACA-3′ | A and B |
| VB21-A | 5′-NNNNTACCCTTTACTGGTACCGGCAG-3′ | A and B |
| VB21-B | 5′-NNNNNTACCCTTTACTGGTACCGGCAG-3′ | A and B |
| VB22-A | 5′-NNNNATACTTCTATTGGTACAGACAAATCT-3′ | A and B |
| VB22-B | 5′-NNNNNATACTTCTATTGGTACAGACAAATCT-3′ | A and B |
| VB23/8b-A | 5′-NNNNCACGGTCTACTGGTACCAGCA-3′ | A and B |
| VB23/8b-B | 5′-NNNNNCACGGTCTACTGGTACCAGCA-3′ | A and B |
| VB24-A | 5′-NNNNCGTCATGTACTGGTACCAGCA-3′ | A and B |
| VB24-B | 5′-NNNNNCGTCATGTACTGGTACCAGCA-3′ | A and B |
| Sequence of primer (reverse) | ||
|---|---|---|
| JB1.1-A | 5′-NNNNNCTTACCTACAACTGTGAATCTGGTG-3′ | A |
| JB1.1-B | 5′-NNNNNNCTTACCTACAACTGTGAATCTGGTG-3′ | A |
| JB1.1-C | 5′-NNNNNNNCTTACCTACAACTGTGAATCTGGTG-3′ | A |
| JB1.2-A | 5′-NNNNNCTTACCTACAACGGTTAACCTGGTC-3′ | A |
| JB1.2-B | 5′-NNNNNNCTTACCTACAACGGTTAACCTGGTC-3′ | A |
| JB1.2-C | 5′-NNNNNNNCTTACCTACAACGGTTAACCTGGTC-3′ | A |
| JB1.3-A | 5′-NNNNNCTTACCTACAACAGTGAGCCAACTT-3′ | A |
| JB1.3-B | 5′-NNNNNNCTTACCTACAACAGTGAGCCAACTT-3′ | A |
| JB1.3-C | 5′-NNNNNNNCTTACCTACAACAGTGAGCCAACTT-3′ | A |
| JB1.4-A | 5′-NNNNNCATACCCAAGACAGAGAGCTGGGTTC-3′ | A |
| JB1.4-B | 5′-NNNNNNCATACCCAAGACAGAGAGCTGGGTTC-3′ | A |
| JB1.4-C | 5′-NNNNNNNCATACCCAAGACAGAGAGCTGGGTTC-3′ | A |
| JB1.5-A | 5′-NNNNNCTTACCTAGGATGGAGAGTCGAGTC-3′ | A |
| JB1.5-B | 5′-NNNNNNCTTACCTAGGATGGAGAGTCGAGTC-3′ | A |
| JB1.5-C | 5′-NNNNNNNCTTACCTAGGATGGAGAGTCGAGTC-3′ | A |
| JB1.6-A | 5′-NNNNNCATACCTGTCACAGTGAGCCTG-3′ | A |
| JB1.6-B | 5′-NNNNNNCATACCTGTCACAGTGAGCCTG-3′ | A |
| JB1.6-C | 5′-NNNNNNNCATACCTGTCACAGTGAGCCTG-3′ | A |
| JB2.2-A | 5′-NNNNNCTTACCCAGTACGGTCAGCCT-3′ | A |
| JB2.2-B | 5′-NNNNNNCTTACCCAGTACGGTCAGCCT-3′ | A |
| JB2.2-C | 5′-NNNNNNNCTTACCCAGTACGGTCAGCCT-3′ | A |
| JB2.6-A | 5′-NNNNNCTCGCCCAGCACGGTCAGCCT-3′ | A |
| JB2.6-B | 5′-NNNNNNCTCGCCCAGCACGGTCAGCCT-3′ | A |
| JB2.6-C | 5′-NNNNNNNCTCGCCCAGCACGGTCAGCCT-3′ | A |
| JB2.7-A | 5′-NNNNNCTTACCTGTAACCGTGAGCCTG-3′ | A |
| JB2.7-B | 5′-NNNNNNCTTACCTGTAACCGTGAGCCTG-3′ | A |
| JB2.7-C | 5′-NNNNNNNCTTACCTGTAACCGTGAGCCTG-3′ | A |
| JB2.1-A | 5′-NNNNNCCTTCTTACCTAGCACGGTGA-3′ | B |
| JB2.1-B | 5′-NNNNNNCCTTCTTACCTAGCACGGTGA-3′ | B |
| JB2.1-C | 5′-NNNNNNNCCTTCTTACCTAGCACGGTGA-3′ | B |
| JB2.3-A | 5′-NNNNNCCCGCTTACCGAGCACTGTCA-3′ | B |
| JB2.3-B | 5′-NNNNNNCCCGCTTACCGAGCACTGTCA-3′ | B |
| JB2.3-C | 5′-NNNNNNNCCCGCTTACCGAGCACTGTCA-3′ | B |
| JB2.4-A | 5′-NNNNNCCAGCTTACCCAGCACTGAGA-3′ | B |
| JB2.4-B | 5′-NNNNNNCCAGCTTACCCAGCACTGAGA-3′ | B |
| JB2.4-C | 5′-NNNNNNNCCAGCTTACCCAGCACTGAGA-3′ | B |
| JB2.5-A | 5′-NNNNNCGCGCACACCGAGCAC-3′ | B |
| JB2.5-B | 5′-NNNNNNCGCGCACACCGAGCAC-3′ | B |
| JB2.5-C | 5′-NNNNNNNCGCGCACACCGAGCAC-3′ | B |
N indicates a degenerate nucleotide position to generate sequence diversity.
Bioinformatics and Statistical Analysis
The overlapped paired-end reads (read 1 and read 2) from the MiSeq were joined using the publicly available FLASH software version 1.2.11 (Fast Length Adjustment of SHort reads, https://sourceforge.net/projects/flashpage/files).27 After removing primer sequences, DNA reads were submitted to IMGT/HighV-Quest for TRB rearrangement analyses.11 The unique CDR3 amino acid sequences for each sample were summarized based on the IMGT/HighV-Quest results. Single-copy CDR3 clones were removed to reduce the likelihood of artificial CDR3s caused by sequencing errors. A clonotype and a CDR3 clone were referred to a unique CDR3 amino acid sequence. CDR3, CDR3 clone, and clonotype are used interchangeably in this article.
The frequency of a clonotype, which represents the relative abundance of the clonotype present in the original sample, was calculated by the copy number of the clonotype divided by the total number of copies of all clonotypes in a sequencing run. When the CDR3 data files from different biological replicates were pooled after separate sequencing runs, we recalculated each clonotype frequency as the total number of copies of that clonotype divided by the total number of all copies in the pooled file.
The Simpson diversity index, , is commonly used in ecology to assess species diversity, where n is the number of unique clonotypes in a sample and Pi is the frequency of the ith clonotype. The Simpson index measures the probability that two randomly selected CDR3s in a sample are the same, with the range of S between 0 and 1. The higher the value of S, the lower the diversity. The CDR3 diversity of each sample was assessed by the Simpson reciprocal index (1/S). The higher the reciprocal index, the higher the diversity of CDR3 clones in a sample. The single-copy CDR3s with their low frequencies have a limited effect on the values of the Simpson reciprocal index due to the index's mathematical nature.28 The Simpson reciprocal indexes were calculated with either inclusion or exclusion of single-copy CDR3s. They showed little difference, as expected. The values of the Simpson reciprocal indexes based on single-copy CDR3 exclusion are shown herein.
The similarity of clonotypes between two samples (p and q) was measured by the Bhattacharyya coefficient (BC(p,q)) using the equation:
| (1) |
where n is the number of the common clonotypes between two samples (p and q), and pi and qi are the frequencies of the ith shared clonotype of the samples p and q, respectively. The Bhattacharyya coefficient ranges from 0 to 1, with 0 indicating no common clonotypes between two samples.
Technical Reproducibility and Variability of NGS TCR Sequencing
The CDR3 clone frequencies from replicate sequencing runs of the same biological replicate library were compared to assess the reproducibility of the sequencing process. CDR3 clone frequencies from sequencing different biological replicates (different libraries) were compared to evaluate the variability of sampling and library preparation.
Biological Variation
We sequenced seven biological replicates (seven different libraries) (L1 to L7) of a healthy person (HP10) in the range of 0.6 million to 1.4 million reads. The CDR3 clonotypes in each biological replicate were characterized as described in Bioinformatics and Statistical Analysis. The seven biological replicate libraries were classified into two test libraries and five reference libraries. To avoid bias in classifying the individual libraries into test and reference libraries, we constructed three test-versus-reference comparison groups by selecting different test libraries in each group. For each group, we approximated the total TRB repertoire by combining the CDR3 clonotypes of five reference libraries as a reference pool. The common clonotypes between each of the two test libraries and a reference pool (the combination of five separate reference libraries) were identified.
Hive plots were generated with scripts that were a gift from Dan Meliza (Gigagen Inc., San Francisco, CA). The hive plots visually display the common clonotypes between samples in relation to their abundance in each sample.29 In hive plots, three axes represent three individual samples. The nodes on an axis represent the CDR3 clonotypes in a sample. They are positioned according to the clonotype abundances: from the most abundant (distal) to the least abundant (central). The abundance is quantified by the CDR3 frequencies in the sample. The arcs link common clonotypes between two samples. Thickness at an arc's end point is proportional to the CDR3 clone abundance in the repertoire. The arcs in yellow indicate the common clonotypes with frequency ≥0.01 in at least one of the two compared samples, and the arcs in red indicate the common clonotypes with frequency <0.01.
Results
BIOMED-2 Primer–Based NGS System
Based on the BIOMED-2 PCR protocol for detection of human TRB rearrangements, which is widely used in clinical settings,26, 30, 31 we developed an NGS method by using a panel of modified multiplexed TCR Vβ and Jβ subunit-specific T-cell primers adopted from BIOMED-2. We performed amplification and high-throughput sequencing of the peripheral blood mononuclear cell genomic DNA samples from patients with GVHD and healthy individuals. We achieved optimal cluster density (850 to 1200 K/mm2) of MiSeq sequencing runs (250 × 2 cycles per run), with more than 90% of clusters passing filter, approximately 20 million reads per end (approximately 40 million reads for both ends in a paired-end run), and >86% of sequences with >Q30 quality metric for read 1 and >81% of sequences with >Q30 for read 2. In lower-depth sequencing runs, we normally obtained 3 million to 4 million DNA reads, translating to 5.6 to 16.2 K unique CDR3 clones per library. In higher-depth sequencing runs, we obtained 35 million to 40 million DNA reads and 38.4 to 46.7 K unique CDR3 clones per library.
Reproducibility of Sequencing Process in TRB Repertoire Estimates
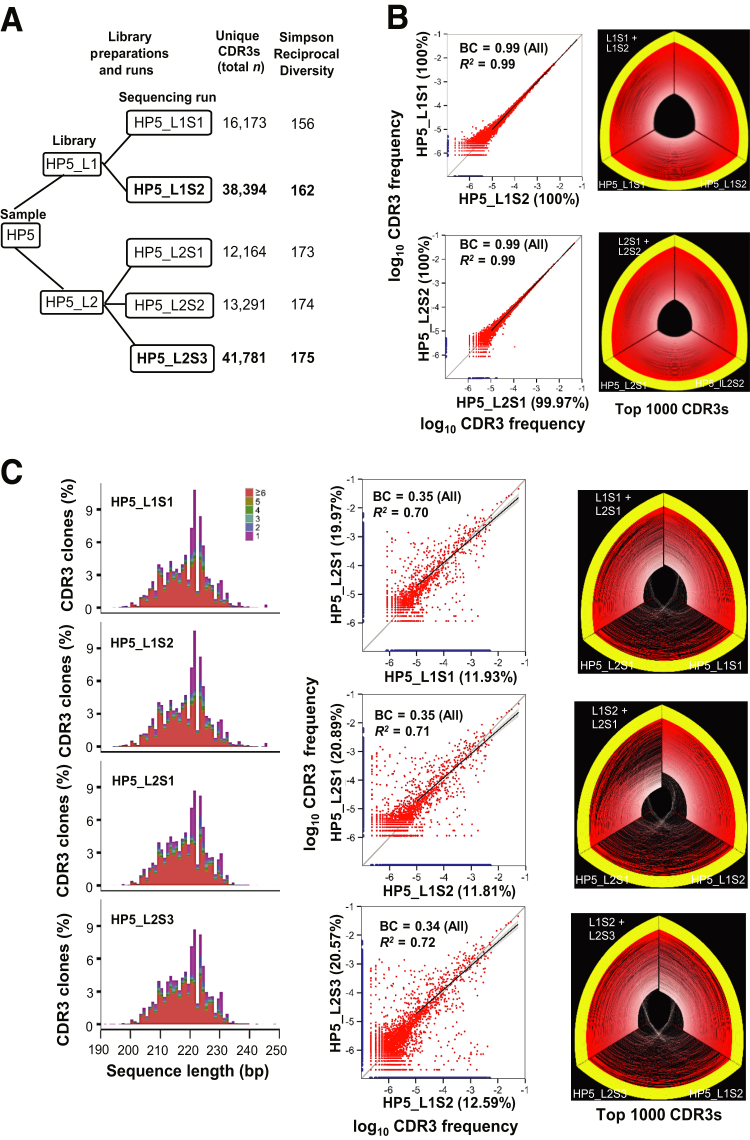
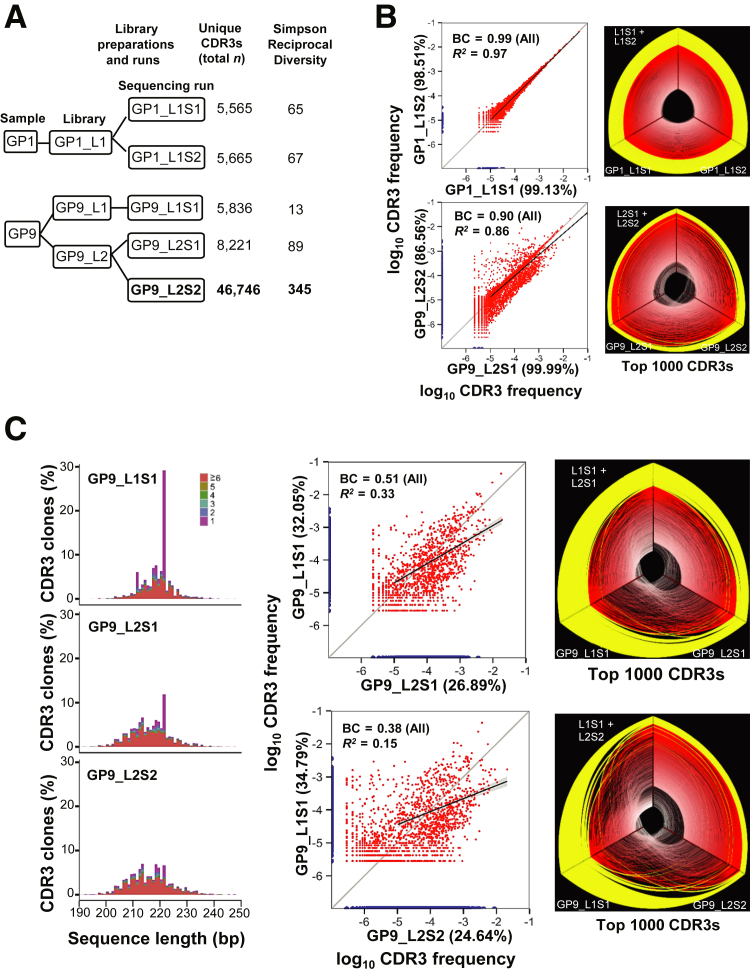
To evaluate the reproducibility of the sequencing process, we compared the results of different sequencing runs of the same biological replicate library prepared from a single individual. One library (GP1_L1) of a patient with GVHD (GP1) and one library (HP5_L2) of a healthy individual (HP5) were prepared. Each library was sequenced twice, each time in a separate sequencing run at similar read depths (1.6 million to 2.6 million reads for GVHD libraries and 4.0 million to 5.1 million reads for healthy person libraries). To determine what role the read depths play in sequencing reproducibility and clonotype identification, we prepared another biological replicate library (HP5_L1) from the same healthy individual (HP5) and a library (GP9_L2) from another patient with GVHD (GP9) also sequenced twice, each time in a separate sequencing run at different sequencing depths; we also sequenced HP5_L2 at different sequencing depth (5.3 million to 17.6 million reads) (Figures 1A and 2A). For a given biological replicate library, a higher number of the sequencing reads resulted in a greater number of clonotypes, and the number of clonotypes was approximately proportional to the number of sequencing reads (Figures 1A and 2A).
Figure 1.
Representative healthy individual (HP5): CDR3 clone comparison between different sequencing runs of the same library and between different libraries. A: Schematic diagram of library preparation and sequencing runs. Higher-depth sequencing runs are in bold. B: Comparison of the CDR3 clones captured in different sequencing runs of the same library. In the scatterplots, the x and y axes represent the CDR3 frequencies in log10 scale in two separate sequencing runs. Red dots are the CDR3 clones detected in both runs. Blue dots on the x or y axis represent the CDR3 clones detected in only one of the two runs. In the hive plots, the three axes represent the CDR3 clones of two individual sequencing runs and the union of these two runs. The yellow arcs indicate the common clonotypes with frequency ≥0.01 on at least one of the two compared axes. The red arcs are secondary clonotypes with frequency <0.01. All the CDR3 clones are used for analysis, and only the top 1000 clones are displayed. C: CDR3 clones captured in different biological replicate libraries made from the same genomic DNA stock. In the histogram plots, the components of CDR3 clones at different sizes of DNA sequences encoding TRB V/D/J regions are shown. The x axis indicates the sizes of the DNA sequences, and the y axis represents the percentage of the translated CDR3 clones in a run. The top five high-frequency CDR3 clones (1 to 5) and the sum of the remaining clones (≥6) at a specific size are represented by different colors. BC, Bhattacharyya coefficient.
Figure 2.
Two representative patients with graft-versus-host disease (GP1 and GP9): CDR3 clone comparison between different sequencing runs of the same library and between different libraries. A: Schematic diagram of library preparation and sequencing runs. B: Comparison of the CDR3 clones captured in different sequencing runs of the same library. C: CDR3 clones captured in different biological replicate libraries made from the same genomic DNA stock. Refer to Figure 1 for the description of each panel. BC, Bhattacharyya coefficient.
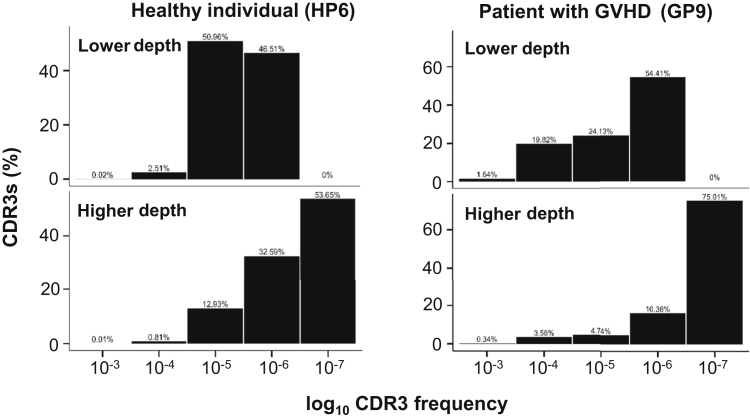
For each of four libraries (GP1_L1, GP9_L2, HP5_L1, and HP5_L2), the shared clonotypes and their frequencies between two sequencing runs were compared (Figures 1B and 2B). Overall, the clonotypes with higher frequency showed greater reproducibility. In all the comparisons except GP9_L2, at least 98.5% of the clonotypes with frequency >10−5 in one run could be detected by the other run (Figures 1B and 2B). For each library comparison, a linear regression was performed on the frequencies of the common clonotypes (frequency >10−5) detected in two separate sequencing runs. The values of R2, which measures how well the data points are fit to the regression line, for the GP1_L1, GP9_L2, HP5_L1, and HP5_L2 libraries were 0.97, 0.86, 0.99, and 0.99, respectively (Figures 1B and 2B). For each library comparison, the Bhattacharyya coefficient was also calculated from all CDR3 clones. The values of Bhattacharyya coefficients, which measure the correlation of CDR3 frequencies, for the GP1_L1, GP9_L2, HP5_L1, and HP5_L2 libraries were 0.99, 0.90, 0.99, and 0.99, respectively (Figures 1B and 2B). In addition, the additional CDR3 clones captured by higher sequencing depth were primarily low-abundance CDR3 clones with frequency <10−7 for healthy people and patients with GVHD (Figure 3), suggesting that higher sequencing depth provided little advantage in reliably detecting a higher number of clonotypes.
Figure 3.
Unique CDR3 distribution profiles of the same biological replicate libraries detected with different sequencing depths in a representative healthy individual (HP6) and a representative patient with graft-versus-host disease (GVHD) (GP9).
These results demonstrated that the NGS technology reproducibly identifies abundant clonotypes with frequency ≥10−5, with little variation across sequencing runs from a single biological replicate library.
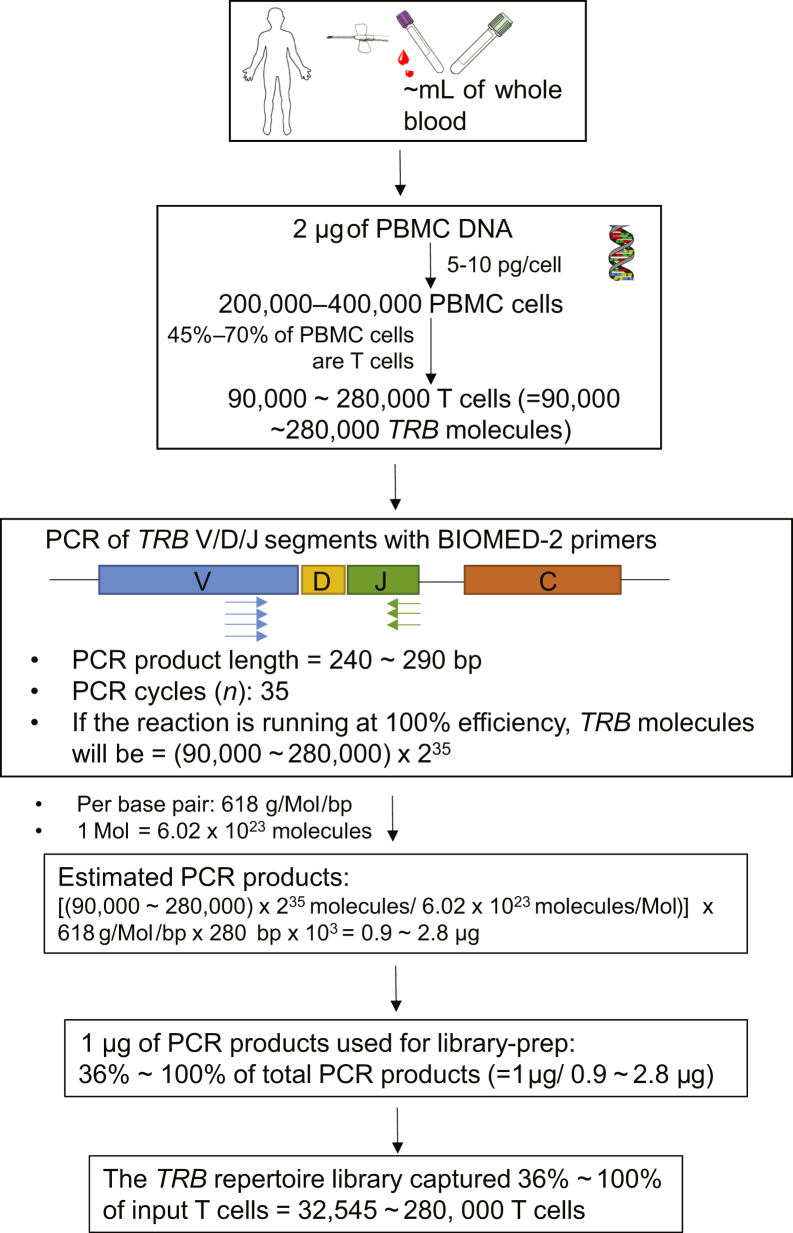
Variability Across Different Biological Replicate Libraries of TRB Repertoire Estimates
Because of the high diversity of human immune systems and the limited depth of sequencing performed, we reasoned that sampling of T cells from the human peripheral blood mononuclear cell captures only a fraction of the true TRB repertoire (Figure 4). To better understand the sampling issues, we compared CDR3 clones from sequencing different biological replicates (different libraries) prepared from the same genomic DNA extraction of a healthy individual (Figure 1A). The range of the percentages of clonotypes with frequency >10−5 in one biological replicate library that could be detected by the other biological replicate library was much lower than that obtained by repeated sequencing of the same biological replicate library (12% to 21%) (Figure 1C). For each comparison, linear regression was also performed on the frequencies of the common clonotypes, with frequency >10−5 detected in two different biological replicate libraries (Figure 1C). The values of R2 for the comparisons were 0.7. Their Bhattacharyya coefficients calculated from all the CDR3 clones were 0.35 (Figure 1C).
Figure 4.
Flowchart of the capture rate of T cells in the BIOMED-2 primer–based NGS system. PBMC, peripheral blood mononuclear cell.
We also performed different biological replicate library comparisons for a GVHD sample (GP9). One biological replicate library (GP9_L2) was sequenced twice in different depths (1.8 million to 16.1 million reads). The clonotypes from sequencing a different biological replicate library (GP9_L1) were compared with both runs of the GP9_L2 library. The results showed that 32% of CDR3 clonotypes with frequency >10−5 in the GP9_L1S1 library existed in the lower-depth run of GP9_L2S1. When GP9_L1S1 was compared with the higher-depth run of GP9_L2S2, the percentage increased slightly to 35%, although the number of clones in the higher-depth run of GP9_L2S2 was approximately six times greater than that in the lower-depth run (Figure 2, A and C).
These results demonstrated the lower reproducibility between different biological replicate libraries made from the same DNA stock and were consistent with the notion that the TRB repertoire detected in each biological replicate library is only a fraction of the true repertoire. These results are consistent with the findings reported previously by other groups.32, 33
Spectratyping assays have been widely used in clinical settings for more than a decade to detect the TCR clonality of patients. This method separates V/D/J DNA clones based on the DNA amplicon size in the TCR repertoire. This method has been reported to be highly reproducible from genomic DNA stock of the same patients.30 However, a limitation of spectratyping assays is that only the highest-frequency clonotypes are observed, and no sequence information is provided; therefore, amplicons of the same length but different sequences could be falsely interpreted as a single clonal population. Similar to spectratyping assays, the V/D/J DNA sequence size distribution of TRB repertoires detected with NGS in different biological replicate libraries made from the same DNA stock were highly similar (Figures 1C and 2C). However, we observed that each DNA clonotype based on DNA size frequently comprised multiple different CDR3 sequences, which should be recorded as many different clonotypes. Therefore, detection of TRB repertoires with NGS technology provides more accurate and more detailed information about clonotype frequency than do spectratyping assays.
Detection Thresholds of Clonotype Frequencies for TRB Repertoires
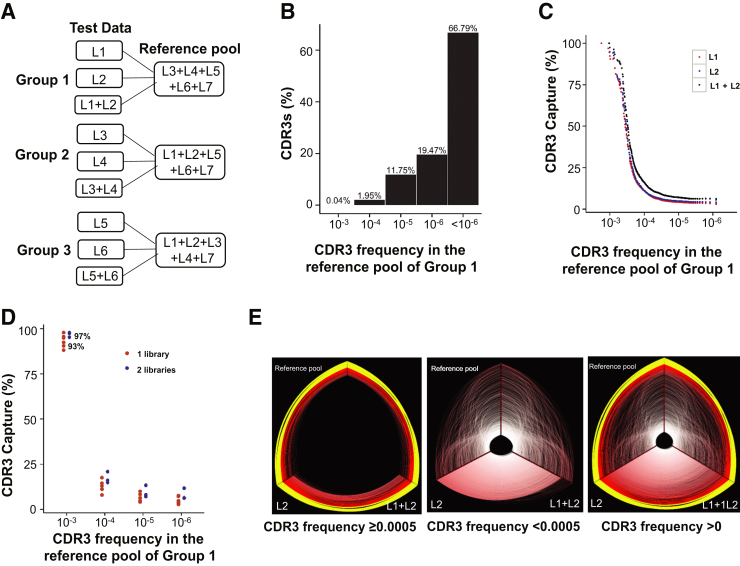
To approximate the diverse TRB repertoires in a single sample, we prepared seven biological replicate libraries (L1 to L7) from seven different PCRs using the same genomic DNA stock of a healthy person (HP10) as the amplification template. The libraries were sequenced separately using the MiSeq instrument. The clonotypes in each biological replicate library were subsequently characterized as described in Materials and Methods.
The seven sets of sequencing data from seven biological replicate libraries were separated into two test libraries and five reference libraries. The clonotypes from the sequencing data of five reference libraries were pooled together as a reference pool to approximate the TRB repertoire of the DNA sample, and the frequencies of the clonotypes in the reference pool were calculated. To assess the threshold of clonotypes in a TRB repertoire that can be detected by a single biological replicate library or by two biological replicate libraries, we compared the clonotypes in each of two test libraries and the clonotypes combined from the sequencing data of two test libraries with the clonotypes in the reference pool, respectively. To avoid bias in classifying the biological replicate libraries into the test and reference pools, we constructed three test-versus-reference comparison groups by selecting different test libraries in each group: tests L1 and L2 versus references L3, L4, L5, L6, and L7 (Group 1); L3 and L4 versus L1, L2, L5, L6, and L7 (Group 2); and L5 and L6 versus L1, L2, L3, L4, and L7 (Group 3) (Figure 5A). We observed that in the reference pool, nearly all the CDR3 clones (99.6%) were low-abundant clones, with frequency <10−3 (Figure 5B). In each test-reference comparison group, the percentages of the clonotypes at different frequencies in the reference pool that were detected by a single test library or two test libraries were evaluated. The results for Group 1 are shown in Figure 5C [Groups 2 and 3 (data not shown) demonstrated similar patterns]. Overall, for clonotypes at any given frequency in the reference pool, the two biological replicate libraries could always detect more than one single biological replicate library. Neither an individual biological replicate library nor two biological replicate libraries could reliably detect low-abundance CDR3 clones (Figure 5E). For each of the three comparison groups, the percentages of the CDR3 clones at several fixed frequencies detected by a single biological replicate library and two biological replicate libraries were determined. The detection rates by one library and two libraries from all three comparison groups were compared and plotted (Figure 5D). A single random biological replicate library identified, on average, >93% of the clones with frequency ≥10−3 in the reference pool, and two biological replicate libraries increased the average detection rate to >97%. The likelihood of the CDR3s to be captured by either single or double biological replicate libraries dropped sharply for clones with frequency <10−3 (Figure 5, C and D).
Figure 5.
Capture of CDR3 clones in different biological replicate libraries made from the same genomic DNA stock of a healthy person (HP10). A: Schematic diagram of three analysis groups. Seven different biological replicate libraries were made from seven different PCRs from the same genomic DNA stock and were sequenced separately. Three two-versus-five (two different biological replicate libraries versus the combination of the other five biological replicate libraries) analytical groups were set up from the sequencing data of these seven biological replicate libraries. In each group, the CDR3 clones of two different libraries were individually or jointly compared with those of the other five libraries combined (the reference pool). B: Percentage of CDR3 clones at different ranges of frequency in the reference pool of Group 1. The results for Groups 2 and 3 (data not shown) show a similar pattern. C: Percentage of CDR3s in the reference pool of Group 1 captured by each of two biological replicate libraries (L1 and L2) and the combination of the two biological replicates (L1 + L2). The x axis represents CDR3 frequency in the reference pool. Each dot is the sum of all the CDR3 clones with the same frequency. The results of Groups 2 and 3 (data not shown) display a similar pattern. D: Representation of likelihood of a given library or two replicate libraries containing a clonotype present in the reference pool (the combination of the other five replicate libraries as indicated in A) of a healthy individual, as % capture on the y axis versus clonotype frequency on the x axis. Only higher frequency clones are reliably detected. E: Hive plots of shared CDR3 clonotypes among different biological replicate libraries made from the same DNA stock (Group 1). In each hive plot, the three axes represent the CDR3 clones of L2, the union of L1 and L2, and the reference pool. The yellow arcs indicate the common clonotypes with frequency ≥0.01 on at least one of the two compared axes, and the red arcs are secondary clonotypes with frequency <0.01. Only the top 1000 clones by frequency in the repertoire on each axis are displayed.
Differences in TRB Repertoires of Healthy Individuals and Patients with GVHD
Patients with GVHD after allogeneic transplant had significantly lower hemoglobin levels, lymphocyte counts, percentages of lymphocytes, and platelet counts compared with healthy individuals (P < 0.0005) (Table 1). These results were consistent with previous reports that post-HSCT patients and patients with acute GVHD have lower cell counts during reconstitution of the immune system, further impaired by the presence of GVHD.34, 35, 36, 37, 38
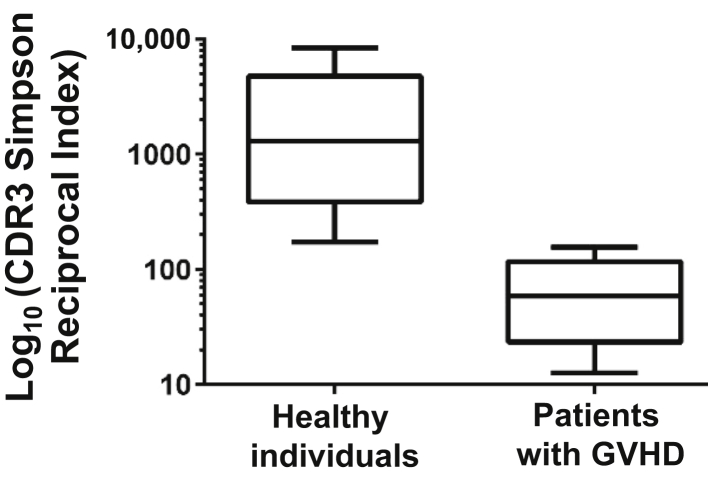
We used several methods to assess the size and diversity of TRB repertoires. As shown in Figure 6, Table 3, and Supplemental Table S1, the total number of CDR3 clones and the total number of unique CDR3 clones in the GVHD group were significantly lower than those in the control group (P = 0.03 and P = 0.003), indicating the significantly smaller size of the TRB repertoires of patients with GVHD than those of healthy individuals. The Simpson reciprocal index was also significantly lower than that of the control group (P = 0.02), indicating that the TRB repertoires of patients with GVHD were smaller and less diverse than those of healthy individuals. Because the number of lymphocytes in the healthy group is approximately four times more than that in the GVHD group, we addressed the possibility that the decreased diversity observed was due to the difference in lymphocyte number between these two groups. To exclude this possibility, we generated three independent CDR3 subsets by randomly selecting one-quarter of the CDR3s for each healthy sample and calculated the Simpson reciprocal indexes. The results demonstrated that the Simpson reciprocal indexes of the subsets were not different from each other or from the original group (data not shown). Thus, the different number of input lymphocytes between the healthy individuals and the patients with GVHD did not affect the diversity differences observed between the two sample groups. The difference in average age between the GVHD and healthy groups was not statistically significantly different. These results were consistent with previous reports that patients with GVHD after HSCT and stem cell transplant recipients had a significantly smaller and restricted T-cell repertoire.7, 14
Figure 6.
Difference in TRB repertoire diversity between healthy individuals and patients with graft-versus-host disease (GVHD). In this box-and-whisker plot, the top of the box is the first quartile, indicating 25% of data greater than this value; the band inside the box is the second quartile (the median), indicating 50% of data is greater than this value; the bottom of the box is the third quartile, indicating 25% of data less than this value. The top and bottom whiskers represent the maximum and minimum values of all the data. The CDR3 Simpson reciprocal index of each sample is calculated as described in Materials and Methods. n = 10 healthy individuals; n = 8 patients with GVHD. P = 0.02 (unpaired t-test).
Table 3.
Summary of the TRB Repertoire Diversity Index in the Control and GVHD Groups
| Group | Total CDR3s, n | Total unique CDR3s, n | Normalized unique CDR3s, n | CDR3 Simpson reciprocal Index |
|---|---|---|---|---|
| GVHD | 448,981 | 5027 | 1.2 | 83 |
| Control | 760,122 | 23,660 | 3.9 | 2341 |
| P value∗ | 0.03 | 0.003 | 0.1 | 0.02 |
Data are expressed as mean values of all individuals in each group.
∗Unpaired t-test.
GVHD, graft-versus-host disease; TRB, T-cell receptor β.
We found that there was little similarity among the TRB repertoires of different healthy individuals with Bhattacharyya coefficients <0.10, and there was even less similarity among the TRB repertoires of different patients with GVHD with Bhattacharyya coefficients typically <0.08 (data not shown). The top 100 CDR3 sequences and their V/D/J gene rearrangements in healthy individuals and patients with GVHD are shown in Supplemental Tables S2 and S3.
Discussion
Traditional methods, including Sanger sequencing and spectratyping assays, have been widely used to study TCR repertoires. However, these methods have important limitations. Sanger sequencing can identify single clone sequences. However, it is not scalable, and it requires enormous effort to isolate individual clones. Owing to low sensitivity, this technology can characterize only highly dominant clones with biased frequency. Spectratyping assays based on the BIOMED-2 PCR protocol for the detection of human TRB rearrangements have been widely used for clinical assessment of clonality. This method assesses only amplicon sizes, without the ability to consider any sequence (clonotype) information. This leads to low sensitivity because only high-frequency clones can be reliably identified, and this method has lower specificity owing to inability to resolve different clonotypes of the same size.
In contrast, NGS offers opportunities to identify and characterize millions of clonotypes in TCR repertoires simultaneously, and it has emerged as an effective tool to study MRD in lymphoid neoplasms. This technology offers the throughput and sensitivity that the traditional methods, including Sanger sequencing and spectratyping assays, cannot match. We developed the BIOMED-2 PCR primer–based NGS protocol to study TRB gene rearrangements. This method allows massively parallel sequencing of the targeted regions of a sample, which covers Vβ-Dβ-Jβ junctions of TRB genes. Using the online high-throughput tool IMGT/HighV-QUEST,11 the sequences are analyzed for information such as rearrangements, V/D/J gene use, and CDR3 clones and their frequencies.
It is estimated that the total number of T cells in the human is approximately 1012, and the TRB repertoire consists of 106 different clones.39 Thus, the higher variability among the sequencing results of different biological replicate libraries is likely a reflection of the enormous number of low-frequency clonotypes. We estimated that 99.6% of the TRB clones were in low abundance, with frequency <10−3. Thus, although sequencing at a higher depth increased the number of low-frequency clonotypes detected, because most clontypes detected were present at frequency <10−3, increasing sequencing depth did not statistically significantly improve the overall reproducibility of the assay. These data are consistent with previous work demonstrating that the T-cell repertoire consists primarily of a diverse set of low-frequency unexpanded memory clones.40, 41 These low-frequency clones cannot be reliably captured or characterized by sequencing depths commonly used to assess TRB repertoires.
Despite these limitations, the NGS technology is robust, with excellent reproducibility with respect to replicates of a given biological replicate library. Its sensitivity of reliably detecting clones in a random biological replicate library is approximately 10−5. Focusing on high-abundance clones and sequencing on more than one separate biological replicate library of an individual sample can mitigate the sampling issues in the library preparation. We showed that one random biological replicate library can detect >93% of the clones in high abundance, with frequency ≥10−3 in a repertoire. Sequencing on two different biological replicate libraries can increase the detection rate to >97%. It is expected that sequencing more than two biological replicate libraries will achieve an even higher detection rate. Herein, we demonstrated that sequencing multiple biological replicates with lower sequencing depth is a cost-effective strategy to increase power and accuracy in detection and study of T- and B-cell receptor repertories. The present result is also consistent with previous work using RNA-seq differential expression studies.42
Assessment of diversity seems to be a reproducible classifier with this method. In the present cohorts of healthy people and patients with GVHD, the latter had significantly smaller size and remarkably lower diversity of TRB repertoires. These results are consistent with previous reports that patients with GVHD after HSCT and stem cell transplant recipients have a significantly smaller and restricted T-cell repertoire.7, 14
NGS technology has emerged as a tool to quantify TCR repertoires that is quickly moving from a research tool to commercial and clinical testing for assessment of clonality and assessment of immune interventions. Defining standards and limitations of library preparation, sequencing and data analysis for detection of TCR repertoires with NGS is a recognized need in the field.28 Toward that end, this work demonstrates that BIOMED-2 primer–based NGS reliably characterizes TRB populations in healthy individuals and patients with GVHD with frequency >10−3 and provides a framework for understanding the limitations of applying NGS immune repertoire methods to clinical testing.
Footnotes
Supported by NIH grant NCI 2PO 1CA49605.
Disclosures: None declared.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.jmoldx.2016.07.009.
Supplemental Data
References
- 1.Wu D., Sherwood A., Fromm J.R., Winter S.S., Dunsmore K.P., Loh M.L., Greisman H.A., Sabath D.E., Wood B.L., Robins H. High-throughput sequencing detects minimal residual disease in acute T lymphoblastic leukemia. Sci Transl Med. 2012;4:1–7. doi: 10.1126/scitranslmed.3003656. [DOI] [PubMed] [Google Scholar]
- 2.Weng W.K., Armstrong R., Arai S., Desmarais C., Hoppe R., Kim Y.H. Minimal residual disease monitoring with high-throughput sequencing of T cell receptors in custaneous T cell lymphoma. Cancer. 2013;5:1–9. doi: 10.1126/scitranslmed.3007420. [DOI] [PubMed] [Google Scholar]
- 3.Logan A.C., Vashi N., Faham M., Carlton V., Kong K., Buño I., Zheng J., Moorhead M., Klinger M., Zhang B., Waqar A., Zehnder J.L., Miklos D.B. Immunoglobulin and T cell receptor gene high-throughput sequencing quantifies residual disease in acute lymphoblastic leukemia and predicts post-transplantation relapse and survival. Biol Blood Marrow Transplant. 2014;20:1307–1313. doi: 10.1016/j.bbmt.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mehr R., Sternberg-Simon M., Michaeli M., Rckman Y. Models and methods for analysis of lymphocyte repertoire generation, development, selection and evolution. Immunol Lett. 2012;148:11–22. doi: 10.1016/j.imlet.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 5.van Dongen J.J.M., van der Velden V.H.J., Bruggemann M., Orfao A. Minimal residual disease diagnostics in acute lymphoblastic leukemia: need for sensitive, fast, and standardized technologies. Blood. 2015;125:3996–4009. doi: 10.1182/blood-2015-03-580027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qi Q., Liu Y., Cheng Y., Glanville J., Zhang D., Lee J.Y., Olshen R.A., Weyand C.M., Boyd S.D., Goronzy J.J. Diversity and clonal selection in the human T-cell repertoire. Proc Natl Acad Sci U S A. 2014;111:13139–13144. doi: 10.1073/pnas.1409155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meier J., Roberts C., Avent K., Hazlett A., Berrie J., Payne K., Hamm D., Desmarais C., Sanders C., Hogan K.T., Archer K.J., Manjili M.H., Toor A.A. Fractal organization of the human T cell repertoire in health and after stem cell transplantation. Biol Blood Marrow Transplant. 2013;19:366–377. doi: 10.1016/j.bbmt.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Bonnet M., Ferrier P., Spicuglia S. Molecular genetics at the T-cell receptor β locus: insights into the regulation of V(D)J recombination. Adv Exp Med Biol. 2009;650:116–132. doi: 10.1007/978-1-4419-0296-2_10. [DOI] [PubMed] [Google Scholar]
- 9.Meyer E.H., Hsu A.R., Liliental J., Löhr A., Florek M., Zehnder J.L., Strober S., Lavori P., Miklos D.B., Johnson D.S., Negrin R.S. A distinct evolution of the T-cell repertoire categorizes treatment refractory gastrointestinal acute graft-versus-host disease. Blood. 2013;121:4955–4962. doi: 10.1182/blood-2013-03-489757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mamedov I.Z., Britanova O.V., Zvyagin I.V., Turchaninova M.A., Bolotin D.A., Putintseva E.V., Lebedev Y.B., Chudakov D.M. Preparing unbiased T-cell receptor and antibody cDNA libraries for the deep next generation sequencing profiling. Front Immunol. 2013;4:1–9. doi: 10.3389/fimmu.2013.00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li S., Lefranc M.P., Miles J.J., Alamyar E., Giudicelli V., Duroux P., Freeman J.D., Corbin V.D., Scheerlinck J.P., Frohman M.A., Cameron P.U., Plebanski M., Loveland B., Burrows S.R., Papenfuss A.T., Gowans E.J. IMGT/High V QUEST paradigm for T cell receptor IMGT clonotype diversity and next generation repertoire immunoprofiling. Nat Commun. 2013;4:1–13. doi: 10.1038/ncomms3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dziubianau M., Hecht J., Kuchenbecker L., Sattler A., Stervbo U., Rodelsperger C., Nickel P., Neumann A.U., Robinson P.N., Mundlos S., Volk H.D., Thiel A., Reinke P., Babel N. TCR repertoire analysis by next generation sequencing allows complex differential diagnosis of T cell-related pathology. Am J Transplant. 2013;13:2842–2854. doi: 10.1111/ajt.12431. [DOI] [PubMed] [Google Scholar]
- 13.Mori A., Deola S., Xumerle L., Mijatovic V., Malerba G., Monsurro V. Next generation sequencing: new tools in immunology and hematology. Blood Res. 2013;48:242–249. doi: 10.5045/br.2013.48.4.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yew P.Y., Alachkar H., Yamaquchi R., Kiyotani K., Fang H., Yap K.L., Liu H.T., Wickrema A., Artz A., van Besien K., Imoto S., Miyano S., Bishop M.R., Stock W., Nakamura Y. Quantitative characterization of T-cell repertoire in allogeneic hematopoietic stem cell transplant recipients. Bone Marrow Transplant. 2015;50:1227–1234. doi: 10.1038/bmt.2015.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sufficool K.E., Lockwood C.M., Abel H.J., Hagemann I.S., Schumacher J.A., Kelley T.W., Duncavage E.J. T-cell clonality assessment by next-generation sequencing improves detection sensitivity in mycosis fungoides. J Am Acad Dermatol. 2015;73:228–236. doi: 10.1016/j.jaad.2015.04.030. [DOI] [PubMed] [Google Scholar]
- 16.Niklas N., Proll J., Weinberger J., Zopf A., Wiesinger K., Krismer K., Bettelheim P., Gabriel C. Qualifying high-throughput immune repertoire sequencing. Cell Immunol. 2014;288:31–38. doi: 10.1016/j.cellimm.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Calis J.J.A., Rosenberg B. Characterizing immune repertoires by high throughput sequencing: strategies and applications. Trends Immunol. 2014;35:581–590. doi: 10.1016/j.it.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robins H. Immunosequencing: applications of immune repertoire deep sequencing. Curr Opin Immunol. 2013;25:646–652. doi: 10.1016/j.coi.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 19.Bolotin D.A., Mamedov I.Z., Britanova O.V., Zvyagin I.V., Shagin D., Ustyugova S.V., Turchaninova M.A., Lukyanov S., Lebedev Y.B., Chudakov D.M. Next generation sequencing for TCR repertoire profiling: platform-specific features and correction algorithms. Eur J Immuol. 2012;42:3073–3083. doi: 10.1002/eji.201242517. [DOI] [PubMed] [Google Scholar]
- 20.Mamedov I.Z., Britanova O.V., Bolotin D.A., Chkalina A.V., Staroverov D.B., Zvyagin I.V., Kotlobay A.A., Turchaninova M.A., Fedorenko D.A., Novik A.A., Sharonov G.V., Lukyanov S., Chudakov D.M., Lebedev Y.B. Quantitative tracking of T cell clones after hematopoietic stem cell transplantation. EMBO Mol Med. 2011;3:201–207. doi: 10.1002/emmm.201100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.lnifro E.M., Ashshi A.M., Cooper R.J., Klapper P.E. Multiplex PCR: optimization and application in diagnostic virology. Clin Microbiol Rev. 2000;13:559–570. doi: 10.1128/cmr.13.4.559-570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petersen S.L. Alloreactivity as therapeutic principle in the treatment of hematologic malignancies. Dan Med Bull. 2007;54:112–139. [PubMed] [Google Scholar]
- 23.Fang H., Yamaguchi R., Liu X., Daigo Y., Yew P.Y., Tanikawa C., Matsuda K., Imoto S., Miyano S., Nakamura Y. Quantitative T cell repertoire analysis by deep cDNA sequencing of T cell receptor α and β chains using next-generation sequencing (NGS) Oncoimmunology. 2015;3:e968467. doi: 10.4161/21624011.2014.968467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freeman J.D., Warren R.L., Webb J.R., Nelson B.H., Holt R.A. Profiling the T-cell receptor beta-chain repertoire by massively parallel sequencing. Genome Res. 2009;19:1817–1824. doi: 10.1101/gr.092924.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robins H.S., Srivastava S.K., Campregher P.V., Turtle C.J., Andriesen J., Riddell S.R., Carlson C.S., Warren E.H. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci Transl Med. 2010;2:1–9. doi: 10.1126/scitranslmed.3001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Dongen J.J.M., Langerak A.W., Bruggemann M., Evans P.A.S., Hummel M., Lavender F.L., Delabesse E., Davi F., Schuuring E., Garcia-Sanz R., van Krieken J.H.J.M., Droese J., Gonzalez D., Bastard C., White H.E., Spaargaren M., Gonzalez M., Parreira A., Smith J.L., Morgan G.J., Kneba M., Macintyre E.A. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 27.Magoc T., Salzberg S. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greiff V., Miho E., Menzel U., Reddy S.T. Bioinformatic and statistical analysis of adaptive immune repertoires. Trends Immunol. 2015;36:738–749. doi: 10.1016/j.it.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 29.Krzywinski M., Birol I., Jones S.J., Marra M.A. Hive plots: rational approach to visualizing networks. Brief Bioinform. 2012;13:627–644. doi: 10.1093/bib/bbr069. [DOI] [PubMed] [Google Scholar]
- 30.Groenen P.J., Langerak A.W., van Dongen J.J., van Krieken J. Pitfalls in TCR gene clonality testing: teaching cases. J Hematopathol. 2008;1:97–109. doi: 10.1007/s12308-008-0013-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandberg Y., van Gastel-Mol E.J., Verhaaf B., Lam K.H., van Dongen J.J.M., Langerak A. Biomed-2 multiplex immunoglobin/T-cell receptor polymerase chain reaction protocols can reliably replace southern blot analysis in routine clonality diagnostics. J Mol Diagn. 2005;7:495–497. doi: 10.1016/S1525-1578(10)60580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laydon D.J., Bangham C.R.M., Asqith B. Estimating T-cell repertoire diversity: limitations of classical estimators and a new approach. Philos Trans R Soc Lond B Biol Sci. 2015;370:1–11. doi: 10.1098/rstb.2014.0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warren R.L., Freeman J.D., Zeng T., Choe G., Munro S., Moore R., Webb J.R., Holt R.A. Exhaustive T-cell repertoire sequencing of human peripheral blood samples reveals signatures of antigen selection and a directly measured repertoire size of at least 1 million clonetypes. Genome Res. 2011;21:790–797. doi: 10.1101/gr.115428.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klyuchnikov E., Asenova S., Kern W., Kilinc G., Ayuk F., Wiedemann B., Lioznov M., Freiberger P., Zalyalov Y., Zander A.R., Kröger N., Bacher U. Post-transplant immune reconstitution after unrelated allogeneic stem cell transplant in patients with acute myeloid leukemia. Leuk Lymphoma. 2010;51:1450–1463. doi: 10.3109/10428194.2010.496015. [DOI] [PubMed] [Google Scholar]
- 35.Kanda J., Chiou L.W., Szabolcs P., Sempowski G.D., Rizzieri D.A., Long G.D., Sullivan K.M., Gasparetto C., Chute J.P., Morris A., McPherson J., Hale J., Livingston J.A., Broadwater G., Niedzwiecki D., Chao N.J., Horwitz M.E. Immune recovery in adult patients after myeloablative dual umbilical cord blood, matched sibling, and matched unrelated donor hematopoietic cell transplantation. Biol Blood Marrow Transplant. 2012;18:1664–1676. doi: 10.1016/j.bbmt.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Z.R., Zhu H.Y., DA W.M., Gao C.J., Yu L., Fu L.Y., Li M. Change and clinical significance of peripheral blood T-lymphocyte functional subsets in acute graft-versus-host disease. J Exp Hematol. 2015;23:190–194. doi: 10.7534/j.issn.1009-2137.2015.01.036. [DOI] [PubMed] [Google Scholar]
- 37.Bayraktar U.D., Milton D.R., Guindani M., Rondon G., Chen J., Al-Atrash G., Rezvani K., Champlin R., Ciurea S.O. Optimal threshold and time of absolute lymphocyte count assessment for outcome prediction after bone marrow transplantation. Biol Blood Marrow Transplant. 2016;22:505–513. doi: 10.1016/j.bbmt.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toor A.A., Sabo R.T., Roberts C.H., Moore B.L., Salman S.R., Scalora A.F., Aziz M.T., Shubar Ali A.S., Hall C.E., Meier J., Thorn R.M., Wang E., Song S., Miller K., Rizzo K., Clark W.B., McCarty J.M., Chung H.M., Manjili M.H., Neale M.C. Dynamical system modeling of immune reconstitution after allogeneic stem cell transplantation identifies patients at risk for adverse outcomes. Biol Blood Marrow Transplant. 2015;21:1237–1245. doi: 10.1016/j.bbmt.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arstila T.P., Casrouge A., Baron V., Even J., Kanellopoulos J., Kourilsky P. A direct estimate of the human αβ T cell receptor diversity. Science. 1999;286:958–961. doi: 10.1126/science.286.5441.958. [DOI] [PubMed] [Google Scholar]
- 40.Klarenbeek P.L., Tak P.P., van Schaik B.D., Zwinderman A.H., Jakobs M.E., Zhang Z., van Kampen A.H., van Lier R.A., Baas F., de Vries N. Human T-cell memory consists mainly of unexpanded clones. Immunol Lett. 2010;133:42–48. doi: 10.1016/j.imlet.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 41.van Heijst J.W., Ceberio I., Lipuma L.B., Samilo D.W., Wasilewski G.D., Gonzales A.M., Nieves J.L., van den Brink M.R., Perales M.A., Pamer E.G. Quantitative assessment of T cell repertoire recovery after hematopoietic stem cell transplantation. Nat Med. 2013;19:372–377. doi: 10.1038/nm.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y., Zhou J., White K.P. RNA-seq differential expression studies: more sequence or more replication? Bioinformatics. 2014;30:301–304. doi: 10.1093/bioinformatics/btt688. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.