Abstract
Fibroblast growth factor receptor 1 (FGFR1) amplification drives poor prognosis and is an emerging therapeutic target. We sought to construct a multigene mRNA expression signature to efficiently identify FGFR1-amplified estrogen receptor–positive (ER+) breast tumors. Five independent breast tumor series were analyzed. Genes discriminative for FGFR1 amplification were screened transcriptome-wide by receiver operating characteristic analyses. The METABRIC series was leveraged to construct/evaluate four approaches to signature composition. A locked-down signature was validated with 651 ER+ formalin-fixed, paraffin-embedded tissues (the University of British Columbia–tamoxifen cohort). A NanoString nCounter assay was designed to profile selected genes. For a gold standard, FGFR1 amplification was determined by fluorescent in situ hybridization (FISH). Prognostic effects of FGFR1 amplification were assessed by survival analyses. Eight 8p11-12 genes (ASH2L, BAG4, BRF2, DDHD2, LSM1, PROSC, RAB11FIP1, and WHSC1L1) together with the a priori selected FGFR1 gene, highly discriminated FGFR1 amplification (area under the receiver operating characteristic curve ≥0.82, all genes and all cohorts). The nine-gene signature Call-FGFR1-amp accurately identified FGFR1 FISH-amplified ER+ tumors in the University of British Columbia–tamoxifen cohort (specificity, 0.94; sensitivity, 0.96) and exhibited prognostic effects (disease-specific survival hazard ratio, 1.57; 95% CI, 1.14–2.16; P = 0.005). Call-FGFR1-amp includes several understudied 8p11-12 amplicon-driven oncogenes and accurately identifies FGFR1-amplified ER+ breast tumors. Our study demonstrates an efficient approach to diagnosing rare amplified therapeutic targets with FISH as a confirmatory assay.
Currently, the diagnosis of amplified therapeutic targets in breast cancer heavily relies on fluorescence in situ hybridization (FISH), a laborious test requiring visual confirmation by a pathologist. In the case of human epidermal growth factor receptor 2 (HER2) gene amplification, immunohistochemistry is frequently used to screen samples for extremes of HER2 expression, with FISH reserved for cases that are equivocal.1, 2, 3, 4 Immunohistochemistry depends on a highly specific HER2 monoclonal antibody, but even in this well-established HER2 amplification diagnosis, immunohistochemistry is worrisomely subjective and exhibits preanalytical variability to the point that some pathologists recommend HER2 FISH for all cases.5 Herein, we consider the diagnosis of amplification events affecting an emerging therapeutic target, fibroblast growth factor receptor 1 (FGFR1). This membrane tyrosine kinase receptor is amplified in a variety of solid tumors,6, 7, 8, 9 including breast,10, 11, 12, 13, 14, 15 lung,16, 17, 18, 19, 20, 21 esophageal,22 bladder,23 and ovarian cancers.24 In breast cancer, FGFR1 is amplified in approximately 10% of estrogen receptor–positive (ER+) patients, corresponding to poor prognosis25 and endocrine resistance.26 FGFR1-targeting agents have shown activity, inhibiting FGFR1 signaling in preclinical studies27, 28, 29, 30 with early signs of activity in clinical trials.25, 31, 32, 33, 34, 35
The diagnosis of FGFR1 amplification is a greater challenge than HER2 amplification because a monoclonal antibody for formalin-fixed, paraffin-embedded (FFPE)–based FGFR1 protein detection has yet to be successfully developed. FGFR1 amplification incidence is lower than HER2, further increasing the cost and time for testing, with only 1 in 10 cases producing a positive result. DNA copy number assays, such as array comparative genomic hybridization (aCGH) and single-nucleotide polymorphism (SNP), are more high throughput than FISH and have been widely used to profile DNA copy number aberrations in the setting of research studies with high-quality DNA.13, 36 However, DNA extracted from FFPE tissues is partially degraded, and these techniques have not, to date, become a diagnostic standard.
Overexpression of FGFR1 at the mRNA level is strongly correlated with FGFR1 amplification21, 26, 37 and, thus, prescreening for FISH positivity based on FGFR1 mRNA levels is an approach to consider. However, the 8p11-12 amplicon is complex, with several peaks of amplification that encompass multiple potential oncogenes.37, 38 FGFR1 is usually coamplified with other genes in this amplicon region, leading to their correlated mRNA overexpression. To develop a comprehensive approach to the diagnosis of the FGFR1/8p11-12 amplicon, we therefore conducted a transcriptome-wide screening and identified nine 8p11-12 genes (FGFR1, LSM1, BAG4, ASH2L, BRF2, DDHD2, PROSC, RAB11FIP1, and WHSC1L1) whose mRNA expression greatly distinguished FGFR1-amplified ER+ breast tumors from nonamplified tumors. We subsequently leveraged a public data set [Molecular Taxonomy of Breast Cancer International Consortium (METABRIC)]36 to optimize an integrated signature for determination of FGFR1 positivity. The favored signature, Call-FGFR1-amp, for accurate identification of FGFR1 FISH-positive ER+ breast tumors was preliminarily confirmed in the setting of Agilent microarray gene expression data. To complete the clinical translation process, we developed a custom NanoString nCounter assay (a gold standard technical platform for gene expression) to specifically profile the target genes and validated the signature Call-FGFR1-amp in a prospective experiment by assaying a well-annotated cohort [University of British Columbia (UBC)–tamoxifen (TAM)] with long follow-up.39 Call-FGFR1-amp was assessed for diagnostic accuracy against FGFR1 FISH amplification and for association with survival outcomes to ensure that it captured the poor outcomes previously associated with the 8p11 amplicon observed in other entirely independent breast cancer studies.9, 14, 26
Materials and Methods
Patients and Specimens
We sought to develop and validate an mRNA gene expression–based signature to accurately triage FGFR1 nonamplified samples so that a FISH assay, required for FGFR1 amplification confirmation, would only be needed in a much smaller number of cases. ER+ breast cancer patients from five independent breast tumor series were used to identify FGFR1-amplification discriminative genes and to construct and validate the signature referred to herein as Call-FGFR1-amp. In brief, the discovery cohort encompassing 64 ER+ breast cancer patients treated by endocrine therapies from the preoperative letrozole40 and the American College of Surgeons Oncology Group (now part of the Alliance for Clinical Trials in Oncology) Z1031 trials (https://clinicaltrials.gov, identifier NCT00265759)41 (Figure 1 and Supplemental Table S1) was used to select genes discriminating FGFR1 amplification at the mRNA level. The clinical trial protocols were approved by the Institutional Review Board at Washington University (St. Louis, MO). Data from 1508 ER+ breast cancer patients (Figure 2A and Supplemental Table S2) from the METABRIC study36 were analyzed to confirm the individual discriminative ability of the selected genes and to develop/evaluate four informatics approaches to combining the nine genes to produce a composite signature. Data from the METABRIC study were accessed with permission from the European Genome-Phenome Archive (EGA; http://www.ebi.ac.uk/ega). A final signature, Call-FGFR1-amp, was locked down before validation efforts in two independent studies, the Strategic Partnering to Evaluate Cancer Signatures (SPECS) cohort and the UBC/British Columbia Cancer Agency (UBC-TAM) cohort. A total of 138 breast cancer samples from the SPECS cohort were used for FISH testing, whereas 104 had paired Agilent whole-genome microarray data, among which 47 were ER+42 (Figure 3A and Supplemental Table S3). The UBC-TAM cohort39 includes 1276 ER+ breast cancer patients treated by tamoxifen alone, with a median follow-up of nearly 10 years. FFPE RNA samples were available from 651 cases of this cohort, and these cases were prospectively subjected to FISH testing (Figure 4A and Supplemental Table S4). Specimen collection for SPECS and for UBC-TAM was approved by the Washington University Institutional Review Board and the UBC/British Columbia Cancer Agency Clinical Research Ethics Board, respectively.
Figure 1.
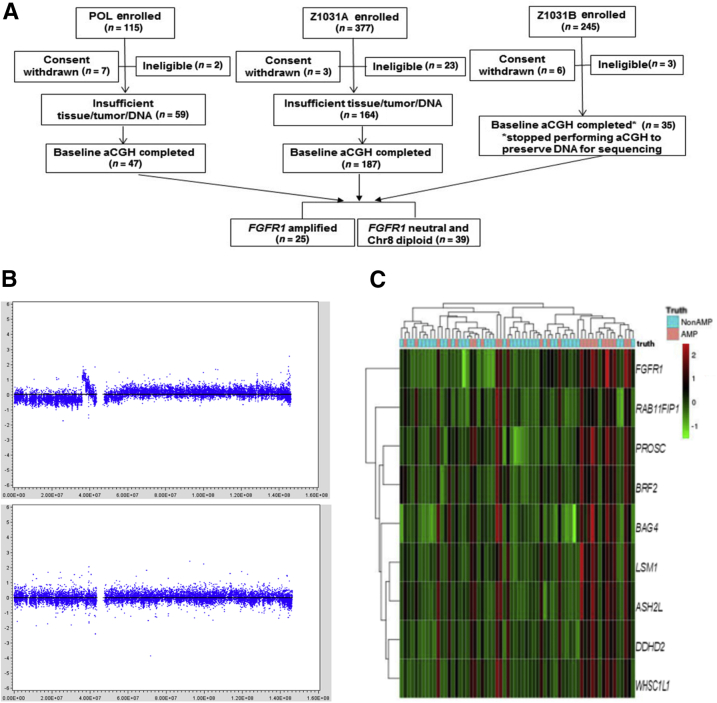
Identification of genes discriminating FGFR1 amplification from FGFR1 neutral in the discovery cohort [ie, the preoperative letrozole (POL)-Z1031 cohort]. A: The REMARK diagram shows 25 FGFR1-amplified estrogen receptor–positive (ER+) tumors were chosen to be compared against 39 FGFR1 neutral and chromosome 8 diploid ER+ tumors for genes discriminative of FGFR1 amplification. B: Firestorm analysis. The segmented array comparative genomic hybridization (aCGH) copy number signals from the circular binary segmentation algorithm (in log2 ratio, y axis) were plotted against chromosome 8 genome locations (x axis). The top panel shows a FGFR1 firestorm amplified tumor example, and the bottom panel shows a FGFR1 neutral and chromosome 8 diploid tumor example. C: Heat map of the mRNA gene expression levels (in red/green color scheme) of the nine genes (at row). Overexpression of the genes corresponds well to the FGFR1-amplified cases among the 64 samples (at column), as determined by the firestorm analysis (indicated in the top image bar as truth: AMP in pink for amplification and NonAMP in blue for nonamplification).
Figure 2.
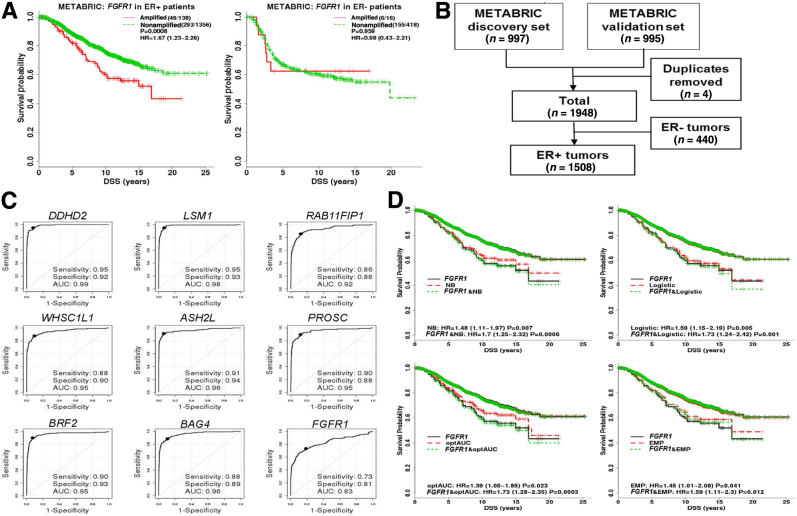
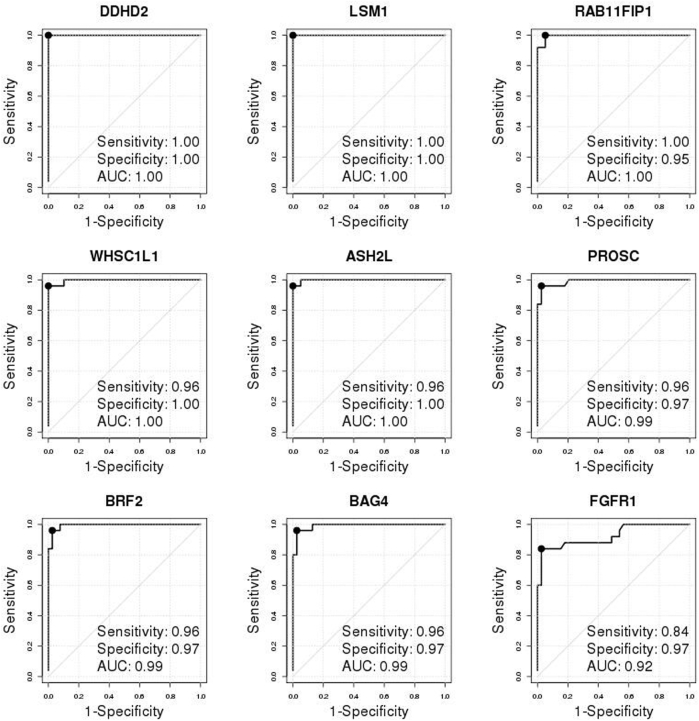
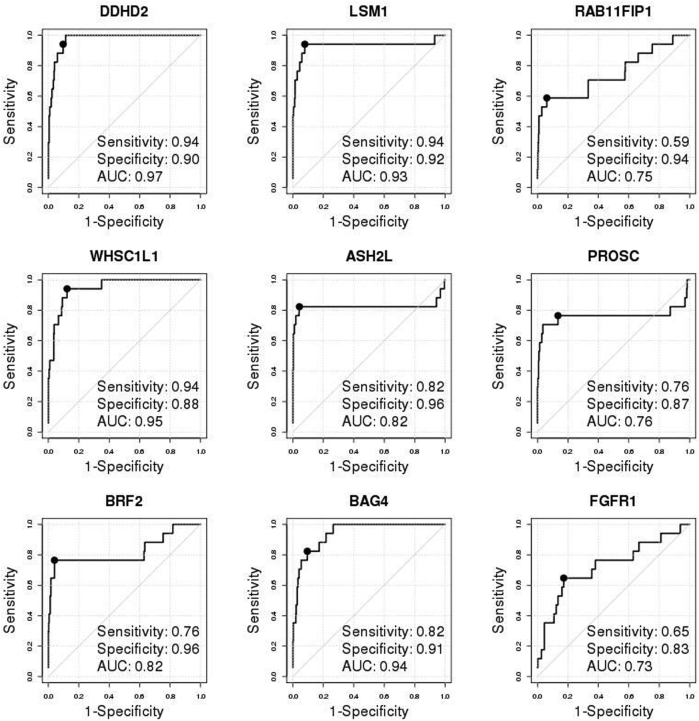
Validation of individual genes and development/evaluation of multigene signatures in the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohort. A: The REMARK diagram; 1508 estrogen receptor–positive (ER+) METABRIC samples were used for FGFR1 amplification analyses. B: Disease-specific survival (DSS) Kaplan-Meier (KM) curves of true FGFR1 amplification status, as determined by single-nucleotide polymorphism (SNP) array copy number analysis stratified by ER status, demonstrated that FGFR1 amplification is prognostic in ER+ tumors but not in ER- tumors. In parentheses are total number of events/total number of samples with valid survival and FGFR1 amplification data. The hazard ratios (HRs) with associated 95% CIs and the log-rank test P values (P) are reported. C: Individual receiver operating characteristic (ROC) curve confirms the discriminative ability of each individual gene for FGFR1 amplification. The solid black point indicates the optimal cutoff point on each ROC curve. The legend shows area under the ROC curve (AUC), sensitivity, and specificity corresponding to the optimal cutoff. D: Overlaid KM curves for DSS indicate that each of the four composite signatures is prognostic and tracks the true nonamplified curve well. KM curves of amplification statuses, as determined by each of the four multigene signatures from 10-fold cross-validations (red dashed curves: upper/lower red curves for nonamplified/amplified patients, respectively), were overlaid by gold standard FGFR1 amplification status, as determined by SNP array copy number analysis (black solid curves, upper/lower curves for nonamplified/amplified patients, respectively), and amplification status, as determined by both a signature and the true status (green dotted curves, upper/lower curve for nonamplified cases determined by both/amplified cases determined by both, respectively). The HRs with associated 95% CIs and the log-rank test P values (P) are reported. The signature Call-FGFR1-amp based on the optimal AUC (optAUC) algorithm was carried forward locked down for validation. EMP, empirical method; Logistic, logistic regression method; NB, naïve Bayes method.
Figure 3.
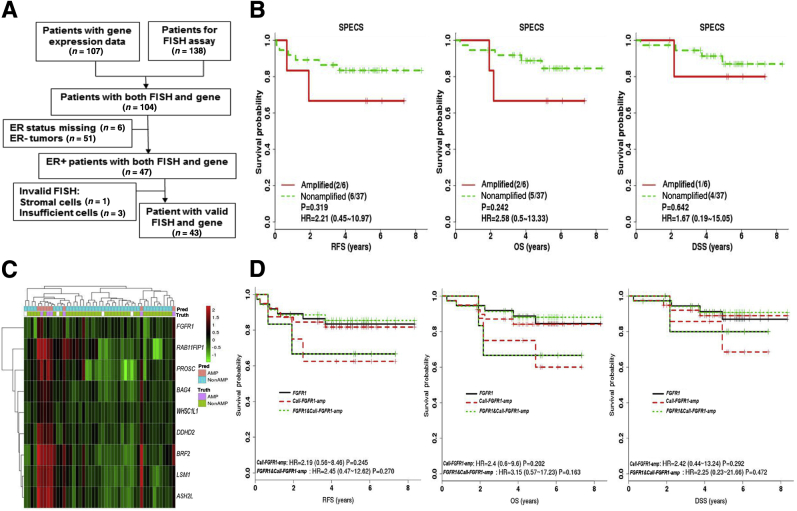
Validation of FGFR1 amplification calls by Call-FGFR1-amp in the Strategic Partnering to Evaluate Cancer Signatures (SPECS) cohort. A: The REMARK diagram; 47 estrogen receptor–positive (ER+) SPECS samples were analyzed for FGFR1 amplification analyses. B: Kaplan-Meier (KM) curves by true FGFR1 amplification status, as determined by fluorescent in situ hybridization (FISH) for relapse-free survival (RFS), overall survival (OS), and disease-specific survival (DSS), show prognostic trends. Indicated in the legend are the total number of events/total number of patients corresponding to the amplified and nonamplified patients in the parenthesis, hazard ratios (HRs), and 95% CIs and log-rank test P values (P). C: Heat map of the nine selected genes (at row) shows concordance between the genes' overexpression, the amplification statuses called by Call-FGFR1-amp, and the FGFR1 FISH amplification statuses of the 47 ER+ tumors (at column), indicated in the top image bar by pred (for Call-FGFR1-amp) and truth (for FISH), respectively. AMP in pink/purple for amplification and NonAMP in blue for nonamplification. White bars are for cases with nonreadable FISH results. D: Overlaid KM curves exhibit close tracking of Call-FGFR1-amp for FGFR1 FISH, showing similar prognostic effects. KM curves for RFS, OS, and DSS by FGFR1 amplification based on Call-FGFR1-amp (Call-FGFR1-amp: red dashed curves, upper/lower red curves corresponding to nonamplified versus amplified patients, as determined by Call-FGFR1-amp, respectively), overlaid with curves by the FISH gold standard (FGFR1: black solid curves, upper/lower curves corresponding to nonamplified versus amplified patients, as determined by FISH, respectively) and FGFR1 status, as determined by both Call-FGFR1-amp and FISH (FGFR1 and Call-FGFR1-amp: green dotted curves, the upper green dotted curve representing patients who were FGFR1 FISH amplified and called so by Call-FGFR1-amp versus the lower green dotted curves representing the remaining patients who were called nonamplified by both or had inconsistent calls between FISH and Call-FGFR1-amp). The HRs with 95% CIs and log-rank test P values (P) were indicated in the legend.
Figure 4.
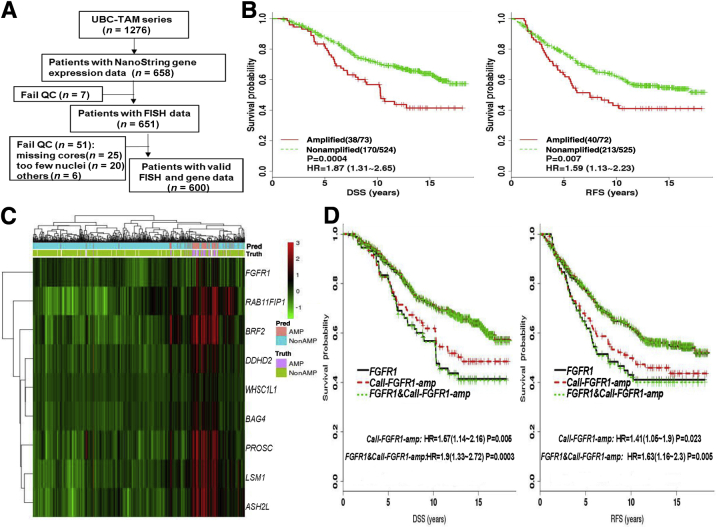
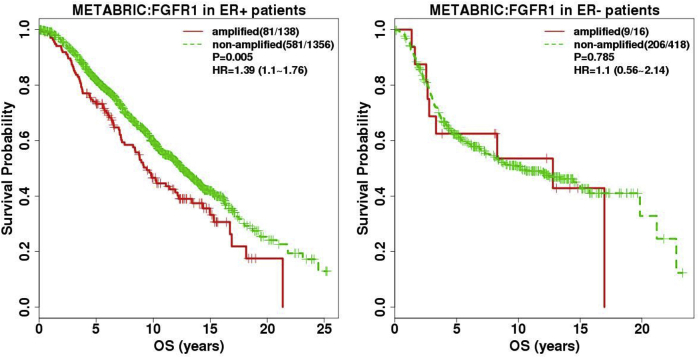
Validation of FGFR1 amplification calls by Call-FGFR1-amp in the University of British Columbia (UBC)–tamoxifen (TAM) cohort. A: The REMARK diagram; 651 estrogen receptor–positive (ER+) UBC-TAM samples were analyzed for FGFR1 amplification analyses. B: Kaplan-Meier (KM) curves by true FGFR1 amplification status, as determined by fluorescent in situ hybridization (FISH), show prognostic effects for disease-specific survival (DSS) and relapse-free survival (RFS). C: Heat map of the nine selected genes (at row) shows concordance between the genes' overexpression, the amplification statuses called by Call-FGFR1-amp, and the FGFR1 FISH amplification statuses of the 651 ER+ tumors (at column), indicated in the top image bar by pred (for Call-FGFR1-amp) and truth (for FISH), respectively. AMP in pink/purple for amplification and NonAMP in blue for nonamplification. White bars are for cases with nonreadable FISH results. D: Overlaid KM curves exhibit close tracking of Call-FGFR1-amp for FGFR1 FISH, showing similar prognostic effects. KM curves for DSS and RFS by FGFR1 amplification based on Call-FGFR1-amp (Call-FGFR1-amp: red dashed curves, upper/lower red curves corresponding to nonamplified versus amplified patients, as determined by Call-FGFR1-amp, respectively), overlaid with curves by the FISH gold standard (FGFR1: black solid curves, upper/lower curves corresponding to nonamplified versus amplified patients, as determined by FISH, respectively) and FGFR1 status, as determined by both Call-FGFR1-amp and FISH (FGFR1 and Call-FGFR1-amp: green dotted curves, the upper green dotted curve representing patients who were FGFR1 FISH amplified and called so by Call-FGFR1-amp versus the lower green dotted curves representing the remaining patients who were called nonamplified by both or had inconsistent calls between FISH and Call-FGFR1-amp). The hazard ratios (HRs) with 95% CIs and log-rank test P values (P) were indicated in the legend. QC, quality control.
FGFR1 Amplification at the DNA Level
The aCGH method was performed on the discovery cohort for DNA copy number profiling and the method was described previously.43 In brief, tumor DNA was extracted from five to seven FFPE tissue sections (50 μm thick) that contained a minimum of 70% tumor cellularity, using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA). Using cell pellets or whole blood, germline DNA was extracted using the QIAamp DNA Mini Kit (Qiagen) or QIAamp DNA Blood Mini/Maxi Kit (Qiagen), according to manufacturer's instructions. Genomic DNAs from both a tumor and a common germline reference sample were restriction digested (AluI/RsaI), fluorescently labeled with either a Cy5 dye (tumor) or a Cy3 dye (germline reference), and column purified using the Bioprime Total Genomic Labeling System (18097-011; Invitrogen, Carlsbad, CA). Both tumor and germline reference purified samples were cohybridized to an Agilent (Santa Clara, CA) Human Genome 244 K CGH array and washed according to Agilent protocols. Log ratios (tumor to reference) were then extracted for each array feature using an Agilent scanner and Feature Extraction software version 9.5.3.1. Circular binary segmentation44, 45 was applied to divide the genome into regions of equal copy numbers, and whole-genome DNA copy number status was assessed by CGHcall.46 The amplification status of FGFR1 in the discovery cohort was determined based on CGHcall results and by observation of a firestorm peak in the FGFR1 region using chromosome 8 scatter plots of segmented copy number signals (Figure 1). Samples were identified as FGFR1 amplified if any genes within the FGFR1 genomic region exhibited amplification. Samples exhibiting chromosome 8 diploidy and FGFR1 neutral copy number status were defined as FGFR1 nonamplified.
For the METABRIC study, whole genome copy number statuses processed from Affymetrix 6.0 human whole genome SNP array data, as previously described,36 were downloaded from the EGA (http://www.ebi.ac.uk/ega) under study accession number EGAS00000000098. To determine FGFR1 gene DNA amplification status in the METABRIC cohort, copy number status for the segmented regions in the FGFR1 gene region (chromosome 8, location 38268656-38326352) was extracted. A tumor was defined to be FGFR1 amplified if any of segmented regions in the gene region demonstrated amplification because the copy number data were rigorously segmented and amplification statuses of segmented regions were called rigorously (see Supplemental Methods of Curtis et al36) and, moreover, the amplification rate (approximately 9%) estimated based on this simple rule in METABRIC is close to published data.
FGFR1 amplification was determined by FISH on both the SPECS and the UBC-TAM cohorts. FISH on the SPECS cohort was conducted on tissue sections (5 or 6 μm thick) after deparaffinization and target retrieval using steam cooking in citrate buffer for 20 minutes, followed by a 20-minute cool-down period and a 5-minute wash with distilled water, then pepsin digestion (37°C, 30 minutes) and a subsequent wash in 2× standard saline citrate. FGFR1 (8p12)/CEN8q FISH probes (Abbott Molecular, Des Plaines, IL) were codenatured with the tissues at 90°C for 13 minutes and hybridized at 37°C overnight. After hybridization, slides were washed in 50% formamide/1× standard saline citrate (5 minutes) and 2× standard saline citrate (5 minutes) at room temperature, air dried, counterstained with DAPI (0.5 L/mL), and examined on an Olympus BX60 fluorescent microscope with appropriate filters (Olympus, Melville, NY). The number of FGFR1 (red) and chromosome 8 (green) signals per cell was scored on a minimum of 100 nonoverlapping tumor nuclei by two technologists blinded to gene expression and patient clinical data. The average gene copy number and FGFR1/CEP8 ratio was calculated for each sample. A region was considered amplified if the number of FGFR1 signals was greater than five relative to the centromeric probe, and a tumor sample was defined as amplified when 10% or more of tumor cells showed such a region amplification. All other readouts were regarded as FGFR1 nonamplified. FISH on the UBC-TAM cohort used FFPE tissue sections (4 μm thick) from tissue microarray blocks that were baked overnight at 60°C, deparaffinized using xylene (5 minutes each, ×3), and dehydrated in 100% ethanol (10 minutes each, ×2). The slides were then pretreated in 10 mmol/L citric acid buffer at 80°C for 50 minutes, followed by pepsin digestion (37°C, 7 to 10 minutes) and dehydration in an ethanol series (70%, 80%, 100%; 1 minute each). FGFR1 (8p12)/CEN8q FISH probes (Abnova, Taipei, Taiwan) were codenatured with the tissues at 73°C for 5 minutes and hybridized at 37°C for 16 to 18 hours. After hybridization, slides were washed with 2× standard saline citrate/0.3% NP40 at 73°C for 2 minutes, dehydrated in the ethanol series, and counterstained with DAPI. The number of FGFR1 (red) and chromosome 8 (green) signals per cell was scored on a minimum of 50 tumor cells from each tissue core by two experienced cytogeneticists who had no access to gene expression or patient clinical data. The average FGFR1 gene copy number and FGFR1/CEP8 ratio was calculated for each core. A sample was defined as FGFR1 amplified if the FGFR1 copy number was >6, or the FGFR1/CEP8 ratio >2.2.14
mRNA Gene Expression Data
The whole-transcriptome mRNA gene expression levels on the fresh frozen tumor tissues that contained a minimum of 50% tumor cellularity in the discovery cohort and the SPECS cohort were each profiled on Agilent Human Gene Expression 4 × 44 K and 244 K Microarrays. RNA was extracted from frozen biopsy tumor tissues. Five to seven sections (50 μm thick) of a tumor tissue were homogenized in Trizol, and total RNA was extracted using the Qiagen RNA extraction reagents. High-quality total RNA from both a tumor biopsy and a universal reference sample1 (100 to 500 ng) was used to synthesize cDNA, followed by T7 polymerase in vitro transcription with either a cy5-CTP (tumor) or cy3-CTP (reference) from Perkin Elmer incorporated during in vitro transcription using Agilent's Low Input Linear Amplification Kit. Amplified, labeled tumor and reference cRNA (825 ng) was then cohybridized to an Agilent 4 × 44 K Whole Human Genome microarray (G4112F), washed, and dried according to Agilent's Two-Color Microarray-Based Gene Expression Analysis protocol (version 5.0.1). Processed arrays were then scanned with an Agilent Microarray Scanner (G2505B), and probe data were extracted from the scanned image using Agilent's Feature Extraction software version 9.5.3.1. Feature Extraction data for each patient were preprocessed through the University of North Carolina Microarray Database (https://genome.unc.edu, registration required),42, 47 where probes of poor quality, as determined by Feature Extraction algorithms, were removed under the following conditions: spot was not found in either channel, spot or background was a nonuniform outlier, spot or background was a nonuniform outlier for the population, or the spot was not positive and significant in either channel. Log2 ratio (tumor/reference) was then calculated for the probes that passed the above filters, and Lowess was used for data normalization.48 Spots with a Lowess normalized net (mean) <10 in either channel were filtered in the final data set. The Agilent microarray raw files and the normalized gene expression data of the whole transcriptomes of the discovery cohort were deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under the accession number GSE77626. Supplemental Tables S5 and S6 provide the gene expression data of the selected genes.
For the METABRIC study, the processed whole genome gene expression data from the Illumina (San Diego, CA) Human HT-12 array version 3, as previously described,36 were downloaded from EGA (http://www.ebi.ac.uk/ega) under study accession number EGAS00000000098.
Total RNA from the FFPE tumor tissues of the UBC-TAM samples was extracted, as previously described,39 and the mRNA expression (Supplemental Table S7) of the nine targeted genes was profiled using the NanoString nCounter analysis system. A custom capture and reporter CodeSet (Supplemental Table S7) for the target genes was designed according to the manufacturer's guidelines (NanoString Technologies, Inc., Seattle, WA) using methods previously described.49 The RNA samples were hybridized with the code set and processed using the NanoString nCounter Analysis system, as described in our previously published studies.47, 50 Six positive control genes at six concentrations, eight negative control genes, and three housekeeping genes (MRPL19, SF3CA1, and PUM1) were profiled together with the nine candidate genes. Quality control and preprocessing of the NanoString nCounter raw counts on the UBC-TAM cohort samples were performed according to the manufacturer's instruction guide, through the following sequential processing steps: i) remove samples with a binding density >2.25 or <0.1; ii) remove samples with three or more positive control genes having zero counts; iii) calculate a positive control normalization factor per sample as the mean summed counts of the six positive control genes across all samples divided by the summed counts of the six positive control genes per sample; iv) remove samples with the positive control normalization factor >3 or <0.3; v) calculate the mean and SD across negative control genes per sample and subtract each gene expression value by the background, defined as mean + 2 × SD; vi) calculate the housekeeping genes geometric mean per sample and the averaged geometric mean across samples, and the housekeeper gene normalization factor is calculated as the averaged geometric mean divided by per-sample geometric mean; vii) gene expression values per sample, multiply the sample-specific housekeeper gene normalization factor; viii) take log2 transformation of the gene expression.
Statistical Analysis
Missing values in gene expression data were imputed using the R impute package.51 Descriptive statistics (mean, SD for continuous variables, and count/frequency for categorical variables) were used to summarize patient characteristics. Fisher's exact test was used to assess associations between categorical patient characteristics. Disease-specific survival (DSS) was defined as the time interval between the date of initial diagnosis of breast cancer and the date of death due to the disease. Overall survival and relapse-free survival (RFS) were defined as the time interval between the date of initial diagnosis of breast cancer and the date of any type of death and relapse of the disease, respectively. Survival analyses were performed to assess prognostic effects of FGFR1 amplification on survival end points. The Kaplan-Meier (KM) product limit method was used to estimate empirical survival probabilities, whereas survival difference was compared by log-rank test. Cox proportional hazard regression modeling was used to estimate the hazard ratio (HR) of FGFR1-amplified cases to nonamplified cases and the associated 95% CI. The proportional hazard assumption was examined by plotting and testing the Schoenfeld residuals.
The FGFR1 amplification status, as determined at the DNA level (aCGH, SNP array, or FISH), was regarded as the true gold standard. The FGFR1 amplification status determined by a signature at the mRNA level was evaluated against these gold standards. For assessment of the discriminative ability of an individual gene for FGFR1 amplification, the receiver operating characteristic (ROC) analyses52 were performed. The area under the ROC curve (AUC) was estimated using the nonparametric trapezoidal method with 95% CI by the method of DeLong et al53 to gauge the discriminative ability across all possible cutoff points. To make binary FGFR1 amplification calls, an optimal cut point was adopted as corresponding to the coordinate on an ROC curve with the minimum distance to the coordinate with a perfect sensitivity and a perfect specificity. By the optimal cut point, various diagnostic measures can be derived, including sensitivity, specificity, positive and negative predictive values, overall accuracy, the ratio of false positives (FPs) to true positives (TPs), labeled as FP/TP, the ratio of false negatives (FNs) to true negatives (TNs), labeled as FN/TN, the Youden index, which does not consider population prevalence and uses equal weight on sensitivity and specificity (ie, FPs and FNs were equally penalized and essentially the summation of sensitivity and specificity), and the Youden index (labeled as Youden*), which considers population prevalence and allows different penalty/cost on FP and FN.54 Herein, we chose a large penalty/cost on FN relative to FP as 5:1 because the objective was to initially screen out FGFR1 nonamplified tumors such that only the remaining tumors would need to be confirmed by more labor-intensive FISH assays for FGFR1 amplification. More detailed definitions on these diagnostic characteristics are as follows: i) sensitivity was defined as the proportion of amplified cases as called by a signature among the true FGFR1-amplified cases; ii) specificity was defined as the proportion of nonamplified cases as called by a signature among the true FGFR1 nonamplified cases; iii) overall accuracy was defined as the proportion of correctly classified amplified and nonamplified cases among all cases; iv) positive predictive value was defined as the proportion of true amplified cases among those called as amplified by a signature; v) negative predictive value was defined as the proportion of true nonamplified cases among those called as nonamplified by a signature; vi) Youden index was calculated as: sensitivity + specificity − 1, essentially the summation of sensitivity and specificity. Youden index does not incorporate population prevalence and uses equal weights on sensitivity and specificity, in another word, FPs and FNs were equally penalized; and vii) Youden* was calculated as:
| (1) |
with CFP and CFN referring to the cost/penalties for FP and FN, respectively. Herein, we chose a large penalty on FN relative to FP (with CFP/CFN = 1/5) because the objective was to initially screen out FGFR1 nonamplified patients by a signature using mRNA gene expression data, whereas the remaining patients will be subjected to more labor-intensive FISH for confirmation. In situations of equal CFP and CFN and a 1:1 case-control study, Youden* reduces to the Youden index as defined above. FP/TP referred to the ratio of FPs to TPs (ie, the ratio of total number of true FGFR1 nonamplified cases, which were called amplified by a signature, to the total number of true FGFR1 amplified cases, which were called amplified by a signature). FN/TN referred to the ratio of FNs to TNs (ie, the ratio of total number of true FGFR1 amplified cases, which were called nonamplified by a signature, to the total number of true FGFR1 nonamplified cases, which were called nonamplified by a signature).
We considered four algorithms to construct a multigene signature during the development stage: the empirical method (labeled as EMP), the naïve Bayes method (NB), the logistic regression method (Logistic), and the optimal AUC method (optAUC).55
The empirical method (EMP). EMP was essentially a majority voting algorithm. The optimal cut point of each candidate gene is the parameter required by the algorithm. The optimal cut point on each gene was adopted as corresponding to the coordinate on the ROC curve with the minimum distance to the coordinate with perfect sensitivity and perfect specificity. By each of the nine candidate genes, each new sample was categorized into being FGFR1 amplified or nonamplified by comparing the gene's expression level to the corresponding optimal cutoff point of the gene. A new sample would be finally called FGFR1 amplified if it was classified as so by at least eight of the nine candidate genes.
The naive Bayes method (NB). The NB method was used to calculate the probability of FGFR1 amplification given the gene expression of all of the candidate genes via the Bayes theorem, where each gene was assumed to be independently normally distributed in the true amplified population and in the true nonamplified population separately. The mean and SD of each candidate gene in the true amplified population and in the true nonamplified population were the required parameters and were separately estimated using the sample mean and SD of the true amplified cases and true nonamplified cases in the training cohort. NB has shown great performance for classification in machine learning despite its simple independence assumption.56, 57 NB estimated the FGFR1 amplification probability in the range of 0 to 1, whereas a new sample was called amplified if the estimated probability was ≥0.5.
The multivariate logistic regression method (Logistic). A multivariate logistic regression model incorporating the gene expression of all of the candidate genes was fitted using the training samples to estimate the regression coefficients (the parameters). A linear score was constructed for a new sample in an independent validation cohort based on the logistic regression model, with the estimated regression coefficients and probabilities of amplification calculated. A new sample was called amplified by Logistic if its associated probability of amplification was ≥0.5.
The optimal AUC method (optAUC). The candidate genes were linearly combined to achieve an optimal AUC. The linear coefficients were estimated by a nonparametric search using the R package optAUC (version 1.0)55, 58 with default arguments in the R function optAUC, namely, the smoothing parameter “λ = 5” and the variables (genes) were standardized by setting scale = T. To dichotomize the resulting linear score, the optimal cut point was adopted as corresponding to the coordinate on the ROC curve (generated for the linear score), with the minimum distance to the coordinate with perfect sensitivity and perfect specificity. A new sample was called amplified if its linear score was above the derived optimal cut point. Thus, the linear coefficients and the optimal cut point were the required parameters.
The performance of the four algorithms was evaluated in the METABRIC cohort via a 10-fold cross-validation (CV) procedure. For the 10-fold CV, the METABRIC cohort was randomly divided into 10 subsets of approximately equal size. At each CV, one subset was held while the algorithms were implemented on the other nine subsets to estimate algorithm-specific parameters. The FGFR1 amplification statuses of the held-out samples were called by each algorithm using the estimated parameters. The performance at each CV was assessed against the true status of the held-out samples based on the diagnostic measures described earlier. According to the averaged performance across all 10 CVs, a locked-down signature, Call-FGFR1-amp, was carried forward to the validation stage. For independent validation analyses in the SPECS and the UBC-TAM cohorts, the expression of each candidate gene in a validation cohort was adjusted to have the same mean and SD as those in the METABRIC cohort to remove any obvious batch effect. The sigclust approach59, 60 was used to test if a major study-wise batch effect was removed such that a validation cohort merged well with the training cohort. A small P value (usually <0.05) from sigclust indicated the existence of major study-wise batch effect because the samples from the validation cohort and the METABRIC cohort could be regarded as from two different multivariate normal distributions, whereas a large P value indicated all of the samples were possibly from one multivariate normal distribution and thus no obvious study-wise batch effect existed. κ Coefficients were calculated to gauge agreement between FGFR1 amplification calls by a signature and the gold standards, and their association was tested by Fisher's exact test, whereas the previously described diagnostic characteristics were also calculated. Hierarchical clustering was used to cluster samples or genes, based on the average linkage and the dissimilarity metric of one minus Pearson correlation coefficient. All tests were two sided at a 5% significance level. All analyses were performed in R 2.15.2 (http://cran.r-project.org).61
Results
Identification of Discriminative Genes for FGFR1 Amplification in the Discovery Cohort
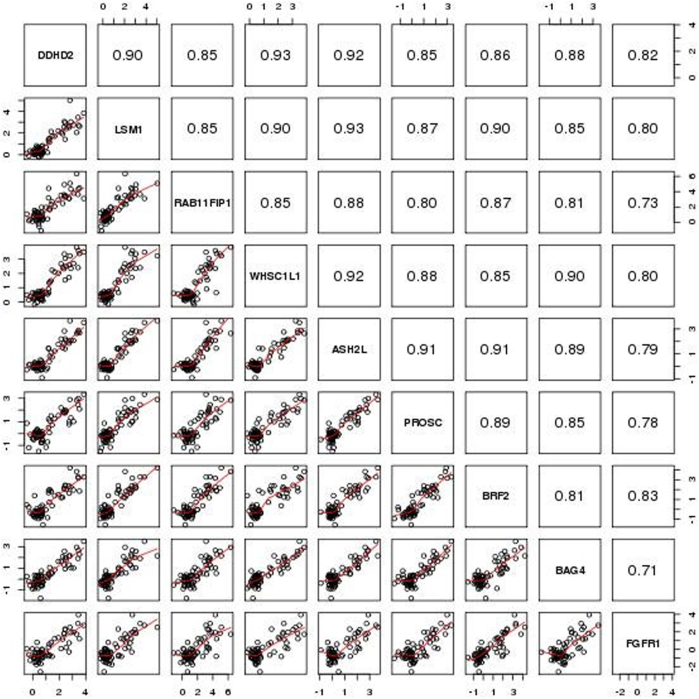
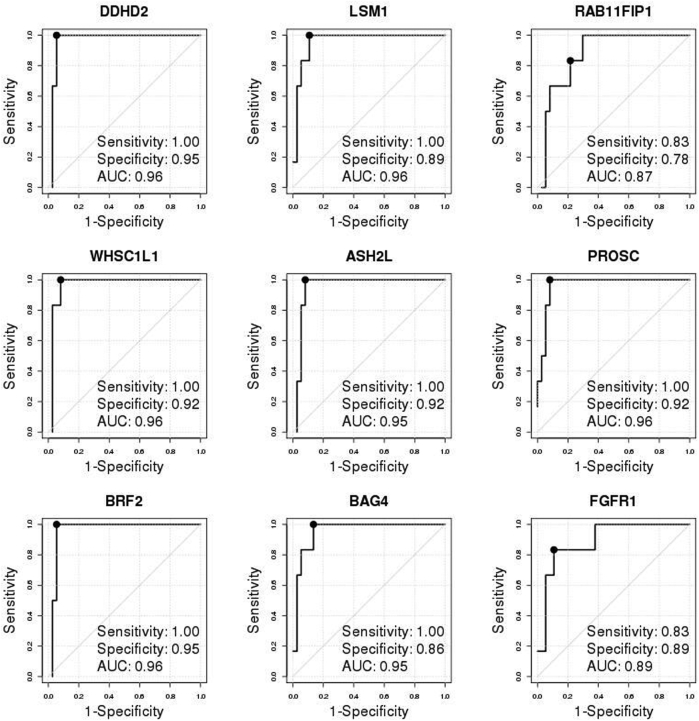
Firestorm analysis (Materials and Methods and Figure 1) of the aCGH DNA copy number profiles of tumors resulted in 25 FGFR1-amplified and 39 FGFR1 neutral ER+ breast tumors (Figure 1) from the POL/Z1031 trials. Patient characteristics of this discovery cohort were briefly summarized in Supplemental Table S1. A total of 22,962 whole-transcriptome probes were analyzed by the ROC method to identify genes discriminative for FGFR1 amplification at the mRNA level (Supplemental Table S5). From ROC analyses, eight genes (ASH2L, BAG4, BRF2, DDHD2, LSM1, PROSC, RAB11FIP1, and WHSC1L1), all located in the same chromosomal region as FGFR1, were selected because their diagnostic test measures, including AUC, sensitivity, specificity, positive predictive value, negative predictive value, and overall accuracy, corresponding to the optimal cut points were all >0.9 (Table 1 and Supplemental Figure S1). The FGFR1 gene itself, with an AUC of 0.92, a high specificity of 0.97 but a relatively low sensitivity of 0.8, was also selected a priori (Table 1 and Supplemental Figure S1). All of the nine genes were highly overexpressed in the FGFR1-amplified cases in this discovery cohort (Supplemental Figure S2). The mRNA expression levels of the selected genes showed high correlation (Pearson correlation coefficient range, 0.71 to 0.93) (Supplemental Figure S3).
Table 1.
Selection of the Candidate Genes in the Discovery Cohort (POL-Z1031)
| Gene | AUC (95% CI) | Specificity | Sensitivity | NPV | PPV | Accuracy | Cutoff |
|---|---|---|---|---|---|---|---|
| ASH2L | 1 (0.99–1) | 1.00 | 0.92 | 0.95 | 1.00 | 0.97 | 0.74 |
| BAG4 | 0.99 (0.98–1) | 0.97 | 0.92 | 0.95 | 0.96 | 0.95 | 0.45 |
| BRF2 | 0.99 (0.98–1) | 0.97 | 0.92 | 0.95 | 0.96 | 0.95 | 0.72 |
| DDHD2 | 1 (1–1) | 1.00 | 0.96 | 0.98 | 1.00 | 0.98 | 1.19 |
| FGFR1 | 0.92 (0.85–1) | 0.97 | 0.80 | 0.88 | 0.95 | 0.91 | 0.09 |
| LSM1 | 1 (1–1) | 1.00 | 0.96 | 0.98 | 1.00 | 0.98 | 1.14 |
| PROSC | 0.99 (0.97–1) | 0.97 | 0.92 | 0.95 | 0.96 | 0.95 | 0.50 |
| RAB11FIP1 | 1.0 (0.99–1) | 0.95 | 0.96 | 0.97 | 0.92 | 0.95 | 1.64 |
| WHSC1L1 | 1.0 (0.99–1) | 1.00 | 0.92 | 0.95 | 1.00 | 0.97 | 0.91 |
The eight selected genes were selected based on all listed receiver operating characteristic diagnostic characteristics and overall accuracy (accuracy column) corresponding to the optimal cutoff point (cutoff column), together with the a priori selected FGFR1 gene.
AUC, area under the receiver operating characteristic curve; NPV, negative predictive value; POL, preoperative letrozole; PPV, positive predictive value.
Development and Evaluation of Multigene Based Signatures in the METABRIC Cohort
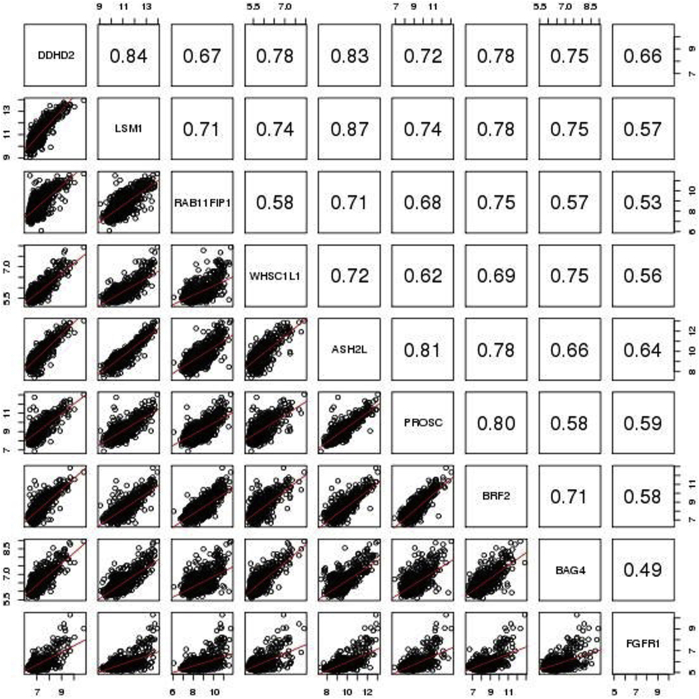
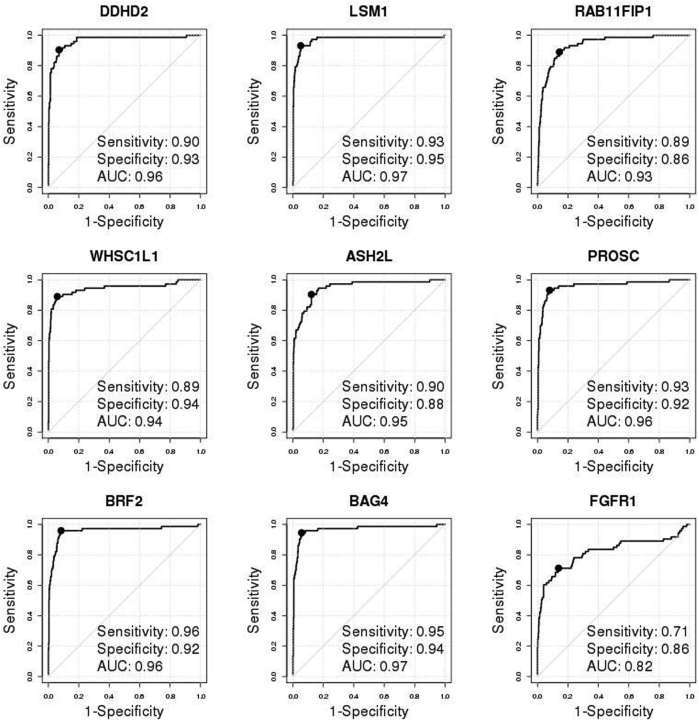
In the METABRIC study,36 the true FGFR1 amplification status of tumors was determined by SNP array copy number analysis (as described in Materials and Methods). FGFR1 was amplified in 138 (approximately 9%) of 1508 ER+ breast patients, significantly higher than in ER- tumors (approximately 4%, 17 of 440; odds ratio, 2.5; 95% CI, 1.49–4.48; Fisher's exact test P = 0.0002). FGFR1 amplification was associated with noticeably worse DSS among ER+ (HR, 1.67; 95% CI, 1.23–2.26; log-rank test P = 0.0008) (Figure 2B) but not among ER- patients (HR, 0.98; 95% CI, 0.43–2.21; log-rank test P = 0.96) (Figure 2B), which was similarly observed for overall survival (Supplemental Figure S4). Thereafter, we focused on the 1508 ER+ breast cancer patients in the METABRIC study for FGFR1 amplification analysis (Figure 2B and Supplemental Table S2). The nine selected genes were all highly overexpressed in the FGFR1-amplified tumors (Supplemental Figure S5) and showed moderate to high correlation (Pearson correlation coefficient range, 0.49 to 0.87) (Supplemental Figure S6), indicating that expression of these genes may provide complementary information for FGFR1 amplification. Consistent with the discovery cohort, all of the eight selected genes well discriminated between FGFR1-amplified and FGFR1 copy neutral ER+ tumors, with AUCs all ≥0.92 and high sensitivities and specificities corresponding to the optimal cutoff points, whereas expression of the FGFR1 gene itself had a comparatively lower AUC of 0.83 (Figure 2C).
Although each single gene already exhibited a discriminative ability for FGFR1 amplification, multigene based signatures may render robust results across studies and may improve FGFR1 amplification calling. We continued to develop a multigene signature (Call-FGFR1-amp) in consideration of four potential classification algorithms (as described in Materials and Methods). The performance of these methods was evaluated in a 10-fold CV setting (as described in Materials and Methods) using the METABRIC cohort. Diagnostic measures from the 10 CVs were averaged for final evaluation of the algorithms (Table 2). The high specificities indicated that all of the algorithms performed well in accurately identifying FGFR1 truly copy neutral cases. The signatures by each of the classification algorithms resulted in a high specificity, a high negative predictive value, and a high overall accuracy (all >0.96) (Table 2). The EMP method yielded the highest specificity of 99.4% but at the cost of a lower sensitivity (70.7%), whereas Logistic performed similarly, with the highest overall accuracy of 97.1%. In contrast, the NB method and the optAUC method showed a high specificity, the highest sensitivity, and the highest negative predictive values (99.4% and 99.5%, respectively) under a cost of slightly more false positives and lower positive predictive values. NB and optAUC also corresponded to the highest Youden index, which summarized the sensitivity and specificity, as well as highest Youden* with incorporation of FGFR1 amplification rate and a greater penalty on FN than on FP (FN/FP cost ratio, 5:1). FGFR1 amplification, as determined by the signature from all of the algorithms, was prognostic of DSS (Figure 2D). The KM curves representing the nonamplified patients, as determined by the signatures, overlapped the true curve of the FGFR1 nonamplified patients. The survival curves of the FGFR1-amplified patients, as determined by each algorithm, showed a slight deviation from the true curve of the FGFR1-amplified patients (Figure 2D). The final signature, Call-FGFR1-amp, determined by the optAUC algorithm was carried forward to the next stage for independent validation because it resulted in both a high specificity and a high sensitivity, a smallest FN/TN ratio, and a highest Youden* when imposing a greater penalty on FN than FP, and also produced slightly better stratifying KM curves when in combination with the FGFR1 true status (Figure 2D). All of the 1508 METABRIC ER+ tumors were used to train the optimal linear coefficients (Supplemental Table S8) of the nine genes using the optAUC algorithm by which the resulting linear score of the nine genes attained an optimal AUC, whereas for binary amplification calls, the optimal cut point (Supplemental Table S8) on the ROC curve was derived (as described in Materials and Methods).
Table 2.
Evaluate the Performance of the Multigene Signature by Four Algorithms Averaged Across 10-Fold CVs in the METABRIC Cohort
| Measures | EMP | NB | Logistic | optAUC |
|---|---|---|---|---|
| Specificity | 0.99 | 0.97 | 0.99 | 0.96 |
| Sensitivity | 0.71 | 0.94 | 0.79 | 0.96 |
| NPV | 0.97 | 0.99 | 0.98 | 1.00 |
| PPV | 0.91 | 0.74 | 0.88 | 0.71 |
| Accuracy | 0.97 | 0.97 | 0.97 | 0.96 |
| Youden | 0.70 | 0.91 | 0.78 | 0.92 |
| Youden* | 0.71 | 0.94 | 0.79 | 0.96 |
| FP/TP | 0.11 | 0.40 | 0.16 | 0.44 |
| FN/TN | 0.03 | 0.01 | 0.02 | 0.00 |
As evaluated against FGFR1 amplification by diagnostic measures, all algorithms perform well; optAUC led to the lowest FN/TN and the highest sensitivity and NPV.
CV, cross-validation; EMP, empirical method; FN, false negative; FP, false positive; Logistic, logistic regression; METABRIC, Molecular Taxonomy of Breast Cancer International Consortium; NB, naïve Bayes method; NPV, negative predictive value; optAUC, optimal area under the receiver operating characteristic curve; PPV, positive predictive value; TN, true negative, TP, true positive; Youden, does not consider population prevalence and uses equal weight on sensitivity and specificity; Youden*, considers population prevalence and allows different penalty/cost on FP and FN.
Independent Validation of the Locked-Down Signature
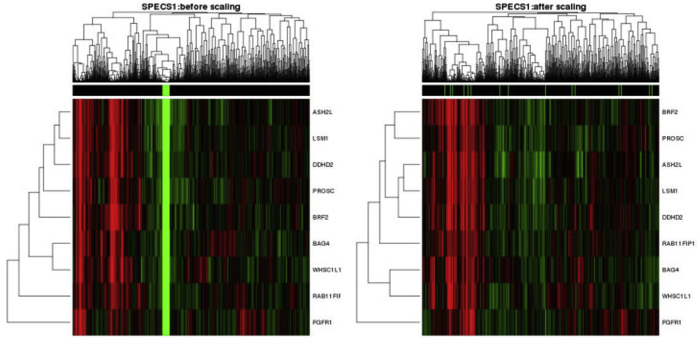
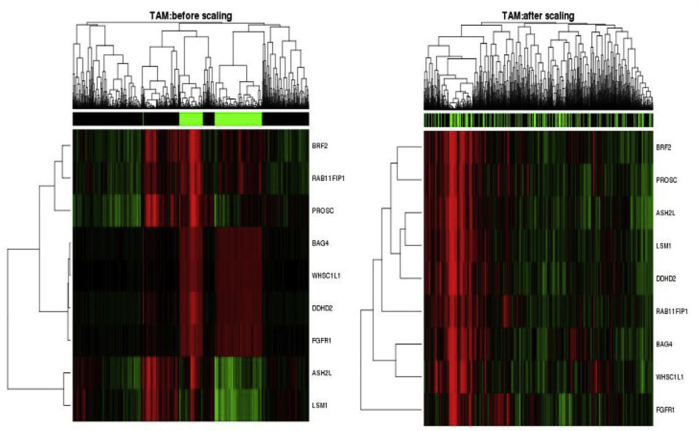
We prospectively evaluated the individual discriminative ability of each selected gene and, more important, the performance of the optimized and locked-down nine-gene–based signature Call-FGFR1-amp from the optAUC algorithm in ER+ patients from two additional independent studies (the SPECS and the UBC-TAM cohorts). As mRNA expression in these series was generated using different platforms (as described in Materials and Methods), the gene expression data of each validation cohort were separately transformed such that each gene had the same mean and SD as in the METABRIC training cohort. For both validation cohorts, the transformation removed major study batch effects between the training and a validation cohort, as visualized by the heat map of the merged data after transformation in contrast to that before transformation (Supplemental Figure S7 and Supplemental Figure S8) and as tested by sigclust59 (both validation cohorts: sigclust P = 0 before transformation and P = 1 after transformation).
The SPECS Cohort
A total of 138 breast tumors from the SPECS study42 were tested by FISH assay as FGFR1 amplification gold statuses, yielding an overall FGFR1 amplification rate of 9.92%. A total of 104 patients had available paired FISH and mRNA gene expression profiled on the Agilent custom 244 K gene expression microarray platform (Figure 3A). Among the 47 ER+ tumors (Supplemental Table S3 and Supplemental Table S6), complete agreement was observed between the readouts by two pathologists. Six (13.95%) of 43 ER+ tumors with a valid FISH readout were amplified. FGFR1-amplified ER+ tumors exhibited a trend toward shorter RFS (HR, 2.21; 95% CI, 0.45–10.97; log-rank test P = 0.319) (Figure 3B), overall survival (HR, 2.58; 95% CI, 0.50–13.33; log-rank test P = 0.242) (Figure 3B), and DSS (HR, 1.67; 95% CI, 0.19–15.05; log-rank test P = 0.642) (Figure 3B), although statistically not significant because of few samples. The individual discriminative performances of the selected genes were consistently confirmed (AUC range, 0.87 to 0.96) (Supplemental Figure S9). Call-FGFR1-amp captured all of the six FGFR1-amplified tumors (sensitivity, 1; and specificity, 0.95) (Figure 3C and Table 3), and rendered KM curves closely tracking those by FISH (Figure 3D).
Table 3.
Contingency Table of FGFR1 Amplification Calls as Determined by Call-FGFR1-amp against the FISH Gold Standards in the SPECS Cohort
| Call-FGFR1-amp | Total no. | FISH |
P value | κ | |
|---|---|---|---|---|---|
| Nonamplified | Amplified | ||||
| Non-amplified | 35 | 35 | 0 | 4.59E-06 | 0.83 |
| Amplified | 8 | 2 | 6 | ||
| Total no. | 43 | 37 | 6 | ||
| Specificity | Sensitivity | NPV | PPV | Accuracy | Youden | Youden* | FP/TP | FN/TN |
|---|---|---|---|---|---|---|---|---|
| 0.95 | 1 | 1 | 0.75 | 0.95 | 0.95 | 1 | 0.33 | 0 |
Call-FGFR1-amp captures all FGFR1-amplified cases and results in a high sensitivity, specificity, NPV, and overall accuracy.
FISH, fluorescent in situ hybridization; FN, false negative; FP, false positive; NPV, negative predictive value; PPV, positive predictive value; SPECS, Strategic Partnering to Evaluate Cancer Signatures; TN, true negative, TP, true positive; Youden, does not consider population prevalence and uses equal weight on sensitivity and specificity; Youden*, considers population prevalence and allows different penalty/cost on FP and FN.
The UBC-TAM Cohort
As a final validation exercise for this study, the NanoString nCounter platform was used to generate new mRNA expression data for the nine candidate genes on 651 FFPE ER+ tumors from the UBC-TAM cohort (Supplemental Table S4 and Supplemental Table S7). FISH was then used to diagnose FGFR1 amplification for all these 651 tumor samples; results were readable in 600 patients, among which 73 (12.17%) were FGFR1 amplified. FGFR1 amplification corresponded to shorter DSS and RFS (HR, 1.87; 95% CI, 1.31–2.65; P = 0.0004 for DSS; HR, 1.59; 95% CI, 1.13–2.23; P = 0.007 for RFS) (Figure 4B) in the ER+ tumors. The individual 8p11-12 amplicon genes were again validated to be overexpressed in FGFR1-amplified patients (Supplemental Figure S10) and were highly discriminative for FGFR1 amplification, with their AUCs ranging from 0.93 to 0.97 (Supplemental Figure S11); again, FGFR1 alone had a lower AUC (0.82). The heat map of the nine genes in the UBC-TAM cohort showed that the amplification calls from Call-FGFR1-amp corresponded well with the overexpression of these genes, in agreement with the true FGFR1 amplification status by FISH (Figure 4C). Call-FGFR1-amp identified 70 of the 73 amplified tumors, yielding a negative predictive value of 99.4%, an overall accuracy of 94.5%, a specificity of 94.3%, and a sensitivity of 95.9% (Figure 4C and Table 4). The call-FGFR1-amp amplification calls were again validated as prognostic for both DSS and RFS (HR, 1.57; 95% CI, 1.14–2.16; P = 0.005 for DSS; HR, 1.41; 95% CI, 1.05–1.9; P = 0.023 for RFS) (Figure 4D).
Table 4.
Contingency Table of FGFR1 Amplification Calls as Determined by Call-FGFR1-Amp against the FISH Gold Standards in the UBC-TAM Cohort
| Call-FGFR1-amp | Total N | FISH |
P value | κ | |
|---|---|---|---|---|---|
| Nonamplified | Amplified | ||||
| Nonamplified | 500 | 497 | 3 | 4.11E-63 | 0.78 |
| Amplified | 100 | 30 | 70 | ||
| Total N | 600 | 527 | 73 | ||
| Specificity | Sensitivity | NPV | PPV | Accuracy | Youden | Youden* | FP/TP | FN/TN |
|---|---|---|---|---|---|---|---|---|
| 0.94 | 0.96 | 0.99 | 0.7 | 0.95 | 0.9 | 0.96 | 0.43 | 0.01 |
Call-FGFR1-amp captures almost all of the FGFR1-amplified cases and results in a high sensitivity, specificity, NPV, and overall accuracy (Materials and Methods provides definitions of the diagnostic measures).
FISH, fluorescent in situ hybridization; FN, false negative; FP, false positive; NPV, negative predictive value; PPV, positive predictive value; TN, true negative, TP, true positive; UBC-TAM, University of British Columbia–tamoxifen; Youden, does not consider population prevalence and uses equal weight on sensitivity and specificity; Youden*, considers population prevalence and allows different penalty/cost on FP and FN.
Discussion
Gene expression signatures have been commonly used for cancer subtyping and for prognostic assays. However, gene expression profiling is not routinely used as an aide to the diagnosis of gene amplification, but would be a valuable approach if cut points and methods for FFPE tissues can be established. We chose the FGFR1 amplicon to address this challenge because of promising therapeutic investigations against this target. Coamplification of multiple genes in the FGFR1 chromosome region 8p11-8p12 has been previously reported.37, 38, 62 Furthermore, mRNA level overexpression of the genes in the region has been shown to correlate with gene copy number individually.13, 37, 38 However, an algorithm for using this information toward a clinical diagnosis had not been clearly established. Through a whole-transcriptome screening, this study identified eight genes that, at the mRNA expression level, prominently discriminate FGFR1 DNA amplification in ER+ breast tumors. The individual discriminative abilities of these genes were evident in all cohorts analyzed in this study. All candidate genes were in the 8p11-12 region, where FGFR1 resides, and are therefore potentially interacting oncogenes.37, 38, 63, 64, 65 Some have transforming properties when overexpressed63, 65 (eg, LSM1, BAG4, WHSC1L1, and DDHD2). BRF2 encodes a RNA polymerase III transcription initiation factor, and RAB11FIP1 is a regulator of RAB GTPases, which regulate virtually all steps of membrane traffic, whereas DDHD2 and WHSC1L1 may play a role in cell proliferation and survival.36, 37 Interestingly, the a priori selected FGFR1 gene consistently shows inferior performance in calling FGFR1 amplification in comparison to the other eight selected genes. The 8p11-12 region is a complex region, and the underlying amplification structure is yet to be defined. Recent studies discovered multiple new putative oncogenes in this region, suggesting that FGFR1 may not be the sole driver gene or even FGFR1 may not be the driver at all.36, 37 Even if these speculations were true, it might still be useful to measure FGFR1 amplification because being a kinase and a transmembrane protein, FGFR1 can be targeted with either kinase inhibitors or antibodies.37
The use of multiple genes in Call-FGFR1-amp has the potential to be more robust considering that the signal from one or more genes may be lost because of mRNA degradation. In addition, even if there are no technical issues, the best single discriminative gene may not be consistent from cohort to cohort. Indeed, in our analyses of the patient cohorts, multiple gene-based classifiers were never inferior to any single gene in any aspect of the diagnostic test measures. From performance averaged across CVs in the METABRIC cohort, the four multigene based signatures had their own strength and weakness. Overall, optAUC and NB consistently provided the best sensitivity (ie, least FN) and also a high specificity, thus the highest Youden index, Youden*, and negative predictive value, whereas the EMP and Logistic method showed slightly better specificity but much worse sensitivity. We carried forward the locked-down Call-FGFR1-amp signature based on the optAUC algorithm to be independently validated in two independent cohorts. In both validation cohorts, Call-FGFR1-amp identified almost all FGFR1 FISH-amplified tumors. Moreover, Call-FGFR1-amp exhibited similar prognostic effects to the DNA-based methods (FISH or SNP chip). Therefore, Call-FGFR1-amp could be used to triage cases, as determined to be nonamplified with only positive samples requiring testing by FISH to identify false positives from true positives.
We have used microarray gene expression data for discovery. Toward clinical diagnosis, it is critical to analytically validate candidate genes and signatures using a proper technical platform. More important, we have demonstrated the applicability of the signature to FFPE samples in the UBC-TAM study using the NanoString nCounter digital system, which is a gold standard analytical platform for gene expression and can be used as the analytical technical platform for future diagnostic test development. Data transformation was necessary herein to apply Call-FGFR1-amp trained in METABRIC to each independent validation cohort profiled on a different platform to remove systematic study/batch effects. However, the chosen informatics approach can be readily applied to diagnose a new sample that has been profiled using the same CodeSet as for the UBC-TAM samples on the NanoString nCounter system, after manufacturer-recommended normalization procedures through built-in negative control, positive control, and housekeeping genes. In this sense, a single sample predictor, where the FGFR1 amplification status of each new patient is determined one at a time separately, can be easily implemented. Yet, the diagnostic accuracy of Call-FGFR1-amp in the clinical diagnosis setting requires further evaluation.
FGFR1 amplification is prognostic of patient survival in ER+ tumors but not ER- tumors,14 also confirmed by our analyses (Figure 2B and Figure 4B). Thus, we focused on ER+ breast tumors for the diagnosis of FGFR1 amplification in this study. However, FGFR1 amplification is also present in ER- tumors and so the question arises as to whether Call-FGFR1-amp will be as accurate in this setting. Box plots of the selected genes individually in the METABRIC ER- patients indicated that the candidate genes selected using ER+ tumors were also overexpressed in FGFR1-amplified ER- tumors (Supplemental Figure S12). ROC analyses showed that DDHD2, LSM1, WHSC11L1, and BAG4 were still highly discriminative of FGFR1 amplification among ER- tumors, although the other genes were less discriminative than in ER+ tumors (Supplemental Figure S13). Tentative application of the locked-down Call-FGFR1-amp signature, trained using the METABRIC ER+ patients, showed that all FGFR1-amplified ER- tumors in METABRIC were identified but with more FPs (sensitivity, 1; specificity, 0.884) (Supplemental Table S9), underlining the importance of FISH confirmation. Call-FGFR1-amp might therefore be applicable to screen for FGFR1-amplified ER- tumors, but additional development and optimization would be required. FGFR1 amplification also occurs frequently in several other cancers, including lung cancer, kidney cancer, prostate cancer, and leukemia, so the testing approach has the potential for more widespread applications, as does the concept that gene expression measurements from the amplified region (readily detected on FFPE tissue blocks by NanoString technology) can reduce the need for FISH testing.
In summary, FGFR1 amplification has been associated with poor prognosis in ER+ breast tumors. We demonstrate the use of an efficient prescreening signature (Call-FGFR1-amp) based on the mRNA gene expression of a set of regionally amplified genes to reduce the need for laborious FISH tests. Ultimately, the validation of Call-FGFR1-amp requires the analysis of a trial where the efficacy of an FGFR1 inhibitor has been investigated.
Acknowledgment
We thank Amy Lum (Huntsman's Laboratory, British Columbia Cancer Agency) for assisting in primary FGFR1 fluorescent in situ hybridization data generation.
Footnotes
Supported in part by National Cancer Institute (NCI) Cancer Center support grant P30CA091842 (J.L.); NIH/NCIU01CA114722 (M.J.E. and T.O.N.); NIH/NCI U10CA180860 and Komen grant PG 12220321 (J.L. and M.J.E.); and NCI/NIH awards U10CA180821 and U10CA180882 (the Alliance for Clinical Trials in Oncology) and U10CA180833. M.J.E. is also a McNair Foundation Scholar.
Disclosures: T.O.N. and M.J.E. have patents and royalty to declare on Prosigna (Nanostring) PAM50 breast cancer subtyping test. S.R.D. has stock ownership in Nanostring, Inc.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Current address of A.A.G., Department of Pathology, University of Virginia, Charlottesville, VA; of D.C.A., Clarient-GE Health Care, Aliso Viejo, CA.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.jmoldx.2016.09.007.
Supplemental Data
Supplemental Figure S1.
Receiver operating characteristic (ROC) curves of the nine selected genes in the discovery cohort [ie, the preoperative letrozole (POL)-Z1031 cohort]. The solid black point indicates the optimal cutoff point on each ROC curve. Indicated in the legend are area under the ROC curve (AUC), sensitivity, and specificity corresponding to the optimal cutoff point.
Supplemental Figure S2.
Boxplot of mRNA gene expression (y axis) of the nine selected genes by FGFR1 amplification status in the discovery cohort [ie, the preoperative letrozole (POL)-Z1031 cohort]. The blue star dots indicate individual data points. The bottom and top edges of a box show the 25% and 75% quantile of the data, respectively. The black horizontal line shows the median, and the bottom and top extend from the edge to 1.5 multiplication of the corresponding quantile. The red dots correspond to the means, with downward and upward arrows representing the SD. Amp, amplification; NonAmp, no amplification.
Supplemental Figure S3.
Pairwise scatter plots (bottom triangle) and Pearson correlation coefficient (top triangle) of the nine genes in the discovery cohort [ie, the preoperative letrozole (POL)-Z1031 cohort]. The pairwise scatter plot was fitted by a loess line in red.
Supplemental Figure S4.
Overall survival (OS) Kaplan-Meier curves by FGFR1 amplification status (as determined by single-nucleotide polymorphism array copy number analysis) stratified by estrogen receptor (ER) status in the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) study. HR, hazard ratio.
Supplemental Figure S5.
Boxplot of mRNA gene expression (y axis) of the nine selected genes by FGFR1 amplification status in the 1508 estrogen receptor–positive tumors in the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) study. The blue stars indicate individual data points. The bottom and top edges of a box show the 25% and 75% quantile of the data, respectively. The black horizontal line shows the median, and the bottom and top extend from the edge to 1.5× the corresponding quantile. The red dots correspond to the means, with downward and upward arrows representing the SD. Amp, amplification; NonAmp, no amplification.
Supplemental Figure S6.
Pairwise scatter plots (bottom triangle) and Pearson correlation coefficient (top triangle) of the nine genes in the 1508 estrogen receptor–positive tumors in the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) study. The pairwise scatter plot was fitted by a loess line in red.
Supplemental Figure S7.
Heat map of merged gene expression data of the nine selected genes across 47 Strategic Partnering to Evaluate Cancer Signatures (SPECS) estrogen receptor–positive (ER+) samples (indicated by green in the top image bar) and the 1508 ER+ Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) samples (indicated by black in the top image bar) before (left panel) and after (right panel) data transformation (as described in Materials and Methods).
Supplemental Figure S8.
Heat map of the merged gene expression data of nine selected genes across the 651 University of British Columbia–tamoxifen series samples (indicated by green in the top image bar) and the 1508 Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) estrogen receptor–positive samples (indicated by black in the top image bar) before (left panel) and after (right panel) data transformation (as described in Materials and Methods).
Supplemental Figure S9.
Receiver operating characteristic (ROC) curves of the nine individual genes in the 47 estrogen receptor–positive tumors of the Strategic Partnering to Evaluate Cancer Signatures cohort. The solid black point indicates the optimal cutoff point on each ROC curve. Indicated in the legend are area under the ROC curve (AUC), sensitivity, and specificity corresponding to the optimal cutoff point.
Boxplot of individual genes by FGFR1 fluorescent in situ hybridization amplification in the University of British Columbia–tamoxifen cohort. The blue stars indicate individual data points. The bottom and top edges of a box show the 25% and 75% quantile of the data, respectively. The black horizontal line shows the median, and the bottom and top extend from the edge to 1.5× the corresponding quantile. The red dots correspond to the means, with downward and upward arrows representing the SD. Amp, amplification; NonAmp, no amplification.
Receiver operating characteristic (ROC) curves of individual genes in the University of British Columbia–tamoxifen cohort. The solid black point indicates the optimal cutoff point on each ROC curve. Indicated in the legend are area under the ROC curve (AUC), sensitivity, and specificity corresponding to the optimal cutoff point.
Individual boxplots of the nine genes by estrogen receptor (ER) status and FGFR1 amplification status in the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohort (0 for nonamplified and 1 for amplified, with sample size in parenthesis). The blue stars indicate individual data points. The bottom and top edges of a box show the 25% and 75% quantile of the data, respectively. The black horizontal line shows the median, and the bottom and top extend from the edge to 1.5* the corresponding quantile. The red dots correspond to the means, with downward and upward arrows representing the SD.
Receiver operating characteristic (ROC) curves of the nine genes in estrogen receptor–negative breast tumors in the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohort. The solid black point indicates the optimal cutoff point on each ROC curve. Indicated in the legend are area under the ROC curve (AUC), sensitivity, and specificity corresponding to the optimal cutoff point.
References
- 1.Wolff A.C., Hammond M.E., Hicks D.G., Dowsett M., McShane L.M., Allison K.H., Allred D.C., Bartlett J.M., Bilous M., Fitzgibbons P., Hanna W., Jenkins R.B., Mangu P.B., Paik S., Perez E.A., Press M.F., Spears P.A., Vance G.H., Viale G., Hayes D.F., American Society of Clinical Oncology; College of American Pathologists Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31:3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 2.Wolff A.C., Hammond M.E., Hicks D.G., Dowsett M., McShane L.M., Allison K.H., Allred D.C., Bartlett J.M., Bilous M., Fitzgibbons P., Hanna W., Jenkins R.B., Mangu P.B., Paik S., Perez E.A., Press M.F., Spears P.A., Vance G.H., Viale G., Hayes D.F., American Society of Clinical Oncology; College of American Pathologists Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med. 2014;138:241–256. doi: 10.5858/arpa.2013-0953-SA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolff A.C., Hammond M.E., Schwartz J.N., Hagerty K.L., Allred D.C., Cote R.J., Dowsett M., Fitzgibbons P.L., Hanna W.M., Langer A., McShane L.M., Paik S., Pegram M.D., Perez E.A., Press M.F., Rhodes A., Sturgeon C., Taube S.E., Tubbs R., Vance G.H., van de Vijver M., Wheeler T.M., Hayes D.F., American Society of Clinical Oncology/College of American Pathologists American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med. 2007;131:18–43. doi: 10.5858/2007-131-18-ASOCCO. [DOI] [PubMed] [Google Scholar]
- 4.Wolff A.C., Hammond M.E., Schwartz J.N., Hagerty K.L., Allred D.C., Cote R.J., Dowsett M., Fitzgibbons P.L., Hanna W.M., Langer A., McShane L.M., Paik S., Pegram M.D., Perez E.A., Press M.F., Rhodes A., Sturgeon C., Taube S.E., Tubbs R., Vance G.H., van de Vijver M., Wheeler T.M., Hayes D.F., American Society of Clinical Oncology; College of American Pathologists American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 5.Sauter G., Lee J., Bartlett J.M., Slamon D.J., Press M.F. Guidelines for human epidermal growth factor receptor 2 testing: biologic and methodologic considerations. J Clin Oncol. 2009;27:1323–1333. doi: 10.1200/JCO.2007.14.8197. [DOI] [PubMed] [Google Scholar]
- 6.Dienstmann R., Rodon J., Prat A., Perez-Garcia J., Adamo B., Felip E., Cortes J., Iafrate A.J., Nuciforo P., Tabernero J. Genomic aberrations in the FGFR pathway: opportunities for targeted therapies in solid tumors. Ann Oncol. 2014;25:552–563. doi: 10.1093/annonc/mdt419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turner N., Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 8.Helsten T., Elkin S., Arthur E., Tomson B.N., Carter J., Kurzrock R. The FGFR landscape in cancer: analysis of 4,853 tumors by next-generation sequencing. Clin Cancer Res. 2016;22:259–267. doi: 10.1158/1078-0432.CCR-14-3212. [DOI] [PubMed] [Google Scholar]
- 9.Chang J.J., Liu X.Y., Wang S.S., Zhang Z., Wu Z., Zhang X.W., Li J. Prognostic value of FGFR gene amplification in patients with different types of cancer: a systematic review and meta-analysis. PLoS One. 2014;9:e105524. doi: 10.1371/journal.pone.0105524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elbauomy Elsheikh S., Green A.R., Lambros M.B., Turner N.C., Grainge M.J., Powe D., Ellis I.O., Reis-Filho J.S. FGFR1 amplification in breast carcinomas: a chromogenic in situ hybridisation analysis. Breast Cancer Res. 2007;9:R23. doi: 10.1186/bcr1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andre F., Cortes J. Rationale for targeting fibroblast growth factor receptor signaling in breast cancer. Breast Cancer Res Treat. 2015;150:1–8. doi: 10.1007/s10549-015-3301-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bollig-Fischer A., Michelhaugh S.K., Wijesinghe P., Dyson G., Kruger A., Palanisamy N., Choi L., Alosh B., Ali-Fehmi R., Mittal S. Cytogenomic profiling of breast cancer brain metastases reveals potential for repurposing targeted therapeutics. Oncotarget. 2015;6:14614–14624. doi: 10.18632/oncotarget.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chin K., DeVries S., Fridlyand J., Spellman P.T., Roydasgupta R., Kuo W.L., Lapuk A., Neve R.M., Qian Z., Ryder T., Chen F., Feiler H., Tokuyasu T., Kingsley C., Dairkee S., Meng Z., Chew K., Pinkel D., Jain A., Ljung B.M., Esserman L., Albertson D.G., Waldman F.M., Gray J.W. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 14.Jang M.H., Kim E.J., Choi Y., Lee H.E., Kim Y.J., Kim J.H., Kang E., Kim S., Kim I.A., Park S.Y. FGFR1 is amplified during the progression of in situ to invasive breast carcinoma. Breast Cancer Res. 2012;14:R115. doi: 10.1186/bcr3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moelans C.B., de Weger R.A., Monsuur H.N., Vijzelaar R., van Diest P.J. Molecular profiling of invasive breast cancer by multiplex ligation-dependent probe amplification-based copy number analysis of tumor suppressor and oncogenes. Mod Pathol. 2010;23:1029–1039. doi: 10.1038/modpathol.2010.84. [DOI] [PubMed] [Google Scholar]
- 16.Monaco S.E., Rodriguez E.F., Mahaffey A.L., Dacic S. FGFR1 amplification in squamous cell carcinoma of the lung with correlation of primary and metastatic tumor status. Am J Clin Pathol. 2016;145:55–61. doi: 10.1093/ajcp/aqv013. [DOI] [PubMed] [Google Scholar]
- 17.Jiang T., Gao G., Fan G., Li M., Zhou C. FGFR1 amplification in lung squamous cell carcinoma: a systematic review with meta-analysis. Lung Cancer. 2015;87:1–7. doi: 10.1016/j.lungcan.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 18.Heist R.S., Mino-Kenudson M., Sequist L.V., Tammireddy S., Morrissey L., Christiani D.C., Engelman J.A., Iafrate A.J. FGFR1 amplification in squamous cell carcinoma of the lung. J Thorac Oncol. 2012;7:1775–1780. doi: 10.1097/JTO.0b013e31826aed28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cihoric N., Savic S., Schneider S., Ackermann I., Bichsel-Naef M., Schmid R.A., Lardinois D., Gugger M., Bubendorf L., Zlobec I., Tapia C. Prognostic role of FGFR1 amplification in early-stage non-small cell lung cancer. Br J Cancer. 2014;110:2914–2922. doi: 10.1038/bjc.2014.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schultheis A.M., Bos M., Schmitz K., Wilsberg L., Binot E., Wolf J., Buttner R., Schildhaus H.U. Fibroblast growth factor receptor 1 (FGFR1) amplification is a potential therapeutic target in small-cell lung cancer. Mod Pathol. 2014;27:214–221. doi: 10.1038/modpathol.2013.141. [DOI] [PubMed] [Google Scholar]
- 21.Weiss J., Sos M.L., Seidel D., Peifer M., Zander T., Heuckmann J.M. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2:62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Loga K., Kohlhaussen J., Burkhardt L., Simon R., Steurer S., Burdak-Rothkamm S., Jacobsen F., Sauter G., Krech T. FGFR1 amplification is often homogeneous and strongly linked to the squamous cell carcinoma subtype in esophageal carcinoma. PLoS One. 2015;10:e0141867. doi: 10.1371/journal.pone.0141867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischbach A., Rogler A., Erber R., Stoehr R., Poulsom R., Heidenreich A., Schneevoigt B.S., Hauke S., Hartmann A., Knuechel R., Veeck J., Gaisa N.T. Fibroblast growth factor receptor (FGFR) gene amplifications are rare events in bladder cancer. Histopathology. 2015;66:639–649. doi: 10.1111/his.12473. [DOI] [PubMed] [Google Scholar]
- 24.Mayr D., Kanitz V., Anderegg B., Luthardt B., Engel J., Lohrs U., Amann G., Diebold J. Analysis of gene amplification and prognostic markers in ovarian cancer using comparative genomic hybridization for microarrays and immunohistochemical analysis for tissue microarrays. Am J Clin Pathol. 2006;126:101–109. doi: 10.1309/n6x5mb24bp42kp20. [DOI] [PubMed] [Google Scholar]
- 25.Jain V.K., Turner N.C. Challenges and opportunities in the targeting of fibroblast growth factor receptors in breast cancer. Breast Cancer Res. 2012;14:208. doi: 10.1186/bcr3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turner N., Pearson A., Sharpe R., Lambros M., Geyer F., Lopez-Garcia M.A., Natrajan R., Marchio C., Iorns E., Mackay A., Gillett C., Grigoriadis A., Tutt A., Reis-Filho J.S., Ashworth A. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 2010;70:2085–2094. doi: 10.1158/0008-5472.CAN-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shiang C.Y., Qi Y., Wang B., Lazar V., Wang J., Fraser Symmans W., Hortobagyi G.N., Andre F., Pusztai L. Amplification of fibroblast growth factor receptor-1 in breast cancer and the effects of brivanib alaninate. Breast Cancer Res Treat. 2010;123:747–755. doi: 10.1007/s10549-009-0677-6. [DOI] [PubMed] [Google Scholar]
- 28.Bello E., Colella G., Scarlato V., Oliva P., Berndt A., Valbusa G., Serra S.C., D'Incalci M., Cavalletti E., Giavazzi R., Damia G., Camboni G. E-3810 is a potent dual inhibitor of VEGFR and FGFR that exerts antitumor activity in multiple preclinical models. Cancer Res. 2011;71:1396–1405. doi: 10.1158/0008-5472.CAN-10-2700. [DOI] [PubMed] [Google Scholar]
- 29.Zhang J., Zhang L., Su X., Li M., Xie L., Malchers F., Fan S., Yin X., Xu Y., Liu K., Dong Z., Zhu G., Qian Z., Tang L., Schottle J., Zhan P., Ji Q., Kilgour E., Smith P.D., Brooks A.N., Thomas R.K., Gavine P.R. Translating the therapeutic potential of AZD4547 in FGFR1-amplified non-small cell lung cancer through the use of patient-derived tumor xenograft models. Clin Cancer Res. 2012;18:6658–6667. doi: 10.1158/1078-0432.CCR-12-2694. [DOI] [PubMed] [Google Scholar]
- 30.Gavine P.R., Mooney L., Kilgour E., Thomas A.P., Al-Kadhimi K., Beck S., Rooney C., Coleman T., Baker D., Mellor M.J., Brooks A.N., Klinowska T. AZD4547: an orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res. 2012;72:2045–2056. doi: 10.1158/0008-5472.CAN-11-3034. [DOI] [PubMed] [Google Scholar]
- 31.Andre F., Bachelot T., Campone M., Dalenc F., Perez-Garcia J.M., Hurvitz S.A., Turner N., Rugo H., Smith J.W., Deudon S., Shi M., Zhang Y., Kay A., Porta D.G., Yovine A., Baselga J. Targeting FGFR with dovitinib (TKI258): preclinical and clinical data in breast cancer. Clin Cancer Res. 2013;19:3693–3702. doi: 10.1158/1078-0432.CCR-13-0190. [DOI] [PubMed] [Google Scholar]
- 32.Gozgit J.M., Wong M.J., Moran L., Wardwell S., Mohemmad Q.K., Narasimhan N.I., Shakespeare W.C., Wang F., Clackson T., Rivera V.M. Ponatinib (AP24534), a multitargeted pan-FGFR inhibitor with activity in multiple FGFR-amplified or mutated cancer models. Mol Cancer Ther. 2012;11:690–699. doi: 10.1158/1535-7163.MCT-11-0450. [DOI] [PubMed] [Google Scholar]
- 33.Dutt A., Ramos A.H., Hammerman P.S., Mermel C., Cho J., Sharifnia T., Chande A., Tanaka K.E., Stransky N., Greulich H., Gray N.S., Meyerson M. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS One. 2011;6:e20351. doi: 10.1371/journal.pone.0020351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soria J.C., DeBraud F., Bahleda R., Adamo B., Andre F., Dienstmann R., Delmonte A., Cereda R., Isaacson J., Litten J., Allen A., Dubois F., Saba C., Robert R., D'Incalci M., Zucchetti M., Camboni M.G., Tabernero J. Phase I/IIa study evaluating the safety, efficacy, pharmacokinetics, and pharmacodynamics of lucitanib in advanced solid tumors. Ann Oncol. 2014;25:2244–2251. doi: 10.1093/annonc/mdu390. [DOI] [PubMed] [Google Scholar]
- 35.Pearson A., Smyth E., Babina I.S., Herrera-Abreu M.T., Tarazona N., Peckitt C., Kilgour E., Smith N.R., Geh C., Rooney C., Cutts R., Campbell J., Ning J., Fenwick K., Swain A., Brown G., Chua S., Thomas A., Johnston S.R., Ajaz M., Sumpter K., Gillbanks A., Watkins D., Chau I., Popat S., Cunningham D., Turner N.C. High-level clonal FGFR amplification and response to FGFR inhibition in a translational clinical trial. Cancer Discov. 2016;6:838–851. doi: 10.1158/2159-8290.CD-15-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Curtis C., Shah S.P., Chin S.F., Turashvili G., Rueda O.M., Dunning M.J., Speed D., Lynch A.G., Samarajiwa S., Yuan Y., Graf S., Ha G., Haffari G., Bashashati A., Russell R., McKinney S., Group M., Langerod A., Green A., Provenzano E., Wishart G., Pinder S., Watson P., Markowetz F., Murphy L., Ellis I., Purushotham A., Borresen-Dale A.L., Brenton J.D., Tavare S., Caldas C., Aparicio S. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia M.J., Pole J.C., Chin S.F., Teschendorff A., Naderi A., Ozdag H., Vias M., Kranjac T., Subkhankulova T., Paish C., Ellis I., Brenton J.D., Edwards P.A., Caldas C. A 1 Mb minimal amplicon at 8p11-12 in breast cancer identifies new candidate oncogenes. Oncogene. 2005;24:5235–5245. doi: 10.1038/sj.onc.1208741. [DOI] [PubMed] [Google Scholar]
- 38.Gelsi-Boyer V., Orsetti B., Cervera N., Finetti P., Sircoulomb F., Rouge C., Lasorsa L., Letessier A., Ginestier C., Monville F., Esteyries S., Adelaide J., Esterni B., Henry C., Ethier S.P., Bibeau F., Mozziconacci M.J., Charafe-Jauffret E., Jacquemier J., Bertucci F., Birnbaum D., Theillet C., Chaffanet M. Comprehensive profiling of 8p11-12 amplification in breast cancer. Mol Cancer Res. 2005;3:655–667. doi: 10.1158/1541-7786.MCR-05-0128. [DOI] [PubMed] [Google Scholar]
- 39.Nielsen T.O., Parker J.S., Leung S., Voduc D., Ebbert M., Vickery T., Davies S.R., Snider J., Stijleman I.J., Reed J., Cheang M.C., Mardis E.R., Perou C.M., Bernard P.S., Ellis M.J. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res. 2010;16:5222–5232. doi: 10.1158/1078-0432.CCR-10-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olson J.A., Jr., Budd G.T., Carey L.A., Harris L.A., Esserman L.J., Fleming G.F., Marcom P.K., Leight G.S., Jr., Giuntoli T., Commean P., Bae K., Luo J., Ellis M.J. Improved surgical outcomes for breast cancer patients receiving neoadjuvant aromatase inhibitor therapy: results from a multicenter phase II trial. J Am Coll Surg. 2009;208:906–914. doi: 10.1016/j.jamcollsurg.2009.01.035. discussion 915–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ellis M.J., Suman V.J., Hoog J., Lin L., Snider J., Prat A., Parker J.S., Luo J., DeSchryver K., Allred D.C., Esserman L.J., Unzeitig G.W., Margenthaler J., Babiera G.V., Marcom P.K., Guenther J.M., Watson M.A., Leitch M., Hunt K., Olson J.A. Randomized phase II neoadjuvant comparison between letrozole, anastrozole, and exemestane for postmenopausal women with estrogen receptor-rich stage 2 to 3 breast cancer: clinical and biomarker outcomes and predictive value of the baseline PAM50-based intrinsic subtype–ACOSOG Z1031. J Clin Oncol. 2011;29:2342–2349. doi: 10.1200/JCO.2010.31.6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parker J.S., Mullins M., Cheang M.C., Leung S., Voduc D., Vickery T., Davies S., Fauron C., He X., Hu Z., Quackenbush J.F., Stijleman I.J., Palazzo J., Marron J.S., Nobel A.B., Mardis E., Nielsen T.O., Ellis M.J., Perou C.M., Bernard P.S. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding L., Ellis M.J., Li S., Larson D.E., Chen K., Wallis J.W. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olshen A.B., Venkatraman E.S., Lucito R., Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 45.Venkatraman E.S., Olshen A.B. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 46.van de Wiel M.A., Kim K.I., Vosse S.J., van Wieringen W.N., Wilting S.M., Ylstra B. CGHcall: calling aberrations for array CGH tumor profiles. Bioinformatics. 2007;23:892–894. doi: 10.1093/bioinformatics/btm030. [DOI] [PubMed] [Google Scholar]
- 47.Wallden B., Storhoff J., Nielsen T., Dowidar N., Schaper C., Ferree S., Liu S., Leung S., Geiss G., Snider J., Vickery T., Davies S.R., Mardis E.R., Gnant M., Sestak I., Ellis M.J., Perou C.M., Bernard P.S., Parker J.S. Development and verification of the PAM50-based Prosigna breast cancer gene signature assay. BMC Med Genomics. 2015;8:54. doi: 10.1186/s12920-015-0129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y.H., Dudoit S., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geiss G.K., Bumgarner R.E., Birditt B., Dahl T., Dowidar N., Dunaway D.L., Fell H.P., Ferree S., George R.D., Grogan T., James J.J., Maysuria M., Mitton J.D., Oliveri P., Osborn J.L., Peng T., Ratcliffe A.L., Webster P.J., Davidson E.H., Hood L., Dimitrov K. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 50.Nielsen T., Wallden B., Schaper C., Ferree S., Liu S., Gao D., Barry G., Dowidar N., Maysuria M., Storhoff J. Analytical validation of the PAM50-based Prosigna Breast Cancer Prognostic Gene Signature Assay and nCounter Analysis System using formalin-fixed paraffin-embedded breast tumor specimens. BMC Cancer. 2014;14:177. doi: 10.1186/1471-2407-14-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hastie T, Tibshirani R, Narasimhan B, Chu G: Impute: imputation for microarray data. R packages version 1.38.1. 2008. Available at http://cran.r-project.org/package=impute.
- 52.Hanley J., McNeil B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 53.DeLong E., DeLong D.M., Clarke-Pearson D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 54.Zweig M.H., Campbell G. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem. 1993;39:561–577. [PubMed] [Google Scholar]
- 55.Huang X., Qin G., Fang Y. Optimal combinations of diagnostic tests based on AUC. Biometrics. 2011;67:568–576. doi: 10.1111/j.1541-0420.2010.01450.x. [DOI] [PubMed] [Google Scholar]
- 56.Rish I: An empirical study of the naive Bayes classifier. In Proceedings of IJCAI-01 Workshop on Empirical Methods in Artificial Intelligence. New York, NY: IBM, 2001.
- 57.Lewis D: Naive (Bayes) at Forty: the independence assumption in information retrieval. In Proceedings 10th European Conference on Machine Learning (ECML). Berlin, Germany: Springer, 1998.
- 58.Huang X, Qin G, Fang Y, optAUC: Optimal Combinations of Diagnostic Tests Based on AUC, R package version 1.0. 2013. Available at http://cran.r-project.org/package=optAUC. [DOI] [PubMed]
- 59.Liu Y., Hayes D.N., Nobel A., Marron J. Statistical significance of clustering for high dimension low sample size data. J Am Stat Assoc. 2008;103:1281–1293. [Google Scholar]
- 60.Huang H, Liu Y, Marron JS: Sigclust: Statistical Significance of Clustering, R package version 1.1.0. 2014. Available at http://cran.r-project.org/package=sigclust.
- 61.R Core Team: A Language and Environment for Statistical Computing. 2015.
- 62.Kwek S.S., Roy R., Zhou H., Climent J., Martinez-Climent J.A., Fridlyand J., Albertson D.G. Co-amplified genes at 8p12 and 11q13 in breast tumors cooperate with two major pathways in oncogenesis. Oncogene. 2009;28:1892–1903. doi: 10.1038/onc.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang Z.Q., Liu G., Bollig-Fischer A., Giroux C.N., Ethier S.P. Transforming properties of 8p11-12 amplified genes in human breast cancer. Cancer Res. 2010;70:8487–8497. doi: 10.1158/0008-5472.CAN-10-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ray M.E., Yang Z.Q., Albertson D., Kleer C.G., Washburn J.G., Macoska J.A., Ethier S.P. Genomic and expression analysis of the 8p11-12 amplicon in human breast cancer cell lines. Cancer Res. 2004;64:40–47. doi: 10.1158/0008-5472.can-03-1022. [DOI] [PubMed] [Google Scholar]
- 65.Yang Z.Q., Streicher K.L., Ray M.E., Abrams J., Ethier S.P. Multiple interacting oncogenes on the 8p11-p12 amplicon in human breast cancer. Cancer Res. 2006;66:11632–11643. doi: 10.1158/0008-5472.CAN-06-2946. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Boxplot of individual genes by FGFR1 fluorescent in situ hybridization amplification in the University of British Columbia–tamoxifen cohort. The blue stars indicate individual data points. The bottom and top edges of a box show the 25% and 75% quantile of the data, respectively. The black horizontal line shows the median, and the bottom and top extend from the edge to 1.5× the corresponding quantile. The red dots correspond to the means, with downward and upward arrows representing the SD. Amp, amplification; NonAmp, no amplification.
Receiver operating characteristic (ROC) curves of individual genes in the University of British Columbia–tamoxifen cohort. The solid black point indicates the optimal cutoff point on each ROC curve. Indicated in the legend are area under the ROC curve (AUC), sensitivity, and specificity corresponding to the optimal cutoff point.
Individual boxplots of the nine genes by estrogen receptor (ER) status and FGFR1 amplification status in the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohort (0 for nonamplified and 1 for amplified, with sample size in parenthesis). The blue stars indicate individual data points. The bottom and top edges of a box show the 25% and 75% quantile of the data, respectively. The black horizontal line shows the median, and the bottom and top extend from the edge to 1.5* the corresponding quantile. The red dots correspond to the means, with downward and upward arrows representing the SD.
Receiver operating characteristic (ROC) curves of the nine genes in estrogen receptor–negative breast tumors in the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohort. The solid black point indicates the optimal cutoff point on each ROC curve. Indicated in the legend are area under the ROC curve (AUC), sensitivity, and specificity corresponding to the optimal cutoff point.