Abstract
Our previous studied indicated that eukaryotic translation initiation factor 3a (eIF3a) increases the sensitive of platinum-based chemotherapy in lung cancer. MiRNAs play an important role in lung carcinogenesis and drug response. In this study, we aimed to identify potential endogenous miRNAs that inhibit eIF3a expression and determine their influence of this inhibition on cisplatin resistance. Using bioinformatics analysis prediction and confirmation with dual-luciferase reporter assays, we found that miRNA-488 inhibited eIF3a expression by directly binding to the 3’UTR of eIF3a. In addition, the overexpression of miRNA-488 inhibited cell migration and invasion in A549 cells, and also inhibited cell proliferation, cell cycle progression by elevated P27 expression. Compared to the parental cell line, A549/cisplatin (DDP) resistant cells exhibited a higher level of miRNA-488. Moreover, we found that miRNA-488 was associated with cisplatin resistance in three NSCLC cells (A549, H1299 and SK-MES-1). The mechanism of miRNA-488 induced cisplatin resistance was that miRNA-488 activated nucleotide excision repair (NER) by increasing the expression of Replication Protein A (RPA) 14 and Xeroderma pigmentosum group C (XPC). In conclusion, our results demonstrated that miRNA-488 is a tumor suppressor miRNA that acts by targeting eIF3a. Moreover, miRNA-488 also participates in eIF3a mediated cisplatin resistance in NSCLC cells.
Lung cancer, which is characterized by uncontrolled cell growth in lung tissues, is still the most common malignant cancer worldwide1,2. It can be classified into non-small-cell lung cancer (NSCLC) and small-cell lung cancer (SCLC), and NSCLC counts more than 85% of lung cancer3. Platinum-based chemotherapy is the basic therapy in advanced NSCLC4,5, but the continuous use of these agents often causes chemotherapy resistance in the clinic, which is one of the key factors affecting prognosis6. Therefore, a better understanding of the mechanisms of platinum resistance in NSCLC will be important for the development of more reasonable therapeutic approaches for lung cancer treatment.
Micro RNAs (MiRNAs) are small non-coding RNA molecules (containing approximately 22 nucleotides) found in plants, animals, and some viruses. They function in RNA silencing and the post-transcriptional regulation of gene expression by perfectly or imperfectly pairing to the 3’ untranslated region (UTR) of target messenger RNAs (mRNAs)7,8. Bioinformatics analysis estimated that miRNAs regulate ∼30% of human genes9. Notably, miRNA deregulation in cancer could partly result from genomic deletion, mutation, or amplification10.
The eukaryotic translation initiation factor 3a (eIF3a) is the largest and core subunit of translation initiation complex 3; it serves as a bridge in the formation of the translation initiation complex and is responsible for ribosomal subunit joining and mRNA recruitment11. It is known that eIF3a plays critical roles in the regulation of various gene products, influencing cell growth and proliferation12,13, differentiation14, DNA repair pathways15, and cell cycle progression16. Recent studies have revealed that eIF3a expression is elevated in several cancer cell lines, while a comparison of the expression levels in human ovary, kidney, lung, breast and colon cancer tissue to normal tissue showed specific high eIF3a expression in lung cancer17. Our previous studies found that genotype variation in the eIF3a gene contributes to platinum-based chemotherapy resistance and severe toxicity in lung cancer patients18,19.
In recent years, ample evidences have revealed that the epigenetic regulation of miRNA alters the pathological progression and prognosis of lung cancer20,21,22. Our latest studies indicated that altered eIF3a expression correlates with the prognosis of non-small lung cancer23 and that eIF3a expression was associated with the response of lung cancer patients to platinum-based chemotherapy through the regulation of DNA repair pathways24. Based on these works, we sought to further identify the relationship between endogenous miRNAs and the inhibition of eIF3a gene expression. Moreover, we also sought to elucidate how the regulation of eIF3a affects cisplatin resistance in NSCLC. The aim of this study was to provide a new explanation and further understanding of eIF3a action in cisplatin resistance in NSCLC and provide new scientific evidences for eIF3a as a molecular target for personalized pharmacotherapy in NSCLC.
Results
A cisplatin sensitive cell line exhibits high eIF3a expression and low miRNA-488 expression, whereas miRNA-488 inhibits eIF3a expression
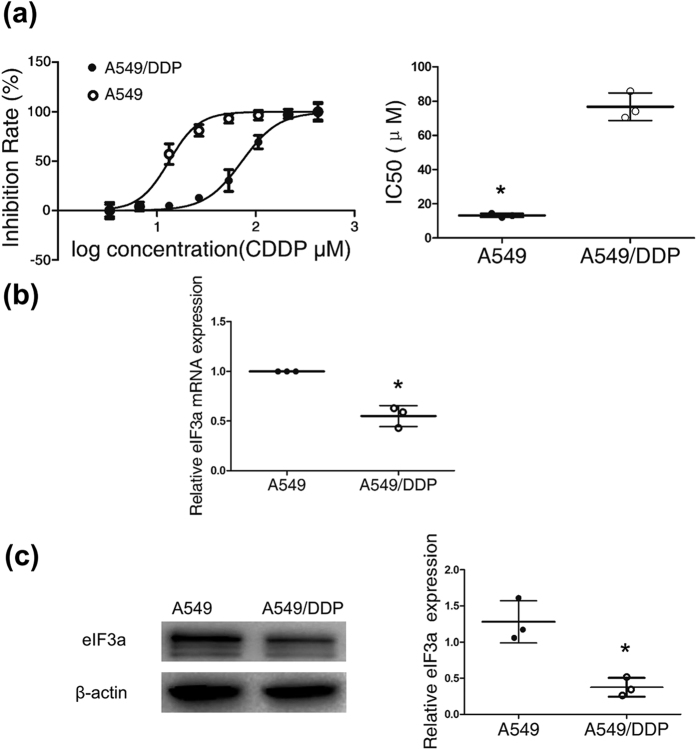
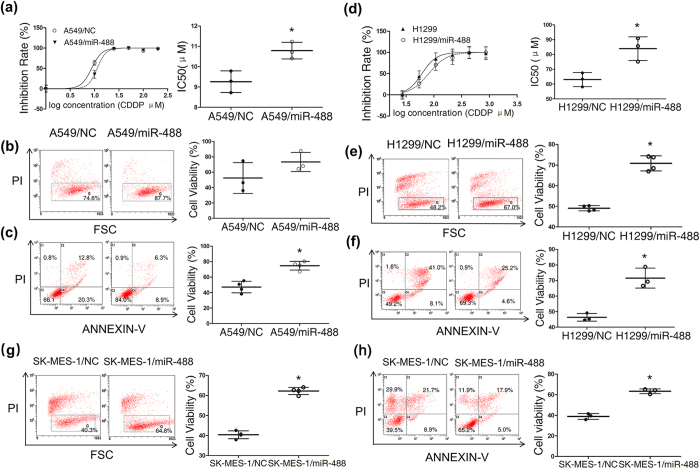
Firstly, we chose the cisplatin-resistant A549/DDP lung adenocarcinoma cell line and its parental cell line as the research models. The resistance index of A549/DDP was identified by evaluating the half-maximal inhibitory concentration (IC50) value of cisplatin in A549/DDP cells relative to that in the A549 cell line. The IC50 of cisplatin in the A549/DDP cell line was significantly higher than that in the A549 cell line (Fig. 1a).
Figure 1. EIF3a showed high expression in a cisplatin sensitive cell line.
(a) Cells were treated with increasing concentrations of cisplatin (3 μM to 384 μM). Forty-eight hours later, cell viability was tested with MTS. The half maximal inhibitory concentration (IC50) was calculated from 3 independent experiments using GraphPad 5.0 software. The A549/DDP cell line showed a higher IC50 than the A549 cell line. (b) The relative expression of eIF3a mRNA in the A549/DDP cell line compared with the parental cell line was measured with qRT-PCR. (c) The eIF3a protein expression in the A549/DDP and A549 cell lines was determined by western blot. The bands cropped for the representative images are shown in Fig. 1c, and full-length blots are presented in Supplementary Fig. S3a. EIF3a was highly expressed in the cisplatin sensitive cell line (A549), and the data from three independent experiments is given as the mean ± SD (*P < 0.05).
To confirm that eIF3a is associated with cisplatin chemotherapy resistance in lung cancer, we tested eIF3a mRNA and protein expression in A549 (cisplatin sensitive cell line) and A549/DDP cells (cisplatin resistant cell line). As predicted, eIF3a showed low expression in the A549/DDP cell line and high expression in the A549 cell line at both the mRNA and the protein level (Fig. 1b,c, respectively).
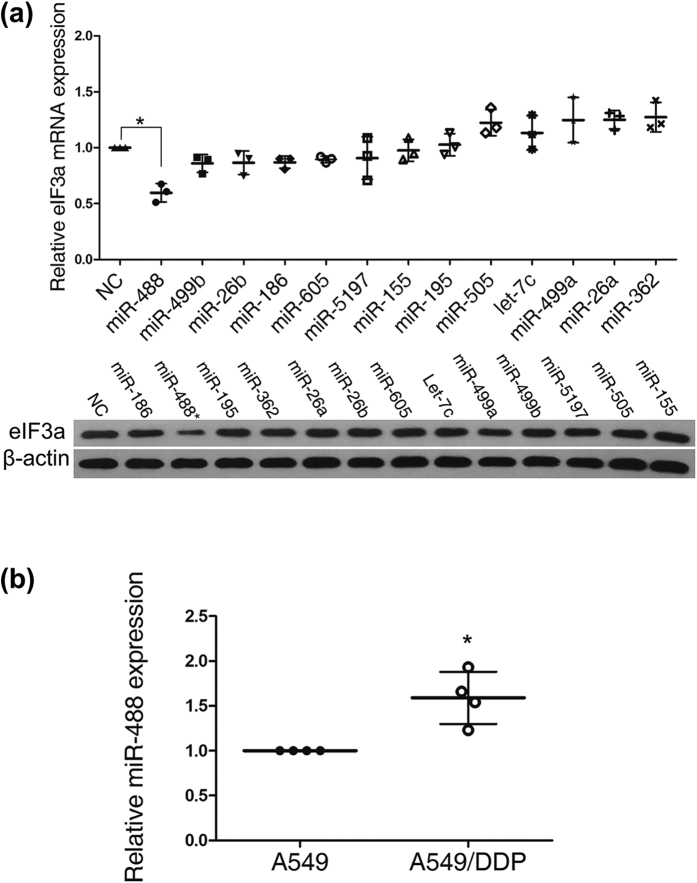
To determine the potential microRNAs regulating the gene expression of eIF3a, the 3’ untranslated region (UTR) of eIF3a was used as a query in the NCBI GenBank database. Subsequently, the potential microRNAs that may regulate eIF3a gene expression were analyzed using TargetScan (http://www.targetscan.org/vert_61), miRBase (http://www.mirbase.org), miRanda (http://www.microrna.org/), miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/index.php) and miRWalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk/index.html). Thirteen microRNAs were selected based on these predictions, including miRNA-186, miRNA-488, miRNA-195, miRNA-362, miRNA-26a, miRNA-26b, miRNA-605, let-7c, miRNA-499a, miRNA-499b, miRNA-5197, miRNA-505 and miRNA-155. Then, we tested eIF3a expression in the A549 cell line after the transfection of miRNA mimics. As shown in Fig. 2a, miR-488 significantly inhibited eIF3a expression at both the mRNA level and the protein level.
Figure 2. MiR-488 inhibited eIF3a expression and is highly expressed in A549/DDP cells.
(a) Relative eIF3a mRNA and protein expression levels in the A549 cell line after the transfection of different miR mimics (75 nM). MiR-488 significantly decreased eIF3a mRNA and protein expression. The representative cropping bands are present, and full-length blots are presented in Supplementary Fig. S3b. (b) Endogenous miR-488 showed higher expression in the A549/DDP cell line than the A549 cell line. The data from 3–4 independent experiments is given as the mean ± SD (*P < 0.05).
Showing a negative correlation with eIF3a expression, miR-488 was highly expressed in the A549/DDP cell line and minimally expressed in the A549 cell line (Fig. 2b). These data indicated that miR-488 may be involved in cisplatin resistance in lung cancer by regulating eIF3a expression.
MiRNA-488 directly targets eIF3a
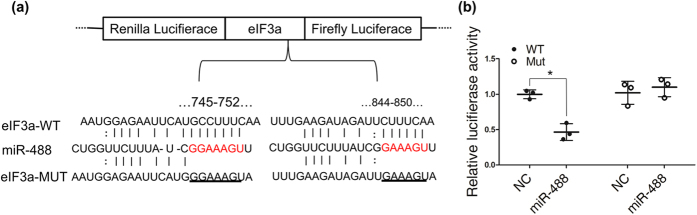
The bioinformatics-based target prediction analysis found two binding and interaction regions for miR-488 and eIF3a mRNA (Fig. 3a). Accumulating evidences indicate that miRNAs can inhibit target gene expression through perfect or nearly perfect complementarity to the 3’UTR of the target mRNA. We performed a dual-luciferase reporter assay to detect the potential regulation of eIF3a by miR-488. A miR-488 mimic and a luciferase reporter plasmid containing a wild-type or mutant 3’UTR binding site of human eIF3a were co-transfected into the A549 cell line. MiR-488 significantly inhibited the luciferase activity in A549 cells containing the eIF3a wild-type 3’UTR but failed to suppress this activity in cells with the mutated eIF3a 3’UTR, suggesting that eIF3a is a specific target of miR-488 (Fig. 3b). Combined with the fact that the mRNA and protein expression of eIF3a was suppressed by miRNA-488 over-expression (Fig. 2a), these results indicate that eIF3a is indeed a direct target of miRNA-488.
Figure 3. MiRNA-488 directly targeted eIF3a.
(a) The pmiR-RB-REPORT™ (vector) containing wild-type or mutant eIF3a 3’UTRs was constructed using the matching seed sequence of miR-488 and eIF3a. (b) The relative luciferase activity of the indicated eIF3a reporter construct was analyzed in A549 cells. The Renilla luciferase activity of each sample was normalized to the firefly luciferase value and plotted as relative luciferase activity.
MiRNA-488 inhibits A549 cell proliferation, migration and invasion
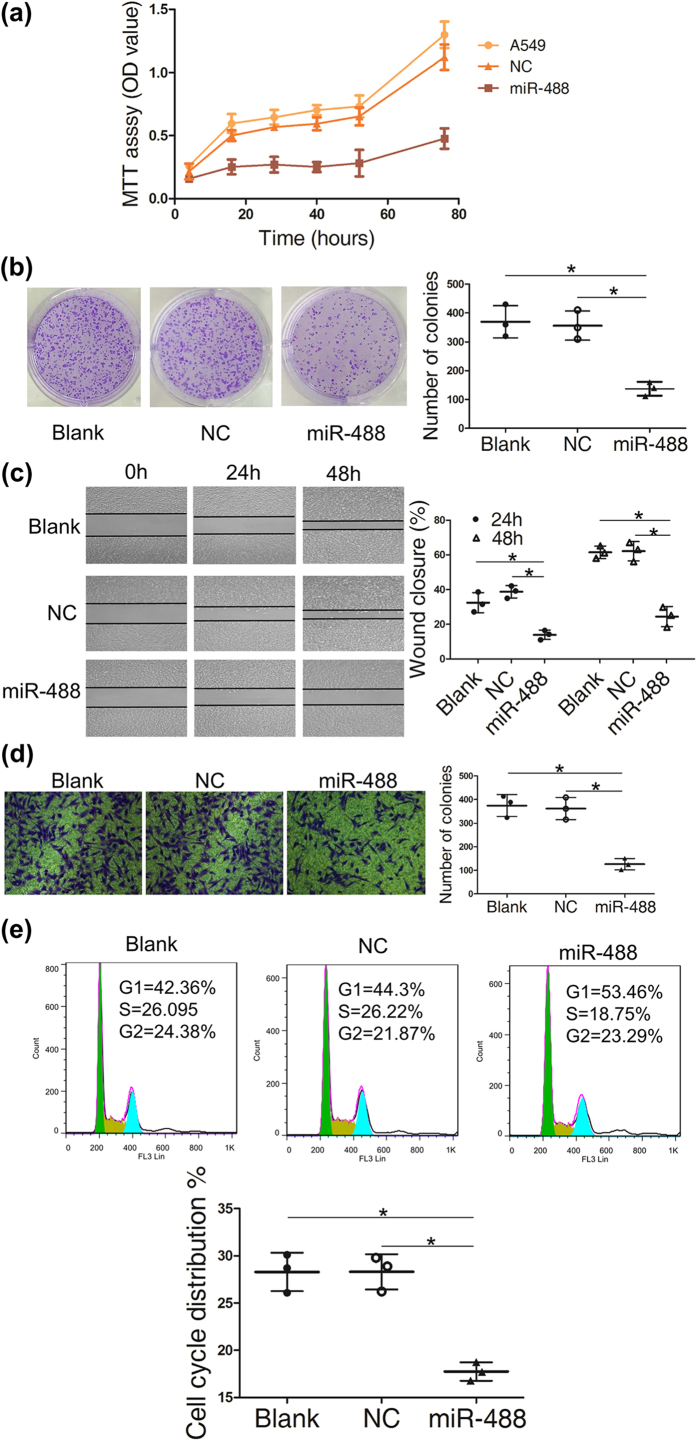
EIF3a is known as a tumor promoting factor, which induced us to investigate the influence of miRNA-488 on the malignant phenotypes in lung cancer cells. Therefore, we investigated the influence of miR-488 on cell proliferation, migration and invasion by transfecting miR-488 mimics into the A549 cell line. Compared with the negative control and un-transfected group, we found that miR-488 significantly inhibited cell proliferation (Fig. 4a), colony formation (Fig. 4b), cell migration (Fig. 4c) and the invasion (Fig. 4d) ability of the A549 cell line. Cell cycle analysis showed that miRNA-488 reduced the proportion of A549 cells in S phase (Fig. 4e), confirming the result in Fig. 4a that miRNA-488 could slow the rate of cell proliferation.
Figure 4. MiR-488 inhibited cell proliferation, migration and invasion in the A549 cell line.
The A549 cell line and cells transfected with negative control (NC) and the miR-488 mimic were used in the following assays. Cell viability (a) was tested with an MTT assay, and colony formation (b) was measured with crystal violet staining. All these showed that miR-488 could inhibit the proliferation of A549 cells. Wound healing (c) and transwell assays with Matrigel (d) were tested in A549 cells with miR-488 overexpression. The percent of wounds closed or number of cells migrating through the membrane were counted and are compared in the diagrams. The cell cycle (e) was evaluated with flow cytometry. The data from three independent experiments are given as the mean ± SD (*P < 0.05).
MiRNA-488 decrease cisplatin sensitivity in NSCLC cell lines
Previous research has shown that eIF3a down-regulation causes cellular resistance to cisplatin in lung cancer24, as shown above, we found that miR-488 regulates eIF3a expression via binding to its 3’UTR. Therefore, we evaluated the association between miR-488 expression and cisplatin sensitivity in three NSCLC cell lines. The MTT experiment showed that A549 and H1299 cells were more resistant to cisplatin after miRNA-488 transfection (Fig. 5a,d, respectively); this finding was confirmed with a dye exclusion assay (Fig. 5b,e). A cell apoptosis assay also showed fewer apoptotic cells in the miRNA-488 group compared to the vehicle group after cisplatin treatment (Fig. 5c,f). Additionally, another NSCLC cell line, the SK-MES-1 human lung squamous carcinoma cell also showed the similar results, in which miR-488 transfection increased cisplatin resistance in SK-MES-1 cells detected by dye exclusion and cell apoptosis assay (see Fig. 5g,h). The reduction efficiency of miR-488 to eIF3a in H1299 and SK-MES-1 cells was demonstrated in the supplementary material (see Supplementary Fig. S1a,b, respectively).
Figure 5. MiR-488 increased cisplatin resistance in three NSCLC cell lines.
(a) A549 and (d) H1299 cells transfected with miR-488 or NC were treated with different concentrations of DDP. Cell viability was tested with an MTS assay. The miR-488 group exhibited a higher IC50 of cisplatin than the NC group. Cell viability was also evaluated using a dye exclusion assay with flow cytometry (b,e,g) after the treatment with 10 μM DDP (for A549 cell line) or 60 μM DDP (for H1299 cell line) or 30 μM DDP (for SK-MES-1 cell line). Representative flow cytometry results showing miR-488 effects on cisplatin induced cell apoptosis in A549 (c) cells, H1299 (f) cells and SK-MES-1 cells (h), the right panels of cell viability refer to cells gated in the C3 quadrants. The results of 3–4 independent experiments are given as the mean ± SD (*P < 0.05).
MiRNA-488 increased cisplatin resistance through NER pathway in the A549 cell line
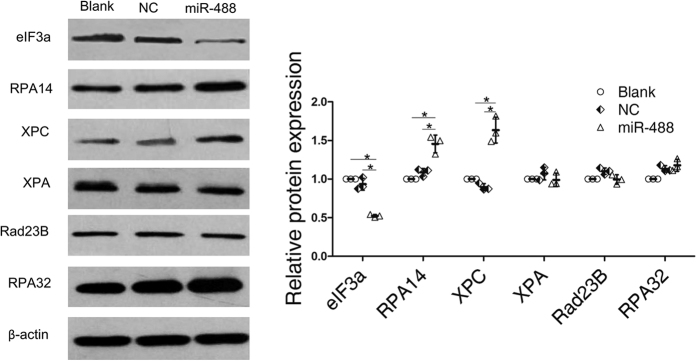
Increased DNA repair is considered one of the major mechanisms of platinum resistance. The DNA damage induced by platinum drugs is primarily repaired by NER25. The NER pathway is a complicated multistep process involving multiple proteins, including replication protein A (RPA) and xeroderma pigmentosum group proteins such as XPA and XPC26. Substantial evidence has suggested that the aberrant expression of these NER proteins was associated with cisplatin resistance27,28. Although we observed that miRNA-488 inhibited eIF3a expression and decreased cisplatin sensitivity, the mechanism was unknown. In this study, we hypothesized that miR-488 activated NER signaling via directly targeting eIF3a, which increased DNA repair ability and decreased cisplatin sensitivity. Here, we chose A549 cell line as our cell model, western blot showed that the expression of RPA14 and XPC increased after miR-488 transfection (Fig. 6). These results suggested that miRNA-488 may increase RPA14 and XPC expression by targeting eIF3a, resulting in cisplatin resistance.
Figure 6. Regulation of NER pathway related proteins by miR-488.
Total protein was collected from A549 cells and cells transfected with NC or miR-488. The eIF3a and NER pathway associated proteins XPA, XPC, RPA14, RPA32 and Rad 23B were detected by western blot. The representative cropping bands are present, and full-length blots are presented in Supplementary Fig. S3c. The relative expression levels were determined with ImageJ software. Actin was used as a loading control. The data shown are from three independent experiments (*P < 0.05).
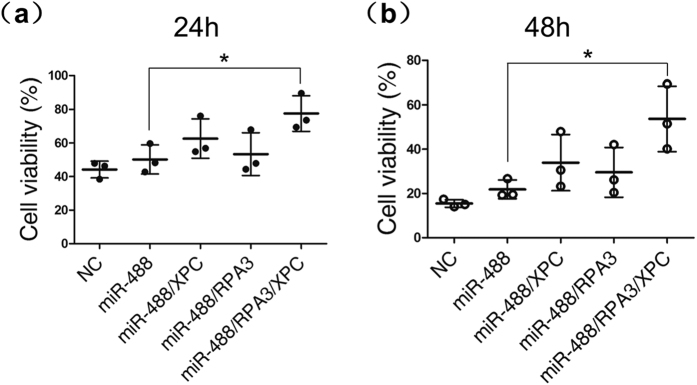
We found that A549 cells transfected with miR-488 could elevated the expression of RPA14 and XPC and increased resistance to cisplatin. Further, miR-488 co-transfected with RPA14 or XPC or both in A549 cells all showed variant rise in cisplatin resistance. Notably, the miR-488/RPA14/XPC transfection group had the greatest resistance to cisplatin compared with miR-488 only group (Fig. 7a,b). These results suggested that XPC and RPA14 enhance cisplatin resistance of cells and further confirmed that miR-488-mediating NER pathway in the cisplatin resistance of NSCLC cells. (The protein expressions after transfection were given in the supplement data (see Supplementary Fig. S1c)).
Figure 7. The overexpression of RPA14 and XPC enhanced miR-488 mediated cisplatin resistance.
Different combinations of NC, miR-488, expression vector of XPC and RPA14 were transfected in A549 cells, cell viability was tested with an MTS assay at 24 h (a) and 48 h (b) after treatment with 15 μM DDP. The results of three independent experiments are given as the mean ± SD (*P < 0.05).
Discussion
In this study, we noted that eIF3a was expressed at a lower level in the A549/DDP cell line compared with parental A549 cells, and identified miR-488 as a tumor suppressor that inhibited cell proliferation, migration and invasion in the A549 lung cancer cell line. The mechanism was to lower expression of the eIF3a oncogene. Our study found that miR-488 increased cisplatin resistance in A549 cells, and this result further conformed in other NSCLC cells, H1299 and SK-MES-1. The mechanism of miR-488 induces cisplatin resistance in NSCLC cells is by enhancing NER pathway proteins expression (Fig. 8).
Figure 8. Roles of miR-488 in lung cancer.
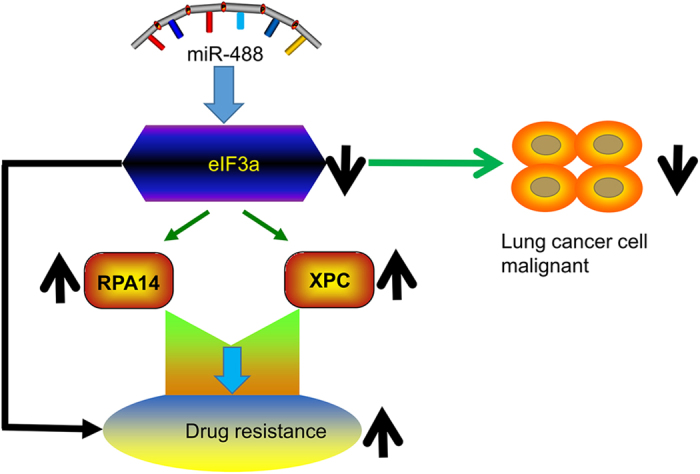
MiR-488 can directly target eIF3a and down-regulate its expression, which is involved in both the malignant phenotype and the cisplatin resistance of lung cancer.
EIF3a was first purified from rabbit reticulocyte lysate29 as one of the largest conserved subunits involved in the core functional domain of eIF3, which is the most complex compound involved in messenger RNA translation initiation. Furthermore, eIF3a can interact with many eIF3 subunits, as exemplified by its binding to eIF3b to form a bridge between eIF3b and eIF3c30,31,32. Tian-Rui Xu et al. revealed that eIF3a regulated the ERK pathway by binding to Raf-1, and the downregulation of eIF3a resulted in prolonged ERK activation33. However, any up-regulators of eIF3a are still unidentified.
It is well known that miRNAs participate in various biological processes, including cell proliferation, cellular differentiation, signal transduction and carcinogenesis34,35. Herein, based on bio-informatics analysis, we sought to identify the potential miRNA(s) in response to eIF3a regulation. Firstly, by detecting the endogenous eIF3a expression level in A549 cells transfected with different miRNA mimics, we focused on miRNA-488 as a regulator of eIF3a. Then, dual-luciferase reporter assay results showed the reduction of luciferase activity in the miRNA-488 transfected A549 cell line. All these results indicated that eIF3a is targeted by miR-488.
Studies have shown that eIF3a is unregulated in several human cancers, including lung cancer36,37,38, and eIF3a mRNA is also highly expressed in adult proliferating tissues (e.g., bone marrow, thymus and digestive tissues) and in tissues during development (e.g., fetal tissues)17. As shown by the evidences above, eIF3a is considered a tumor promoting factor because of its potential role in malignant transformation and the control of cell growth. Moreover, recent studies have found that decreasing eIF3a expression significantly reversed the malignant growth phenotype both in a human lung cancer cell line and in a breast cancer cell line13. An increasing number of studies have shown that miR-488 plays critical roles in several diseases39,40,41,42. In prostate carcinoma cells, it functions as tumor suppressor by inhibiting cancer cell proliferation and promoting cancer cell apoptosis39. Moreover, it suppresses cell migration during chondrogenic differentiation43. In the present study, we found that the over expression of miR-488 inhibited cell proliferation, migration and invasion in a lung cancer cell line by directly downregulating eIF3a expression.
Additionally, our lab found that chemotherapy sensitive patients showed higher eIF3a expression in their lung cancers, which was partly ascribed to the role of eIF3a in the regulation of NER pathway proteins24. Accordingly, we found lower eIF3a expression in the A549/DDP cell line (a drug resistant lung adenocarcinoma cell line) compared with parental A549 cells. Our results of three NSCLC cells lines suggested that miR-488 increased cell resistance to cisplatin in a mild way, which may be partly due to the increased expression of XPC and RPA14 of the NER pathway when eIF3a is down regulated by miRNA-488.
NER is the most important repair system among several DNA repair pathways in organisms and can prevent gene mutation and repair DNA distortion44. RPA is a single-stranded DNA binding protein required for DNA replication and NER; this protein is a heterotrimeric complex consisting of three subunits: RPA1 (RPA70), RPA2 (RPA32) and RPA3 (RPA14), with RPA14 being the smallest subunit45. The structure of the RPA-ssDNA complex is essential to DNA damage repair46. Salas, T.R. et al. provided evidence that RPA3 interacted directly with single-stranded DNA (ssDNA) at the 3′-end of a 31 nt ssDNA molecule47. XPC is a conserved DNA repair enzyme involved in the early stages of the global genome repair (GGR) NER pathway with the primary function of DNA damage recognition48. Polymorphisms in XPC were reported to be associated with response to platinum-based chemotherapy49. Our study showed that RPA14 and XPC levels were elevated after eIF3a was down-regulated by miRNA-488, which was consistent with our previous research in ovarian cancer50.
However, we found the miR-488 has inconsistent effect that suppresses eIF3a oncogene expression but increases cisplatin resistant in cells. The chemo-resistant effect by eIF3a inhibition is according to our previous study that cell downregulated eIF3a was resistant to platinum treatment24. Further investigation of eIF3a action on cell quiescent are requires to accurate the chemotherapy in NSCLC cells since studies have shown that quiescent cells tend to be quite resistant to all drugs51 and our research revealed that A549 cells with miR-488 overexpression would raise more proportion of G1 status but this result could not tell cells between G1 or G0 phase. Studies have shown that maximum expression of P27 is associating to G0 phase cells, implying a role for p27 in maintaining a quiescent state52,53,54. In addition, Dong et al. found that the decreased expression of eIF3a could elevate translation of P2755, and in our study, we found that the downregulation of eIF3a by miR-488 could increase the expression of P27 (see Supplementary Fig. S2). These may explain why cell proliferation was suppressed and cisplatin resistance was elevated after eIF3a downregulated by miR-488.
Our study has several limitations. First, as the endogenous expression level of miRNA-488 is not high, only the overexpression of miRNA-488 was investigated in the entire study. Second, as one miRNA could target various genes, the direct regulation of the NER pathway proteins by miR-488 was not researched. More studies of miRNA-488 are needed in this area.
In summary, we are first time to revealed that miR-488 directly targets eIF3a and decreases the expression of eIF3a, which play as tumor suppressor to reduce cell proliferation, migration and invasion in a lung cancer cell line. Meanwhile, we confirmed that this regulation functions in cisplatin resistance by increasing the expression of NER pathway proteins, such as XPC and RPA14. The current study provides new clues for NSCLC development and progression under miRNA regulation and novel issues should pay attention in NSCLC treatments.
Materials and Methods
Cell culture
The A549 and H1299 human lung adenocarcinoma cell lines, SK-MES-1 human lung squamous carcinoma cell line were purchased from the Chinese Academy of Sciences (Shanghai, China), and the A549/DDP cell line was obtained from the cell biology research laboratory and Modern Analysis Testing Center of Central South University (Changsha, China). All the cell lines were cultured in RPMI 1640 medium (Gibco, Life Technologies) supplemented with 10% fetal bovine serum (FBS, Gibco, Grand Island, NY, USA). Moreover, the A549/DDP cell line was cultured in medium with 2 mg/L cisplatin (Sigma, P4394) to maintain the drug-resistant phenotype before experimentation. All the cell lines were maintained at 37 °C in a humidified atmosphere of 5% CO2.
Cell viability/proliferation and cell cycle and apoptosis assays
Cell viability was tested with the Cell Titer 96® AQueous One Solution Assay (Promega), which was performed according to the instructions after transfection. Cell apoptosis was detected with flow cytometry using an Annexin V FITC Apoptosis Detection kit (BD, USA). The cell cycle was evaluated by flow cytometry with propidium iodide (PI) (Sigma) staining. For dye exclusion assays, cells were analyzed through flow cytometry. PI-positive and -negative cells were considered dead and living cells, respectively. X-axis is Forward-scattered light (FSC) that means proportional to cell-surface area or size.
Cell clone formation, migration and invasion assays
For clone formation experiments, 1000 cells were seeded into 6-well plates and then cultured for two weeks in medium containing 1% FBS. The cell clusters were stained with crystal violet. The wound healing assay was performed to evaluate cell migration. Cells were cultured for 48 h after being scratched with a pipette tip. For the invasion assay, 2 × 104 cells in serum-free medium were placed into the upper chamber (Costar, Corning, Inc., Corning), which was covered with Matrigel (BD Biosciences, Franklin Lakes). After 48 hours of incubation at 37 °C, the number of cells adhering to the lower membrane of the chambers was counted after staining with a solution containing 0.1% crystal violet and 20% methanol.
QRT-PCR of miRNA
Total RNA was extracted using a miRNeasy kit (Qiagen), and cDNA was synthesized with a Mir-X™ miRNA First-Strand Synthesis kit (Clontech Laboratories, Inc.). Gene expression was assessed by qRT-PCR using SYBR Premix Dimer Eraser (Perfect Real Time) assay kits. Real-time PCR was performed using the Roche LightCycler 480 PCR System.
Dual luciferase reporter assay
The recombinant pmir-REPORT Dual-Luciferase vector (Ambion) was used for eIF3a 3’UTR luciferase assays. Cells were cultured in 24-well plates, and wild-type or mutated eIF3a plasmids were co-transfected with miR-488 mimics or NC mimics using Lipofectamine 2000. Forty-eight hours after transfection, firefly and Renilla luciferase activities were measured using the Dual-Luciferase® Reporter Assay System (Promega).
Western blot analysis
Total protein was collected from cells in RIPA lysis buffer, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto PVDF membrane (Millipore, Bedford, MA). Then, the membranes were incubated in blocking solution (5% non-fat milk) and probed with primary antibody at 4 °C overnight. The primary antibodies used in this study were as follows: rabbit monoclonal antibodies for eIF3a, P27, Rad23B, RPA32, and XPA (Cell Signaling Technology, USA) and mouse monoclonal antibodies for RPA14 and XPC (Abcam, UK). Beta-actin (Sigma, St. Louis, MO) was used as a loading control.
Statistical analysis
The statistical analyses were performed using SPSS 18.0 for Windows (IBM, Inc., Chicago, IL, USA). One-way ANOVA was performed for comparison of more than two groups, and the differences between two groups were compared using an Independent Samples T-test. A value of P<0.05 was considered statistically significant. All values were expressed as the mean ± SD.
Additional Information
How to cite this article: Fang, C. et al. MiR-488 inhibits proliferation and cisplatin sensibility in non-small-cell lung cancer (NSCLC) cells by activating the eIF3a-mediated NER signaling pathway. Sci. Rep. 7, 40384; doi: 10.1038/srep40384 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by the National High-tech R&D Program of China (863 Program) (2012AA02A517), the National Natural Science Foundation of China (81373490, 81573508, 81573463), the Hunan Provincial Science and Technology Plan of China (2015TP1043), and the Open Foundation of Innovative Platform in University of Hunan Province of China (2015–14).
Footnotes
Author Contributions C.F., J.Y.Y. and Z.Q.L. wrote the manuscript. C.F., J.Y.Y., H.S.H. and Z.Q.L. conceived and designed the experiments. C.F., Y.X.C., N.Y.Y.W. and X.P.L. and performed the experiments. C.F., Y.X.C., N.Y.Y.W., J.Y., X.P.L. and W.Z. analyzed the results. C.F., Y.X.C., N.Y.Y.W., X.P.L. and H.H.Z. contributed new reagents/analytical tools.
References
- Losanno T. & Gridelli C. Safety profiles of first-line therapies for metastatic non-squamous non-small-cell lung cancer. Expert opinion on drug safety, doi: 10.1517/14740338.2016.1170116 (2016). [DOI] [PubMed] [Google Scholar]
- Kadara H., Scheet P., Wistuba, II & Spira, A. E. Early events in the molecular pathogenesis of lung cancer. Cancer prevention research (Philadelphia, Pa.), doi: 10.1158/1940-6207.capr-15-0400 (2016). [DOI] [PubMed] [Google Scholar]
- Siegel R., Ma J., Zou Z. & Jemal A. Cancer statistics, 2014. CA: a cancer journal for clinicians 64, 9–29, doi: 10.3322/caac.21208 (2014). [DOI] [PubMed] [Google Scholar]
- Spira A. & Ettinger D. S. Multidisciplinary management of lung cancer. The New England journal of medicine 350, 379–392, doi: 10.1056/NEJMra035536 (2004). [DOI] [PubMed] [Google Scholar]
- Chemotherapy in addition to supportive care improves survival in advanced non-small-cell lung cancer. a systematic review and meta-analysis of individual patient data from 16 randomized controlled trials. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 26, 4617–4625, doi: 10.1200/jco.2008.17.7162 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannetti E., Toffalorio F., De Pas T. & Peters G. J. Pharmacogenetics of conventional chemotherapy in non-small-cell lung cancer: a changing landscape? Pharmacogenomics 13, 1073–1086, doi: 10.2217/pgs.12.91 (2012). [DOI] [PubMed] [Google Scholar]
- Perron M. P. & Provost P. Protein interactions and complexes in human microRNA biogenesis and function. Frontiers in bioscience: a journal and virtual library 13, 2537–2547 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature 431, 350–355, doi: 10.1038/nature02871 (2004). [DOI] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004). [DOI] [PubMed] [Google Scholar]
- Garzon R., Calin G. A. & Croce C. M. MicroRNAs in Cancer. Annual review of medicine 60, 167–179, doi: 10.1146/annurev.med.59.053006.104707 (2009). [DOI] [PubMed] [Google Scholar]
- Damoc E. et al. Structural characterization of the human eukaryotic initiation factor 3 protein complex by mass spectrometry. Molecular & cellular proteomics: MCP 6, 1135–1146, doi: 10.1074/mcp.M600399-MCP200 (2007). [DOI] [PubMed] [Google Scholar]
- Dong Z. & Zhang J. T. Initiation factor eIF3 and regulation of mRNA translation, cell growth, and cancer. Crit Rev Oncol Hematol 59, 169–180, doi: 10.1016/j.critrevonc.2006.03.005 (2006). [DOI] [PubMed] [Google Scholar]
- Dong Z., Liu L. H., Han B., Pincheira R. & Zhang J. T. Role of eIF3 p170 in controlling synthesis of ribonucleotide reductase M2 and cell growth. Oncogene 23, 3790–3801, doi: 10.1038/sj.onc.1207465 (2004). [DOI] [PubMed] [Google Scholar]
- Liu Z. et al. Role of eIF3a (eIF3 p170) in intestinal cell differentiation and its association with early development. Differentiation; research in biological diversity 75, 652–661, doi: 10.1111/j.1432-0436.2007.00165.x (2007). [DOI] [PubMed] [Google Scholar]
- Yin J. Y. et al. Translational regulation of RPA2 via internal ribosomal entry site and by eIF3a. Carcinogenesis 34, 1224–1231, doi: 10.1093/carcin/bgt052 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z. et al. Role of eIF3a in regulating cell cycle progression. Experimental cell research 315, 1889–1894, doi: 10.1016/j.yexcr.2009.03.009 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincheira R., Chen Q. & Zhang J. T. Identification of a 170-kDa protein over-expressed in lung cancers. British journal of cancer 84, 1520–1527, doi: 10.1054/bjoc.2001.1828 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X. et al. The A/G allele of eIF3a rs3740556 predicts platinum-based chemotherapy resistance in lung cancer patients. Lung Cancer 79, 65–72, doi: 10.1016/j.lungcan.2012.10.005 (2013). [DOI] [PubMed] [Google Scholar]
- Xu X. et al. Association between eIF3alpha polymorphism and severe toxicity caused by platinum-based chemotherapy in non-small cell lung cancer patients. British journal of clinical pharmacology 75, 516–523, doi: 10.1111/j.1365-2125.2012.04379.x (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorkova Z., Skarda J. & Andel J. The role of microRNA in metastatic processes of non-small cell lung carcinoma. A review. Biomedical papers of the Medical Faculty of the University Palacky, Olomouc, Czechoslovakia, doi: 10.5507/bp.2016.021 (2016). [DOI] [PubMed] [Google Scholar]
- Langsch S. et al. miR-29b mediates NF-kB signaling in KRAS-induced non-small cell lung cancers. Cancer research, doi: 10.1158/0008-5472.can-15-2580 (2016). [DOI] [PubMed] [Google Scholar]
- Jiang L. P., Zhu Z. T. & He C. Y. Expression of miRNA-26b in the diagnosis and prognosis of patients with non-small-cell lung cancer. Future oncology (London, England) 12, 1105–1115, doi: 10.2217/fon.16.21 (2016). [DOI] [PubMed] [Google Scholar]
- Shen J. et al. The prognostic value of altered eIF3a and its association with p27 in non-small cell lung cancers. PloS one 9, e96008, doi: 10.1371/journal.pone.0096008 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J. Y. et al. Effect of eIF3a on response of lung cancer patients to platinum-based chemotherapy by regulating DNA repair. Clinical cancer research: an official journal of the American Association for Cancer Research 17, 4600–4609, doi: 10.1158/1078-0432.ccr-10-2591 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabholkar M., Vionnet J., Bostick-Bruton F., Yu J. J. & Reed E. Messenger RNA levels of XPAC and ERCC1 in ovarian cancer tissue correlate with response to platinum-based chemotherapy. The Journal of clinical investigation 94, 703–708, doi: 10.1172/jci117388 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Lindsey-Boltz L. A., Unsal-Kacmaz K. & Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annual review of biochemistry 73, 39–85, doi: 10.1146/annurev.biochem.73.011303.073723 (2004). [DOI] [PubMed] [Google Scholar]
- Wang G., Dombkowski A., Chuang L. & Xu X. X. The involvement of XPC protein in the cisplatin DNA damaging treatment-mediated cellular response. Cell research 14, 303–314, doi: 10.1038/sj.cr.7290375 (2004). [DOI] [PubMed] [Google Scholar]
- Andrews B. J. & Turchi J. J. Development of a high-throughput screen for inhibitors of replication protein A and its role in nucleotide excision repair. Molecular cancer therapeutics 3, 385–391 (2004). [PubMed] [Google Scholar]
- Benne R. & Hershey J. W. Purification and characterization of initiation factor IF-E3 from rabbit reticulocytes. Proceedings of the National Academy of Sciences of the United States of America 73, 3005–3009 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methot N., Rom E., Olsen H. & Sonenberg N. The human homologue of the yeast Prt1 protein is an integral part of the eukaryotic initiation factor 3 complex and interacts with p170. The Journal of biological chemistry 272, 1110–1116 (1997). [DOI] [PubMed] [Google Scholar]
- Asano K., Phan L., Anderson J. & Hinnebusch A. G. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. The Journal of biological chemistry 273, 18573–18585 (1998). [DOI] [PubMed] [Google Scholar]
- Block K. L., Vornlocher H. P. & Hershey J. W. Characterization of cDNAs encoding the p44 and p35 subunits of human translation initiation factor eIF3. The Journal of biological chemistry 273, 31901–31908 (1998). [DOI] [PubMed] [Google Scholar]
- Xu T. R. et al. Eukaryotic translation initiation factor 3, subunit a, regulates the extracellular signal-regulated kinase pathway. Molecular and cellular biology 32, 88–95, doi: 10.1128/mcb.05770-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que T. et al. Decreased miRNA-637 is an unfavorable prognosis marker and promotes glioma cell growth, migration and invasion via direct targeting Akt1. Oncogene 34, 4952–4963, doi: 10.1038/onc.2014.419 (2015). [DOI] [PubMed] [Google Scholar]
- Yu S. L. et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer cell 13, 48–57, doi: 10.1016/j.ccr.2007.12.008 (2008). [DOI] [PubMed] [Google Scholar]
- Bachmann F., Banziger R. & Burger M. M. Cloning of a novel protein overexpressed in human mammary carcinoma. Cancer research 57, 988–994 (1997). [PubMed] [Google Scholar]
- Dellas A. et al. Expression of p150 in cervical neoplasia and its potential value in predicting survival. Cancer 83, 1376–1383 (1998). [DOI] [PubMed] [Google Scholar]
- Chen G. & Burger M. M. p150 expression and its prognostic value in squamous-cell carcinoma of the esophagus. International journal of cancer. Journal international du cancer 84, 95–100 (1999). [DOI] [PubMed] [Google Scholar]
- Sikand K., Slaibi J. E., Singh R., Slane S. D. & Shukla G. C. miR 488* inhibits androgen receptor expression in prostate carcinoma cells. International journal of cancer. Journal international du cancer 129, 810–819, doi: 10.1002/ijc.25753 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik S. K., Kannisto E., Knudsen S. & Yendamuri S. Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. Cancer research 70, 36–45, doi: 10.1158/0008-5472.can-09-3153 (2010). [DOI] [PubMed] [Google Scholar]
- Zhao Y. et al. miR-488 acts as a tumor suppressor gene in gastric cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine, doi: 10.1007/s13277-015-4645-y (2016). [DOI] [PubMed] [Google Scholar]
- Song J. et al. MicroRNA-488 regulates zinc transporter SLC39A8/ZIP8 during pathogenesis of osteoarthritis. Journal of biomedical science 20, 31, doi: 10.1186/1423-0127-20-31 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Kim D. & Jin E. J. MicroRNA-488 suppresses cell migration through modulation of the focal adhesion activity during chondrogenic differentiation of chick limb mesenchymal cells. Cell biology international 35, 179–185, doi: 10.1042/cbi20100204 (2011). [DOI] [PubMed] [Google Scholar]
- Wood R. D. DNA repair in eukaryotes. Annual review of biochemistry 65, 135–167, doi: 10.1146/annurev.bi.65.070196.001031 (1996). [DOI] [PubMed] [Google Scholar]
- Iftode C., Daniely Y. & Borowiec J. A. Replication protein A (RPA): the eukaryotic SSB. Crit Rev Biochem Mol Biol 34, 141–180, doi: 10.1080/10409239991209255 (1999). [DOI] [PubMed] [Google Scholar]
- Shechter D., Costanzo V. & Gautier J. Regulation of DNA replication by ATR: signaling in response to DNA intermediates. DNA repair 3, 901–908, doi: 10.1016/j.dnarep.2004.03.020 (2004). [DOI] [PubMed] [Google Scholar]
- Salas T. R., Petruseva I., Lavrik O. & Saintome C. Evidence for direct contact between the RPA3 subunit of the human replication protein A and single-stranded DNA. Nucleic acids research 37, 38–46, doi: 10.1093/nar/gkn895 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl T., Hanaoka F. & Egly J. M. The comings and goings of nucleotide excision repair factors on damaged DNA. The EMBO journal 22, 5293–5303, doi: 10.1093/emboj/cdg489 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y. et al. Modulation of repair of ultraviolet damage in the host-cell reactivation assay by polymorphic XPC and XPD/ERCC2 genotypes. Carcinogenesis 23, 295–299 (2002). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. eIF3a improve cisplatin sensitivity in ovarian cancer by regulating XPC and p27Kip1 translation. Oncotarget 6, 25441–25451, doi: 10.18632/oncotarget.4555 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Zhang C. M. & Wu Y. S. Research progress in cancer stem cells and their drug resistance. Chinese journal of cancer 29, 261–264 (2010). [DOI] [PubMed] [Google Scholar]
- Hengst L. & Reed S. I. Translational control of p27Kip1 accumulation during the cell cycle. Science (New York, N.Y.) 271, 1861–1864 (1996). [DOI] [PubMed] [Google Scholar]
- Susaki E., Nakayama K. & Nakayama K. I. Cyclin D2 translocates p27 out of the nucleus and promotes its degradation at the G0-G1 transition. Molecular and cellular biology 27, 4626–4640, doi: 10.1128/mcb.00862-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson A. et al. A pathway in quiescent cells that controls p27Kip1 stability, subcellular localization, and tumor suppression. Genes & development 20, 47–64, doi: 10.1101/gad.1384406 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z. & Zhang J. T. EIF3 p170, a mediator of mimosine effect on protein synthesis and cell cycle progression. Molecular biology of the cell 14, 3942–3951, doi: 10.1091/mbc.E02-12-0784 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.